Abstract
It remains an enigma how the nervous system of different animal species produces different behaviors. We studied the neural circuitry for mating behavior in Drosophila subobscura, a species that displays unique courtship actions not shared by other members of the genera including the genetic model D. melanogaster, in which the core courtship circuitry has been identified. We disrupted the D. subobscura fruitless (fru) gene, a master regulator for the courtship circuitry formation in D. melanogaster, resulting in complete loss of mating behavior. We also generated frusoChrimV, which expresses the optogenetic activator Chrimson fused with a fluorescent marker under the native fru promoter. The fru-labeled circuitry in D. subobscura visualized by frusoChrimV revealed differences between females and males, optogenetic activation of which in males induced mating behavior including attempted copulation. These findings provide a substrate for neurogenetic dissection and manipulation of behavior in non-model animals, and will help to elucidate the neural basis for behavioral diversification.
SIGNIFICANCE STATEMENT How did behavioral specificity arise during evolution? Here we attempted to address this question by comparing the parallel genetically definable neural circuits controlling the courtship behavior of Drosophila melanogaster, a genetic model, and its relative, D. subobscura, which exhibits a courtship behavioral pattern unique to it, including nuptial gift transfer. We found that the subobscura fruitless circuit, which is required for male courtship behavior, was slightly but clearly different from its melanogaster counterpart, and that optogenetic activation of this circuit induced subobscura-specific behavior, i.e., regurgitating crop contents, a key element of transfer of nuptial gift. Our study will pave the way for determining how and which distinctive cellular elements within the fruitless circuit determine the species-specific differences in courtship behavior.
Keywords: courtship, CRISPR/Cas9, Drosophila, fruitless
Introduction
The species difference in mating behavior patterns is crucial for premating isolation (Seehausen et al., 2008; Kitano et al., 2009; van Doorn et al., 2009). However, little is known about how the mating circuitry of different species produces different behavioral patterns and what change in the genome underlies the species difference in the neural circuitry for the behavior (but see Tootoonian et al., 2012).
As an attempt to determine the circuitry basis for species differences in mating behavior, we focused on Drosophila subobscura, which possesses unique elements in the courtship ritual, most notably the phenomenon of the nuptial gift, in which a courting male extends his proboscis to make contact with the proboscis of his potential mate and gives her a regurgitated drop of his crop contents (Spieth, 1952; Steele, 1986; Immonen et al., 2009). We recently succeeded in targeted mutagenesis with the CRISPR/Cas9 system and piggyBac-mediated transgenesis in D. subobscura (Tanaka et al., 2016), paving the way to the genetic dissection of behavior in this non-model species.
In D. melanogaster, the gene fruitless (fru) has been shown to play a master regulator-like role in the formation of the circuitry for male courtship behavior (Dickson, 2008). The important features of the fru gene revealed by the studies in D. melanogaster are as follows. The most distal fru gene promoter (the P1 promoter) is dedicated to neuronal sexual differentiation (Ryner et al., 1996). In males, the P1 promoter generates several transcripts, each encoding isoforms with distinct C-termini (Goodwin et al., 2000; Song et al., 2002), whereas, in females, fru transcripts derived from the P1 promoter are not translated (Lee et al., 2000; Usui-Aoki et al., 2000). Thus, the P1 promoter contributes exclusively to the generation of FruM (M denotes male-specific), which are male-specific Fruitless proteins (Ferri et al., 2008; Neville et al., 2014; von Philipsborn et al., 2014). FruM is a member of the BTB-zinc finger protein family (Ito et al., 1996; Ryner et al., 1996), forms a complex with the chromatin regulators, and binds to ∼140 genomic target sites (Ito et al., 2012). FruM thus functions as a transcription factor, likely orchestrating a large set of target genes required for producing the male-typical characteristics of a neuron (Ito et al., 2016). Indeed, many of the fru-expressing neurons display sexual dimorphisms (Kimura et al., 2005, 2008; Cachero et al., 2010; Ruta et al., 2010; Yu et al., 2010; Ren et al., 2016). In loss-of-function fru mutant males, the majority of sexually dimorphic fru-neurons convert their structures into female-typical ones (Kimura et al., 2005; Datta et al., 2008), resulting in a reduction in male-to-female courtship and an increase in male-to-male courtship (Hall, 1978; Villella et al., 1997). Artificial formation of one particular group of fru-positive male-specific neurons (the P1 cluster neurons) in females causes them to court other females with a display typical of male courtship (Kimura et al., 2008), and artificial activation of the P1 cluster neurons in a male provokes courtship behavior even in the absence of a target mate (Kohatsu et al., 2011; von Philipsborn et al., 2011). It has been suggested that fru-positive neurons connect to each other, forming a core circuitry for generating male courtship behavior (Stockinger et al., 2005), and thus seem to compose a “fru-labeled circuitry” dedicated to male courtship behavior. We infer that the cellular composition of the fru-labeled circuitry might vary from species to species, and as a result, every species could possess distinct elements of courtship behavior. Using newly generated knock-in fru mutants of D. subobscura, here we demonstrate that artificial activation of fru-expressing neurons induces mating behavior in these males. Interestingly, the fru-labeled circuitry in D. subobscura has similar but distinct anatomical features from that in D. melanogaster. Our results suggest that the neural substrates for D. subobscura-specific mating behavior are embodied in the fru-labeled circuitry of this species, which function differently from the counterpart in D. melanogaster.
Materials and Methods
Fly strain and rearing conditions.
D. subobscura was obtained from the Drosophila Species Stock Center in San Diego, CA. The flies were reared on a cornmeal yeast medium at 22°C for D. subobscura or 25°C for D. melanogaster. A Canton-S fly line was used as wild-type D. melanogaster. w;; fruNP21/TM3 Ser was generated by the NP consortium (Hayashi et al., 2002). w;; 20XUAS-CsChrimson::mVenus was obtained from the Bloomington stock center (stock #55136).
Determination of a partial sequence of the targeted gene and gRNA design.
Genomic DNA was extracted from four D. subobscura males with a DNeasy Blood & Tissue Kit (Qiagen). A portion of the second exon of the fru gene was sequenced. The primers for sequencing were designed based on a sequence conserved across three Drosophila species: D. melanogaster, D. pseudoobscura, and D. persimilis (Drosophila 12 Species Consortium et al., 2007). Amplified DNA fragments were sequenced directly with a 3500 Genetic Analyzer (Applied Biosystems). For the sequence analysis of the modified fru locus, genomic DNAs were extracted from mutant flies in 50 μl of a squishing buffer (10 mm Tris-HCl, pH 8.2, 1 mm EDTA, 25 mm NaCl, 200 mg/ml proteinase K). We chose the sequences flanking the conserved splicing donor site in males as the targets for mutagenesis and transgenesis in D. subobscura after confirming the presence of the protospacer adjacent motif (PAM) there. PAM guides Cas9 to the nearby target for cleavage.
Construction of a knock-in vector and the generation of knock-in fru alleles.
To construct a donor vector for generating the frusoDR allele, 698 bp (HAL) and 759 bp (HAR) sequences flanking the predicted cut site of gRNA were amplified by PCR and incorporated, with an In-Fusion HD Cloning kit (Takara), into the EcoRI and XhoI sites of pDsRed-attP (Addgene plasmid; 51019), respectively.
To prepare the donor vector for generating the frusoChrimV allele, genomic DNA was extracted from the fly stock P{20XUAS-IVS-CsChrimson.mVenus}attP2 (Bloomington Stock Center, 55136). The sequence containing UAS-Chrimson-mVenus was amplified by PCR and incorporated into the SphI site of the donor vector.
Generation of Cas9 mRNA and gRNA.
Cas9 mRNA and gRNA were generated as described previously by Bassett et al. (2013). Briefly, the plasmid MLM3613 (Addgene plasmid 42251; Hwang et al., 2013) was linearized with PmeI (New England Biolabs) and in vitro transcription of a 1 μg linearized template was performed with an mMESSAGE mMACHINE T7 Ultra Transcription kit (Ambion) to produce the Cas9 mRNA. The Cas9 mRNA was then purified with an RNeasy Mini Kit (Qiagen) and frozen at −80°C until being injected into eggs. Templates for gRNA transcription were produced by a PCR with an oligonucleotide encoding the gRNA target sequence (CRISPR_fru-F; GAAATTAATACGACTCACTATAGGGGCTTACCTCGGCGATGTGTTTTAGAGCTAGAAATAGC) and a common oligonucleotide encoding the remainder of the gRNA sequence(gRNA-R;AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC) with an Ex Taq Hot Start version kit (Takara Bio). gRNAs were generated with a Megascript T7 kit (Ambion) by in vitro transcription of 300 ng DNA templates, which had been purified with a PCR purification kit (Qiagen). gRNAs were then purified by means of phenol chloroform extraction and frozen at −80°C until being injected into eggs.
Preparation of the injection solution for fru locus mutagenesis and transgenesis.
The solution used for the injection of gRNA and Cas9 mRNA into eggs for the generation of fru-deletion mutants was prepared from an RNA mixture containing 12 μg Cas9 mRNA and 1 μg gRNA in 30 μl of pure water. Three microliters of 3M sodium acetate, pH 5.2, and 90 μl of 99.5% ethanol were added to the mixture to precipitate RNAs. The RNA pellet was washed twice in 70% ethanol and resuspended in 11 μl of pure water. In the case of CRISPR-mediated knock-in, 9 μl of a donor vector was mixed with 12 μg Cas9 mRNA and 1 μg gRNA in 30 μl of pure water. A mixture of the donor vector, gRNA, and Cas9 mRNA was recovered by the same procedure as used for the generation of fru-deletion mutants.
Microinjection.
Fertilized eggs (embryos) were collected within 30 min after egg laying. The embryos were washed with distilled water and kept on a coverslip anterior side-up for several minutes to remove water on the surface. Lined-up embryos on the coverslip were coated with silicone oil (Shin-Etsu). Microinjection was performed at 20°C with Femtotip II micropipettes (Eppendorf) in conjunction with a 5246 Transjector system (Eppendorf).
PCR analysis for genotyping.
To ascertain the fruso3 and frusoDR fly genotypes, PCR was performed with an Ex Taq Hot Start version kit (Takara Bio). The genotype determination was achieved by 10% PAGE (for fruso3) or 1% agarose gel electrophoresis (for frusoDR).
Fly preparation for optogenetic activation.
Optogenetic activation of neurons was performed on flies that were collected ∼8 h after eclosion and kept in a vial with the food medium for 6–8 d. At 2 d before their use in the experiments, the flies were transferred to a new vial with the retinal-containing food medium; an aliquot of all-trans retinal (R2500; Sigma-Aldrich) at 50 mg/ml in 100% ethanol was dropped on the food medium so as to form a uniform layer of 0.3 mg/cm2 on the medium surface. To avoid the possible activation of fru-labeled neurons by environmental light, the retinal-fed flies were kept in the dark until they were used in the experiments. The control fly groups were handled in the same way as the experimental groups, except that they were transferred to a new vial containing the food medium without retinal 2 d before use. Under tethered conditions, a short copper wire of ∼150 μm in diameter and 3 mm in length was attached to the dorsal thorax of the male using UV glue (1773E; ThreeBond) under cold anesthesia. Then the males were kept singly in individual plastic chambers of 35 mm in diameter and 10 mm in height, which were humidified by placing a piece of filter paper soaked with 120 μl of distilled water onto the bottom of each. These chambers were kept in a humid container and flies were allowed to recover within them for ∼3 h in the dark. After the resting period, the free end of the copper wire (the end that was not affixed to the thorax) was attached to the fly holder, and then the fly was carefully positioned over an air-supported Styrofoam ball on the locomotion simulator so that the ball rotated smoothly along with the locomotion of the fly.
Light stimulation of the fru-labeled circuitry in free-moving and tethered males.
To evoke behavior in a fly under tethered conditions, the LED light source (M625L3; peak wavelength: 632 nm; Thorlabs) in front of a test male on the locomotion simulator was used to deliver excitation light to activate CsChrimson. LED lights were collimated using an aspheric lens mount (SM1V05, Thorlabs) in an adjustable lens tube (ACL2520U-A, Thorlabs), then projected onto the test male. The light on/off and lighting duration were controlled by electrical signals fed to the LED driver (LEDD1B, Thorlabs).
To evoke behavior in an unrestrained fly, a halogen light source (KTS-150, Kenko) in conjunction with a light-pad was used to deliver excitation light to activate CsChrimson.
Analysis of abdominal bending and wing extension under tethered conditions.
To quantify the behavior, the tethered frusoChrimV heterozygous males were video recorded from the lateral side with a CMOS camera (Point Gray, FL3-U3-13S2C-CS). An imaginary line connecting the halter and the abdominal tip was drawn on movie frames, and the angle between this line and the thoracic midline was measured: when this angle was 15° or larger, we judged that the fly was performing abdominal bending. To quantify wing extension, an imaginary line was drawn along the anteroposterior midline of the fly body on movie frames, and the angle between this line and the anterior margin of a wing was measured: when this angle was 15° or larger, we judged that the fly was performing wing extension. For this analysis, the ImageJ software (v1.40 r) was used. These recorded events were displayed in a raster plot and frequency histogram as a function of time.
Analysis of courtship behavior in fruso3 and frusoDR males.
For the analysis of courtship behavior in fruso3 and frusoDR males, the test flies were kept singly for 10–12 d after eclosion in food vials unless specifically mentioned. In the behavioral assays, a male was placed in a chamber of 14 mm in diameter and 4 mm in height; the floor of the chamber was covered with a slide glass (S9213, Matsunami Glass Industries), The flies were video recorded under white light illumination for 60 min by using an overhead CMOS camera (DFK 23U445, Imaging Source Asia). Each courtship element was counted with the video annotation software vCode (Hagedorn et al., 2008).
Dissection, immunohistochemistry, and imaging of the CNS and SEM observation of compound eyes.
The CNS of the fruso3 and frusoDR mutants and respective controls were dissected in PBS and fixed in 4% paraformaldehyde for 60 min on ice. Immunostaining was performed using the anti-FruMale antibody (at a dilution of 1:500) for FruM in combination with the AlexaFluor 546 anti-Guinea Pig IgG antibody (Invitrogen; 1:200). Counter-staining of the CNS was performed with a mouse nc82 antibody (Developmental Studies Hybridoma Bank; 1:10) in combination with the AlexaFluor 647 anti-mouse IgG antibody (Invitrogen; 1:200). For immunostaining of the frusoChrimV CNS, the anti-GFP rabbit polyclonal serum (Invitrogen; 1:500) was used for CsChrimson-mVenus in combination with the AlexaFluor 488 anti-rabbit IgG antibody (Invitrogen; 1:200), in addition to the anti-FruMale and nc82 antibodies. In some experiments, the CNSs of wild-type D. subobscura and D. melanogaster were additionally labeled by the anti-GABA rabbit polyclonal serum (Sigma-Aldrich; 1:500), which was visualized by the AlexaFluor 488 anti-rabbit IgG antibody (Invitrogen; 1:200). Images were acquired with a LSM 510 META confocal microscope (Carl Zeiss) using LSM Image Browser software. Stacks of optical sections at 0.84 μm were obtained with a LSM 510 META confocal microscope. Images of the compound eye surface were obtained with a scanning electron microscope (SU8000; Hitachi High-Technologies).
Experimental design and statistical analysis.
All statistical analyses were performed using GraphPad Prism7. The data shown in Figures 2H, and 7B,C were statistically analyzed using the Fisher exact test. Other data were statistically analyzed using the Mann–Whitney test. Exact p values were reported in the legend for each figure.
Figure 2.
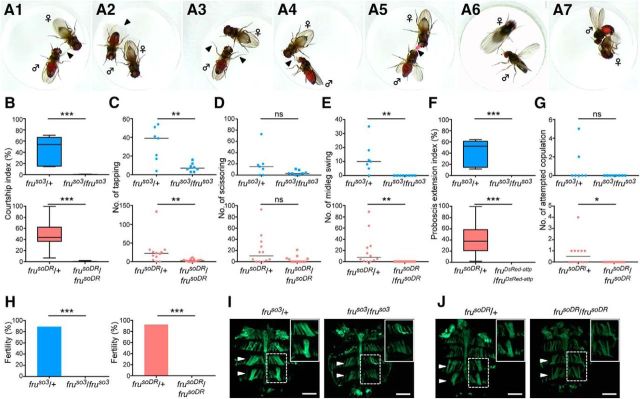
D. subobscura fru mutant phenotypes. A, The steps of mating behavior in D. subobscura include tapping (A1), scissoring (A2), midleg swing (A3), proboscis extension (A4), nuptial gift (A5), wing extension (A6), and attempted copulation (A7). All these steps except for the nuptial gift were included in estimating the courtship index. Shown are representative behavioral acts recorded with a wild-type D. subobscura male. The courtship indices (B; top: p = 0.0003, bottom: p < 0.0001), number of tappings (C; top: p = 0.0084, bottom: p = 0.0063), number of scissorings (D; top: p = 0.1228, bottom: p = 0.0511), number of midleg swings (E; top; p = 0.0014, bottom: p < 0.0001), proboscis extension indices (F; top; p = 0.0002, bottom: p < 0.0001), and number of attempted copulations (G; top; p = 0.2, bottom: p = 0.0137) are shown for fruso3 heterozygous males (left, bars in the top: n = 7), fruso3 homozygous males (right, bars in the top: n = 8), frusoDR heterozygous males (left, bars in the bottom: n = 12), and frusoDR homozygous males (righ, bars in the bottom: n = 11). The statistical significance of differences was evaluated by the Mann–Whitney's U test. ns, Not significant; *p < 0.05, **p < 0.01, ***p < 0.001. H, Comparisons of fertility between heterozygotes (left bars) and homozygotes (right bars) of fruso3(left graph; p < 0.0001) and frusoDR(right graph; p < 0.0001) males. The statistical significance of differences was evaluated by the Fisher exact test. ***p < 0.001. I, Dorsal abdominal musculature in a fruso3 heterozygous male (left) and a fruso3 homozygous male (right). J, Dorsal abdominal musculature in a frusoDR heterozygous male (left) and a frusDR3 homozygous male (right). Inset, Enlarged views of a MOL (dotted white line) for each genotype. Scale bars, 500 μm.
Figure 7.
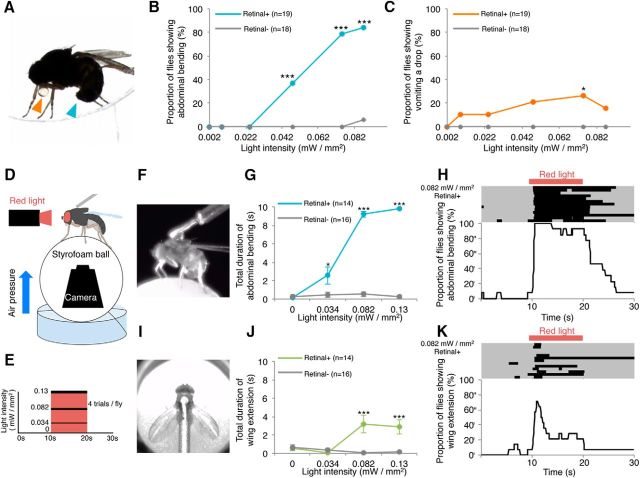
Mating motor acts induced by CsChrimson-mediated activation of the fru-labeled circuitry in D. subobscura. A, Abdominal bending (blue arrowhead) and regurgitating a droplet (orange arrowhead) as induced by CsChromson activation via light illumination in an unrestrained male placed alone in a circular chamber. B, C, The proportion of flies showing abdominal bending (B) and regurgitating a droplet (C) under unrestrained conditions in the fly groups fed on diets with or without retinal. D–K, Activation of the fru-labeled circuitry in a tethered male on the treadmill. A schematic drawing of the experimental setup (D) and a stimulation protocol for CsChrimson activation (E). Snapshots of a male fly displaying abdominal bending (F) and wing extension (I) on the treadmill. Cumulative durations of abdominal bending (G) and wing extension (J) in tethered male flies during a 10 s observation period as a function of the intensity of activation light in the fly groups fed on diets with or without retinal. Raster plots showing responses to the light stimulation in 14 test males (top) and changes in the proportion of flies exhibiting abdominal bending (H) and wing extension (K) over time, with reference to the period of red light illumination for CsChrimson activation (bottom). In the raster plot, each row represents an action record, in which a black bar appears when the male fly exhibits a behavioral act (i.e., abdominal bending and wing extension).
Results
Mutagenesis at the subobscura fru locus
In the D. melanogaster fru locus, sex-specific splicing in the second exon is decisive in determining whether FruM proteins are produced and thus whether the cell adopts the male-fate or female-fate (Fig. 1A): in the male, default splicing occurs at GT immediately after the coding sequence corresponding to the male-specific N-terminal extension, whereas, in the female, this GT is ignored as a result of the binding of a female determinant splicing accelerator, Transformer, to a more 3′ site, leading to the inclusion of a termination codon in the mRNA and thus to a failure in translation (Ryner et al., 1996; Heinrichs et al., 1998). We therefore targeted this conserved GT for the deletion by CRISPR/Cas9 to obtain mutants in which no splicing occurs at this site not only in females but also in males, and consequently, the resulting fru mRNA has a stop codon that prevents its translation regardless of the fly's sex (Fig. 1B). We recovered three deletion alleles (fruso1, fruso2, and fruso3; “so” stands for subobscura), one of which (fruso3) lost the GT, whereas the other two retained it (Fig. 1C). fruso3 is thus a promising fru mutant candidate. Encouraged by our success in generating deletions in the fru locus, we further endeavored to knock-in the attP site within the fru locus (Fig. 1D). attP is the landing site for phiC31-integrase-mediated gene insertions, allowing one to integrate into the fru locus any sequence of interest, provided that it has been cloned into the donor vector with attB. The construct to be inserted also carried the fluorescent marker gene 3xP3DsRed-p(A) and targeted the same site as used in deletion mutagenesis (Fig. 1D). The DsRed fluorescence allowed us to recover the genome-edited offspring simply by selecting “glowing” larvae among those derived from eggs injected with the donor vector under a fluorescent microscope (Fig. 1E). We chose this target site because we expected that the vector integration into the fru open reading frame would intervene in the translation of FruM, and since the target site worked well in mutagenesis (Fig. 1A) by the CRISPR/Cas9 system for genome editing. We recovered the DsRed-positive offspring, leading to the establishment of a fru knock-in allele, frusoDsRed (frusoDR) (Fig. 1, E–G).
Figure 1.
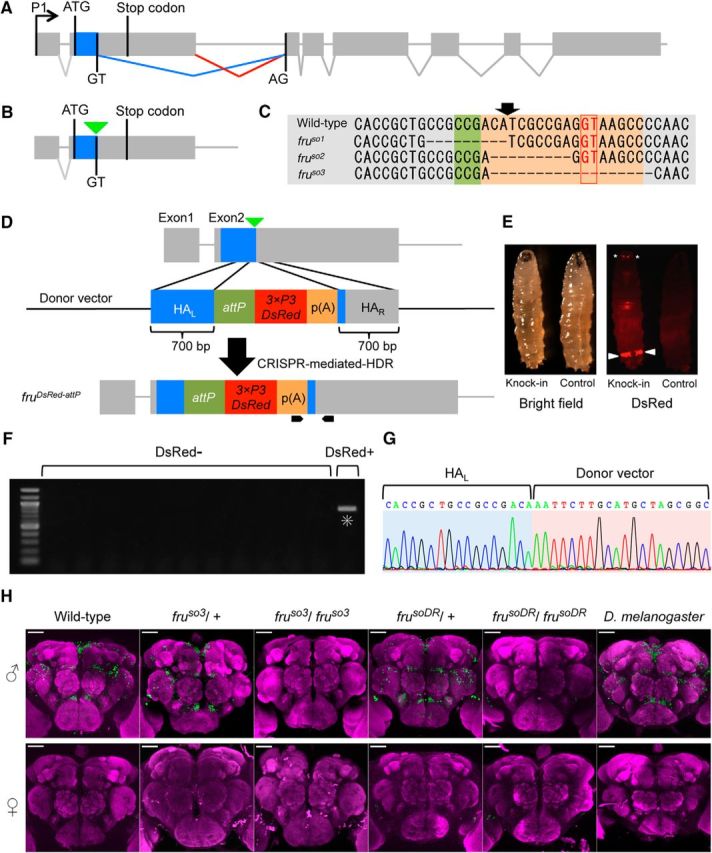
Generation of fru mutants in D. subobscura. A, The exon-intron organization of the fru gene in D. melanogaster, with the initiation and stop codons in the second exon as well as the splicing donor and acceptor sites for the conjunction of exon 2 and exon 3, highlighting the sex difference in fru splicing, which underlies the male-specific production of the full-length FruM protein. B, A schematic representation of the targeted site for gRNA (indicated by an inverted triangle) used for mutagenesis. C, The nucleotide sequence around the targeted region (shaded) that includes the splice donor site (boxed) in the wild-type genome (WT) and the induced deletion in the mutants fruso1, fruso2, and fruso3. The planned position for cutting is indicated with an arrow. D, A schematic representation of CRISPR-mediated knock-in targeting of the fru locus. The black pointed bars at the bottom indicate the position of the primers used for screening. HAL and HAR represent left and right homology arms, respectively. attP indicates the landing site for phiC31-integrase-mediated gene insertions. p(A) indicates a poly(A) signal sequence for the fluorescent marker gene 3xP3DsRed. E, Bright-field (left) and fluorescent (right) images of a third-instar larva expressing the DsRed protein (Knock-in) and a non-injected larva (Control). Arrowheads indicate anal pads and asterisks indicate Bolwig organs. F, Screening for CRISPR-mediated knock-in events by PCR. The left-most lane representing a sample derived from a DsRed+ fly but none of the other lanes from DsRed- flies had a band (highlighted with an asterisk) of the predicted size (905 bp) indicative of successful knock-in. G, The genomic sequence around the 5′ junction of HAL and the transgene of the DsRed+ fly. H, Anti-FruMale antibody immunoreactivity (green) of the male (top) and female (bottom) brains of D. melanogaster wild-type flies, D. subobscura wild-type and variants, i.e., fruso3 heterozygotes, fruso3 homozygotes, frusoDR heterozygotes, frusoDR homozygotes, and frusoDR homozygotes (from left to right), counterstained with nc82 (magenta). Scale bars, 50 μm.
By immunostaining the brain tissue of D. subobscura males that are heterozygous for fruso3 or frusoDR with the anti-Fru antibody raised against D. melanogaster Fru, we successfully labeled a subset of D. subobscura neurons (Fig. 1H), the location and number of which were very similar to those of fru-expressing neurons in wild-type D. melanogaster (Fig. 1H; Cachero et al., 2010; Yu et al., 2010). We consider that the anti-Fru antibody recognized FruM expressed in the male brain of D. subobscura. In keeping with this idea, the anti-Fru antibody revealed no immunoreactive material in the brain of D. subobscura females that are heterozygous for fruso3 or frusoDR or in the brain of wild-type D. melanogaster females (Fig. 1H). Importantly, no discernible immunoreactivity to the anti-Fru antibody was detected in the male brain homozygous of fruso3 or frusoDR (Fig. 1H). We conclude that fruso3 and frusoDR are protein null fru alleles.
Subobscura fru mutant phenotypes
In D. melanogaster, males of fru protein-null mutants exhibit little or no courtship activity toward a female (Ito et al., 1996; Villella et al., 1997). We quantified the courtship activity in D. subobscura males with the courtship index (CI), which represents the percentage of time spent by a male courting a female within a 15 min observation period in a mating chamber. The following male motions directed to a female were judged to constitute courtship behavior: tapping the female with a leg (Fig. 2A1), wing scissoring (Fig. 2A2), midleg swing (Fig. 2A3), proboscis extension (Fig. 2A4), nuptial gift (Fig. 2A5), wing extension (Fig. 2A6), and attempted copulation (Fig. 2A7). We also included in the male courtship behavior the “dance” (Maynard Smith, 1956), which refers to the male movement to maintain his position facing the female upon her sidestepping (Movie 1). fru heterozygous males exhibited a high CI of 47% for fruso3 and 48.2% for frusoDR, whereas homozygous males exhibited an almost negligibly low CI (0.6% for fruso3 and 0.7% for frusoDR; Fig. 2B). The incidence of occurrence of three elementary courtship actions, i.e., tapping (Fig. 2C), midleg swing (Fig. 2E), and proboscis extension (Fig. 2F), decreased in mutants homozygous for fruso3 or frusoDR, compared with the respective heterozygous controls. The incidence of occurrence of scissoring in fru homozygotes was indistinguishable from that of heterozygotes in both fru alleles (Fig. 2D) and that of attempted copulation was reduced in frusoDR homozygotes but not fruso3 homozygotes compared with the respective controls (Fig. 2G). In parallel with the difference in the courtship activity, males heterozygous for fruso3 or frusoDR were fertile, whereas homozygous males were infertile in both alleles (Fig. 2H).
Movie 1.
Courtship behavior in D. subobscura. Male courtship behavior is composed of tapping, scissoring, proboscis extension, midleg swing, nuptial gift, dance, wing extension, and attempted copulation.
In addition to the behavioral phenotypes, fru mutant males in D. melanogaster manifest a gross-anatomical deficit, i.e., loss of the muscle of Lawrence (MOL), a pair of dorsal longitudinal muscles running beneath the fifth abdominal (A5) tergite that are specifically present in the adult wild-type male (Gailey et al., 1991). The MOL is produced by the inductive action of an innervating neuron (Lawrence and Johnston, 1986; Currie and Bate, 1995), which needs to have the male identity as a result of the expression of the masculinizer protein FruM (Nojima et al., 2010). Therefore, lack of the MOL signifies neural feminization due to the fru deficiency in the motoneuron. In D. subobscura, wild-type males are known to have two pairs of MOL, one in A4 and the other in A5 (Gailey et al., 1997; Takayanagi et al., 2015). We found that males heterozygous for fruso3 or frusoDR invariably had two pairs of MOL (Fig. 2I,J). In homozygous males, the MOL was present but its shape and orientation were markedly distorted in both alleles (Fig. 2I,J). We conclude that the fru mutations disturb neural masculinization in D. subobscura as does the fru mutation in D. melanogaster.
The fru-labeled circuitry in D. subobscura
The finding that the fru gene functions are conserved leads to the question of how this gene contributes to the production of distinctly different mating behaviors in the two species of Drosophila. In an effort to answer this question, we first attempted to determine whether fru-expressing neurons in D. subobscura are able to generate mating behavior elements unique to this species, by optogenetically activating these neurons via the channelrhodopsin Chrimson (csChrimson; Klapoetke et al., 2014). Unlike for the genetic model of D. melanogaster (Venken et al., 2011), no genetic tools for this approach are available in D. subobscura. In D. melanogaster, a fru-specific GAL4 is used to drive a UAS-csChrimson transgene for the activation of fru-expressing neurons. For the purpose of visualizing activated fru-expressing cells, a visible marker such as UAS-GFP is also required. Because transforming flies per se is quite difficult in D. subobscura, we endeavored to edit the native fru locus so that it expresses mVenus-tagged csChrimson instead of FruM by means of the CRISPR/Cas9 system, rather than generating three transformant lines respectively carrying fru-GAL4, UAS-csChrimson, and UAS-GFP. To confer GAL4-responsiveness onto the edited fru allele, UAS-csChrimson-mVenus, rather than the coding region for csChrimson-mVenus alone, was knocked into the fru locus of D. subobscura (Fig. 3A). To leave room for further modifications of the edited fru locus, we once again included attP in the donor vector, along with 3xP3DsRed-p(A) (Fig. 3A).
Figure 3.
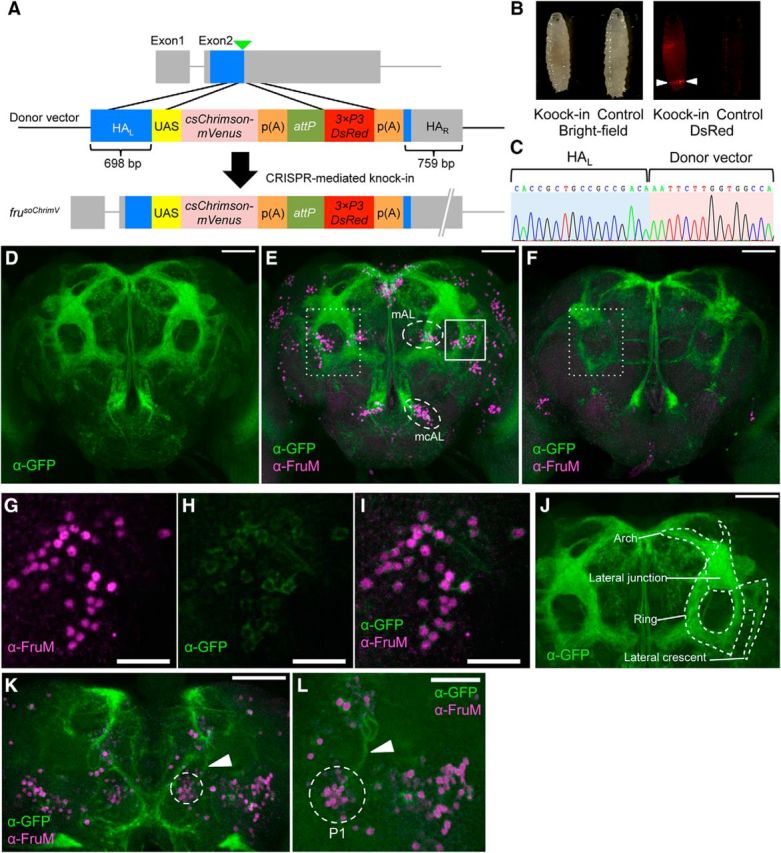
Generation of a fru allele that expresses csChrimson-mVenus under the native fru-P1 promoter in D. subobscura. A, Top, A schematic representation of the minigene knock-in site (indicated by an inverted triangle). Middle, The structure of the donor vector that contains homologous arms for recombination (HAL and HAR), UAS, the coding region of csChrimson-mVenus, attP and 3xP3-DsRed. Bottom, An expected genomic organization after the successful integration of the construct. B, Larvae with and without the minigene integration as viewed under a light field (left) and a dark field (right). 3xP3-DsRed expression in the anal pad indicates the minigene integration (arrowheads). C, Sequence around the 5′ junction of HAL and the transgene in frusoChrimV. D–F, Anterior view of the fru-labeled circuitry in the brain of frusoChrimV males (D, E) and females (F) stained for mVenus (green) by the anti-GFP antibody alone (D) or together with the anti-FruMale antibody (red; E, F). Scale bars: D–F, 50 μm. The somata of mAL and mcAL neurons are encircled with a white broken line (E). The ring region is boxed with a white dotted line in E and F. G–I, Magnified images of the region boxed with a solid white line in E, showing the immunoreactivity to the anti-FruM antibody (G) and anti-GFP antibody (H), and merged images (I). Scale bars, 20 μm. J, Anterior view of the fru-labeled circuitry. The arch, lateral junction, ring, and lateral crescent are indicated. Scale bar, 50 μm. K, Posterior view of the fru-labeled circuitry in male. P1 cell bodies are encircled with a white dotted line and the primary neurites are indicated by an arrowhead, respectively. Scale bar, 50 μm. L, Enlarged image of P1 cell bodies (encircled with a white dotted line) and neurites (indicated by an arrowhead). Scale bar, 30 μm.
In this attempt, we succeeded in recovering frusoChrimV, a fru allele (Fig. 3B,C) that expresses CsChrimson-mVenus in a large proportion of neurons positive for anti-Fru antibody immunoreactivity (Fig. 3D,E,G–I). Surveying across the entire brain region in several males, we estimated that 96.4% of brain neurons with endogenous FruM have CsChrimson-mVenus expression, whereas only a minor fraction of FruM-negative neurons (2.9%) expressed CsChrimson-mVenus in frusoChrimV heterozygous males (Table 1). The neurons singly positive for GFP (i.e., Venus), those singly positive for FruM, and those doubly positive for GFP and FruM were intermingled in a given neural cluster (Fig. 3J). mVenus visualized both cell bodies and neurites, highlighting the entire fru-labeled circuitry in D. subobscura (Fig. 3D–F,K,L). Overall, the fru-labeled circuitry in the male brain of D. subobscura appeared similar to that of D. melanogaster (Fig. 3E,F). Neural subdomains characteristic of the fru-labeled circuitry in D. melanogaster (Yu et al., 2010), i.e., dorsally locating fiber bundles connecting two hemispheres called the arch, circular fiber tracts in the lateral protocerebrum called the ring, the lateral junction that connects the arch and ring, and fiber extensions aside the loop called the lateral crescent, were all recognizable in the male brain of D. subobscura (Fig. 3J). These four subdomains are enriched with interconnections among fru-expressing neurons, likely facilitating the information processing relevant to courtship activities in D. melanogaster (Yu et al., 2010). The lateral crescent receives inputs from the lateral horn (LH), a higher olfactory processing center for innate behavior (Yu et al., 2010; Kohl et al., 2013). LH outputs also impinge onto the lateral junction, where some ascending neurons from the SOG terminate (Yu et al., 2010) to convey contact chemical pheromone inputs. Higher sensory interneurons with different sensory modalities converge at the ring, from which some descending fibers originate to control motor centers (Yu et al., 2010). The arch represents a dorsal commissure connecting two brain halves but nonetheless is decorated with synapses along nearly the entire length (Kimura et al., 2008). Thus, it appears that the fundamental architecture of the neural center for courtship behavior is conserved between D. subobscura and D. melanogaster.
Table 1.
The number of brain neurons that are positive for the anti-FruM antibody and for theVenus reporter in frusoChrimV heterozygous males
| Cluster | FruM[+]a | Venus[+]a | Double[+]/FruM[+], %b | Venus[+]-double[+]/Venus[+], %c |
|---|---|---|---|---|
| aSP1 | 16.0 ± 0.7 | 15.3 ± 0.5 | 95.3 | 0 |
| aSP2 | 53.0 ± 2.7 | 52.0 ± 2.0 | 98.1 | 0 |
| aSP3 | 50.3 ± 1.5 | 48.8 ± 1.3 | 96.5 | 0 |
| AL | 63.8 ± 1.9 | 64.0 ± 1.7 | 98.8 | 1.6 |
| mAL | 28.0 ± 1.7 | 26.8 ± 1.7 | 94.6 | 0.9 |
| mcAL | 31.8 ± 1.1 | 32.5 ± 1.6 | 97.6 | 4.6 |
| SG | 15.5 ± 1.3 | 14.8 ± 1.0 | 88.7 | 6.8 |
| Lo | 45.5 ± 1.0 | 45.5 ± 0.6 | 98.9 | 1.1 |
| pSP1 + pSP2 + P + pL | 131.3 ± 3.2 | 124.8 ± 3.6 | 94.7 | 0.4 |
| Lateral-AL | 0 | 7.5 ± 0.3 | NA | 100 |
| Total | 435.0 ± 5.3 | 431.8 ± 6.6 | 96.4 | 2.9 |
aThe mean ± SEM values of the counts in four brains are indicated.
bThe proportion of cells that are Venus-positive (Venus[+]) in anti-FruM-labeled cells (FruM[+]) is indicated.
cThe proportion of cells that are anti-FruM negative in Venus-labeled cells is indicated.
Some of the fru-expressing neurons in D. melanogaster have conspicuous features that allow us to reasonably assign their possible homologs in D. subobscura. These include: posteriorly located somata of P1 cluster neurons with a dorsally extending primary neurite that turns ventrally to form a U-shaped tract (Fig. 3K,L; Hall, 1978), mAL cluster neurons with bilateral neurites with tufts in the SOG (Fig. 3E; Kimura et al., 2005), and mcAL cluster neurons with cell bodies alongside the esophagus and the ascending neurite along the midline (Fig. 3E; Yu et al., 2010). The roles of these three groups of neurons in male courtship behavior have been well established in D. melanogaster (Yamamoto and Koganezawa, 2013). The P1 cluster is composed of 20 male-specific neurons that initiate courtship behavior (Kimura et al., 2008). The mAL cluster is involved in contact chemical pheromone processing, likely making contact with P1 neurons as a presynaptic neural element (Koganezawa et al., 2010; Clowney et al., 2015; Kallman et al., 2015). The mcAL cluster regulates overall courtship activity levels (Manoli and Baker, 2004). We therefore consider that some of the neurons composing the core courtship circuitry are conserved between D. subobscura and D. melanogaster.
As in D. melanogaster, the fru P1 promoter is active in both the female and male, even though FruM is produced only in the male, and therefore, we can compare the fru-labeled circuitry in the two sexes by relying on mVenus staining in frusoChrimV heterozygotes (Figs. 3D–I, Fig. 4). Most of the fiber tracts composing the fru-labeled circuitry in the female brain appeared thinner than the male counterparts (compare Figs. 3E vs F, Fig. 4A–C). In D. melanogaster, the tip of the contralateral neurite in the SOG of mAL neurons bifurcates in females but not in males (Kimura et al., 2005). This sex difference in mAL neurons was similarly detected in mAL neurons of D. subobscura (Fig. 4D–I). We conclude that the fru-labeled circuitry in D. subobscura manifests sexual dimorphisms.
Figure 4.
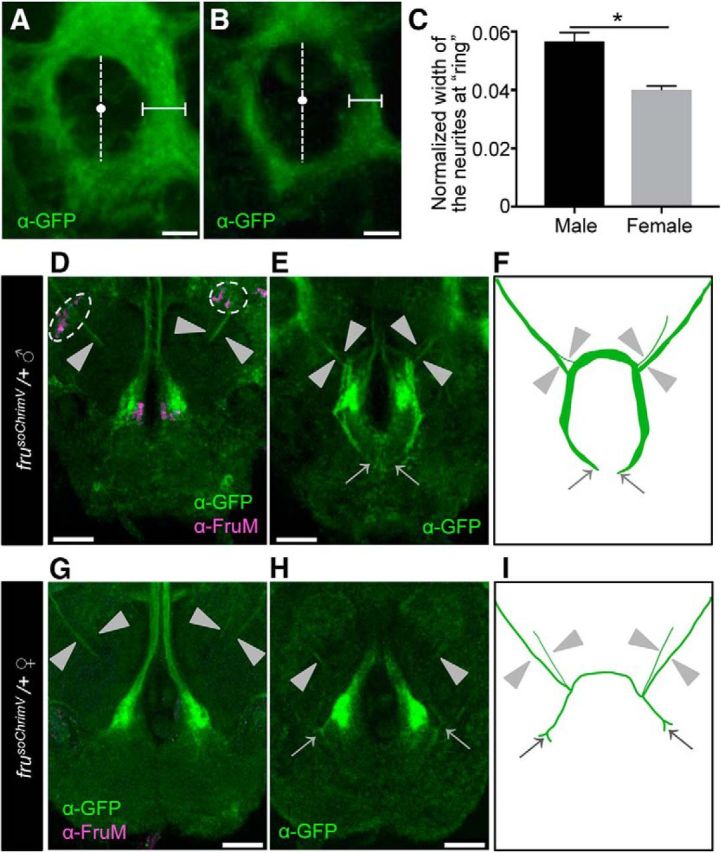
Sexual dimorphisms in the fru-labeled circuitry in D. subobscura. A, B, Magnified images of the ring region boxed by a dotted white line in Figure 3E and F, respectively. The width of the medial portion of the inner fiber tract in the ring (indicated by horizontal bars) was measured along the lateral axis at the midpoint, which was defined as the midpoint (white dots) of the inner ring diameter along the dorsal-ventral axis (indicated by vertical broken lines). The width of the ring measured was normalized by the largest width of the dorsal central brain. Scale bars, 15 μm. C, A comparison of the normalized width of the ring fiber tract between the male (n = 5) and female (n = 3) frusoChrimV heterozygotes. The statistical significance of differences was evaluated by the Mann–Whitney U test: p = 0.0357. D–I, Sexual dimorphisms of the mAL neurites in the frusoChrimV heterozygous male and female. Scale bars, 30 μm. D, G, Magnified images of mAL cell bodies and neurites in the male (D) and female (G) brain. Cell bodies are encircled by a broken white line, and neurites are indicated by arrowheads. E, H, mAL neurites observed at a different plane, highlighting their tip portions (indicated by arrows) in the subesophageal ganglion (SOG) of the male (E) and female (H). The thick bilateral fiber tracts along the midline and large terminals in the SOG are of mcAL neurons. F, I, Schematic representation of the neurite projection pattern in the male (F) and female (I). Neurites are indicated by arrowheads. The dark oblique structure at the center represents the esophagus. Scale bars, 30 μm.
Although the fru-labeled circuitry in D. subobscura appeared similar to the counterpart in D. melanogaster, there were discernible differences between them (Fig. 5). The most obvious difference was found in the male optic lobe; ascending fibers of fru-positive M neurons in the medulla are densely distributed in the ventral half of the optic lobe in D. melanogaster (Fig. 5A,C), whereas the corresponding M-neuron fibers occupy the entire width of the optic lobe in D. subobscura (Fig. 5B,D). In accord with the apparent expansion of fiber projections, the counts of somata in the male brain revealed that D. subobscura contains ∼2.5 times more M neurons than D. melanogaster does (Fig. 5E–G). Interestingly, the number of ommatidia composing a compound eye was ∼2.5 times larger in D. subobscura than D. melanogaster (Fig. 5H–J). This suggests that D. subobscura has the larger number of M neurons as the third order visual interneurons than D. melanogaster and this is correlated with the larger number of primary sensory neurons in the retina in the former species. The fru-expressing lobula plate Lo neurons, on which M-neuron fibers likely terminate (Yu et al., 2010), were expanded in number in D. subobscura compared to D. melanogaster, though in a lesser extend than the increase in M neurons (Fig. 5E–G). As aforementioned, mAL neurons, second-order interneurons in the pheromone processing pathway (Koganezawa et al., 2010; Clowney et al., 2015; Kallman et al., 2015), and mcAL neurons with a negative regulator role in initiating male courtship behavior (Manoli and Baker, 2004), were similar in number between D. melanogaster and D. subobscura (Fig. 5E). These observations imply that the sensory pathway for visual information processing is overrepresented in the fru-labeled circuitry of D. subobscura relative to that of D. melanogaster. We have carefully examined the optic lobe in the female, and we indeed found a clear difference between D. melanogaster and D. subobscura. The fru-positive “M neurons” in the optic lobe are male-specific in D. melanogaster, and we confirmed this is true (Fig. 5A,C). Unexpectedly, however, D. subobscura females did have the M-neurons (Fig. 5D). This might imply that vision is important in female courtship behavior in D. subobscura.
Figure 5.
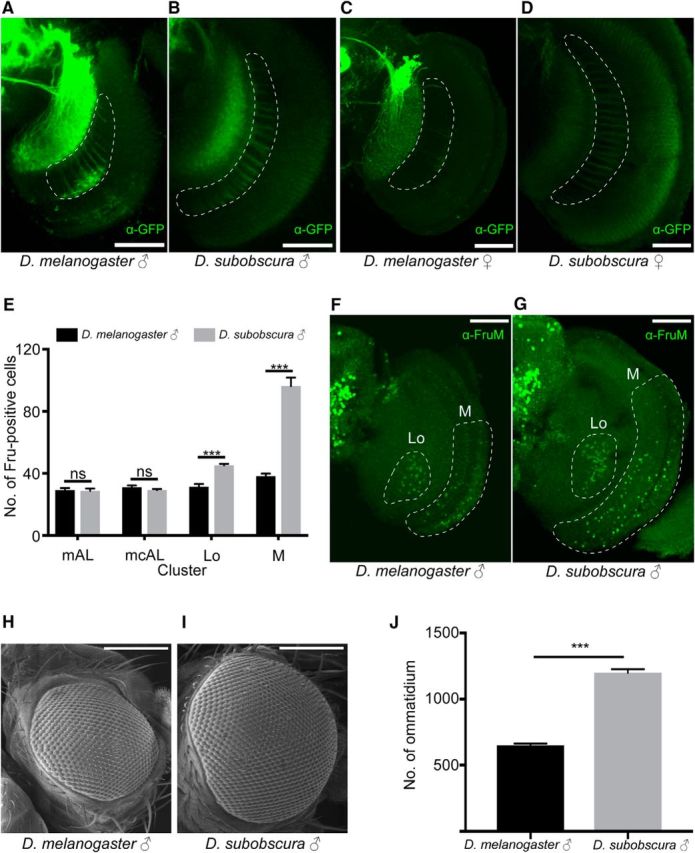
Differences of the fru-labeled circuitry in the optic lobe between D. subobscura and D. melanogaster. A–D, The fru-labeled fibers of M neruons in the optic lobe (encircled with white broken lines) in D. melanogaster (A, C) and D. subobscura (B, D) in male (A, B) and (C, D). Scale bars, 50 μm. E, The number of anti-FruM antibody-immunoreactive cells in the mAL: p = 0.9213; mcAL: p = 0.6702; Lo: p = 0.0006; and M: p = 0.0006 clusters compared between D. melanogaster (n = 7) and D. subobscura (n = 7). The statistical significance of differences was evaluated by the Mann–Whitney's U test. ns, Not significant. ***p < 0.001. Error bars show SEM. F, G, Images of the optic lobe stained with the anti-FruM antibody in D. melanogaster (F) and D. subobscura (G) males. The region with Lo and M neuron somata is encircled with a white broken line. Scale bars, 50 μm. H, I, Scanning electron micrographs of the compound eye in D. melanogaster (H) and D. subobscura (I) males. J, The number of ommatidia composing a compound eye in D. melanogaster (n = 8) and D. subobscura (n = 8) males. The statistical significance of differences was evaluated by the Mann–Whitney's U test. p = 0.0006. Error bars show SEM.
mAL neurons are the best-characterized fru-expressing neurons, and are known to be GABAergic in D. melanogaster (Koganezawa et al., 2010; Fig. 6A–D). We have performed an additional experiment to examine whether mAL neurons in D. subobscura are also GABAergic. Our anti-GABA antibody staining of mAL neurons unambiguously demonstrated this is indeed the case (Fig. 6E–H).
Figure 6.
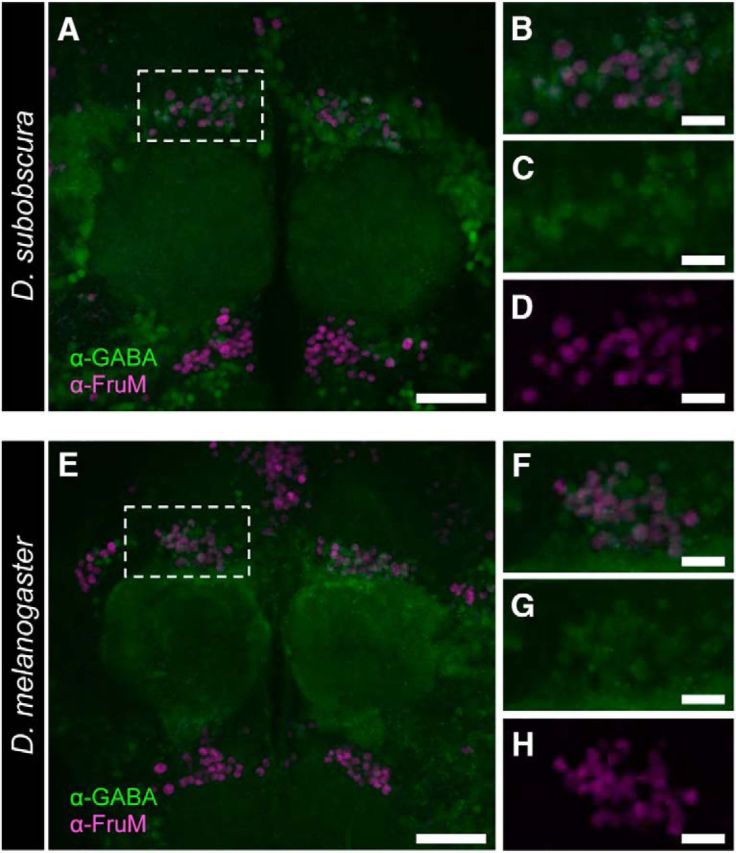
The GABAergic nature of mAL-cluster neurons in D. melanogaster and D. subobscura. A, E, Anti-GABA antibody immunoreactivity in male brains of D. subobscura (A) and D. melanogaster (E) compared with anti-FruMale-antibody immunoreactivity. B–D, F–H, Magnified images of the mAL cluster, showing the merged images (B, F) highlighting immunoreactivity to the anti-GABA antibody (C, G) and the anti-FruMale antibody (D, H).
The subobscura fru circuitry produces mating behavior
To explore the functions of the fru-labeled circuitry in D. subobscura, we artificially activated it via CsChrimson. We placed a frusoChrimV heterozygous adult in a round observation chamber to irradiate it with strong visible light to activate CsChrimson. Intriguingly, isolated males, in response to the irradiated light, repetitively, and persistently bent the abdomen in a manner reminiscent of attempted copulation in normal mating behavior (Fig. 7A,B). Although less frequently, some of the isolated frusoChrimV heterozygous males regurgitated a drop in response to light (Fig. 7A,C), as did courting wild-type males for a nuptial gift (Fig. 2A6). Both elementary courtship acts were induced in an increasing number of flies as the intensity of light increased up to ∼0.075 mW/mm2 in a retinal-dependent manner (Fig. 7B,C). Although these results demonstrated that optogenetic activation of the fru-labeled circuitry induces elementary acts composing mating behavior in D. subobscura, quantitative analysis of the stimulus–response relationship was difficult because the position of the test fly relative to the light source changes time-to-time and thus the stimulus efficacy may not be constant throughout the experiment. To circumvent this difficulty, we used a treadmill-aided tethered male preparation (Kohatsu et al., 2011; Kohatsu and Yamamoto, 2015), in which the test male is stationary but can voluntarily walk on the treadmill (Fig. 7D). With this system, we found that the proportion of flies exhibiting abdominal bending increased as the stimulus intensity increased (Fig. 7E,G). Likewise, the cumulative duration of abdominal bending increased as the stimulus intensity increased (Fig. 7G). Practically no abdominal bending was observed in isolated males without light-activation of CsChrimson (Fig. 7H), or without retinal feeding (Fig. 7G). Our careful examination of fly behavior during optogenetic activation of the fru-circuit has unraveled that bilateral wing extension is reproducibly induced in response to light illumination (Fig. 7I). This response was retinal-dependent and thus judged to be mediated by CsChrimson activation (Fig. 7J,K). We conclude that the fru-labeled circuitry generates elements of mating behavior, including those unique to D. subobscura, such as the nuptial gift.
Discussion
Based on the proposition that fru plays a master regulator role in organizing the courtship circuitry in D. melanogaster (Dickson, 2008), evolutionary changes in the structure and function of this gene have been implicated as the bases for the species difference in mating behavior patterns (Yamamoto and Ishikawa, 2013). A quantitative trait loci (QTL) analysis for a courtship song difference between D. simulans and D. sechellia highlighted six chromosomal intervals that revealed significant QTLs, and the interval found to have the largest effect contained the fru locus (Gleason and Ritchie, 2004). On the other hand, a recent exhaustive QTL mapping investigating the differences in sine songs among the strains of D. simulans and D. mauritiana revealed that a transposon insertion into the K+ channel gene Slowpoke was responsible for the variations in sine song carrier frequency (Ding et al., 2016). Yet another study examined whether a complete wild-type fru locus introduced into the D. melanogaster fru mutant genome from D. yakuba, D. ananassae, or D. persimilis could restore male courtship behavior, and if so, whether the rescued flies would exhibit the courtship behavior pattern of the donor species, rather than that of the host species, i.e., D. melanogaster (Cande et al., 2014). The results of this comprehensive work revealed that the heterospecific wild-type fru locus perfectly rescued the courtship behavior that had been lost by the fru mutation in the D. melanogaster host, with no sign of cross-species transfer of courtship behavior patterns, allowing the authors to claim that fru is not a significant source of evolutionary variation in courtship behavior (Cande et al., 2014). However, in that study, only quantitative differences in the parameters of courtship songs were analyzed, i.e., the elements of courtship behavior that were shared by all the tested species. It is theoretically possible that a qualitative difference in courtship behavior among species, such as the addition of a novel elementary act to the ritual, might arise from a master regulator gene mutation, which could recruit to the courtship circuitry a set of neurons that have been used for behavior unrelated to courtship. It remains an open question as to whether “microevolution” of the song pattern and “macroevolution” of the courtship ritual are causally related to each other (Reznick and Ricklefs, 2009). Yet another question here is whether a gene transplanted from the donor species genome to the host species genome operates normally even under a different trans environment. FruM functions as a transcription factor that orchestrates >100 target genes, whose cis elements have likely coevolved with the trans environment, which is variable from species to species (Wray et al., 2003). Moreover, specific connections among neurons are not entirely determined by the genetic program, but rather, are heavily dependent on cell-to-cell interactions during development (Luo, 2016). In fact, FruM is required in the innervating motoneuron for induction of the target MOL, revealing a “noncell autonomous” effect of FruM. Interestingly, a genomic fragment 5′ to the P1 promoter of the D. subobscura fru gene (∼30 kb in length) was able to rescue the MOL-less phenotype of D. melanogaster fru mutant males, leading to the formation of a single MOL pair in A5 when introduced into the D. melanogaster genome, but this fragment was not able to recapitulate the MOL formation pattern typical of D. subobscura, which has two MOL pairs, one in A5 and the other in A4 (Gailey et al., 1997). On the other hand, our present results clearly documented the fru requirement for the proper induction of MOL pairs in both the A4 and A5 segments in D. subobscura males, as the fruso3 and frusoDR mutations affected not only the MOL in A5 but also that in A4, which is D. subobscura-specific. These considerations inevitably lead to the notion that it is crucial to analyze the fru gene function and fru-labeled circuitry in every non-model species (e.g., D. subobscura) to elucidate the molecular and cellular underpinnings of the species difference in mating behavior.
In this study, we observed a profound enlargement of the fru-labeled visual pathway in D. subobscura compared to the D. melanogaster counterpart (Fig. 5). This is intriguing because, unlike D. melanogaster, vision is known to be indispensable for successful mating in D. subobscura (Milani, 1950). The fact that D. subobscura males have ∼100 M neurons and ∼40 Lo neurons, whereas D. melanogaster males have ∼40 M neurons and ∼30 Lo neurons suggests that convergence of visual inputs at postsynaptic lobular neurons is more extensive in D. subobscura than in D. melanogaster. It remains to be examined whether this quantitative difference in the neural connectivity found in the visual pathway between the two species has any relevance to the species difference in the vision dependence of male courtship activities. In this study, we successfully induced regurgitation of crop contents and bilateral wing extension, two courtship elements unique to D. subobscura that are not observed in D. melanogaster. The tantalizing possibility is that neurons involved in these behavioral actions acquired fru expression in D. subobscura and thus were recruited to the courtship circuitry in this species, whereas their counterparts in D. melanogaster are fru-negative and dedicated to other behavioral actions. This hypothesis will become testable when roles of individual neurons or neural groups in these behaviors are determined in D. subobscura upon the establishment of a genetic tool kit that allows clonal analysis of cellular functions. Our success in generating fru mutants and the fru knock-in fly line for optogenetics in D. subobscura opens an avenue for the systematic comparisons of functions of the fru gene and fru-labeled circuitry between D. melanogaster and D. subobscura, thereby paving the way for understanding the mechanistic basis of diversification in behavior.
Footnotes
This work was supported, in part, by Grant-in-Aids for Scientific Research (Nos. 17K19371, 17H05935, and 16H06371) from The Ministry of Education, Culture, Sports, Science and Technology (MEXT) to D.Y., and a Grant-in-Aid for JSPS fellows (15J05088) to R.T. We thank A. Utsumi for secretarial assistance.
The authors declare no competing financial interests.
References
- Bassett AR, Tibbit C, Ponting CP, Liu JL (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4:220–228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachero S, Ostrovsky AD, Yu JY, Dickson BJ, Jefferis GS (2010) Sexual dimorphism in the fly brain. Curr Biol 20:1589–1601. 10.1016/j.cub.2010.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande J, Stern DL, Morita T, Prud'homme B, Gompel N (2014) Looking under the lamp post: neither fruitless nor doublesex has evolved to generate divergent male courtship in Drosophila. Cell Rep 8:363–370. 10.1016/j.celrep.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Iguchi S, Bussell JJ, Scheer E, Ruta V (2015) Multimodal chemosensory circuits controlling male courtship in Drosophila. Neuron 87:1036–1049. 10.1016/j.neuron.2015.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie DA, Bate M (1995) Innervation is essential for the development and differentiation of a sex-specific adult muscle in Drosophila melanogaster. Development 121:2549–2557. [DOI] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, Flores J, Balonze K, Dickson BJ, Axel R (2008) The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 452:473–477. 10.1038/nature06808 [DOI] [PubMed] [Google Scholar]
- Dickson BJ. (2008) Wired for sex: the neurobiology of Drosophila mating decisions. Science 322:904–909. 10.1126/science.1159276 [DOI] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, Stern DL (2016) Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 536:329–332. 10.1038/nature19093 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, et al. (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Ferri SL, Bohm RA, Lincicome HE, Hall JC, Villella A (2008) fruitless Gene products truncated of their male-like qualities promote neural and behavioral maleness in Drosophila if these proteins are produce in the right places at the right times. J Neurogenet 22:17–55. 10.1080/01677060701671947 [DOI] [PubMed] [Google Scholar]
- Gailey DA, Taylor BJ, Hall JC (1991) Elements of the fruitless locus regulate development of the muscle of Lawrence, male-specific structure in the abdomen of Drosophila melanogaster adults. Development 113:879–890. [DOI] [PubMed] [Google Scholar]
- Gailey DA, Ohshima S, Santiago SJ, Montez JM, Arellano AR, Robillo J, Villarimo CA, Roberts L, Fine E, Villella A, Hall JC (1997) The muscle of lawrence in Drosophila: a case of repeated evolutionary loss. Proc Natl Acad Sci U S A 94:4543–4547. 10.1073/pnas.94.9.4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JM, Ritchie MG (2004) Do quantitative trait loci (QTL) for a courtship song difference between Drosophila simulans and D. sechellia coincide with candidate genes and intraspecific QTL? Genetics 166:1303–1311. 10.1534/genetics.166.3.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SF, Taylor BJ, Villella A, Foss M, Ryner LC, Baker BS, Hall JC (2000) Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics 154:725–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn J, Hailpern J, Karahalios KG (2008) VCode and VData: illustrating a new framework for supporting the video annotation workflow [abstract]. Working Conference on Advanced Visual Interfaces, AVI 08, pp 317–321. Naples, Italy, May 10.1145/1385569.1385622 [DOI] [Google Scholar]
- Hall JC. (1978) Courtship among males due to a male-sterile mutation in Drosophila melanogaster. Behav Genet 8:125–141. 10.1007/BF01066870 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, Ueda R, Uemura T, Yoshihara M, Goto S (2002) GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34:58–61. 10.1002/gene.10137 [DOI] [PubMed] [Google Scholar]
- Heinrichs V, Ryner LC, Baker BS (1998) Regulation of sex-specific selection of fruitless 5′ splice sites by Transformer and Transformer-2. Mol Cell Biol 18:450–458. 10.1128/MCB.18.1.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen E, Hoikkala A, Kazem AN, Ritchie MG (2009) When are vomiting males attractive? Sexual selection on condition-dependent nuptial feeding in Drosophila subobscura. Behav Ecol 20:289–295. 10.1093/beheco/arp008 [DOI] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D (1996) Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci U S A 93:9687–9692. 10.1073/pnas.93.18.9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Sato K, Koganezawa M, Ote M, Matsumoto K, Hama C, Yamamoto D (2012) Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell 149:1327–1338. 10.1016/j.cell.2012.04.025 [DOI] [PubMed] [Google Scholar]
- Ito H, Sato K, Kondo S, Ueda R, Yamamoto D (2016) Fruitless represses robo1 transcription to shape male-specific neural morphology and behavior in Drosophila. Curr Biol 26:1532–1542. 10.1016/j.cub.2016.04.067 [DOI] [PubMed] [Google Scholar]
- Kallman BR, Kim H, Scott K (2015) Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. eLife 4:e11188. 10.7554/eLife.11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ote M, Tazawa T, Yamamoto D (2005) Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438:229–233. 10.1038/nature04229 [DOI] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D (2008) Fruitless and Doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron 59:759–769. 10.1016/j.neuron.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, Kingsley DM, Peichel CL (2009) A role for a neo-sex chromosome in stickleback speciation. Nature 461:1079–1083. 10.1038/nature08441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK, et al. (2014) Independent optical excitation of distinct neural populations. Nat Methods 11:338–346. 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koganezawa M, Haba D, Matsuo T, Yamamoto D (2010) The shaping of male courtship posture by lateralized gustatory inputs to male-specific interneurons. Curr Biol 20:1–8. 10.1016/j.cub.2009.11.038 [DOI] [PubMed] [Google Scholar]
- Kohatsu S, Yamamoto D (2015) Visually induced initiation of Drosophila innate courtship-like following pursuit is mediated by central excitatory state. Nat Commun 6:6457. 10.1038/ncomms7457 [DOI] [PubMed] [Google Scholar]
- Kohatsu S, Koganezawa M, Yamamoto D (2011) Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron 69:498–508. 10.1016/j.neuron.2010.12.017 [DOI] [PubMed] [Google Scholar]
- Kohl J, Ostrovsky AD, Frechter S, Jefferis GS (2013) A bidirectional circuit switch reroutes pheromone signals in male and female brains. Cell 155:1610–1623. 10.1016/j.cell.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P (1986) The muscle pattern of a segment of Drosophila may be determined by neurons and not by contributing myoblasts. Cell 45:505–513. 10.1016/0092-8674(86)90282-5 [DOI] [PubMed] [Google Scholar]
- Lee G, Foss M, Goodwin SF, Carlo T, Taylor BJ, Hall JC (2000) Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol 43:404–426. [DOI] [PubMed] [Google Scholar]
- Luo L. (2016) Principles of neurobiology. Abingdon, UK: Garland Science. [Google Scholar]
- Manoli DS, Baker BS (2004) Median bundle neurons coordinate behaviours during Drosophila male courtship. Nature 430:564–569. 10.1038/nature02713 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. (1956) Fertility, mating behavior and sexual selection in Drosophila subobscura. J Genetics 54:261–279. 10.1007/BF02982781 [DOI] [PubMed] [Google Scholar]
- Milani R. (1950) Sexual behavior of D. subobscura, ambigua, bidasciata, tristis, obscuroides. Dros Inf Service No 24:88–89. [Google Scholar]
- Neville MC, Nojima T, Ashley E, Parker DJ, Walker J, Southall T, Van de Sande B, Marques AC, Fischer B, Brand AH, Russell S, Ritchie MG, Aerts S, Goodwin SF (2014) Male-specific Fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr Biol 24:229–241. 10.1016/j.cub.2013.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Kimura K, Koganezawa M, Yamamoto D (2010) Neuronal synaptic outputs determine the sexual fate of postsynaptic targets. Curr Biol 20:836–840. 10.1016/j.cub.2010.02.064 [DOI] [PubMed] [Google Scholar]
- Ren Q, Awasaki T, Huang YF, Liu Z, Lee T (2016) Cell class-lineage analysis reveals sexually dimorphic lineage compositions in the Drosophila brain. Curr Biol 26:2583–2593. 10.1016/j.cub.2016.07.086 [DOI] [PubMed] [Google Scholar]
- Reznick DN, Ricklefs RE (2009) Darwin's bridge between microevolution and macroevolution. Nature 457:837–842. 10.1038/nature07894 [DOI] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R (2010) A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature 468:686–690. 10.1038/nature09554 [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA (1996) Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87:1079–1089. 10.1016/S0092-8674(00)81802-4 [DOI] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HD, Miyagi R, van der Sluijs I, Schneider MV, Maan ME, Tachida H, Imai H, Okada N (2008) Speciation through sensory drive in cichlid fish. Nature 455:620–626. 10.1038/nature07285 [DOI] [PubMed] [Google Scholar]
- Song HJ, Billeter JC, Reynaud E, Carlo T, Spana EP, Perrimon N, Goodwin SF, Baker BS, Taylor BJ (2002) The fruitless gene is required for the proper formation of axonal tracts in the embryonic central nervous system of Drosophila. Genetics 162:1703–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth HT. (1952) Mating behavior within the genus Drosophila (Diptera). Bull AMNH 99:399–474. [Google Scholar]
- Steele RH. (1986) Courtship feeding in Drosophila subobscura: I. The nutritional significance of courtship feeding. Animal Behav 34:1087–1098. 10.1016/S0003-3472(86)80168-3 [DOI] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ (2005) Neural circuitry that governs Drosophila male courtship behavior. Cell 121:795–807. 10.1016/j.cell.2005.04.026 [DOI] [PubMed] [Google Scholar]
- Takayanagi S, Toba G, Lukacsovich T, Ote M, Sato K, Yamamoto D (2015) A fruitless upstream region that defines the species specificity in the male-specific muscle patterning in Drosophila. J Neurogenet 29:23–29. 10.3109/01677063.2014.983101 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Murakami H, Ote M, Yamamoto D (2016) Clustered regulatory interspaced short palindromic repeats (CRISPR)-mediated mutagenesis and phenotype rescue by piggyBac transgenesis in a nonmodel Drosophila species. Insect Mol Biol 25:355–361. 10.1111/imb.12232 [DOI] [PubMed] [Google Scholar]
- Tootoonian S, Coen P, Kawai R, Murthy M (2012) Neural representations of courtship song in the Drosophila brain. J Neurosci 32:787–798. 10.1523/JNEUROSCI.5104-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui-Aoki K, Ito H, Ui-Tei K, Takahashi K, Lukacsovich T, Awano W, Nakata H, Piao ZF, Nilsson EE, Tomida J, Yamamoto D (2000) Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat Cell Biol 2:500–506. 10.1038/35019537 [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Edelaar P, Weissing FJ (2009) On the origin of species by natural and sexual selection. Science 326:1704–1707. 10.1126/science.1181661 [DOI] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ (2011) Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72:202–230. 10.1016/j.neuron.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Gailey DA, Berwald B, Ohshima S, Barnes PT, Hall JC (1997) Extended reproductive roles of the fruitless gene in Drosophila melanogaster revealed by behavioral analysis of new fru mutants. Genetics 147:1107–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Philipsborn AC, Liu T, Yu JY, Masser C, Bidaye SS, Dickson BJ (2011) Neuronal control of Drosophila courtship song. Neuron 69:509–522. 10.1016/j.neuron.2011.01.011 [DOI] [PubMed] [Google Scholar]
- von Philipsborn AC, Jörchel S, Tirian L, Demir E, Morita T, Stern DL, Dickson BJ (2014) Cellular and behavioral functions of fruitless isoforms in Drosophila courtship. Curr Biol 24:242–251. 10.1016/j.cub.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20:1377–1419. 10.1093/molbev/msg140 [DOI] [PubMed] [Google Scholar]
- Yamamoto D, Ishikawa Y (2013) Genetic and neural bases for species-specific behavior in Drosophila species. J Neurogenet 27:130–142. 10.3109/01677063.2013.800060 [DOI] [PubMed] [Google Scholar]
- Yamamoto D, Koganezawa M (2013) Genes and circuits of courtship behaviour in Drosophila males. Nat Rev Neurosci 14:681–692. 10.1038/nrn3567 [DOI] [PubMed] [Google Scholar]
- Yu JY, Kanai MI, Demir E, Jeffferis GS, Dickson BJ (2010) Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol 20:1602–1614. 10.1016/j.cub.2010.08.025 [DOI] [PubMed] [Google Scholar]