Abstract
Circular RNAs (circRNAs) are highly expressed in the CNS and regulate physiological and pathophysiological processes. However, the potential role of circRNAs in stroke remains largely unknown. Here, we show that the circRNA DLGAP4 (circDLGAP4) functions as an endogenous microRNA-143 (miR-143) sponge to inhibit miR-143 activity, resulting in the inhibition of homologous to the E6-AP C-terminal domain E3 ubiquitin protein ligase 1 expression. circDLGAP4 levels were significantly decreased in the plasma of acute ischemic stroke patients (13 females and 13 males) and in a mouse stroke model. Upregulation of circDLGAP4 expression significantly attenuated neurological deficits and decreased infarct areas and blood–brain barrier damage in the transient middle cerebral artery occlusion mouse stroke model. Endothelial-mesenchymal transition contributes to blood–brain barrier disruption and circDLGAP4 overexpression significantly inhibited endothelial-mesenchymal transition by regulating tight junction protein and mesenchymal cell marker expression. Together, the results of our study are illustrative of the involvement of circDLGAP4 and its coupling mechanism in cerebral ischemia, providing translational evidence that circDLGAP4 serves as a novel therapeutic target for acute cerebrovascular protection.
SIGNIFICANCE STATEMENT Circular RNAs (circRNAs) are involved in the regulation of physiological and pathophysiological processes. However, whether circRNAs are involved in ischemic injury, particularly cerebrovascular disorders, remains largely unknown. Here, we demonstrate a critical role for circular RNA DLGAP4 (circDLGAP4), a novel circular RNA originally identified as a sponge for microRNA-143 (miR-143), in ischemic stroke outcomes. Overexpression of circDLGAP4 significantly attenuated neurological deficits and decreased infarct areas and blood–brain barrier damage in the transient middle cerebral artery occlusion mouse stroke model. To our knowledge, this is the first report describing the efficacy of circRNA injection in an ischemic stroke model. Our investigation suggests that circDLGAP4 may serve as a novel therapeutic target for acute ischemic injury.
Keywords: blood–brain barrier integrity, circular RNA DLGAP4, endothelial-mesenchymal transition, ischemic stroke, miR-143, transient middle cerebral artery occlusion
Introduction
Stroke is a leading cause of death and long-term disability worldwide (Baltan et al., 2013; Vemuganti, 2013; Acosta et al., 2015; Denorme and De Meyer, 2016; Hu et al., 2017). Despite intensive research regarding neuroprotective agents designed to block one or more steps of the neuronal ischemia cascade, no drugs have been found to yield unequivocal improvements in clinical outcomes (Fisher and Schaebitz, 2000; Catanese et al., 2017). Currently, stroke therapies aim to establish early reperfusion through thrombolytic and/or mechanical recanalization of obstructed blood vessels. Ischemic stroke thrombolytic therapy efficacy is limited by a narrow time window and thrombolytic recombinant tissue plasminogen activator (rt-PA) side effects (Denorme et al., 2016; Snow, 2016; Alberts, 2017). Although several clinical trials aimed at identifying effective therapies have been performed (Malhotra et al., 2017), no pharmacological therapies that protect brain tissues are available. Therefore, novel treatment approaches that may enable clinicians and researchers to overcome the aforementioned side effects and extend the therapeutic window of time for existing treatments are urgently needed.
Cerebral ischemia that disrupts blood–brain barrier (BBB) integrity has been extensively studied in cellular and animal stroke models (Shi et al., 2017). Inflammatory mediators are potent contributors to endothelial inflammation and mediate changes in gene expression in endothelial cells such that the cells shift toward displaying an “inflamed” phenotype, which is accompanied by their consequent dedifferentiation into mesenchymal cells, a process known as endothelial-mesenchymal transition (EndoMT) (Potenta et al., 2008). Under these pathological conditions, these specialized brain endothelial cells lose their protective function, leading to neuroinflammation and neurodegeneration. Dedifferentiation of endothelial cells into mesenchymal cells has also been observed in brain disorders, particularly multiple sclerosis, which lends credence to speculation that EndoMT may contribute to BBB damage during multiple sclerosis pathogenesis (Troletti et al., 2016). However, none of these studies investigated the potential role of EndoMT in BBB dysfunction during stroke pathogenesis.
MicroRNAs (miRNAs) are involved in a number of CNS pathologies, including brain tumors, neurodegeneration, and multiple sclerosis (Liu et al., 2012; Amin et al., 2015; Tominaga et al., 2015; Wu et al., 2017). Moreover, miRNAs have been envisioned as a potential therapeutic strategy for treating neurovascular-dependent brain diseases because a number of miRNAs have been implicated in the regulation of endothelial cell function under normal and inflammatory conditions (Reijerkerk et al., 2013). A previous study demonstrated that miR-31 acts as a positive modulator of EndoMT in mouse pancreatic microvascular endothelial cells (Katsura et al., 2016). Our previous work indicated that silencing microRNA-143 (miR-143) protects BBB integrity by targeting p53-upregulated modulator of apoptosis (Bai et al., 2016). Mounting evidence indicates that miR-143 is associated with ischemic stroke and has clinical utility as an early diagnostic marker (Wei et al., 2016; Tiedt et al., 2017; Zeng et al., 2017). Moreover, miR-143 is involved in regulating cerebral vasculature in a rat experimental subarachnoid hemorrhage model (Müller et al., 2015). However, whether the involvement of miR-143 in BBB integrity occurs via regulation of EndoMT remains largely unknown.
Genome-wide bioinformatic analysis has revealed that the circular RNA DLGAP4 (circDLGAP4), which is derived from exons 8, 9, and 10 of the DLGAP4 gene, acts as an miR-143 sponge. Circular RNAs (circRNAs), generated from back-spliced exons, have recently been identified as a naturally occurring family of noncoding RNAs that are highly represented in the eukaryotic transcriptome (Jeck and Sharpless, 2014). circRNAs have been shown to be involved in the regulation of physiological and pathophysiological processes, including neural development and plasticity, Alzheimer's disease, heart senescence, hypertrophy and failure, and cell growth (You et al., 2015; Gruner et al., 2016; Wu et al., 2016; Zhao et al., 2016a; Floris et al., 2017). However, whether circDLGAP4 is involved in cerebral ischemic injury, particularly BBB damage, remains largely unknown. We hypothesize that circDLGAP4/miR-143 plays physiological and pathological roles at the cellular and tissue levels. Therefore, in this context, we aimed to investigate whether circDLGAP4/miR-143 is mechanistically responsible for stroke outcomes.
Materials and Methods
Reagents.
The circDLGAP4 lentivirus and the miR-143 lentivirus were purchased from Hanbio. TRIzol reagent was purchased from Takara (9109). 2,3,5-Triphenyltetrazolium chloride (TTC) was obtained from Sigma-Aldrich (T8877). HiScript Q RT SuperMix for qPCR (+gDNA wiper) (R123-01), HiScript Q Select RT SuperMix for qPCR (+gDNA wiper) (R133-01), and AceQ qPCR SYBR Green Master Mix (High ROX Premixed) (Q141-02) were purchased from Vazyme Biotech. The oligonucleotide primers used for RT-PCR were synthesized by Invitrogen.
Standard protocol approval and patient consent.
The ethics committee of the Affiliated Jiangsu Province Hospital approved this research protocol (approval ID: 2016-SR-235), and the participants or their legally authorized representatives provided written informed consent to participate in the study.
Human plasma collection.
Acute ischemic stroke (AIS) patients were recruited from among those admitted to the Emergency Department of the Affiliated Jiangsu Province Hospital between January 2016 and December 2016. Either MRI or CT of the brain confirmed the diagnosis of ischemic stroke. Experienced neurologists determined the severity of patients' neurological deficits using the National Institute of Health Stroke Scale within 24 h after stroke onset. Patients with intracerebral hemorrhages or unknown diseases were excluded from the study. The mean time of the enrollment blood draw was 16.5 ± 2.6 h after stroke onset. Nonstroke controls were recruited from among those patients who underwent an annual medical examination at the hospital. The demographic and clinical characteristics of the 26 stroke patients and 26 nonstroke controls enrolled in the study are provided in Table 1.
Table 1.
Baseline participant characteristicsa
| Characteristic | Control (n = 26) | AIS (n = 26) | p |
|---|---|---|---|
| Age (yr) | 65.4 ± 2.03 | 66.9 ± 2.30 | 0.627 |
| Gender (male), n (%) | 13 (50.0) | 13 (50.0) | 1.000 |
| BMI (kg/m2) | 23.9 ± 0.58 | 25.4 ± 0.50 | 0.055 |
| Smoking, n (%) | 6 (23.1) | 4 (15.4) | 0.482 |
| Drinking, n (%) | 3 (11.5) | 4 (15.4) | 1.000 |
| Hypertension, n (%) | 5 (19.2) | 13 (50.0) | 0.020 |
| Diabetes mellitus, n (%) | 5 (19.2) | 4 (15.4) | 1.000 |
| Total cholesterol (mm) | 4.28 ± 0.18 | 4.45 ± 0.20 | 0.526 |
| Triglycerides (mm) | 1.55 ± 0.15 | 1.49 ± 0.12 | 0.763 |
| LDL (mm) | 2.61 ± 0.13 | 2.72 ± 0.16 | 0.579 |
| HDL (mm) | 1.16 ± 0.06 | 1.30 ± 0.09 | 0.197 |
| Lpa (g/L)b | 0.20 (0.13–0.30) | 0.18 (0.11–0.34) | 0.634 |
| NIHSS score | |||
| 1–4 | 13 (50.0%) | ||
| 5–15 | 11 (42.3%) | ||
| 15–20 | 0 (0%) | ||
| 21–42 | 2 (7.7%) |
aBMI, Body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein; Lpa, lipoprotein (a); NIHSS, National Institute of Health Stroke Scale.
bLpa is presented as the median and interquartile range; other data are mean ± SEM
Animals and treatment.
All animal procedures were performed in strict accordance with the ARRIVE guidelines, and all animal protocols were approved by the Institutional Animal Care and Use Committee of the Medical School of Southeast University (approval ID: SYXK-2010.4987). Adult male C57BL/6J mice (25.0–30.0 g, 8–10 weeks old, RRID:IMSR_JAX:000664) were randomly assigned to experimental groups. The Mir-143+/− mice (034438-UCD, RRID:MMRRC_034438-UCD, Mutant Mouse Resource and Research Centers supported by the National Institutes of Health; https://www.mmrrc.org/catalog/cellLineSDS.php?mmrrc_id=34438) used herein were described in detail in our previous study (Zhang et al., 2016). Additionally, Tie2 promoter-driven GFP mice (N000274, Stock Tg (TIE2GFP)/Nju, Biomedical Research Institute of Nanjing University) were used in this study. All animals were housed under a constant temperature and humidity and under a 12 h light/12 h dark cycle, with the lights on at 7:00 A.M. Food and water were available ad libitum.
Transient middle cerebral artery (MCA) occlusion (tMCAO).
tMCAO was performed according to a previous report (Longa et al., 1989). Briefly, anesthesia was induced with 3% isoflurane mixed with 30% oxygen and 70% nitrous oxide in an anesthetic chamber and maintained with 1.5% isoflurane via a facemask. Cannulation of the femoral artery allowed monitoring of mean arterial blood pressure and arterial blood gases. Rectal temperature was maintained at 37.0 ± 0.5°C during surgery and the recovery period using a temperature-controlled heating pad. A 1-cm-long midline skin incision was made in the neck area, and the right common carotid artery was carefully dissected free from the surrounding nerves under a stereo dissecting microscope and tied off using 4-0 silk suture. The right external carotid artery was exposed and isolated from its small artery branches. The external carotid artery was ligated with 6-0 silk suture ∼3 mm distal to its origin, and an arteriotomy was then performed in the external carotid artery. Next, a silicone-rubber-coated 6-0 nylon filament (602356PK5Re, Doccol) was inserted into the external carotid artery and advanced for 9–10 mm, to the carotid bifurcation along the internal carotid artery and to the origin of the MCA. The incision in the neck was subsequently sutured, and the mouse was placed in a 35°C nursing box to recover from the anesthesia. After 1 h of occlusion, the filament was removed to restore the blood flow of the MCA region. In sham-operated mice, the internal carotid artery was surgically prepared for filament insertion, but the filament was not inserted. Monitoring of physiologic variables was performed in companion cohorts for all groups before tMCAO and 1 h after reperfusion. Sham-operated mice served as controls.
Cortical cerebral blood flow (CBF) measurements.
Cortical CBF was monitored using a moorFLPI-2 Full-Field Laser Perfusion Imager, following the manufacturer's instructions (Moor Instruments). Briefly, a charge-coupled device camera was placed above the head of an anesthetized mouse, and the intact skull surface was illuminated with a laser diode (785 nm) to allow laser penetration through the brain in a diffuse manner. CBF was measured in both cerebral hemispheres and recorded 15 min before tMCAO and throughout the ischemic period until 15 min after the onset of reperfusion. There was no direct sunlight or infrared radiation, and room temperature was maintained at 26°C. To evaluate changes in CBF, the region of interest (ROI) included the right cortical infarct region, which is posterior to the coronal suture and medial to the linear temporalis. Animals that did not show a reduction in CBF by at least 75% of the baseline level or died after ischemia induction (<10%) were excluded from further experiments (Shi et al., 2016).
In vivo MRI scanning.
In vivo MRI was performed using a 7.0 tesla small animal magnetic resonance scanner (PharmaScan 7T, Bruker). T2-weighted images were acquired to calculate the percentage of the infarct volume (Bai et al., 2014; Chang et al., 2015). The mice were anesthetized with 2% isoflurane delivered through a nose cone, and their respiratory rate and body temperature were monitored via a physiology monitor. T2-weighted imaging was conducted at 24 h after tMCAO using a 2D fast-spin echo sequence (2500/33 ms of repetition time/echo time, 1 average). Twelve 1-mm-thick axial slices, each with a 256 × 256 matrix and a 20 × 20 mm FOV, were positioned over the brain, excluding the olfactory bulb. The total scan time was 5 min. The percentage of the infarct volume was calculated based on T2-weighted imaging. Briefly, the percentage of the infarct volume was presented as the lesion volume/contralateral hemisphere volume covering the entire slices of T2-weighted images. The lesion volume was acquired based on the high-signal area of T2-weighted images from which the artifact of brain edema was subtracted. The infarct volume was traced and calculated using ImageJ software (RRID:SCR_003070, National Institutes of Health).
Measurements of neurological deficits.
Neurological deficits were evaluated by an experimenter blinded to the experimental groups 24 h after tMCAO. Measurements of neurological function were performed using the modified neurological severity score test (Li et al., 2000). The score was graded on a scale of 0–14 (normal score, 0; maximum point score, 14). A score of 1–4 indicates mild injury, 5–9 indicates moderate injury, and 10–14 indicates severe injury. For determining the scores for the severity of impairment, one score point is awarded for the inability to perform the test or for the lack of a tested reflex; thus, higher scores indicate more severe injuries.
TTC staining and measurement of cerebral infarction.
Infarct volume was evaluated at 24 h after tMCAO. The mice were anesthetized with 1% pentobarbital sodium and perfused with 0.01 m PBS. The mice were then decapitated, and their brains were collected and immediately frozen at −20°C for 6 min. The brains were subsequently sectioned coronally into 1-mm-thick slices using a brain matrix on ice. Next, the brain slices were incubated in 1% TTC at 37°C for 10 min and fixed in 4% PFA to determine the size and extent of the infarction. Images were analyzed using ImageJ software (RRID:SCR_003070, National Institutes of Health, Bethesda, MD). To correct for brain swelling, the infarct area was determined by subtracting the area of noninfarcted tissue in the ipsilateral hemisphere from that in the intact contralateral hemisphere. Infarct volume was calculated via the integration of infarct areas for all slices of each brain (Arumugam et al., 2006).
Brain endothelial cell culture.
The mouse brain endothelial cell line bEnd.3 was obtained from ATCC (CRL-2299, RRID:CVCL_0170) (Lee et al., 2012) and cultured in humidified 5% CO2 at 37°C in DMEM (30-2002, ATCC) supplemented with 10% (v/v) FBS (10099-141, Invitrogen), penicillin (100 U/ml), and streptomycin (100 U/ml) (10378-016, Invitrogen). The mouse brain endothelial bEnd.3 cells were used in experiments after 4–10 passages.
Microinjection of the circDLGAP4 lentivirus.
The lateral ventricles of 8-week-old C57BL/6J mice were microinjected with either a circControl-GFP lentivirus or circDLGAP4-GFP lentivirus (5 μl of 2 × 108 viral genome/μl, Hanbio) at the following microinjection coordinates: 0.3 mm behind the bregma and 1.0 mm lateral from the sagittal midline, at a depth of 2.2 mm from the skull surface. To evaluate the effects of circDLGAP4 on the tMCAO mouse stroke model, mice were divided into the following four groups 2 weeks after lentivirus microinjection: circControl+Sham, circDLGAP4+Sham, circControl+tMCAO, and circDLGAP4+tMCAO.
Immunofluorescence staining.
Mice were microinjected with the circControl or circDLGAP4 lentivirus for 2 weeks, followed by tMCAO surgery. Mouse brain sections that encompassed the MCA region were cut into 30 μm slices, and every sixth section was spaced 180 μm apart with a cryostat. To detect blood vessels, frozen sections were permeabilized with 0.3% Triton X-100 in PBS for 15 min. After blocking with 10% normal goat serum (NGS) in 0.3% Triton X-100 for 1 h, sections were incubated overnight with a mouse anti-CD31 antibody (ab24590, RRID:AB_448167, 1:500, Abcam). Finally, sections were incubated with AlexaFluor-594 goat anti-rabbit IgG (A-11037, RRID:AB_2534095, 1:1000, Thermo Fisher Scientific) and mounted on poly-L-lysine-coated glass slides.
Western blot (WB) analysis.
Proteins were extracted in RIPA lysis buffer (P0013B, Beyotime), separated on SDS polyacrylamide gels (8% and 12%), and electrophoretically transferred onto PVDF membranes according to our previously described methods (Zhang et al., 2017). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with Tween 20, probed with antibodies recognizing claudin-5 (ab15106, RRID:AB_301652, 1:1000, Abcam), occludin (33-1500, RRID:AB_87033, 1:1000, Invitrogen), ZO-1 (40-2300, RRID:AB_2533457, 1:1000, Thermo Fisher Scientific), collagen I (Col I; BS1530, RRID:AB_1662101, 1:1000, Bioworld), collagen III (Col III; 22734-1-AP, 1:1000, Proteintech), α-smooth muscle actin (α-SMA; 14395-1-AP, RRID:AB_2223009, 1:1000, Proteintech), and homologous to the E6-AP C-terminal (HECT) domain E3 ubiquitin protein ligase 1 (HECTD1; 20605-1-AP, RRID:AB_10732804, 1:1000, Proteintech) overnight at 4°C before incubation with an HRP-conjugated goat anti-mouse IgG secondary antibody (7076P2, RRID:AB_330924, 1:2000, Cell Signaling Technology) and an HRP-conjugated goat anti-rabbit IgG secondary antibody (7074P2, 1:2000, RRID:AB_2099233, Cell Signaling Technology). Signals were detected by chemiluminescence and imaged using a Microchemi 4.2 (DNR Bio-Imaging Systems) digital image scanner, as described in our previous study (Zhang et al., 2016). Individual protein bands were quantified by densitometry using ImageJ software (RRID:SCR_003070, National Institutes of Health).
RT-PCR.
RT-PCR analysis of mature miR-143 was performed using an Applied Biosystems Real-Time PCR System. First, total RNA was extracted using the TRIzol reagent and treated with gDNA wiper. Then, the RNA was reverse transcribed with the stem-loop RT primer (RiboBio) using the HiScript Q Select RT SuperMix for qPCR Kit (R133-01, Vazyme Biotech). Next, the RT products were quantified using AceQ qPCR SYBR Green Master Mix (Q141-02, Vazyme Biotech). The levels of miR-143 determined via qRT-PCR were normalized to the level of U6. Specific primers for mature miR-143 and U6 were obtained from RiboBio. circRNAs and mRNAs were reverse transcribed using the HiScript Q RT SuperMix for qPCR Kit (R123-01, Vazyme Biotech) and quantified via SYBR Green RT-PCR. The results were standardized to the control values of 18S. The following primers were used: human circDLGAP4 (forward primer: 5′-ACGGCTACTGGTTCCTAAAGC-3′; reverse primer: 5′-GGGGTCTTCTTATACGCCACT-3′), mouse circDLGAP4 (forward primer: 5′-TGCCAGATGGACAAGGAGACC-3′; reverse primer: 5′-CTGGACGGTGACTGAGATGAA-3′), and 18S (forward primer: 5′-CAGCCACCCGAGATTGAGCA-3′; reverse primer: 5′-TAGTAGCGACGGGCGGTGTG-3′).
Northern blots.
Total RNA was extracted from the brain tissues of mice treated with tMCAO using Trizol reagent (9109, Takara). Northern blotting was performed according to the manufacturer's protocol (DIG Northern Starter Kit, 12039672910, Roche). In brief, prepared total RNA (50 μg) was subjected to electrophoresis in a 5% urea denatured PAGE gel. The RNA was then transferred to a Hybond-N+ nylon transfer membrane (RPN303B, GE Healthcare). The membranes were UV-cross-linked and prehybridized at 38°C for at least 2 h with DIG Easy Hyb Granules (11603558001, Roche), followed by hybridization with a circDLGAP4 digoxigenin-labeled probe at 20 nm (Invitrogen) at 38°C overnight. A DIG Northern Starter Kit (12039672910, Roche) was used to detect the bound RNA molecules.
FISH.
According our previously described methods (Yao et al., 2014), mouse brain endothelial bEnd.3 cells cultured on coverslips were fixed with 4% PFA for 20 min and incubated in PBS overnight at 4°C, followed by processing to detect circDLGAP4 or miR-143 expression. Next, the cells were permeabilized with 0.25% Triton X-100 in PBS for 15 min and prehybridized in hybridization buffer (50% formamide, 10 mm Tris-HCl, pH 8.0, 200 μg/ml yeast tRNA, 1× Denhardt's solution, 600 mm NaCl, 0.25% SDS, 1 mm EDTA, and 10% dextran sulfate) for 1 h at 37°C. Then hybridization buffer containing a biotin-labeled circDLGAP4 probe (50 nm, Invitrogen) and a digoxigenin-labeled miR-143 probe (38515-15, 25 nm, Exiqon) was heated to 65°C for 5 min and dripped onto the coverslips, followed by hybridization at 37°C overnight. The next day, the coverslips were washed three times in 2× SSC and twice in 0.2× SSC at 42°C and then blocked with 1% BSA and 3% NGS in PBS for 1 h at room temperature. The coverslips were subsequently incubated with an HRP-conjugated anti-digoxigenin antibody (11207733910, RRID:AB_514500, 1:200, Roche Diagnostics) and FITC-streptavidin (434311, 1:200, Invitrogen) overnight at 4°C. After the coverslips were washed three times with TBS, they were incubated with a TSA Cy5 kit (NEL74500KT, PerkinElmer) for 10 min at room temperature. Then the coverslips were washed twice with PBS and incubated with Hoechst 33342 (H1399, Invitrogen) for 10 min at room temperature to visualize nuclei. The sections were finally washed once with DEPC water and mounted with 30% glycerine. Immunofluorescence images were captured via microscopy (DP73, Olympus). The circDLGAP4 probe sequence, biotinylated at the 5′ end, was 5′-AAAACTAGGCATGATGAACCTTCTTCAGAGAGGTT-3′; and the digoxigenin-labeled miR-143 probe sequence was 5′-GAGCTACAGTGCTTCATCTCA-3′.
Pull-down assay with biotinylated miR-143.
A total of 2 × 106 HEK293T or mouse endothelial bEnd.3 cells were seeded 1 d before transfection. On the following day, the cells were transfected with 3′-end-biotinylated miR-143 or control RNA (GenePharma) at a final concentration of 50 nm for 36 h. Then, the cells were washed with PBS, briefly vortexed, and incubated in lysis buffer [20 mm Tris, pH 7.5, 200 mm NaCl, 2.5 mm MgCl2, 0.05% Igepal (18896, Sigma-Aldrich) containing 60 U/ml Superase-In (AM2694, Invitrogen) and 1 mm DTT (43816, Sigma-Aldrich); and protease inhibitors] on ice for 10 min. The lysates were precleared by centrifugation, and 50 μl aliquots of the samples were prepared for input. The remaining lysates were incubated with M-280 streptavidin magnetic beads (11205D, Invitrogen), which were coated with yeast tRNA (15401011, Invitrogen) in advance to prevent nonspecific binding of RNA and protein complexes. The beads were incubated at 4°C for 2.5 h and then washed twice with ice-cold lysis buffer, twice with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 150 mm NaCl), and once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 500 mm NaCl). The bound RNA was purified using TRIzol to measure circDLGAP4 levels.
Evans blue extravasation assay.
As described in our previous study (Yao et al., 2011), we evaluated cerebrovascular integrity in C57BL/6J mice. To assess cerebrovascular permeability, we injected a 2% Evans blue solution (4 ml/kg) into the lateral tail vein. After 4 h, the mice were anesthetized and perfused through the left ventricle with ice-cold PBS, pH 7.4, to remove the intravascular Evans blue. The brains were harvested, sliced, and scanned before being homogenized in PBS (1:10 g/v). The homogenates were precipitated in 15% trichloroacetic acid (1:1 v/v) and centrifuged at 1000 × g for 10 min. The pH was adjusted by adding 125 μl of 5 m sodium hydroxide to 500 μl of each supernatant aliquot. Evans blue concentrations were measured spectrophotometrically at 620 nm.
Oxygen glucose deprivation and reoxygenation (OGD/R) treatment.
Mouse brain endothelial bEnd.3 cells were cultured with deoxygenated DMEM without glucose and FBS (11966-025, Invitrogen) in a sealed chamber (MIC-101, Billups-Rothenburg) loaded with mixed gas consisting of 5% CO2 and 95% N2 for 5 min at 25 L/min. The chamber was placed in a water-jacketed incubator (Forma, Thermo Fisher Scientific). The cells were incubated at 37°C for 4 h before reperfusion. For reperfusion, the cells were refreshed with normal DMEM (30-2002, ATCC) containing 10% FBS and placed in a CO2 incubator (95% air and 5% CO2) for 24 h. The cells in the control group were cultured with normal DMEM and 10% FBS for the same amount of time.
Colocalization analysis.
The sections that encompassed the MCA region were cut into 30 μm slices, and every sixth section was spaced 180 μm apart with a cryostat. The sections were subsequently incubated with 0.3% Triton X-100 in PBS for 15 min and blocked with 10% NGS in 0.3% Triton X-100 for 1 h at room temperature. Next, the sections were incubated with a rabbit anti-α-SMA antibody (14395-1-AP, RRID:AB_2223009, 1:50, Proteintech) overnight at 4°C. On the following day, the sections were washed and incubated with AlexaFluor-594 goat anti-rabbit IgG (A-11037, RRID:AB_2534095, 1:250, Thermo Fisher Scientific) for 1 h. After a final wash step with PBS, the sections were mounted on poly-L-lysine-coated glass slides and numerically coded. The colocalization of green (Tie2-GFP: endothelial cell marker) and red (α-SMA: mesenchymal cell marker) fluorescence was evaluated by experimenters blinded to the experimental conditions for each sample. Immunofluorescence images were captured via microscopy in the ROI (LSM700, Carl Zeiss). Three sections were analyzed for each brain, and five ROIs were randomly selected from the inner infarct border in each section. Fifteen vessels (diameter size 5–10 μm) were randomly selected for colocalization analysis in each group. Quantification was accomplished by acquiring grayscale images and integrating fluorescence intensities into the images using ImageJ software (RRID:SCR_003070, National Institute of Health). GFP and α-SMA colocalization was quantified using Pearson correlation coefficient and Manders overlap coefficient, as described in detail in a previous study (Costes et al., 2004; Zinchuk et al., 2007; French et al., 2008; Shi et al., 2016). Descriptive data are presented for the colocalization coefficient in each group.
Luciferase activity assays.
The 3′-untranslated region (UTR) of the 829 bp human Hectd1 gene containing the putative miR-143 target site was PCR-amplified from human genomic DNA using forward (5′-GCGGCTCGAGGAGCTTTGAAGTGCAATGGG-3′) and reverse (5′-AATGCGGCCGCATGACACAGTTCTGTACGCT-3′) primers, and the DNA fragment was cloned into the XhoI and NotI sites on the 3′ end of the Rluc gene in a pmiR-RB-REPORT vector. For the pmiR-RB-Hectd1-3′-UTR-miR-143-target-mutant vector, the miR-143 target site (CATCTCA) within the Hectd1 3′-UTR was changed to CTAGAGT via PCR mutagenesis with the primers Hectd1 miR-143T-F (5′-TTAATGAGGTAGAGTAGCTTTTTCTTTTCCTTT-3′) and Hectd1 miR-143T-R (5′-AAAAAGCTACTCTACCTCATTAATTCTCTATGT-3′). Briefly, HEK293T cells were transfected with an miR-143 OE pLV-[hsa-miR-143] vector and a target plasmid, pmiR-RB-Hectd1-3′-UTR or pmiR-RB-Hectd1-3′-UTR-miR-143-target-mutant. An miR control pLV-[miR-control] plasmid was used as a negative control. Luciferase activity was determined at 24 h after transfection, and a reporter assay was performed using Dual-Glo Luciferase Assay System (Promega, E2920). Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as a percentage of the control (n > 3, determined from 5 wells per sample).
Upregulation of target DNA using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technologies.
Mouse brain endothelial bEnd.3 cells were transiently transfected with Control CRISPR Activation Plasmid (sc-437275, Santa Cruz Biotechnology) and Hectd1 CRISPR Activation Plasmid (m) (sc-431500-CRISPR Activation Plasmid [ACT], Santa Cruz Biotechnology) according to the CRISPR Activation Plasmid Transfection Protocol (Santa Cruz Biotechnology) to examine downstream effects. The transfection efficiency was determined by WB.
Experimental design and statistical analysis.
For all comparisons between WT and Mir-143+/− mice, littermates were used. All mice were male, and experimental groups were age-matched. Saple size is reported in the figure legends. All statistical analyses were performed using SigmaPlot 12.0 (Systat Software, RRID:SCR_003210).
For comparison between two groups, the Shapiro–Wilk Test and Equal Variance Test were used to evaluate the normality and equality of the variances, respectively. If the Shapiro–Wilk Test and Equal Variance Test passed, the two groups were compared using Student's t test. The differences between groups with respect to neurological deficit (Mir-143+/− vs WT) and infarct size were examined by Student's t test. The expression of circDLGAP4 in circControl and circDLGAP4-microinjected groups in vivo as well as circControl and circDLGAP4 lentivirus-transduced groups in vitro was examined by Student's t test. The same analysis was also performed to examine differences between the Sham and tMCAO groups, control, and OGD/R-treated groups for HECTD1 expression and the effects of mimic miR-143/anti-miR-143 or Hectd1 ACT on the expression of HECTD1. The same analysis was performed to analyze the colocalization of GFP and α-SMA (Manders overlap coefficient) as well as the result about the luciferase activity assay and pull-down assay. If the Shapiro–Wilk Test or Equal Variance Test failed, the Mann–Whitney Test was used for the statistical analysis of two groups. The analysis of differences in miR-143 and circDLGAP4 levels between control and AIS patients was performed with the Mann–Whitney Test. The same analysis was performed to analyze differences between groups with respect to neurological deficits (circDLGAP4-microinjected group vs and circControl-microinjected group) and the colocalization of GFP and α-SMA (Pearson correlation coefficient).
For the comparison of multiple groups, the Shapiro–Wilk Test for normal distribution and the Equal Variance Test for equal variances were performed to determine the variances between multiple groups, and one-way ANOVA, followed by the Holm–Sidak Test, was used to test statistical significance. Analysis of differences in BBB damage between Sham and tMCAO mice in WT or Mir-143+/− mice was performed with a one-way ANOVA followed by the Holm–Sidak Test. The same analysis was performed to analyze differences between groups in terms of tight junction proteins (TJPs) (claudin-5, occluding, and ZO-1) and mesenchymal cell markers (Col I, Col III, and α-SMA) as well as differences between groups with respect to miR-143 and circDLGAP4 levels in tMCAO mice at 12 and 24 h after reperfusion.
Results
miR-143 is upregulated in the plasma of AIS patients and tMCAO mice
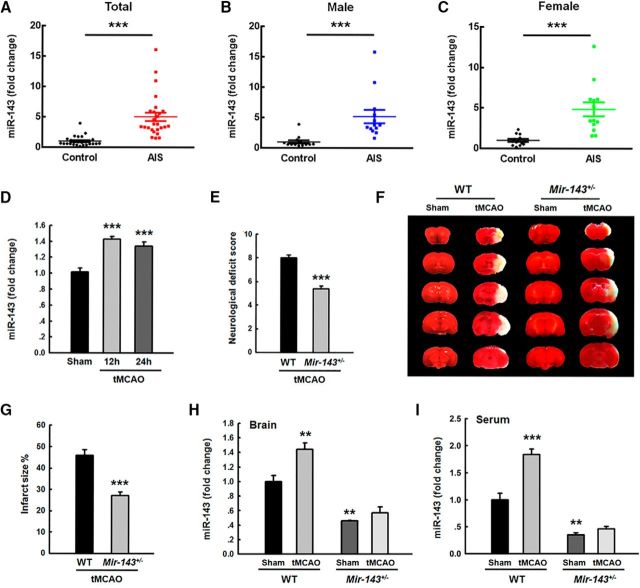
Our previous work demonstrated that silencing of miR-143 exerted a protective effect against cerebrovascular damage induced by methamphetamine (Bai et al., 2016). However, the potential role of miR-143 in stroke remains largely unknown. To determine whether miR-143 is involved in stroke, we examined miR-143 levels in AIS patients. Thus, plasma samples were collected from these patients (age 66.9 ± 2.30 years), whose socio-demographic and clinical characteristics and National Institute of Health Stroke Scale scores are listed in Table 1. Twenty-six patients with AIS were enrolled in the study, and 26 nonstroke volunteers served as controls. As shown in Figure 1A, miR-143 levels were significantly increased in these subjects (p < 0.001, n = 26) compared with those of age- and gender-matched cognitively healthy controls (n = 26). There were no gender-based differences in miR-143 levels, as shown in Figure 1B, C. Consistent with this finding, we noted increased ipsilateral miR-143 expression in mice subjected to tMCAO compared with that in sham-treated mice (Fig. 1D). To further confirm the role of miR-143 in cerebral ischemia-induced injury, we examined the brain infarct volumes of both WT and Mir-143+/− mice in a TTC assay at 24 h after tMCAO surgery. Consistent with our findings regarding brain damage, neurological deficit scores were significantly decreased in Mir-143+/− mice compared with those in WT mice at 24 h after tMCAO surgery (Fig. 1E). Infarct sizes were also significantly decreased in Mir-143+/− mice compared with those in WT mice (Fig. 1F,G). As expected, there were significant decreases in miR-143 levels in Mir-143+/− mice in the brain and serum compared with the level in the WT group under sham conditions. Interestingly, tMCAO treatment significantly increased the level of miR-143 levels in WT mice but not in Mir-143+/− mice (Fig. 1H,I). These results indicate that silencing of miR-143 exerts protective effects against cerebral ischemia-induced injury in tMCAO mice.
Figure 1.
miR-143 is upregulated in the plasma of AIS patients and tMCAO mice. A, miR-143 expression levels in the serum of AIS patients were increased 5.01-fold compared with that in healthy controls. n = 26 individuals/group. ***p < 0.001, AIS versus control (Mann–Whitney Test). B, C, miR-143 expression levels in the serum of male (B) and female (C) AIS patients were increased compared with that in male and female healthy controls. n = 13 individuals/group. ***p < 0.001, male AIS versus male control. ***p < 0.001, female AIS versus female control (Mann–Whitney Test). D, Examination of miR-143 expression in brain tissues isolated from sham and tMCAO mice at 12 and 24 h after reperfusion (ischemia/reperfusion); n = 6 mice/group. F(2,16) = 21.754: ***p < 0.001, 12 h tMCAO versus Sham. ***p < 0.001, 24 h tMCAO versus Sham (one-way ANOVA followed by the Holm–Sidak Test). E, Neurological deficit scores were significantly decreased in Mir-143+/− mice compared with those in WT mice at 24 h after tMCAO surgery. n = 8 mice/group. ***p < 0.001, Mir-143+/− versus WT (Student's t test). F, G, Infarct volumes were significantly decreased in Mir-143+/− tMCAO mice compared with those in WT tMCAO mice. Representative TTC-stained sections of mouse brains from WT or Mir-143+/− mice at 24 h after tMCAO surgery (F). Viable tissue is stained red, whereas damaged ischemic brain tissue appears unstained/white. Infarct volumes were quantified via TTC staining using ImageJ software (G). n = 5 mice/group. ***p < 0.001, Mir-143+/− versus WT (Student's t test). H, Expression of miR-143 in the ipsilateral side of Mir-143+/− mice under Sham and tMCAO conditions. n = 6 mice/group. F(3,8) = 38.341: **p = 0.008, WT tMCAO versus WT Sham. **p = 0.003, Mir-143+/− Sham versus WT Sham (one-way ANOVA followed by the Holm–Sidak Test). I, Expression of miR-143 in the serum of Mir-143+/− mice under sham and tMCAO conditions. F(3,8) = 64.673: ***p < 0.001, WT tMCAO versus WT Sham. **p = 0.002, Mir-143+/− Sham versus WT Sham (one-way ANOVA followed by the Holm–Sidak Test).
Silencing of miR-143 ameliorates the disruption of cerebrovascular integrity in tMCAO mice
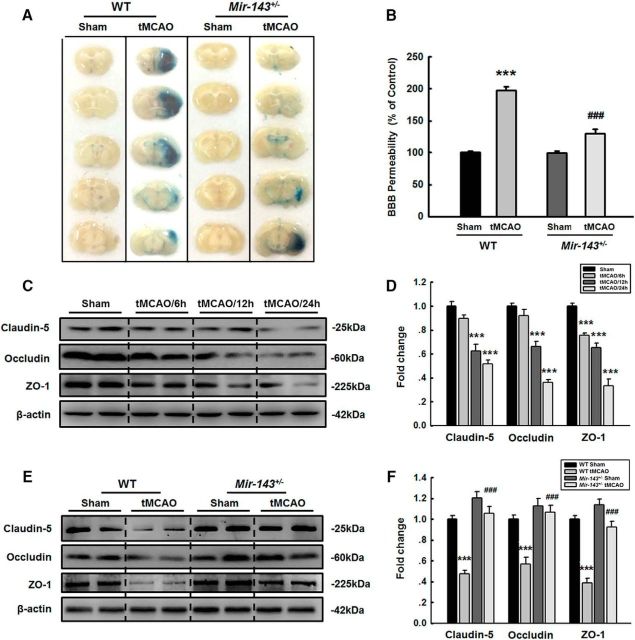
Given that cerebral ischemia and stroke induced damage to cerebrovascular integrity, we next examined the role of miR-143 in cerebrovascular injury in vivo by studying Evans blue extravasation in tMCAO mice. As shown in Figure 2A, B, Evans blue extravasation was decreased in Mir-143+/− mice subjected to tMCAO treatment but not in WT mice subjected to tMCAO treatment, demonstrating that miR-143 plays a role in regulating cerebrovascular integrity in vivo. We then examined how miR-143 regulates the ipsilateral expression of TJPs, including claudin-5, occludin, and ZO-1. WT mice subjected to tMCAO treatment displayed decreased TJP expression (Fig. 2C,D), and this change was significantly ameliorated in Mir-143+/− mice at 24 h after tMCAO surgery (Fig. 2E,F).
Figure 2.
Silencing of miR-143 ameliorates the disruption of cerebrovascular integrity in tMCAO mice. A, Representative images of extravasated Evans blue staining of the entire brains of WT and Mir-143+/− mice. B, Cerebrovascular permeability in WT or Mir-143+/− mice was determined by measuring the concentrations of brain-extracted Evans blue using spectrophotometry at 620 nm. n = 6 mice/group. F(3,20) = 97.964: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, Mir-143+/− tMCAO versus WT tMCAO (one-way ANOVA followed by the Holm–Sidak Test). C, D, Cortical TJP expression levels decreased significantly at 12 and 24 h after tMCAO surgery. TJP expression levels were determined via WB analysis (C) and quantified through densitometric analysis (D). n = 6 mice/group. Claudin-5, F(3,20) = 32.327: p = 0.088, tMCAO/6 h versus Sham. ***p < 0.001, tMCAO/12 h versus Sham. ***p < 0.001, tMCAO/24 h versus Sham. Occludin, F(3,20) = 61.330: p = 0.150, tMCAO/6 h versus Sham. ***p < 0.001, tMCAO/12 h versus Sham. ***p < 0.001, tMCAO/24 h versus Sham. ZO-1, F(3,20) = 55.018: ***p < 0.001, tMCAO/6 h versus Sham. ***p < 0.001, tMCAO/12 h versus Sham. ***p < 0.001, tMCAO/24 h versus Sham (one-way ANOVA followed by the Holm–Sidak Test). Two representative immunoblots were presented from 6 mice/group. E, F, The decrease in ipsilateral TJP expression observed at 24 h after tMCAO surgery was ameliorated in Mir-143+/− mice. TJP expression levels were determined via WB analysis (E) and quantified through densitometric analysis (F). n = 6 mice/group. Claudin-5, F(3,20) = 38.884: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, Mir-143+/− tMCAO versus WT tMCAO. Occludin, F(3,20) = 17.630: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, Mir-143+/− tMCAO versus WT tMCAO. ZO-1, F(3,20) = 49.511: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, miR-143+/− tMCAO versus WT tMCAO (one-way ANOVA followed by the Holm–Sidak Test). Two representative immunoblots were presented from 6 mice/group.
circDLGAP4 functions to bind miR-143 and is downregulated in the plasma of AIS patients and tMCAO mice
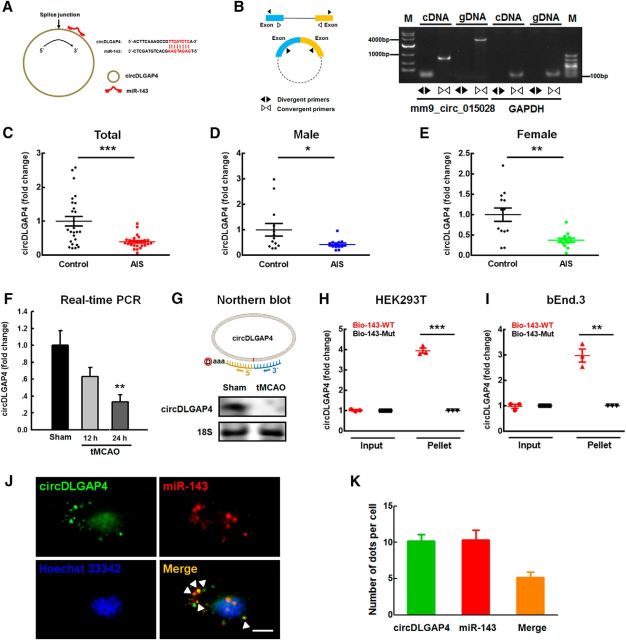
Accumulating evidence indicates that the circRNAs that are highly expressed in mouse tissues and in human cell lines act as ceRNA sponges that interact with miRNAs and influence the expression of their target proteins (Hansen et al., 2013). We next sought to determine which circRNA might act as an endogenous miR-143 sponge to inhibit miR-143 activity. We found that circDLGAP4 contains one miR-143 target site using the bioinformatics program RNAhybrid (Fig. 3A). By using divergent primers specific to mm9_circ_015028, which was designated circDLGAP4 (Fig. 3B), we found that circDLGAP4 levels in these AIS subjects were significantly decreased (p < 0.01, n = 26) compared with those in age- and gender-matched cognitively healthy controls (n = 26) (Fig. 3C). There were no gender-based differences in circDLGAP4 levels, as shown in Figure 3D, E. Consistent with these clinical findings, circDLGAP4 expression levels were observed to be decreased in the ipsilateral hemispheres of mice with tMCAO compared with those in the sham group at 24 h after reperfusion (Fig. 3F). This was further confirmed by Northern blotting (Fig. 3G). Furthermore, biotin-coupled miR-143 mimics were used to assess whether miR-143 could pull down circDLGAP4. More circDLGAP4 enrichment was detected in the miR-143-captured fraction than in the fractions in which mutations disrupted base-pairing between circDLGAP4 and miR-143 in both HEK293T and bEnd.3 cells (Fig. 3H,I). This finding was further confirmed via FISH, demonstrated by colocalization between circDLGAP4 and miR-143 in cytoplasm (Fig. 3J,K).
Figure 3.
circDLGAP4 binds miR-143 and is downregulated in the plasma of AIS patients and tMCAO mice. A, Left, circDLGAP4 contains one site that is complementary to miR-143 according to the bioinformatics program RNA hybrid. Right, A biotin-coupled miR-143 mutant. B, Divergent primers amplified circDLGAP4 from cDNA, but not genomic DNA (gDNA). GAPDH, linear control; M, marker. C, Examination of circDLGAP4 levels in the plasma of healthy controls and AIS patients via RT-PCR. circDLGAP4 levels were decreased in AIS patients by 0.6-fold the levels observed in healthy controls. n = 26 individuals/group. ***p < 0.001, AIS versus control (Mann–Whitney Test). D, E, circDLGAP4 levels in expression the plasma of male (D) and female (E) AIS patients were decreased compared with those in male and female healthy controls. n = 13 individuals/group. *p = 0.046, male AIS versus male control (Mann–Whitney Test). **p = 0.003, female AIS versus female control (Mann–Whitney Test). F, Effects of stroke on circDLGAP4 expression in the brain tissues of tMCAO mice. circDLGAP4 expression was determined at 12 and 24 h after tMCAO surgery via qRT-PCR. n = 8 mice/group. F(2,21) = 6.907, p = 0.102: 12 h tMCAO versus Sham. **p = 0.004, 24 h tMCAO versus Sham (one-way ANOVA followed by the Holm–Sidak Test). G, The expression of circDLGAP4 decreased in the mouse tMCAO mouse stroke model compared with that in sham group according to Northern blotting. H, I, circDLGAP4 was pulled down with biotinylated WT miR-143 (Bio-miR-143-WT) or mutant miR-143 (Bio-miR-143-mut). Biotinylated WT miR-143 (Bio-143-WT) or mutant miR-143 (Bio-143-mut) was transfected into HEK293T cells (H) and bEnd.3 cells (I). After streptavidin capture, circDLGAP4 and GAPDH mRNA levels were quantified via RT-PCR, and the relative immunoprecipitate/input ratios were plotted. Data are mean ± SEM of three independent experiments. HEK293T cells: ***p < 0.001, Bio-143-Mut Pellet versus Bio-143-WT Pellet. bEnd.3 cells: **p = 0.002, Bio-143-Mut Pellet versus Bio-143-WT Pellet (Student's t test). J, K, FISH hybridization of mature miR-143 and circDLGAP4 in brain endothelial cells (J) and quantitation of colocalization (K). White arrowheads indicate the colocalization of miR-143 and circDLGAP4. Green represents circDLGAP4. Red represents miR-143. Blue represents Hoechst 33342. Scale bar, 5 μm.
Overexpression of circDLGAP4 levels in the brains of stroke mice via delivery of recombinant circDLGAP4 RNA using a lentivirus vector
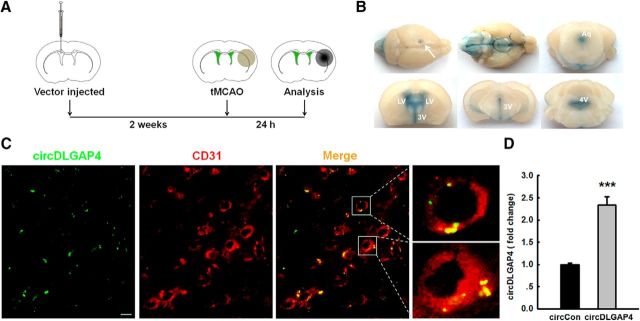
We next sought to verify the role of circDLGAP4 in vivo by microinjecting circDLGAP4 lentiviruses into the lateral ventricles of C57BL/6J mice. The lateral ventricles of C57BL/6J mice were microinjected with either the circControl-GFP lentivirus or the circDLGAP4-GFP lentivirus, and the mice were monitored to determine the role of circDLGAP4 in stroke pathogenesis, as illustrated in Figure 4A. To track the distribution of the lentiviruses, we injected the same volume of Evans blue into the left lateral ventricles of the mice, and the injected dye diffused throughout both hemispheres and the brainstem (Fig. 4B). We also examined the efficacy of circDLGAP4-GFP lentivirus transduction in vivo. As shown in Figure 4C, GFP was widely expressed around blood vessels and colocalized with CD31-positive cells. As expected, increased expression of circDLGAP4 was observed in circDLGAP4-injected mice compared with that in circControl-injected mice (Fig. 4D).
Figure 4.
Overexpression of circDLGAP4 via the delivery of recombinant circDLGAP4 RNA using lentiviral vectors. A, Illustration of the in vivo locations of GFP-labeled circDLGAP4 microinjection and the experimental procedure. Mice were microinjected with the circControl or circDLGAP4 lentivirus for 2 weeks, followed by tMCAO surgery. B, Distribution and infiltration of Evans blue, which was injected into the lateral ventricle (LV). A total of 5 μl of Evans blue was injected into the left lateral ventricles of the mice, and its distribution was observed at 2 h after injection. Evans blue was distributed throughout the entire brain after lateral ventricle injection. Arrow indicates the injection site. The infiltration and appearance of Evans blue in the bilateral ventricles, third ventricle (3V), aqueduct (Aq), fourth ventricle (4V), and adjacent regions. C, GFP lentiviruses were distributed in the microvessels of C57BL/6J mice after lentivirus microinjection. Representative images with CD31 immunostaining obtained 2 weeks after the microinjection of circDLGAP4 into the lateral ventricle. Green represents circDLGAP4. Red represents CD31. Scale bar, 5 μm. D, LV-circDLGAP4-intraventricular injection efficiency. circDLGAP4 expression was examined by RT-PCR. n = 5 mice/group. ***p < 0.001, circDLGAP4 versus circControl (Student's t test).
Overexpression of circDLGAP4 exerts protective effects against cerebral ischemia-induced injury in tMCAO mice
Two weeks after lentivirus injection, mice were subjected to tMCAO. After 24 h of reperfusion, neurological deficit scores were significantly decreased in circDLGAP4-injected mice compared with those in circControl-injected mice (Fig. 5A). We evaluated infarct sizes using MRI to determine the extent of brain damage within the circControl-injected injury territory (50.6% infarct volume) and the circDLGAP4-injected injury territory (34.8% infarct volume) to determine the initial damage suffered by each mouse (Fig. 5B). Compared with circControl-injected mice, circDLGAP4-injected mice exhibited significantly decreased infarct sizes that were 31.2% smaller than those of their counterparts (Fig. 5C). To verify these observations further, we determined brain infarct volumes by TTC assay at 24 h after tMCAO surgery. Infarct sizes were significantly decreased in circDLGAP4-injected mice in the tMCAO group compared with those in circControl-injected mice (Fig. 5D,E). We next examined the role of circDLGAP4 in cerebrovascular injury in vivo by subjecting mice to tMCAO. The procedure disrupted cerebrovascular injury in the circControl-injected group, but its effects were significantly ameliorated in the circDLGAP4-injected group (Fig. 5F,G). We then examined how circDLGAP4 regulated the ipsilateral expression of TJPs, including claudin-5, occludin, and ZO-1. circControl mice subjected to tMCAO treatment displayed decreased TJP expression, and this change was significantly ameliorated in circDLGAP4-injected mice (Fig. 5H,I).
Figure 5.
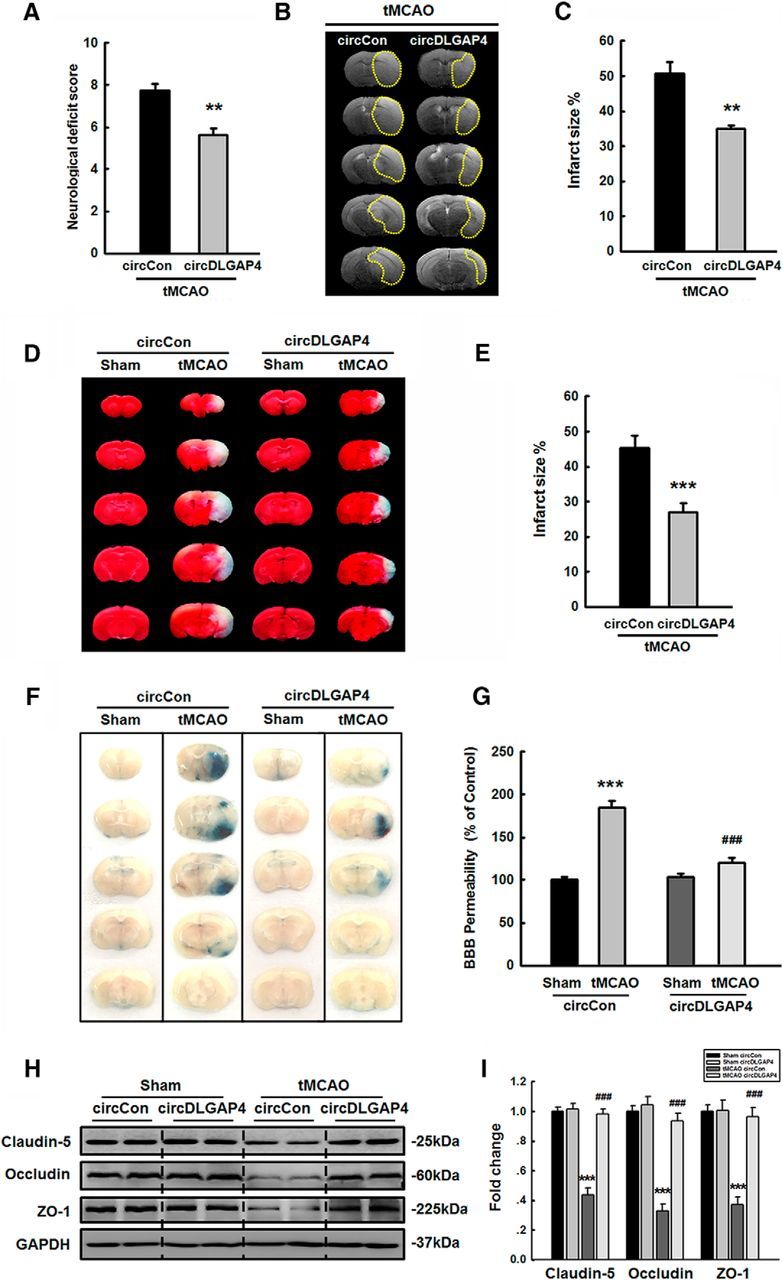
Overexpression of circDLGAP4 results in neuroprotective effects against cerebral ischemia-induced injury in tMCAO mice. A, Microinjection of circDLGAP4 reduced neurological deficit scores at 24 h after tMCAO. n = 8 mice/group. **p = 0.001, circDLGAP4 tMCAO versus circControl tMCAO (Mann–Whitney Test). B, Representative T2-weighted MRI images of circControl-microinjected (left) and circDLGAP4-microinjected (right) tMCAO mice. C, Effects of circDLGAP4 on infarct volumes in tMCAO mice. Infarct volumes were decreased in circDLGAP4-microinjected tMCAO mice compared with those in circControl-microinjected tMCAO mice, as demonstrated by MRI. n = 6 mice/group. **p = 0.001, circDLGAP4 versus circControl (Student's t test). D, Representative TTC-stained sections from mice microinjected with circControl or circDLGAP4 at 24 h after tMCAO surgery. Viable tissue is stained red, whereas damaged ischemic brain tissue appears unstained/white. E, Microinjection of circDLGAP4 significantly reduced infarct volumes at 24 h after tMCAO surgery compared with those in the group microinjected with circControl. n = 8 mice/group. ***p < 0.001, circDLGAP4 versus circControl (Student's t test). F, G, circDLGAP4 overexpression ameliorated the cerebrovascular damage induced by tMCAO. circControl- and circDLGAP4-microinjected mice were administered a 2% Evans blue solution via tail vein injections. After 4 h, cerebrovascular permeability was evaluated by visualizing Evans blue extravasation following vigorous heart perfusion. Representative images of extravasated Evans blue staining of the brain (F). Cerebrovascular permeability in circControl- and circDLGAP4-microinjected mice was determined by measuring the concentrations of brain-extracted Evans blue by spectrophotometry at 620 nm (G). n = 6 mice/group. F(3,20) = 54.834: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO (one-way ANOVA followed by the Holm–Sidak Test). H, I, Microinjection of circDLGAP4 ameliorated the decreased ipsilateral TJP expression observed in the tMCAO group. TJP expression levels were determined through WB analysis (H) and quantified via densitometric analysis (I). n = 6 mice/group. Claudin-5, F(3,20) = 55.852: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO. Occludin, F(3,20) = 47.798: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO. ZO-1, F(3,20) = 27.872: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO (one-way ANOVA followed by the Holm–Sidak Test). Two representative immunoblots were presented from 6 mice/group.
circDLGAP4/miR-143 regulates EndoMT in tMCAO mice
Because EndoMT is characterized by the loss of endothelial markers, and we determined that endothelial marker expression levels were significantly decreased at 24 h after tMCAO surgery, we hypothesized that EndoMT contributes to cerebrovascular damage. To confirm this hypothesis, we subsequently investigated whether endothelial cells acquired the ability to express mesenchymal cell markers, such as Col I, Col III, and α-SMA. As shown in Figure 6A, B, Col I, Col III, and α-SMA expression levels were increased in mice subjected to tMCAO. To verify this observation, we examined EndoMT using a Tie2-GFP genetic mouse model combined with quantitative colocalization analysis, in which colocalization is estimated using specially developed algorithms that calculate two coefficients, such as Pearson correlation coefficient and Manders overlap coefficient. As shown in Figure 6C, D, the tMCAO group displayed a significantly increased colocalization rate, which was 4.13-fold higher according to Pearson correlation coefficient and 2.66-fold higher according to Manders overlap coefficient than that in the sham group, suggesting that the brain microvessel endothelial cells exposed to tMCAO had differentiated from an endothelial to a mesenchymal phenotype. To examine the role of circDLGAP4/miR-143 in EndoMT, we first examined WT and Mir-143+/− mice at 24 h after tMCAO surgery. As illustrated in Figure 6E, F, WT mice subjected to tMCAO showed increases in Col I, Col III, and α-SMA expression in the infarct core. These changes were significantly ameliorated in Mir-143+/− mice subjected to tMCAO. Next, circDLGAP4-GFP lentivirus was microinjected into the left lateral ventricles of the mice to examine the effects of circDLAP4 on the expression of mesenchymal cell markers. As illustrated in Figure 6G, H, circControl mice subjected to tMCAO treatment displayed increases in Col I, Col III, and α-SMA expression in the infarct core, and these changes were significantly ameliorated in the circDLGAP4-injected group.
Figure 6.
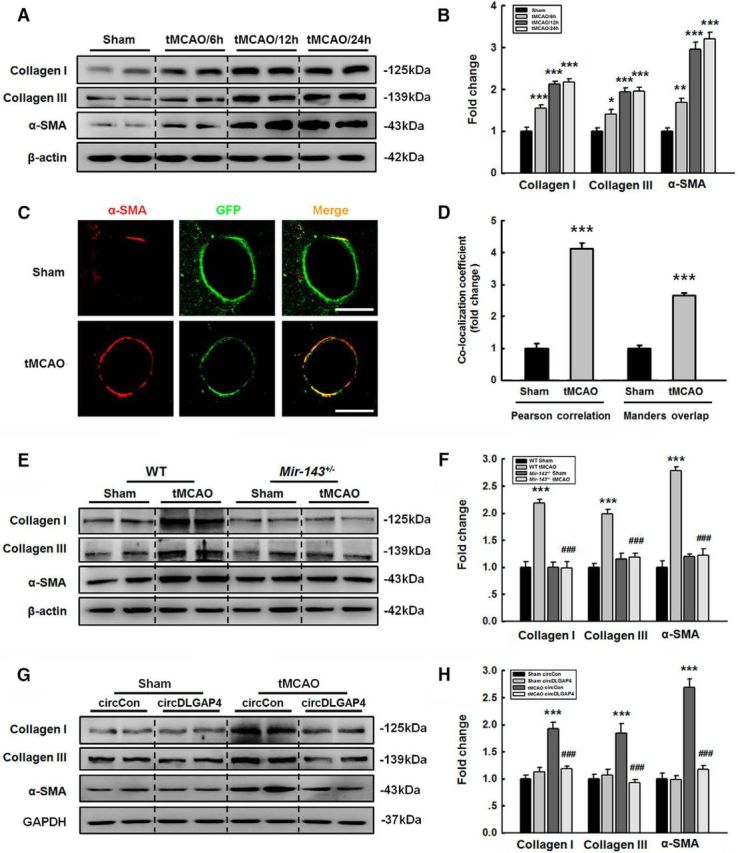
circDLGAP4/miR-143 regulates EndoMT in tMCAO mice. A, B, Ipsilateral mesenchymal cell marker expression levels were significantly increased at 12 and 24 h after tMCAO. The expression of mesenchymal cell markers was determined through WB analysis (A) and quantified via densitometric analysis (B); n = 6 mice/group. Col I, F(3,20) = 45.997: ***p < 0.001, tMCAO/6 h versus Sham. ***p < 0.001, tMCAO/12 h versus Sham. ***p < 0.001, tMCAO/24 h versus Sham. Col III, F(3,20) = 23.784: p = 0.015, tMCAO/6 h versus Sham. ***p < 0.001, tMCAO/12 h versus Sham. ***p < 0.001, tMCAO/24 h versus Sham. α-SMA, F(3,20) = 68.767: **p = 0.002, tMCAO/6 h versus Sham. ***p < 0.001, tMCAO/12 h versus Sham. ***p < 0.001, tMCAO/24 h versus Sham (one-way ANOVA followed by the Holm–Sidak Test). Two representative immunoblots were presented from 6 mice/group. C, D, Analysis of GFP+ and α-SMA colocalization in vivo. Immunostaining for α-SMA in the cortex of the Tie2-GFP genetic mouse model. Z-stack images were acquired on a Carl Zeiss confocal laser-scanning microscope (C). Pearson correlation coefficient and Manders overlap coefficient of GFP+ and α-SMA immunofluorescence were calculated (D). Scale bar, 5 μm. n = 15 microvessels/group. Pearson correlation coefficient: ***p < 0.001, tMCAO versus Sham (the Mann–Whitney Test). Manders overlap coefficient: ***p < 0.001, tMCAO versus Sham (Student's t test). E, F, Mir-143+/− mice ameliorated the increases in cortical mesenchymal cell marker expression induced by tMCAO. The expression of mesenchymal cell markers was determined via WB analysis (E) and quantified via densitometric analysis (F). n = 6 mice/group. Col I, F(3,20) = 35.557: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, Mir-143+/− tMCAO versus WT tMCAO. Col III, F(3,20) = 28.210: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, Mir-143+/− tMCAO versus WT tMCAO. α-SMA, F(3,20) = 81.617: ***p < 0.001, WT tMCAO versus WT Sham. ###p < 0.001, Mir-143+/− tMCAO versus WT tMCAO (one-way ANOVA followed by the Holm–Sidak Test). Two representative immunoblots were presented from 6 mice/group. G, H, Microinjection of circDLGAP4 ameliorated the decreases in ipsilateral mesenchymal cell marker expression induced by tMCAO. The expression of mesenchymal cell markers was determined via WB analysis (G) and quantified via densitometric analysis (H). n = 6 mice/group. Col I, F(3,20) = 26.338: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO. Col III, F(3,20) = 13.132: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO. α-SMA, F(3,20) = 64.707: ***p < 0.001, circControl tMCAO versus circControl Sham. ###p < 0.001, circDLGAP4 tMCAO versus circControl tMCAO (one-way ANOVA followed by the Holm–Sidak Test). Two representative immunoblots were presented from 6 mice/group.
circDLGAP4 inhibits EndoMT targeting of miR-143 in mouse endothelial cells
Because EndoMT was found to be involved in the observed cerebrovascular damage, we next sought to study the effects of circDLGAP4 on EndoMT. Mouse brain endothelial bEnd.3 cells were transduced with circDLGAP4; and as shown in Figure 7A, the circDLGAP4 group displayed increased circDLGAP4 expression compared with that in the circControl group based on the results of qRT-PCR. We also detected the effects of circDLGAP4 on OGD/R-induced EndoMT. As shown in Figure 7B, transducing cells with circDLGAP4 significantly ameliorated the decreases in TJP expression induced by OGD/R, according to the WB analysis of claudin-5, occludin, and ZO-1 expression. In contrast, transducing mouse brain endothelial bEnd.3 cells with circDLGAP4 significantly inhibited the increases in mesenchymal cell marker expression induced by OGD/R, as determined via WB analysis of Col I, Col III, and α-SMA expression (Fig. 7C).
Figure 7.
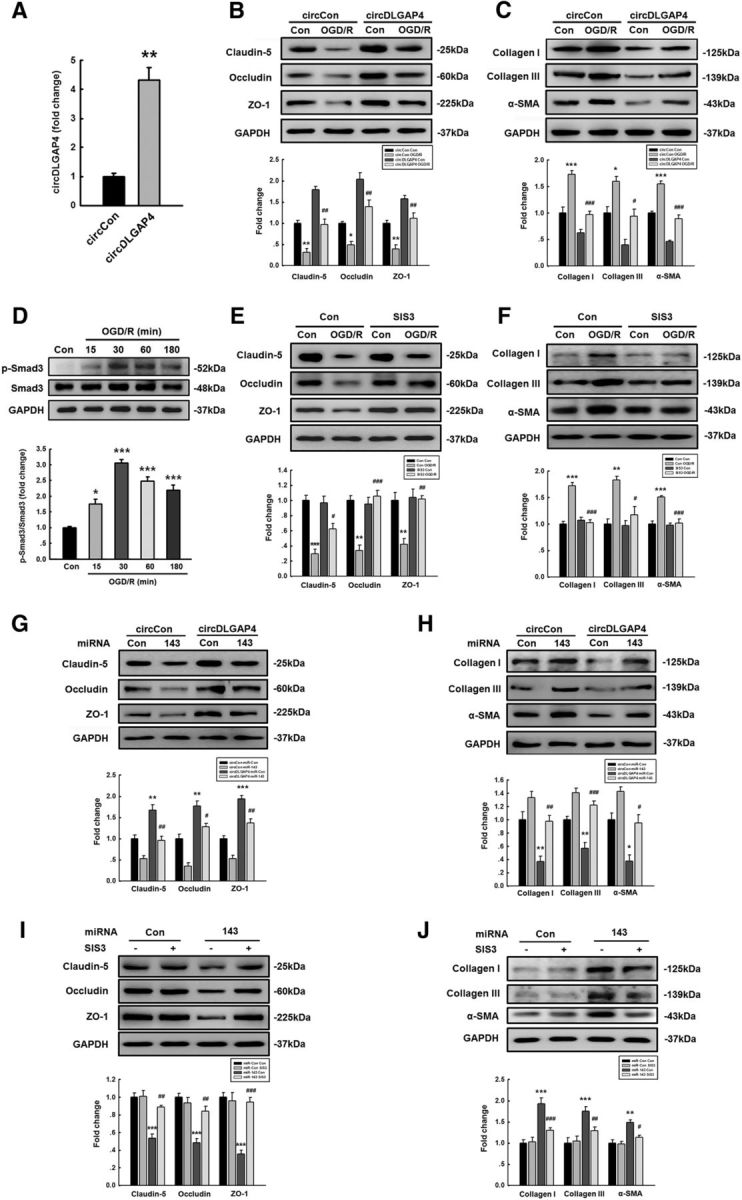
circDLGAP4 inhibits EndoMT targeting of miR-143 in mouse brain endothelial cells. A, Transduction of circDLGAP4 lentiviruses increased circDLGAP4 expression in mouse brain endothelial bEnd.3 cells, as determined via RT-PCR. Data are mean ± SEM of three independent experiments. **p = 0.002, circDLGAP4 versus circControl (Student's t test). B, Effects of circDLGAP4 overexpression on TJP expression in mouse brain endothelial bEnd.3 cells treated with OGD/R. Transduction of cells with circDLGAP4 lentiviruses significantly inhibited the decrease in the expression of TJP induced by OGD/R. Claudin-5, F(3,8) = 41.632: **p = 0.002, circControl OGD/R versus circControl Control. ##p = 0.002, circDLGAP4 OGD/R versus circControl OGD/R. Occludin, F(3,8) = 30.396: *p = 0.031, circControl OGD/R versus circControl Control. ##p = 0.003, circDLGAP4 OGD/R versus circControl OGD/R. ZO-1, F(3,8) = 24.985: **p = 0.009, circControl OGD/R versus circControl Control. ##p = 0.004, circDLGAP4 OGD/R versus circControl OGD/R (one-way ANOVA followed by the Holm–Sidak Test). C, Effects of circDLGAP4 overexpression on mesenchymal cell marker expression in mouse brain endothelial bEnd.3 cells treated with OGD/R. Transduction of cells with circDLGAP4 lentiviruses significantly inhibited the increased expression of mesenchymal cell markers induced by OGD/R. Col I, F(3,8) = 32.075: ***p < 0.001, circControl OGD/R versus circControl Control. ###p < 0.001, circDLGAP4 OGD/R versus circControl OGD/R. Col III, F(3,8) = 19.027: *p = 0.022, circControl OGD/R versus circControl Control. #p = 0.015, circDLGAP4 OGD/R versus circControl OGD/R. α-SMA, F(3,8) = 81.157: ***p < 0.001, circControl OGD/R versus circControl Control. ###p < 0.001, circDLGAP4 OGD/R versus circControl OGD/R (one-way ANOVA followed by the Holm–Sidak Test). D, Activation of Smad3 signaling in mouse brain endothelial bEnd.3 cells treated with OGD/R. Data are mean ± SEM of three independent experiments. F(4,10) = 34.500: *p = 0.012, OGD/R (15 min) versus Control. ***p < 0.001, OGD/R (30 min) versus Control. ***p < 0.001, OGD/R (60 min) versus Control. ***p < 0.001, OGD/R (180 min) versus Control (one-way ANOVA followed by the Holm–Sidak Test. E, Effects of Smad3 inhibitor-SIS3 on TJP expression in mouse brain endothelial bEnd.3 cells treated with OGD/R. Cells pretreated with SIS3 significantly inhibited the decreased expression of TJPs induced by OGD/R. Claudin-5, F(3,8) = 20.550: ***p < 0.001, OGD/R versus Control. #p = 0.026, SIS3-treated OGD/R versus OGD/R. Occludin, F(3,8) = 19.420: **p = 0.001, OGD/R versus Control. ###p < 0.001, SIS3-treated OGD/R versus OGD/R. ZO-1, F(3,8) = 11.638: **p = 0.006, OGD/R versus Control. ##p = 0.007, SIS3-treated OGD/R versus OGD/R (one-way ANOVA followed by the Holm–Sidak Test). F, Effects of the Smad3 inhibitor SIS3 on mesenchymal cell marker expression in mouse brain endothelial bEnd.3 cells treated with OGD/R. Cells pretreated with SIS3 significantly inhibited the increased expression of mesenchymal cell markers induced by OGD/R. Col I, F(3,8) = 37.289: ***p < 0.001, OGD/R versus Control. ###p < 0.001, SIS3-treated OGD/R versus OGD/R. Col III, F(3,8) = 14.258: **p = 0.003, OGD/R versus Control. #p = 0.010, SIS3-treated OGD/R versus OGD/R. α-SMA, F(3,8) = 23.199: ***p < 0.001, OGD/R versus Control. ###p < 0.001, SIS3-treated OGD/R versus OGD/R (one-way ANOVA followed by the Holm–Sidak Test). G, Transduction of miR-143 significantly inhibited the increased TJP expression levels in mouse brain endothelial bEnd.3 cells cotransduced with circDLGAP4 lentiviruses. Claudin-5, F(3,8) = 23.414: **p = 0.005, circDLGAP4 miR-Control versus circControl miR-Control. ##p = 0.004, circDLGAP4 miR-143 versus circDLGAP4 miR-Control. Occludin, F(3,8) = 39.606: **p = 0.002, circDLGAP4 miR-Control versus circControl miR-Control. #p = 0.013, circDLGAP4 miR-143 versus circDLGAP4 miR-Control. ZO-1, F(3,8) = 53.654: ***p < 0.001, circDLGAP4 miR-Control versus circControl miR-Control. ##p = 0.004, circDLGAP4 miR-143 versus circDLGAP4 miR-Control (one-way ANOVA followed by the Holm–Sidak Test). H, Transduction of miR-143 lentiviruses significantly inhibited the decreases in mesenchymal cell marker expression induced by circDLGAP4 lentiviruses. Expression of TJPs and mesenchymal cell markers was determined via WB analysis and quantified via densitometric analysis. Data are mean ± SEM of three independent experiments. Col I, F(3,8) = 17.506: **p = 0.008, circDLGAP4 miR-Control versus circControl miR-Control. ##p = 0.008, circDLGAP4 miR-143 versus circDLGAP4 miR-Control. Col III, F(3,8) = 27.008: **p = 0.009, circDLGAP4 miR-Control versus circControl miR-Control. ###p < 0.001, circDLGAP4 miR-143 versus circDLGAP4 miR-Control. α-SMA, F(3,8) = 19.070: *p = 0.010, circDLGAP4 miR-Control versus circControl miR-Control. #p = 0.013, circDLGAP4 miR-143 versus circDLGAP4 miR-Control (one-way ANOVA followed by the Holm–Sidak Test). I, SIS3 pretreatment significantly inhibited the decreases in TJP expression levels in mouse brain endothelial bEnd.3 cells transduced with miR-143 lentiviruses. Claudin-5, F(3,8) = 21.241: ***p < 0.001, miR-143 versus miR-Control. ##p = 0.003, SIS3-treated miR-143 versus miR-143. Occludin, F(3,8) = 19.707: ***p < 0.001, miR-143 versus miR-Control. ##p = 0.005, SIS3-treated miR-143 versus miR-143. ZO-1, F(3,8) = 23.728: ***p < 0.001, miR-143 versus miR-Control. ###p < 0.001, SIS3-treated miR-143 versus miR-143 (one-way ANOVA followed by the Holm–Sidak Test). J, SIS3 pretreatment significantly inhibited the increases in mesenchymal cell marker expression levels in mouse brain endothelial bEnd.3 cells cotransduced with miR-143 lentiviruses. The expression of mesenchymal cell markers was determined via WB analysis and quantified via densitometric analysis. Data are mean ± SEM of three independent experiments. Col I, F(3,8) = 57.599: ***p < 0.001, miR-143 versus miR-Control. ###p < 0.001, SIS3-treated miR-143 versus miR-143. Col III, F(3,8) = 30.331: ***p < 0.001, miR-143 versus miR-Control. ##p = 0.003, SIS3-treated miR-143 versus miR-143. α-SMA, F(3,8) = 14.701: **p = 0.003, miR-143 versus miR-Control. #p = 0.014, SIS3-treated miR-143 versus miR-143 (one-way ANOVA followed by the Holm–Sidak Test).
To dissect the causal relationship between circDLGAP4 binding miR-143 and the observed effects on TJPs and EndoMT, we first examined Smad3 activation in mouse brain endothelial bEnd.3 cells treated with OGD/R based on a previous study showing that the Smad3 inhibitor SIS3 blocks EndoMT (Li et al., 2010). As shown in Figure 7D, OGD/R treatment induced the phosphorylation of Smad3 with a peak response at OGD/R 30 min. Then, endothelial cells were treated with SIS3 to examine the causal relationship between TJPs and EndoMT. As shown in Figure 7E, F, treatment of cells with SIS3 significantly inhibited the decreased expression of TJPs as well as the increased mesenchymal cell markers induced by OGD/R treatment.
To verify that miR-143 is a mediator of circDLGAP4, we cotransfected mouse brain endothelial bEnd.3 cells with mimic-miR-143 and circDLGAP4. circDLGAP4 overexpression enhanced TJP expression, and this change was significantly attenuated by miR-143 (Fig. 7G). In contrast, miR-143 reversed the decreases in mesenchymal cell marker expression induced by circDLGAP4 overexpression (Fig. 7H). These results indicate that circDLGAP4 acts as an endogenous miR-143 sponge to regulate EndoMT. Next, we examined the role of SIS3 in the process of EndoMT induced by miR-143. As shown in Figure 7I, J, treatment of bEnd.3 cells with SIS3 significantly inhibited the decreased expression of TJPs as well as the increased mesenchymal cell markers induced by miR-143 overexpression.
miR-143 enhances EndoMT by targeting HECTD1
Given that miR-143 plays a critical role in cerebrovascular integrity, we examined the mechanisms underlying the function of miR-143. We first predicted that a consensus binding site of miR-143 is present in the 3′-UTR of Hectd1 using the TargetScan algorithm. As shown in Figure 8A, Hectd1 exhibits a conserved miR-143 binding site within its 3′-UTR in most species. An in vivo experiment showed that tMCAO mice exhibited significantly decreased expression of HECTD1 compared with that in the sham control group (Fig. 8B). These findings were further confirmed in mouse brain endothelial bEnd.3 cells treated with OGD/R (Fig. 8C). Intriguingly, cotransfection of an miR-143-overexpressing vector and the pmiR-GLO plasmid with the Hectd1 WT 3′-UTR resulted in the downregulation of luciferase activity, and this effect was reversed by transfection with a mutated Hectd1 3′-UTR in both HEK293T and mouse brain endothelial bEnd.3 cells (Fig. 8D,E). In line with these findings, miR-143 decreased HECTD1 expression, whereas anti-miR-143 increased its expression in mouse brain endothelial bEnd.3 cells (Fig. 8F). Having determined that miR-143 regulates HECTD1 expression, we next examined the role of circDLGAP4 in regulating the expression of HECTD1. As shown in Figure 8G, overexpression of circDLGAP4 significantly ameliorated the decreased expression of HECTD1 induced by OGD/R treatment in mouse brain endothelial bEnd.3 cells. In contrast, miR-143 reversed the increase in HECTD1 expression induced by circDLGAP4 overexpression (Fig. 8H). These results indicate that circDLGAP4 acts as an endogenous miR-143 sponge to regulate HECTD1 expression.
Figure 8.
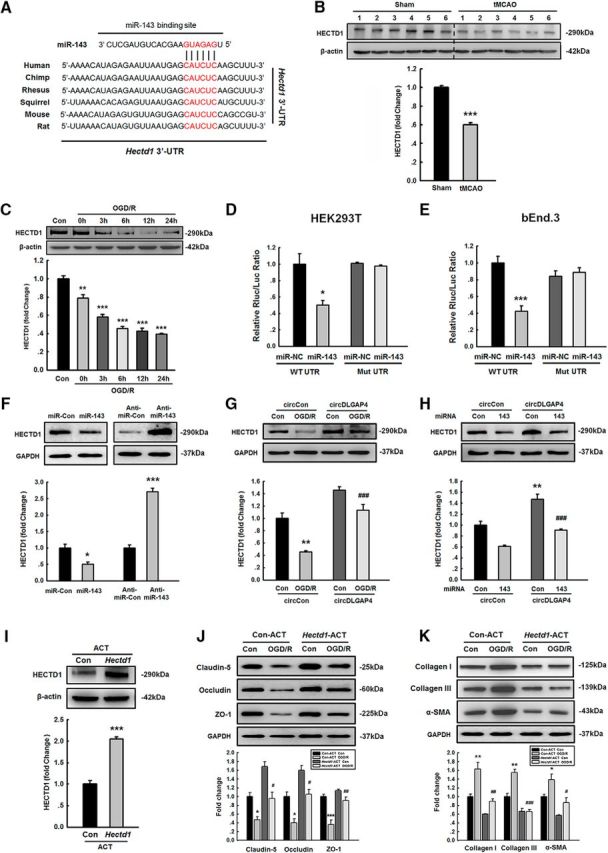
miR-143 enhances EndoMT by targeting HECTD1. A, Putative miR-143 binding sites in Hectd1 (HECTD1 gene). Red represents the potential complementary residues. B, The expression of HECTD1 was significantly decreased in the tMCAO mouse stroke model compared with that in the sham group. The expression of HECTD1 was determined via WB analysis and quantified via densitometric analysis. n = 6 mice/group. ***p < 0.001, tMCAO versus Sham (Student's t test). C, OGD/R treatment resulted in a significant decrease in the expression of HECTD1 in mouse brain endothelial bEnd.3 cells compared with that in the control group in a time-dependent manner. Data are mean ± SEM of three individual experiments. F(5,12) = 68.299: **p = 0.002, OGD/R 0 h versus Control. ***p < 0.001, OGD/R 3 h versus Control. ***p < 0.001, OGD/R 6 h versus Control. ***p < 0.001, OGD/R 12 h versus Control. ***p < 0.001, OGD/R 24 h versus Control (one-way ANOVA followed by the Holm–Sidak Test). D, E, Relative luciferase activity of wild-type and 3′-UTR mutant constructs of Hectd1 cotransfected with an miR-143 OE vector and the pmiR-GLO plasmid in both HEK293T cells (D) and bEnd.3 cells (E). Data are mean ± SEM of three individual experiments. HEK293T cells: *p = 0.016, WT UTR miR-143 versus WT UTR miR-NC. bEnd.3 cells: ***p < 0.001, WT UTR miR-143 versus WT UTR miR-NC (Student's t test). F, HECTD1 expression was evaluated at the protein level in mouse brain endothelial bEnd.3 cells transduced with the miR-control/miR-143 and the anti-miR-control/anti-miR-143 lentiviruses. Data are mean ± SEM of three individual experiments. *p = 0.02, miR-143 versus miR-Control (Student's t test). ***p < 0.001, anti-miR-143 versus anti-miR-Control (Student's t test). G, Transduction of cells with circDLGAP4 lentiviruses significantly inhibited the decreased expression of HECTD1 induced by OGD/R. Data are mean ± SEM of three individual experiments. F(3,8) = 34.032: **p = 0.003, circControl OGD/R versus circControl Control. ###p < 0.001, circDLGAP4 OGD/R versus circControl OGD/R (one-way ANOVA followed by the Holm–Sidak Test). H, Transduction of miR-143 significantly inhibited the increased HECTD1 expression levels in mouse brain endothelial bEnd.3 cells cotransduced with circDLGAP4 lentiviruses. Data are mean ± SEM of three individual experiments. F(3,8) = 34.104: **p = 0.002, circDLGAP4 miR-Control versus circControl miR-Control. ###p < 0.001, circDLGAP4 miR-143 versus circDLGAP4 miR-Control (one-way ANOVA followed by the Holm–Sidak Test). I, Transfection of mouse brain endothelial bEnd.3 cells with Hectd1-ACT increased the expression of HECTD1. Data are mean ± SEM of three individual experiments. ***p < 0.001, Hectd1-ACT versus Con-ACT (Student's t test). J, Transfection of mouse brain endothelial bEnd.3 cells with Hectd1-ACT significantly inhibited the decrease in TJP expression compared with that in the Con-ACT group treated with OGD/R. Claudin-5, F(3,8) = 23.159: *p = 0.020, Con-ACT OGD/R versus Con-ACT Control. #p = 0.021, Hectd1-ACT OGD/R versus Con-ACT OGD/R. Occludin, F(3,8) = 22.148: *p = 0.014, Con-ACT OGD/R versus Con-ACT Control. #p = 0.011, Hectd1-ACT OGD/R versus Con-ACT OGD/R. ZO-1, F(3,8) = 25.717: ***p < 0.001, Con-ACT OGD/R versus Con-ACT Control. ##p = 0.002, Hectd1-ACT OGD/R versus Con-ACT OGD/R (one-way ANOVA followed by the Holm–Sidak Test). K, Transfection of bEnd.3 cells with Hectd1-ACT significantly inhibited the increase of mesenchymal cell marker expression compared with that in the Con-ACT group treated with OGD/R. The expression of TJPs and mesenchymal cell markers was determined via WB analysis and quantified via densitometric analysis. Data are mean ± SEM of three independent experiments. Col I, F(3,8) = 26.032: **p = 0.003, Con-ACT OGD/R versus Con-ACT Control. ##p = 0.001, Hectd1-ACT OGD/R versus Con-ACT OGD/R. Col III, F(3,8) = 42.527: **p = 0.001, Con-ACT OGD/R versus Con-ACT Control. ###p < 0.001, Hectd1-ACT OGD/R versus Con-ACT OGD/R. α-SMA, F(3,8) = 15.712: *p = 0.037, Con-ACT OGD/R versus Con-ACT Control. #p = 0.012, Hectd1-ACT OGD/R versus Con-ACT OGD/R (one-way ANOVA followed by the Holm–Sidak Test). Hectd1-ACT, Hectd1 CRISPR Activation Plasmid (ACT); Con-ACT, Control CRISPR Activation Plasmid (ACT).
Having determined that miR-143 regulates HECTD1 expression, we next examined the role of HECTD1 in EndoMT induced by OGD/R. As shown in Figure 8I, transfection with the Hectd1 ACT upregulated the expression of HECTD1 in mouse brain endothelial bEnd.3 cells. Moreover, transfection with Hectd1-ACT resulted in significant amelioration of the decreased expression of TJPs (claudin-5, occluding, and ZO-1) and increased expression of mesenchymal cell markers (Col I, Col III, and α-SMA) induced by OGD/R, as shown in Figure 8J, K.
Discussion
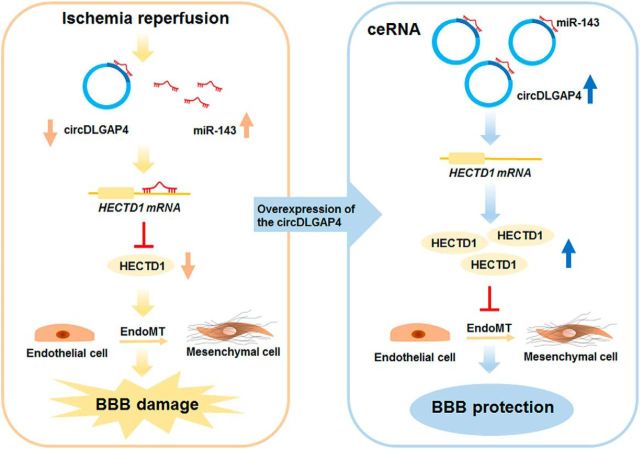
Our study provides new insights into the function of circDLGAP4, which was found to be involved in EndoMT in the context of stroke via binding with miR-143 and subsequently targeting HECTD1 (Fig. 9). Specific upregulation of circDLGAP4 may be a potential therapeutic target for the treatment of cerebral ischemia.
Figure 9.
circDLGAP4/miR-143 affects the ischemic stroke outcome by regulating EndoMT associated with cerebrovascular integrity. circDLGAP4 levels were significantly decreased in the plasma of AIS patients and in the mouse stroke model. circDLGAP4, identified as ceRNA, functions as an endogenous miR-143 sponge to sequester miR-143, resulting in the inhibition of HECTD1 expression. Decreased HECTD1 expression enhanced EndoMT by decreasing TJP expression with concomitant increased expression of mesenchymal cell markers. circDLGAP4 overexpression significantly inhibited EndoMT and attenuated BBB damage in the tMCAO mouse stroke model.
Although miRNAs are considered a potential therapeutic strategy for the treatment of neurovascular-dependent brain diseases (Reijerkerk et al., 2013), the specific roles of miRNAs in EndoMT in the brain are largely unknown. In addition to the involvement of miR-31 in EndoMT, other miRNAs, including miR-21, miR-23, miR-20a, and miR-302c, are known to be involved in EndoMT in peripheral cells, such as cardiac endothelial cells and hepatocellular carcinoma cells (Lagendijk et al., 2011; Ghosh et al., 2012; Kumarswamy et al., 2012; Laurila and Kallioniemi, 2013; Zhu et al., 2014). The findings of our previous work indicated that miR-143 regulates cerebrovascular integrity (Bai et al., 2016); however, whether miR-143 is involved in EndoMT in the pathology of stroke remained unknown. In the present study, downregulation of miR-143 significantly decreased ischemic injury via the regulation of EndoMT in the tMCAO mouse stroke model using Mir-143+/− mice. Our finding was consistent with a recent study showing that circulating miR-143 is upregulated in ischemic patients compared with its expression level in healthy controls, indicating that miR-143 is associated with AIS and has clinical utility as an early diagnostic marker (Tiedt et al., 2017).
Consistent with the findings of previous studies (Hansen et al., 2013; Wang et al., 2016), the current study showed that circDLGAP4 acts as an miR-143 sponge to decrease the sizes of infarct areas and attenuate neuronal deficits and BBB damage in tMCAO mice. The interaction between miR-143 and circDLGAP4 was confirmed via miRNA pull-down and FISH assays. Thus, circRNAs may act as endogenous sponges that interact with miRNAs and regulate the expression of miRNA target genes. A recent study demonstrated that the circRNA HRCR acts as an miR-223 sponge to regulate cardiac hypertrophy and heart failure (Wang et al., 2016). Another well-known circSRY expressed in mouse tissues is known to act as a sponge for miR-138 (Hansen et al., 2013). Moreover, another circRNA, HIPK3, has multiple miRNA binding sites, and miR-124 was found to bind with this circRNA most prominently (Zheng et al., 2016). In addition to exerting their biological functions by regulating miRNAs, circRNAs also exert their biological functions by other mechanisms. For example, circFox3 was found to participate in cell cycle progression by forming of a ternary complex with p21 and CDK2 (Du et al., 2017). Another complex, circRNA-cZNF292, is regulated by hypoxia and displays proangiogenic activity in endothelial cells. However, it has also been suggested that cZNF292 does not act as miRNA sponge, indicating that different circRNAs have different features and contribute to regulatory networks in distinct ways (Boeckel et al., 2015). We demonstrated that miR-143 is regulated by circDLGAP4; however, we could not rule out the possibility that other mechanisms may underlie the functions of circDLGAP4.
Our study has provided us with new insights regarding the function of circDLGAP4, which was found to significantly promote the maintenance of BBB integrity in the tMCAO mouse stroke model. This promotion resulted in decreased infarct sizes and improved neurological deficits. Our results indicated that plasma circDLGAP4 is a promising biomarker for diagnosing AIS and evaluating the degree of damage caused by ischemic insults. Our study elucidated the involvement of circDLGAP4 and its coupling mechanism in cerebral ischemia, specifically its involvement in maintaining BBB integrity, and provided translational evidence indicating that circDLGAP4 may serve as a novel biomarker for and therapeutic target in acute cerebrovascular protection.
Accumulating evidence indicates that circRNAs have important functions in development and in heart senescence, hypertrophy and failure, as well as in cell growth (Boeckel et al., 2015; Hong et al., 2016; Zheng et al., 2016; Du et al., 2017). Despite the biological importance of circRNAs, whether they are involved in the pathogenesis of stroke has not yet been determined. In this study, plasma circDLGAP4 levels were found to be decreased in AIS patients compared with those in controls. Consistent with this finding, circDLGAP4 levels were also significantly decreased in the tMCAO mouse stroke model, suggesting a correlation between circDLGAP4 expression in patient plasma samples and that in the tMCAO mouse stroke model. Universality, conserved expression, defined specificity, high stability, and abundance are the main characteristics of circRNAs that render them very attractive as diagnostic tools for assessing disease. Thus far, circRNAs have been shown to be potential biomarkers of gastric and hepatocellular carcinoma (Li et al., 2015; Qin et al., 2016). A previous study suggested that circRNAs also have the potential to be used as biomarkers of neurodegenerative diseases (Kumar et al., 2016). Our findings indicate that the expression of circDLGAP4 is a valuable biomarker of cerebral ischemia.
Because damage to BBB integrity is not only a common consequence of but also a contributor to the progression of stroke (Hu et al., 2017), and EndoMT is thought to be involved in the disruption of BBB integrity, we examined the role of circDLGAP4 in EndoMT. Mounting evidence from studies of peripheral body systems indicates that EndoMT is involved in cardiac development and a variety of pathological conditions (LeBleu et al., 2013; Maddaluno et al., 2013; Cooley et al., 2014; Petrini et al., 2017). However, it should also be noted that most of the research regarding EndoMT has been performed in the context of studying embryonic development or evaluating non–brain-derived endothelial cells. To our knowledge, this is the first study to demonstrate that EndoMT contributes to BBB damage in the context of stroke, as we noted decreases in endothelial cell-specific marker expression and increases in mesenchymal cell product expression. Using a Tie2-GFP genetic mouse model colocalization analysis, we clearly demonstrated that the mesenchymal cell marker-α-SMA colocalizes with GFP. Multiple pathways, including the TGF-β signaling pathway and other signaling pathways, such as the Wnt, Notch, and Hh pathways, are involved in EndoMT (Aisagbonhi et al., 2011; Piera-Velazquez et al., 2016; Zhao et al., 2016b; Wermuth et al., 2017). circRNAs are regulated by the RNA binding protein Quaking and are known to play a specific role in epithelial-mesenchymal transition (Conn et al., 2015), although this role remains enigmatic. Our study shed light on this unknown area by showing that Smad3 activation was involved in circDLGAP4/miR-143 axis-mediated EndoMT. Our findings were consistent with a previous study in which a Smad3 inhibitor inhibited EndoMT, thus contributing to the delayed development of diabetic nephropathy (Li et al., 2010).
Our findings demonstrated that circDLGAP4/miR-143 regulated EndoMT during cerebral ischemia; thus, we deemed it necessary to identify the downstream target of miR-143 and gain further insights into the mechanism underlying miR-143-mediated EndoMT regulation. Computational algorithms, such as TargetScan, have been used to identify evolutionarily conserved sequences in Hectd1 that are targeted by miR-143. Consistent with these predictions, the current study indicated that HECTD1 was a target of miR-143 based on the results of a Luciferase activity assay. The gene encoding HECTD1 has been mapped to a region of chromosome 14q12 encoding a novel protein HECT domain-containing E3 ubiquitin ligase, which regulates the selective ubiquitination of Hsp90 client proteins (Sarkar and Zohn, 2012; Shen et al., 2017). Hectd1 knock-out mice exhibit perinatal lethality, impaired neural fold elevation, and an abnormal head mesenchyme morphology (Sarkar and Zohn, 2012). A previous study indicated that HECTD1 enhances adenomatous polyposis coli-axin interactions to negatively regulate Wnt signaling (Tran et al., 2013). Moreover, HECTD1 ubiquitinates phosphatidylinositol 4 phosphate 5-kinase Type I γ at K97, resulting in its degradation and focal adhesion assembly/disassembly during cell migration (Li et al., 2013; Deng and Huang, 2014). However, the involvement of HECTD1 in endothelial cell permeability has remained elusive. To the best of our knowledge, our study is the first to demonstrate that HECTD1, as a target of miR-143, functions in the EndoMT of endothelial cells induced by OGD/R, lending credence to our speculation that miR-143/HECTD1 contributes to EndoMT during stroke. The detailed mechanism underlying the regulation of EndoMT by HECTD1, and especially whether TJPs ubiquitinated by HECTD1 are involved in this process, must be further confirmed in future studies.
Together, the findings of the current study have demonstrated, for the first time, that circDLGAP4 binds with miR-143 and acts as an endogenous miR-143 sponge to inhibit miR-143 activity, resulting in the increased expression of HECTD1, a target of miR-143. Restoration of circDLGAP4 attenuated neuronal deficits, decreased infarct areas, and ameliorated BBB damage through the inhibition of EndoMT. Thus, circDLGAP4 may be important both as a target for therapeutic interventions in stroke and as a biomarker for disease activity.
Footnotes
This work was supported by Major State Basic Research Development Program of China 973 Program Grants 2013CB733800 and 2013CB733803, National Natural Science Foundation of China Grants 81673410, 81603090, and 81473190, Scientific Research Foundation of the Graduate School of Southeast University YBJJ1745, China Scholarship Council, and Jiangsu Specially Appointed Professor Grants.
The authors declare no competing financial interests.
References
- Acosta SA, Tajiri N, Hoover J, Kaneko Y, Borlongan CV (2015) Intravenous bone marrow stem cell grafts preferentially migrate to spleen and abrogate chronic inflammation in stroke. Stroke 46:2616–2627. 10.1161/STROKEAHA.115.009854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisagbonhi O, Rai M, Ryzhov S, Atria N, Feoktistov I, Hatzopoulos AK (2011) Experimental myocardial infarction triggers canonical Wnt signaling and endothelial-to-mesenchymal transition. Dis Model Mech 4:469–483. 10.1242/dmm.006510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts MJ. (2017) Stroke treatment with intravenous tissue-type plasminogen activator: more proof that time is brain. Circulation 135:140–142. 10.1161/CIRCULATIONAHA.116.025724 [DOI] [PubMed] [Google Scholar]
- Amin ND, Bai G, Klug JR, Bonanomi D, Pankratz MT, Gifford WD, Hinckley CA, Sternfeld MJ, Driscoll SP, Dominguez B, Lee KF, Jin X, Pfaff SL (2015) Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 350:1525–1529. 10.1126/science.aad2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam TV, Chan SL, Jo DG, Yilmaz G, Tang SC, Cheng A, Gleichmann M, Okun E, Dixit VD, Chigurupati S, Mughal MR, Ouyang X, Miele L, Magnus T, Poosala S, Granger DN, Mattson MP (2006) Gamma secretase-mediated Notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med 12:621–623. 10.1038/nm1403 [DOI] [PubMed] [Google Scholar]
- Bai YY, Gao X, Wang YC, Peng XG, Chang D, Zheng S, Li C, Ju S (2014) Image-guided pro-angiogenic therapy in diabetic stroke mouse models using a multi-modal nanoprobe. Theranostics 4:787–797. 10.7150/thno.9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Hua J, Yang X, Zhang X, Duan M, Zhu X, Huang W, Chao J, Zhou R, Hu G, Yao H (2016) Silencing microRNA-143 protects the integrity of the blood–brain barrier: implications for methamphetamine abuse. Sci Rep 6:35642. 10.1038/srep35642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltan S, Morrison RS, Murphy SP (2013) Novel protective effects of histone deacetylase inhibition on stroke and white matter ischemic injury. Neurotherapeutics 10:798–807. 10.1007/s13311-013-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S (2015) Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res 117:884–890. 10.1161/CIRCRESAHA.115.306319 [DOI] [PubMed] [Google Scholar]
- Catanese L, Tarsia J, Fisher M (2017) Acute ischemic stroke therapy overview. Circ Res 120:541–558. 10.1161/CIRCRESAHA.116.309278 [DOI] [PubMed] [Google Scholar]
- Chang D, Wang YC, Bai YY, Lu CQ, Xu TT, Zhu L, Ju S (2015) Role of P38 MAPK on MMP activity in photothrombotic stroke mice as measured using an ultrafast MMP activatable probe. Sci Rep 5:16951. 10.1038/srep16951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Cooley BC, et al. (2014) TGF-beta signaling mediates endothelial-to-mesenchymal transition (EndMT) during vein graft remodeling. Sci Transl Med 6:227ra234. 10.1126/scitranslmed.3006927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S (2004) Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86:3993–4003. 10.1529/biophysj.103.038422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Huang C (2014) E3 ubiquitin ligases in regulating stress fiber, lamellipodium, and focal adhesion dynamics. Cell Adh Migr 8:49–54. 10.4161/cam.27480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denorme F, De Meyer SF (2016) The VWF-GPIb axis in ischaemic stroke: lessons from animal models. Thromb Haemost 116:597–604. 10.1160/TH16-01-0036 [DOI] [PubMed] [Google Scholar]
- Denorme F, Langhauser F, Desender L, Vandenbulcke A, Rottensteiner H, Plaimauer B, François O, Andersson T, Deckmyn H, Scheiflinger F, Kleinschnitz C, Vanhoorelbeke K, De Meyer SF (2016) ADAMTS13-mediated thrombolysis of t-PA-resistant occlusions in ischemic stroke in mice. Blood 127:2337–2345. 10.1182/blood-2015-08-662650 [DOI] [PubMed] [Google Scholar]
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412. 10.1093/eurheartj/ehw001 [DOI] [PubMed] [Google Scholar]
- Fisher M, Schaebitz W (2000) An overview of acute stroke therapy: past, present, and future. Arch Intern Med 160:3196–3206. 10.1001/archinte.160.21.3196 [DOI] [PubMed] [Google Scholar]
- Floris G, Zhang L, Follesa P, Sun T (2017) Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol 54:5156–5165. 10.1007/s12035-016-0055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3:619–628. 10.1038/nprot.2008.31 [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Nagpal V, Covington JW, Michaels MA, Vaughan DE (2012) Molecular basis of cardiac endothelial-to-mesenchymal transition (EndMT): differential expression of microRNAs during EndMT. Cell Signal 24:1031–1036. 10.1016/j.cellsig.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner H, Cortés-López M, Cooper DA, Bauer M, Miura P (2016) circRNA accumulation in the aging mouse brain. Sci Rep 6:38907. 10.1038/srep38907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hong Y, Wang X, Sun S, Xue G, Li J, Hou Y (2016) Progesterone exerts neuroprotective effects against Abeta-induced neuroinflammation by attenuating ER stress in astrocytes. Int Immunopharmacol 33:83–89. 10.1016/j.intimp.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Hu X, De Silva TM, Chen J, Faraci FM (2017) Cerebral vascular disease and neurovascular injury in ischemic stroke. Circ Res 120:449–471. 10.1161/CIRCRESAHA.116.308427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura A, Suzuki HI, Ueno T, Mihira H, Yamazaki T, Yasuda T, Watabe T, Mano H, Yamada Y, Miyazono K (2016) MicroRNA-31 is a positive modulator of endothelial-mesenchymal transition and associated secretory phenotype induced by TGF-beta. Genes Cells 21:99–116. 10.1111/gtc.12323 [DOI] [PubMed] [Google Scholar]
- Kumar L, Shamsuzzama, Haque R, Baghel T, Nazir A (2016) Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol 54:7224–7234. 10.1007/s12035-016-0213-8 [DOI] [PubMed] [Google Scholar]
- Kumarswamy R, Volkmann I, Jazbutyte V, Dangwal S, Park DH, Thum T (2012) Transforming growth factor-beta-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler Thromb Vasc Biol 32:361–369. 10.1161/ATVBAHA.111.234286 [DOI] [PubMed] [Google Scholar]
- Lagendijk AK, Goumans MJ, Burkhard SB, Bakkers J (2011) MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ Res 109:649–657. 10.1161/CIRCRESAHA.111.247635 [DOI] [PubMed] [Google Scholar]
- Laurila EM, Kallioniemi A (2013) The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosomes Cancer 52:1103–1113. 10.1002/gcc.22107 [DOI] [PubMed] [Google Scholar]
- LeBleu VS, Taduri G, O'Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H, Kalluri R (2013) Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19:1047–1053. 10.1038/nm.3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kim HS, Choi HY, Oh TH, Yune TY (2012) Fluoxetine inhibits matrix metalloprotease activation and prevents disruption of blood-spinal cord barrier after spinal cord injury. Brain 135:2375–2389. 10.1093/brain/aws171 [DOI] [PubMed] [Google Scholar]
- Li J, Qu X, Yao J, Caruana G, Ricardo SD, Yamamoto Y, Yamamoto H, Bertram JF (2010) Blockade of endothelial-mesenchymal transition by a Smad3 inhibitor delays the early development of streptozotocin-induced diabetic nephropathy. Diabetes 59:2612–2624. 10.2337/db09-1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J (2015) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132–136. 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- Li X, Zhou Q, Sunkara M, Kutys ML, Wu Z, Rychahou P, Morris AJ, Zhu H, Evers BM, Huang C (2013) Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I gamma by HECTD1 regulates focal adhesion dynamics and cell migration. J Cell Sci 126:2617–2628. 10.1242/jcs.117044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chopp M, Chen J, Wang L, Gautam SC, Xu YX, Zhang Z (2000) Intrastriatal transplantation of bone marrow nonhematopoietic cells improves functional recovery after stroke in adult mice. J Cereb Blood Flow Metab 20:1311–1319. 10.1097/00004647-200009000-00006 [DOI] [PubMed] [Google Scholar]
- Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM (2012) The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482:519–523. 10.1038/nature10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longa EZ, Weinstein PR, Carlson S, Cummins R (1989) Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke 20:84–91. 10.1161/01.STR.20.1.84 [DOI] [PubMed] [Google Scholar]
- Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, Tournier-Lasserve E, Chapon F, Richichi C, Retta SF, Lampugnani MG, Dejana E (2013) EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 498:492–496. 10.1038/nature12207 [DOI] [PubMed] [Google Scholar]
- Malhotra K, Rayi A, Khunger M, Thompson S, Liebeskind DS (2017) Reporting compliance of stroke trials: cross-sectional analysis. J Stroke Cerebrovasc Dis 26:1472–1480. 10.1016/j.jstrokecerebrovasdis.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Müller AH, Povlsen GK, Bang-Berthelsen CH, Kruse LS, Nielsen J, Warfvinge K, Edvinsson L (2015) Regulation of microRNAs miR-30a and miR-143 in cerebral vasculature after experimental subarachnoid hemorrhage in rats. BMC Genomics 16:119. 10.1186/s12864-015-1341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini I, Barachini S, Carnicelli V, Galimberti S, Modeo L, Boni R, Sollini M, Erba PA (2017) ED-B fibronectin expression is a marker of epithelial-mesenchymal transition in translational oncology. Oncotarget 8:4914–4921. 10.18632/oncotarget.13615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piera-Velazquez S, Mendoza FA, Jimenez SA (2016) Endothelial to mesenchymal transition (EndoMT) in the pathogenesis of human fibrotic diseases. J Clin Med 5:E45. 10.3390/jcm5040045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenta S, Zeisberg E, Kalluri R (2008) The role of endothelial-to-mesenchymal transition in cancer progression. Br J Cancer 99:1375–1379. 10.1038/sj.bjc.6604662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W (2016) Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 16:161–169. 10.3233/CBM-150552 [DOI] [PubMed] [Google Scholar]
- Reijerkerk A, Lopez-Ramirez MA, van Het Hof B, Drexhage JA, Kamphuis WW, Kooij G, Vos JB, van der Pouw Kraan TC, van Zonneveld AJ, Horrevoets AJ, Prat A, Romero IA, de Vries HE (2013) MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. J Neurosci 33:6857–6863. 10.1523/JNEUROSCI.3965-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AA, Zohn IE (2012) Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. J Cell Biol 196:789–800. 10.1083/jcb.201105101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Jia Z, D'Alonzo D, Wang X, Bruder E, Emch FH, De Geyter C, Zhang H (2017) HECTD1 controls the protein level of IQGAP1 to regulate the dynamics of adhesive structures. Cell Commun Signal 15:2. 10.1186/s12964-016-0156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang L, Pu H, Mao L, Hu X, Jiang X, Xu N, Stetler RA, Zhang F, Liu X, Leak RK, Keep RF, Ji X, Chen J (2016) Rapid endothelial cytoskeletal reorganization enables early blood–brain barrier disruption and long-term ischaemic reperfusion brain injury. Nat Commun 7:10523. 10.1038/ncomms10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Jiang X, Zhang L, Pu H, Hu X, Zhang W, Cai W, Gao Y, Leak RK, Keep RF, Bennett MV, Chen J (2017) Endothelium-targeted overexpression of heat shock protein 27 ameliorates blood–brain barrier disruption after ischemic brain injury. Proc Natl Acad Sci U S A 114:E1243–E1252. 10.1073/pnas.1621174114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow SJ. (2016) Stroke and t-PA-triggering new paradigms of care. N Engl J Med 374:809–811. 10.1056/NEJMp1514696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedt S, Prestel M, Malik R, Schieferdecker N, Duering M, Kautzky V, Stoycheva I, Böck J, Northoff BH, Klein M, Dorn F, Krohn K, Teupser D, Liesz A, Plesnila N, Holdt LM, Dichgans M (2017) Seq identifies circulating miR-125a-5p, miR-125b-5p and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res 121:970–980. 10.1161/CIRCRESAHA.117.311572 [DOI] [PubMed] [Google Scholar]
- Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, Lötvall J, Nakagama H, Ochiya T (2015) Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood–brain barrier. Nat Commun 6:6716. 10.1038/ncomms7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Bustos D, Yeh R, Rubinfeld B, Lam C, Shriver S, Zilberleyb I, Lee MW, Phu L, Sarkar AA, Zohn IE, Wertz IE, Kirkpatrick DS, Polakis P (2013) HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J Biol Chem 288:3753–3767. 10.1074/jbc.M112.415240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troletti CD, de Goede P, Kamermans A, de Vries HE (2016) Molecular alterations of the blood–brain barrier under inflammatory conditions: the role of endothelial to mesenchymal transition. Biochim Biophys Acta 1862:452–460. 10.1016/j.bbadis.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Vemuganti R. (2013) All's well that transcribes well: non-coding RNAs and post-stroke brain damage. Neurochem Int 63:438–449. 10.1016/j.neuint.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37:2602–2611. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- Wei YS, Xiang Y, Liao PH, Wang JL, Peng YF (2016) An rs4705342 T>C polymorphism in the promoter of miR-143/145 is associated with a decreased risk of ischemic stroke. Sci Rep 6:34620. 10.1038/srep34620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermuth PJ, Carney KR, Mendoza FA, Piera-Velazquez S, Jimenez SA (2017) Endothelial cell-specific activation of transforming growth factor-beta signaling in mice induces cutaneous, visceral, and microvascular fibrosis. Lab Invest 97:806–818. 10.1038/labinvest.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Zhang CY, Zhang S, Chang M, Wang HY (2016) Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell Physiol Biochem 39:205–216. 10.1159/000445617 [DOI] [PubMed] [Google Scholar]
- Wu R, He Q, Chen H, Xu M, Zhao N, Xiao Y, Tu QQ, Zhang W, Bi X (2017) MicroRNA-448 promotes multiple sclerosis development through induction of Th17 response through targeting protein tyrosine phosphatase non-receptor type 2 (PTPN2). Biochem Biophys Res Commun 486:759–766. 10.1016/j.bbrc.2017.03.115 [DOI] [PubMed] [Google Scholar]
- Yao H, Duan M, Buch S (2011) Cocaine-mediated induction of platelet-derived growth factor: implication for increased vascular permeability. Blood 117:2538–2547. 10.1182/blood-2010-10-313593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, Kook Y, Niu F, Liao K, Fu M, Kolattukudy P, Buch S (2014) MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun 5:4386. 10.1038/ncomms5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Liu N, Zhang J, Wang L, Zhang Z, Zhu J, Li Q, Wang Y (2017) Inhibition of miR-143 during ischemia cerebral injury protects neurones through recovery of the hexokinase 2-mediated glucose uptake. Biosci Rep 37:BSR20170216. 10.1042/BSR20170216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shen K, Bai Y, Lv X, Huang R, Zhang W, Chao J, Nguyen LK, Hua J, Gan G, Hu G, Yao H (2016) Mir143-BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy: implications for methamphetamine-mediated neurotoxicity. Autophagy 12:1538–1559. 10.1080/15548627.2016.1191723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Bai Y, Chao J, Hu G, Chen X, Yao H (2017) Involvement of PUMA in pericyte migration induced by methamphetamine. Exp Cell Res 356:28–39. 10.1016/j.yexcr.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ (2016a) Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel) 7:E116. 10.3390/genes7120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Qiao X, Wang L, Tan TK, Zhao H, Zhang Y, Zhang J, Rao P, Cao Q, Wang Y, Wang Y, Wang YM, Lee VW, Alexander SI, Harris DC, Zheng G (2016b) Matrix metalloproteinase 9 induces endothelial-mesenchymal transition via Notch activation in human kidney glomerular endothelial cells. BMC Cell Biol 17:21. 10.1186/s12860-016-0101-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Pan Q, Jia LQ, Dai Z, Ke AW, Zeng HY, Tang ZY, Fan J, Zhou J (2014) MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci Rep 4:5524. 10.1038/srep05524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V, Zinchuk O, Okada T (2007) Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem 40:101–111. 10.1267/ahc.07002 [DOI] [PMC free article] [PubMed] [Google Scholar]