Abstract
Sleep is highly conserved across animal species. Both wake- and sleep-promoting neurons are implicated in the regulation of wake–sleep transition at dusk in Drosophila. However, little is known about how they cooperate and whether they act via different mechanisms. Here, we demonstrated that in female Drosophila, sleep onset was specifically delayed by blocking the Shaker cognate L channels [Shal; also known as voltage-gated K+ channel 4 (Kv4)] in wake-promoting cells, including large ventral lateral neurons (l-LNvs) and pars intercerebralis (PI), but not in sleep-promoting dorsal neurons (DN1s). Delayed sleep onset was also observed in males by blocking Kv4 activity in wake-promoting neurons. Electrophysiological recordings show that Kv4 channels contribute A-type currents in LNvs and PI cells, but are much less conspicuous in DN1s. Interestingly, blocking Kv4 in wake-promoting neurons preferentially increased firing rates at dusk ∼ZT13, when the resting membrane potentials and firing rates were at lower levels. Furthermore, pigment-dispersing factor (PDF) is essential for the regulation of sleep onset by Kv4 in l-LNvs, and downregulation of PDF receptor (PDFR) in PI neurons advanced sleep onset, indicating Kv4 controls sleep onset via regulating PDF/PDFR signaling in wake-promoting neurons. We propose that Kv4 acts as a sleep onset controller by suppressing membrane excitability in a clock-dependent manner to balance the wake–sleep transition at dusk. Our results have important implications for the understanding and treatment of sleep disorders such as insomnia.
SIGNIFICANCE STATEMENT The mechanisms by which our brains reversibly switch from waking to sleep state remain an unanswered and intriguing question in biological research. In this study, we identified that Shal/Kv4, a well known voltage-gated K+ channel, acts as a controller of wake–sleep transition at dusk in Drosophila circadian neurons. We find that interference of Kv4 function with a dominant-negative form (DNKv4) in subsets of circadian neurons specifically disrupts sleep onset at dusk, although Kv4 itself does not exhibit circadian oscillation. Kv4 preferentially downregulates neuronal firings at ZT9–ZT17, supporting that it plays an essential role in wake–sleep transition at dusk. Our findings may help understand and eventually treat sleep disorders such as insomnia.
Keywords: circadian, Drosophila, Kv4, Shaker, sleep
Introduction
Sleep is an essential process that is believed to be regulated by circadian clock and other factors. Drosophila melanogaster has become an ideal and well accepted model for sleep and circadian rhythms research (Kayser and Biron, 2016; Dubowy and Sehgal, 2017). In Drosophila, groups of neurons that regulate sleep were found in mushroom body neurons (MBNs), fan-shaped body (FB), the ellipsoid body (EB), pars intercerebralis (PI), large ventral lateral neurons (l-LNvs), dorsal neurons (DN1s), pars lateralis, and dorsolateral neurons (LNds; Strauss and Heisenberg, 1993; Joiner et al., 2006; Pitman et al., 2006; Foltenyi et al., 2007; Shang et al., 2008; Sheeba et al., 2008a; Keene et al., 2010; Afonso et al., 2015; Artiushin and Sehgal, 2017). Among them, l-LNvs are a group of wake-promoting neurons, which express neuropeptide pigment-dispersing factor (PDF), and regulate night-time sleep (Parisky et al., 2008). PDF, released by LNvs (also named PDF neurons), regulates circadian and sleep by acting on PDF receptor (PDFR)-expressing neurons, including LNds, DN1s, and PI neurons (Im and Taghert, 2010; Dubowy and Sehgal, 2017). DN1s, a group of potent sleep-promoting neurons at dusk, show effect on transition from wake to sleep via feedback mechanisms and may also relay signals from PDF neurons to PI (Cavanaugh et al., 2014; Kunst et al., 2014; Guo et al., 2016). PI, analogous to the mammalian hypothalamus, is an important output brain region for rest–activity rhythms (Foltenyi et al., 2007; Cavanaugh et al., 2014; Park et al., 2014; Cavey et al., 2016), and activation of PI neurons reduced sleep amount (Crocker et al., 2010). Although previous studies have implicated various types of circadian neurons in regulating sleep behavior, it remains to be determined whether these sleep- and wake-promoting neurons control wake/sleep amount and transition via different mechanisms, and the precise mechanisms by which they cooperate in the brain.
Insomnia is a condition characterized by difficulty initiating or maintaining sleep and dissatisfaction with sleep quantity or quality (Rosenberg, 2006). Although the biological basis of classification of initiating and maintaining sleep is unknown, sleep initiation is suggested to be uncoupled from sleep maintenance in Drosophila. In particular, hyperactivity of l-LNvs resulted in increased sleep latency and GABAA receptor played a critical role in the regulation (Agosto et al., 2008; Chung et al., 2009; Liu et al., 2014; Li et al., 2017). Another study shows that sleep initiation is dependent on the amnesiac gene expression (Liu et al., 2008). However, our understanding of the key molecules and neurons that control wake–sleep transition remains rudimentary.
Compared with wake state, there are large-scale changes in neuronal activity during sleep. Reports have shown that different types of ion channels, including Shaker and sandman, are required for normal wake–sleep cycles (Cirelli et al., 2005; Pimentel et al., 2016). Only two genes, Shaker/Kv1 and Shal/Kv4, encode voltage-gated A-type K+ channels in Drosophila neurons, whereas mammals contain multiple genes (Salkoff and Wyman, 1981; Wei et al., 1990; Covarrubias et al., 1991). In most Drosophila neurons, Shaker expression is extensively restricted to axons and terminals (Rogero et al., 1997; Ueda and Wu, 2006). In contrast, Kv4 channels are localized exclusively to soma and dendrites (Gasque et al., 2005; Bergquist et al., 2010; Diao et al., 2010; Srinivasan et al., 2012). In mammals, neurons in suprachiasmatic nucleus (SCN) function as central pacemaker and their firing activity exhibits circadian oscillation (Colwell, 2011). Loss of Shaker results in dysregulated sleep patterns in both Drosophila and mammals, and most Kv channels contributing to A-type currents (IA) regulate activity of neurons in SCN (Cirelli et al., 2005; Douglas et al., 2007; Granados-Fuentes et al., 2012). However, little is known about the role of Kv4 playing in sleep.
In this study, we found that Shal495 mutants exhibited delayed sleep onset at dusk, which could be rescued by restoring Shal/Kv4 expression. Our results further demonstrated that Kv4 controls sleep onset in wake-promoting neurons, but not in subsets of sleep-promoting neurons. These results support that Kv4 has an important role in controlling wake–sleep transition.
Materials and Methods
Animals.
Flies were kept at 23°C on cornmeal, yeast, and sucrose and agar food. w1118 (5905; RRID:BDSC_5905), elav-GAL4 (458; RRID:BDSC_8760), pdf-GAL4 (6900, 6899; RRIDs:BDSC_69002, BDSC_6899, 50y-GAL4 (30820; RRID:BDSC_30820), tub-GAL80ts (7019), UAS-dTrpA1 (26263), pdf01 (26654; RRID:BDSC_26654), Shmns (22837), UAS-EKO (40973; RRID:BDSC_40974), UAS-Shal-RNAi (31879), and clk4.1M-GAL4 (36316; RRID:BDSC_36316) were obtained from the Bloomington Stock Center. UAS-CD8GFP (RRID:BDSC_5137), Shal495 (Bergquist et al., 2010), R18H11-GAL4 (RRID:BDSC_48832), UAS-white-RNAi, and pdf-GAL80 (Stoleru et al., 2004) were obtained from Drs. Susan Tsunoda, Bingwei Lu, and Yufeng Pan. UAS-DNKv4 and UAS-Kv4 transgenic lines were generated previously (Ping and Tsunoda, 2011; Ping et al., 2011). Flies were outcrossed for more than five generations with the w1118 strain to standardize the background.
Sleep and activity assays.
All behavior was done on female flies unless otherwise specified. Three- to 5-d-old virgin flies were monitored at 25°C in glass tube containing 5% sucrose and 2% agar using the Drosophila Activity Monitoring System (Trikinetics). Flies were raised in behavior tubes for >72 h at 25°C in 12 h light/dark (LD) conditions for acclimation before data collection. Flies were entrained at least 3 d in LD before switching to constant darkness (DD). Activity counts were collected in 1 min bins in LD for 3 d. Sleep was defined as 5 consecutive minutes of inactivity. Sleep latency was measured from the time lights-off to the onset of first sleep episode. Sleep parameters were analyzed using R software. For analysis of sleep in DD, sleep latency was calculated from CT12. For UAS-dTrpA1 experiments, data were recorded at 29°C to activate the relevant neurons. For conditional expression experiments using Tub-GAL80ts, animals containing UAS-DNKv4, driven by Tub-GAL80ts and PDF-GAL4, were reared at 18°C to prevent DNKv4 expression at developmental stages. The temperature was then raised to 29°C to induce expression for 14 d post-eclosion, followed by conventional sleep assays. Free-running activity was measured over 3 d entrainment for LD and over 7 d for DD, as reported previously (Song et al., 2017). Rhythmic flies were defined by χ2 periodogram analysis (ClockLab software), and power is a measure of rhythm amplitude and corresponds to the height of the periodogram peak above the significance line. More than 20 flies were examined for each genotype and condition, and dead flies were excluded from the data analysis.
Electrophysiology.
Flies were kept at 25°C in LD cycles. Whole-cell recordings were performed in perforated patch-clamp configuration by adding 400–800 μg/ml amphotericin-B (Sigma-Aldrich) in the pipette, as reported previously (Feng et al., 2018). For voltage-dependent K+ current recordings from GFP-labeled PDF (l-LNvs), DN1s, and PI neurons in the isolated brain, we first dissected brain in the Drosophila culture medium (Invitrogen) and then we exchanged culture medium to the recording external solution (in mm): 110 NaCl, 2 KCl, 6 MgCl2, 1 CaCl2, 5 glucose, 20 NaHCO3, 2 NaH2PO4. Solution was continuously bubbled with carbogen (5% oxygen and 95% carbon dioxide) throughout recording, pH was set at 7.2. TTX (1 μm) and nifedipine (10 μm) were added to the external solution to block Na+ and Ca2+ currents (for IA recordings). Electrodes were filled with internal solution (in mm): K-gluconate, 120; KCl, 20; Mg-ATP, 4; Na-GTP, 0.5; HEPEs, 10; EGTA, 1.1; MgCl2, 2; CaCl2, 0.1, with pH 7.2. The l-LNvs were distinguished from the sLNvs by their size and anatomical location. We performed all recordings at room temperature. Electrode resistances for all recordings were 5–10 MΩ. Gigaohm seals were obtained for whole-cell recordings. Cells were clamped at −80 mV. The Kv2–Kv3-mediated delayed rectifier (DR) component was recorded using a pre-pulse of −45 mV to completely inactivate Kv4 channels, before stepping to a test potential of +50 mV; the Kv4 current or IA was then isolated by subtracting this DR component from the total whole-cell current elicited from a pre-pulse of −125 mV, stepping to a test potential of +50 mV. In current-clamp experiments, we clamped the membrane current at 0 pA unless otherwise specified (Fig. 5-1B). Resting potential was determined until stabilization of the membrane potential after the transition from voltage-clamp to current-clamp mode. 4-AP (Sigma-Aldrich) was diluted at the appropriate concentration in the external saline solution, and superfused to the recording chamber. Phrixotoxin-2 (PaTx2; Alomone Labs) was kept frozen for <2 months as 10× stock (10 μm) aliquots and thawed immediately before experiments.
For cell-attached recordings, recording pipettes were filled with internal solution, with additional 200 mm BaCl2 to prevent perforated patch effect mediated by high density of Kir channels in the pipettes. Cell attached configuration was achieved by gentle suction through a syringe. Recordings were performed in voltage-clamp mode without holding. Action potential number was quantified using MiniAnalysis software for at least 1 min.
Experimental design and statistical analysis.
The main objective of the present study was to investigate the sleep deficits in DNKv4 mutants of both sexes and the effects of Shal/Kv4 on neuronal excitability. Therefore, sleep measurements and electrophysiological recordings of GFP-labeled circadian neurons were performed on at least six flies for each genotype. The n number and gender of flies used in this study were described in the corresponding figure legends or results. For sleep measurements, as shown in Figures 1, 1-1, 2, 2-1, 3, 5-1A, 6A–C, 7, and 7-1, each n corresponds to one animal. For electrophysiological recordings, as shown in Figures 4, 4-1, 5, 5-1B, and 6D,E, each n corresponds to one cell, with one cell recorded per animal. We required an experimental result to be significantly different from both genetic controls (if applicable).
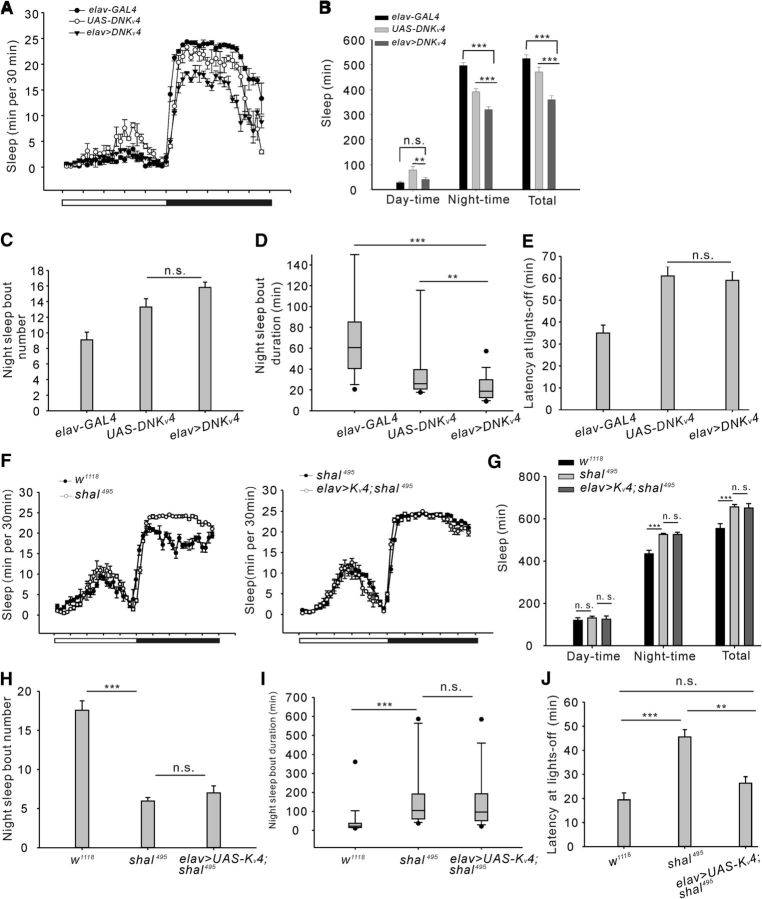
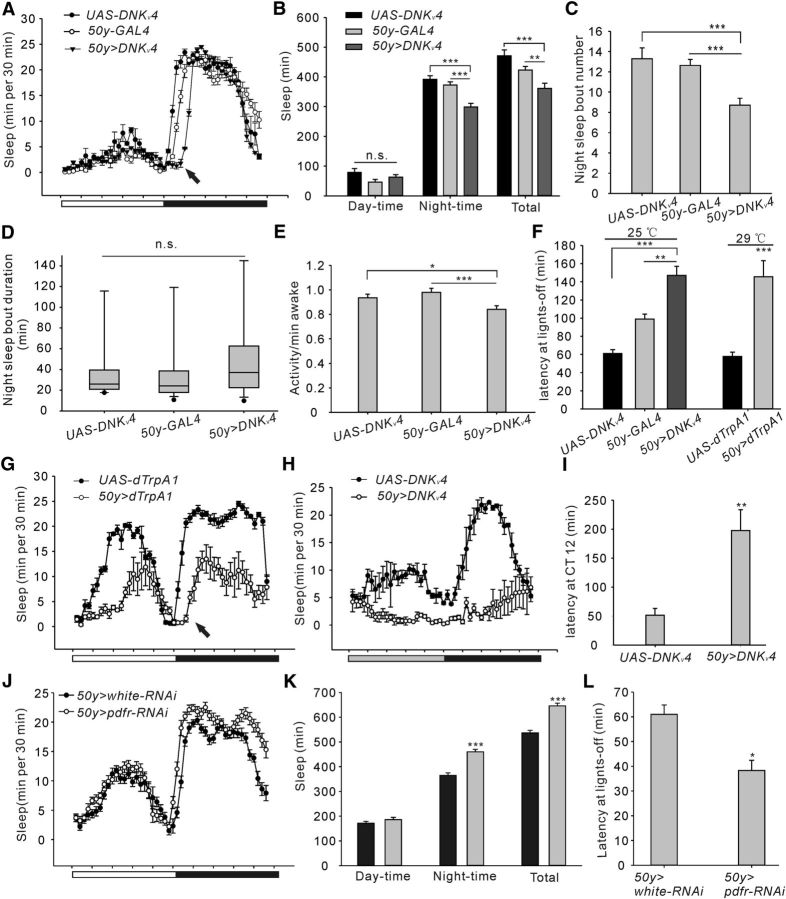
Figure 1.
Sleep phenotypes in two different types of voltage-gated K+ channel 4 (Kv4) mutants. A, Conventional sleep plots of controls (elav-GAL4 and UAS-DNKv4, n = 27) and experimental flies (elav > DNKv4, n = 69) in 12 h LD. DN, dominant negative. White and black bars indicate 12 h light/dark periods, respectively. B, Total sleep amount (in 24 h) and sleep during daytime and night-time in controls and experimental lines as described in A. C, Histograms of the number of night-time sleep bouts during night-time for controls and experimental flies. D, Box plots for night-time sleep bout duration. As the data are not normally distributed, box plots were used. The line inside the box indicates the median; the upper and lower box limits the 75 and 25% quantiles; vertical lines above and below the box represent the 90 and 10% quantiles; points show the 95% and 5% outliers. E, Sleep latency after lights-off at ZT12. Sleep latency was measured from the time lights-off to the onset of first sleep episode. F, Conventional sleep plots of w1118 (n = 35), Shal495 (n = 95), and Shal495 with Kv4 overexpression lines (elav> Kv4;Shal495) (n = 33). G–J, Histograms of total sleep amount and sleep during daytime and night-time (G), night-time sleep bout number (H), night-time sleep bout duration (I), and sleep latency at lights-off (J). Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant. Data are compared by Student's t test, except Mann–Whitney U test for comparison of nonparametrically distributed data. For multiple comparisons, one-way ANOVAs followed by post hoc Tukey were performed. For the quantification of Shaker mRNA expression levels in Shal/Kv4 mutants and sleep deficits in Shaker mutants (Figure 1-1).
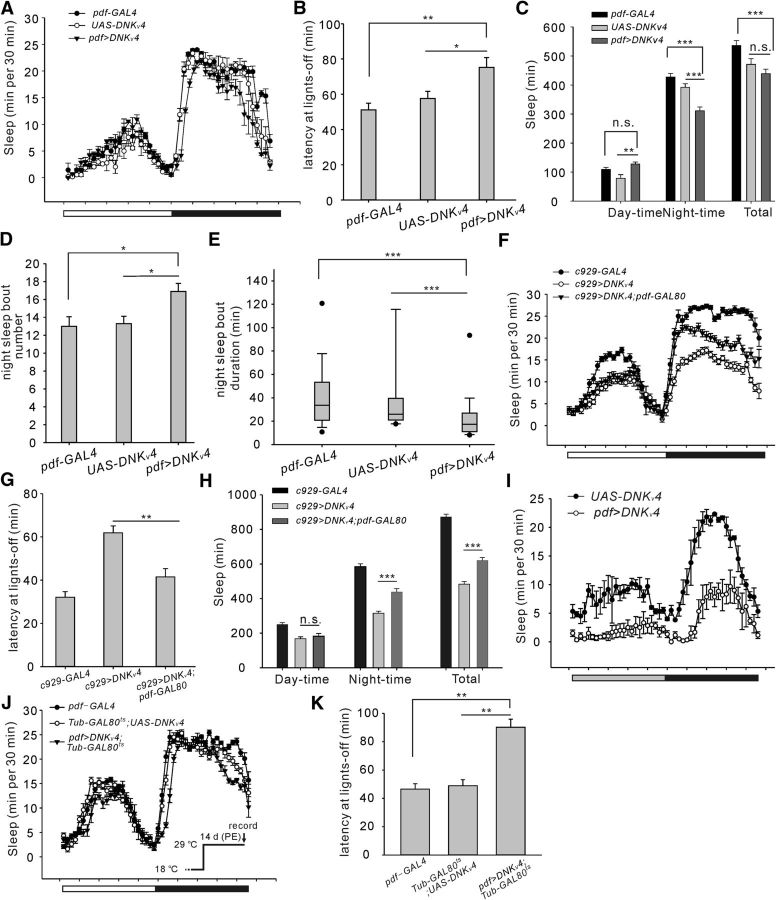
Figure 2.
Kv4 in pigment-dispersing factor (PDF) cells regulates sleep onset. A, Conventional sleep plots of controls (pdf-GAL4 and UAS-DNKv4, n = 32) and experimental flies (pdf>DNKv4, n = 47) in LD. B–E, Quantification of sleep latency after lights-off (B), and other sleep parameters showing total sleep amount and sleep during daytime and night-time (C), night-time sleep bout number (D) and night-time sleep duration (E). F, Conventional sleep plots of the transgenic lines, including c929-GAL4 (n = 31), c929>DNKv4 (n = 33), and c929>DNKv4;pdf-GAL80 (n = 28). G, H, Quantification of sleep latency and sleep amount of flies shown in F. I, Conventional sleep plots in DD of control (UAS-DNKv4, n = 30) and experimental (pdf>DNKv4, n = 31) flies. Gray and black bars indicate 12 h subjective day and night, respectively. Note that sleep latency at subjective night was increased by expressing DNKv4 in DD. J, K, Conventional sleep plots of controls (pdf-GAL4 and Tub-GAL80ts;UAS-DNKv4, n = 22, and 27, respectively) and experimental flies (pdf>DNKv4;Tub-GAL80ts, n = 27) at 29°C in LD (J) and quantification of sleep latency (K; inset procedure). Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant. Data are compared by Student's t test, except Mann–Whitney U test for comparison of nonparametrically distributed data. For multiple comparisons, one-way ANOVAs followed by post hoc Tukey were performed. For the quantification of sleep onset in males, Shal-RNAi mutants and circadian period (Figure 2-1).
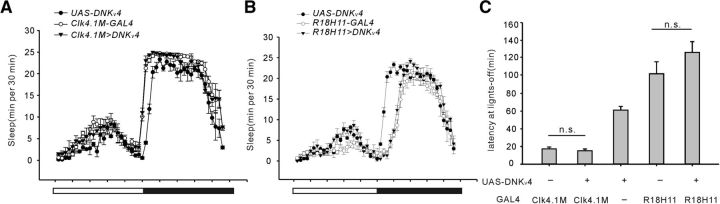
Figure 3.
Interference of Kv4 in dorsal neurons (DN1s) does not significantly regulate sleep onset. A, B, Conventional sleep plots of controls (Clk4.1M-GAL4, UAS-DNKv4, and R18H11-GAL4; n = 27–32) and experimental flies (Clk4.1M>DNKv4 and R18H11>DNKv4; n = 30) in LD cycles. C, Quantification of sleep latency after lights-off. Mean ± SEM is shown. n.s., not significant. One-way ANOVAs followed by post hoc Tukey were performed. Data are compared by one-way ANOVAs followed by post hoc Tukey.
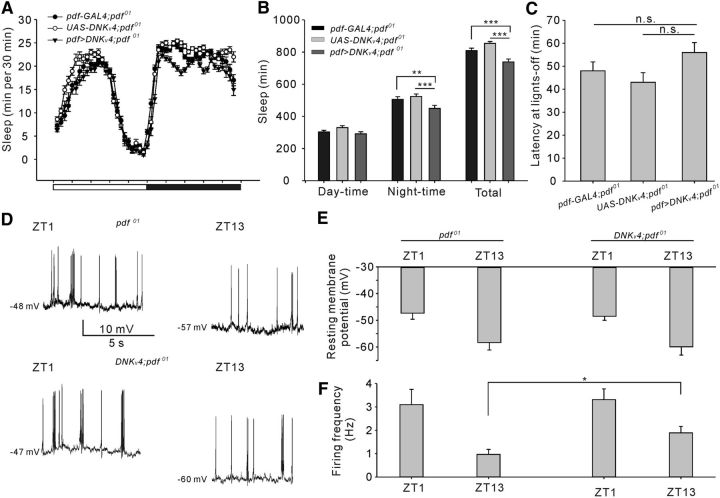
Figure 6.
PDF is required for DNKv4-mediated delayed sleep onset in PDF neurons. A, Conventional sleep plots of indicated transgenic lines, including pdf-GAL4;pdf01 (n = 33), UAS-DNKv4;pdf01 (n = 27), and pdf>DNKv4;pdf01 (n = 27). B, C, Quantification of sleep amount (B) and sleep latency (C) of flies shown in A. D, Representative spontaneous firing traces showing RMP values of GFP-labeled l-LNvs in pdf01 background flies by current-clamp recordings. Recordings were performed at ZT1 and ZT13 and two transgenic lines were used (pdf>CD8GFP;pdf01 and pdf>CD8GFP;DNKv4;pdf01). E, F, Quantification of RMPs and firing frequency of l-LNvs recorded in D. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant. One-way ANOVAs followed by post hoc Tukey were performed.
Figure 7.
Function-loss of Kv4 in PI also leads to delayed sleep onset. A, Conventional sleep plots of controls (UAS-DNKv4, n = 27; 50y-GAL4, n = 32) and experimental (50y>DNKv4, n = 49) flies in LD. White and black bars indicate 12 h light/dark periods, respectively, and the arrow indicates delayed sleep onset in experimental flies. B–E, Quantification of sleep parameters showing total sleep amount (24 h), sleep during daytime and night-time (B), night-time sleep bout number (C), night-time sleep duration (D), and waking activity (E). F, Sleep latency after lights-off at ZT12 for controls versus experimental flies at 25°C (left), and UAS-dTrpA1 vs. 50y>dTrpA1 at 29°C (right). G, Conventional sleep plots of control (UAS-dTrpA1, n = 30) and experimental (50y>dTrpA1, n = 39) flies in LD. Flies were raised at 29°C to activate dTrpA1 during test period. The arrows indicates delayed sleep onset in experimental flies. Quantifications are shown in F. H, Conventional sleep plots in DD of control (UAS-DNKv4, n = 30) and experimental (50y>DNKv4, n = 25) flies. Gray and black bars indicate 12 h subjective day and night, respectively. I, Quantification of sleep latency at subjective night in DD for the genotypes as indicated in H. J, Conventional sleep plots of control (50y>white-RNAi, n = 26) and experimental (50y>pdfr-RNAi, n = 28) flies. K, L, Quantification of sleep amount (K) and sleep latency (L) for the genotypes as indicated in J. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant. Data are compared by Student's t test, except Mann–Whitney U test for comparison of nonparametrically distributed data. For multiple comparisons, one-way ANOVAs followed by post hoc Tukey were performed. For the quantification of sleep onset in males, see Figure 7-1.
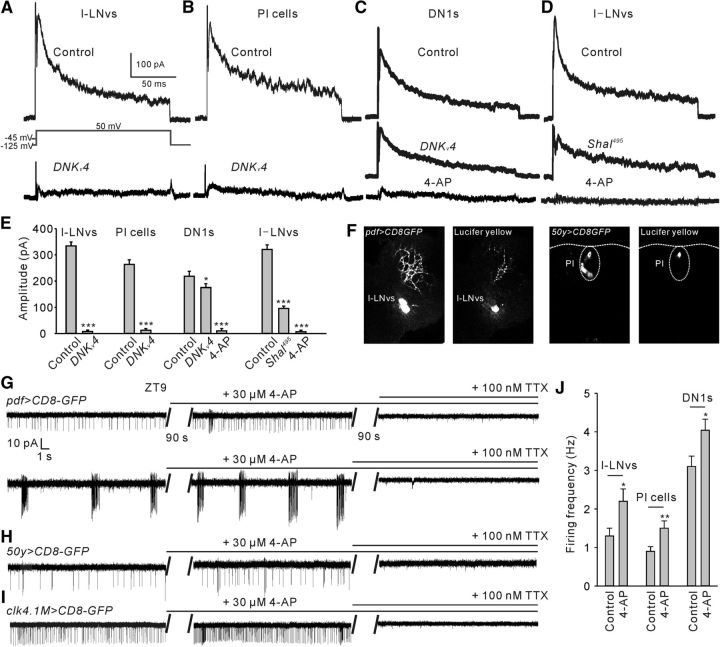
Figure 4.
Kv4 contributes to A-type currents (IA) and regulates activities in subsets of circadian neurons. A, Shown are representative voltage-clamp recordings of IA from l-LNvs in two genotypes: pdf>CD8GFP (Control) and pdf>CD8GFP;DNKv4 (DNKv4). We first recorded total whole-cell K+ current, which was elicited by a 500 ms prepulse of −125 mV with a voltage jump to +50 mV. Then Kv4 (or IA) is completely inactivated and the total delayed rectifier (DR) current remains with a prepulse of +45 mV. IA trace obtained by subtracting the DR current trace from the total K+ current trace. B, Shown are representative recordings of IA from PI neurons in two genotypes: 50y>CD8GFP and 50y>CD8GFP;DNKv4. C, Shown are representative recordings of IA from DN1s in two genotypes: clk4.1M>CD8GFP and clk4.1M>CD8GFP;DNKv4, in the presence or absence of 2 mm 4-AP. D, Shown are representative recordings of IA from large ventral lateral neurons (l-LNvs) in two genotypes: pdf>CD8GFP and pdf>CD8GFP;Shal495, in the presence or absence of 4-AP. E, Quantification of IA amplitudes recorded from the genotypes shown in A–D. Note that there is still remaining IA (∼100 pA) in clk4.1M>CD8GFP;DNKv4 (DN1s) and Shal495 (l-LNvs) mutants; n = 6–9 for each case. F, GFP-labeled PDF cells and a typical Lucifer yellow-filled l-LNv during recording (left two). GFP labeled pars intercerebralis (PI) cells and a typical Lucifer yellow-filled PI neuron during recording (right two). G, Two typical firing patterns (tonic and burst firing) as shown in the up and down panels in GFP-labeled l-LNvs. 4-AP, a specific IA blocker, increased firing rate in both firing patterns at ZT9, and all the AP currents could be completely inhibited by TTX. H, I, 4-AP also increased firing rate in PI neurons and DN1s, and all the AP currents were blocked by TTX. J, Quantification of firing frequency in the cells as shown in G–I; n = 6–12 for each case. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001. Data are compared by Student's t test. For multiple comparisons, one-way ANOVAs followed by post hoc Tukey were performed. For the comparison of RMPs and firing rate in DN1s between control and DNKv4 mutants (Figure 4-1).
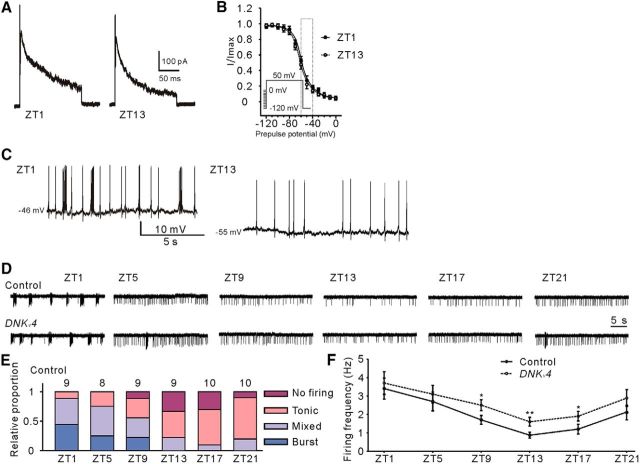
Figure 5.
Kv4 controls membrane excitability at dusk. A, Shown are representative recordings of IA from GFP-labeled l-LNvs at ZT1 and ZT13. Note that there is no significant change in IA amplitudes. n = 12 for ZT1 and n = 13 for ZT13. B, Steady-state inactivation properties of Kv4 at ZT1 and ZT13. We used a pre-pulse from −120 to 0 mV, in 10 mV intervals, then stepped to a test potential of +50 mV (inset, procedure). Steady-state inactivation curves were fitted with Boltzmann function: I/Imax = 1/(1 + exp[(V − V1/2)/k]). There was no significant difference in half-maximal inactivation potential values (V1/2). (ZT1: V1/2 = −59.3 ± 1.9 mV, n = 11; ZT13: V1/2 = −61.5 ± 3.1 mV, n = 11). C, Representative spontaneous firing traces (at ZT1 and ZT13) showing resting membrane potential (RMP) values of GFP-labeled l-LNvs by current-clamp recordings. Note that RMP is significantly depolarized at ZT1 compared with ZT13; n = 6 for ZT1 and n = 7 for ZT13. D, Representative spontaneous firing patterns (cell-attached) are shown at the indicated time of recordings in control (pdf>CD8GFP) or DNKv4-expressing l-LNvs (pdf>DNKv4;CD8GFP). Note that l-LNvs exhibit circadian oscillation of firing, with higher firing frequency at dawn and lower firing frequency at dusk. E, Relative proportions of firing phenotypes from l-LNvs at the indicated time of the circadian day. F, Quantification of firing rates at the indicated time of the circadian day as shown in E. Loss of Kv4 resulted in increased firing rate preferentially at dusk, when the firing frequency and RMP are relatively lower in l-LNvs; n = 8–10 for each case. Mean ± SEM is shown. *p < 0.05, **p < 0.01. Data are compared by Student's t test. For multiple comparisons, one-way ANOVAs followed by post hoc Tukey were performed. For the comparison of sleep onset by activating PDF cells between ZT9–12 and ZT12–15 (Figure 5-1A). For the comparison of firing rate between control and PaTx2-treated l-LNvs at different membrane potentials, (Figure 5-1B).
We used Student's t test for comparisons of two groups of normally distributed data using Sigma-AldrichPlot 10.0 software (RRID:SCR_003210), as shown in Fig. 1-1D,G,J; 2J; 2-1A,B,E,F; 4E,J; 5A,C,F; 5-1A; and 7I,K,L. For multiple comparisons, one-way ANOVA followed by post hoc Tukey were performed using SPSS software (RRID:SCR_002865), as shown in Figures 1B,C,E,G,H,J; 1-1A; 2B–D,G,H,L; 2-1D,H; 3C; 4E; 4-1C,D; 5-1B; 6B,C,E,F; 7B,C,E,F; and 7-1B. For comparisons of non-normally distributed data (sleep bout duration), Mann–Whitney U tests were performed using SPSS software as shown in Figures 1D,I, 2E, and 7D. See Results and figure legends for specifics on statistical tests for each experiment. Statistical significance was set at *p < 0.05 and data are reported as mean ± SEM.
Results
Kv4 is important for sleep in Drosophila
To determine whether Kv4 is important for regulating sleep in Drosophila, we first selected dominant-negative mutation, UAS-DNKv4, and expressed it with the pan-neuronal driver (elav/+;+;UAS-DNKv4/+, DNKv4), which could completely eliminate Kv4 function (Ping and Tsunoda, 2011; Ping et al., 2015). Total and night-time sleep amounts were reduced in DNKv4 female flies (Fig. 1A,B; daytime: F(2,120) = 7.4, **p = 0.0046, p = 0.1056; night-time: F(2,120) = 38.89, elav-GAL4 vs elav>DNKv4, ***p < 0.0001, UAS-DNKv4 vs elav>DNKv4, ***p = 0.0009; total: F(2,120) = 43.27, ***p < 0.0001; one-way ANOVA). Daytime sleep was not significantly reduced (Fig. 1A,B), and it might be due to a floor effect, because the control lines exhibited very little daytime sleep. The night-time sleep phenotypes were largely due to a decrease in sleep-bout duration (Fig. 1D; elav-GAL4 vs elav>DNKv4, U = 164, p < 0.0001; UAS-DNKv4 vs elav>DNKv4, U = 556.5, p = 0.001. Mann–Whitney U test), because no significant change was found in sleep number and latency (Fig. 1C,E; sleep number: F(2,120) = 14.12, p = 0.1167; latency: F(2,120) = 11.54, p = 0.6774; one-way ANOVA). These results indicate that Kv4 can improve sleep maintenance and has sleep-promoting effects.
Shaker (Sh) mRNA expression levels in Shal/Kv4 mutants and sleep phenotypes in Sh mutants. A, Sh mRNA expression was measured in DNKv4 (elav/+;+;UAS-DNKv4/+), Shal495 and Kv4 + Shal495 (elav/+;UAS-Kv4/+;Shal495) animals. For quantitative RT-PCR, brains were isolated and total RNA was isolated from each sample using the standard Trizol Protocol. A DNase digestion was then done to remove potential DNA contamination and RT was performed. Each experimental animal sample was compared to each wild-type (wt) sample. All bars are represented as percent of wt animals. We noted that increased Shaker RNA expression in Shal495 mutants was not rescued by restoration of Shal/Kv4 in Shal495 mutants using the GAL4/UAS system. B, Conventional sleep plots of w1118 (n = 24) and experimental flies (Shmns/+, n = 35) in 12h:12h light:dark (LD). C, Total sleep amount (24h) and sleep during day-time and night-time in wt and experimental lines were analyzed as described in B. These data show reduced sleep amount in heterozygous female Shmns flies. D, Sleep latency after lights-off at ZT12. E, Conventional sleep plots of control (Shmns;+;+, n = 26) and experimental flies (Shmns;+;Shal495, n = 25) in 12h:12h light:dark (LD). F, Total sleep amount (24h) and sleep during day-time and night-time in wt and experimental lines. These data show reduced night-time sleep amount in Shal495 flies, in the presence of Shmns. Note that these flies exhibited very little day-time sleep. G, Sleep latency after lights-off at ZT12. H, Conventional sleep plots of control (w;UAS-Eko/+;+, n = 33) and experimental flies (pdf-GAL4/+;UAS-Eko/+;+, n = 27) in 12h:12h light:dark (LD). I, Total sleep amount (24h) and sleep during day-time and night-time in wt and experimental lines. J, Sleep latency after lights off at ZT12. Download Figure 1-1, TIF file (10.6MB, tif)
Next, we tested whether another null mutation of Shal495 could lead to sleep phenotypes observed in DNKv4 flies. Surprisingly, compared with background control (w1118), night-time sleep was markedly increased in Shal495, without significant changes in daytime sleep (Fig. 1F,G; daytime: F(2,160) = 0.5637, w1118 vs Shal495, p = 0.1732, Shal495 vs elav>Kv4;Shal495, p = 0.3318; night-time: F(2,160) = 26.47, w1118 vs Shal495, ***p < 0.0001, Shal495 vs elav>Kv4;Shal495, p = 0.2351; total: F(2,160) = 26.89, w1118 vs Shal495, ***p < 0.0001, Shal495 vs elav>Kv4;Shal495, p = 0.1879; one-way ANOVA). Further analysis showed the increased sleep was due to lengthened sleep bout duration (Fig. 1H; sleep number: F(2,160) = 8.69, w1118 vs Shal495, ***p < 0.0001; Shal495 vs elav>Kv4;Shal495, p = 0.2004, one-way ANOVA; I, sleep bout duration: w1118 vs Shal495, U = 432.8, ***p = 0.0003, Shal495 vs elav>Kv4;Shal495, U = 641.3, p = 0.1374, Mann–Whitney U test), suggesting that Shal495 mutation enhances sleep maintenance. Additionally, sleep latency in Shal495 mutant was increased significantly compared with control (Fig. 1J; F(2,160) = 11.45, w1118 vs Shal495, ***p = 0.0005, Shal495 vs elav>Kv4;Shal495, **p = 0.0034; w1118 vs elav>Kv4;Shal495, p = 0.0894, one-way ANOVA). Studies have shown that loss of Shal/Kv4 could induce compensatory upregulation of Shaker/Kv1 (Bergquist et al., 2010; Parrish et al., 2014). Moreover, Shaker mutants (Shmns) exhibited reduced sleep in Drosophila, even in heterozygous flies (Fig. 1-1B–D; sleep: F(5,171) = 32.56, ***p < 0.0001, **p = 0.0035 for night-time, **p = 0.0054 for total, one-way ANOVA; latency: T(60) = 3.25, *p = 0.0142, t test). Thus, we assume that the increased sleep in Shal495 mutants could be caused by the upregulation of Shaker. Indeed, Shal495 mutant exhibited significant increase in Shaker expression as detected using qPCR (Fig. 1-1A; F(2,14) = 74.53, ***p < 0.0001, one-way ANOVA), whereas Shaker expression was not increased in DNKv4 mutants (Fig. 1-1A). Moreover, when Shaker function was inhibited in shal495 mutants, the animals exhibited reduced night-time sleep, similar as observed in DNKv4 expression animals (Fig. 1-1E–G; F(5, 147) = 15.76, ***p < 0.0001, one-way ANOVA; T(49) = 3.87, p = 0.3155, t test).
Subsequently, we tested whether the sleep phenotypes in Shal495 mutant could be rescued by overexpression of Shal/Kv4 via the GAL4/UAS system. However, Shal495 mutant flies with Shal overexpression (elav/+; UAS-Kv4/+;Shal495) showed similar sleep time, as well as sleep number, sleep duration (Fig. 1F–I; statistics were shown above) and Shaker expression (Fig. 1-1A) compared with Shal495 mutants. Interestingly, there was a significant reduction in sleep latency after overexpressing Kv4 (Fig. 1J), indicating that the delayed sleep onset in Shal495 could be attributed to loss of Kv4, and not compensated by Shaker overexpression. Whereas DNKv4-expressing flies did not show significant change in timing of sleep onset (Fig. 1E). We presume that the sleep phenotypes induced by pan-neuronal expression of UAS-DNKv4 may reflect an integrated effect of multiple regulatory mechanisms operating in different neuronal populations, as we will show later that DNKv4 expression in wake-promoting neurons delays sleep onset, but without effects on sleep onset when expressed in subsets of sleep-promoting neurons.
Loss of Kv4 function in l-LNvs delays sleep onset
We next tested sleep-regulating effects of Kv4 in subsets of circadian neurons. Studies have shown that l-LNvs, a group of wake-promoting cells, regulate night-time sleep via GABA receptors (Agosto et al., 2008; Parisky et al., 2008; Liu et al., 2014). Thus, we first selected pdf-GAL4 to drive expression of UAS-DNKv4 to eliminate Kv4 function (pdf-GAL4/+;+;UAS-DNKv4/+) in small- (s-) and l-LNvs. Strikingly, sleep latency was markedly increased in pdf>DNKv4 female flies compared with controls, accompanied by decreased night-time sleep (Fig. 2A–C; latency: F(2,108) = 26.34, **p = 0.0076, *p = 0.0328; sleep: F(2,108) = 8.31, p = 0.0631, **p = 0.0039 for daytime, F(2,108) = 25.78, ***p < 0.0001 for night-time, F(2,108) = 9.62, p = 0.0733, ***p = 0.0003 for total, one-way ANOVA). Nevertheless, total and daytime sleep amounts did not exhibit a significant change (Fig. 2C). There was an increase in night sleep bout number, but night sleep bout duration was decreased (Fig. 2D; F(2,108) = 18.22, pdf-GAL4 vs pdf>DNKv4, *p = 0.0276, UAS-DNKv4 vs pdf>DNKv4, *p = 0.0289, one-way ANOVA, E, pdf-GAL4 vs pdf>DNKv4, U = 223.5, UAS-DNKv4 vs pdf>DNKv4, U = 336, ***p < 0.0001, Mann–Whitney U test). Delayed sleep onset latency was further confirmed using another pdf-GAL4 on chromosome 2 (Fig. 2-1A; T(72) = 6.15, ***p < 0.0001, t test), and was also observed in male flies (Fig. 2-1B; T(56) = 5.46, *p = 0.0219, t test). Moreover, increase or decrease in wakefulness was found in the early night by activating or inhibiting PDF cells, respectively (Fig. 1-1H–J; F(5, 174) = 12.44, ***p = 0.0003 for daytime, ***p < 0.0001 for night-time and total, one-way ANOVA; T(57) = 2.92, **p = 0.0051, t test; Parisky et al., 2008). Restored Kv4 expression only in PDF cells partially rescued delayed sleep onset in Shal495 flies (Fig. 2-1E,F; T(53) = 2.16, *p = 0.033, t test). We did not observe significant change in circadian period in pdf>DNKv4 flies (Fig. 2-1C; pdf-GAL4: period = 23.7 ± 0.10 h; UAS-DNKv4: period = 23.8 ± 0.12 h; pdf>DNKv4: 24.1 ± 0.11 h; F(2,93) = 11.03, pdf-GAL4 vs pdf>DNKv4, p = 0.0842, UAS-DNKv4 vs pdf>DNKv4, p = 0.1037, one-way ANOVA). Downregulation of Kv4 expression in PDF cells (pdf>Shal-RNAi) also delayed sleep onset (Fig. 2-1G,H; F(2,76) = 51.76, UAS-Shal-RNAi vs pdf>Shal-RNAi, **p = 0.0039, pdf-GAL4 vs pdf>Shal-RNAi, **p = 0.0054, one-way ANOVA), consistent with the results of DNKv4.
DNKv4 and Shal-RNAi expression in PDF neurons delay sleep onset, but without effects on circadian period by interference of Kv4 in PDF and DN1s. A, Conventional sleep plots of control (w;UAS-DNKv4/+;+, n = 27) and experimental flies (w;UAS-DNKv4/pdf-GAL4;+, n = 49) in 12h:12h light:dark (LD) (left), and Quantification of sleep latency after lights-off at ZT12 (right). Our data show that blocking Kv4 function in PDF cells delayed sleep onset. B, Quantification of sleep latency at lights-off at ZT12 in male flies (n = 24 for UAS-DNKv4, n = 34 for pdf>DNKv4). C, Activity records of representative controls (pdf-GAL4, and UAS-DNKv4) and experimental flies (pdf>DNKv4) for 3 d in LD and following 7 d in DD. Quantification of circadian periods and rhythmicity is presented in the top panel. Our data show that there is no significant difference in circadian periods. D, Quantification of circadian periods for indicated genotypes, including UAS-DNKv4, R18H11-GAL4, R18H11>DNKv4, 50y-GAL4, and 50y>DNKv4. Rhythmicity is also shown in the bar graph. Our data show that DNKv4 expression in subsets of DN1s does not change the circadian periods (p > 0.05) and rhythmicity, while most (over 80%) flies became arrhythmic when expressing DNKv4 in PI cells and the remaining rhythmic ones had ∼25.3-hour period phenotype. E, F, Conventional sleep plots of two genotypes (Shal495 , n = 26, and pdf>Kv4;Shal495, n = 29) in LD (E), and sleep latency after lights-off at ZT12 (F). Our data indicate that restoring Kv4 expression selectively in PDF cells partially rescues delayed sleep onset in Shal495 mutants. G, H, Conventional sleep plots of controls (UAS-Shal-RNAi and pdf-GAL4, n = 24-28) and experimental (pdf>Shal-RNAi, n = 27) flies (G), and sleep latency after lights-off at ZT12 (H). Our data show that down-regulation of Kv4 expression by RNA interference (RNAi) also delays sleep onset, similar to those of DNKv4. Download Figure 2-1, TIF file (16.8MB, tif)
To examine the specific roles of s- and l-LNvs, we overexpressed DNKv4 with the c929-GAL4 driver, which drives expression in l-LNvs, but not s-LNvs. We found that sleep latency was increased in c929>DNKv4 flies, accompanied by decreased daytime and night-time sleep amounts. Since pdf-GAL80 would suppress the transcription activity of GAL4 in s/l-LNvs, we assayed the sleep phenotype of c929>DNKv4 flies expressing pdf-GAL80 (c929-GAL4/+;UAS-DNKv4/+;pdf-GAL80/+). These flies exhibited decreased sleep latency and increased night-time sleep, compared with c929>DNKv4 flies (Fig. 2F,G; F(2,89) = 91.29, **p = 0.0063; H, daytime: F(2,89) = 2.31, p = 0.2318, night-time: F(2,89) = 65.49, ***p < 0.0001; total: F(2,89) = 78.39, ***p < 0.0001, one-way ANOVA). These results further support that Kv4 functions to control sleep onset in l-LNvs, but not s-LNvs.
To rule out the possibility that the increased sleep latency was caused by an enhanced startle response to lights-off, we examined sleep in pdf>DNKv4 flies under constant darkness (Day 1 of DD), and assayed the time it took flies to fall asleep from CT12. The increase of sleep latency persisted in DD (Fig. 2I; latency at CT12, 51 ± 10.4 min for UAS-DNKv4, 163 ± 19.7 min for pdf>DNKv4, T(59) = 5.23, ***p < 0.0001, t test). Next, to bypass possible complications from the persistent Kv4 expression during development, we induced pdf-GAL4 along with tub-GAL80ts, which inhibits GAL4 function in a temperature-dependent manner, to silence Kv4 expression during development and switch the expression on in the adulthood. Compared with control lines, sleep latency of pdf>DNKv4;tub-GAL80ts was significantly increased (Fig. 2J,K; F(2,73) = 107.5, pdf-GAL4 vs pdf>DNKv4;tub-GAL80ts, **p = 0.0021, tub-GAL80ts; UAS-DNKv4 vs pdf>DNKv4;tub-GAL80ts, **p = 0.0028, one-way ANOVA).
No significant change in sleep onset was observed by blocking Kv4 activity in DN1s
Wake–sleep transition is expected to be determined by integration of signals from both wake- and sleep-promoting neurons. Next we examined whether Kv4 in sleep-promoting neurons also regulates sleep onset. Because reports show activation of DN1s promotes sleep at dusk (Kunst et al., 2014; Guo et al., 2016), we first selected clk4.1M-GAL4 to drive expression of UAS-DNKv4 to eliminate Kv4 function in DN1s. Sleep latency at lights-off was not changed by inhibiting Kv4 function in DN1s (Fig. 3A,C; F(2,84) = 47.78, p = 0.1068, one-way ANOVA). We then used another DN1s driver, R18H11-GAL4, and further confirmed no significant change in sleep latency by inhibiting Kv4 function compared with controls (Fig. 3B,C; F(2,84) = 24.37, p = 0.0927, one-way ANOVA). Moreover, we did not observe any significant change in circadian period in R18H11>DNKv4 flies compared with controls (Fig. 2-1D).
Kv4 channels regulate excitability of circadian neurons
Next, we investigated how DNKv4 regulates sleep onset. We first tested whether DNKv4 expression eliminates IA in the circadian neurons. We chose pdf-GAL4, clk4.1M-GAL4 and 50y-GAL4 to drive DNKv4 and GFP expression in l-LNvs (UAS-CD8GFP; pdf-GAL4/+;UAS-DNKv4/+), DN1s (UAS-CD8GFP; clk4.1M-GAL4/+;UAS-DNKv4/+), and PI (UAS-CD8GFP; 50y-GAL4/+;UAS-DNKv4/+) neurons, respectively. IA was almost completely inhibited by expressing DNKv4 in l-LNvs and PI cells (Fig. 4A,B,E; l-LNvs: T(12) = 54.33; PI cells: T(12) = 54.33, ***p < 0.0001, t test, F). However, we still detected remaining IA component in DN1s, and all the IA components were completely blocked by a specific IA inhibitor, 4-AP (Fig. 4C,E; F(2,19) = 33.42, *p = 0.0239, ***p < 0.0001, one-way ANOVA), indicating that Shaker/Kv1 mediates the majority of IA in DN1s, compared with Kv4. Subsequently, specific interference of Kv4 function did not significantly change the excitability of DN1s (Fig. 4-1). A remaining IA component was also detected in l-LNvs in Shal495 mutants (UAS-CD8GFP; pdf-GAL4/+;Shal495) (Fig. 4D,E; F(2,20) = 55.72, ***p < 0.0001, one-way ANOVA). As previously stated in motor neurons, remaining IA currents could be contributed by overexpression of Shaker (Bergquist et al., 2010).
Blocking Kv4 activity in GFP-labelled DN1s does not change RMPs and firing rate. A, Representative spontaneous firing traces showing RMP values and firing in DN1s (clk4.1M>CD8GFP) by current-clamp recordings at ZT1 and ZT13. B, Spontaneous firing traces showing RMP values and firing with interference of Kv4 function in DN1s (clk4.1M>CD8GFP;DNKv4) at ZT1 and ZT13. C, D, Quantification of RMPs and firing frequency of DN1s recorded in A and B. Our data show that RMP values declined from -49.2 mV at ZT1 to -63.6 mV at ZT13, and spontaneous firing rate was higher at ZT1 compared to firing at at ZT13 in control DN1s. Specific interference of Kv4 has little effects on RMPs and firing rate at ZT1 or ZT13. Download Figure 4-1, TIF file (4.2MB, tif)
To confirm IA regulates membrane excitability of circadian neurons, we monitored spontaneous neuronal activity in the presence of 4-AP. We used cell-attached voltage-clamp of neuronal activity without changing the cytoplasmic milieu to record action potential (AP) currents. As reported previously (Cao and Nitabach, 2008; Sheeba et al., 2008b; Depetris-Chauvin et al., 2011), l-LNvs exhibited tonic and burst AP currents at ZT9 (Fig. 4G). 4-AP significantly increased frequency of AP currents, which were completely blocked by TTX in l-LNvs (Fig. 4G,J; T(13) = 4.96, *p = 0.0143, t test). Similarly, 4-AP also increased neuronal AP currents in GPF-labeled PI and DN1s (Fig. 4H–J; T(13) = 3.66, **p = 0.0029, T(21) = 2.80, *p = 0.0107, t test).
DNKv4 induces neuronal hyperactivity in a time-of-day-dependent manner
Since Kv4 in l-LNvs specifically controls sleep onset at dusk, we next asked whether IA varies throughout the day in l-LNvs. However, our results showed that there was no difference in IA amplitudes between ZT1 (dawn) and ZT13 (dusk; Fig. 5A; 318 ± 17.3 pA at ZT1, 309 ± 15.1 pA at ZT13, T(22) = 0.75, p = 0.5549, t test). We also compared steady-state inactivation properties of Kv4 currents in neurons. No difference was observed between ZT1 and ZT13 in half-maximal inactivation potential values (Fig. 5B; T(20) = 1.36, p = 0.1873, t test).
Selectively activating PDF cell at ZT12-15 delayed sleep onset. A, Continuous sleep data from control (UAS-dTrpA1, n = 20) and a fly expressing the temperature-gated cation channel dTrpA1 in PDF cells (pdf-GAL4>dTrpA1, n = 20) (top panel). The temperature changes are shown in the middle panel. Quantification of sleep latency after lights off at ZT12 is shown at bottom panel. Our data show that activating PDF cells at ZT12-15 significantly increased sleep latency as compared to activating at ZT9-12. B, Quantification of firing frequency of l-LNvs at ZT1. Our data show that perfusion of 1 µM Phrixotoxin-2 (PaTx2) does not change the firing frequency at RMP (without current injection), but significantly increased firing frequency after hyperpolarization of ∼10 mV by current injection (∼-4 pA). Our data confirm that Kv4 may not regulate neuronal activity at depolarizing membrane potentials (for example at ZT1). Download Figure 5-1, TIF file (11.5MB, tif)
The membrane excitability and Ca2+ waves of l-LNvs and PI neurons was reported to be strongly rhythmic (Cao and Nitabach, 2008; Sheeba et al., 2008b; Barber et al., 2016; Liang et al., 2016). Our cell-attached recordings of GFP-labeled l-LNvs (pdf >CD8GFP) revealed a pattern of circadian regulation of spontaneous AP currents, with higher firing rate during daytime than night-time (Fig. 5D,F; F(5,41) = 141.40, ***p < 0.0001, one-way ANOVA). We detected different kinds of firing properties of l-LNvs, including no firing cells, tonic firing cells, bursting cells, and cells mixed with tonic firing and bursting (Fig. 5E). Blocking Kv4 function in l-LNvs by DNKv4 expression (pdf >CD8GFP;DNKv4) increased the frequency of AP currents during dusk (∼ZT13; Fig. 5D,F; ZT9: T(14) = 2.55, *p = 0.0230; ZT13: T(13) = 4.07, **p = 0.0053; ZT17: T(14) = 2.50, *p = 0.0306, t test), but without effects on firing frequency during dawn (∼ZT1). It appears that loss of Kv4 function did not upregulate the firing rate when activity was relatively high (ZT1, 5, 17, and 21). Moreover, we found that the resting membrane potential (RMP) of l-LNvs exhibited circadian oscillations, with more depolarized RMPs at dawn, declining to lower levels at dusk (from −45 mV at ZT1 to −54 mV at ZT13; Fig. 5C; −45.6 ± 2.0 mV at ZT1, −54.2 ± 1.8 mV at ZT13, T(11) = 2.83, *p = 0.0162, t test). Kv4-mediated IA was sensitive to the pre-pulse window between −60 mV and −40 mV (Fig. 5B), indicating that more Kv4 channels were inactivated at ZT1 when RMPs were relatively depolarized. To confirm membrane potential oscillation contributes to time-of-day–dependent regulation of excitability by Kv4, l-LNvs were artificially hyperpolarized from ∼−45 to ∼−55 mV at ZT1 by current injection during recordings in Figure 5C. Firing frequency was not significantly increased at RMPs by PaTx2 (Kv4-specific blocker) perfusion at ZT1, but it was significantly increased when the membrane potential was hyperpolarized to −55 mV by current injection (Fig. 5-1B; F(3, 27) = 199.5, p = 0.1657, **p = 0.0026, one-way ANOVA). Because IA was mainly contributed by Shaker/Kv1 in DN1s, DNKv4 expression in DN1s did not change the frequency and oscillation of AP currents (Clk4.1M>CD8GFP;DNKv4) (Fig. 4-1). Thus, although Kv4 maintains its expression during the day and night, it may still preferentially control excitability of neurons at dusk due to oscillation of RMPs in wake-promoting neurons.
To demonstrate that the activity of PDF neurons at dusk is crucial for sleep onset, we selectively activated PDF neurons at dusk (ZT9–12 and ZT12–15) by dTrpA1 expression. Our results show that activating PDF neurons during ZT12–15 significantly delayed sleep onset, compared with activation during ZT9–12 (Fig. 5-1A; T(38) = 8.84, ***p < 0.0001, T(38) = 1.95, p = 0.1076, t test). These results indicate that control of the firing of l-LNvs at dusk by Kv4 is essential for normal sleep onset.
PDF is necessary for delayed sleep onset in DNKv4 mutants
To determine whether neuropeptide PDF is required for the delayed sleep onset in pdf>DNKv4 flies, we examined sleep behavior in pdf01 flies that lack PDF. Compared with PDF-null background controls, pdf>DNKv4;pdf01 flies showed decreased total sleep time, but no difference was found in sleep latency (Fig. 6A–C; daytime: F(2,84) = 0.34, p > 0.05; night-time: F(2,84) = 23.65, **p = 0.0034, ***p = 0.0008; total: F(2,84) = 23.96, ***p < 0.0001, one-way ANOVA). Furthermore, we determined whether DNKv4 would still induce circadian-dependent hyperactivity in l-LNvs in PDF-null background. Excitability and RMPs of l-LNvs still exhibited circadian oscillation in the absence of PDF (Fig. 6D–F; F(3,27) = 203.5, *p = 0.0260, one-way ANOVA). More importantly, compared with PDF-null background control, loss of Kv4 function still preferentially increased frequency of AP currents at dusk (ZT13), suggesting that lack of PDF has little effect on the regulation of neuronal excitability by Kv4. In summary, l-LNvs hyperactivity induced by DNKv4 expression delayed sleep onset in a PDF-dependent mechanism.
Kv4 also regulates timing of sleep onset in circadian output area
We set out to identify possible downstream neurons involved in Kv4-mediated sleep onset delay. PI is developmentally and functionally analogous to the hypothalamus in vertebrates (Foltenyi et al., 2007; de Velasco et al., 2007), and has been established as an important component of the circadian output pathway for rest: activity rhythms under circadian control (Cavanaugh et al., 2014; Cavey et al., 2016). We next chose 50y-GAL to drive the expression of DNKv4 in subsets of PI neurons (w;50y-GAL4/+;UAS-DNKv4/+), which has been shown to regulate sleep (Foltenyi et al., 2007). Interestingly, compared with controls (w;+;UAS-DNKv4/+, and w;50y-GAL4/+;+), 50y-GAL4>UAS-DNKv4 showed difficulty in falling asleep after lights-off as well, accompanied with significantly reduced night-time sleep amount and bout number (Fig. 7A,B; daytime: F(2,105) = 2.357, UAS-DNKv4 vs 50y-GAL4>DNKv4, p = 0.0807, 50y-GAL4 vs 50y-GAL4>DNKv4, p = 0.1094, night-time: F(2,105) = 123.0, ***p < 0.0001; total: F(2,105) = 89.77, UAS-DNKv4 vs 50y-GAL4>DNKv4, ***p < 0.0001, 50y-GAL4 vs 50y-GAL4>DNKv4, **p = 0.0034, one-way ANOVA; C, F(2,105) = 44.97, UAS-DNKv4 vs 50y-GAL4>DNKv4, ***p < 0.0001, 50y-GAL4 vs 50y-GAL4>DNKv4, ***p = 0.0008, one-way ANOVA; D, UAS-DNKv4 vs 50y-GAL4>DNKv4, U = 619, p = 0.2139, 50y-GAL4 vs 50y-GAL4>DNKv4, U = 716.5, p = 0.1082, Mann–Whitney U test; F, F(2,105) = 10.54, ***p = 0.0002, **p = 0.0038, one-way ANOVA). These flies exhibited very little daytime sleep in LD (Fig. 7A,B) and lengthened sleep latency in DD (Fig. 7H,I; T(54) = 11.88, ***p < 0.0001, t test). Most (>80%) of the mutants became arrhythmic under DD and the rhythmic ones had a ∼25.4 h period phenotype (Fig. 2-1D; F(2,59) = 5.92, *p = 0.0106, ***p = 0.0004, one-way ANOVA). To exclude the possibility that the reduction of sleep amount is due to animals' hyperactivity, we calculated waking activity (activity counts per minute awake) and no hyperactivity was found, but with a mild hypoactivity (Fig. 7E; F(2,105) = 7.29, *p = 0.0268, ***p = 0.0007, one-way ANOVA). We also observed delayed sleep onset in male flies (Fig. 7-1; F(2, 79) = 49.9, **p = 0.0026, ***p < 0.0001, one-way ANOVA). All these results suggest that Kv4 in PI neurons is also required to control sleep onset, but without effects on the sleep maintenance.
DNKv4 expression in subsets of PI cells delayed sleep onset in males. A, Conventional sleep plots of controls (50y-GAL4, n = 26, and UAS-DNKv4, n = 26) and experimental flies (50y>DNKv4, n = 30). B, Sleep latency after lights off at ZT12. Our data show that blocking Kv4 function by DNKv4 expression in PI cells delayed sleep onset in males. Download Figure 7-1, TIF file (2.7MB, tif)
We next asked whether the defective sleep initiation was due to the hyperactivity of PI neurons. Expression of the temperature-gated nonspecific cation channel dTrpA1 driven by 50y-GAL4 can impose a fast firing pattern on the cell at 29°C. Similar to 50y>DNKv4 mutants, 50y>dTrpA1 flies also exhibited markedly longer sleep latency (Fig. 7F,G; T(67) = 4.27, ***p < 0.0001, t test). This result verified the notion that function loss of Kv4 could lead to hyperactivity of neurons and subsequent delayed sleep onset.
We examined whether PDF receptors in PI neurons regulate sleep onset. Interestingly, downregulation of PDFR by expressing pdfr-RNAi in PI cells (50y>pdfr-RNAi) significantly advanced sleep onset, accompanied with increased night-time sleep amount (Fig. 7J,K; daytime: T(52) = 0.18, p = 0.2541, night-time: T(52) = 5.85, ***p < 0.0001, T(52) = 5.89, ***p < 0.0001, t test; L, T(52) = 3.19, *p = 0.0114, t test). These results suggest that PDF neurons may act on downstream PI neurons to regulate night-time sleep, and Kv4 in these neurons regulates sleep onset likely through regulating PDF/PDFR signaling.
Discussion
To the best of our knowledge, this is the first demonstration of the function of Kv4 in sleep regulation. In this study, we showed that Kv4 is important for night-time sleep in Drosophila, and is especially crucial to normal sleep onset. Pan-neuronal expression of DNKv4 leads to decreased night-time sleep, indicating a general sleep-promoting function of Kv4. The increased night-time sleep in Shal495-null mutant reveals a compensatory overexpression of Shaker. A transcription factor named Kruppel (Kr) was identified to be a central regulator of this process (Parrish et al., 2014), consistent with our conclusion that the compensatory modulation occurs at the transcriptional level. However, the Kr expression is suggested to be a result from detecting Kv4 or Shaker/Kv1 conductance, since 4-AP also increased Shaker RNA expression (Parrish et al., 2014). In our study, we did not observe significant increase in Shaker RNA expression in DNKv4 mutants suggesting that other transcription factor(s), rather than Kr, may be involved in regulating Shal/Kv4 and Shaker balance.
Previous studies have shown that cyclin A and GABAA receptors (or RDL) in circadian neurons also regulate sleep latency (Agosto et al., 2008; Rogulja and Young, 2012; Afonso et al., 2015). A molecule named WAKE interacts with RDL, and the cycling manner of WAKE promotes excitability oscillation of l-LNvs (Liu et al., 2014). Moreover, DN1s may regulate wake–sleep transition at dusk in a clock-dependent manner (Kunst et al., 2014; Guo et al., 2016). Our results verified that Kv4-mediated IA in l-LNvs, and function-loss of Kv4 preferentially upregulated membrane excitability at dusk, although Kv4 expression was not rhythmic. Moreover, RMP values exhibit circadian oscillation in l-LNvs, with depolarized RMPs at dawn. Thus, less Kv4 channels would be available for opening when RMPs are depolarized. This supports our data showing that blocking Kv4 did not significantly increased firing rate when circadian neurons were hyperexcitable at dawn. Although RMPs and firing rate also exhibit circadian oscillation in DN1s (Fig. 4-1), blocking Kv4 activity has no effects on sleep latency. We further demonstrated that this may be caused by the near absence of Kv4-mediated IA in DN1s. Thus, Kv4 controls wake–sleep transition at dusk in subsets wake-promoting cells, but not in DN1s. The phases of Ca2+ waves recorded with a fine temporal resolution were approximately coincident with membrane excitability and RMPs oscillations in l-LNvs, but with slight (∼1–2 h) delay in peaks (Cao and Nitabach, 2008; Liang et al., 2016). In this study, DNKv4 preferentially reduced excitability at dusk without causing daily excitability oscillation shifts (Fig. 5F), and presumably would reduce Ca2+ peaks as well. But how Kv4 exactly regulates intracellular Ca2+ waves and coordination of electrical excitability and Ca2+ waves need further study.
PI exerts its effects as downstream of clock neurons and is one of the key targets (direct or indirect) of PDF neurons, although PI cells do not express molecular clock machinery (Jaramillo et al., 2004; Chung et al., 2009; Cavanaugh et al., 2014). Previous studies have suggested that EGFR/ERK signal in insulin-producing PI neurons plays important roles in regulating the consolidation and maintenance of sleep in Drosophila, indicating that PI cells act as wake-promoting neurons (Foltenyi et al., 2007; Cong et al., 2015; Barber et al., 2016). Our results are the first to demonstrate that the neuronal activities and PDF/PDFR signaling in subsets of PI cells (50y-GAL4 driver) participate in the regulation of sleep onset.
In this study, defects in sleep initiation were presumed to be due to the hyperactivity of specific circadian neurons caused by blocking Kv4 activity. dTrpA1 experiments further supported this notion. However, a recent study concluded that reducing the Shaker/Kv1 current would decrease, rather than increase, the action potential discharge in dFB (dorsal FB; Pimentel et al., 2016). Depleting Shaker from dFB neurons could shift the interspike interval distribution toward longer values, making it more difficult to generate next spike. We suppose that the opposite effect may be due to diverse types of Kv channels and different discharge and biophysical properties of neurons. For example, Kv4 is required for maintaining excitability in cultured neurons (Ping et al., 2011), but not in groups of neurons from dissected brain in this study (Figs. 4, 5). Whether driving DNKv4 expression in other sleep-promoting or wake-promoting neurons would cause parallel or opposite effects on sleep needs to be investigated. Neurons in EB (Liu et al., 2016), FB (Donlea et al., 2014; Pimentel et al., 2016), or a subset MBNs driven by 201y-GAL4 (Cavanaugh et al., 2016) could be tested in further studies.
Previous work has provided a strong link between circadian periods with habitual sleep timing in human, and CRY1 is suggested to be associated with a familial form of delayed sleep phase disorder (Wright et al., 2005; Patke et al., 2017). However, we did not detect significant change in circadian periods by blocking Kv4 in LNvs and DN1s, indicating that delayed sleep onset in the mutants was not due to changes in circadian periods. Blocking Kv4 in PI cells disrupted rhythmic activities in most flies and we did detect a lengthened circadian period in the rhythmic ones. Because PI is a circadian output region and does not express clock machinery, it is not likely that intrinsic circadian rhythm disorder contributes to the sleep onset phenotype caused by blocking Kv4.
Studies have provided evidence that links types of potassium channels to sleep phenotypes in Drosophila and human (Cornelius et al., 2011; Allebrandt et al., 2013; Barone and Krieger, 2016). For example, insomnia has been delineated in patients with neurologic disorders of Voltage-gated K+ channels (Kv) autoimmunity (or named Kv antibody syndrome). Sleep onset insomnia, defined as inability to fall asleep at the desired time, is observed in patients with neurodegenerative diseases and psychiatric disorders (Wulff et al., 2010). Abnormal sleep timing and pattern have also been observed in Drosophila disease models (Wulff et al., 2010; Tabuchi et al., 2015; Gonzales et al., 2016; Song et al., 2017). Our study may provide a potential alternative therapy of sleep onset insomnia by targeting Kv4 channels.
Footnotes
This work was supported by a National Natural Science Foundation of China Grant (81371482), a National Science and Technology Ministry Major Project Grant (2016YFC0906400), and a Science and Technology Commission of Shanghai Municipality Grant (18ZR1419400). We thank Hai Huang, Bingwei Lu, and Xiong-Li Yang for critical reading of the paper and helpful feedback.
The authors declare no competing financial interests.
References
- Afonso DJ, Liu D, Machado DR, Pan H, Jepson JE, Rogulja D, Koh K (2015) TARANIS functions with cyclin A and Cdk1 in a novel arousal center to control sleep in Drosophila. Curr Biol 25:1717–1726. 10.1016/j.cub.2015.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC (2008) Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 11:354–359. 10.1038/nn2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allebrandt KV, Amin N, Müller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, Hayward C, van Mill J, Vogelzangs N, Green EW, Melville SA, Lichtner P, Wichmann HE, Oostra BA, Janssens AC, Campbell H, Wilson JF, Hicks AA, Pramstaller PP, Dogas Z, et al. (2013) A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry 18:122–132. 10.1038/mp.2011.142 [DOI] [PubMed] [Google Scholar]
- Artiushin G, Sehgal A (2017) The Drosophila circuitry of sleep–wake regulation. Curr Opin Neurobiol 44:243–250. 10.1016/j.conb.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber AF, Erion R, Holmes TC, Sehgal A (2016) Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev 30:2596–2606. 10.1101/gad.288258.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone DA, Krieger AC (2016) Sleep disturbances in voltage-gated potassium channel antibody syndrome. Sleep Med 21:171–173. 10.1016/j.sleep.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Bergquist S, Dickman DK, Davis GW (2010) A hierarchy of cell intrinsic and target-derived homeostatic signaling. Neuron 66:220–234. 10.1016/j.neuron.2010.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN (2008) Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci 28:6493–6501. 10.1523/JNEUROSCI.1503-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A (2014) Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell 157:689–701. 10.1016/j.cell.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Vigderman AS, Dean T, Garbe DS, Sehgal A (2016) The Drosophila circadian clock gates sleep through time-of-day dependent modulation of sleep-promoting neurons. Sleep 39:345–356. 10.5665/sleep.5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Collins B, Bertet C, Blau J (2016) Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci 19:587–595. 10.1038/nn.4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R (2009) The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 19:386–390. 10.1016/j.cub.2009.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G (2005) Reduced sleep in Drosophila shaker mutants. Nature 434:1087–1092. 10.1038/nature03486 [DOI] [PubMed] [Google Scholar]
- Colwell CS. (2011) Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12:553–569. 10.1038/nrn3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X, Wang H, Liu Z, He C, An C, Zhao Z (2015) Regulation of sleep by insulin-like peptide system in Drosophila melanogaster. Sleep 38:1075–1083. 10.5665/sleep.4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius JR, Pittock SJ, McKeon A, Lennon VA, Aston PA, Josephs KA, Tippmann-Peikert M, Silber MH (2011) Sleep manifestations of voltage-gated potassium channel complex autoimmunity. Arch Neurol 68:733–738. 10.1001/archneurol.2011.106 [DOI] [PubMed] [Google Scholar]
- Covarrubias M, Wei AA, Salkoff L (1991) Shaker, Shal, Shab, and Shaw express independent K+ current systems. Neuron 7:763–773. 10.1016/0896-6273(91)90279-9 [DOI] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A (2010) Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65:670–681. 10.1016/j.neuron.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF (2011) Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol 21:1783–1793. 10.1016/j.cub.2011.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V (2007) Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol 302:309–323. 10.1016/j.ydbio.2006.09.035 [DOI] [PubMed] [Google Scholar]
- Diao F, Chaufty J, Waro G, Tsunoda S (2010) SIDL interacts with the dendritic targeting motif of shal (K(v)4) K+ channels in Drosophila. Mol Cell Neurosci 45:75–83. 10.1016/j.mcn.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Miesenböck G (2014) Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81:860–872. 10.1016/j.neuron.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CL, Vyazovskiy V, Southard T, Chiu SY, Messing A, Tononi G, Cirelli C (2007) Sleep in Kcna2 knockout mice. BMC Biol 5:42. 10.1186/1741-7007-5-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy C, Sehgal A (2017) Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205:1373–1397. 10.1534/genetics.115.185157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Pang J, Yi X, Song Q, Zhang J, Li C, He G, Ping Y (2018) Down-regulation of KV4 channel in Drosophila mushroom body neurons contributes to Abeta42-induced courtship memory deficits. Neuroscience 370:236–245. 10.1016/j.neuroscience.2017.06.008 [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW (2007) Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 10:1160–1167. 10.1038/nn1957 [DOI] [PubMed] [Google Scholar]
- Gasque G, Labarca P, Reynaud E, Darszon A (2005) Shal and shaker differential contribution to the K+ currents in the Drosophila mushroom body neurons. J Neurosci 25:2348–2358. 10.1523/JNEUROSCI.4384-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ED, Tanenhaus AK, Zhang J, Chaffee RP, Yin JC (2016) Early-onset sleep defects in Drosophila models of Huntington's disease reflect alterations of PKA/CREB signaling. Hum Mol Genet 25:837–852. 10.1093/hmg/ddv482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Norris AJ, Carrasquillo Y, Nerbonne JM, Herzog ED (2012) IA channels encoded by Kv1.4 and Kv4.2 regulate neuronal firing in the suprachiasmatic nucleus and circadian rhythms in locomotor activity. J Neurosci 32:10045–10052. 10.1523/JNEUROSCI.0174-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M (2016) Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536:292–297. 10.1038/nature19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Taghert PH (2010) PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol 518:1925–1945. 10.1002/cne.22311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, Sehgal A, Levitan IB (2004) Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci 5:3. 10.1186/1471-2202-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A (2006) Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441:757–760. 10.1038/nature04811 [DOI] [PubMed] [Google Scholar]
- Kayser MS, Biron D (2016) Sleep and development in genetically tractable model organisms. Genetics 203:21–33. 10.1534/genetics.116.189589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Duboué ER, McDonald DM, Dus M, Suh GS, Waddell S, Blau J (2010) Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol 20:1209–1215. 10.1016/j.cub.2010.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, Nitabach MN (2014) Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 24:2652–2664. 10.1016/j.cub.2014.09.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH (2016) Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351:976–981. 10.1126/science.aad3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li Y, Wang X, Qi J, Jin X, Tong H, Zhou Z, Zhang ZC, Han J (2017) Fbxl4 serves as a clock output molecule that regulates sleep through promotion of rhythmic degradation of the GABAA receptor. Curr Biol 27:3616–3625.e5. 10.1016/j.cub.2017.10.052 [DOI] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S, Lloyd TE, Montell C, Sehgal A, Koh K, Wu MN (2014) WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82:151–166. 10.1016/j.neuron.2014.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, Wu MN (2016) Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165:1347–1360. 10.1016/j.cell.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Guo F, Lu B, Guo A (2008) amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophys Res Commun 372:798–803. 10.1016/j.bbrc.2008.05.119 [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60:672–682. 10.1016/j.neuron.2008.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Sonn JY, Oh Y, Lim C, Choe J (2014) SIFamide and SIFamide receptor defines a novel neuropeptide signaling to promote sleep in Drosophila. Mol Cells 37:295–301. 10.14348/molcells.2014.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Kim CC, Tang L, Bergquist S, Wang T, Derisi JL, Jan LY, Jan YN, Davis GW (2014) Kruppel mediates the selective rebalancing of ion channel expression. Neuron 82:537–544. 10.1016/j.neuron.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke A, Murphy PJ, Onat OE, Krieger AC, Özçelik T, Campbell SS, Young MW (2017) Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169:203–215.e13. 10.1016/j.cell.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, Miesenböck G (2016) Operation of a homeostatic sleep switch. Nature 536:333–337. 10.1038/nature19055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping Y, Tsunoda S (2011) Inactivity-induced increase in nAChRs upregulates shal K(+) channels to stabilize synaptic potentials. Nat Neurosci 15:90–97. 10.1038/nn.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping Y, Waro G, Licursi A, Smith S, Vo-Ba DA, Tsunoda S (2011) Shal/K(v)4 channels are required for maintaining excitability during repetitive firing and normal locomotion in Drosophila. PLoS One 6:e16043. 10.1371/journal.pone.0016043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping Y, Hahm ET, Waro G, Song Q, Vo-Ba DA, Licursi A, Bao H, Ganoe L, Finch K, Tsunoda S (2015) Linking Aβ42-induced hyperexcitability to neurodegeneration, learning and motor deficits, and a shorter lifespan in an Alzheimer's model. PLoS Genet 11:e1005025. 10.1371/journal.pgen.1005025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R (2006) A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441:753–756. 10.1038/nature04739 [DOI] [PubMed] [Google Scholar]
- Rogero O, Hämmerle B, Tejedor FJ (1997) Diverse expression and distribution of shaker potassium channels during the development of the Drosophila nervous system. J Neurosci 17:5108–5118. 10.1523/JNEUROSCI.17-13-05108.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Young MW (2012) Control of sleep by cyclin A and its regulator. Science 335:1617–1621. 10.1126/science.1212476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg RP. (2006) Sleep maintenance insomnia: strengths and weaknesses of current pharmacologic therapies. Ann Clin Psychiatry 18:49–56. 10.1080/10401230500464711 [DOI] [PubMed] [Google Scholar]
- Salkoff L, Wyman R (1981) Genetic modification of potassium channels in Drosophila shaker mutants. Nature 293:228–230. 10.1038/293228a0 [DOI] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M (2008) Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A 105:19587–19594. 10.1073/pnas.0809577105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC (2008a) Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18:1537–1545. 10.1016/j.cub.2008.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O'Dowd DK, Holmes TC (2008b) Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol 99:976–988. 10.1152/jn.00930.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Feng G, Huang Z, Chen X, Chen Z, Ping Y (2017) Aberrant axonal arborization of PDF neurons induced by Abeta42-mediated JNK activation underlies sleep disturbance in an Alzheimer's model. Mol Neurobiol 54:6317–6328. 10.1007/s12035-016-0165-z [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Lance K, Levine RB (2012) Segmental differences in firing properties and potassium currents in Drosophila larval motoneurons. J Neurophysiol 107:1356–1365. 10.1152/jn.00200.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431:862–868. 10.1038/nature02926 [DOI] [PubMed] [Google Scholar]
- Strauss R, Heisenberg M (1993) A higher control center of locomotor behavior in the Drosophila brain. J Neurosci 13:1852–1861. 10.1523/JNEUROSCI.13-05-01852.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Lone SR, Liu S, Liu Q, Zhang J, Spira AP, Wu MN (2015) Sleep interacts with Aβ to modulate intrinsic neuronal excitability. Curr Biol 25:702–712. 10.1016/j.cub.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Wu CF (2006) Distinct frequency-dependent regulation of nerve terminal excitability and synaptic transmission by IA and IK potassium channels revealed by Drosophila Shaker and Shab mutations. J Neurosci 26:6238–6248. 10.1523/JNEUROSCI.0862-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A, Covarrubias M, Butler A, Baker K, Pak M, Salkoff L (1990) K+ current diversity is produced by an extended gene family conserved in Drosophila and mouse. Science 248:599–603. 10.1126/science.2333511 [DOI] [PubMed] [Google Scholar]
- Wright KP Jr, Gronfier C, Duffy JF, Czeisler CA (2005) Intrinsic period and light intensity determine the phase relationship between melatonin and sleep in humans. J Biol Rhythms 20:168–177. 10.1177/0748730404274265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Gatti S, Wettstein JG, Foster RG (2010) Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci 11:589–599. 10.1038/nrn2868 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shaker (Sh) mRNA expression levels in Shal/Kv4 mutants and sleep phenotypes in Sh mutants. A, Sh mRNA expression was measured in DNKv4 (elav/+;+;UAS-DNKv4/+), Shal495 and Kv4 + Shal495 (elav/+;UAS-Kv4/+;Shal495) animals. For quantitative RT-PCR, brains were isolated and total RNA was isolated from each sample using the standard Trizol Protocol. A DNase digestion was then done to remove potential DNA contamination and RT was performed. Each experimental animal sample was compared to each wild-type (wt) sample. All bars are represented as percent of wt animals. We noted that increased Shaker RNA expression in Shal495 mutants was not rescued by restoration of Shal/Kv4 in Shal495 mutants using the GAL4/UAS system. B, Conventional sleep plots of w1118 (n = 24) and experimental flies (Shmns/+, n = 35) in 12h:12h light:dark (LD). C, Total sleep amount (24h) and sleep during day-time and night-time in wt and experimental lines were analyzed as described in B. These data show reduced sleep amount in heterozygous female Shmns flies. D, Sleep latency after lights-off at ZT12. E, Conventional sleep plots of control (Shmns;+;+, n = 26) and experimental flies (Shmns;+;Shal495, n = 25) in 12h:12h light:dark (LD). F, Total sleep amount (24h) and sleep during day-time and night-time in wt and experimental lines. These data show reduced night-time sleep amount in Shal495 flies, in the presence of Shmns. Note that these flies exhibited very little day-time sleep. G, Sleep latency after lights-off at ZT12. H, Conventional sleep plots of control (w;UAS-Eko/+;+, n = 33) and experimental flies (pdf-GAL4/+;UAS-Eko/+;+, n = 27) in 12h:12h light:dark (LD). I, Total sleep amount (24h) and sleep during day-time and night-time in wt and experimental lines. J, Sleep latency after lights off at ZT12. Download Figure 1-1, TIF file (10.6MB, tif)
DNKv4 and Shal-RNAi expression in PDF neurons delay sleep onset, but without effects on circadian period by interference of Kv4 in PDF and DN1s. A, Conventional sleep plots of control (w;UAS-DNKv4/+;+, n = 27) and experimental flies (w;UAS-DNKv4/pdf-GAL4;+, n = 49) in 12h:12h light:dark (LD) (left), and Quantification of sleep latency after lights-off at ZT12 (right). Our data show that blocking Kv4 function in PDF cells delayed sleep onset. B, Quantification of sleep latency at lights-off at ZT12 in male flies (n = 24 for UAS-DNKv4, n = 34 for pdf>DNKv4). C, Activity records of representative controls (pdf-GAL4, and UAS-DNKv4) and experimental flies (pdf>DNKv4) for 3 d in LD and following 7 d in DD. Quantification of circadian periods and rhythmicity is presented in the top panel. Our data show that there is no significant difference in circadian periods. D, Quantification of circadian periods for indicated genotypes, including UAS-DNKv4, R18H11-GAL4, R18H11>DNKv4, 50y-GAL4, and 50y>DNKv4. Rhythmicity is also shown in the bar graph. Our data show that DNKv4 expression in subsets of DN1s does not change the circadian periods (p > 0.05) and rhythmicity, while most (over 80%) flies became arrhythmic when expressing DNKv4 in PI cells and the remaining rhythmic ones had ∼25.3-hour period phenotype. E, F, Conventional sleep plots of two genotypes (Shal495 , n = 26, and pdf>Kv4;Shal495, n = 29) in LD (E), and sleep latency after lights-off at ZT12 (F). Our data indicate that restoring Kv4 expression selectively in PDF cells partially rescues delayed sleep onset in Shal495 mutants. G, H, Conventional sleep plots of controls (UAS-Shal-RNAi and pdf-GAL4, n = 24-28) and experimental (pdf>Shal-RNAi, n = 27) flies (G), and sleep latency after lights-off at ZT12 (H). Our data show that down-regulation of Kv4 expression by RNA interference (RNAi) also delays sleep onset, similar to those of DNKv4. Download Figure 2-1, TIF file (16.8MB, tif)
Blocking Kv4 activity in GFP-labelled DN1s does not change RMPs and firing rate. A, Representative spontaneous firing traces showing RMP values and firing in DN1s (clk4.1M>CD8GFP) by current-clamp recordings at ZT1 and ZT13. B, Spontaneous firing traces showing RMP values and firing with interference of Kv4 function in DN1s (clk4.1M>CD8GFP;DNKv4) at ZT1 and ZT13. C, D, Quantification of RMPs and firing frequency of DN1s recorded in A and B. Our data show that RMP values declined from -49.2 mV at ZT1 to -63.6 mV at ZT13, and spontaneous firing rate was higher at ZT1 compared to firing at at ZT13 in control DN1s. Specific interference of Kv4 has little effects on RMPs and firing rate at ZT1 or ZT13. Download Figure 4-1, TIF file (4.2MB, tif)
Selectively activating PDF cell at ZT12-15 delayed sleep onset. A, Continuous sleep data from control (UAS-dTrpA1, n = 20) and a fly expressing the temperature-gated cation channel dTrpA1 in PDF cells (pdf-GAL4>dTrpA1, n = 20) (top panel). The temperature changes are shown in the middle panel. Quantification of sleep latency after lights off at ZT12 is shown at bottom panel. Our data show that activating PDF cells at ZT12-15 significantly increased sleep latency as compared to activating at ZT9-12. B, Quantification of firing frequency of l-LNvs at ZT1. Our data show that perfusion of 1 µM Phrixotoxin-2 (PaTx2) does not change the firing frequency at RMP (without current injection), but significantly increased firing frequency after hyperpolarization of ∼10 mV by current injection (∼-4 pA). Our data confirm that Kv4 may not regulate neuronal activity at depolarizing membrane potentials (for example at ZT1). Download Figure 5-1, TIF file (11.5MB, tif)
DNKv4 expression in subsets of PI cells delayed sleep onset in males. A, Conventional sleep plots of controls (50y-GAL4, n = 26, and UAS-DNKv4, n = 26) and experimental flies (50y>DNKv4, n = 30). B, Sleep latency after lights off at ZT12. Our data show that blocking Kv4 function by DNKv4 expression in PI cells delayed sleep onset in males. Download Figure 7-1, TIF file (2.7MB, tif)