Abstract
It was proposed that the Drosophila amnesiac gene (amn) is required for consolidation of aversive memory in the dorsal paired medial (DPM) neurons, a pair of large neurons that broadly innervate the mushroom bodies (MB), the fly center for olfactory learning and memory (Waddell et al., 2000). Yet, a conditional analysis showed that it was not possible to rescue the memory deficit of amnX8 null mutant flies when amn expression was restored only in the adult (DeZazzo et al., 1999), which led the authors to suggest that amn might be involved in the development of brain structures that normally promote adult olfactory memory. To further investigate temporal and spatial requirements of Amnesiac (AMN) peptide in memory, we used RNA interference in combination with conditional drivers. Experiments were conducted either in both sexes, or in either sexes. Our data show that acute modulation of amn expression in adult DPM neurons does not impact memory. We further show that amn expression is required for normal development of DPM neurons. Detailed enhancer trap analyses suggest that amn transcription unit contains two distinct enhancers, one specific of DPM neurons, and the other specific of α/β MB neurons. This prompted us to investigate extensively the role of AMN in the adult MB. Together, our results demonstrate that amn is acutely required in adult α/β MB neurons for middle-term and long-term memory. The data thus establish that amn plays two distinct roles. Its expression is required in DPM neurons for their development, and in adult MB for olfactory memory.
SIGNIFICANCE STATEMENT The Drosophila amnesiac gene encodes a neuropeptide whose expression was proposed to be required for consolidation of aversive memory in the dorsal paired medial (DPM) neurons, a pair of large neurons that broadly innervate the mushroom bodies (MB), the olfactory memory center. Here, we investigated amnesiac temporal and spatial requirement using conditional tools that allowed us to manipulate its expression in selected neurons. This work leads to a complete reassessment of the role of amnesiac in brain development and memory. We show that amnesiac is required for two distinct processes: for normal development of DPM neurons, and in adult MB for memory.
Keywords: amnesiac, dorsal paired medial neurons, Drosophila, mushroom body, olfactory memory
Introduction
Drosophila is well suited to studying memory formation. The mushroom bodies (MB) constitute the central integrative brain structure for olfactory memory (de Belle and Heisenberg, 1994). They are composed of 4000 intrinsic neurons, called Kenyon cells (KC), classed into three subtypes whose axons form two vertical (α and α′) and three medial (β, β′, and γ) lobes (Crittenden et al., 1998). Using a classical conditioning paradigm in which an odorant is paired with the delivery of electric shocks, the fly is capable of forming six discrete memory phases reflected at the neural network level (Bouzaiane et al., 2015). Memory measured 2 h after a single conditioning is a composite memory formed of labile middle-term memory (MTM) encoded in the α/β neurons, and middle-term anesthesia-resistant memory encoded in γ KC. The fly can also produce two exclusive forms of consolidated memory measured 24 h after training (Isabel et al., 2004): long-term anesthesia-resistant memory (LT-ARM) is formed after five massed cycles of conditioning, whereas the robust long-term memory (LTM) is only formed after five spaced cycles. Crucially, LTM is the only memory phase dependent on de novo protein synthesis (Tully et al., 1994). LT-ARM and LTM are encoded in α′/β ′ and α/β KC, respectively.
The Drosophila amnesiac (amn) mutant was first isolated in a behavioral screen for memory defects (Quinn et al., 1979). The amn gene encodes a predicted neuropeptide precursor with a signal sequence (Feany and Quinn, 1995). Several reports have implicated AMN in activation of the cAMP pathway (Moore et al., 1998; Bhattacharya et al., 2004). Although the mature products have not been identified, sequence analyses point to three neuropeptides, one of which is homologous to the mammalian pituitary adenylate cyclase-activating polypeptide (PACAP) (Feany and Quinn, 1995; Hashimoto et al., 2002). PACAP is a neuropeptide that is widely expressed throughout the brain and acts as a neuromodulator or neurotrophic factor through activation of G-protein-linked receptors. It has been shown to regulate a variety of physiological processes through stimulation of cAMP production (Miyata et al., 1989; Arimura, 1998; Vaudry et al., 2000). As in mammals, the fly cAMP/PKA pathway plays a key role in associative memory, the adenylate cyclase Rutabaga being thought to act as a coincidence detector in the MB to associate the olfactory and electric-shock pathways (Levin et al., 1992; Tomchik and Davis, 2009; Gervasi et al., 2010).
The Amnesiac (AMN) peptide is expressed in dorsal paired medial (DPM) neurons, a pair of large serotoninergic neurons that intensively innervate all lobes of the MB (Waddell et al., 2000; Lee et al., 2011). DPM output was shown to be required during the consolidation phase for MTM, prompting the notion that DPM neurons might release the AMN modulatory neuropeptide which alters the physiology of MB neurons to help stabilize or consolidate odor memories (Keene et al., 2004). Despite many studies addressing AMN role in memory, its function has remained unclear. Noteworthy, memory rescue experiments succeeded when a wild-type amn transgene was expressed throughout development including the adult stage (DeZazzo et al., 1999; Waddell et al., 2000), but not when its expression was limited to the adult stage (DeZazzo et al., 1999), suggesting that amn is required for development.
Here, to clarify the role of amn in memory, we used conditional RNA interference (RNAi) to manipulate amn expression with the spatial and temporal control allowed by Gal4 drivers. As expected, constitutive expression of amn-specific RNAi in DPM neurons mimicked memory phenotypes previously described for amn mutants. In sharp contrast, conditional expression in adult DPM neurons did not impact memory. Furthermore, we observed abnormal DPM morphology in amn-deficient flies. Together, the data show that amn expression in DPM neurons is required for neuronal development. Strikingly, we next show that AMN plays a key role in adult α/β neurons for MTM and LTM formation. Thus AMN plays two distinct roles: it is required for DPM neuronal development, and in the adult MB for memory.
Materials and Methods
Drosophila stocks.
Drosophila melanogaster wild-type strain Canton-Special (Canton-S) and mutant flies were raised on standard medium at 18°C in 60% humidity in a 12 h light/dark cycle. All strains used for memory experiments were outcrossed to the Canton-S background. amn-RNAi lines were obtained from the Vienna Drosophila Resource Center (VDRC; amn-RNAi1, 5606) and from the Bloomington Drosophila Stock Center (Indiana University; amn-RNAi2, 25797). GAL4 drivers used to achieve RNAi expression were VT64246 (VDRC, 264246) and VT26149 (VDRC, 204563) for expression in DPM neurons, 238Y, c747 and VT30559 (VDRC, 206077) for expression in the MB, c739 for expression in α/β neurons, VT30604 (VDRC, 200228) for expression in α′/β ′ neurons, and VT49483 (VDRC, 206419) for expression in γ neurons. To specifically induce RNAi expression in adults, we took advantage of the TARGET system (McGuire et al., 2003). To achieve RNAi induction, flies were kept at 30°C for 3 or 5 d before conditioning, and also until memory test for LTM and LT-ARM analyses. To specifically block RNAi induction in the MB, MB247-Gal80 (MB-Gal80; Krashes et al., 2007) was used. Rescue experiments were conducted with the UAS-amn construct described by Waddell et al. (2000).
Behavioral experiments.
Flies were trained with classical olfactory aversive conditioning protocols as described by Pascual and Préat (2001). Training and testing were performed at 25°C in 80% humidity. Conditioning was performed on samples of 25–35 flies aged between 3 and 4 d with 3-octanol (∼95% purity; Sigma-Aldrich) and 4-methylcyclohexanol (99% purity; Sigma-Aldrich) at 0.360 and 0.325 mm, respectively. Odors were diluted in paraffin oil (VWR International). Memory tests were performed with a T-maze apparatus (Tully and Quinn, 1985). Flies were given 1 min to choose between two arms, each delivering a distinct odor. An index was calculated as the difference between the numbers of flies in each arm divided by the sum of flies in both arms. The average of two reciprocal experiments gave a performance index (PI). For analyses of immediate memory, flies were tested immediately after a single training cycle (1 min memory). To assess MTM, flies were submitted to one-cycle training, and memory was tested 2 h later. Cold anesthesia was achieved by a 2 min cold shock performed 1 h before the test. For LTM analyses, flies were trained with five cycles spaced at 15 min rest intervals, and tested 24 h later, and for LT-ARM analyses, flies were submitted to five massed conditioning cycles and memory was tested 24 h later. For odor avoidance tests after electric shock and response to electric shock, flies were treated as described by Pascual and Préat (2001).
Immunohistochemistry experiments.
Gal80ts;VT64246/UAS-mCD8::GFP and Gal80ts;VT26149/UAS-mCD8::GFP flies were incubated for 3 d at 30°C before brain dissection. For amn-RNAi analyses, VT64246;UAS-mCD8::GFP female flies were crossed to amn-RNAi1 males, and brains of female F1 progenies were analyzed. For amnX8 analyses, amnX8;MB-Gal80 were crossed to VT64246;UAS-mCD8::GFP or VT26149;UAS-mCD8::GFP males, and brains of male F1 progenies were analyzed. For amnX8 rescue analyses, amnX8;UAS-amn female flies were crossed to c739 males, and brains of male F1 progenies were analyzed. Flies were collected 3–4 d after hatching (at 25°C). Brains were dissected and fixed for 20 min at room temperature in 4% formaldehyde in PBT (PBS containing 1% Triton X-100). Samples were then rinsed three times for 20 min in PBT, blocked with 2% bovine serum albumin in PBT for 2 h, and further incubated at 4°C overnight with primary antibodies [rabbit anti-GFP at 1:400 (ThermoFisher Scientific) or rabbit anti-5HT at 1:500 (Sigma-Aldrich) and mouse anti-nc82 at 1:100 (DSHB)] in the blocking solution. Brains were washed three times for 20 min in PBT and then incubated at 4°C overnight with secondary antibodies at 1:400 [anti-rabbit conjugated to AlexaFluor 488 (ThermoFisher Scientific) and anti-mouse IgG (H+L) conjugated to AlexaFluor 488 (Life Technologies, A11029), or anti-mouse IgG (H+L) conjugated to AlexaFluor 633 (Life Technologies, A21126) for amnX8 analyses] in the blocking solution. After three washes (20 min), brains were mounted in the Prolong Mounting Medium (Life Technologies) for microscopy analysis. Images were acquired with a Nikon A1R confocal microscope. Confocal Z-stacks were acquired in 1 μm (amnX8 analyses) or 1.5 μm slices (amn-RNAi1 analyses), and imported into NIH ImageJ for analyses. For axonal length Fiji's plugin Simple Neurite Tracer (Longair et al., 2011) was used to determine the trajectory and length of the axone along all plans in stack images. For soma volume, we used Fiji's plugin Volumest (Merzin, 2008).
Quantitative PCR analyses.
Flies were raised at 25°C. Total RNA was extracted from 60 female heads using the RNeasy Plant Mini Kit (Qiagen). Preparations underwent a DNaseI treatment (BioLabs) for 15 min at 37°C. DNase was heat-inactivated with EDTA (10 mm). Samples were cleaned with the RNA Minielute Cleanup kit (Qiagen), and reverse-transcribed with oligo(dT)20 primers using the SuperScript III First-Strand kit (Life Technologies) according to the manufacturer's instructions. The amn oligonucleotides used were as follows: forward: 5′-ACATGCGCAGTTTTTGTTGT-3′; reverse: 5′-AACAGTAATACGCAGTGCAACG-3′. Level of the target cDNA was compared against level of α-Tub84B (Tub; CG1913) cDNA used as a reference. Amplification was performed using a LightCycler 480 (Roche) and the SYBR Green I Master mix (Roche). Reactions were performed in triplicate. Specificity and size of amplification products were assessed by melting curve analyses and agarose gel electrophoresis, respectively. Expression relative to reference is expressed as a ratio (2−ΔCp, where Cp is the crossing point).
Statistical analyses.
Comparisons of the data series between two conditions were achieved by a two-tailed unpaired Student's t test. Results of t tests are given as the value t(x) of the t distribution with x degrees of freedom obtained from the data. Comparisons between more than two distinct groups were made using one-way ANOVA followed, if significant at p ≤ 0.05, by Newman–Keuls multiple-comparisons tests. Overall ANOVA p value is given in the legends along with the value of the corresponding Fisher distribution F(x,y), where x is number of degrees of freedom for groups and y is total number of degrees of freedom for the distribution. Asterisks on the figure denote the least significant of the pairwise post hoc comparisons between the genotype of interest and its controls, following the usual nomenclature. Statistical tests were performed using GraphPad Prism 5 software. All data are displayed as mean ± SEM.
Results
Acute modulation of amn expression in adult DPM neurons does not impact memory
Amn memory studies conducted to date have mainly used constitutive amn mutants, and therefore have not been able to address temporal requirement. To study the temporal involvement of amn in olfactory memory, we used RNAi and conditional tissue-specific drivers. We targeted amn expression with a UAS-RNAi construct (amn-RNAi1) that was previously shown to phenocopy the thermal nociceptive defect of amn mutants (Aldrich et al., 2010). Memory was analyzed using the classical conditioning paradigm of odor-avoidance response. In this paradigm, groups of flies are successively exposed to two distinct odors, only one of which is associated with electric shocks. Following a single cycle of conditioning, we measure immediate memory (1 min memory) and 2 h memory. For consolidated memory phases, LT-ARM is assessed 24 h after five massed conditioning cycles whereas LTM is assessed 24 h after five spaced conditioning cycles.
We first analyzed the 2 h memory of flies constitutively expressing amn-RNAi1 in DPM neurons. To this end, we used the VT64246 GAL4-driver that specifically labels DPM neurons (Lee et al., 2011). As expected, VT64246/amn-RNAi1 flies showed a significant 2 h memory deficit (Fig. 1A), and this result was confirmed with a second non-overlapping amn-RNAi construct (amn-RNAi2; Fig. 1A). To address the specificity of the memory phenotype, we next analyzed LTM and observed a strong LTM deficit (Fig. 1B). In contrast, VT64246/amn-RNAi1 and VT64246/amn-RNAi2 flies displayed normal scores when memory was assessed immediately after a single cycle of conditioning (Fig. 1C). The ability of these flies to avoid electric shocks and their olfactory acuity to each odor after electric shock was unaffected (Table 1), showing that they displayed normal perception of the conditioning stimuli. We conclude that amn-RNAi expression specifically in DPM neurons during development and in adulthood mimics memory phenotypes described for amn-mutant flies (Quinn et al., 1979; Yu et al., 2006; Lee et al., 2011), thus confirming amn-RNAi is an efficient tool for studying the effect of amn inhibition on memory.
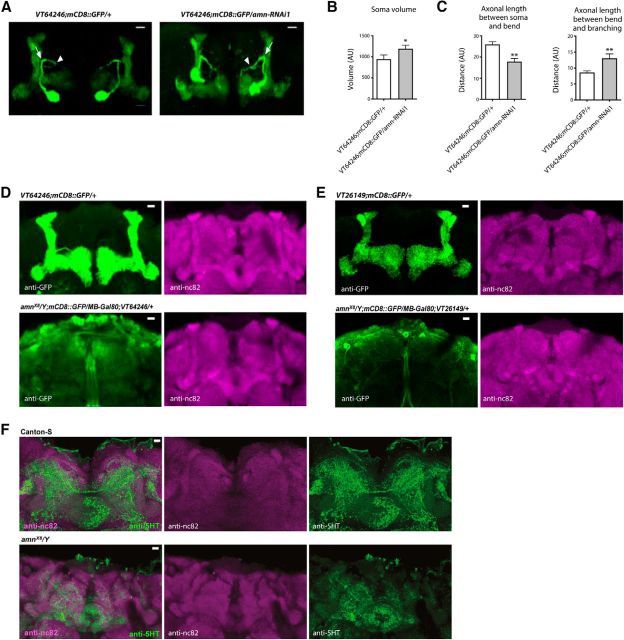
Figure 1.
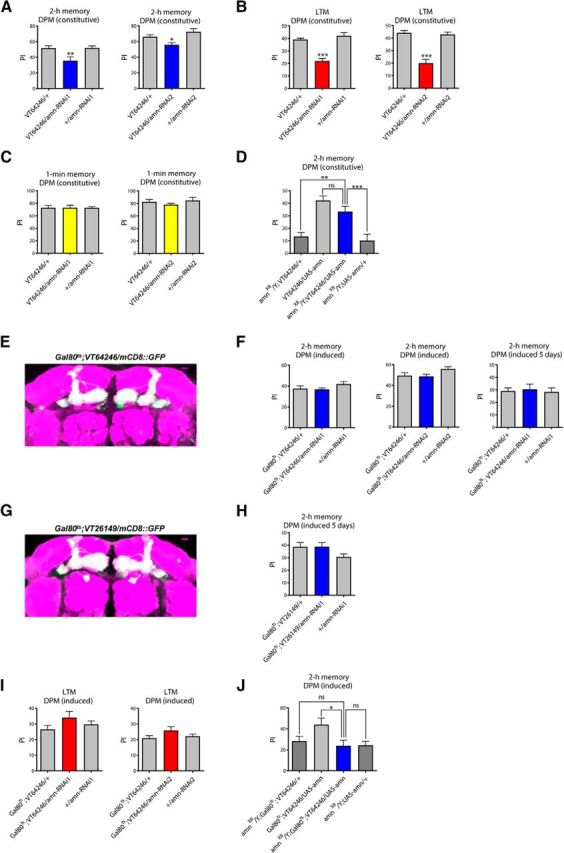
Acute modulation of amn expression in adult DPM neurons does not impact memory. A, Constitutive expression of amn-RNAi in DPM neurons leads to a 2 h memory deficit. Flies expressing amn-RNAi1 or amn-RNAi2 under the control of the constitutive VT64246 driver exhibit significantly lower scores than genetic controls (amn-RNAi1: F(2,29) = 6.793, **p = 0.004, n = 10; amn-RNAi2: F(2,39) = 7.415, **p = 0.002, n ≥ 13). B, Constitutive expression of amn-RNAi in DPM neurons leads to a LTM deficit. Flies expressing amn-RNAi1 or amn-RNAi2 under the control of the VT64246 driver exhibit significantly lower scores than genetic controls (amn-RNAi1: F(2,34) = 29.670, ***p < 0.0001, n ≥ 11; amn-RNAi2: F(2,34) = 35.47, ***p < 0.0001, n ≥ 11). C, Immediate memory is not altered by constitutive expression of amn-RNAi in DPM neurons. VT64246/amn-RNAi1 and VT64246/amn-RNAi2 flies show similar scores to genetic controls (amn-RNAi1: F(2,29) = 0.0003, p = 0.100, n = 10; amn-RNAi2: F(2,14) = 0.778, p = 0.481, n = 5). D, amn expression in DPM neurons rescues amnX8 memory defects. amnX8/Y;VT64246/UAS-amn flies expressing amn in DPM neurons show higher MTM scores than their amnX8/Y genetic controls, and similar scores to normal flies expressing amn in DPM neurons (F(3,57) = 13.79, ***p < 0.0001, n ≥ 13). Only males were analyzed. E, Analysis of induced expression of the Gal80ts;VT64246 driver. Gal80ts;VT64246/mCD8::GFP flies were incubated for 3 d at 30°C. Dissected brains were used for immunostaining (green, mCD8::GFP; magenta, nc82). Scale bar, 10 μm. F, Acute inhibition of amn expression in DPM neurons does not impact 2 h memory. After 3 d of induction, amn-RNAi-expressing flies show similar scores to genetic controls (amn-RNAi1: F(2,47) = 1.479, p = 0.239, n = 16; amn-RNAi2: F(2,46) = 2.906, p = 0.065, n ≥ 15). After 5 d of induction, Gal80ts;VT64246/amn-RNAi1 flies show normal 2 h memory (F(2,38) = 0.103, p = 0.903, n = 8). G, Analysis of induced expression of the Gal80ts;VT26149 driver. Gal80ts;VT26149/mCD8::GFP flies were incubated for 3 d at 30°C. Dissected brains were used for immunostaining (green, mCD8::GFP; magenta, nc82). Scale bar, 10 μm. H, Acute inhibition of amn expression for 5 d using the Gal80ts;VT26149 driver does not impact 2 h memory (F(2,38) = 0.119, p = 0.102, n = 8). I, Acute inhibition of amn expression in DPM neurons does not affect LTM. Flies were incubated for 3 d at 30°C before conditioning, and also until memory test. amn-RNAi-expressing flies show similar scores to genetic controls (amn-RNAi1: F(2,24) = 1.693, p = 0.2070, n ≥ 8; amn-RNAi2: F(2,29) = 1.711, p = 0.200, n = 10). J, amn expression in adult DPM neurons does not rescue amnX8 memory defects. After 3 d of induction, amnX8/Y;Gal80ts;VT64246/UAS-amn flies show similar MTM scores to their amnX8/Y genetic controls, and lower scores than normal flies expressing amn in adult DPM neurons (F(3,50) = 3.49, *p = 0.023, n ≥ 12). Only males were analyzed. Error bars indicate SEM.
Table 1.
Shock reactivity and olfactory acuity of flies expressing amn-RNAi in DPM neurons
| Genotype | Shock reactivity | Olfactory acuity |
|
|---|---|---|---|
| Octanol | Methylcyclohexanol | ||
| VT64246/+ | 71.80 ± 4.36 | 58.30 ± 2.77 | 60.50 ± 5.15 |
| VT64246/amn-RNAi1 | 65.30 ± 2.87 | 73.30 ± 4.60 | 59.80 ± 4.78 |
| +/amn-RNAi1 | 69.70 ± 2.83 | 61.90 ± 4.53 | 51.20 ± 4.01 |
| VT64246/+ | 69.50 ± 4.11 | 59.80 ± 4.45 | 67.70 ± 4.39 |
| VT64246/amn-RNAi2 | 67.20 ± 3.65 | 60.40 ± 3.35 | 63.60 ± 5.19 |
| +/amn-RNAi2 | 74.00 ± 4.39 | 58.40 ± 5.31 | 62.20 ± 5.10 |
Data are shown as mean ± SEM. VT64246/amn-RNAi1 and VT64246/amn-RNAi2 flies exhibit normal shock reactivity (amn-RNAi1: F(2,29) = 0.938, p = 0.406, n = 10; amn-RNAi2: F(2,29) = 0.725, p = 0.493, n = 10) and normal olfactory acuity for octanol (amn-RNAi1: F(2,29) = 3.726, p = 0.037, n = 10, post hoc Newman–Keuls test; VT64246/amn-RNAi versus VT64246/+, *p < 0.05; amn-RNAi2: F(2,29) = 0.053, p = 0.948, n = 10) and methylcyclohexanol (amn-RNAi1: F(2,29) = 1.230, p = 0.308, n = 10; amn-RNAi2: F(2,29) = 0.352, p = 0.707, n = 10).
Waddell et al. (2000) previously showed that reestablishing amn expression in DPM neurons was sufficient to rescue the memory of amn mutant flies. However, the c316 driver used in their study was later shown to also label, although to a lesser extent, other cells including KC (Wu et al., 2011). To better assess the spatial requirement of amn in memory, we aimed to reestablish amn expression specifically in DPM neurons of amnX8 null mutant flies. As expected, amnX8/Y control flies expressing the driver (amnX8/Y;VT64246/+) or containing the UAS-amn construct alone (amnX8/Y;UAS-amn/+) showed strong 2 h memory deficits (Fig. 1D). amnX8/Y;VT64246/UAS-amn flies expressing amn specifically in DPM neurons showed significantly higher MTM scores than amnX8/Y control groups (Fig. 1D), indicating a functional rescue.
We next investigated amn function specifically in adult DPM neurons. To restrict amn-RNAi expression to adulthood, we took advantage of the TARGET system (McGuire et al., 2003) that relies on a conventional GAL4 activation system and a temperature-sensitive Gal80 inhibitor (Gal80ts). Gal80ts represses GAL4 transcriptional activity at low temperature (18°C) but this inhibition is abolished at high temperature (30°C), allowing GAL4-mediated transgene expression. Experiments were performed with a tub-Gal80ts;VT64246 line that ubiquitously expresses Gal80ts (Gal80ts;VT64246). Using an UAS-mCD8::GFP transgene (mCD8::GFP), we first verified that the Gal80ts;VT64246 driver generated a high level of expression after 3 d of induction at 30°C (Fig. 1E), consistently with recent data showing that it leads to efficient RNAi expression after 3 d of induction (Turrel et al., 2016). Surprisingly, Gal80ts;VT64246/amn-RNAi1 and Gal80ts;VT64246/amn-RNAi2 induced flies both showed normal 2 h memory (Fig. 1F). Even after 5 d of induction, Gal80ts;VT64246/amn-RNAi1 flies displayed normal 2 h memory (Fig. 1F). To confirm these data, we repeated the experiment with tub-Gal80ts;VT26149, another DPM-specific line that drives efficient expression after induction (Fig. 1G). After 5 d of RNAi induction, Gal80ts;VT26149/amn-RNAi1 flies displayed normal 2 h memory (Fig. 1H). We next analyzed LTM and found that neither amn-RNAi1 nor amn-RNAi2 expression in adult DPM neurons were able to induce a LTM deficit (Fig. 1I). In conclusion, constitutive inhibition of amn expression in DPM neurons leads to memory impairment whereas acute inhibition in adult DPM does not.
We next restored amn expression in DPM neurons of adult amnX8 null mutant flies. amnX8/Y;Gal80ts;VT64246/UAS-amn flies showed 2 h memory that was not different from amnX8/Y mutant genotypes, and significantly lower than control flies (Fig. 1J). Thus restoration of amn expression in adult DPM neurons does not rescue amnX8 memory impairment. Together, the findings suggest that amn expression is required in DPM neurons during developmental stages, and not acutely during adulthood.
amn is involved in the development of DPM neurons
To investigate the potential influence of amn on the development of DPM neurons, we analyzed the morphology of these neurons in flies expressing mCD8::GFP and amn-RNAi1 under the control of the VT64246 driver. As expected, we observed extensive arborization of DPM neurons onto the MB lobes. However, flies expressing amn-RNAi1 in DPM neurons exhibited DPM neurons that were morphologically abnormal on several aspects (Fig. 2). DPM cell bodies exhibited irregular shape and less accurate localization relative to the MB (Fig. 2A) and they were bigger in flies expressing amn-RNAi1 in DPM neurons compared with controls (Fig. 2B). In addition, the DPM axonal path was modified in amn-deficient flies (Fig. 2C). Together, the data indicate that amn expression in DPM neurons is required for their own normal development.
Figure 2.
amn is involved in the development of DPM neurons. A–C, Constitutive expression of amn-RNAi1 in DPM neurons alters their morphology. DPM neurons were revealed using anti-GFP staining. AU, Arbitrary unit. A, A representative brain of each genotype is shown. Scale bar, 10 μm. B, DPM cell body volume is increased in VT64246;mCD8::GFP/amn-RNAi1 compared with VT64246;mCD8::GFP/+ control flies (t test, t(22) = 2.177, *p = 0.041, n = 12). C, Axon measurements. Axonal length between the cell body and the major axonal bend (arrow) is reduced in VT64246;mCD8::GFP/amn-RNAi1 compared with VT64246;mCD8::GFP/+ control flies (t test, t(22) = 3.756, **p = 0.001, n = 12), whereas axonal length between the bend and the DPM axonal bifurcation point (arrowhead) is increased (t test, t(22) = 2.952, **p = 0.007, n = 12). D–F, Analyses of amnX8 null mutant fly brains. D, VT64246;mCD8::GFP/+ control brains (n = 11) and amnX8/Y;mCD8::GFP/MB-Gal80;VT64246/+ mutant brains (n = 13) were analyzed with anti-GFP (left) and anti-nc82 (right) primary antibodies. Identical Z-stack projections are shown on the left and right. A representative brain of each genotype is shown. Scale bar, 10 μm. E, VT26149;mCD8::GFP/+ control brains (n = 7) and amnX8/Y;mCD8::GFP/MB-Gal80;VT26149/+ mutant brains (n = 10) were analyzed with anti-GFP (left) and anti-nc82 (right) primary antibodies. Identical Z-stack projections are shown on the left and right. A representative brain of each genotype is shown. Scale bar, 10 μm. F, Canton-S control brains (n = 8) and amnX8/Y mutant brains (n = 8) were analyzed with anti-nc82 and anti-5HT primary antibodies. Identical Z-stack projections are shown for each panel. A representative brain of each genotype is shown. Scale bar, 10 μm. Error bars indicate SEM.
To further assess amn involvement in the development of DPM neurons, we next aimed at analyzing the morphology of DPM neurons in amn null mutant flies. The amnX8 null mutant line results from the excision of a GAL4-encoding P-element inserted in the amn28A line (Moore et al., 1998). Yet, the amn28A line contained two insertions and only one was excised in the amnX8 flies (Moore et al., 1998). It was later mentioned that amnX8 flies had retained GAL4 activity localized in the MB (Keene et al., 2004). To be able to analyze GFP expression specifically driven in DPM neurons, we constructed a line expressing Gal80 in the MB of amnX8 flies using the MB247-Gal80 transgene (MB-Gal80; Krashes et al., 2007). Strikingly, we did not observe immunostaining on the MB lobes in amnX8/Y;mCD8::GFP/MB-Gal80;VT64246/+ fly brains (Fig. 2D). We verified that MB did not display any gross alteration of their morphology using the nc82 antibody to label synapses (Fig. 2D). To rule out the possibility that the loss of expression might be due to a change of the VT64246 expression pattern in an amn background, we analyzed GFP expression with the DPM-specific driver VT26149. We did not observe immunostaining on the MB lobes in amnX8/Y;mCD8::GFP/MB-Gal80;VT26149/+ fly brains (Fig. 2E). Finally, to further confirm that the absence of staining was due to an abnormal development of the DPM neurons, we took advantage of the fact that DPM neurons can be revealed by serotoninergic (5HT) staining (Lee et al., 2011). In Canton-S control fly brains, we observed numerous ramifications coming from DPM neurons onto the MB (Fig. 2F). In contrast, in amnX8 mutant flies, these ramifications were mostly absent, confirming an abnormal development of DPM neurons (Fig. 2F). We conclude that in the absence of amn expression, DPM neurons do not develop normally.
amn expression in the MB
As we could not evidence any memory impact upon acute modulation of amn expression in the adult, and as we observed that amn is required for the proper development of neurons involved in memory, the next question to address was whether AMN expression plays a role in the adult brain for memory.
As mentioned in the previous section, Keene et al. (2004) reported as data not shown that amnX8 flies expressed GAL4 activity in the MB. Indeed, when we analyzed GFP expression driven by amnX8, we observed immunostaining of the MB α and β lobes (Fig. 3A), confirming that amnX8 flies had retained a GAL4-coding sequence, and suggesting the presence of a MB-specific enhancer in the amn gene. Analysis of GAL4-expression patterns in the adult brain points, in addition to a DPM enhancer-sensitive sequence, to the presence of a second enhancer located upstream of the amn transcript that leads to specific expression in the MB (Fig. 3B). GAL4 inserted in a region located at the start of the amn coding sequence (blue box) lead to expression in DPM neurons (amn651 and amnCHPD; Waddell et al., 2000). A fragment of genomic DNA covering a region upstream of the amn transcription unit is linked to GAL4 expression in the MB α/β neurons (GMR65A01; Jenett et al., 2012; expression pattern available from http://flybase.org/reports/FBtp0061966.html). Similarly, the Vienna Tile VT64511 covering a region upstream and at the start of the amn transcription unit leads to GAL4 expression in the MB α/β neurons.
Figure 3.
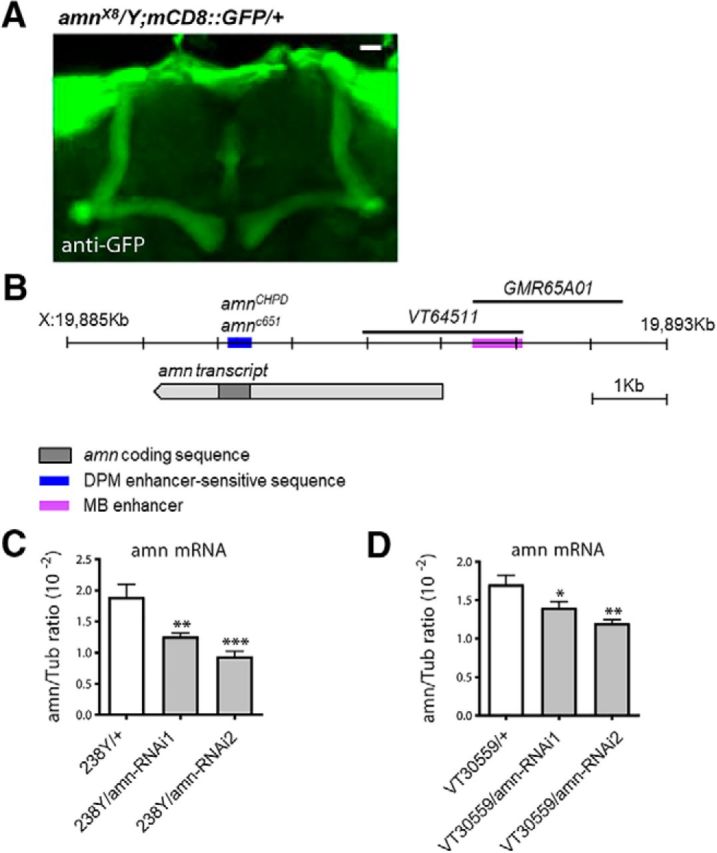
amn expression in the MB. A, amnX8 flies express GAL4 in the MB α/β neurons. Dissected brains were used for immunostaining (green, mCD8::GFP). B, Schematic representation of the amn transcript with the localization of a MB enhancer sequence and of DPM enhancer-sensitive sequence. C, D, qPCR analyses of amn expression levels. Total RNA extracted from female heads were reverse-transcribed and further quantified by PCR. Expression relative to reference is expressed as a ratio (2−ΔCp, where Cp is the crossing point). C, 238Y/amn-RNAi1 and 238Y/amn-RNAi2 show 33% and 50% decrease in amn mRNA compared with the 238Y/+ control. Each bar corresponds to six measures from three independent experiments (F(2,17) = 13.16, ***p = 0.0005, n = 6). D, VT30559/amn-RNAi1 and VT30559/amn-RNAi2 show 18% and 30% decrease in amn mRNA compared with the VT30559/+ control. (F(2,23) = 8.522, **p = 0.002, n = 8). Each bar corresponds to eight measures from four independent experiments. Error bars indicate SEM.
We could not directly assess amn expression in the MB with immunochemistry experiments because AMN antibodies are not available. We tried to perform in situ experiments, but probably because amn RNA is expressed at very low levels (Feany and Quinn, 1995; Chintapalli et al., 2007; Affymetrix data available from http://flyatlas.org/atlas.cgi?name=FBgn0000076), we did not observe specific staining. Thus, to assess amn expression in MB neurons, we analyzed the effect of amn-RNAi expression under the control of the 238Y driver known to label MB neurons (Aso et al., 2009). qPCR analyses revealed a decrease of amn mRNA level in 238Y/amn-RNAi1 and 238Y/amn-RNAi2 fly heads compared with controls (Fig. 3C). We next used the VT30559 driver that labels KC more specifically than the 238Y driver (Plaçais et al., 2017). VT30559/amn-RNAi1 and VT30559/amn-RNAi2 fly heads also expressed less amn mRNA than controls (Fig. 3D). Together, the data strongly suggest that amn is indeed expressed in the adult MB.
Acute inhibition of amn expression in adult MB neurons leads to memory defects
We then investigated the involvement of amn in memory in the MB. Flies expressing amn-RNAi in all adult MB neuronal types with a conditional tub-Gal80ts;238Y driver (Gal80ts;238Y) showed abnormal 2 h memory (Fig. 4A), whereas non-induced flies showed normal scores (Fig. 4B), establishing that the memory deficit is caused by RNAi induction.
Figure 4.
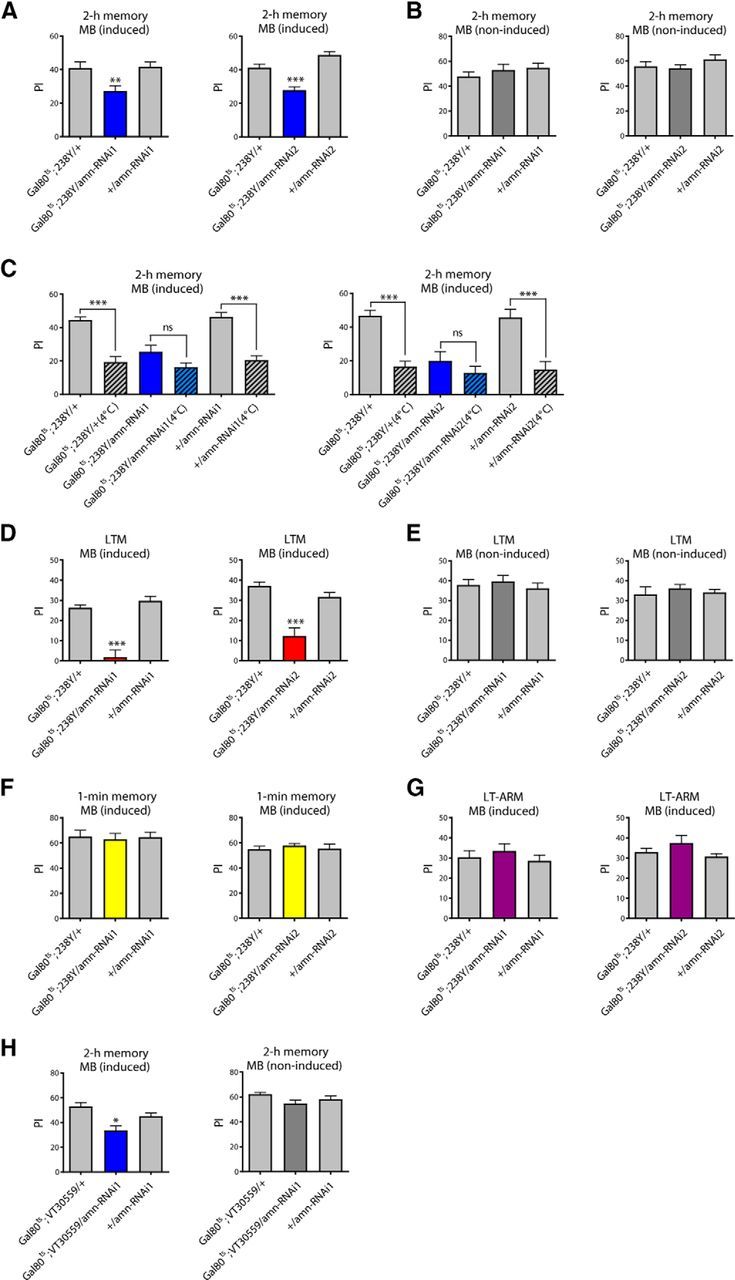
Acute inhibition of amn expression in adult MB neurons leads to memory defects. A, B, amn inhibition in adult MB neurons induces MTM defects. A, Flies expressing amn-RNAi in the adult MB exhibit a 2 h memory deficit (amn-RNAi1: F(2,35) = 7.348, **p = 0.002, n = 12; amn-RNAi2: F(2,39) = 27.81, ***p < 0.0001, n ≥ 13). B, Non-induced flies display normal 2 h memory scores (amn-RNAi1: F(2,29) = 0.822, p = 0.451, n = 10; amn-RNAi2: F(2,34) = 1.242, p = 0.302, n ≥ 11). C, When indicated (4°C), flies were submitted to 2 min cold shock 1 h before the memory test. Cold-shock treatment induces a memory decrease in control flies but not in Gal80ts;238Y/amn-RNAi flies (amn-RNAi1: F(5,75) = 21.99, ***p < 0.0001, n ≥ 12; amn-RNAi2: F(5,71) = 12.98, ***p < 0.0001, n = 12). D, E, amn inhibition in adult MB neurons induces LTM defects. D, Gal80ts;238Y/amn-RNAi-induced flies show LTM impairment (amn-RNAi1: F(2,24) = 35.70, ***p < 0.0001, n ≥ 8; amn-RNAi2: F(2,34) = 20.22, ***p < 0.0001, n ≥ 11). E, In the absence of induction, Gal80ts;238Y/amn-RNAi flies show normal LTM (amn-RNAi1: F(2,29) = 0.393, p = 0.679, n = 10; amn-RNAi2: F(2,29) = 0.363, p = 0.699, n = 10). F, Immediate memory is not affected by amn inhibition in the adult MB (amn-RNAi1: F(2,23) = 0.060, p = 0.942, n = 8; amn-RNAi2: F(2,29) = 0.320, p = 0.729, n = 10). G, LT-ARM is not affected by amn inhibition in the adult MB (amn-RNAi1: F(2,29) = 0.565, p = 0.575, n = 10; amn-RNAi2: F(2,46) = 1.709, p = 0.193, n ≥ 15). H, MTM analyses with the Gal80ts;VT30559 driver. Gal80ts;VT30559/amn-RNAi1 flies show impaired MTM (F(2,35) = 9.953, ***p = 0.0004, n = 12). In absence of induction, Gal80ts;VT30559/amn-RNAi1 flies show normal memory (F(2,35) = 2.547, p = 0.936, n = 12). Error bars indicate SEM.
Memory measured 2 h after training is composed of two distinct phases: MTM, which is a labile memory sensitive to cold anesthesia, and MT-ARM which is resistant to anesthesia (Quinn and Dudai, 1976). To examine which memory phase was affected by amn inhibition, we applied cold-shock treatments to the flies. The memory scores of control flies dropped whereas the memory scores of amn-RNAi-expressing flies remained unaffected (Fig. 4C), indicating that MTM is absent. Furthermore, after cold shock treatment, Gal80ts;238Y/amn-RNAi flies exhibited similar memory scores to control flies, showing that MT-ARM is not affected. We next examined LTM after spaced training and observed that Gal80ts;238Y/amn-RNAi flies showed a strong LTM impairment (Fig. 4D,E) but normal 1 min memory (Fig. 4F) and LT-ARM (Fig. 4G). The sensory-motor capacities of these flies were normal (Table 2). We next used a second MB driver, VT30559, to knockdown amn expression specifically in KC. After 3 d of induction, Gal80ts;VT30559/amn-RNAi1 flies displayed impaired 2 h memory whereas noninduced flies displayed normal 2 h memory (Fig. 4H). We verified that the ability of induced Gal80ts;VT30559/amn-RNAi1 flies to perceive conditioning stimuli was normal (Table 2). Strikingly, we conclude that inhibition of amn expression specifically in the adult MB recapitulates memory phenotypes previously reported for amn-mutant flies (Quinn et al., 1979; Yu et al., 2006; Lee et al., 2011). In short, MTM and LTM are altered, whereas STM and ARM remain unaffected.
Table 2.
Shock reactivity and olfactory acuity of flies expressing amn-RNAi in adult MB neurons
| Genotype | Shock reactivity | Olfactory acuity |
|
|---|---|---|---|
| Octanol | Methylcyclohexanol | ||
| Gal80ts;238Y/+ | 79.75 ± 3.92 | 75.10 ± 3.79 | 50.70 ± 0.92 |
| Gal80ts;238Y/amn-RNAi1 | 73.13 ± 3.83 | 70.50 ± 3.09 | 57.00 ± 4.10 |
| +/amn-RNAi1 | 72.00 ± 4.67 | 73.50 ± 3.62 | 52.00 ± 1.65 |
| Gal80ts;238Y/+ | 83.70 ± 4.42 | 53.50 ± 5.43 | 55.50 ± 6.57 |
| Gal80ts;238Y/amn-RNAi2 | 81.20 ± 2.84 | 58.50 ± 6.82 | 46.10 ± 4.15 |
| +/amn-RNAi2 | 71.40 ± 1.82 | 44.20 ± 4.59 | 49.40 ± 2.90 |
| Gal80ts;VT30559/+ | 74.88 ± 5.11 | 69.38 ± 4.56 | 66.25 ± 7.01 |
| Gal80ts;VT30559/amn-RNAi1 | 77.38 ± 2.10 | 66.38 ± 3.82 | 70.88 ± 5.27 |
| +/amn-RNAi1 | 77.88 ± 3.74 | 60.88 ± 3.57 | 55.00 ± 4.83 |
Data are shown as mean ± SEM. Gal80ts;238Y/amn-RNAi1 and Gal80ts;238Y/amn-RNAi2-induced flies exhibit normal shock reactivity (amn-RNAi1: F(2,23) = 1.015, p = 0.380, n = 8; amn-RNAi2: F(2,29) = 0.992, p = 0.384, n = 10) and normal olfactory acuity for octanol (amn-RNAi1: F(2,29) = 0.442, p = 0.647, n = 10; amn-RNAi2: F(2,29) = 4.113, p = 0.028, n = 10) and methylcyclohexanol (amn-RNAi1: F(2,29) = 1.627, p = 0.215, n = 10; amn-RNAi2: F(2,29) = 1.628, p = 0.215, n = 10). Gal80ts;VT30559/amn-RNAi1-induced flies exhibit normal shock reactivity (F(2,23) = 0.174, p = 0.842, n = 8) and normal olfactory acuity for octanol (F(2,23) = 1.156, p = 0.334, n = 8) and methylcyclohexanol (amn-RNAi1: F(2,23) = 1.993, p = 0.161, n = 8).
Acute inhibition of amn expression in adult α/β neurons leads to memory defects
Because MTM and LTM rely on MB α/β neurons (Bouzaiane et al., 2015), we next aimed to specifically restrict amn inhibition to α/β neurons using the inducible tub-Gal80ts;c739 driver (Gal80ts;c739). Flies expressing amn-RNAi in adult α/β neurons exhibited deficits in 2 h memory (Fig. 5A,B) and LTM (Fig. 5C,D) but normal 1 min memory (Fig. 5E). We verified that their responses to the stimuli used for training were unaffected (Table 3). To further characterize in which KC amn is required for memory, we analyzed the effect of amn inhibition in adult α′/β ′ KC using a tub-Gal80ts;VT30604 driver (Wu et al., 2013), and in adult γ KC using a tub-Gal80ts;VT49483 driver (see Materials and Methods). Neither α′/β ′ KC-driven amn-RNAi1 expression (Fig. 5F) nor γ KC-driven amn-RNAi1 expression (Fig. 5G) did impact MTM or LTM.
Figure 5.
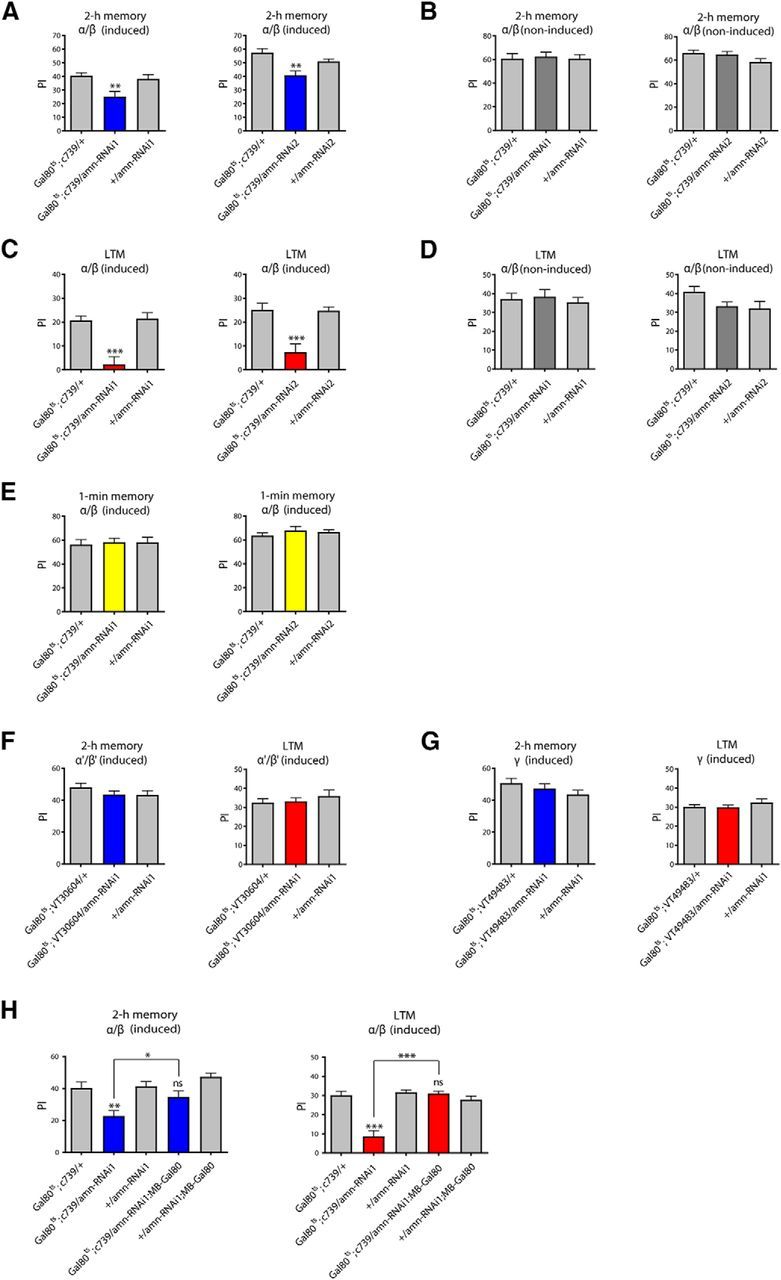
Acute inhibition of amn expression in adult α/β neurons leads to memory defects. A, B, amn expression in adult α/β neurons is required for MTM. A, After induction, Gal80ts;c739/amn-RNAi flies show lower MTM scores than genetic controls (amn-RNAi1: F(2,29) = 6.600, **p = 0.0046, n = 10; amn-RNAi2: F(2,42) = 10.17, ***p = 0.0003, n ≥ 14). B, In absence of induction, Gal80ts;c739/amn-RNAi flies show normal MTM (amn-RNAi1: F(2,29) = 0.064, p = 0.939, n = 10; amn-RNAi2: F(2,29) = 2.141, p = 0.137, n = 10). C, D, amn expression in the adult α/β neurons is required for LTM. C, Gal80ts;c739/amn-RNAi-induced flies show LTM deficits (amn-RNAi1: F(2,29) = 16.56, ***p < 0.0001, n ≥ 9; amn-RNAi2: F(2,29) = 13.88, ***p < 0.0001, n = 10). D, In absence of induction, Gal80ts;c739/amn-RNAi flies show normal LTM (amn-RNAi1: F(2,29) = 0.220, p = 0.804, n = 10; amn-RNAi2: F(2,29) = 2.450, p = 0.105, n = 10). E, amn expression is not required in adult α/β neurons for immediate memory. Gal80ts;c739/amn-RNAi induced flies show similar scores to genetic controls (amn-RNAi1: F(2,29) = 0.087, p = 0.917, n = 10; amn-RNAi2: F(2,29) = 0.719, p = 0.496, n = 10). F, amn expression in adult α′/β′ neurons is not required for MTM nor LTM. After induction Gal80ts;VT30604/amn-RNAi1 flies show normal MTM (F(2,35) = 1.175, p = 0.321, n = 12) and normal LTM (F(2,38) = 0.602, p = 0.553, n = 13). G, amn expression in adult γ neurons is not required for MTM nor LTM. After induction Gal80ts;VT49483/amn-RNAi1 flies show normal MTM (F(2,35) = 1.463, p = 0.246, n = 12) and normal LTM (F(2,38) = 0.856, p = 0.433, n = 13). H, Intersectional analyses. Gal80ts;c739/amn-RNAi1-induced flies show MTM and LTM deficits whereas Gal80ts;c739/amn-RNAi1;MB-Gal80-induced flies show normal MTM and LTM (MTM: F(4,72) = 7.672, ***p < 0.0001, n ≥ 14; LTM: F(4,59) = 24.12, ***p < 0.0001, n = 12). Error bars indicate SEM.
Table 3.
Shock reactivity and olfactory acuity of flies expressing amn-RNAi in adult α/β neurons
| Genotype | Shock reactivity | Olfactory acuity |
|
|---|---|---|---|
| Octanol | Methylcyclohexanol | ||
| Gal80ts;c739/+ | 74.25 ± 4.86 | 52.70 ± 3.82 | 45.40 ± 3.78 |
| Gal80ts;c739/amn-RNAi1 | 78.75 ± 4.46 | 56.80 ± 4.96 | 48.30 ± 4.84 |
| +/amn-RNAi1 | 70.25 ± 5.36 | 59.50 ± 6.58 | 44.50 ± 3.55 |
| Gal80ts;c739/+ | 70.60 ± 5.56 | 52.20 ± 4.36 | 48.80 ± 4.67 |
| Gal80ts;c739/amn-RNAi2 | 69.10 ± 4.78 | 57.60 ± 5.12 | 56.30 ± 3.92 |
| +/amn-RNAi2 | 68.50 ± 4.14 | 60.90 ± 5.19 | 49.80 ± 5.33 |
Data are shown as mean ± SEM. Gal80ts;238Y/amn-RNAi1 and Gal80ts;238Y/amn-RNAi2-induced flies exhibit normal shock reactivity (amn-RNAi1: F(2,23) = 0.752, p = 0.484, n = 8; amn-RNAi2: F(2,29) = 0.050, p = 0.952, n = 10) and normal olfactory acuity for octanol (amn-RNAi1: F(2,29) = 0.427, p = 0.657, n = 10; n = 10; amn-RNAi2: F(2,29) = 0.744, p = 0.485, n = 10) and methylcyclohexanol (amn-RNAi1: F(2,29) = 0.236, p = 0.792, n = 10; amn-RNAi2 F(2,29) = 0.759, p = 0.478, n = 10).
Together, the data show that amn is specifically required in the adult α/β KC for MTM and LTM formation. To strengthen these results, we performed intersectional experiments using the MB-Gal80 transgene to inhibit RNAi expression in the MB. Gal80ts;c739/amn-RNAi1-induced flies showed mutant 2 h memory, as previously observed, whereas Gal80ts;c739/amn-RNAi1;MB-Gal80-induced flies displayed normal 2 h memory (Fig. 5H). Similarly, Gal80ts;c739/amn-RNAi1 flies showed a strong LTM deficit whereas Gal80ts;c739/amn-RNAi1;MB-Gal80 flies showed normal LTM (Fig. 5H). We conclude that preventing GAL4-mediated amn-RNAi expression in the MB with Gal80 abolishes memory deficits. In conclusion, the results demonstrate that AMN is acutely required in MB α/β neurons for MTM and LTM formation.
Expression of the wild-type amn gene in MB neurons of amnX8 mutant flies rescues their memory deficits
The next question we addressed was whether reestablishing amn gene expression in MB neurons of amn null mutant flies could rescue their memory deficits. First, we showed that overexpression of the amn gene in MB neurons with the 238Y, c739, or c747 driver did not affect MTM nor LTM (Fig. 6A). We then performed rescue experiments. Interestingly, amnX8/Y;238Y/UAS-amn MTM scores were indistinguishable from 238Y/UAS-amn control fly scores (Fig. 6B), indicating a full rescue of the MTM deficit. Similarly, we also observed complete rescue of the LTM deficit (Fig. 6B). To confirm these results, we used the c739 α/β-specific driver and found that amn expression driven in the α/β KC allows complete rescue of both the MTM and LTM deficit in amnX8/Y flies (Fig. 6C). In conclusion, amn constitutive expression in either DPM neurons or α/β KC is capable of restoring functional memory.
Figure 6.
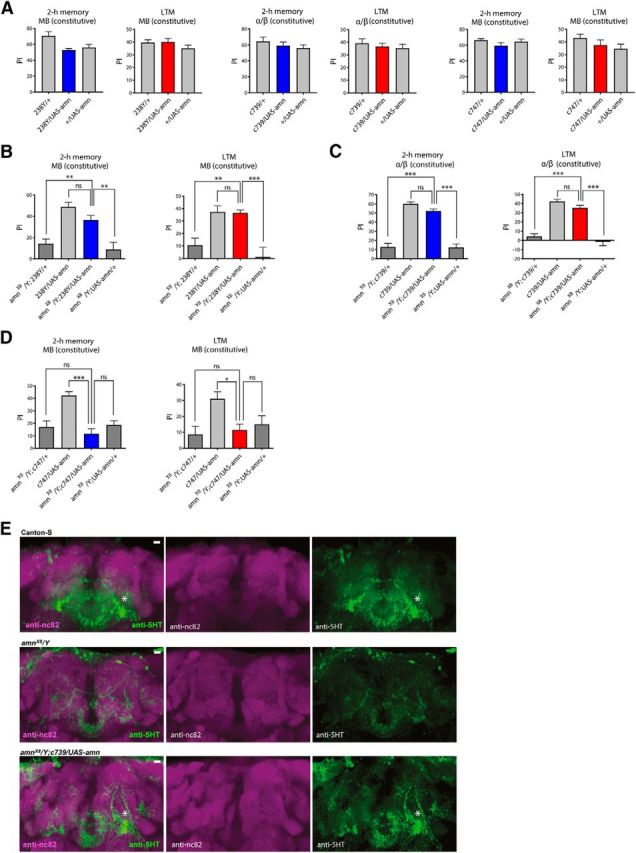
Expression of the wild-type amn gene in MB neurons of amnX8 mutant flies rescues their memory deficits. A, MTM and LTM scores are not affected by expressing amn in the MB. MTM and LTM scores are not affected by expressing amn in the different subsets of the MB with the 238Y driver (MTM: F(2,23) = 5.967, **p = 0.0089, n = 8; LTM: F(2,33) = 1.178, p = 0.3205, n = 12), the c739 driver (MTM: F(2,23) = 0.8268, p = 0.4512, n = 8; LTM: F(2,33) = 0.5209, p = 0.5988, n = 12) or the c747 driver (MTM: F(2,23) = 1.425, p = 0.2629, n = 8; LTM: F(2,33) = 1.419, p = 0.2563, n = 12). B, Analyses with the 238Y driver. amnX8/Y flies expressing amn show higher MTM and LTM scores than their amnX8/Y genetic controls and similar MTM and LTM scores to flies expressing amn in the MB (MTM: F(3,39) = 14.14, ***p < 0.0001, n = 10; LTM: F(3,39) = 10.94, ***p < 0.0001, n = 10). C, Analyses with the c739 driver. amnX8/Y flies expressing amn in the α/β KC display higher MTM and LTM scores than their amnX8/Y genetic controls and similar MTM and LTM scores to flies expressing amn in the α/β KC (MTM: F(3,59) = 66.52, ***p < 0.0001, n = 15; LTM: F(3,59) = 53.87, ***p < 0.0001, n = 15). D, c747 GAL4-driven amn expression does not rescue memory deficits of amnX8 mutant flies. amnX8/Y;c747/UAS-amn flies show similar MTM and LTM scores to amnX8/Y genotypes (MTM: F(3,39) = 11.83, ***p < 0.0001, n = 10; LTM: F(3,23) = 4.449, *p = 0.015, n = 6). Only males were analyzed in these experiments. E, Expression of the wild-type amn gene rescues normal 5HT staining. Canton-S control (n = 8), amnX8/Y mutant (n = 8) and amnX8/Y;c739/UAS-amn (n = 5) brains were analyzed with anti-nc82 and anti-5HT primary antibodies. DPM neuron staining is indicated by a star. Identical Z-stack projections are shown for each panel. A representative brain of each genotype is shown. Scale bar, 10 μm. Error bars indicate SEM.
These results seem to contradict a previous study (Waddell et al., 2000) reporting that amn expression in the MB failed to rescue the olfactory memory phenotypes of amnX8 flies (Waddell et al., 2000). To resolve this apparent discrepancy, we also conducted rescue experiments with the same c747 driver that Waddell et al. (2000) used. We observed that c747 GAL4-driven amn expression had no effect on the memory deficits of amn null mutant flies (Fig. 6D). Thus, amn expression driven by either 238Y or c739 rescues amnX8 memory deficits whereas expression driven by c747 does not, indicating that these drivers are functionally distinct. The fact that c747 shows a broad brain expression (Rodan et al., 2002) may explain this difference.
Finally, to analyze whether amn expression in the MB of amnX8 flies could also rescue the abnormal development of DPM neurons, we performed 5HT staining of amnX8/Y;c739/UAS-amn fly brains. As previously observed (Fig. 2F), amnX8 mutant flies show a loss of specific staining of the ramifications and the soma of DPM neurons compared with Canton-S control flies (Fig. 6E). Strikingly, in amnX8/Y;c739/UAS-amn flies, the 5HT staining of DPM neurons was restored (Fig. 6E), indicating that expression of the amn gene in α/β neurons during development and adulthood is sufficient to restore normal development of DPM neurons in amnX8 mutants.
Discussion
Here, we show that amn expression in DPM neurons is required for their development. Next, using inducible inhibition of amn expression, we establish that AMN peptide is specifically required in adult α/β MB neurons for MTM and LTM.
It was previously shown that acute ubiquitous amn expression in the adult was inefficient to rescue memory phenotypes of amnX8 null mutant flies (DeZazzo et al., 1999). Here, induced amn expression in adult DPM neurons using the TARGET system also failed to rescue amnX8 memory deficits, suggesting that amn expression in DPM neurons may be required for a developmental stage. We analyzed the morphology of DPM neurons in fly coexpressing amn-RNAi and GFP in these neurons. Although DPM arborization onto MB lobes is still visible, their abnormal cell body and axon morphology indicate that amn expression in DPM neurons is required to achieve their normal development. Keene et al. (2004) analyzed DPM morphology in amnex1 mutant fly brains using c316 to drive GFP expression in DPM neurons. They concluded that amn is not essential for DPM targeting to the MB during development. Interestingly, although they did not describe it in detail, they showed a typical amnex1 brain that apparently displayed DPM neurons with similar subtle morphology alterations to those described in our study. We further analyzed DPM development in an amn knock-out context. Imaging DPM neurons in amnX8 brains is hampered by the fact that amnX8 flies express GAL4 in the MB (Keene et al., 2004). To circumvent this issue, we took advantage of the MB-Gal80 transgene to inhibit GAL4 activity in the MB. Strikingly, using VT64246 or VT26149 to drive GFP expression in DPM neurons, we did not observe DPM arborization in amnX8 brains, despite the presence of all MB lobes. Together the results show that amn plays an essential role in the development of DPM neurons, a pair of neurons involved in olfactory memory; thus raising the question of whether amn is involved in memory in the adult brain.
Although constitutive expression of amn-RNAi in DPM neurons mimics memory deficits described for amn mutants, amn inhibition restricted to adult DPM neurons does not induce any memory impairment. The absence of a memory phenotype using conditional RNAi does not demonstrate that the target gene is not involved in memory processes, as we cannot rule out that conditional RNAi expression does not drive sufficient inhibition of the target gene expression to generate a loss-of-function phenotype. However, several lines of evidence suggest that amn expression should be efficiently inhibited by RNAi in adult DPM neurons: (1) each of the two amn-RNAi constructs used here induces memory deficits when acutely expressed in the adult MB; (2) to achieve amn inhibition in adult DPM neurons, two distinct DPM drivers were used after up to 5 d of GAL4-induction; and (3) one of these specific drivers has been previously shown to generate strong memory deficits upon RNAi-mediated silencing (Turrel et al., 2016). Also, restoring amn expression in amnX8 mutant flies during development and adulthood rescues their memory deficits, whereas expression restricted to adulthood fails to restore memory. Together, the data suggest that amn expression in DPM neurons is not required during adulthood for memory processes. When amn expression is constitutively inhibited in DPM neurons, these neurons do not form properly. As a consequence, memory is affected because DPM neurons cannot fulfill their physiological role (Keene et al., 2004), independently of amn adult function. The fact that constitutive amn expression in either MB or DPM neurons can rescue memory deficits of amnX8 flies suggests that the AMN peptide is secreted and can act non-autonomously to fulfill its roles during development and in the adult.
Analysis of GAL4-expression patterns indicates the presence of a MB enhancer located upstream of the amn transcription unit. Consistently, qPCR experiments show that amn-RNAi expression in the MB leads to a reduced level of amn RNA. Thus, similar to genes encoding components of the cAMP cascade, rutabaga, dunce, and DCO, and consistent with a role in this pathway, amn is expressed in the MB. Surprisingly, Waddell et al. (2000) did not report AMN expression in adult KC. They analyzed reporter-gene staining in amnchpd and amnc651 fly brains, both lines that appear to contain an insertion in the DPM enhancer located in the amn coding sequence. Immunohistology experiments revealed AMN staining in DPM neurons at the site of protein synthesis, i.e., cell bodies (Waddell et al., 2000). AMN concentration in KC may be much lower than that observed in DPM large cell bodies, thus making difficult its observation with immunohistology experiments. Several reports indicate that amn is expressed at very low levels (Feany and Quinn, 1995; Chintapalli et al., 2007). A low level of expression is consistent with the fact that neuropeptides are known to display a higher receptor binding affinity than classical neurotransmitters (Merighi et al., 2011), thus eliciting their biological effects when released at lower quantities.
Remarkably, conditional inhibition of amn expression in the adult MB recapitulates memory phenotypes previously reported for amn mutant flies (Quinn et al., 1979; Yu et al., 2006; Lee et al., 2011). MTM and LTM are impaired, whereas MT-ARM and LT-ARM are normal, which is consistent with a requirement for amn in the α/β neurons known to sustain MTM and LTM processes (Bouzaiane et al., 2015). In agreement with amn requirement in the MB, we show that amn expression in α/β KC rescues amnX8 memory deficits. These results are in apparent contradiction with a previous study showing that restoring AMN expression in the MB of amnX8 flies did not restore normal MTM (Waddell et al., 2000). However, we also observed a lack of memory rescue when using one of the MB drivers that Waddell et al. (2000) had used, indicating that using distinct drivers to achieve AMN expression generates different results. Supporting these observations, several reports have previously described similar discrepancies among these MB drivers (Chen et al., 2008; Pavlopoulos et al., 2008; Bushey et al., 2009), probably reflecting different levels of expression and/or expression in different KC subpopulations (Rodan et al., 2002).
We report here that AMN is required for two distinct processes: its expression in DPM neurons is required for their development, and its expression in the adult MB is required for memory formation. Strikingly, amn memory phenotypes are rescued by AMN constitutive expression in either DPM or MB neurons. Therefore, AMN overexpression in MB neurons during development is able to compensate the absence of AMN in DPM neurons and conversely, AMN overexpression in adult DPM neurons is capable of rescuing the functional deficit in the MB. In other words, because AMN is secreted (Feany and Quinn, 1995), it could act on a nearby structure, thereby fulfilling both functions.
One of the three peptides encoded by amn is homologous to PACAP which is also known to be involved in brain development and function (Shen et al., 2013). Interestingly, several reports in mammals have suggested that PACAP is implicated in learning and memory (Sacchetti et al., 2001; Matsuyama et al., 2003; Schmidt et al., 2015; Cabezas-Llobet et al., 2018). An attractive hypothesis is that AMN neuropeptide synthesized by KC would act autocrine-like to activate Rutabaga, the fly adenylate cyclase that mediates coincidence detection sustaining memory formation in the MB lobes (Levin et al., 1992; Tomchik and Davis, 2009; Gervasi et al., 2010). When overexpressed, AMN could act as a paracrine signal from a close structure. This would be possible at DPM/MB synapses, or in a longer range signaling process as most neuropeptides have multiple functions and are likely to also act as hormones (Nässel and Winther, 2010).
In conclusion, the observations reported here probably reflect tight anatomical and functional links between DPM neurons and MB lobes. In addition to its physiological role in memory, AMN is involved in the development of neuronal structures required for memory processes. Added to the fact that when overexpressed, AMN can act on a nearby structure, these features probably explain why areas of darkness have persisted for so long regarding the function of AMN. However, AMN function in adult DPM neurons remains to be determined.
Footnotes
This work was supported by the French national agency for research (Agence Nationale de la Recherche; ANR-15-CE32-0008-01, Memomap) and the French foundation for medical research (Fondation pour la Recherche Médicale; DEQ20140329540) to T.P. We thank Aurélie Lampin-Saint-Amaux, Lisa Scheunemann, and Yann Dromard for help with the immunohistology experiments, Alice Pavlowsky and Julia Minatchy for help with the behavioral experiments, Pierre-Yves Plaçais for characterization of some tub-Gal80ts GAL4-lines and statistical expertise, and all the members of our laboratory and Nicolas Gervasi (Inserm/UPMC, Institut du Fer à Moulin) for valuable discursive input and for critically reading the paper.
The authors declare no competing financial interests.
References
- Aldrich BT, Kasuya J, Faron M, Ishimoto H, Kitamoto T (2010) The amnesiac gene is involved in the regulation of thermal nociception in Drosophila melanogaster. J Neurogenet 24:33–41. 10.3109/01677060903419751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura A. (1998) Perspectives on pituitary adenylate cyclase activating polypeptide (PACAP) in the neuroendocrine, endocrine, and nervous systems. Jpn J Physiol 48:301–331. 10.2170/jjphysiol.48.301 [DOI] [PubMed] [Google Scholar]
- Aso Y, Grübel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H (2009) The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet 23:156–172. 10.1080/01677060802471718 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Lakhman SS, Singh S (2004) Modulation of L-type calcium channels in Drosophila via a pituitary adenylyl cyclase-activating polypeptide (PACAP)-mediated pathway. J Biol Chem 279:37291–37297. 10.1074/jbc.M403819200 [DOI] [PubMed] [Google Scholar]
- Bouzaiane E, Trannoy S, Scheunemann L, Plaçais PY, Preat T (2015) Two independent mushroom body output circuits retrieve the six discrete components of Drosophila aversive memory. Cell Rep 11:1280–1292. 10.1016/j.celrep.2015.04.044 [DOI] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C (2009) The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci 29:1948–1961. 10.1523/JNEUROSCI.4830-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Llobet N, Vidal-Sancho L, Masana M, Fournier A, Alberch J, Vaudry D, Xifró X (2018) Pituitary adenylate cyclase-activating polypeptide (PACAP) enhances hippocampal synaptic plasticity and improves memory performance in Huntington's disease. Mol Neurobiol 55:8263–8277. 10.1007/s12035-018-0972-5 [DOI] [PubMed] [Google Scholar]
- Chen G, Li W, Zhang QS, Regulski M, Sinha N, Barditch J, Tully T, Krainer AR, Zhang MQ, Dubnau J (2008) Identification of synaptic targets of Drosophila pumilio. PLoS Comput Biol 4:e1000026. 10.1371/journal.pcbi.1000026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39:715–720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL (1998) Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem 5:38–51. [PMC free article] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M (1994) Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science 263:692–695. 10.1126/science.8303280 [DOI] [PubMed] [Google Scholar]
- DeZazzo J, Xia S, Christensen J, Velinzon K, Tully T (1999) Developmental expression of an amn(+) transgene rescues the mutant memory defect of amnesiac adults. J Neurosci 19:8740–8746. 10.1523/JNEUROSCI.19-20-08740.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Quinn WG (1995) A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science 268:869–873. 10.1126/science.7754370 [DOI] [PubMed] [Google Scholar]
- Gervasi N, Tchénio P, Preat T (2010) PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron 65:516–529. 10.1016/j.neuron.2010.01.014 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Shintani N, Baba A (2002) Higher brain functions of PACAP and a homologous Drosophila memory gene amnesiac: insights from knockouts and mutants. Biochem Biophys Res Commun 297:427–431. 10.1016/S0006-291X(02)02144-7 [DOI] [PubMed] [Google Scholar]
- Isabel G, Pascual A, Preat T (2004) Exclusive consolidated memory phases in Drosophila. Science 304:1024–1027. 10.1126/science.1094932 [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, et al. (2012) A GAL4-driver line resource for Drosophila neurobiology. Cell Rep 2:991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Stratmann M, Keller A, Perrat PN, Vosshall LB, Waddell S (2004) Diverse odor-conditioned memories require uniquely timed dorsal paired medial neuron output. Neuron 44:521–533. 10.1016/j.neuron.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S (2007) Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron 53:103–115. 10.1016/j.neuron.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Lin HW, Chang YH, Fu TF, Dubnau J, Hirsh J, Lee T, Chiang AS (2011) SI serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc Natl Acad Sci U S A 108:13794–13799. 10.1073/pnas.1019483108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, Reed RR (1992) The Drosophila learning and memory gene rutabaga encodes a Ca2+/calmodulin-responsive adenylyl cyclase. Cell 68:479–489. 10.1016/0092-8674(92)90185-F [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD (2011) Simple neurite tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27:2453–2454. 10.1093/bioinformatics/btr390 [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Matsumoto A, Hashimoto H, Shintani N, Baba A (2003) Impaired long-term potentiation in vivo in the dentate gyrus of pituitary adenylate cyclase-activating polypeptide (PACAP) or PACAP type 1 receptor-mutant mice. Neuroreport 14:2095–2098. 10.1097/00001756-200311140-00017 [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302:1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Merighi A, Salio C, Ferrini F, Lossi L (2011) Neuromodulatory function of neuropeptides in the normal CNS. J Chem Neuroanat 42:276–287. 10.1016/j.jchemneu.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Merzin M. (2008) Applying stereological method in radiology: volume measurement. Bachelor's thesis University of Tartu. [Google Scholar]
- Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, Culler MD, Coy DH (1989) Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun 164:567–574. 10.1016/0006-291X(89)91757-9 [DOI] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U (1998) Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell 93:997–1007. 10.1016/S0092-8674(00)81205-2 [DOI] [PubMed] [Google Scholar]
- Nässel DR, Winther AM (2010) Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol 92:42–104. 10.1016/j.pneurobio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Pascual A, Préat T (2001) Localization of long-term memory within the Drosophila mushroom body. Science 294:1115–1117. 10.1126/science.1064200 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Anezaki M, Skoulakis EMC (2008) Neuralized is expressed in the alpha/beta lobes of adult Drosophila mushroom bodies and facilitates olfactory long-term memory formation. Proc Natl Acad Sci U S A 105:14674–14679. 10.1073/pnas.0801605105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaçais PY, de Tredern É, Scheunemann L, Trannoy S, Goguel V, Han KA, Isabel G, Preat T (2017) Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nat Commun 8:15510. 10.1038/ncomms15510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn WG, Dudai Y (1976) Memory phases in Drosophila. Nature 262:576–577. 10.1038/262576a0 [DOI] [PubMed] [Google Scholar]
- Quinn WG, Sziber PP, Booker R (1979) The Drosophila memory mutant amnesiac. Nature 277:212–214. 10.1038/277212a0 [DOI] [PubMed] [Google Scholar]
- Rodan AR, Kiger JA Jr, Heberlein U (2002) Functional dissection of neuroanatomical loci regulating ethanol sensitivity in Drosophila. J Neurosci 22:9490–9501. 10.1523/JNEUROSCI.22-21-09490.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B, Lorenzini CA, Baldi E, Bucherelli C, Roberto M, Tassoni G, Brunelli M (2001) Pituitary adenylate cyclase-activating polypeptide hormone (PACAP) at very low dosages improves memory in the rat. Neurobiol Learn Mem 76:1–6. 10.1006/nlme.2001.4014 [DOI] [PubMed] [Google Scholar]
- Schmidt SD, Myskiw JC, Furini CR, Schmidt BE, Cavalcante LE, Izquierdo I (2015) PACAP modulates the consolidation and extinction of the contextual fear conditioning through NMDA receptors. Neurobiol Learn Mem 118:120–124. 10.1016/j.nlm.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Shen S, Gehlert DR, Collier DA (2013) PACAP and PAC1 receptor in brain development and behavior. Neuropeptides 47:421–430. 10.1016/j.npep.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Tomchik SM, Davis RL (2009) Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron 64:510–521. 10.1016/j.neuron.2009.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG (1985) Classical-conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 157:263–277. 10.1007/BF01350033 [DOI] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M (1994) Genetic dissection of consolidated memory in Drosophila. Cell 79:35–47. 10.1016/0092-8674(94)90398-0 [DOI] [PubMed] [Google Scholar]
- Turrel O, Lampin-Saint-Amaux A, Préat T, Goguel V (2016) Drosophila neprilysins are involved in middle-term and long-term memory. J Neurosci 36:9535–9546. 10.1523/JNEUROSCI.3730-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry D, Gonzalez BJ, Basille M, Yon L, Fournier A, Vaudry H (2000) Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol Rev 52:269–324. [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG (2000) The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell 103:805–813. 10.1016/S0092-8674(00)00183-5 [DOI] [PubMed] [Google Scholar]
- Wu CL, Shih MF, Lai JS, Yang HT, Turner GC, Chen L, Chiang AS (2011) Heterotypic gap junctions between two neurons in the Drosophila brain are critical for memory. Curr Biol 21:848–854. 10.1016/j.cub.2011.02.041 [DOI] [PubMed] [Google Scholar]
- Wu CL, Shih MF, Lee PT, Chiang AS (2013) An octopamine-mushroom body circuit modulates the formation of anesthesia-resistant memory in Drosophila. Curr Biol 23:2346–2354. 10.1016/j.cub.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Yu DB, Akalal DB, Davis RL (2006) Drosophila α/β mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron 52:845–855. 10.1016/j.neuron.2006.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]