Abstract
PDGFRα, specifically expressed by immature oligodendrocyte progenitor cells (OPCs) in the CNS, plays a critical role in OPC proliferation and migration. However, it has been uncertain whether all cells of oligodendrocyte lineage are derived from the PDGFRα-expressing OPCs. In the present study, we uncovered a PDGFRα-independent oligodendrocyte lineage in the developing cortex. This OPC subpopulation originates from the local ventricular/subventricular zone after birth and contributes to the earliest mature oligodendrocytes in the cortex. PDGFRα signaling does not regulate the generation and differentiation of cortical OPCs. Fate-mapping studies in the PDGFRαCreER; Sox10-GFP/tdTom double-transgenic mice of either sex have further corroborated the PDGFRα-independent oligodendrocyte lineage. This study provides additional missing genetic evidence for PDGFRα-independent oligodendrocyte lineage in the developing hindbrain.
SIGNIFICANCE STATEMENT This is the first report of a subpopulation of oligodendrocyte lineage in the developing mouse cortex independent of PDGFRα signaling. These oligodendrocyte progenitor cells are generated from the local ventral ventricular zone/subventricular zone after birth, and contribute to the earliest mature oligodendrocytes in the cortex.
Keywords: analysis, gene expression, lineage, oligodendrocytes, PDGFRα, transgenic mice
Introduction
Myelin sheaths are critical structures that enable rapid conduction of nerve impulses and brain functioning. In the CNS, myelin sheaths are elaborated by oligodendrocytes (OLs) that differentiate from immature oligodendrocyte progenitor cells (OPCs). During development, early OPCs are generated from the ventral ventricular zone (VZ) under the influence of Sonic hedgehog (SHH) morphogen, although a subpopulation of OPCs can also be produced from the dorsal neural progenitor cells at later stages independent of SHH signaling (Cai et al., 2005; Fogarty et al., 2005; Vallstedt et al., 2005). The newly generated OPCs undergo rapid proliferation and migration when they spread into the surrounding parenchymal regions. In the developing forebrain, there are three waves of OPC production in a ventral-to-dorsal sequence (Kessaris et al., 2006). OPCs are first produced from the VZ of the (MGE)-(AEP) area at embryonic day (E) 12.5, then from the VZ of the (LGE)-(CGE) at ∼E15, and finally from the VZ of the cortex after birth. Cell fate-tracing studies have demonstrated that the dorsal cortical OPCs gradually replace the ventrally derived OPCs at later postnatal stages (Kessaris et al., 2006; Richardson et al., 2006). The multiorigin feature of brain OPCs suggests the possible heterogeneity of this glial population (Richardson et al., 2006).
PDGFRα receptor tyrosine kinase is one of the earliest OPC markers starting at ∼E12.5 in the mouse spinal cord and forebrain (Pringle et al., 1992; Pringle and Richardson, 1993; Miller et al., 1999). Upon stimulation by PDGF-A, PDGFRα undergoes autophosphorylation on tyrosine residues and phosphorylates the downstream molecules to deliver and amplify downstream signals (Sultzman et al., 1991). Astrocytes and neurons in the mouse CNS secrete PDGF-A, which functions as the major in vivo mitogen for OPC proliferation and migration (Yeh et al., 1991; Fruttiger et al., 1999). Overexpression of PDGF-A in the embryonic mouse spinal cord leads to a dramatic increase in the number of OPCs (van Heyningen et al., 2001). Recently, we reported that PDGFRα signaling also regulates the timing of OL differentiation and maturation downstream of Nkx2.2 transcription factor (Fu et al., 2002; Zhu et al., 2014). These and other studies indicate that PDGFRα signaling plays a critical role in OPC proliferation, migration, and differentiation.
Interestingly, previous studies have reported a small population of OLs that are PDGFRα-negative but PLP/DM-20-positive in the hindbrain and olfactory bulb during early glial development (Timsit et al., 1995; Spassky et al., 2000, 2001), suggesting the possibility of a separate OL lineage independent of PDGFRα signaling. However, the evidence for the PDGFRα lineage in these studies is indirect and mostly based on gene-expression analyses and pharmacological assays (Spassky et al., 1998, 2001). Given that PLP is normally considered a major myelin protein expressed in mature OLs, an alternative explanation proposes that the PLP/DM-20+ OLs observed in the embryonic hindbrain may represent the mature OLs that have originated from the PDGFRα+ OPCs but have lost the receptor expression during differentiation stage (Richardson et al., 2006). Thus, it remains uncertain whether all cells of OL lineage are derived from PDGFRα-expressing OPCs during development.
In the present study, we report the identification of a separate PDGFRα-independent OPC population in the developing cerebral cortex. In Olig1+/crePDGFRαflox/flox conditional knock-out (cko) mice, the generation of early OPCs from the ventral forebrain is largely inhibited at embryonic stages, but a small number of Olig1+/Sox10+ OPCs are still produced in the dorsal telencephalon after birth. Despite their relatively late birthdate during development, the dorsally derived PDGFRα-independent OPCs differentiate earlier than the ventrally derived PDGFRα-dependent OPCs, and their maturation is not affected by PDGFRα mutation. The concept of PDGFRα-independent OPC lineages in the cerebral cortex and hindbrain is further verified by the Cre/LoxP-mediated fate-mapping studies.
Materials and Methods
Knock-out and transgenic mice.
PDGFRαflox mice were obtained from Jackson Laboratories (Tallquist and Soriano, 2003) and mated to Olig1+/cre knock-in mice (Lu et al., 2002) to produce the Olig1+/cre; PDGFRαfl/+ double-heterozygous mice. Conditional PDGFRα mutants (cko) were generated by interbreeding of double heterozygotes. The PDGFRα cko mice die at ∼2 weeks after birth and display obvious characteristics of hypomyelination, such as trembling and shaking. PDGFRαcre-ER mice obtained from Jackson Laboratories (De Biase et al., 2011) were mated to Sox10-GFP/tdTom transgenic line mice (Tripathi et al., 2011) to obtain the PDGFRαcre-ER; Sox10-GFP/tdTom double-hemizygous mice of either sex. Tamoxifen was administered from E11.5 to postnatal day (P) 0 by intraperitoneal injection in pregnant female mice. When Cre is activated by tamoxifen in OPCs, the GFP gene is excised out and tdTomato protein is subsequently expressed.
In situ RNA hybridization and immunofluorescent staining.
Brain tissues were isolated from E15.5–P15 mouse embryos and then fixed in 4% paraformaldehyde at 4°C overnight. Following fixation, tissues were transferred to 20% sucrose in PBS at 4°C overnight, embedded in optimum cutting temperature media, and then sectioned [18 μm thickness for in situ hybridization (ISH) and 14 μm thickness for GFP and tdTomato red observations] on a cryostat. Adjacent sections from the control and mutant embryos were subjected to ISH or immunofluorescent staining. Regular ISH was performed as described in Schaeren-Wiemers and Gerfin-Moser (1993) with minor modifications. Double immunofluorescent procedures were described previously (Qi et al., 2001). The dilution ratio of antibodies is as follows: anti-Olig2, 1:6000; anti-CRE (Millipore), 1:500; anti-Sox10, 1:3000 (Stolt et al., 2002); and anti-PDGFRα (Santa Cruz Biotechnology), 1:200.
Experimental design and statistical analyses.
For each analysis, OPCs or OLs detected by immunostaining or ISH were counted from three independent mouse tissues. Statistical analyses were performed with two-tailed Student's t test. Error bars represented the SDs. Statistical significance was considered to be at *p < 0.05 and **p < 0.01 (ns: no significant difference). The exact p values are listed in the figure legends.
Results
PDGFRα is essential for the generation and migration of ventral OPCs in the forebrain
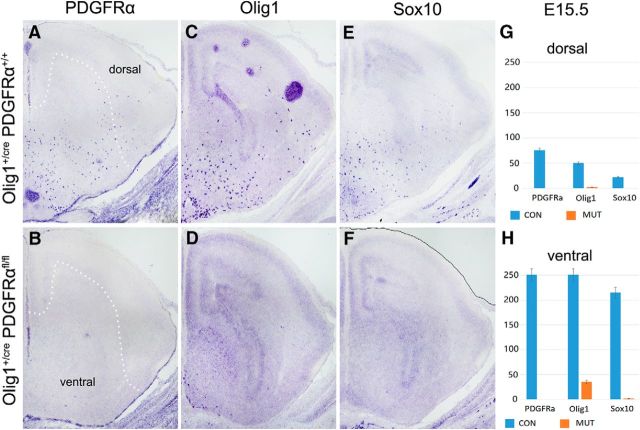
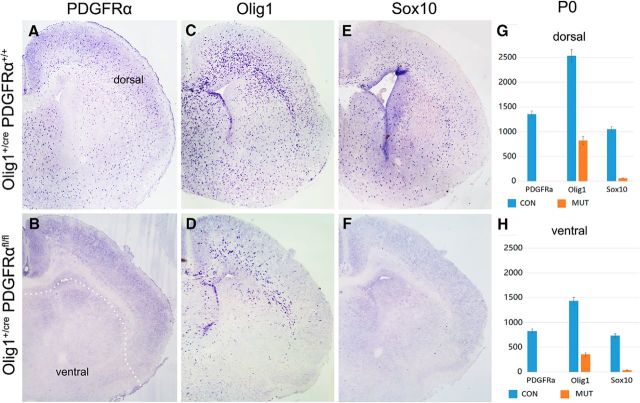
Previous studies revealed that early OPCs, defined by the expression of PDGFRα, emerge from the Olig2+ ventral neuroepithelium in the MGE and AEP after E12 in mouse embryos (Richardson et al., 2006). As a first step to investigate the role of PDGFRα signaling in subpallial oligodendrogenesis, we compared the expression of several OPC markers in embryonic control and Olig1+/crePDGFRαflox/flox cko mice (PDGFRα cko). At E15.5, numerous PDGFRα+/Olig1+/Sox10+ OPCs were produced in the ventral forebrain of the wild-type embryos, and some of these ventral OPCs started to migrate into the dorsal cortical region (Fig. 1A,C,E). Consistent with the observation that Olig1 is expressed slightly earlier than PDGFRα in the ventral telencephalon, including the MGE and AEP (Lu et al., 2002), PDGFRα expression was lost in the PDGFRα cko (Fig. 1B). Expression of another OPC marker, Sox10, was also absent in the mutants at this stage (Fig. 1D), although a few Olig1+ cells were detected along the ventral midline of the hypothalamus (Fig. 1F). At P0, expression of PDGFRα, Sox10, and Olig1 in wild-type mice was significantly increased and widely distributed throughout the entire telencephalon, including the cortex (Fig. 2A,C,E). By contrast, only a few PDGFRα+ and Sox10+ OPCs were detected in the ventral region of the mutants, but none in the cortex (Fig. 2B,F). Thus, in the absence of PDGFRα signaling, few OPCs were generated from the subpallium, and they failed to multiply and spread a long distance to reach the neocortex, indicating that PDGFRα signaling is crucial for the proliferation and migration of ventrally derived OPCs in the telencephalon. Surprisingly, many Olig1-positive cells of the mutants emerged at this stage in the dorsal forebrain, including the corpus callosum and the septal and striatal regions surrounding the lateral ventricles (Fig. 2D). Given the lack of contribution from migratory ventral OPCs, these dorsal OPCs must have originated from the local VZ/subventricular zone (SVZ) of the cortex.
Figure 1.
Inhibition of OPC generation and migration in the ventral forebrain of PDGFRα cko mutants. A–F, E15.5 forebrain tissues from control (A, C, E) and Olig1crePDGFRαflox/flox cko (B, D, F) mice were examined for expression of PDGFRα, Olig1, and Sox10 by RNA ISH. G, H, Statistical analyses of PDGFRα+, Olig1+, and Sox10+ OPCs in the dorsal and ventral forebrain. OPCs were produced in the ventral forebrain and migrated dorsally in the control but not cko tissues. White dot line indicates the boundary of dorsal and ventral forebrain.
Figure 2.
Local generation of OPCs in the dorsal forebrain of PDGFRα mutants. A–F, P0 forebrain tissues from control and PDGFRα cko mice were examined for PDGFRα, Olig1, and Sox10 by RNA ISH. G, H, Statistical analyses of various OPCs in the dorsal and ventral forebrain. While Olig1 expression in the mutants was mostly detected in the SVZ surrounding the lateral ventricle, expression of Sox10 was nearly undetectable at this stage. White dot line indicates the boundary of dorsal and ventral forebrain.
PDGFRα is not required for generation and differentiation of dorsally derived cortical OPCs
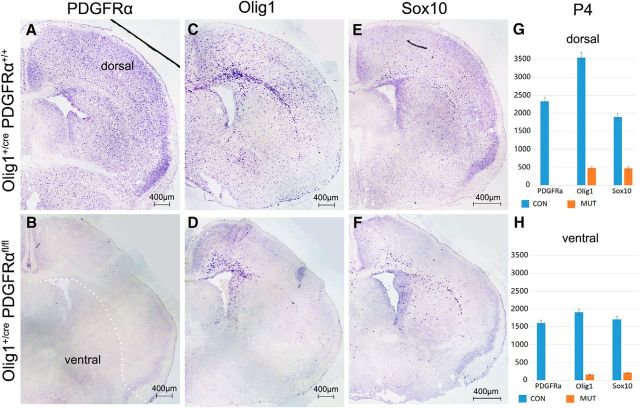
To investigate the importance of PDGFRα signaling in the development of dorsally derived OPCs, we next examined the expression of several well defined OPC and OL markers in the postnatal forebrain of PDGFRα cko mutants. While the expression of PDGFRα remained absent in the mutants at P4 as expected (Fig. 3A,B), Olig1+ OPCs in the mutant cortex were mostly confined to the regions adjacent to the ventricle (Fig. 3C,D). Moreover, Sox10+ cells were also detected with a distribution pattern similar to that of Olig1+ cells (Fig. 3E,F), suggesting that the dorsal OPCs had acquired Sox10 expression by this stage.
Figure 3.
Dorsal distribution of OPCs in P4 PDGFRα mutant brain. A–F, P4 forebrain tissues from control and PDGFRα cko mice were examined for expression of OPC markers. G, H, Statistical analyses of various OPCs in the dorsal and ventral forebrain. At this stage, Olig1+ and Sox10+ cells in the mutants were mostly confined to the dorsal cortex adjacent to the lateral ventricle. White dot line indicates the boundary of dorsal and ventral forebrain.
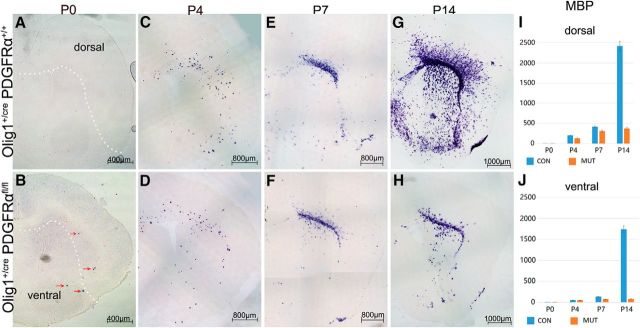
The terminal differentiation of dorsally derived OPCs was analyzed in PDGFRα cko mice by the expression of myelin basic protein (MBP) gene. At P0, expression of MBP was not detected throughout the entire forebrain in normal tissues (Fig. 4A). Similarly, no MBP expression was detected in the neocortex of the PDGFRα cko mutants. However, a few were found in the ventral forebrain (Fig. 4B, arrows), resulting from the premature differentiation of ventrally derived OPCs in the absence of PDGFRα signaling, as previously reported in the spinal cord tissues (Zhu et al., 2014). From P4 to P7, many MBP+ OLs were found in the ventral forebrain of the control tissues, but very few in that of the mutants (Fig. 4C–F) due to the reduced proliferation of ventral OPCs and their premature differentiation when PDGFRα was conditionally deleted. Surprisingly, MBP+ cells were observed in the corpus callosum and striatum in both control and mutant tissues with a similar density and distribution pattern (Fig. 4C–F), suggesting that early MBP+ OLs in these regions are likely to originate from dorsal OPCs, and their differentiation is not regulated by PDGFRα signaling. At P14 stage, MBP+ cells in the control cortical tissues were further increased in number and widely distributed throughout the entire forebrain as the ventrally derived OPCs underwent terminal differentiation in the cortex (Fig. 4G,H). By contrast, the density and distribution of MBP+ cells in the mutant cortex and striatum remained unchanged over time, because of the lack of contribution from ventrally derived OPCs. Together, these studies suggest that dorsal cortical OPCs that arise in the absence of PDGFRα mature earlier than their ventral counterparts despite their later birth date and their generation and differentiation are not regulated by PDGFRα signaling.
Figure 4.
Expression of MBP in the cortex at early postnatal stages. A–H, Cortical tissues from P0–P14 control and PDGFRα cko mice are examined for MBP expression by ISH. I, J, Statistical analyses of OPCs in the dorsal and ventral forebrain. Red arrows in B indicated fewer MBP+ cells in the mutant forebrain. A similar number and distribution of MBP+ oligodendrocytes in the dorsal cortex were observed between control and mutant tissues from P0–P7 mice. White dot line indicates the boundary of dorsal and ventral forebrain.
Genetic evidence for PDGFRα-independent lineage in the cortex
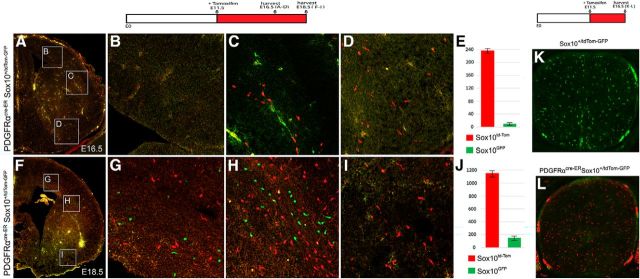
To further test the hypothesis of the PDGFRα-independent OPC lineage in the cerebral cortex, we generated the PDGFRαCre-ER; Sox10-GFP/tdTOM double-transgenic mice (Tripathi et al., 2011) in which all Sox10+ OPCs that ever express PDGFRα are permanently labeled for Tomato red, whereas those that never express PDGFRα remain GFP green. To ensure that all PDGFRα-expressing OPCs are labeled successfully, pregnant mice were injected daily with tamoxifen starting at E11.5 before PDGFRα expression in brain OPCs (Spassky et al., 2001; Richardson et al., 2006). Double-labeling studies revealed that the vast majority of Sox10-positive OPCs in E16.5 embryos were Tomato red+ (Fig. 5A,B,D), as expected. Consistent with our hypothesis for a PDGFRα -independent lineage, a small fraction of Sox10+ OPCs appeared as GFP+ green cells at the boundary region between the cortex and striatum (Fig. 5C,E). At E18.5, the number of GFP+ OPCs was significantly higher in the dorsal forebrain, including the corpus callosum (Fig. 5F–J). Unlike the PDGFRα-expressing Tomato+ OPCs, which were widely distributed in the forebrain, the GFP+ OPCs were largely confined to the dorsal cortical tissues, and careful examination revealed no GFP+ green cells in the most ventral region (Fig. 5I). To evaluate the efficiency and specificity of Cre induction, we also examined the percentage of GFP+ OPCs in E16.5 spinal cords and found that all OPCs were tdTom+ red cells (Fig. 5K,L), indicating that the Cre-mediated recombination in OPCs is highly efficient and reliable. Together, these results provide additional evidence for the dorsal origin of this PDGFRα-independent OPC subpopulation.
Figure 5.
A–J, Expression of GFP and tdTomato in the PDGFRα-cre-ER; Sox10-GFP/tdTOM double-transgenic embryos. Embryos were treated with tamoxifen from E11.5 to E16.5 (A–E) or from E11.5 to E18.5 (F–J). A, GFP+ OPCs were observed in dorsal cortex at E16.5. B–D, Higher magnifications of three regions outlined in A. E, Statistical analysis of tdTomato+ OPCs (average, 238/section) and GFP+ OPCs (average, nine/section) in the cortex (p = 0.0004608). F, GFP+ OPCs were observed in the dorsal cortex at E18.5. G–I, Higher magnifications of regions outlined in F. J, Statistical analysis of tdTomato+ OPCs (average, 1151/section) and GFP+ OPCs (average, 172/section) in the cortex (p = 0.0000661). K, L, Expression of GFP and tdTomato in E16.5 control (K) and DTG (L) embryos following tamoxifen treatment at E11.5.
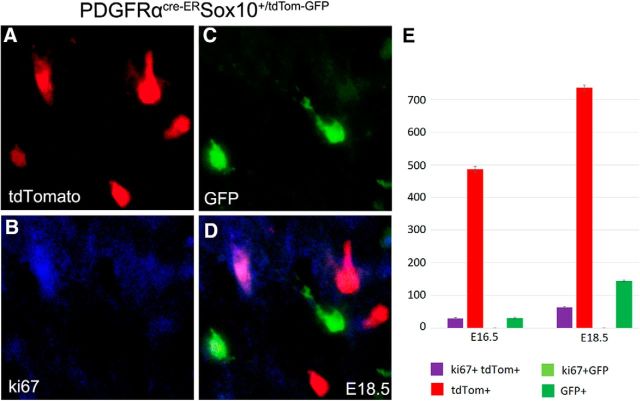
We next examined whether these PDGFRα-independent cells can proliferate in the forebrain of PDGFRαCreER;Sox10-GFP/tdTom double transgenic mice at E16.5 and E18.5. It was found that the GFP+ cells did not express Ki67, a marker for proliferating cells, and the tdTom+ cells also showed weak proliferative capacity (Fig. 6). These results suggest that PDGFRα-independent cells do not proliferate after expressing Sox10.
Figure 6.
Expression of Ki-67 in GFP+ and tdTom+ cells in the forebrain of PDGFRαCreER;Sox10-GFP/tdTom embryos at E16.5 and E18.5. A–D, Representative images of colabeling from E18.5 double transgenic mouse embryos. E, Statistical analysis showing the lack of Ki67 expression in GFP+ cells and a low level of coexpression in tdTom+ cells.
PDGFRα-independent OPCs in the hindbrain
Previous studies suggested that in the developing hindbrain, one type of OL expresses PDGFRα and another expresses PLP/DM-20 (Timsit et al., 1995; Peyron et al., 1997; Fruttiger et al., 1999). They were hypothesized to represent two separate populations of OLs with distinct origins (Spassky et al., 2000). However, the evidence was mostly indirect and based on gene-expression analyses, and it was argued that the PLP/DM-20+ OLs are early-differentiated cells derived from PDGFRα-expressing OPCs (Richardson et al., 2000). This controversy had been difficult to resolve due to the lack of appropriate mouse lines for genetic and fate-mapping studies.
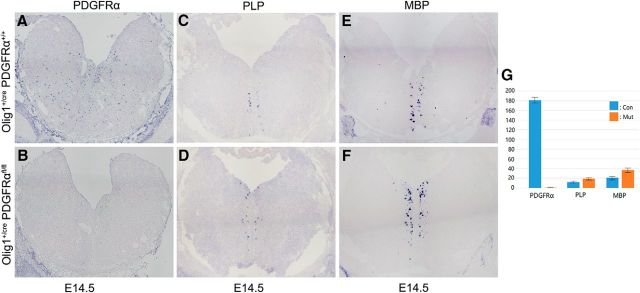
To reinvestigate this issue, we recently examined the development of OLs in the embryonic hindbrain of PDGFRα cko mutants. As expected, PDGFRα expression was absent in the hindbrain of the E14.5 mutants (Fig. 7A,B), confirming the complete inactivation of PDGFRα gene in OPC cells. In agreement with the previous finding, PLP/DM20+ OLs were discovered in the ventral midline region with a similar number and distribution pattern in both genotypes (Fig. 7C,D). Moreover, MBP+ cells were also found at the same position on immediately adjacent slides (Fig. 7E,F), indicating that the PLP+ and MBP+ cells represent mature OLs whose generation and differentiation are independent of PDGFRα signaling. These findings also strongly suggest that the early PLP+/MBP+ OLs observed in the E14.5 hindbrain originate from the PDGFRα-independent OL lineage.
Figure 7.
Expression of PLP/DM20 and MBP in the hindbrain of PDGFRα cko mutants. A–F, E14.5 hindbrain tissues from control and PDGFRα cko mice were examined for expression of PDGFRα (A, B), PLP (C, D), and MBP (E, F) by ISH. G, Statistical analysis showing the lack of PDGFRα+ OPCs and the slightly increased number of MBP+/PLP + oligodendrocytes in the mutant hindbrain. PPDGFRα = 0.000509, PMBP = 0.0261483, PPLP = 0.010093.
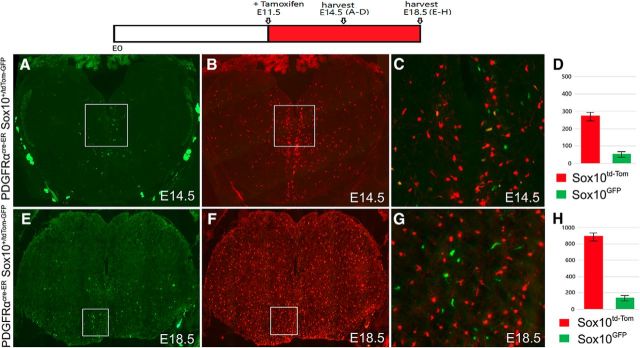
The PDGFRα-independent OL lineage in the hindbrain was further corroborated by the lineage study with the PDGFRαCreER; Sox10-GFP/tdTOM double-transgenic mice as described earlier. Direct immunofluorescence demonstrated that ∼20% of OLs in the E14.5–E18.5 hindbrain were green cells that never expressed PDGFRαCreER, whereas the other 80% Sox10-derived OLs were tdTomato red after PDGFRα-mediated activation of tdTomato transcription (Fig. 8). Similar to PLP+/MBP+ cells, the GFP+ green cells predominantly localized in the ventromedial region of the hindbrain (Fig. 8). Collectively, these results provide strong support for the existence of the PDGFRα-independent population of OLs in the caudal brain region as well.
Figure 8.
Detection of GFP+ cells in the hindbrain. PDGFRαcre-ER Sox10-GFP/tdTOM double-transgenic embryos were treated with tamoxifen from E11.5 to E14.5 (A–D) or from E11.5 to E18.5 (E–H). The numbers of GFP+ oligodendrocytes (PDGFRα-independent) and Tomato+ oligodendrocytes (PDGFRα-dependent) in the hindbrain at E14.5 (average of 61 green cells vs 283 red cells per section, p = 0.0003818) and E18.5 (171 green vs 915 red, p = 0.0000124) are depicted in D and H, respectively. (C) Higher magnifications of regions outlined in A and B, and (G) Higher magnifications of regions outlined in E and F respectively.
Discussion
PDGFRα is a widely used molecular marker for progenitor cells of OL lineage. During development, it is exclusively expressed in immature OPCs in the developing CNS and functions to stimulate OPC proliferation and migration. Recently, we showed that PDGFRα and its downstream effector Shp2 also negatively regulate the terminal differentiation of OLs in the spinal cord and therefore the timing of OL differentiation (Zhu et al., 2010, 2014). In this study, we demonstrate that PDGFRα signaling also regulates the proliferation and differentiation of OPCs derived from the ventral forebrain (Figs. 1, 2). In Olig1+/crePDGFRαflox/flox cko mice, a few precocious MBP+ OLs were detected in the newborn ventral forebrain ∼4 d earlier than normal (Fig. 4B). However, the number of MBP+ cells in the ventral region did not increase significantly with time in the mutants (Fig. 4D,F), probably due to the reduced OPC proliferation and premature differentiation in the absence of PDGFRα signaling. Together, these findings indicate that PDGFRα plays important roles in regulating the proliferation, migration, and terminal differentiation of ventrally derived OPCs in the developing forebrain.
The lack of contribution of migratory OPCs from the ventral forebrain in PDGFRα cko mice helped uncover the PDGFRα-independent OPC population generated in the dorsal cortex. Olig1+ OPCs began to emerge in the mutant cortical region surrounding the lateral ventricle at birth (Figs. 1, 2), and later gained the expression of the critical differentiation factor Sox10 (Fig. 3). MBP+ mature OLs were initially detected in the corpus callosum and striatum in both control and mutant mice, and there was no apparent difference in their numbers and distribution patterns in both genotypes from P4 to P7 (Fig. 4). These observations imply that the earliest group of mature OLs in the cortex originate from the dorsally or locally derived OPCs. This implication is somewhat surprising, given that ventral OPCs from the MGE and LGE migrate into the cortex several days before dorsal cortical OPCs are born (Kessaris et al., 2006).
The relatively normal generation and differentiation of early cortical OPCs in PDGFRα mutants also argue for a PDGFRα-independent OPC lineage arising from the local VZ/SVZ at neonatal stages. Considering that PDGFRα signaling promotes OPC proliferation and migration, the PDGFRα-independent OPCs do not appear to proliferate as they fail to costain Ki67 (Fig. 6) and are likely to have limited capacity to migrate. This may explain the lack of visible increase in the number of MBP+ OLs from P7 to P14 in PDGFRα cko mutants (Fig. 4) and the confinement of early MBP+ OLs to the corpus callosum and striatum adjacent to the VZ/SVZ in both normal mice and mutants. In the normal tissues, MBP+ OLs increased dramatically after P7 due to the continuous proliferation and maturation of PDGFRα-dependent OLs derived from the ventral forebrain and possibly from the cortex as well at later postnatal stages.
The existence of PDGFRα-independent lineage in the forebrain was further tested by the fate-mapping study with the PDGFRαCreER; Sox10-GFP/tdTOM double-transgenic mice. The Sox10+ green OPCs that have never expressed PDGFRα first appeared in dorsal cortex at ∼E16.5, and the number of this subpopulation was significantly increased at E18.5 (Fig. 5). Based on the restricted location in the dorsal region of the forebrain, these PDGFRα-independent OPCs are likely to have originated from the dorsal and lateral VZ/SVZ neural progenitor cells during the third wave of oligodendrogenesis in the mouse forebrain (Kessaris et al., 2006). Nevertheless, at this stage, we cannot exclude the possibility that the GFP green cells in the double transgenic mouse forebrain may represent a small population of unrecombined cells. However, this alternative appears unlikely, as the efficiency of PDGFRα-cre-ER-mediated recombination is extremely high, and all the Sox10+ cells in E16.5 double transgenic mouse spinal cord tissues were tdTom red cells and no green cells were found (Fig. 5K,L).
Previous studies have suggested that one type of OPCs precursor cells in the developing hindbrain expresses PDGFRα and another expresses PLP/DM-20 (Timsit et al., 1995; Spassky et al., 1998, 2000). However, there is uncertainty about the origin and identity of these PLP/DM-20+-lineage cells because of the alternative explanation that they may represent the early-differentiated PDGFRα+ OPCs (Richardson et al., 2000; Kessaris et al., 2006). Our studies found that the PLP+ and MBP+ OLs are indeed present in the E14.5 hindbrain, despite the absence of PDGFRα+ OPCs in the PDGFRα cko mutants (Fig. 7). These PLP+/MBP+ cells are in the ventromedial regions of hindbrain adjacent to the VZ that produces OPCs. Moreover, even at E12.5 stage, MBP/PLP+ OLs are found in the ventral VZ/SVZ before they emigrate out (data not shown). These observations provide support for the existence of a PDGFRα-independent OL lineage in the hindbrain that differentiates early during development, similar to the scenario in the forebrain. The direct evidence for the PDGFRα-independent OL lineage in the hindbrain comes from the cell fate-tracing study with the use of the PDGFRαCreER; Sox10-GFP/tdTOM transgenic mice. It has been shown that a small number of GFP+ green cells are present in the same ventrolateral positions overlapping with those of PLP+/MBP+ OLs (Fig. 8), suggesting that they may represent the same group of cells.
In summary, our studies provide the long-sought genetic evidence that a small population of OPCs in the developing cortex and hindbrain do not express PDGFRα, and they differentiate early during development without being regulated by the PDGFRα-signaling pathway. It would be of great interest and importance to define the functional importance of these PDGFRα-independent OLs in mouse brain development in future studies.
Footnotes
This work was supported by National Natural Science Foundation of China (31771621, 31572224) and by the Zhejiang Provincial Natural Science Foundation of China (LY14C090005, LQ18C090005, LY18H090014). We thank Dr. Bill Richardson for providing the Sox10-GFP/tdTomato transgenic mouse line.
The authors declare no competing financial interests.
References
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M (2005) Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and shh signaling. Neuron 45:41–53. 10.1016/j.neuron.2004.12.028 [DOI] [PubMed] [Google Scholar]
- De Biase LM, Kang SH, Baxi EG, Fukaya M, Pucak ML, Mishina M, Calabresi PA, Bergles DE (2011) NMDA receptor signaling in oligodendrocyte progenitors is not required for oligodendrogenesis and myelination. J Neurosci 31:12650–12662. 10.1523/JNEUROSCI.2455-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Richardson WD, Kessaris N (2005) A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development 132:1951–1959. 10.1242/dev.01777 [DOI] [PubMed] [Google Scholar]
- Fruttiger M, Karlsson L, Hall AC, Abramsson A, Calver AR, Boström H, Willetts K, Bertold CH, Heath JK, Betsholtz C, Richardson WD (1999) Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development 126:457–467. [DOI] [PubMed] [Google Scholar]
- Fu H, Qi Y, Tan M, Cai J, Takebayashi H, Nakafuku M, Richardson W, Qiu M (2002) Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development 129:681–693. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD (2006) Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci 9:173–179. 10.1038/nn1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH (2002) Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell 109:75–86. 10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Miller RH, Hayes JE, Dyer KL, Sussman CR (1999) Mechanisms of oligodendrocyte commitment in the vertebrate CNS. Int J Dev Neurosci 17:753–763. [DOI] [PubMed] [Google Scholar]
- Peyron F, Timsit S, Thomas JL, Kagawa T, Ikenaka K, Zalc B (1997) In situ expression of PLP/DM-20, MBP and CNP during embryonic and postnatal development of the jimpy mutant and of transgenic mice overexpressing PLP. J Neurosci Res 50:190–201. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Richardson WD (1993) A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development 117:525–533. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Mudhar HS, Collarini EJ, Richardson WD (1992) PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development 115:535–551. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M (2001) Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128:2723–2733. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Smith HK, Sun T, Pringle NP, Hall A, Woodruff R (2000) Oligodendrocyte lineage and the motor neuron connection. Glia 29:136–142. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N (2006) Oligodendrocyte wars. Nat Rev Neurosci 7:11–18. 10.1038/nrn1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A (1993) A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100:431–440. [DOI] [PubMed] [Google Scholar]
- Spassky N, Goujet-Zalc C, Parmantier E, Olivier C, Martinez S, Ivanova A, Ikenaka K, Macklin W, Cerruti I, Zalc B, Thomas JL (1998) Multiple restricted origin of oligodendrocytes. J Neurosci 18:8331–8343. 10.1523/JNEUROSCI.18-20-08331.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Olivier C, Perez-Villegas E, Goujet-Zalc C, Martinez S, Thomas Jl, Zalc B (2000) Single or multiple oligodendroglial lineages: a controversy. Glia 29:143–148. [DOI] [PubMed] [Google Scholar]
- Spassky N, Heydon K, Mangatal A, Jankovski A, Olivier C, Queraud-Lesaux F, Goujet-Zalc C, Thomas JL, Zalc B (2001) Sonic hedgehog-dependent emergence of oligodendrocytes in the telencephalon: evidence for a source of oligodendrocytes in the olfactory bulb that is independent of PDGFRα signaling. Development 128:4993–5004. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16:165–170. 10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultzman L, Ellis C, Lin LL, Pawson T, Knopf J (1991) Platelet-derived growth factor increases the in vivo activity of phospholipase C-gamma 1 and phospholipase C-gamma 2. Mol Cell Biol 11:2018–2025. 10.1128/MCB.11.4.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P (2003) Cell autonomous requirement for PDGFRα in populations of cranial and cardiac neural crest cells. Development 130:507–518. 10.1242/dev.00241 [DOI] [PubMed] [Google Scholar]
- Timsit S, Martinez S, Allinquant B, Peyron F, Puelles L, Zalc B (1995) Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J Neurosci 15:1012–1024. 10.1523/JNEUROSCI.15-02-01012.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi RB, Clarke LE, Burzomato V, Kessaris N, Anderson PN, Attwell D, Richardson WD (2011) Dorsally and ventrally derived oligodendrocytes have similar electrical properties but myelinate preferred tracts. J Neurosci 31:6809–6819. 10.1523/JNEUROSCI.6474-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J (2005) Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron 45:55–67. 10.1016/j.neuron.2004.12.026 [DOI] [PubMed] [Google Scholar]
- van Heyningen P, Calver AR, Richardson WD (2001) Control of progenitor cell number by mitogen supply and demand. Curr Biol 11:232–241. [DOI] [PubMed] [Google Scholar]
- Yeh HJ, Ruit KG, Wang YX, Parks WC, Snider WD, Deuel TF (1991) PDGF A-chain gene is expressed by mammalian neurons during development and in maturity. Cell 64:209–216. 10.1016/0092-8674(91)90222-K [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zhao X, Zheng K, Li H, Huang H, Zhang Z, Mastracci T, Wegner M, Chen Y, Sussel L, Qiu M (2014) Genetic evidence that Nkx2.2 and PDGFRα are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development 141:548–555. 10.1242/dev.095323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Park J, Hu X, Zheng K, Li H, Cao Q, Feng GS, Qiu M (2010) Control of oligodendrocyte generation and proliferation by Shp2 protein tyrosine phosphatase. Glia 58:1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]