Abstract
Angelica dahurica is a widely grown plant species with multiple uses, especially in the medical field. However, the frequent introduction of A. dahurica to new areas has made it difficult to distinguish between varieties. Simple sequence repeats (SSRs) detected based on transcriptome analyses are very useful for constructing genetic maps and analyzing genetic diversity. They are also relevant for the molecular marker-assisted breeding of A. dahurica. We identified 33,724 genic SSR loci based on transcriptome sequencing data. A total of 114 primer pairs were designed for the SSR loci and were tested for their specificity and diversity. Ten SSR loci in untranslated regions were ultimately selected. Subsequently, 56 A. dahurica ecotypes collected from different regions were analyzed. The SSR loci comprised 2–8 alleles, with a mean of 5.2 alleles per locus. The polymorphic information content value and Shannon’s information index were 0.6274–0.2702 (average of 0.4091) and 1.3040–0.5618 (average of 0.8475), respectively. Thus, the 10 novel SSRs identified in this study were almost in accordance with Harvey-Weinberg equilibrium and will be useful for analyzing A. dahurica genetic relationships. The results of this study confirm the potential value of transcriptome databases for the development of new SSR markers.
Introduction
Angelica dahurica [1, 2], which is known as ‘Baizhi’ in China, is a perennial dicotyledonous herb of the family Apiaceae. It originated in Taiwan and is widely grown in Korea, China, Japan, and Russia [3]. A. dahurica has been used in traditional Chinese medicine [4] because of its anti-proliferative [5], anti-inflammatory [6], anti-depressive [7], anti-oxidative [8], and anti-microbial [9] effects. It has also been applied to regulate hormones [10], decrease blood glucose levels [11], and treat headaches, ulcers, toothaches, abscesses [12], and other diseases. Angelica dahurica root extracts contain furanocoumarin compounds [13], phenolic compounds, and volatile oils, all of which account for the multiple pharmacological effects of this plant species. A. dahurica ecotypes may vary in their root surface texture and color, stem color, active ingredient compositions and abundances, and other characteristics. However, it remains unknown whether these differences are the result of phenotypic plasticity or ecotypic variation. Analyses of molecular data may clarify this issue.
Angelica dahurica has a functionally diploid genome (2n = 22) [14], and is cultivated in Sichuan, Zhejiang, Henan, Hebei, Zhejiang, Anhui, and other provinces in China. Additionally, there are many wild A. dahurica populations. The abundance of available A. dahurica germplasm resources is ideal for genetic studies as the core of germplasm resource diversity is genetic diversity. The development of molecular genetic markers may be useful for clarifying the genetic variation among varieties. Eukaryotic genes are broken up by introns [15]. In the process of eukaryotic gene expression, introns are spliced out to form different mRNAs that guide protein expression and ultimately produce phenotypic diversity. Therefore, SSR markers can be divided into two types: genic markers, such as those in mRNA coding regions and untranslated regions (UTRs), and genomic markers, including markers in non-functional genes and intron regions. SSR markers are usually developed through transcriptome sequencing. Compared with genomic markers, genic markers are more practical and may be closely associated with traits because variations in the nucleotide sequences of functional genes often result in phenotypic diversity.
Simple sequence repeats (SSRs), which are also known as microsatellites, are one of the most efficient types of genetic markers because of their reproducibility, multiallelic nature, codominant inheritance, relative abundance, and high genome coverage[16]. SSR loci are generally divided into genomic SSRs and genic SSRs based on their genomic locations. Genic SSRs occur in gene coding regions, making them suitable for explaining the phenotypic and functional diversity in various populations, gene mapping, and analyses of evolution [17].
The cross-species transferability of SSRs, which is dependent on phylogenetic closeness, is greater for genic SSRs than for genomic SSRs. Advances in next-generation (i.e., high-throughput) sequencing technology have improved the efficiency and reliability of transcriptome sequencing experiments[18]. Consequently, many expressed sequence tag (EST)-SSR markers exhibiting cross-species transferability have been developed via the transcriptome sequencing of various plant species, such as Euphorbiaceae family members [19], Lolium multiflorum [20], and Saccharina japonica [21]. The frequent introduction of A. dahurica to new regions in China has made it difficult to distinguish between different populations, which is problematic for the development of A. dahurica-specific EST-SSR markers. Thus, A. dahurica-specific EST-SSR markers have not been reported. We herein present the first report of the development of genic SSR markers in A. dahurica. These markers may provide researchers with new opportunities for assessing the molecular phylogeny and genetic diversity among A. dahurica varieties.
Materials and methods
2.1 Plant materials
Our research object was A. dahurica, which is not an endangered or protected species. No specific permission was required for the materials we collected because they are ecotypes that have been planted for a long period, and the exchange of planting areas occurs frequently. We collected them with the owners’ permission. We collected 56 A. dahurica ecotypes from Sichuan, Zhejiang, Hebei, Jiangsu, Anhui, and Guizhou provinces as well as the Chongqing municipality (Table 1) for the subsequent development of genic SSR markers.
Table 1. 56 A. dahurica ecotypes collected from different regions.
| No. | Material ID | Types | Origins | No. | Material ID | Types | Origins |
|---|---|---|---|---|---|---|---|
| 1 | 1801 | cultivar | Suining city, Sichuan province, China | 29 | BZC035 | cultivar | Suining city, Sichuan province, China |
| 2 | 1802 | cultivar | Suining city, Sichuan province, China | 30 | BZC036 | cultivar | Suining city, Sichuan province, China |
| 3 | 1804 | cultivar | Ziyang city, Sichuan province, China | 31 | BZC037 | cultivar | Suining city, Sichuan province, China |
| 4 | 1805 | cultivar | Ziyang city, Sichuan province, China | 32 | BZC040 | cultivar | Dazu district, Chongqing municipality, China |
| 5 | 1806 | cultivar | Nanchong city, Sichuan province, China | 33 | BZC048 | cultivar | Suining city, Sichuan province, China |
| 6 | 1807 | cultivar | Luzhou city, Sichuan province, China | 34 | BZC052 | cultivar | Suining city, Sichuan province, China |
| 7 | 1808 | cultivar | Suining city, Sichuan province, China | 35 | BZC056 | cultivar | Suining city, Sichuan province, China |
| 8 | 1809 | cultivar | Suining city, Sichuan province, China | 36 | BZC057 | cultivar | Suining city, Sichuan province, China |
| 9 | 1810 | cultivar | Suining city, Sichuan province, China | 37 | BZC059 | cultivar | Suining city, Sichuan province, China |
| 10 | 1811 | cultivar | Suining city, Sichuan province, China | 38 | BZC060 | cultivar | Suining city, Sichuan province, China |
| 11 | 1812 | cultivar | Neijiang city, Sichuan province, China | 39 | BZC061 | cultivar | Suining city, Sichuan province, China |
| 12 | 1814 | cultivar | Dazhou city, Sichuan province, China | 40 | BZC062 | cultivar | Suining city, Sichuan province, China |
| 13 | 1815 | cultivar | Guang’an city, Sichuan province, China | 41 | BZC063 | cultivar | Suining city, Sichuan province, China |
| 14 | 1816 | cultivar | Guang’an city, Sichuan province, China | 42 | BZC064 | cultivar | Suining city, Sichuan province, China |
| 15 | 1817 | cultivar | Zunyi city, Guizhou province, China | 43 | BZC067 | cultivar | Suining city, Sichuan province, China |
| 16 | BZA001 | cultivar | Bred by Sichuan Agricultural University | 44 | BZC069 | cultivar | Dazu district, Chongqing municipality, China |
| 17 | BZA002 | cultivar | Bred by Sichuan Agricultural University | 45 | BZC070 | cultivar | The specimen garden of Sichuan Agricultural University |
| 18 | BZA003 | cultivar | Bred by Sichuan Agricultural University | 46 | BZC071 | cultivar | Suining city, Sichuan province, China |
| 19 | BZA004 | cultivar | Bred by Sichuan Agricultural University | 47 | BZC072 | cultivar | Dazu district, Chongqing municipality, China |
| 20 | BZB002 | cultivar | Bred by Sichuan Agricultural University | 48 | BZC073 | cultivar | Suining city, Sichuan province, China |
| 21 | BZB003 | cultivar | Bred by Sichuan Agricultural University | 49 | BZC074 | cultivar | Suining city, Sichuan province, China |
| 22 | BZC014 | cultivar | Suining city, Sichuan province, China | 50 | BZC079 | cultivar | Suining city, Sichuan province, China |
| 23 | BZC015 | cultivar | Suining city, Sichuan province, China | 51 | BZC080 | cultivar | Suining city, Sichuan province, China |
| 24 | BZC017 | cultivar | Nanchong city, Sichuan province, China | 52 | BZC081 | cultivar | Anhui province, China |
| 25 | BZC018 | cultivar | Anguo city, Hebei province, China | 53 | BZC082 | cultivar | Sichuan province, China |
| 26 | BZC019 | cultivar | Suining city, Sichuan province, China | 54 | BZC084 | cultivar | Jiangsu province, China |
| 27 | BZC024 | cultivar | Suining city, Sichuan province, China | 55 | BZC086 | cultivar | Hangzhou city, Zhejiang province, China |
| 28 | BZC033 | cultivar | Suining city, Sichuan province, China | 56 | BZC089 | cultivar | Anguo city, Hebei province, China |
Leaves, stems, and phloem and xylem tissues of A. dahurica cultivar ‘Chuan zhi No. 2’ were collected at the seedling and bolting stages for transcriptome sequencing. The samples were immediately frozen in liquid nitrogen and stored at −80°C for subsequent RNA extraction. Three biological replicates were prepared for each sample.
2.2 RNA extraction, sequencing, and analysis of the A. dahurica transcriptome and SSR locus identification
Total RNA was extracted with the mirVana miRNA ISOlation Kit (Thermo Fisher, Carlsbad, CA, USA). The 2100 Bioanalyzer RNA Nanochip (Agilent, Santa Clara, CA, USA) was used to verify RNA quality. All samples had an RNA Integrity Number > 7 and a 28S/18S ratio > 2. The TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) was used to purify and fragment the mRNA, after which double-stranded cDNA was synthesized with SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). Short fragments were modified with an A-tail on the 3′ end and adapters were ligated via PCR amplification. The resulting modified fragments were sequenced with the Illumina Genome Analyzer HiSeq X Ten sequencing platform. Data analyses and base calling were performed with the Illumina instrument software. The transcriptome datasets are available in the NCBI Sequence Read Archive (accession number SRX247927).
The Trinity software (version: trinityrnaseq_r20131110) [1] was used to splice and obtain transcript sequences. Additionally, the MISA software (http://pgrc.ipk-gatersleben.de/misa/) was used to detect genic SSR markers based on the de novo transcriptome sequencing data [22]. The SSR loci containing repeat units of 1–6 nucleotides were identified, and the minimum SSR length was set at six iterations. SSR primers were designed using the Primer3 software (http://pgrc.ipk-gatersleben.de/misa/) with the following parameters: primer length of 18–22 bp, with an optimum length of 20 bp; an annealing temperature of 60°C; a PCR product size of 100–280 bp; and a GC content of 40–60%, with an optimum of 50% [23].
2.3 Germplasm resource DNA extraction
The seeds of the various ecotypes were sown in pots and germinated in a growth chamber under the following conditions: 22°C, 75% relative humidity, and a 16-h light/8-h dark cycle. Leaves were collected at the seedling stage, and genomic DNA was extracted according to a modified cetyltrimethylammonium bromide protocol, with polysaccharides removed using high salt concentrations and polyphenols removed using polyvinyl pyrrolidone, an extended RNase treatment and phenol-chloroform extraction [24]. The DNA quality was assessed by 1% agarose gel electrophoresis (m/v) and OD260/OD280 with a NanoDrop 2000 spectrophotometer (Wilmington, DE, USA). The DNA quantity was assessed with a NanoDrop 2000 spectrophotometer.
2.4 Development and validation of SSR markers
A total of 114 SSR primer pairs were synthesized by Chengdu TsingKe Biotechnology Co., Ltd. (Chengdu, China). PCR amplifications were completed with the T100 Thermal Cycler (Foster City, CA, USA). The PCR program was as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s, 55–65°C for 30 s, and 72°C for 2 min; 72°C for 10 min. The specificity and ideal annealing temperature of the PCR products were determined by 1% agarose gel electrophoresis. Primers that amplified a single band of the expected size were selected.
The forward primers were labeled with HEX fluorescent probe by Chengdu TsingKe Biotechnology Co., Ltd. (Chengdu, China) to analyze fragments on the Applied Biosystems 3730xl DNA Analyzer (Carlsbad, CA, USA). For genotyping, 0.5 μL PCR amplification products in 10 µL ABI highly deionized (Hi-Di) formamide and ABI GeneScan 500 LIZ Size standard mixture (130:1) were first pre-degenerated at 95℃ for 5 min and then separated by capillary electrophoresis. Alleles were detected with the GeneMapper software (version 4.1) [25]. Markers with a strong tendency to form stutter peaks were excluded in this step. The sequences of SSR products were determined by Chengdu TsingKe Biotechnology Co., Ltd. (Chengdu, China).
2.5 Data analysis
We used the GenAlEx software (version 6.501) [26] to edit the data and transform it the codominant format automatically. The number of alleles (Na), number of effective alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), Shannon’s information index (I), and Hardy-Weinberg equilibrium (HWE) were calculated with the Popgene software (version 1.32) [27]. Additionally, the polymorphic information content (PIC) was calculated with the PIC_Calc program.
Results
3.1 Transcriptome sequencing and de novo assembly
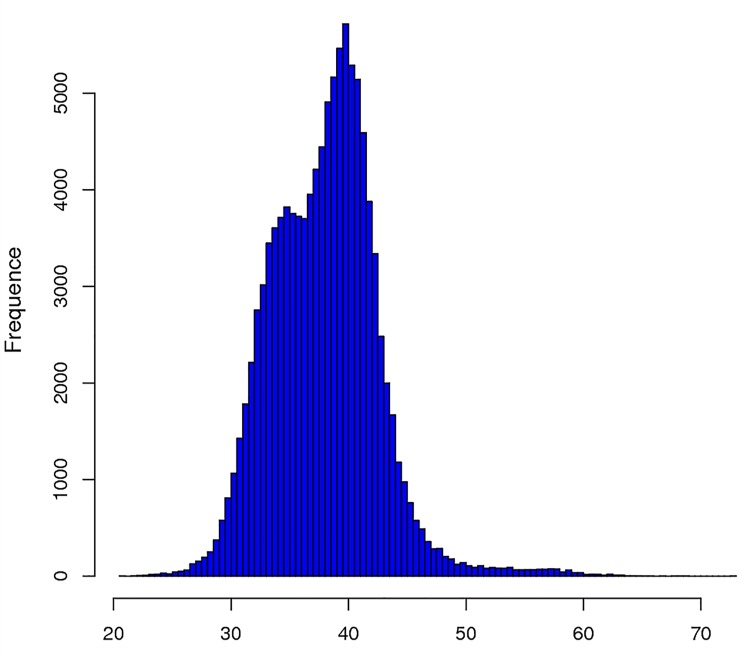
The quality of the extracted total RNA was appropriate for transcriptome sequencing (see S1 Table, S1 Fig). After a stringent quality assessment and data filtering step, 49,580,458.67 raw reads were selected for further analyses and deposited in the NCBI SRA database (PRJNA523076). An overview of the sequencing results is presented in Table 2. All high-quality reads were assembled with the Trinity software, which produced a contig N50 length of 1,858 bp and a mean length of 1,299.83 bp. After sequencing, 110,251 unigenes were identified in the A. dahurica transcriptome based on the de novo assembly of clean raw reads comprising a total length of 143,307,954 bp. The GC content of the unigenes was mostly 30–50% (Fig 1). The mean A. dahurica transcriptome GC content was 43.33%.
Table 2. Transcriptome sequencing data quality and splicing results statistics.
| Projects | Numbers | Projects | Numbers |
|---|---|---|---|
| Raw reads | 49580458.67 | Total number of Unigene >300 (bp) | 110251 |
| Raw bases | 7437068800.00 | Total number of Unigene≥500 (bp) | 86958 |
| Clean reads | 47550772.74 | Total number of Unigene≥1000 (bp) | 50559 |
| Clean bases | 7128569462.00 | N50 | 1858 |
| Valid bases | 95.86% | The total length of unigene (bp) | 143307954 |
| Q30 | 91.97% | The length of the longest unigene (bp) | 20496 |
| GC | 43.33% | The length of the shortest unigene (bp) | 290 |
| Raw Bases(G) | 6.93 | The average length of unigene (bp) | 1299.83 |
| Clean bases(G) | 6.64 |
Fig 1. GC Content frequence distribution of unigene.
3.2 Identification of simple sequence repeats from the A. dahurica transcriptome
A total of 33,724 potential SSR loci were identified across 26,455 unigenes from the transcriptome of A. dahurica. The total size of the examined unigene sequences was 143,307,954 bp, meaning one SSR site was detected per 4,249 bp. Moreover, 20,734 unigenes (78.37%) contained a single SSR, 5,723 unigenes (21.63%) had more than one SSR, and 1,993 unigenes (7.53%) contained compound SSRs (Table 3). Thus, A. dahurica contains many types of SSRs, and all of the repeats comprising 1–6 nucleotides were detected.
Table 3. A. dahurica transcriptome SSR general statistics.
| Items | Number |
|---|---|
| Total number of unigene sequences examined | 110251 |
| Total size of examined unigene sequences (bp) | 143307954 |
| Total number of identified SSRs | 33724 |
| The number of sequences containing SSR loci | 26455 |
| The number of sequences with 1 SSR loci | 20734 |
| The number of unigene sequences with more than 1 SSR locus | 5723 |
| The number of SSRs present in compound formation | 1993 |
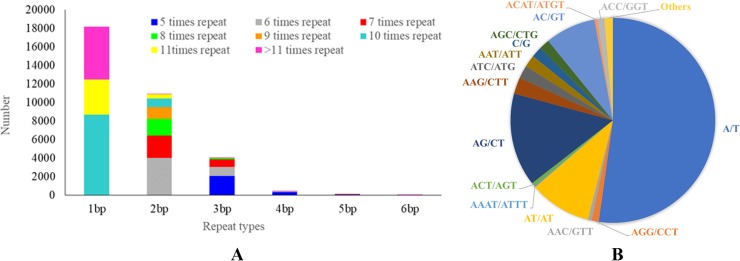
In total, 18,179 mononucleotide repeat motifs with more than 10 repeats (53.91%) were identified, which was the most frequent type, followed by dinucleotide repeat motifs with more than six repeats (10,975; 32.54%), trinucleotide repeat motifs with more than five repeats (4,022; 11.93%), tetranucleotide repeat motifs with more than five repeats (436; 1.29%), pentanucleotide repeat motifs with more than five repeats (77; 0.23%), and hexanucleotide repeat motifs with more than five repeats (35; 0.1%). Additionally, A/T was the most abundant motif among the two possible types of mononucleotide repeats, representing 52.51% of the total SSRs, followed by the AG/CT (14.73%), AT/AT (9.80%), and AC/GT (7.99%) motifs. Among the trinucleotide repeat motifs, the two most frequent repeats were AAG/TTC (2.58% of the total SSRs) and ATC/ATG (2.06%), followed by AAT/ATT (1.92%) and AGC/CTG (1.66%) (Table 4). Quad-, penta-, and hexanucleotide repeats represented 1.62% of the total SSRs (Fig 2A).
Table 4. The most abundant SSR repeat types in the transcriptome.
| Mononucleotide | Dinucleotide | Trinucleotide | Tetranucleotide |
|---|---|---|---|
| A/T (52.21%) | AG/CT (14.73%) | AAG/CTT (2.58%) | ACAT/ATGT (0.45%) |
| C/G (1.69%) | AT/AT (9.80%) | ATC/ATG (2.06%) | AAAT/ATTT (0.20%) |
| AC/GT (7.99%) | AAT/ATT (1.92%) | AGAT/ATCT (0.10%) | |
| CG/CG (0.03%) | AGC/CTG (1.66%) | AAAG/CTTT (0.07%) | |
| ACC/GGT (1.13%) | AGGG/CCCT (0.07%) | ||
| AGG/CCT (1.13%) | AAAC/GTTT (0.06%) | ||
| AAC/GTT (0.59%) | AGCT/AGCT (0.05%) | ||
| ACT/AGT (0.56%) | AACC/GGTT (0.04%) | ||
| ACG/CGT (0.18%) | ACAG/CTGT (0.03%) | ||
| CCG/CGG (0.13%) | ACTC/AGTG (0.03%) | ||
| AGGC/CCTG (0.03%) |
Fig 2. Repeat motif type distribution in A. dahurica SSR.
A:1–6 nucleotides repeats statistical results, B: types and quantities of dinucleotides and trinucleotide repeats.
3.3 Development and validation of identified SSR markers
A total of 114 primer pairs were designed and synthesized (S1 Table). After PCR amplification, 14 primer pairs that produced a single band of the expected size and many polymorphisms were selected for subsequent analysis. All of the forward primers were labeled with different HEX fluorescent dyes and then used to analyze the fragments from the 56 A. dahurica ecotypes collected from different locations in China (Table 1). Loci that failed to provide clear signals in the expected size range or that lacked polymorphisms were eliminated. Finally, 10 novel SSR primer pairs were developed for A. dahurica (Table 5). A total of 45 alleles were identified, with 2–8 alleles per locus. The observed and expected heterozygosities at each locus were 0.6955–0.3137 (average of 0.5332) and 0.6802–0.3018 (average of 0.4626), respectively. Shannon’s information index was 1.3040–0.5618 (average of 0.8475) and the PIC was 0.6274–0.2702 (average of 0.4091). The loci were moderately polymorphic. HWE tests revealed that only Locus 8 significantly deviated from HWE in this captive population (0.05 ≥ P ≥ 0.01). The results showed Loci 1, 2, 3, 4, 7, 9, and 10 were moderately polymorphic and in accordance with HWE.
Table 5. Characteristics of the 10 polymorphic SSR markers and the novel microsatellite marker system validated in 56 A. dahuricaes.
| loci No. | Motif | Primer Sequence(5'→3') | Tm (℃) |
Allele Size (bp) |
Na | Ne | Ho | He | I | PIC | HWE | Regions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (TA)8 | F: | GCCTCAAGAACTTGCCTCAC | 61 | 229–241 | 4 | 1.4323 | 0.6955 | 0.3018 | 0.5618 | 0.2702 | 0.1328 | 5’UTR |
| R: | GTTTCACCACTTCTGCCACA | ||||||||||||
| 2 | (AG)9 | F: | CAGACCTGAATGGCACTGAA | 60 | 173–181 | 6 | 1.8518 | 0.5359 | 0.46 | 0.9361 | 0.4292 | 0.3709 | 5’UTR |
| R: | TGAAGGAGTCTGCACACCAA | ||||||||||||
| 3 | (TG)7 | F: | GATTGAGCAAGGAGAGGTGC | 60 | 241–250 | 4 | 1.7398 | 0.5709 | 0.4252 | 0.7959 | 0.3835 | 0.3024 | 5’UTR |
| R: | CCATCAAATAAGAGGCACAAAA | ||||||||||||
| 4 | (CA)6 | F: | CAGCCGGCTAAGACAGAATC | 60 | 254–258 | 2 | 1.6228 | 0.6128 | 0.3838 | 0.5719 | 0.3101 | 0.2457 | 5’UTR |
| R: | TGGGATCACTTTCTTGGCATA | ||||||||||||
| 5 | (TA)7 | F: | TTTTCATGGATGCACAATAAGC | 57 | 200–210 | 6 | 3.1266 | 0.3137 | 0.6802 | 1.304 | 0.6272 | 0.1465 | 5’UTR |
| R: | GAAATTGATGGCATTCTCCC | ||||||||||||
| 6 | (TC)9 | F: | TTGCAGCGTCAATTTGCTAC | 59 | 237–243 | 5 | 2.48 | 0.3978 | 0.5968 | 1.1452 | 0.5498 | 0.2736 | 5’UTR |
| R: | AGAAAAAGTCCGGGCAAAGT | ||||||||||||
| 7 | (AT)8 | F: | TTACGCAATTGCTGATTCCA | 58 | 212–220 | 3 | 1.9222 | 0.5159 | 0.4798 | 0.7084 | 0.3729 | 0.3362 | 5’UTR |
| R: | CCCCGCTGGCTTATCTTTAT | ||||||||||||
| 8 | (AT)9 | F: | CCATGGAGAAATTGGCTGTT | 58 | 149–158 | 8 | 1.8909 | 0.5246 | 0.4711 | 1.0793 | 0.4521 | 0.0102 | 5’UTR |
| R: | TTCTTGATCCAGTGATGCCA | ||||||||||||
| 9 | (GT)9 | F: | TTTGGGGCTGCTTTGTATTC | 59 | 269–285 | 3 | 1.7302 | 0.5742 | 0.422 | 0.6872 | 0.3528 | 0.6730 | 5’UTR |
| R: | TCCAGCATCCAGTATCACCA | ||||||||||||
| 10 | (TG)9 | F: | GGTTTGTGAGATCCTTTGGG | 60 | 237–243 | 4 | 1.682 | 0.5909 | 0.4055 | 0.6852 | 0.3427 | 0.6662 | 5’UTR |
| R: | GAACTGACACTTGGGAATCCA | ||||||||||||
Na: Number of Different Alleles, Ne: Number of Effective Alleles, I: Shannon‘s Information Index, Ho: Observed Heterozygosity, He: Expected Heterozygosity, PIC: Polymorphism Information Content. HWE: Hardy-Weinberg equilibrium P-value (≥0.05).
Discussion
4.1 Developing SSRs based on the transcriptome is reliable and efficient
With the advent and rapid development of high-throughput sequencing technology, RNA sequencing and the development of molecular markers has become easy and reliable in many species [28–30]. Multiple SSR loci can be identified by analyzing the transcriptome, which is significant for the localization of genes responsible for specific traits and functions. This is the first report describing the application of high-throughput sequencing to the development of A. dahurica genic SSR markers based on unigenes. A total of 33,724 genic SSR loci were detected, and a set of 10 novel genic SSR markers was developed to provide additional tools for analyzing the genetic diversity of A. dahurica. The 10 polymorphic loci were then validated in 56 individuals. These loci showed abundant polymorphism, implying they are useful for analyzing A. dahurica relationships and confirming the potential value of an A. dahurica transcriptome database for the development of new SSR markers.
4.2 The infrequent observation of GC repeats may be related to CpG clusters and GC contents
GC repeat motifs were the least common among the mononucleotide and dinucleotide repeats, possibly because of the presence of CpG clusters. Previous studies have indicated that CpG dinucleotides often occur in discrete regions, and almost 60% of these CpG-rich clusters are located in or close to genes, mostly at the 5′ end [31, 32]. The GC content and CpG pattern, along with the chromatin condensation and B–Z transition, in a gene and around the promoter can determine gene expression characteristics through transcription factor binding sites [33]. Cell differentiation involving the deamination of mC to T has been proposed to result in CpG deficiency [34], and CpG deficiency is associated with a corresponding TpG (CpA) excess [35]. The mean GC content of the A. dahurica transcriptome unigenes was 43.33% (i.e., less than the AT content), which may be related to the limited number of GC dinucleotide repeats or G/C mononucleotides. Moreover, CpG clusters may be essential for regulating gene expression, and are therefore infrequently observed.
4.3 The presence of 10 novel SSRs in untranslated regions implies SSR diversity is related to different phenotypes
Cis-acting elements involved in post-transcriptional control are generally located in the UTRs of mRNAs [36]. In the current study, 10 novel A. dahurica SSRs were localized to UTRs. UTRs are vital at the interface between mRNAs and proteins. They regulate stability, transport, and translation efficiency, as well as the function and subcellular localization of translated proteins, and they increase the coding capacity of the genome [36–39]. In future studies, the relationship between SSRs and regulatory elements in the UTR should be characterized to clarify phenotypic diversity.
Conclusion
The A. dahurica transcriptome characterization and the substantial body of transcripts reported here will facilitate research to develop the medicinal and nutritional properties of this species. Ten novel A. dahurica SSR markers with polymorphism were developed, which provides a foundation for genetic diversity analysis, genetic mapping and marker breeding in A. dahurica.
Supporting information
(DOCX)
(DOCX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The work is supported by the project of 13th Five-year Plan for Breeding Cooperation in Sichuan Province, grant number: 2016NYZ0036 (http://202.61.89.120/) to WW. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.The European Pharmacopoeia Commission. European pharmacopoeia: Council of Europe; 2013.
- 2.Yakuji Nippo Ltd. The Japanese Pharmacopoeia: Society of Japanese Pharmacopoeia; 2010.
- 3.Zhang X-n, Ma Z-j, Wang Y, Sun B, Guo X, Pan C-q, et al. Angelica Dahurica ethanolic extract improves impaired wound healing by activating angiogenesis in diabetes. PLOS ONE. 2017;12(5):e0177862 10.1371/journal.pone.0177862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang J, Zhou L, Sun J, Han J, Guo D-A. Chromatographic fingerprint analysis and characterization of furocoumarins in the roots of Angelica dahurica by HPLC/DAD/ESI-MSn technique. Journal of Pharmaceutical and Biomedical Analysis. 2008;47(4):778–85. 10.1016/j.jpba.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y-K, Kim YS, Ryu SY. Antiproliferative effect of furanocoumarins from the root of Angelica dahurica on cultured human tumor cell lines. Phytotherapy Research. 2007;21(3):288–90. 10.1002/ptr.2043 . [DOI] [PubMed] [Google Scholar]
- 6.Wei W, Wu X-W, Deng G-G, Yang X-W. Anti-inflammatory coumarins with short- and long-chain hydrophobic groups from roots of Angelica dahurica cv. Hangbaizhi. Phytochemistry. 2016;123:58–68. 10.1016/j.phytochem.2016.01.006 . [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Liu J, Wang Q, Liu M, Cheng Y, Zhang X, et al. Antidepressive-like effect of imperatorin from Angelica dahurica in prenatally stressed offspring rats through 5-hydroxytryptamine system. Neuroreport. 2017;28(8):426–33. 10.1097/WNR.0000000000000778 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G-H, Chen C-Y, Tsai T-H, Chen C-K, Cheng C-Y, Huang Y-H, et al. Evaluation of tyrosinase inhibitory and antioxidant activities of Angelica dahurica root extracts for four different probiotic bacteria fermentations. Journal of Bioscience and Bioengineering. 2017;123(6):679–84. 10.1016/j.jbiosc.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 9.Lechner D, Stavri M, Oluwatuyi M, Pereda-Miranda R, Gibbons S. The anti-staphylococcal activity of Angelica dahurica (Bai Zhi). Phytochemistry. 2004;65(3):331–5. 10.1016/j.phytochem.2003.11.010 [DOI] [PubMed] [Google Scholar]
- 10.Kimura Y, Ohminami H, Arichi H, Okuda H, Baba K, Kozawa M, et al. Effects of various coumarins from roots of Angelica dahurica on actions of adrenaline, ACTH and insulin in fat cells. Planta Med. 1982;45(3):183–7. . [DOI] [PubMed] [Google Scholar]
- 11.Han HS, Jeon H, Kang SC. Phellopterin isolated from Angelica dahurica reduces blood glucose level in diabetic mice. Heliyon. 2018;4:e00577 10.1016/j.heliyon.2018.e00577 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinese Pharmacopeia Committee. Pharmacopoeia of the People's Republic of China Volume 1 Beijing: China Medical Science Press; 2015. 2 p. [Google Scholar]
- 13.Li B, Zhang X, Wang J, Zhang L, Gao B, Shi S, et al. Simultaneous Characterisation of Fifty Coumarins from the Roots of Angelica dahurica by Off-line Two-dimensional High-performance Liquid Chromatography Coupled with Electrospray Ionisation Tandem Mass Spectrometry. Phytochemical Analysis. 2014;25(3):229–40. 10.1002/pca.2496 [DOI] [PubMed] [Google Scholar]
- 14.Wang N, Qin H, Shu P, Huang L, Wang M, Yuan C. study on original plant of tranditional drug 'baizhi' (Radix Angelicae Dahuricae) and Its Closely Related Wild Plants Ⅱ:Karyological and Pollen Morphological Studies On 'baizhi' and Closely Related Wild Plants. China Journal of Chinese Materia Medica. 2001;09:8–12. [PubMed] [Google Scholar]
- 15.Rogozin I, V Sverdlov A, Babenko V, Koonin E. Analysis of evolution of exon-intron structure of eukaryotic genes 2005. 118–34 p. [DOI] [PubMed] [Google Scholar]
- 16.Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: features and applications. Trends in Biotechnology. 2005;23(1):48–55. 10.1016/j.tibtech.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 17.Varshney RK, Grosse I, Hahnel U, Siefken R, Prasad M, Stein N, et al. Genetic mapping and BAC assignment of EST-derived SSR markers shows non-uniform distribution of genes in the barley genome. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2006;113(2):239–50. Epub 2006/06/23. 10.1007/s00122-006-0289-z . [DOI] [PubMed] [Google Scholar]
- 18.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends in Genetics. 2008;24(3):133–41. 10.1016/j.tig.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 19.Sen S, Dehury B, Sahu J, Rathi S, Narain Singh Yadav R. Mining and comparative survey of EST–SSR markers among members of Euphorbiaceae family. 2018;45 10.1007/s11033-018-4181-0 [DOI] [PubMed] [Google Scholar]
- 20.Pan L, Huang T, Yang Z, Tang L, Cheng Y, Wang J, et al. EST-SSR marker characterization based on RNA-sequencing of Lolium multiflorum and cross transferability to related species 2018. [Google Scholar]
- 21.Zhang J, Liu T, Rui F. Development of EST-SSR markers derived from transcriptome of Saccharina japonica and their application in genetic diversity analysis 2017. [Google Scholar]
- 22.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2003;106(3):411–22. Epub 2003/02/18. 10.1007/s00122-002-1031-0 . [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Liu Y-X, Jiang G-F. De novo Assembly and Characterization of the Testis Transcriptome and Development of EST-SSR Markers in the Cockroach Periplaneta americana 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porebski S, Grant Bailey L, Baum B. Modification of CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components 1997. 8–15 p. [Google Scholar]
- 25.Chatterji S, Pachter L. Reference based annotation with GeneMapper. Genome Biology. 2006;7(4):R29 10.1186/gb-2006-7-4-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smouse PE, Peakall R. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28(19):2537–9. 10.1093/bioinformatics/bts460 %J Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh FC, R-C Y, Boyle T. POPGENE Version 1.31. Microsoft windows-based freeware for population genetic analysis 1998. 11–23 p. [Google Scholar]
- 28.Colburn BC, Mehlenbacher SA, Sathuvalli VR. Development and mapping of microsatellite markers from transcriptome sequences of European hazelnut (Corylus avellana L.) and use for germplasm characterization. Molecular Breeding. 2017;37:16 10.1007/s11032-016-0616-2 [DOI] [Google Scholar]
- 29.Chen X, Li J, Xiao S, Liu X. De novo assembly and characterization of foot transcriptome and microsatellite marker development for Paphia textile. Gene. 2016;576:537–43. 10.1016/j.gene.2015.11.001 . [DOI] [PubMed] [Google Scholar]
- 30.Cui X, Huang X, Chen J, Yang X, Rong J. An efficient method for developing polymorphic microsatellite markers from high-throughput transcriptome sequencing: a case study of hexaploid oil-tea camellia (Camellia oleifera). Euphytica. 2018;214:26 10.1007/s10681-018-2114-6 [DOI] [Google Scholar]
- 31.Larsen F, Gundersen G, Lopez R, Prydz H. CpG Islands as Gene Markers in the Human Genome. Genomics. 1992;13(4):1095–107. 10.1016/0888-7543(92)90024-M [DOI] [PubMed] [Google Scholar]
- 32.Cross SH, Bird AP. CpG islands and genes. Current opinion in genetics & development. 1995;5:309–14. 10.1016/0959-437X(95)80044-1 [DOI] [PubMed] [Google Scholar]
- 33.Vinogradov AE. Dualism of gene GC content and CpG pattern in regard to expression in the human genome: magnitude versus breadth. Trends Genet. 2005;21(12):639–43. Epub 2005/10/06. 10.1016/j.tig.2005.09.002 . [DOI] [PubMed] [Google Scholar]
- 34.Scarano E, Iaccarino M, Grippo P, Parisi E. The Heterogeneity of Thymine Methyl Group Origin in DNA Pyrimidine Isostichs of Developing Sea Urchin Embryos 1967. 1394–400 p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jabbari K, Bernardi G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 2004;333:143–9. 10.1016/j.gene.2004.02.043 [DOI] [PubMed] [Google Scholar]
- 36.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biology. 2002;3(3):reviews0004.1-.10. 10.1186/gb-2002-3-3-reviews0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrillo E A. Godoy Herz M, Barta A, Kalyna M, Kornblihtt A. Let there be light: Regulation of gene expression in plants. RNA biology. 2015;11(10):1215–20. 10.4161/15476286.2014.972852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernando CE, Romanowski A, Yanovsky MJ. Transcriptional and post-transcriptional control of the plant circadian gene regulatory network. Biochim Biophys Acta Gene Regul Mech. 2017;1860(1):84–94. 10.1016/j.bbagrm.2016.07.001 . [DOI] [PubMed] [Google Scholar]
- 39.Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J-K. UTR-Dependent Control of Gene Expression in Plants. Trends Plant Sci. 2018;23(3):248–59. 10.1016/j.tplants.2017.11.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.