Abstract
Breast cancer is the most significant cause of cancer-related death in women around the world. The vast majority of breast cancer-associated mortality stems from metastasis, which remains an incurable disease state. Metastasis results from evolution of clones that possess the insidious properties required for dissemination and colonization of distant organs. These clonal populations are descended from breast cancer stem cells (CSCs), which are also responsible for their prolonged maintenance and continued evolution. Telomeres impose a lifespan on cells that can be extended when they are actively elongated, as occurs in CSCs. Thus, changes in telomere structure serve to promote the survival of CSCs and subsequent metastatic evolution. The selection of telomere maintenance mechanism (TMM) has important consequences not only for CSC survival and evolution, but also for their coordination of various signaling pathways that choreograph the metastatic cascade. Targeting the telomere maintenance machinery may therefore provide a boon to the treatment of metastatic breast cancer. Here we review the two major TMMs and the roles they play in the development of stem and metastatic breast cancer cells. We also highlight current and future approaches to targeting these mechanisms in clinical settings to alleviate metastatic breast cancers.
Keywords: breast cancer, cancer evolution, cancer stem cells, metastasis, telomerase
Introduction
Breast cancer is the most common malignancy and most frequent cause of cancer-related death in women globally[1]. The vast majority of breast cancer-related morbidity and mortality can be ascribed to metastasis, which occurs in ~30 percent of cases and underlies ~90 percent of breast cancer deaths[2,3]. Metastasis is a multistage cascade that commences when cancer cells migrate from their primary tumor of origin and undergo hematogenous dissemination that terminates in the seeding and colonization of distant organs[4]. This so-called “invasion-metastasis cascade” serves as an evolutionary bottleneck that requires disseminated cancer cells (DTCs) to: (i) activate migratory and invasive programs; (ii) survive within the vasculature in an anchorage-independent manner; (iii) interact with other circulating cells to facilitate survival and extravasation; and (iv) coordinate tissue-specific signaling inputs to persist in unfamiliar microenvironments[5–7]. Thus, metastasis can be viewed as a process of clonal selection whereby a heterogeneous primary tumor gives rise to subpopulations that are fit to traverse the invasion-metastasis cascade. Following tissue colonization, these disseminated subclones retain growth-permissive features of the original primary tumor and undergo further evolution and clonal expansion within metastatic microenvironments[8,9].
Metastatic evolution occurs via a number of distinct yet spatiotemporally overlapping mechanisms, including linear and parallel progression of monophyletic or polyphyletic founder clones[10]. Cancer stem cells (CSCs) are fundamental components of tumors that enable the maintenance of emergent clonal populations yielded by evolutionary forces[11–13]. CSCs are operationally defined by their self-renewal and tumor-initiating capacities; that is, a single CSC can recapitulate a tumor in its entirety, including a stable CSC pool[14]. Historically, stochastic clonal evolution was believed to be mutually exclusive with a tumor developmental hierarchy built upon a stem cell population[15,16]. More recent evidence suggests that there is a relationship between tumor evolution and CSCs that manifests through at least two mechanisms. First, the CSC population itself becomes highly heterogeneous during tumor development, indicating that CSCs are directly subjected to selective pressures[17,18]. Second, non-stem cancer cells that define unique genetic and epigenetic lineages can be reprogrammed into CSCs[19,20]. Thus, the plasticity that exists within and between stem and non-stem cancer cells provides a bidirectional route to engender clones that harbor distinctive properties, including the ability to metastasize. Of note, the functional significance of CSC evolution in the development and progression of multiple malignancies has been extensively documented[21–23].
Numerous pathways that exert control over the metastatic propensity of cancer cells do so by regulating the production or function of CSCs. For instance, Wnt/β-catenin signaling in both the primary tumor and metastatic microenvironments enhances breast CSC self-renewal and metastatic colonization[24,25]. Likewise, inhibiting Wnt signaling abrogates metastatic outgrowth by depleting the CSC population[26,27]. Similarly, vascular endothelial growth factor (VEGF) activates stem programs in breast cancer cells via VEGF receptor (VEGFR)- and neuropilin (NRP)-dependent cascades[28,29]. VEGF can additionally push breast CSCs to undergo endothelial-like differentiation, thereby promoting tumor vascularization and cancer cell dissemination[30]. The NF-κB transcription factor pathway also acts as a critical regulator of breast CSC function[31]. In particular, microenvironmental stimuli from resident stromal cells, extracellular matrix components, and the local immune milieu activate NF-κB signaling to sustain CSC development[25,32,33]. As a result, NF-κB inhibitors demonstrate potent activity against breast CSCs[34]. Related to these events, CSC expansion is associated with the epithelial-mesenchymal transition (EMT), a process whereby epithelial cells lose their intrinsic polarity and markers of differentiation and adopt features of mesenchymal cells, including enhanced migration and invasiveness[35,36]. Key transcription factors that orchestrate EMT in breast cancer, such as Snail, Slug, and Twist1, simultaneously play a role in the acquisition of stem-like traits[37]. Importantly, both Wnt/β-catenin and NF-κB signaling exert direct transcriptional control over these EMT-associated factors[36,38]. Furthermore, EMT induces upregulation of VEGF, which bolsters the activities of β-catenin and NF-κB and promotes angiogenesis to support CSC self-renewal and permit dissemination[39–41]. In short, breast CSC survival and maturation are determined by a confluence of cell-intrinsic and microenvironment-derived signals that are transduced through parallel EMT-dependent and -independent circuits.
CSCs, like embryonic and tissue stem cells, possess replicative immortality[42], a process achieved in part by activating telomere maintenance mechanisms (TMMs)[43,44]. As outlined below, TMMs function within a network that unites cellular immortalization with processes, including EMT, that drive the development and outgrowth of metastatic cells. Telomeres, therefore, serve as essential mediators of CSC maintenance and consequent metastatic evolution. In addition, the results detailed below implicate telomere homeostasis as an attractive target for novel therapeutics to treat metastatic breast cancer.
Telomeres and Telomere Dynamics in CSCs and Metastatic Cells
Telomeres are nucleoprotein complexes located at the ends of linear chromosomes that safeguard against chromosomal instability and the loss of genetic information during cell division[45]. In humans, the DNA component of telomeres is composed of tandem (TTAGGG)n repeats with a 3’ single-stranded overhang that invades telomeric duplex DNA to form a protective loop[46]. These DNA regions are coated with proteins that collectively constitute the shelterin complex. Shelterin proteins serve to shield telomeres from illicit activation of DNA damage responses (DDRs); they also maintain genome integrity and recruit factors responsible for regulating telomere length[47,48]. In somatic (i.e., non-immortalized) cells, telomeres shorten during iterative rounds of cell division. To combat this event, stem cells and cancer cells maintain their telomeres using one of two TMMs: telomerase or alternative lengthening of telomeres (ALT). Telomerase is a reverse transcriptase enzyme composed of an RNA moiety (TERC, also known as TR) that provides a template for telomeric DNA synthesis and a protein moiety (TERT) that facilitates telomerase recruitment and carries out its polymerase activity[49]. In contrast, ALT relies upon homology-directed, recombination-dependent synthesis of nascent telomeric DNA[50]. ALT requires transient deprotection of telomeres coupled to activation of a DDR that is accompanied by telomere extension in a manner similar to break-induced DNA synthesis[51,52]. DDR activation occurs in response to alterations in telomeric and subtelomeric chromatin structure that are brought about by loss of the chromatin remodelers ATRX and DAXX[53,54]. Notably, evidence of each of these mechanisms has been found in breast cancer and can be correlated with specific histologic subtypes or disease stages[55,56]. These findings support the idea that TMM identity may impact breast cancer progression, including the onset of metastasis.
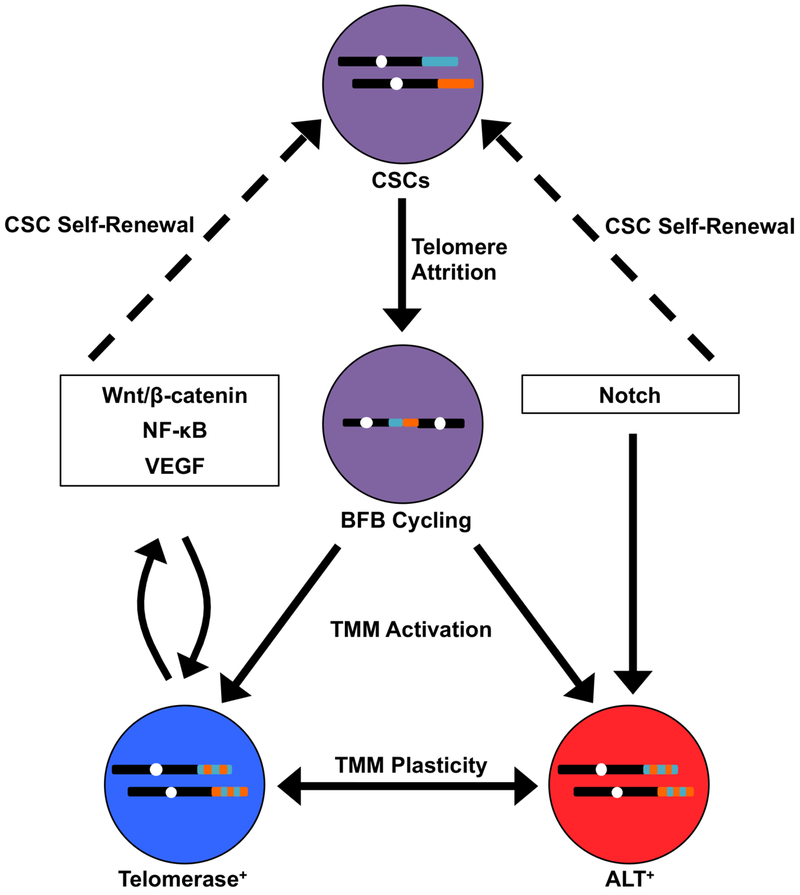
While TMM acquisition has been identified as a feature of both stem and non-stem cancer cells, these processes play an essential role in preferentially sustaining the CSC population[42]. By virtue of their replicative immortality, CSCs function as progenitors that exist over a sufficient timescale for evolution to take place. Remarkably, telomere shortening appears to be a primary driving force underlying tumor evolution. Telomere shortening precedes TMM activation[57], which allows for the formation of critically short telomeres that cannot be adequately capped by shelterin. Cells interpret these short telomeres as free DNA ends, which are temporarily repaired by chromosome end-to-end fusions that ultimately induce breakage-fusion-bridge (BFB) cycles[58,59]. BFB cycling leads to complex genomic rearrangements including deletions, non-reciprocal translocations, and formation of dicentric or circular chromosomes[60]. Telomere catastrophe may also yield chromosomal instability that is resolved via chromothripsis or other forms of chromoanagenesis, an event termed telomere crisis[61,62]. Breast cancer-initiating cells can harbor both short telomeres and telomerase activity[41], consistent with the model that telomere shortening instigates genomic instability and CSC evolution while telomere elongation maintains emergent CSC subpopulations (Fig. 1). Evidence identifying ALT in breast CSCs has not yet been found. However, ALT has a stem cell origin[63], while ALT activity has been observed in non-breast CSCs[44,63,64]. Future studies examining TMMs in breast CSCs and their connection to genome architecture and tumor heterogeneity will be of great value.
Figure 1. Telomere-centric Model of Breast CSC Biogenesis and Metastatic Evolution.
Cancer stem cells (CSCs; purple) harboring telomeres of a given length (shown for two different chromosomes in teal and orange) undergo telomere attrition as a by-product of self-renewal. This ultimately yields critically short telomeres that are temporarily repaired by chromosome end-to-end fusions, resulting in breakage-fusion-bridge (BFB) cycling (represented by dicentric chromosome). BFB cycling or chromoanagenesis (not shown) cause widespread chromosomal instability (represented by dual-colored telomeres) and the acquisition of new genetic features, including those that are advantageous for metastasis. At the same time, telomere maintenance mechanisms (TMMs) are activated in these new clonal populations, which are defined in part by their reliance on telomerase (blue) or ALT (red). In addition, TMMs exhibit a degree of plasticity, such that TMM identity may interconvert between telomerase and ALT. TMM selection is influenced by signaling pathways that simultaneously promote CSC propagation (dashed arrows). In turn, telomere maintenance proteins directly regulate these signaling pathways, establishing reciprocal feedback loops that coordinate TMM activation and CSC maintenance.
Telomere maintenance proteins have been heavily implicated in many of the central signaling pathways in metastasis[65] (Fig. 1). For instance, TERT is capable of regulating Wnt target genes by forming a transcriptional co-activation complex with β-catenin[66]. In addition, TERT directly regulates NF-κB-dependent gene expression by binding to the NF-κB p65 subunit at the promoters of target genes[67,68]. Each of these pathways exerts reciprocal control over TERT[69,70], thereby preserving TMM identity and CSC phenotype. Of note, TERT can also bind to the VEGF promoter to stimulate VEGF expression and neoangiogenesis[71]. Other transcriptional regulators of TERT, such as c-Myc, further serve to induce EMT and stemness in breast cancer cells[72–74]. Compared to telomerase, ALT is less well-characterized at a molecular level; therefore, our understanding of its role in EMT and breast cancer metastasis is presently incomplete. Nevertheless, ALT is most often associated with tumors of mesenchymal origin[75], indicating a possible role for ALT in EMT. Accordingly, carcinoma cells exhibiting telomerase dysfunction were driven to adopt a mesenchymal stem-like phenotype, which was accompanied by activation of ALT and the formation of metastatic tumors[76,77]. In breast cancer cells, TERT expression is mutually exclusive with the mesenchymal state[78]. Similarly to TERT, the expression of ALT-associated proteins, such as the Bloom syndrome protein (BLM), is governed by signaling pathways, such as Notch, that are responsible for CSC fate specification and self-renewal[79,80]. Given these findings, it is paramount that future studies explore the significance of the relationship between stemness and telomere plasticity in breast cancer progression.
Telomere-directed Therapies for Metastatic breast cancer: Current and Future Perspectives
The functions of telomerase in tumorigenesis have been rigorously interrogated over the last several decades, as has the potential to target telomerase therapeutically[65,81]. The telomerase inhibitors BIBR1532 and GRN163L (also known as Imetelstat) display high efficacy in depleting the CSC pool and disrupting breast cancer metastasis[82–85]. Indeed, Imetelstat was assessed in a Phase I clinical trial for recurrent or metastatic breast cancer, although the trial was suspended due to dose-limiting toxicity[81]. In addition to such toxicity concerns, the success of telomerase inhibitors in clinical trials has thus far been moderated by the inherent complexity of telomere homeostasis. First, telomere shortening-induced senescence can be bypassed in the absence of functional p53 or other components of the DDR machinery[86]. Second, the critically short telomeres and chromosomal instability associated with telomere crisis are disproportionately associated with metastasis[87,88]. Thus, the evolution of DTCs that underlie metastatic disease may be enhanced unwittingly by therapies that promote telomere shortening. Despite these challenges, telomerase remains an appealing therapeutic objective in need of innovative targeting approaches in which these evolutionary considerations are taken into account.
Emerging telomerase-targeting strategies include cytotoxic small molecules that act as substrates for telomerase as well as anti-telomerase immunotherapies[89–92]. Current immunotherapeutic platforms are primarily centered on telomerase peptide or dendritic cell vaccines, which can be engineered to elicit either CD4+ or CD8+ T cell antitumor responses[93]. These strategies are being assessed in diverse preclinical settings, including breast cancer. Indeed, the telomerase peptide vaccine Vx-001 is progressing through clinical trials for advanced solid tumors[90]. More recent investigations have examined the feasibility of adoptive transfer of anti-telomerase chimeric antigen receptor (CAR) T cells for treating triple-negative breast cancer[94]. Future studies into the generalizability of anti-telomerase CAR T cell therapy to other breast cancer subtypes, as well as the to efficacy of these diverse immunotherapeutic approaches in clinical settings will be of tremendous value.
Although the functions of specific ALT-associated proteins have been elucidated, their utility as therapeutic targets for ALT-driven cancers has only recently been investigated. For example, the DNA damage-responsive kinase ataxia-telangectasia and Rad3-related (ATR) is activated secondary to depletion of ATRX, which leads to persistent retention of replication protein A (RPA) at telomeres and generation of a recombinogenic substrate. Inhibition of ATR, in turn, triggers apoptosis of ALT-positive cells[95]. BLM, a RecQ DNA helicase, unwinds telomeric G-quadruplex structures and coordinates 5’→3’ end resection during telomere recombination[96,97]. Accordingly, a recently-developed small molecule inhibitor of BLM may possess great potential as an anticancer agent against ALT-driven tumors[98]. Finally, topoisomerase IIIα (Topo IIIα) associates with BLM and regulates the topology of telomeric recombination intermediates. Interestingly, genetic inactivation of Topo IIIα selectively reduces the survival of ALT-positive compared to telomerase-positive cells[99]. Moreover, telomerase activity is enhanced in the surviving fraction of Topo IIIα-deficient cells[100], suggesting that telomerase activation provides a pathway for chemoresistance. Thus, targeting TMMs may best be achieved using a multidrug regimen consisting of multiple anti-TMM agents or an anti-TMM agent in combination with chemotherapy or other targeted agents[101]. The effectiveness of these therapeutic modalities in eliminating breast CSCs and in treating metastatic breast cancers remain intriguing and important open questions.
Conclusion
By overseeing multiple pathways that promote breast cancer stemness, EMT, and metastasis, telomeres function as critical nodes in the nexus between cellular immortalization, tumor evolution, and disease progression. The selection of TMM likely exhibits a high degree of plasticity in different tumor cell types or across disparate stages of breast cancer development, including metastasis. Indeed, TMM selection may itself be subject to evolutionary dynamic forces. In addition, the plasticity inherent in TMM identity has far-reaching prognostic and therapeutic implications. Tumors driven by distinct TMMs may show sensitivity or resistance to specific treatments, which has substantial impact on patient survival. Moreover, different subpopulations within a single tumor (e.g., stem versus non-stem cells) may be reliant upon unique TMMs. Such TMM heterogeneity may beget residual, resistant clones that underlie disease recurrence. In the future, gaining a deeper understanding of telomeres and the pathways controlled by the telomere machinery will provide immense insight into the origin, progression, and eradication of one of the world’s deadliest cancers.
Acknowledgements
Members of the Schiemann Laboratory are thanked for critical comments and reading of the manuscript.
Financial Support and Sponsorship
Research support was provided in part by the National Institutes of Health to W.P.S. (CA236273), to D.J.T. (CA186571), and to N.J.R (T32 GM007250 and F30 CA213892). Additional support was graciously provided by the METAvivor Foundation (W.P.S.), and by pilot funding from the Case Comprehensive Cancer Center’s Research Innovation Fund, which is supported by the Case Council and Friends of the Case Comprehensive Cancer Center (W.P.S.), and from the Case Clinical &Translational Science Collaborative (W.P.S.). Finally, D.J.T. is also supported by the American Cancer Society (RSG-13-211-01-DMC).
Footnotes
Conflict of Interest
All authors declare that there are no conflicts of interest.
Availability of Data and Materials
Not applicable.
Ethical Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist 2004;9:606–16. [DOI] [PubMed] [Google Scholar]
- 3.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011;331:1559–64. [DOI] [PubMed] [Google Scholar]
- 4.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 2003;3:453–8. [DOI] [PubMed] [Google Scholar]
- 5.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell 2017;168:670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YN, Koo KH, Sung JY, Yun UJ, Kim H. Anoikis resistance: an essential prerequisite for tumor metastasis. Int J Cell Biol 2012;2012:306879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie HY, Shao ZM, Li DQ. Tumor microenvironment: driving forces and potential therapeutic targets for breast cancer metastasis. Chin J Cancer 2017;36:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I et al. Cancer: evolution within a lifetime. Annu Rev Genet 2014;48:215–36. [DOI] [PubMed] [Google Scholar]
- 9.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer 2009;9:302–12. [DOI] [PubMed] [Google Scholar]
- 10.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science 2016;352:169–75. [DOI] [PubMed] [Google Scholar]
- 11.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science 2009;324:1670–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bapat SA. Evolution of cancer stem cells. Semin Cancer Biol 2007;17:204–13. [DOI] [PubMed] [Google Scholar]
- 13.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010;140:62–73. [DOI] [PubMed] [Google Scholar]
- 14.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 2008;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23–8. [DOI] [PubMed] [Google Scholar]
- 16.La Porta CAM, Zapperi S. Complexity in cancer stem cells and tumor evolution: Toward precision medicine. Semin Cancer Biol 2017;44:3–9. [DOI] [PubMed] [Google Scholar]
- 17.Poleszczuk J, Hahnfeldt P, Enderling H. Evolution and phenotypic selection of cancer stem cells. PLoS Comput Biol 2015;11:e1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang-Verslues WW, Kuo WH, Chang PH, Pan CC, Wang HH et al. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS One 2009;4:e8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011;146:633–44. [DOI] [PubMed] [Google Scholar]
- 20.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A 2011;108:1397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature 2011;469:362–7. [DOI] [PubMed] [Google Scholar]
- 22.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 2011;469:356–61. [DOI] [PubMed] [Google Scholar]
- 23.Piccirillo SG, Combi R, Cajola L, Patrizi A, Redaelli S et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene 2009;28:1807–11. [DOI] [PubMed] [Google Scholar]
- 24.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 2011;17:867–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011;481:85–9. [DOI] [PubMed] [Google Scholar]
- 26.Jang GB, Kim JY, Cho SD, Park KS, Jung JY et al. Blockade of Wnt/beta-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep 2015;5:12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L, Zhang L, Hu C, Liang S, Fei X et al. WNT pathway inhibitor pyrvinium pamoate inhibits the self-renewal and metastasis of breast cancer stem cells. Int J Oncol 2016;48:1175–86. [DOI] [PubMed] [Google Scholar]
- 28.Zhao D, Pan C, Sun J, Gilbert C, Drews-Elger K et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene 2015;34:3107–19. [DOI] [PubMed] [Google Scholar]
- 29.Elaimy AL, Guru S, Chang C, Ou J, Amante JJ et al. VEGF-neuropilin-2 signaling promotes stem-like traits in breast cancer cells by TAZ-mediated repression of the Rac GAP beta2-chimaerin. Sci Signal 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Li C, Li Y, Zhu Z. Involvement of breast cancer stem cells in tumor angiogenesis. Oncol Lett 2017;14:8150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, Sakamaki T, Casimiro MC, Willmarth NE, Quong AA et al. The canonical NF-kappaB pathway governs mammary tumorigenesis in transgenic mice and tumor stem cell expansion. Cancer Res 2010;70:10464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storci G, Sansone P, Mari S, D’Uva G, Tavolari S et al. TNFalpha up-regulates SLUG via the NF-kappaB/HIF1alpha axis, which imparts breast cancer cells with a stem cell-like phenotype. J Cell Physiol 2010;225:682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinkenbaugh AL, Baldwin AS. The NF-kappaB Pathway and Cancer Stem Cells. Cells 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Zhang H, Gu P, Bai J, Margolick JB et al. NF-kappaB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res Treat 2008;111:419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parvani JG, Schiemann WP. Sox4, EMT programs, and the metastatic progression of breast cancers: mastering the masters of EMT. Breast Cancer Res 2013;15:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor MA, Parvani JG, Schiemann WP. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-beta in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia 2010;15:169–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Micalizzi DS, Ford HL. Epithelial-mesenchymal transition in development and cancer. Future oncology 2009;5:1129–43. [DOI] [PubMed] [Google Scholar]
- 38.Felipe Lima J, Nofech-Mozes S, Bayani J, Bartlett JM. EMT in Breast Carcinoma-A Review. J Clin Med 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo M, Hou L, Li J, Shao S, Huang S et al. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-kappaB and beta-catenin. Cancer Lett 2016;373:1–11. [DOI] [PubMed] [Google Scholar]
- 40.Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res 2014;74:1566–75. [DOI] [PubMed] [Google Scholar]
- 41.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005;65:5506–11. [DOI] [PubMed] [Google Scholar]
- 42.Shay JW, Wright WE. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett 2010;584:3819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannen R, Bartsch JW. Essential roles of telomerase reverse transcriptase hTERT in cancer stemness and metastasis. FEBS Lett 2018;592:2023–31. [DOI] [PubMed] [Google Scholar]
- 44.Bojovic B, Booth RE, Jin Y, Zhou X, Crowe DL. Alternative lengthening of telomeres in cancer stem cells in vivo. Oncogene 2015;34:611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lange T How telomeres solve the end-protection problem. Science 2009;326:948–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A et al. Mammalian telomeres end in a large duplex loop. Cell 1999;97:503–14. [DOI] [PubMed] [Google Scholar]
- 47.de Lange T Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005;19:2100–10. [DOI] [PubMed] [Google Scholar]
- 48.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 2010;11:171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev 2015;29:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 2014;159:108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR et al. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016;539:54–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol 2009;16:1244–51. [DOI] [PubMed] [Google Scholar]
- 53.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011;333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 2012;8:e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subhawong AP, Heaphy CM, Argani P, Konishi Y, Kouprina N et al. The alternative lengthening of telomeres phenotype in breast carcinoma is associated with HER-2 overexpression. Mod Pathol 2009;22:1423–31. [DOI] [PubMed] [Google Scholar]
- 56.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 2011;179:1608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka H, Abe S, Huda N, Tu L, Beam MJ et al. Telomere fusions in early human breast carcinoma. Proc Natl Acad Sci U S A 2012;109:14098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P et al. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci U S A 2001;98:12683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res 2009;19:167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cleal K, Norris K, Baird D. Telomere Length Dynamics and the Evolution of Cancer Genome Architecture. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015;163:1641–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lafferty-Whyte K, Cairney CJ, Will MB, Serakinci N, Daidone MG et al. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene 2009;28:3765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silvestre DC, Pineda JR, Hoffschir F, Studler JM, Mouthon MA et al. Alternative lengthening of telomeres in human glioma stem cells. Stem Cells 2011;29:440–51. [DOI] [PubMed] [Google Scholar]
- 65.Robinson NJ, Schiemann WP. Means to the ends: The role of telomeres and telomere processing machinery in metastasis. Biochim Biophys Acta 2016;1866:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park JI, Venteicher AS, Hong JY, Choi J, Jun S et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009;460:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM et al. Telomerase directly regulates NF-kappaB-dependent transcription. Nat Cell Biol 2012;14:1270–81. [DOI] [PubMed] [Google Scholar]
- 68.Ding D, Xi P, Zhou J, Wang M, Cong YS. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-kappaB-dependent transcription. Faseb j 2013;27:4375–83. [DOI] [PubMed] [Google Scholar]
- 69.Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A et al. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science 2012;336:1549–54. [DOI] [PubMed] [Google Scholar]
- 70.Akiyama M, Hideshima T, Hayashi T, Tai YT, Mitsiades CS et al. Nuclear factor-kappaB p65 mediates tumor necrosis factor alpha-induced nuclear translocation of telomerase reverse transcriptase protein. Cancer Res 2003;63:18–21. [PubMed] [Google Scholar]
- 71.Liu N, Ding D, Hao W, Yang F, Wu X et al. hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res 2016;44:8693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A et al. Direct activation of TERT transcription by c-MYC. Nat Genet 1999;21:220–4. [DOI] [PubMed] [Google Scholar]
- 73.Yin S, Cheryan VT, Xu L, Rishi AK, Reddy KB. Myc mediates cancer stem-like cells and EMT changes in triple negative breast cancers cells. PLoS One 2017;12:e0183578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang A, Qin S, Schulte BA, Ethier SP, Tew KD et al. MYC Inhibition Depletes Cancer Stem-like Cells in Triple-Negative Breast Cancer. Cancer Res 2017;77:6641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett 2010;584:3800–11. [DOI] [PubMed] [Google Scholar]
- 76.Xue Y, Li L, Zhang D, Wu K, Chen Y et al. Twisted epithelial-to-mesenchymal transition promotes progression of surviving bladder cancer T24 cells with hTERT-dysfunction. PLoS One 2011;6:e27748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lagunas AM, Wu J, Crowe DL. Telomere DNA damage signaling regulates cancer stem cell evolution, epithelial mesenchymal transition, and metastasis. Oncotarget 2017;8:80139–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.El-Badawy A, Ghoneim NI, Nasr MA, Elkhenany H, Ahmed TA et al. Telomerase reverse transcriptase coordinates with the epithelial-to-mesenchymal transition through a feedback loop to define properties of breast cancer stem cells. Biol Open 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen GH, Tang W, Robles AI, Beyer RP, Gray LT et al. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc Natl Acad Sci U S A 2014;111:9905–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grudzien P, Lo S, Albain KS, Robinson P, Rajan P et al. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res 2010;30:3853–67. [PubMed] [Google Scholar]
- 81.Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med 2016;8:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hochreiter AE, Xiao H, Goldblatt EM, Gryaznov SM, Miller KD et al. Telomerase template antagonist GRN163L disrupts telomere maintenance, tumor growth, and metastasis of breast cancer. Clin Cancer Res 2006;12:3184–92. [DOI] [PubMed] [Google Scholar]
- 83.Dogan F, Ozates NP, Bagca BG, Abbaszadeh Z, Sogutlu F et al. Investigation of the effect of telomerase inhibitor BIBR1532 on breast cancer and breast cancer stem cells. J Cell Biochem 2018. 10.1002/jcb.27089. [DOI] [PubMed] [Google Scholar]
- 84.Joseph I, Tressler R, Bassett E, Harley C, Buseman CM et al. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res 2010;70:9494–504. [DOI] [PubMed] [Google Scholar]
- 85.Koziel JE, Herbert BS. The telomerase inhibitor imetelstat alone, and in combination with trastuzumab, decreases the cancer stem cell population and self-renewal of HER2+ breast cancer cells. Breast Cancer Res Treat 2015;149:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cosme-Blanco W, Shen MF, Lazar AJ, Pathak S, Lozano G et al. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep 2007;8:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A 2003;100:7737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffith JK, Bryant JE, Fordyce CA, Gilliland FD, Joste NE et al. Reduced telomere DNA content is correlated with genomic instability and metastasis in invasive human breast carcinoma. Breast Cancer Res Treat 1999;54:59–64. [DOI] [PubMed] [Google Scholar]
- 89.Zeng X, Hernandez-Sanchez W, Xu M, Whited TL, Baus D et al. Administration of a Nucleoside Analog Promotes Cancer Cell Death in a Telomerase-Dependent Manner. Cell Rep 2018;23:3031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer Treat Rev 2013;39:444–56. [DOI] [PubMed] [Google Scholar]
- 91.Vonderheide RH. Telomerase as a universal tumor-associated antigen for cancer immunotherapy. Oncogene 2002;21:674–9. [DOI] [PubMed] [Google Scholar]
- 92.Mender I, Gryaznov S, Dikmen ZG, Wright WE, Shay JW. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2’-deoxyguanosine. Cancer Discov 2015;5:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zanetti M A second chance for telomerase reverse transcriptase in anticancer immunotherapy. Nat Rev Clin Oncol 2017;14:115–28. [DOI] [PubMed] [Google Scholar]
- 94.Sandri S, Bobisse S, Moxley K, Lamolinara A, De Sanctis F et al. Feasibility of Telomerase-Specific Adoptive T-cell Therapy for B-cell Chronic Lymphocytic Leukemia and Solid Malignancies. Cancer Res 2016;76:2540–51. [DOI] [PubMed] [Google Scholar]
- 95.Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015;347:273–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol 2015;210:191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pan X, Drosopoulos WC, Sethi L, Madireddy A, Schildkraut CL et al. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc Natl Acad Sci U S A 2017;114:E5940–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nguyen GH, Dexheimer TS, Rosenthal AS, Chu WK, Singh DK et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem Biol 2013;20:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C et al. Topoisomerase IIIalpha is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J 2008;27:1513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai HJ, Huang WH, Li TK, Tsai YL, Wu KJ et al. Involvement of topoisomerase III in telomere-telomere recombination. J Biol Chem 2006;281:13717–23. [DOI] [PubMed] [Google Scholar]
- 101.Ward RJ, Autexier C. Pharmacological telomerase inhibition can sensitize drug-resistant and drug-sensitive cells to chemotherapeutic treatment. Mol Pharmacol 2005;68:779–86. [DOI] [PubMed] [Google Scholar]