Abstract
In an article published recently in Nature Medicine, the authors generate organoid models of liver neoplasia. In doing so, they highlight both the diversity of current organoid methodologies and their application to cancer modeling and therapeutics discovery.
A major goal of cancer precision medicine is to identify the most appropriate treatment modality for the genetic landscape of a patient’s tumor. Despite advances in cancer genomic analysis and discovery of druggable targets, a large number of patients do not respond to therapies as predicted, indicating the need for improved models to predict therapeutic outcome1. The development of 3D organoid culture methods provides an emerging approach for preclinical studies of cancer therapy, complementing established cancer cell lines and patient-derived xenografts. These miniaturized organoid ‘avatars’ of a patient’s tumor recapitulate the histological and gene expression features of the original tumor and can be genetically characterized, manipulated and expanded, rendering them an attractive model for guiding cancer precision medicine2. Organoid models may be particularly applicable to identifying patient-specific drug sensitivities, as is being increasingly demonstrated through treatment of a patient’s miniature tumor avatar in a dish with a myriad of drugs to identify the most effective treatment strategy3,4. Applications of organoids to study diverse or rare tumor types, to model premalignant states such as polyposis and to perform anticancer drug discovery are only now being explored.
A recent study in Nature Medicine highlights the use of patient-specific organoid strategies to model cancer therapeutic responses in patient-specific tissue. In this study Broutier et al5 used patient tumor biopsy tissue to generate and characterize organoid models of three types of primary liver cancer (PLC): hepatocellular carcinoma (HCC), cholangiocarcinoma (CC) and combined HCC/CC cancer (CHC). They demonstrate the feasibility of using the resultant organois, termed ‘tumoroids’, to model tumor subtypes and for anticancer drug discovery (Fig. 1).
Figure 1.
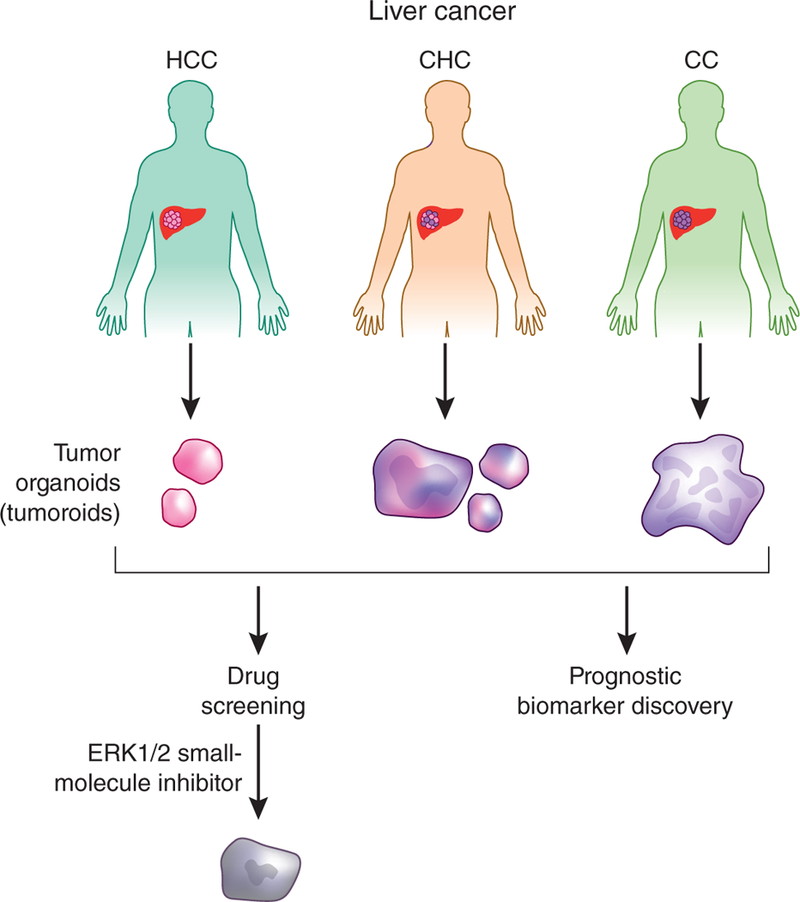
Organoid models of liver cancer. Broutier et al.5 generated tumor organoids, termed tumoroids, from liver cancer tumor biopsies spanning a range of histological types. Screening of these organoids identified a small-molecule ERK1/2 inhibitor as a potential liver cancer therapeutic and also identified potential prognostic biomarkers.
Broutier et al.5 built on previous work demonstrating growth of organoids from clinical tumor biopsies to develop culture models that accurately reflect the histological diversity of PLC. The authors carefully optimized their processing and culture conditions to minimize overgrowth of normal hepatocytes and epithelial cells through omission of the Wnt agonist R-spondin-1 and inclusion of other factors. The resultant protocol allowed successful derivation of organoids from eight different patients with PLC, spanning the HCC, CC and CHC tumor subtypes. These PLC organoids preserved the histology and mutational spectrum of the original tumors, even after long-term passaging, and exhibited in vivo tumorigenicity upon subcutaneous transplantation into mice. Broutier et al.5 also carried out comparative gene expression analysis of PLC-derived organoids versus those derived from healthy liver, identifying several genes that were upregulated among the different PLC subtypes, some of which correlated with poor survival. While future validation studies of these genes are needed to accurately define their role in PLC progression, this study demonstrates the feasibility of using organoid models for biomarker discovery.
Broutier et al.5 then tested the sensitivity of the PLC organoid lines to 29 compounds, finding that the ERK1 and ERK2 (ERK1/2) kinase inhibitor SCH772984 repressed in vitro growth of HCC, CC and CHC organoid lines, including some that were resistant to inhibitors targeting BRAF and MEK within the same pathway. Furthermore, SCH772984 inhibited in vivo tumor growth of representative HCC and CC organoids, suggesting that it might have clinical utility as a novel treatment strategy for patients with liver cancer.
As illustrated by this example, organoid technologies are progressively entering the mainstream for cancer biology and translational research. Certainly, organoids allow the creation of in vitro models from rare subtypes of cancer for which cell lines do not exist, such as for liver cancer in the current study, and facilitate the accumulation of sufficient tumor-derived biomass for deep ‘omics’ study. Importantly, organoids provide a robust method to preserve clinical tumor samples with a high success rate. Furthermore, these organoids represent an early state of the original tumor, in contrast to long-passaged cancer cell lines. As current organoid strategies begin with limiting amounts of biopsy tissue, in vitro expansion is typically required before therapeutic screening. To reach the ultimate goal of cancer precision medicine, it will be important to accelerate contemporary organoid methods, as in this study, to actionable time frames to allow guidance of treatment choices in real time.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Prasad V Nature 537, S63 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Neal JT & Kuo CJ Annu. Rev. Pathol. 11, 199–220 (2016). [DOI] [PubMed] [Google Scholar]
- 3.van de Wetering M et al. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauli C et al. Cancer Discov. 7, 462–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broutier L et al. Nat. Med. 23, 1424–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


