Abstract
OBJECTIVE.
The objective of this article is to show how artificial intelligence (AI) has impacted different components of the imaging value chain thus far as well as to describe its potential future uses.
CONCLUSION.
The use of AI has the potential to greatly enhance every component of the imaging value chain. From assessing the appropriateness of imaging orders to helping predict patients at risk for fracture, AI can increase the value that musculoskeletal imagers provide to their patients and to referring clinicians by improving image quality, patient centricity, imaging efficiency, and diagnostic accuracy.
Keywords: artificial intelligence, deep learning, fast MRI, machine learning, MRI, musculoskeletal imaging
Imaging remains an important tool for the evaluation of patients with musculoskeletal (MSK) conditions, and its value has contributed to increased utilization of common MSK imaging modalities [1]. Increased utilization has had several downstream effects for a radiology department or private practice, including an increased need for achieving operational efficiency while maintaining excellent accuracy and imaging report quality [2].
Artificial intelligence (AI) is an exciting tool that can help radiologists meet these needs. AI has the potential to significantly affect every step in the imaging value chain. In the current early stages of the introduction of AI into radiology, several studies involving MSK imaging have already examined and shown the potential value of AI. The purpose of this article is to give MSK radiologists an introduction to AI by reviewing the most current literature highlighting its use in various stages of image formation and utilization.
Technical Aspects
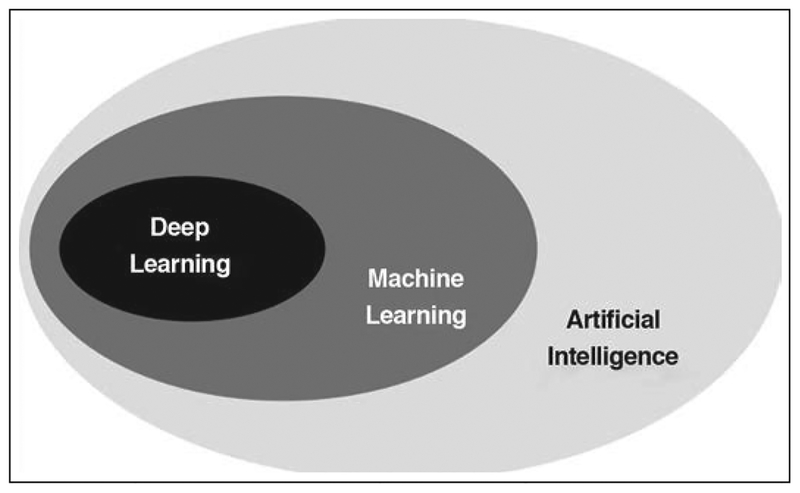
The terms AI, machine learning (ML), and deep learning (DL) are often used interchangeably; however, there are notable differences in what these related terms signify (Fig. 1). AI refers to any technique that enables computers to mimic human intelligence [3]. ML is a more specialized subfield of AI that enables machines to improve performance of tasks with experience, using various tools drawn from statistics, mathematics, and computer science. DL is an even more specialized subfield within ML that studies the use of a certain category of computational models, called deep neural networks, that are exposed to large datasets. DL [4] has led to several breakthrough improvements in areas such as image classification [5], semantic labeling [6], optical flow [7], and gaming [8].
Fig. 1—
Schematic showing machine learning as specialized subcategory of artificial intelligence. Deep learning is another subcategory of machine learning that studies use of certain category of computational models that are fit to large datasets.
The building blocks of all neural networks are nodes, which might be considered the computational analogue of neurons in a biologic brain. These nodes perform elementary mathematic operations, such as weighted addition, on their various inputs, resulting in outputs that are passed on to other nodes to which they are connected. The weighted connections between nodes may be seen as analogues to the synapses between biologic neurons. In addition to linear operations like addition, neural networks also typically perform nonlinear operations such as thresholding or rectification, yielding output only when certain levels of input signal are reached. In this respect, computational neural networks emulate neuronal activation (and, indeed, the functions describing elementary nonlinear operations are usually known as activation functions). In deep neural networks, the nodes are generally organized into layers, with each layer receiving input from previous layers (and, in some specialized cases, also receiving recurrent feedback from subsequent layers). One might be tempted to draw analogies to various stages in the processing of sensory stimuli, such as processing by the retina, the lateral geniculate nucleus, and the primary visual cortex, each of which performs distinct operations essential to the interpretation of visual input by the human brain. A neural network is generally identified as deep when it has more than just a few layers.
This general network structure can be traced back to 1958, when Rosenblatt [9] described what he called the Perceptron, a computational model to describe the function of the human brain. In such a computational network, just as in the brain, the computational capacity of any single neuron is very limited, but the grouping of neurons together into organized layers has a substantially greater computational capacity. This was first shown formally in the late 1980s via the so-called universal approximation theorem [10–12].
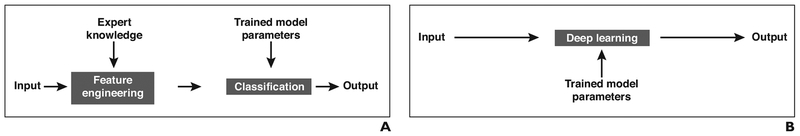
Whereas the basic architecture of artificial neural networks has a rich historical backdrop, the means of training neural networks have undergone a revolution in recent times. For example, until 2012, the predominant paradigm in ML for vision emphasized the careful design of handcrafted features [13], which were then used as the input to a trained classifier that would identify useful combinations of features [14] (Fig. 2). In contrast, the DL paradigm largely removes human experts from the feature discovery process [5]. The input to a typical DL model is the unprocessed data (e.g., image data, in the case of image categorization), and in the course of training, intermediate layers automatically learn salient features of the data while subsequent layers of the same network perform the classification (Fig. 2). Because DL models can be very large, containing many layers and very many nodes and having extremely large total numbers of trainable parameters, the essential prerequisites for DL training are the availability of both large datasets and substantial computational power. In this sense, both the network structures and the required data reserves in DL may be characterized as deep. The advent of the modern Internet, with its vast repositories of easily accessible data, as well as modern advances in computational speed and capacity have therefore been key enablers of DL.
Fig. 2—
Learning paradigms.
A, Schematic shows classic machine learning paradigm.
B, Schematic shows deep learning paradigm.
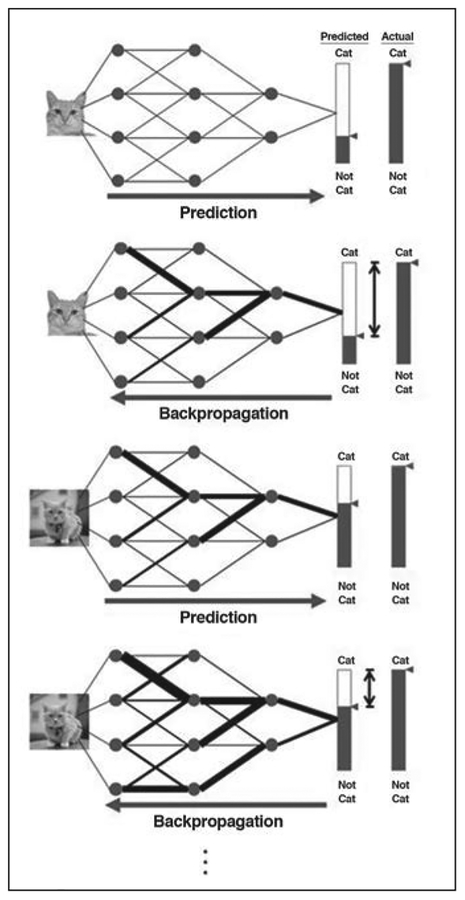
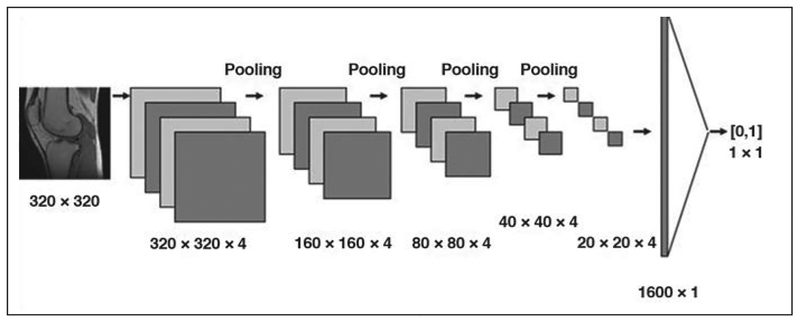
The process of training a deep neural network is as follows for the exemplary concrete task of image classification (Fig. 3). Pixels from a sample image containing items of interest (e.g., a cat or a knee) are fed as input into the nodes comprising the first layer of the network, and the data are propagated through the network using a default set of weights for the weighted addition operations previously described. This results in a numeric classification (e.g., the likelihood of the image con-taining a knee) that is compared with a ground truth label previously provided by human inspection or other suitable means. Any difference from ground truth is then used to adjust the network’s weights (e.g., with use of a procedure known as backpropagation) [15], to increase the likelihood of achieving a correct classification the next time. This process is repeated with numerous additional images from the training dataset, and the weights are progressively refined. For robustness, the training dataset is often configured to include diverse examples containing items of interest as well as counterexamples that do not contain those items. Once the weights have been suitably refined (as measured, for example, by the performance of the network on a validation dataset), they are fixed, and the network is ready to be presented with new data. As we previously implied, given the large number of data points, nodes, and weights involved, the training of a deep neural network may be computationally intensive. However, once effort has been invested up front in training the network, processing of any new data (e.g., classification of a new image) can be remarkably fast and efficient, with data simply flowing through the various layers and being subjected to simple preset operations along the way. When processing image data, one of the most important architecture classes is convolutional neural networks (CNNs). In a CNN architecture, each node is connected to only a comparatively small number of neighboring nodes. Such structures are highly efficient in extracting local features in images (Fig. 4).
Fig. 3—
Schematic shows training of deep neural network for task of image classification. Process of prediction and backpropagation is repeated for numerous examples from training dataset, with progressive refinement of weights to improve future predictions. Circles represent nodes. Black lines represent weighted connections between nodes, with thickness of each line representing magnitude of corresponding weight. Long arrow below each process denotes flow of information through network at this step. Shaded areas of bars and small triangles to right of each process show probability of input image being categorized in certain category. Areas of bars between double-headed arrows denote difference of prediction to actual ground truth that drives refining of weights in training.
Fig. 4—
Schematic of example of convolutional neural network (CNN) architecture that takes input image and performs binary classification on basis of content of image. Dimensions of images, as they are processed by CNN, are shown below images. Please note that choice of four convolutional channels in every convolution layer is arbitrary choice in this didactic example of CNN architecture. Architecture consists of five convolutional layers that alternate with pooling layers, each of which combines its input into output that is four times smaller (i.e., 320 × 320, followed by 160 × 160, 80 × 80, 40 × 40, and then 20 × 20), followed by fully connected layer. Convolutional layers perform task of feature extraction at progressively higher level. Fully connected layer performs classification. Values in brackets denote output of classification obtained by network (1 corresponding to image being corrupted by artifacts, 0 corresponding to uncorrupted image).
Imaging Appropriateness and Protocoling
Selecting the most appropriate imaging examination for a patient can be a difficult decision for a clinician. Although tools such as imaging ordering guidelines, decision support software, and virtual consult platforms are available to make this decision easier, ML can provide a more comprehensive evidence-based resource to help select the best imaging examination [2, 16]. ML algorithms can incorporate various sources of information from a patient’s medical records, including symptoms, laboratory values, physical examination findings, and prior imaging results, to recommend an appropriate patient-specific imaging examination tailored to the clinical question that needs to be answered [17].
Once the appropriate examination is ordered, it is the responsibility of the radiologist to make sure the examination is protocoled and performed correctly. Inappropriately protocoled studies can lead to suboptimal patient care and outcomes, repeat examinations that could involve additional radiation exposure, significant frustration and inconvenience both for patients and for referring physicians, and added cost to the radiology practice.
Recently, two studies have examined the use of DL for natural language classification and its potential use in automatically determining MSK protocols and the necessity of IV contrast medium [18, 19]. The study by Lee [18] showed the feasibility of using deep CNNs to classify MSK MRI examinations as following routine protocols versus tumor protocols with the use of word combinations that included “referring department,” “region,” “contrast media,” “gender,” and “age” [18]. Trivedi et al. [19] used a DL-based natural language classification system (Watson, IBM) to determine the necessity of IV contrast medium for MSK MRI examinations on the basis of the free-text clinical indication. Although both studies showed promising results for potential applications in clinical decision and protocoling support, further research will no doubt explore increasingly complex classifiers to more fully reflect the diversity of available MSK imaging protocols. For example, to determine the most appropriate protocol, ML not only could draw from information on the examination order but could also potentially mine the electronic medical record, prior examination protocols and examination reports, CT or MRI scanner data, the contrast injection system and contrast agent data, the cumulative or annual radiation dose, and other quantitative data [20].
Scheduling
No-shows and same-day cancellations represent a significant opportunity cost for radiology practices. This is especially true for advanced imaging examinations such as MRI and CT [21].
Predictive analysis with AI regression models using electronic medical record data has been used to predict imaging no-shows successfully [22]. ML algorithms have also been used to predict missed appointments in other clinical settings, ranging from diabetes clinics to urban academic centers [23, 24]. Although these prior studies used regression-based analyses of inputs to define the best predictors of the desired outcome (missed appointments), more advanced ML algorithms may identify relationships between inputs that are not presently identified and that could better predict these outcomes.
Image Acquisition and Reconstruction
Increasing the Speed of MRI Data Acquisition
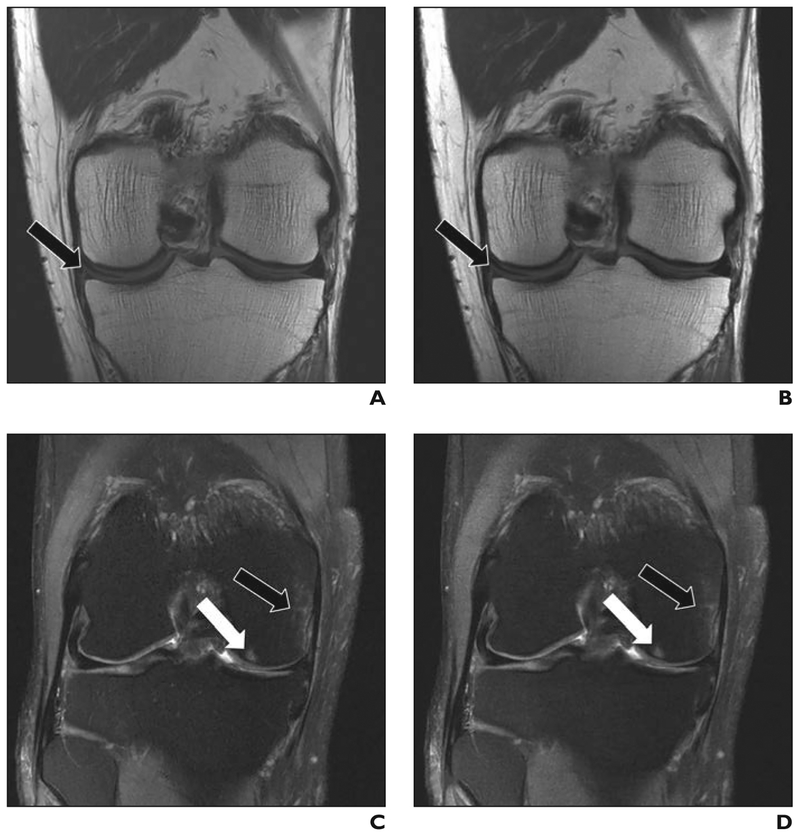
Decreasing imaging acquisition time has been a major ongoing field of research since the invention of MRI. Parallel imaging was one of the most successful developments in this field in the late 1990s and early 2000s [25–28], followed by the introduction of compressed sensing in 2007 [28]. Both of these methods achieve accelerated acquisitions by subsampling k-space, which means that the number of phase-encoding lines that are acquired during a scan is reduced below the Nyquist limit. Although this speeds up the acquisition because less data need to be collected, it also introduces artifacts in the reconstructed images. ML has been proposed as a means of solving the problem of reconstructing MR images from accelerated acquisitions [29–32]. The central idea is to learn the separation of true image content from aliasing artifacts. Early studies have shown promising results in terms of image quality and diagnostic accuracy when ML-accelerated knee MRI is compared to conventional MRI [33]. Figure 5 shows an example in which data acquisition accelerated four times faster with the use of ML reconstruction is compared with a fully sampled reconstruction with traditional reconstruction by Fourier transformation. A systematic comparison of this accelerated ML-based reconstruction of a complete clinical knee protocol with parallel imaging [34], the combination of parallel imaging and compressed sensing [35], and dictionary learning [36] can be found in a recent study by Hammernik et al. [29].
Fig. 5—
Comparison of 4-times-accelerated data acquisition with machine learning (ML) reconstruction to conventional, fully sampled (FS) clinical protocol in two anonymized patients.
A and B, Tear of medial meniscus (arrows) can be easily seen on FS (A) and ML (B) coronal proton density images. C and D, Bone contusion (black arrows) and subchondral edema (white arrows) in medial femoral condyle can be easily seen on FS (C) and ML (D) coronal fat-suppressed T2-weighted images.
Decreasing CT Radiation Dose
Continuous efforts have been made to decrease the amount of radiation that a patient is exposed to when undergoing diagnostic imaging [37, 38]. ML provides an exciting new tool for reducing the radiation dose in CT. The current ML-based techniques for radiation reduction operate in a manner similar to techniques used to increase the speed of MRI acquisition-namely, they aim to reconstruct high-quality images from reduced quantities of raw data or raw data of a reduced quality (e.g., as a result of noisy data obtained at low tube currents). In a recent study by Cross et al. [39], more than 90% of readers found that the quality of low-radiation-dose CT images, which were produced in part with the use of an artificial neural network, was equal to or greater than that of CT images obtained using standard radiation doses.
Image Presentation
Radiologists are facing ever-greater pressure to increase productivity, confronting higher daily volumes of more complex cases than they have been asked to interpret in the past [40]. Radiologists can work more efficiently if the PACS automatically displays each series in the correct preferred position, orientation, and magnification as well as the correct preferred window and level, syncing, and cross-referencing settings. Such hanging protocols should load consistently and should be accurately based on modality, body part, laterality, and time (in the case of prior available imaging). AI has the potential to revolutionize the way in which a PACS displays information for the radiologist, by using smarter tools that process a variety of available data. One PACS vendor uses ML algorithms to learn how radiologists prefer to view examinations, collect contextual data, present layouts for future similar studies, and adapt after any corrections [41]. These intelligent systems could overcome issues related to variable or missing data that may cause traditional hanging protocols to fail and can help radiologists achieve greater efficiency.
Image Interpretation
Currently, the most popular area of research within ML relates to pattern detection and image interpretation. In MSK radiology alone, ML algorithms have been applied to various conditions, including diagnosis of fractures, osteoarthritis, bone age, and bone strength [42–46].
Fractures
Two studies in the orthopedic literature have shown that deep CNNs perform equally as well as or better than orthopedic surgeons in the detection of proximal humerus, hand, wrist, and ankle fractures on radiographs [47, 48]. In a study by Chung et al. [47], the network performed well in detecting the presence of fracture in the proximal humerus, but it did not perform as well in classifying fractures according to the Neer classification. Given that the data-sets for training three- and four-part fractures were smaller, Chung and colleagues suggested that the lower accuracies for more complex proximal humerus fracture types would likely improve with more training.
ML has also been used to detect posterior element fractures and vertebral body compression fractures on CT [49, 50]. Roth et al. [49] established the feasibility of using a deep CNN to facilitate automated detection of posterior element fractures using a relatively small dataset, reporting an AUC value of 0.857 and sensitivities of 71% and 81%. The same group was also able to develop an automated ML system to detect, localize, and classify thoracic and lumbar vertebral compression fractures with the use of support vector machine regression (SVMR), reporting sensitivity of 95.7% and a false-positive rate of 0.29 per patient for the detection and localization of compression fractures [50].
Osteoarthritis
Using a binary classifier (normal vs abnormal), Xue et al. [51] reported that a CNN was able to automatically detect hip osteoarthritis on radiographs with performance comparable to that of an attending radiologist with 10 years of experience. Using a pretrained CNN model with fine-tuning for the final model on a set of 420 radiographs, the authors reported sensitivity of 95%, specificity of 90.7%, and accuracy of 92.8%, compared with a reference standard of chief physicians (defined as physicians with more than 20 years of experience).
Other studies have examined the performance of CNNs in grading the severity of knee osteoarthritis using existing large datasets, such as the Osteoarthritis Initiative (OAI) and Multicenter Osteoarthritis Study (MOST) [52–54]. Tiulpin et al. [52] used a deep CNN to automatically score the severity of knee osteoarthritis on radiographs according to the Kellgren-Lawrence (KL) grading scale, which yielded results comparable to known values of human reader agreement. The authors also provided the probability distribution of KL grades with the images, showing when the CNN may have similar predicted probabilities across two adjacent KL grades. This is not unlike real-life clinical practice, in which the severity of arthritis may not clearly be classified as one grade or another and may reflect the overlap or transition between two grades.
Antony et al. [53] attempted to address the same limitation of using the finite KL grading scale by redefining knee osteoarthritis grading as a regression rather than a classification ML problem (e.g., treating KL grading as a continuous variable, which the authors argued more accurately replicated continuous disease progression). A more recent study by the same group used CNNs to both localize the knee joint and quantify the severity of knee osteoarthritis with both multiclass classification and regression outputs [54].
Bone Age
Several studies have shown promising results of using ML to determine bone age [55–58]. Using datasets from two separate children’s hospitals, Larson et al. [55] found that their deep CNN was able to estimate skeletal maturity with accuracy comparable to that of an expert radiologist as well as to that of existing automated bone age software. Tajmir et al. [59] showed that AI-assisted radiologist interpretation performed better than AI alone, a radiologist alone, or a pooled cohort of experts, by increasing accuracy and decreasing variability and the root-mean-square error. Their findings suggest that the most optimal use of AI for determination of bone age may be in combination with a radiologist’s interpretation.
Bone Fragility
There is increasing interest in how ML can improve quantitative bone imaging for the assessment of bone strength and quality. Several recent studies have attempted to incorporate existing methods of assessing trabecular bone microarchitecture, such as geometric and textural characteristics, with the ML methods of support vector machines and SVMR [60–62].
A study by Yang et al. [60] used SVMR to predict the failure loads of ex vivo proximal femur specimens on the basis of a combination of conventional dual-energy x-ray absorptiometry bone mineral density (BMD) measurements and other methods of capturing trabecular bone microarchitecture on MDCT. These methods included statistical moments of MDCT BMD distribution, morphometric parameters like bone fraction and trabecular thickness, and geometric features derived from the scaling index method. They found that prediction of failure load was significantly improved with the addition of geometric features to supplement conventional dual-energy x-ray absorptiometry BMD distribution.
A study by Huber et al. [61] that used similar techniques showed the ability to predict trabecular bone strength of ex vivo proximal tibia specimens from knee MRI data by use of scaling index method–derived features and SVMR. They showed that the addition of scaling index method features and SVMR to the standard bone volume fraction parameter had the highest prediction accuracy when compared with bone volume fraction and linear multiple regression analysis. Ferizi et al. [63] found that random undersamling-boosted trees, logistic regression, and linear discriminant were the best ML classifiers for predicting osteoporotic fractures. This group also found that the intertrochanteric, greater trochanteric, and femoral head regions contributed the most to ML prediction performance.
These studies have shown that the combination of trabecular bone microarchitecture features and ML techniques can be used to more accurately predict biomechanical strength of trabecular bone and that ML automated segmentation has the capacity to more rapidly translate MRI-based bone structural assessment into clinical practice. Further development and refinement of these prediction and segmentation models will aid the automated and objective assessment of osteoporosis, disease progression, and treatment response.
Quantitative Image Analysis
Segmentation
Current imaging segmentation techniques, including model-, atlas-, and graph-based techniques, are limited for several reasons, including the need for large training sets, user interaction, significant time investment, and accurate robust registration technique [64]. ML can improve quantitative analysis by allowing automatic segmentation of the areas of interest, depending on the region of the body and clinical question. Attempts to use ML for the purposes of MSK image segmentation can be found as far back as 2008, in a study by Gassman et al. [65], who used artificial neural networks to successfully segment hand phalanges. Most of the recent literature has focused on knee cartilage segmentation, with promising initial results noted [66, 67]. Liu et al. [67] successfully used a combination of a deep CNN, a 3D fully connected conditional random field, and 3D simplex deformable modeling to accurately and efficiently segment the different structures of the knee, including cartilage, menisci, and bones.
ML-driven segmentation sets the stage for easier assessment of osteoarthritis progression and degeneration of important stabilizing structures, such as the meniscus, with advanced imaging sequences. One recent study by Norman et al. [68] used a DL-based model to automatically segment knee cartilage and menisci while also determining cartilage relaxometry and morphologic findings with T1-rho and 3D double-echo steady-state imaging. By use of manual segmentation and quantifications of the articular surfaces and menisci as ground truth, the model was found to be strongly accurate for both the segmentation and morphologic tasks. These techniques can also be extremely useful for outcome prediction when applied to other common MSK joint-related conditions, such as femoroacetabular impingement and anterior shoulder instability.
It is necessary to segment the proximal femur when using MRI to assess proximal fem-oral structural bone quality. Most studies have used manual segmentation of the whole proximal femur, but a recent study by Deniz et al. [69] that involved proximal femur MRI segmentation using deep CNN showed a high dice similarity score of 0.95. The dice score is a statistic used to assess the similarity of two samples, with values approaching 1 indicating higher similarity. In the case of segmentation, it is often used to compare the similarity between the algorithm-generated segmentation mask and the ground truth, which is typically manual segmentation by human experts. A dice score of 0.95 is indicative of successful automated segmentation.
Radiomics
Radiomics is an emerging field in medicine that is based on the extraction of diverse quantitative characteristics from images and the use of these characteristics for data mining and pattern identification. These data can then be used with other patient information to better characterize and predict disease processes [70]. ML techniques have led to a rapid expansion of the potential of radiomics to impact clinical care. For instance, the description of a sarcoma diagnosed on MRI will typically include estimates of tumor size, shape, and enhancement pattern. ML-driven algorithms can also identify and collect other characteristics that are not easily appreciated on images (e.g., texture analysis, image intensity histograms, and image voxel relationships) and can lead to more precise treatment [71].
One recent study applied ML-enhanced radiomics to differentiate sacral chordomas from sacral giant cell tumors on 3D CT [72]. Comparing different feature selection and classification methods, the authors found contrast-enhanced CT characteristics more useful than those from unenhanced imaging for differentiation of these two tumor types. This type of works highlights the potential for ML-enhanced radiomics in evaluating other MSK tumors as well as in guiding more precise treatment of this patient population.
Conclusion
The use of AI has the potential to greatly enhance every component of the imaging value chain. From assessing the appropriateness of imaging orders to helping predict patients at risk for fracture, AI can increase the value that MSK imagers provide to their patients and to referring clinicians by improving image quality, patient centricity, imaging efficiency, and diagnostic accuracy.
Acknowledgment
We thank Daniel Sodickson for his guidance and contributions to this manuscript.
Supported in part by grant R01 EB024532 from the National Institutes of Health for F. Knoll’s machine learning research.
References
- 1.Harkey P, Duszak R Jr, Gyftopoulos S, Rosen-krantz AB. Who refers musculoskeletal extremity imaging examinations to radiologists? AJR 2018; 210:834–841 [DOI] [PubMed] [Google Scholar]
- 2.Doshi AM, Moore WH, Kim DC, et al. Informatics solutions for driving an effective and efficient radiology practice. RadioGraphics 2018; 38:1810–1822 [DOI] [PubMed] [Google Scholar]
- 3.Poole DL, Mackworth AK, Goebel R. Computational intelligence and knowledge In: Computational intelligence: a logical approach. New York, NY: Oxford University Press, 1998: 1–22 [Google Scholar]
- 4.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521:436–444 [DOI] [PubMed] [Google Scholar]
- 5.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems, vol. 1 Lake Tahoe, NV: Curran Associates, 2012:1097–1105 [Google Scholar]
- 6.Chen LC, Papandreou G, Kokkinos I, Murphy K, Yullie AL. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 2018; 40:834–848 [DOI] [PubMed] [Google Scholar]
- 7.Dosovitskiy A, Fischer P, Ilg E, et al. FlowNet: learning optical flow with convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision (ICCV) Washington, DC: IEEE, 2015:2758–2766 [Google Scholar]
- 8.Silver D, Huang A, Maddison CJ, et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016; 529:484–489 [DOI] [PubMed] [Google Scholar]
- 9.Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev 1958; 65:386–408 [DOI] [PubMed] [Google Scholar]
- 10.Pinkus A. Approximation theory of the MLP model in neural networks. Acta Numer 1999;8: 143–195 [Google Scholar]
- 11.Cybenko G. Approximation by superpositions of sigmoidal function. MCSS 1989; 2:303–314 [Google Scholar]
- 12.Poggio T, Mhaskar H, Rosasco L, Miranda B, Liao Q. Why and when can deep–but not shallow–networks avoid the curse of dimensionality: a review. International Journal of Automation and Computing 2017; 14:503–519 [Google Scholar]
- 13.Lowe DG. Object recognition from local scale-invariant features. In: Proceedings of the Seventh IEEE International Conference on Computer Vision Kerkyra, Greece: IEEE, 1999:1150–1157 [Google Scholar]
- 14.Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995; 20:273–297 [Google Scholar]
- 15.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986; 323:6533–6536 [Google Scholar]
- 16.American College of Radiology (ACR). Appropriateness criteria. ACR website; acsearch.acr.org/list. Published 2019. Accessed April 1, 2019 [Google Scholar]
- 17.Lakhani P, Prater AB, Hutson RK, et al. Machine learning in radiology: applications beyond image interpretation. J Am Coll Radiol 2018; 15:350–359 [DOI] [PubMed] [Google Scholar]
- 18.Lee YH. Efficiency improvement in a busy radiology practice: determination of musculoskeletal magnetic resonance imaging protocol using deep-learning convolutional neural networks. J Digit Imaging 2018; 31:604–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trivedi H, Mesterhazy J, Laguna B, Vu T, Sohn JH. Automatic determination of the need for intravenous contrast in musculoskeletal MRI examinations using IBM Watson’s natural language processing algorithm. J Digit Imaging 2018; 31:245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohli M, Dreyer KJ, Geis JR. Rethinking radiology informatics. AJR 2015; 204:716–720 [DOI] [PubMed] [Google Scholar]
- 21.Mieloszyk RJ, Rosenbaum JI, Hall CS, Raghavan UN, Bhargava P. The financial burden of missed appointments: uncaptured revenue due to outpatient no-shows in radiology. Curr Probl Diagn Radiol 2018; 47:285–286 [DOI] [PubMed] [Google Scholar]
- 22.Harvey HB, Liu C, Ai J, et al. Predicting no-shows in radiology using regression modeling of data available in the electronic medical record. J Am Coll Radiol 2017; 14:1303–1309 [DOI] [PubMed] [Google Scholar]
- 23.Kurasawa H, Hayashi K, Fujino A, et al. Machine-learning-based prediction of a missed scheduled clinical appointment by patients with diabetes. J Diabetes Sci Technol 2016; 10:730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres O, Rothberg MB, Garb J, Ogunneye O, Onyema J, Higgins T. Risk factor model to predict a missed clinic appointment in an urban, academic, and underserved setting. Popul Health Manag 2015; 18:131–136 [DOI] [PubMed] [Google Scholar]
- 25.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn Reson Med 1997; 38:591–603 [DOI] [PubMed] [Google Scholar]
- 26.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med 1999; 42:952–962 [PubMed] [Google Scholar]
- 27.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002; 47:1202–1210 [DOI] [PubMed] [Google Scholar]
- 28.Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med 2007; 58:1182–1195 [DOI] [PubMed] [Google Scholar]
- 29.Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018; 79:3055–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoll F, Hammernik K, Kobler E, Pock T, Recht MP, Sodickson DK. Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magn Reson Med 2019; 81:116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 2018; 37:491–503 [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Su Z, Ying L. Accelerating magnetic resonance imaging via deep learning. In: Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI) Prague, Czech Republic: Institute of Electrical and Electronics Engineers, 2016:514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoll F, Hammernik A, Garwood K, et al. Accelerated knee imaging using a deep learning based reconstruction. In: Proceedings of the International Society of Magnetic Resonance in Medicine (ISMRM) Honolulu, HI: ISMRM, 2017:645 [Google Scholar]
- 34.Pruessmann KP, Weiger M, Börnert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Magn Reson Med 2001; 46:638–651 [DOI] [PubMed] [Google Scholar]
- 35.Knoll F, Bredies K, Pock T, Stollberger R. Second order total generalized variation (TGV) for MRI. Magn Reson Med 2011; 65:480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravishankar S, Bresler Y. MR image reconstruction from highly undersampled k-space data by dictionary learning. IEEE Trans Med Imaging 2011; 30:1028–1041 [DOI] [PubMed] [Google Scholar]
- 37.Subhas N, Pursyko CP, Polster JM, et al. Dose reduction with dedicated CT metal artifact reduction algorithm: CT phantom study. AJR 2018; 210:593–600 [DOI] [PubMed] [Google Scholar]
- 38.Subhas N, Polster JM, Obuchowski NA, et al. Imaging of arthroplasties: improved image quality and lesion detection with iterative metal artifact reduction, a new CT metal artifact reduction technique. AJR 2016; 207:378–385 [DOI] [PubMed] [Google Scholar]
- 39.Cross N, DeBerry J, Ortiz D, et al. Diagnostic quality of machine learning algorithm for optimization of low dose computed tomography. cdn.ymaws.com/siim.org/resource/resmgr/siim2017/abstracts/posters-Cross.pdf. Published 2017. Accessed April 1, 2019
- 40.McDonald RJ, Schwartz KM, Eckel LJ, et al. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad Radiol 2015; 22:1191–1198 [DOI] [PubMed] [Google Scholar]
- 41.Wang T, Iankoulski A. Intelligent tools for a productive radiologist workflow: how machine learning enriches hanging protocols. GE Healthcare website; www3.gehealthcare.com.sg/~/media/downloads/asean/healthcare_it/rdiology%20solutions/radiology%20solutions%20additional%20resources/smart_hanging_protocol_white_paper_doc1388817_july_2013_kl.pdf. Published 2013. Accessed April 1, 2019 [Google Scholar]
- 42.Roth HR, Yao J, Lu L, Stieger J, Burns JE, Summers RM. Detection of sclerotic spine metastases via random aggregation of deep convolutional neural network classifications In: Yao J, Glocker B, Klinder T, Li S, eds. Recent advances in computational methods and clinical applications for spine imaging. Basel, Switzerland: Springer International Publishing, 2015:3–12 [Google Scholar]
- 43.Golan D, Donner Y, Mansi C, Jaremko J, Ramachandran M. Fully automating Graf’s method for DDH diagnosis using deep convolutional neural networks In: Carneiro G, Mateus D, Peter L, et al. , eds. Deep learning and data labeling for medical applications. Basel, Switzerland: Springer, 2016:130–141 [Google Scholar]
- 44.Shun Miao, Wang ZJ, Rui Liao. A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging 2016; 35:1352–1363 [DOI] [PubMed] [Google Scholar]
- 45.Štern D, Payer C, Lepetit V, Urschler M. Automated age estimation from hand MRI volumes using deep learning. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention Athens, Greece: Springer, 2016:194–202 [Google Scholar]
- 46.Jamaludin A, Lootus M, Kadir T, et al. ; Genodisc Consortium. ISSLS Prize in Bioengineering Science 2017: automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J 2017; 26:1374–1383 [DOI] [PubMed] [Google Scholar]
- 47.Chung SW, Han SS, Lee JW, et al. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop 2018; 89:468–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olczak J, Fahlberg N, Maki A, et al. Artificial intelligence for analyzing orthopedic trauma radio-graphs. Acta Orthop 2017; 88:581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth HR, Wang Y, Yao J, Lu L, Burns JE, Summers RM. Deep convolutional networks for automated detection of posterior-element fractures on spine CT. arXiv website. arxiv.org/abs/1602.00020. Submitted January 29, 2016. Accessed April 4, 2019 [Google Scholar]
- 50.Burns JE, Yao J, Summers RM. Vertebral body compression fractures and bone density: automated detection and classification on CT images. Radiology 2017; 284:788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue Y, Zhang R, Deng Y, Chen K, Jiang T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS One 2017; 12:e0178992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S. Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning-based approach. Sci Rep 2018; 8:1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antony J, McGuinness K, Connor NEO, Moran K. Quantifying radiographic knee osteoarthritis severity using deep convolutional neural networks. arXiv website. arxiv.org/abs/1602.00020. Submitted September 8, 2016. Accessed April 4, 2019 [Google Scholar]
- 54.Antony J, McGuinness K, Moran K, O’Connor NE. Automatic detection of knee joints and quantification of knee osteoarthritis severity using convolutional neural networks. New York, NY: Springer, 2017 [Google Scholar]
- 55.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology 2018; 287:313–322 [DOI] [PubMed] [Google Scholar]
- 56.Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging 2017; 30:427–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JR, Shim WH, Yoon HM, et al. Computerized bone age estimation using deep learning based program: evaluation of the accuracy and efficiency. AJR 2017; 209:1374–1380 [DOI] [PubMed] [Google Scholar]
- 58.Spampinato C, Palazzo S, Giordano D, Aldinucci M, Leonardi R. Deep learning for automated skeletal bone age assessment in X-ray images. Med Image Anal 2017; 36:41–51 [DOI] [PubMed] [Google Scholar]
- 59.Tajmir SH, Lee H, Shailam R, et al. Artificial intelligence-assisted interpretation of bone age radiographs improves accuracy and decreases variability. Skeletal Radiol 2019; 48:275–283 [DOI] [PubMed] [Google Scholar]
- 60.Yang CC, Nagarajan MB, Huber MB, et al. Improving bone strength prediction in human proximal femur specimens through geometrical characterization of trabecular bone microarchitecture and support vector regression. J Electron Imaging 2014; 23:013013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huber MB, Lancianese SL, Nagarajan MB, Ikpot IZ, Lerner AL, Wismuller A. Prediction of biomechanical properties of trabecular bone in MR images with geometric features and support vector regression. IEEE Trans Biomed Eng 2011; 58:1820–1826 [DOI] [PubMed] [Google Scholar]
- 62.Sharma GB, Robertson DD, Laney DA, Gambello MJ, Terk M. Machine learning based analytics of micro-MRI trabecular bone microarchitecture and texture in type 1 Gaucher disease. J Biomech 2016; 49:1961–1968 [DOI] [PubMed] [Google Scholar]
- 63.Ferizi U, Besser H, Hysi P, et al. Artificial intelligence applied to osteoporosis: a performance comparison of machine learning algorithms in predicting fragility fractures from MRI data. J Magn Reson Imaging 2018; 49:1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedoia V, Majumdar S, Link TM. Segmentation of joint and musculoskeletal tissue in the study of arthritis. MAGMA 2016; 29:207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gassman EE, Powell SM, Kallemeyn NA, et al. Automated bony region identification using artificial neural networks: reliability and validation measurements. Skeletal Radiol 2008; 37:313–319 [DOI] [PubMed] [Google Scholar]
- 66.Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018; 80:2759–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2018; 79:2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Norman B, Pedoia V, Majumdar S. Use of 2D UNet convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 2018; 288:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deniz CM, Xiang S, Hallyburton RS, et al. Seg mentation of the proximal femur from MR images using deep convolutional neural networks. Sci Rep 2018; 8:16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278:563–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McBee MP, Awan OA, Colucci AT, et al. Deep learning in radiology. Acad Radiol 2018; 25:1472–1480 [DOI] [PubMed] [Google Scholar]
- 72.Yin P, Mao N, Zhao C, et al. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol 2019; 29:1841–1847 [DOI] [PubMed] [Google Scholar]