Abstract
Mass spectrometry imaging is a powerful tool widely used in biological, clinical and forensic research, but its often poor repeatability limits its application for quantitative and large-scale analysis. A systematic evaluation of infrared matrix-assisted laser desorption electrospray ionization mass spectrometry (IR-MALDESI-MS) repeatability in absolute ion abundances during short- and long-term experiments was carried out on liver slices from the same rat with minimal biological variability to be expected. Results of median %RSDs ranging from 14-45, pooled %RMADs ranging from 11-33 and Pearson correlation coefficients ranging from 0.83 to 1.00 demonstrated an acceptable repeatability of IR-MALDESI-MS. Normalization is commonly applied for the purpose of accounting for analytical variability of spectra generated from different runs so to reveal real biological differences. Nine data normalization strategies were performed on the rat liver data sets to examine their effects on reducing analytical variation, and further on a hen ovary data set containing more morphological features for the investigation of their impact on ion images. Results demonstrated that the majority of normalization approaches benefit data quality to some extent, and local normalization methods significantly outperforms their global counterparts, resulting in a reduction of median %RSD up to 22. Local median normalization was found to be promisingly robust for both homogeneous and heterogeneous samples.
Keywords: IR-MALDESI, Mass Spectrometry Imaging, Repeatability, Normalization
INTRODUCTION
Mass spectrometry imaging (MSI) has been gaining considerable popularity in tissue analysis due to its capability of simultaneous detection of hundreds of biomolecules while retaining their spatial distributions. The development and application of ambient MSI allows for direct tissue characterization with minimal sample pretreatment [1]. One such ambient ionization source is infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI), which combines features of both matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI). IR-MALDESI employs a pulsed mid-infrared (mid-IR) laser at 2940 nm to resonantly excite O-H stretching modes of molecules present in the sample. An exogenous ice layer can be deposited to facilitate the desorption of neutral materials from sample [2]. The high laser fluence enables complete ablation of a 10 μm thick tissue section at each spatial location with two laser pulses, therefore each pixel in the ion images are usually referred to as voxel to represent a volume element [3]. The desorbed neutrals are allowed to partition into an orthogonally oriented electrospray plume, where they are ionized via an ESI-like mechanism [4, 5]. IR-MALDESI, due to its matrix- and label-free nature, has been widely used to study biological samples ranging from single hair strands [6] to complex biological matrices such as whole neonatal mice [7].
High repeatability of analytical techniques is of prime importance for meaningful intra- and inter-sample comparison, which is particularly essential for large-scale studies where measurements are made over long time periods or where accurate quantification is required (e.g., clinical applications). However, variability in ion abundances between replicates is a widely recognized problem with MSI [8, 9]. These uncertainties present in mass spectra may compromise the precision of MSI, pose challenges for quantification, and consequently limit its usefulness as a tool for routine analysis. Attempts have been made to characterize the repeatability of various MSI techniques. The intraexperiment relative standard deviations (RSD) of protein peak abundances were reported to be 2-40% varying among studies using MALDI-MS [8]. The origins of the analytical variation in MALDI-MS are currently poorly understood, but studies have suggested that this issue could be attributed to non-uniform matrix coating, heterogeneous crystallization, variations in laser energy, and detector sensitivity [10-12]. Gurdak et al. reported average absolute abundance repeatability of 30% over 8 separate days using a desorption electrospray ionization mass spectrometry (DESI-MS) platform [13]. Impurities in electrospray solvent was proposed to possibly play an important factor in the day-to-day variation. Our previous studies demonstrated that the raw voxel-to-voxel %RSD of antiretroviral drugs in IR-MALDESI is 41-56% depending on the adducted cations [3], but the variability in ion abundances over longer time periods remains to be characterized. Additionally, the limited number of m/z features selected in some studies may bias the precise estimation of the instrument repeatability. A thorough evaluation with a significant number of features should be undertaken to allow more quantitative statistics to be derived.
Normalization is a crucial data processing step to identify and address experimental variance, hence improving the comparability of spectra generated from different measurements. The process is normally performed by subtracting an offset from a mass spectrum (optional) and then dividing by a normalization factor. The offset and normalization factor can be calculated over the full spectrum, which is known as global normalization, or over segmented m/z windows, which is known as local normalization [14]. The most frequently applied normalization techniques in MSI include total ion current (TIC) normalization, which forces all voxels in a data set to have an identical TIC value [15], and reference normalization, which scales analyte responses by a well-matched reference ion, usually an analogue of the analyte deposited uniformly over the sample (internal standard) [16, 17] or a matrix ion [18, 19]. Although they frequently correct for signal variability [3, 15, 20-22], TIC normalization and reference normalization may be prone to bias due to their inherent limitations. For example, the assumption of TIC normalization may not be fulfiled for heterogeneous samples where high-abundance but localized peaks exist [23] or when multiple classes of samples are compared. Internal standard normalization is impractical for untargeted analysis since analytes with different chemical structures have different ablation and ionization efficiencies. The fact that the optimal normalization method for a certain platform or a data set is not readily clear requires careful exploration of the selection so to ensure a reliable downstream analysis, otherwise severe data skewness can result, causing inaccurate conclusions or interpretation.
This work aims at comprehensively and quantitatively evaluating IR-MALDESI performance on yielding repeatable data from animal tissues across voxels, lines, time points and days. Then detailed comparisons were performed to illustrate the effect of a wide range of commonly used normalization strategies on minimizing data variation between technical measurements and improving ion image quality.
MATERIALS AND METHODS
Materials
HPLC-grade methanol and water were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Formic acid and acetic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). Hematoxylin and Eosin (H&E) reagents were purchased from Electron Microscopy Sciences (Hatfield, PA, USA). All chemicals were used without any purification. Nitrogen gas used for purging the MALDESI sample stage enclosure was obtained from Arc3 Gases (Raleigh, NC, USA).
Samples
Rat liver tissue samples were obtained from NCSU Department of Biological Sciences. Liver was selected for this study as a relatively homogeneous model to provide quasi-technical replicate sections with minimal biological variability. An ovary tissue coming from a healthy white leghorn commercial egg laying hen was obtained from an in-house biorepository. The ovary was an example for a heterogeneous model, of which mass spectrometry images could serve for evaluating the effect of normalization on image quality. Animals were managed in accordance with the Institute for Laboratory Animal Research Guide, and all husbandry practices were approved by North Carolina State University Institutional Animal Care and Use Committee (IACUC). Both tissues were frozen in isopentane/dry ice and stored at −80 °C until sectioning.
Tissue sections of 10 μm thickness were produced using a Leica CM1950 cryostat (Buffalo Grove, IL, USA) at a temperature of −15 or −20 °C for rat liver and hen ovary, respectively. The sections were thaw mounted onto standard glass microscope slides and kept frozen until the time of analysis. A serial section of ovary tissue was Hematoxylin and Eosin (H&E) stained then imaged by Leica LMD7000 microscope (Leica Microsystems, Buffalo Grove, IL, USA). The microscopic image was used to provide an independent evaluation of the effects of normalization by comparing the agreement between raw/ normalized ion images and the histology shown on the optical image.
IR-MALDESI-MS Analysis
Details about the in-house built IR-MALDESI source and the implementation of tissue imaging has been reported elsewhere [2, 24]. Briefly, an ice layer is formed on the surface of the tissue section. Then, a mid-IR laser at 20 Hz pulse rates of 2940 nm incident wavelength (IR-Opolette 2731, Opotek, Carlsbad, CA, USA) is focused on the sample surface to resonantly excite water, desorbing neutrals from sample. Each voxel is subjected to two laser pulses to completely ablate probed sample materials. The laser spot size on tissue was measured to be 150 μm. The desorbed neutrals partition into charged droplets of the electrospray and are converted into gas-phase ions. 50% (v/v) aqueous methanol modified by 0.2% formic acid and 1 mM acetic acid were used for electrospray solvent in positive and negative ionization mode respectively. A Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was coupled to the IR-MALDESI source for accurate mass detection. The automatic gain control function (AGC) was disabled and the injection time (IT) was fixed in imaging experiments to coordinate the laser desorption and ion acquisition events.
The rat liver data sets were acquired at a spatial resolution of 200 μm in both positive and negative ionization mode in the m/z range of 250-1000 with the resolving power of 140,000 (FWHM, m/z = 200). A 10-by-10 voxel region was sampled on each tissue section for each ionization mode, producing 100 mass spectra per section. Regions containing visible vessels and bile channels were avoided to minimize the contribution from possible biological variability. The IT was held constantly throughout the experiments as 75 ms. Lock mass re-calibration was used to achieve parts per million mass accuracy [25]. The peaks of polysiloxane at m/z 371.1012 [M + H]+ and diisooctyl phthalate at m/z 391.2843 [M + H]+ were used as lock masses in positive ionization mode. Two peaks of palmitic acid at 255.2329 [M − H]− and stearic acid at 283.2643 [M − H]− were used as lock masses in negative ionization mode.
The hen ovary-based data was acquired in positive ionization mode in the m/z range of 250-1000 with the resolving power of 140,000 (FWHM, m/z = 200). The spatial resolution of 200 μm-by-100 μm was achieved by applying the over-sampling method. More details about the experimental parameters for ovarian tissue have been described previously [26].
Data Processing
The raw data files were converted into a mzML file using MSConvert from the ProteoWizard toolkit [27], followed by conversion into an imzML image file by imzMLConverter [28]. These files were subsequently loaded, processed and visualized in MatLab (R2018a; MathWorks, Natick, MA, USA) environment using MSiReader [29, 30]. Ion images were generated with ±2.5 ppm m/z tolerance. Peak picking in MSiReader was performed using the MSiPeakfinder tool to generate a list of biological ions which mainly originated from animal tissue sections but barely from the background. Each image was queried for peaks present over a threshold abundance in at least 80% of a user-defined interrogated region (on-tissue) while either exist in less than 20% of a reference region (off-tissue), or present at an average abundance ratio of 2 or higher. The abundance thresholds for peak picking in positive and negative mode were set to be 3000 and 1000 counts⋅s−1, respectively, for the purpose of producing sufficient ion statistics and incorporating consistently existing features.
Repeatability Measurements
Four different levels of variability of rat liver-based data were evaluated in this work, including voxel-to-voxel, line-to-line, intra-day and inter-day. Signal variability across all voxels on one tissue section was assessed at voxel-to-voxel level. Line-to-line variability was measured by comparing mean spectra that were acquired from replicate scan lines on the same tissue section. 10 scan lines were acquired per sample. each line consisting of 10 non-overlapping measurements. For Intra-day and inter-day variability measurements, the mean spectrum across 100 measurements on each tissue section was calculated to create a single data vector. Mean spectra that were acquired at 8am, 12pm and 4pm during the same day were compared for intra-day variability study, while spectra acquired within 11 consecutive days were compared for inter-day variability study.
Considering the linear relationship between the mean and standard deviation with ion abundances, relative standard deviation (RSD)
and relative median absolute deviation (RMAD)
were chosen to estimate scale-independent estimations of variance, where σA is the standard deviation, μA is the mean and Ai is the abundance of an ion in the ith spectrum. %RSD quantifies the amount of variation or dispersion of data, while %RMAD serves as a supplementary estimator since it is more resilient to outliers [31]. %RSDs and %RMADs over all selected tissue-relevant peaks, including 196 cations and 101 anions, were computed, then the median %RSD value with interquartile range and pooled %RMAD at each level were presented. Pearson correlation coefficients were further calculated by
to measure the degree of linear relationship between spectrum X and Y, so to quantify their similarity [32], where Xi and Yi are the ith ion abundance in X and Y, respectively. μA and μB, σA and σB are mean and standard deviation of ion abundances in spectrum X and Y, respectively. cov(A, B) represent the covariance of spectrum X and Y.
Normalization Techniques
Nine commonly used normalization approaches based on different assumptions were specifically selected for adjusting ion abundances between measurements. For those normalization methods with the notable exception of quantile normalization, the mass spectrum is divided by a normalization factor (f). All calculations were completed in MatLab with in-house scripts which are provided in the Electronic Supplementary Material (ESM).
Total ion current normalization (TIC normalization):
where yt is the abundance of the tth peak in the spectrum. This classic and simple method based on the assumption that all measurements have equal TIC value may be limited for ambient MSI, in which numerous ions generated from environment or solvent are also detected and contribute a lot to TIC. These background ions vary over time unpredictably, causing problems performing TIC normalization.
In order to overcome the limitation of involving all detected ions, we tend to only use selected biological ions for estimating normalization factor. For this reason, all normalization methods used in this work except TIC normalization are based on biological ions that are mainly present on animal tissues. The peak picking process was detailed in “Data Processing”.
p-normalization-sum normalization, vector normalization, max normalization:
where yi is the abundance of the ith biological ion in the spectrum. Sum, vector and max normalization are special cases of p-normalization.
For p = 1, the spectrum is normalized to the sum of biological ion abundances. Sum normalization is a variant of traditional TIC normalization. The rationale that the biological constituents are expected to be constant across pixels or samples makes it more robust to the fluctuation in background ions. Sum normalization seems to be more reliable especially for IR laser-based MSI, in which probed tissue areas are completely ablated. For p = 2, the formula leads to vector normalization. It forces all spectra to have the equal length. For p → ∞, it is known as maximum normalization and normalizes spectrum based on the most abundant biological ion.
Median normalization:
Median normalization is the division of the ion abundance by the median value of ion abundances in a spectrum. Using this approach, the normalization factor is less affected by outlying ions from a statistical point of view, so median normalization is thought to be more robust than p-normalization. Although there is no theoretical basis for it, median normalization has been successfully applied on MALDI-MS data [18, 23].
Median fold change normalization (MFC normalization):
MFC normalization scales the spectrum to the median fold change of ion abundances with respect to a reference spectrum. The choice of the reference spectrum is not critical but typically the median spectrum [33]. The rationale behind this method is that non-differentially expressed molecules should have fold changes in abundances approximately to be one across different spectra [34]. However, applying MFC normalization to MSI data may be less appropriate when the distribution of fold-change regarding to the reference spectrum deviates far from symmetrical Gaussian distribution [18].
Standard deviation normalization (Std Normalization) and Median absolute deviation normalization (MAD Normalization):
Std and MAD normalization factor can be computed respectively as
and
The dispersion-based techniques are frequently used to scale microarray data with the assumption of equal spread for all arrays [35, 36]. MAD was also estimated as the noise level in MSI [23], and normalizing to MAD is performed when a consistent noise level is assumed for all spectra.
Quantile Normalization:
This non-parametric technique is motivated by the idea that the distribution of ion abundances is expected to be identical across measurements [37]. Median quantile normalization is performed by first sorting m/z variables in each spectrum by their abundances, then the median value of the highest abundances was substituted for the highest abundance in every spectrum, the median value of the next highest abundances was assigned to the next highest abundance in every spectrum, and so forth. Therefore, quantile normalization as a rank order-based method is thought to be more robust than intensity-based in some respects, but its limitations including discarding potentially informative abundance values and impracticability on tissue with different compositions may decrease its effectiveness [18].
RESULTS AND DISCUSSION
Assessments of Voxel-to-voxel, Line-to-line, Intra- and Inter-day Repeatability
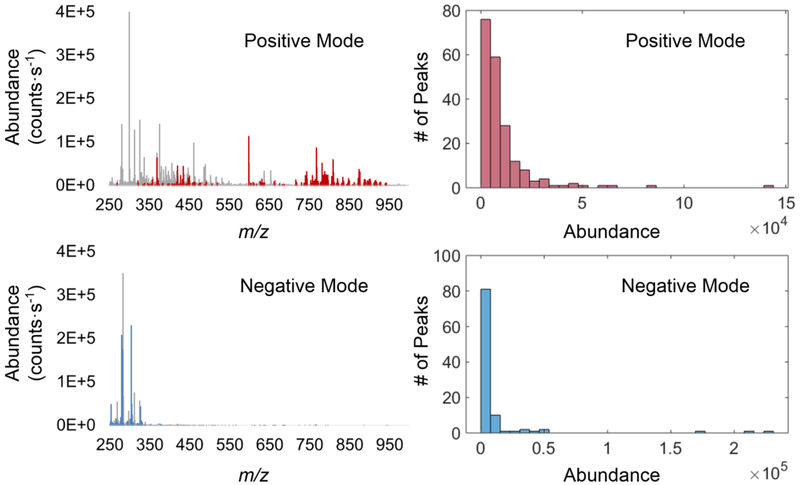
Rat liver was used as a chemically homogeneous model to study the repeatability of IR-MALDESI-MS. Molecular profiles of tissue sections were collected at different voxels (voxel-to-voxel), scan lines (line-to-line), three time points during one day (intra-day) or on 11 continuous days (inter-day). The variations in absolute abundances of selected biological ions were characterized for 196 and 101 peaks in positive and negative mode, respectively. The mean centroid mass spectra of rat liver with selected biological ions and their abundance distributions are presented in Figure 1. Abundances of representative ions which are common lipids or metabolites present in animal tissues were plotted over time to visually display the signal fluctuation (Figure 2). %RSDs and %RMADs in ion abundances of all selected biological ions were computed at different levels to represent scale-independent estimations of variance, and the median %RSDs with interquartile ranges and pooled %RMADs were shown in Table 1. Pearson correlation coefficients were further calculated to quantify the similarity of replicate spectra (ESM Fig. S1). A low variation value and a high correlation score mean a stronger similarity between replicates, thus indicating a better repeatability of the analysis.
Fig. 1.
Left: mean centroid mass spectra of rat liver acquired in positive and negative ionization mode with selected biological peaks marked in red and blue, respectively. Right: abundance (counts⋅s−1) distributions of selected biological peaks in positive and negative mode.
Fig. 2.
The sum ion abundances (counts⋅s−1) of representative ions and TIC values from rat liver sections were monitored over time. Data points within two vertical gridlines are from the same section. Top: m/z 369.3517, 435.3329, 744.5547, 792.5551, 810.6016 in positive mode; Bottom: m/z 279.2330, 303.2330, 306.0766, 329.2487, 379.0829 in negative mode. Solid brown and blue lines are Lowess smoothed lines fitted to the data points.
Table 1.
Analysis of intra-section, intra- and inter-day %RSDs and %RMAD in ion abundances.
| Intra-section | Intra-day | Inter-day | |||
|---|---|---|---|---|---|
| Voxel-to-voxel | Line-to-line | ||||
| 196 Ions in positive mode | Median %RSD | 38 | 17 | 14 | 40 |
| (IQR) | (12) | (8) | (28) | (13) | |
| Pooled %RMAD | 26 | 11 | 21 | 30 | |
| 101 Ions in negative mode | Median %RSD | 45 | 14 | 17 | 29 |
| (IQR) | (28) | (11) | (11) | (7) | |
| Pooled %RMAD | 33 | 11 | 14 | 16 | |
As an important indicator of instrument stability, the TIC values have been monitored during the acquisition period. It can be observed from Figure 2 that the TIC values from the same section remained fairly constant, while they varied from day to day, and the change is not necessarily positively associated with the abundance change of biological ions. Data points of biological ions spread within a single section with median %RSD of approximately 40%, which is in accordance with our previous study [3]. This variation may originate from a range of sources, including laser energy, stability of electrospray and ionization efficiency. Most ions have line-to-line and intra-day %RSD lower than 20%, revealing decent repeatability at these two levels. No significant change in acquisition direction indicates that stage movement does not adversely influence data quality.
For inter-day data, the relatively significant variability in positive mode with median %RSD of 40% and mean correlation coefficient of 0.83 can be possibly attributed to the fluctuation in atmosphere or accumulation of contamination across days. For example, strong signals m/z 637.3060 and m/z 666.3327 were only observed in spectra shown on day 7 (data not shown). These ions might suppress ionization and/or detection of biological signals. This hypothesis is supported by the opposite trend between TIC and biological ions from inter-day data sets in positive mode. Interestingly, data in negative mode revealed fairly constant ion abundances, which could be explained by more stable background ions. Median %RSD of 29% and mean Pearson’s coefficient of 0.99 were achieved in this ionization mode.
Effect of Normalization on Voxel-to-voxel, Line-to-line, Intra-and Inter-day Repeatability
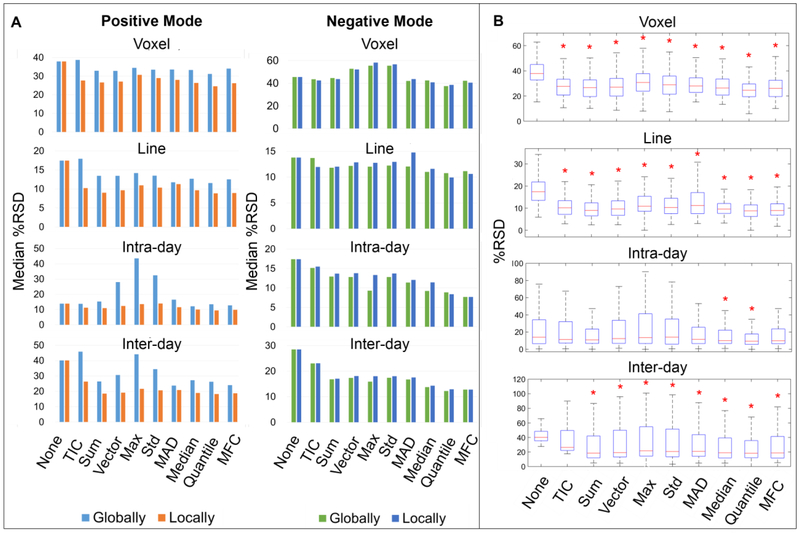
The observed variations in ion abundances emphasize the need for normalization to remove experimental bias while revealing the authentic biological features. A comparison of nine commonly used normalization techniques including TIC, sum, vector, max, std, MAD, median, quantile and MFC normalization were performed on rat liver data. The performances of these normalization methods were judged in terms of the reduction of %RSDs and %RMADs (Figure 3 and Table 2). Additionally, multiple comparison tests with Bonferroni correction were carried out at the 5% significance level to determine whether the mean %RSDs of normalized data are significantly lower than unnormalized data.
Fig. 3.
Box-plots show %RSD distributions of raw or normalized ion abundances acquired in positive (A) and negative (B) mode. From top to bottom, the variability was measured at voxel level, line level, intra-day level and inter-day level. The red asterisk indicates the mean %RSD of the normalized data is significantly lower than the corresponding unnormalized one, while the blue asterisk indicates the opposite.
Table 2.
Variation analysis before and after normalization.
| Measurements | Un-norm | Normalization Methods | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | Sum | Vector | Max | Std | MAD | Median | Quantile | MFC | ||||
| Positive mode | Voxel | Median %RSD | 38 | 39 | 33 | 33 | 34 | 33 | 34 | 33 | 31 | 34 |
| (IQR) | (12) | (14) | (17) | (15) | (12) | (13) | (21) | (17) | (16) | (24) | ||
| Pooled %RMAD | 26 | 26 | 22 | 22 | 24 | 23 | 25 | 22 | 21 | 22 | ||
| Line | Median %RSD | 17 | 18 | 13 | 13 | 14 | 14 | 12 | 13 | 12 | 13 | |
| (IQR) | (8) | (9) | (9) | (8) | (8) | (8) | (15) | (9) | (8) | (9) | ||
| Pooled %RMAD | 11 | 12 | 10 | 10 | 10 | 10 | 14 | 10 | 9 | 10 | ||
| Intra-day | Median %RSD | 14 | 14 | 15 | 28 | 44 | 32 | 17 | 12 | 13 | 13 | |
| (IQR) | (28) | (18) | (17) | (12) | (11) | (10) | (28) | (21) | (16) | (22) | ||
| Pooled %RMAD | 21 | 19 | 18 | 19 | 29 | 20 | 24 | 19 | 11 | 20 | ||
| Inter-day | Median %RSD | 40 | 46 | 26 | 31 | 44 | 34 | 24 | 27 | 26 | 24 | |
| (IQR) | (13) | (9) | (32) | (25) | (16) | (22) | (40) | (33) | (29) | (42) | ||
| Pooled %RMAD | 30 | 33 | 27 | 31 | 40 | 33 | 31 | 26 | 23 | 26 | ||
| Negative mode | Voxel | Median %RSD | 45 | 43 | 45 | 53 | 55 | 55 | 42 | 42 | 38 | 42 |
| (IQR) | (28) | (34) | (40) | (43) | (46) | (46) | (33) | (30) | (33) | (34) | ||
| Pooled %RMAD | 33 | 30 | 28 | 28 | 28 | 28 | 30 | 28 | 27 | 28 | ||
| Line | Median %RSD | 14 | 14 | 12 | 12 | 12 | 12 | 12 | 11 | 11 | 11 | |
| (IQR) | (11) | (12) | (11) | (11) | (11) | (11) | (9) | (10) | (10) | (10) | ||
| Pooled %RMAD | 11 | 11 | 10 | 10 | 10 | 10 | 11 | 10 | 7 | 10 | ||
| Intra-day | Median %RSD | 17 | 15 | 13 | 13 | 9 | 13 | 11 | 9 | 9 | 8 | |
| (IQR) | (11) | (11) | (11) | (11) | (9) | (11) | (11) | (8) | (10) | (9) | ||
| Pooled %RMAD | 14 | 13 | 13 | 13 | 13 | 13 | 14 | 13 | 6 | 13 | ||
| Inter-day | Median %RSD | 29 | 23 | 17 | 17 | 16 | 17 | 17 | 14 | 12 | 13 | |
| (IQR) | (8) | (9) | (9) | (8) | (9) | (8) | (8) | (9) | (12) | (10) | ||
| Pooled %RMAD | 16 | 16 | 14 | 15 | 14 | 15 | 14 | 14 | 10 | 13 | ||
Un-norm, unnormalized data; IQR, interquartile range; TIC, total ion current normalization; Std, standard deviation normalization; MAD, median absolute deviation normalization; MFC, median fold change normalization.
Although normalizing to TIC is one of the most common methods in mass spectrometry field, it did not help decrease signal variability in this work, especially in positive mode. This may be a result of abnormally intense background ions accounting for approximately 65-80% TIC while suppressing the biological ion abundances. Therefore, this normalization approach is not recommended for ambient MSI-based data, especially for processing data sets acquired over a long period.
Normalizing to a sum or vector of biological ions achieved significant %RSD reduction up to 14. Nevertheless, these p-normalization methods may cause dangerous artifacts when applied on chemically heterogeneous samples with highly abundant localized peaks. The artifacts will be amplified with increasing p because abundant peaks have greater impact on normalization factor with larger p. For instance, Deininger et al [23] found that the insulin signal was unusually intense in the islets of Langerhans in mouse pancreas. After normalizing to TIC or vector, the signal at m/z 14,014 which was expected to be ubiquitous in the entire mouse pancreas was decreased in the islets of Langerhans. The artifacts should be taken seriously since the reconstructed image would be in agreement with the histology. The authors suggested a possible solution by excluding these high abundance peaks before normalization, but this step requires manual interaction which is comparatively time-consuming and introduces potential operator error.
Our results show that the median, MFC and quantile normalization approaches led to considerable improvements of inter-day repeatability by reducing the median %RSD by 12-16, whereas the voxel-to-voxel, line-to-line and intra-day %RSDs were less affected. The robustness of normalizing to median signal has been reported previously [18, 23, 38]. It is robust not only to significantly changing background (unlike TIC normalization), but also to abnormally high but localized peaks (unlike p-normalization), hence is able to avoid detrimental normalization artifacts. Quantile normalization works constantly well on all data sets. It is a well-established method in microarray analysis to remove systematic bias between arrays of non-biological origin [39]. However as stated previously, quantile normalization may cause loss of informative abundance values and the assumption may not hold true for chemically heterogeneous tissues where distinct distributions are existent.
Global vs. Local Normalization
One interesting observation was made for inter-day data sets in positive mode, that the boxes of %RSDs in Figure 3A show extended ranges following most of the normalization methods, implying that some ion abundances varied even more after normalization. To characterize the ions of which variation can be reduced by normalization, the relationship between variation reduction ratio and m/z or ion abundance was assessed. Here, the variation reduction ratio of each ion was calculated as:
where a ratio less than 1 indicates normalization improved agreement. It turns out that positive ions with m/z > 600 show an overall %RSD reduction by normalization, while those with m/z < 600 generally obtain larger variability after normalization (Figure 4). The contrary behavior could be tentatively explained in terms of competitive ablation, ionization, and/or detection between these two ion groups, which is suggested by the roughly negative association for the sum of ion abundances with m/z < 600 and > 600 (ESM Fig. S2). No similar pattern was observed in negative mode.
Fig. 4.
Scatter plots of RSD ratios of normalized data to raw data versus m/z in positive mode (top) and negative mode (bottom). A data point below x=1 line indicates the ion achieves reduced inter-day variation after normalization.
It is proposed that local normalization may be more suitable for addressing spectra with non-uniformly distributed noise levels [40]. Based on what we found above, the same normalization methods with locally estimated scaling factors were investigated using m/z segments of 250 < m/z < 600 and 600 < m/z < 1000 for positive mode. Although no similar scenario was observed in negative mode, we still evaluated how local normalization performed on negative mode data, and the m/z window was selected with the criteria of similar number of features and totally different background signal present in the two windows. Therefore, 250 < m/z < 300 and 300 < m/z < 1000 for negative mode were chosen. The comparison between global and local normalization is graphically depicted in Figure 5. Local normalization methods perform generally better on positive mode data than their global counterparts, displaying lower median %RSDs and more normalized data achieving statistically significant %RSD reduction (Figure 5 and Table 3). For example, normalizing to TIC globally deteriorated the signal repeatability, while normalizing to TIC locally reduced the median %RSD from 38 to 28 (voxel-to-voxel), 17 to 10 (line-to-line), 14 to 11 (intra-day), 40 to 26 (inter-day). A similar observation can also be made when using the pooled %RMAD as the variation estimator. More details can be found in Table 3. Local quantile, median and MFC normalization were overall the optimal methods in reducing data variability, bringing the median %RSDs from 38 to 25 (across voxels) and from 40 to 18 (across days), showing comparable performance to normalization on internal standards [3]. No obvious improvements was observed for negative mode data.
Fig. 5.
Comparison of the performance of global and local normalization on reducing signal variability. From top to bottom, the variability was measured at voxel level, line level, intra-day level and inter-day level. (A) Lower median %RSDs of positive mode data were achieved by applying local normalization. No improvement was found on negative mode data. (B) More locally normalized data in positive mode obtained significantly reduced variation (marked by red asterisk) than globally normalized ones (Figure 3A).
Table 3.
Normalizing positive data locally leads to improved repeatability.
| Measurements | Un-norm | Normalization Methods | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | Sum | Vector | Max | Std | MAD | Median | Quantile | MFC | |||
| Voxel | Median %RSD | 38 | 28 | 27 | 27 | 31 | 29 | 28 | 26 | 25 | 26 |
| (IQR) | (12) | (12) | (13) | (14) | (14) | (14) | (12) | (13) | (10) | (13) | |
| Pooled %RMAD | 26 | 19 | 18 | 19 | 21 | 20 | 20 | 18 | 17 | 18 | |
| Line | Median %RSD | 17 | 10 | 9 | 10 | 11 | 10 | 11 | 10 | 9 | 9 |
| (IQR) | (8) | (6) | (6) | (7) | (7) | (7) | (10) | (4) | (5) | (5) | |
| Pooled %RMAD | 11 | 8 | 7 | 8 | 9 | 8 | 9 | 8 | 7 | 7 | |
| Intra-day | Median %RSD | 14 | 11 | 11 | 12 | 14 | 14 | 12 | 10 | 9 | 10 |
| (IQR) | (28) | (25) | (17) | (27) | (35) | (29) | (19) | (16) | (12) | (17) | |
| Pooled %RMAD | 21 | 19 | 14 | 14 | 18 | 15 | 18 | 12 | 9 | 12 | |
| Inter-day | Median %RSD | 40 | 26 | 19 | 19 | 22 | 21 | 21 | 19 | 18 | 19 |
| (IQR) | (13) | (28) | (30) | (37) | (40) | (38) | (30) | (28) | (24) | (30) | |
| Pooled %RMAD | 30 | 26 | 22 | 26 | 30 | 28 | 23 | 22 | 20 | 22 | |
Visual Inspection of Bias between Measurements
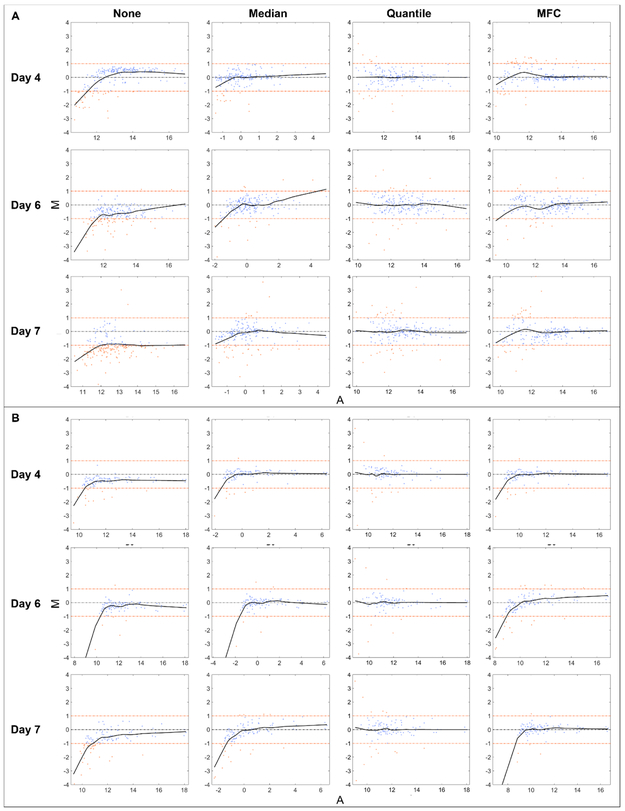
Another way of evaluating normalization procedures is by looking at their effect on minimizing differences in pairwise comparisons between spectra. MA plot is a practical tool for discerning the differences between two runs by presenting a scatter plot of the logged abundance ratios (M) versus the logged abundance products (A). M and A are calculated by
where Xi and Yi are the abundance of the ith ion in mass spectrum X and Y, respectively. [41, 42]. It can be seen that the more closely the data points are centered around M = 0, the more the two spectra are similar. In this work, MA plots of pairwise inter-day data were drawn with each normalization method. Since local normalization was found to be more successful in positive mode, we only compared positive mode data normalized locally, while negative mode data was processed with global normalization. As shown in Figure 6, the clear curvature in MA plot of unnormalized data suggests the abundance-dependent bias, while representative MA plots of normalized data show the point clouds were concentrated more tightly around x axis, the locally weighted scatter plot smooth (Lowess) lines approached M = 0 and less data points had larger than 2-fold change. MA plots of other pairs of data can be found in ESM Figs. S3-S4. These results clearly indicate improved similarities in ion abundances between experiment pairs after most normalization techniques.
Fig. 6.
Representative MA plots in (A) positive and (B) negative mode under different normalization strategies. Spectra from different days were compared with day 1. Two horizontal fold-change lines marked in orange correspond to a ratio of 1 and −1 on a log2 (Ratio) scale, showing a fold-change level of 2. Each data point represents an ion, and those with 2-fold increased or decreased abundances are located outside of the fold-change lines and appear in orange. The black solid line is the Lowess smoothed line fitted to the data points with window size of 0.5 to visually exhibit the abundance-dependent bias.
Effect of Normalization on Ion Images
Normalization was further applied on a hen ovary tissue-based data acquired in positive mode, which contains more morphological features and represents a typical imaging data set, to assess the impact of normalization on reconstructed images. Local normalization with the same m/z windows used previously was mainly evaluated here considering its excellent performance on variation reduction. The ion images for four m/z values prior to or following different normalization strategies (global or local TIC, local median) are displayed in Figure 7. See ESM Fig. S5 for the ion images using other normalization methods. Note that quantile normalization was excluded here since the sampling area included regions outside the tissue section. Applying this method will largely change the image if pixel selection is not performed first.
Fig. 7.
Comparison of ion images in positive mode from a hen ovary section before/after different normalization strategies. (A) H&E staining image of ovary tissue in parallel section; (B) TIC graph for all scans showing a dramatic signal change in the middle of experiment. Example ion images show normalizing m/z (C) 732.5563, (D) 768.5900; (E) 632.6362 and (F) 630.6197 to global/local TIC and median abundance of biological ions. Color bar shows signal from low (bottom) to high abundance (top).
It can be clearly observed that unnormalized images show significant variability in ion abundances, and experimental artifacts coming from changes in background ions, manifested as a dramatic signal change in TIC image (Figure 7B). Normalizing to TIC globally does not compensate for these anomalies, while local normalization approaches such as local TIC and median normalization substantially reduce voxel-to-voxel variation across the entire tissue section, so to allow tissue anatomy to be more readily distinguished (Figure 7C, D), improve the agreement of ion images with the stained optical image (Figure 7A), and minimize experimental artifacts (Figure 7E, F). Indeed, some normalization methods which achieved satisfactory results in rat liver-based data show unfavorable results for ovary-based data. A good example is that local MFC normalization efficiently reduces the spectral variation but leads to less accurate reconstructed images by highlighting certain voxels on the sample edge (ESM Fig. S5), which is clearly undesirable. Local TIC or median normalization are therefore recommended for processing images of heterogeneous tissues in the light of our evaluation.
CONCLUSIONS
In this study, serial rat liver tissue sections as quasi-technical replicates were analyzed by IR-MALDESI-MS to evaluate the short-term and long-term repeatability in absolute ion abundances. Minor median %RSD (<18) at line-to-line/ intra-day level and acceptable median %RSD (~40) at voxel-to-voxel/ inter-day level, along with high Pearson’s coefficient (values ranging from 0.83 to 1.00) demonstrated that IR-MALDESI is essentially a well-behaved technique with decent repeatability for profiling metabolites and lipids in animal tissues. The nonbiological variability can be efficiently reduced with appropriate normalization strategies. Normalization was found to work differently on ions with m/z < 600 and m/z > 600 in positive mode, which could be refined by using locally calculated normalization factors. Quantile, median and MFC normalization outperformed other methods in variation reduction, bringing the median %RSDs from 38 to 25 (across voxels) and from 40 to 18 (across days), showing comparable performance to normalization on internal standards. The same normalization approaches were further compared on hen ovary tissue to investigate their impact on image quality improvement. Local normalization of imaging data led to minimized experimental artifacts and voxel-to-voxel variability, hence constructed more smoothed images showing higher agreement with the optical image. This study can serve to increase confidence in applying IR-MALDESI on clinical research which typically requires high comparability and long acquisition periods.
Supplementary Material
ACKNOWLEDGEMENTS
All mass spectrometry measurements were carried out in the Molecular Education, Technology, and Research Innovation Center (METRIC) at NC State University. The authors gratefully acknowledge the financial support received from the National Institutes of Health (R01GM087964), and North Carolina State University.
Footnotes
Conflicts of Interest
Neither of the authors have any conflicts of interest.
Use of Research Animals
This study utilized tissues sourced from animals managed in accordance with the Institute for Laboratory Animal Research Guide. All husbandry practices were approved by North Carolina State University Institutional Animal Care and Use Committee (IACUC).
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Wu C, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013; 32:218–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robichaud G, Barry JA, Muddiman DC. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom. 2014; 25:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokhart MT, Rosen E, Thompson C, Sykes C, Kashuba ADM, Muddiman DC. Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal Bioanal Chem. 2015; 407:2073–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampson JS, Hawkridge AM, Muddiman DC. Generation and Detection of Multiply-Charged Peptides and Proteins by Matrix-Assisted Laser Desorption Electrospray Ionization (MALDESI) Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. J Am Soc Mass Spectrom. 2006; 17:1712–1716. [DOI] [PubMed] [Google Scholar]
- 5.Dixon RB, Muddiman DC. Study of the ionization mechanism in hybrid laser based desorption techniques. Analyst. 2010; 135:880–882. [DOI] [PubMed] [Google Scholar]
- 6.Rosen EP, Thompson CG, Bokhart MT, Prince HMA, Sykes C, Muddiman DC, et al. Analysis of Antiretrovirals in Single Hair Strands for Evaluation of Drug Adherence with Infrared-Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry Imaging. Anal Chem. 2016; 88:1336–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazari M, Bokhart MT, Muddiman DC. Whole-body Mass Spectrometry Imaging by Infrared Matrix-assisted Laser Desorption Electrospray Ionization (IR-MALDESI). J Vis Exp. 2016; 109:e53942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrethsen J Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem. 2007; 53:852–858. [DOI] [PubMed] [Google Scholar]
- 9.Douglass KA, Jain S, Brandt WR, Venter AR. Deconstructing desorption electrospray ionization: Independent optimization of desorption and ionization by spray desorption collection. J Am Soc Mass Spectrom. 2012; 23:1896–1902. [DOI] [PubMed] [Google Scholar]
- 10.Rohner TC, Staab D, Stoeckli M. MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev. 2005; 126:177–185. [DOI] [PubMed] [Google Scholar]
- 11.Gusev Al, Wilkinson WR, Proctor A, Hercules DM. Improvement of Signal Reproducibility and Matrix/Comatrix Effects in MALDI Analysis. Anal Chem. 1995; 67:1034–1041. [Google Scholar]
- 12.Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, et al. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005; 51:102–112. [DOI] [PubMed] [Google Scholar]
- 13.Gurdak E, Green FM, Rakowska PD, Seah MP, Salter TL, Gilmore IS. VAMAS interlaboratory study for desorption electrospray ionization mass spectrometry (DESI MS) intensity repeatability and constancy. Anal Chem. 2014; 86:9603–9611. [DOI] [PubMed] [Google Scholar]
- 14.Meuleman W, Engwegen JYMN, Gast MCW, Beijnen JH, Reinders MJT, Wessels LFA. Comparison of normalisation methods for surface-enhanced laser desorption and ionisation (SELDI) time-of-flight (TOF) mass spectrometry data. BMC Bioinformatics. 2008; 9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, et al. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int J Mass Spectrom. 2007; 260:212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan KD, Fang R, Yuan J, Chu RK, Dey SK, Burnum-Johnson KE, et al. Quantitative Mass Spectrometry Imaging of Prostaglandins as Silver Ion Adducts with Nanospray Desorption Electrospray Ionization. Anal Chem. 2018; 90:7246–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rzagalinski I, Volmer DA. Quantification of low molecular weight compounds by MALDI imaging mass spectrometry - A tutorial review. Biochim Biophys Acta - Proteins Proteomics. 2017; 1865:726–739. [DOI] [PubMed] [Google Scholar]
- 18.Fonville JM, Carter C, Cloarec O, Nicholson JK, Lindon JC, Bunch J, et al. Robust data processing and normalization strategy for MALDI mass spectrometric imaging. Anal Chem. 2012; 84:1310–1319. [DOI] [PubMed] [Google Scholar]
- 19.Burrell MM, Earnshaw CJ, Clench MR. Imaging Matrix Assisted Laser Desorption Ionization Mass Spectrometry: A technique to map plant metabolites within tissues at high spatial resolution. J Exp Bot. 2007; 58:757–763. [DOI] [PubMed] [Google Scholar]
- 20.Nazari M, Bokhart MT, Loziuk PL, Muddiman D. Quantitative Mass Spectrometry Imaging of Glutathione in Healthy and Cancerous Hen Ovarian Tissue Sections by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). Analyst. 2018; 143:654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Gordaliza E, Esteban-Fernández D, Lázaro A, Humanes B, Aboulmagd S, Tejedor A, et al. MALDI-LTQ-Orbitrap mass spectrometry imaging for lipidomic analysis in kidney under cisplatin chemotherapy. Talanta. 2017; 164:16–26. [DOI] [PubMed] [Google Scholar]
- 22.Lanekoff I, Thomas M, Carson JP, Smith JN, Timchalk C, Laskin J. Imaging nicotine in rat brain tissue by use of nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2013; 85:882–889. [DOI] [PubMed] [Google Scholar]
- 23.Deininger SO, Cornett DS, Paape R, Becker M, Pineau C, Rauser S, et al. Normalization in MALDI-TOF imaging datasets of proteins: Practical considerations. Anal Bioanal Chem. 2011; 401:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom. 2013; 24:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, de Godoy LMF, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per Million Mass Accuracy on an Orbitrap Mass Spectrometer via Lock Mass Injection into a C-trap. Mol Cell Proteomics. 2005; 4:2010–2021. [DOI] [PubMed] [Google Scholar]
- 26.Nazari M, Muddiman DC. Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst. 2016; 141:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics. 2008; 24:2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Race AM, Styles IB, Bunch J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J Proteomics. 2012; 75:5111–5112. [DOI] [PubMed] [Google Scholar]
- 29.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on matlab platform. J Am Soc Mass Spectrom. 2013; 24:718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bokhart MT, Nazari M, Garrard KP, Muddiman DC. MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses. J Am Soc Mass Spectrom. 2018; 29:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eldin AAH, Refaey MA. A novel algorithm for discrimination between inrush current and internal faults in power transformer differential protection based on discrete wavelet transform. Electr Power Syst Res. 2011; 81:19–24. [Google Scholar]
- 32.Puth MT, Neuhäuser M, Ruxton GD. Effective use of Pearson’s product-moment correlation coefficient. Anim Behav. 2014; 93:183–189. [Google Scholar]
- 33.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in1H NMR metabonomics. Anal Chem. 2006; 78:4281–4290. [DOI] [PubMed] [Google Scholar]
- 34.Abbassi-Ghadi N, Jones EA, Veselkov KA, Huang J, Kumar S, Strittmatter N, et al. Repeatability and reproducibility of desorption electrospray ionization-mass spectrometry (DESI-MS) for the imaging analysis of human cancer tissue: a gateway for clinical applications. Anal Methods. 2015; 7:71–80. [Google Scholar]
- 35.Fundel K, Haag J, Gebhard PM, Zimmer R, Aigner T. Normalization strategies for mRNA expression data in cartilage research. Osteoarthr Cartil. 2008; 16:947–955. [DOI] [PubMed] [Google Scholar]
- 36.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003; 31:265–273. [DOI] [PubMed] [Google Scholar]
- 37.Callister SJ, Barry RC, Adkins JN, Johnson ET, Qian WJ, Webb-Robertson BJM, et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res. 2006; 5:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Noo ME, Tollenaar RAEM, Özalp A, Kuppen PJK, Bladergroen MR, Eilers PHC, et al. Reliability of human serum protein profiles generated with C8 magnetic beads assisted Maldi-TOF mass spectrometry. Anal Chem. 2005; 77:7232–7241. [DOI] [PubMed] [Google Scholar]
- 39.Irizarry RA, Hobbs B, Collin F, Beazer-barclay YD, Antonellis KJ, Scherf U, et al. Exploration , Normalization , and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics. 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 40.Colantuoni C, Henry G, Zeger S, Pevsner J. SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics. 2002; 18:1540–1541. [DOI] [PubMed] [Google Scholar]
- 41.Välikangas T, Suomi T, Elo LL. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief Bioinform. 2018; 19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003; 19:185–193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.