Summary
Neuronal subtypes show diverse injury responses, but the molecular underpinnings remain elusive. Using transgenic mice that allow reliable visualization of axonal fate, we demonstrate that intrinsically photosensitive-retinal ganglion cells (ipRGCs) are both resilient to cell death and highly regenerative. Using RNA-seq, we show genes that are differentially expressed in ipRGCs, and that associate with their survival and axon regeneration. Strikingly, thrombospondin-1 (Thbs1) ranked as the most differentially expressed gene, along with the well documented injury-response genes, Atf3 and Jun. THBS1 knockdown in RGCs eliminated axon regeneration. Conversely, RGC-overexpression of THBS1 enhanced regeneration in both ipRGCs and non-ipRGCs, an effect that was dependent on syndecan-1, a known THBS1 binding protein. All structural domains of the THBS1 were not equally effective: the trimerization and C-terminal domains promoted regeneration while the THBS type-1 repeats were dispensable. Our results identify cell-type specific induction of Thbs1 as a novel gene conferring high regenerative capacity.
Keywords: Retinal Ganglion Cells, Axon Regeneration, Axon Growth, Axon Injury, Extracellular Matrix Protein, Thrombospondin, ipRGCs, Retina
eTOC Blurb:
Here, Bray et al. used a variety of transgenic mice to demonstrate high survival and regenerative ability of one neuronal type in the retina, and show several factors within these neurons including thrombospondin-1 and syndecan-1 that confer high regenerative capacity.
Introduction:
Projection neurons’ death and failure in regeneration are hallmark features of axonal injury, posing a major challenge for treating central nervous system (CNS) trauma and neurodegenerative conditions. Nonetheless, not all neurons respond to an injury the same way. In fact, different types of neurons come to their demise at different speeds, while other injured neurons are surprisingly resilient. Moreover, some surviving neurons can regrow their axons, contributing to partial regain of lost functions (Cafferty et al., 2008; Chen and Zheng, 2014). How these select populations of neurons deal with otherwise fatal stress and regenerate axons remains an enigma.
Retinal ganglion cells (RGCs) are a heterogeneous population of cells, grouped into different subtypes based on functional, morphological and molecular features (Sanes and Masland, 2015). RGCs exemplify classic subtype dependent-differences in responses to injury; studies in cats and mice have demonstrated that RGCs with large cell bodies belonging to the alpha type survive and regrow axons better than other RGCs (Duan et al., 2015; Watanabe et al., 1993). In effort to investigate the mechanisms underlying RGCs’ regenerative ability, previous studies have profiled gene expression in RGCs (Fischer et al., 2004; Sun et al., 2011). However, these studies were done using mixed populations of RGCs, thus unable to reveal the transcriptomic differences that are specific to the regeneration competent-RGCs.
A group of RGCs known as ipRGCs express melanopsin (Opn4) (Pickard and Sollars, 2012). They have a stunning resilience to various types of insults including axonal injury (La Morgia et al., 2010; Li et al., 2006; Perez de Sevilla Muller et al., 2014). The regenerative ability of ipRGCs has been examined previously. However, the results have varied, with some studies indicating that ipRGCs do not regenerate axons at all (Duan et al., 2015; Li et al., 2016), but others demonstrating that these neurons can regenerate axons (Lim et al., 2016), though rarely (i.e. 1% of ipRGCs) (Robinson and Madison, 2004). Since the previous studies used antibodies and transgenic lines that fail to detect all ipRGCs, we sought to use multiple Cre driver lines as a more sensitive way to track ipRGCs. To this end, we assessed ipRGC regeneration using mouse lines that have either Cre-recombinase or tamoxifen-inducible Cre (CreERT) gene inserted into the Opn4 locus. We show that ipRGCs not only are resilient to axotomy, but regenerate robustly under different conditions. RNA-seq reveals transcriptomic responses unique to ipRGCs after injury, and uncovers Thbs1 as the most highly upregulated gene compared to non-regenerative RGCs. Functional studies demonstrate that THBS1 is both necessary and sufficient to increase regeneration. In order to gain mechanistic insights into how THBS1-mediated regeneration occurs, we investigated downstream mechanisms that underlie this form of regeneration.
Results
ipRGCs are resilient to axonal injury and their axons make up a large portion of the regenerating fibers.
ipRGCs constitute about 5% of the total RGCs. They are divided into five subtypes (i.e. M1-M5), grouped based on dendrite stratification and Opn4 expression (Schmidt et al., 2011). Previously, studies had examined ipRGC axon regeneration using different approaches. Retrograde labeling of regenerated RGCs in peripheral nerve (PN) grafted-animals had shown that ipRGCs regenerate poorly with 1% of ipRGCs being able to regenerate an axon(Robinson and Madison, 2004). Mice in which GFP expression is driven by the melanopsin promoter (Opn4-GFP) (Schmidt et al., 2008) indicated that ipRGCs do not regenerate axons (Li et al., 2016). These methods detect ipRGCs that have high to moderate levels of Opn4 expression (M1-M3 ipRGCs), but lack the sensitivity to detect the remaining subtypes (Li et al., 2016). An additional limitation is the fluctuation of Opn4 expression in responses to physiological and environmental changes (e.g. circadian rhythm and light availability) (Hannibal, 2006) as well as downregulation after injury (Nadal-Nicolas et al., 2015). Thus, relying on Opn4 expression per se is unlikely to capture all regenerated ipRGCs and their axons.
To comprehensively determine ipRGCs’ axon regenerative capacity, we used Cre driver lines; both non-inducible (Opn4Cre/+) and inducible (Opn4CreERT/+) Opn4 lines crossed to Rosa26 reporter (R26-tdTomatof/f)7,8. About 5,000 tdTomato+ RGCs are labeled in the Opn4Cre/+;R26-tdTomatof/f, and about 350 tdTomato+ RGCs labeled in the Opn4CreERT/+;R26-tdTomatof/f mice (Figures 1A-1D). In the Opn4Cre/+;R26-tdTomatof/f mice, all ipRGCs and any cells that had Opn4 expression during development would be labelled. On the other hand, consistent with a previous study(Chen et al., 2011), ~80% of the RGCs labelled in the Opn4CreERT/+;R26-tdTomatof/f mouse retina are M1 ipRGCs, 12% the M2/M3 types, <1% M4, 2% M5, and the remaining being indistinguishable M1/M2/M3 types (categorized based on dendritic stratification and soma size; data not shown). Adeno-associated virus (AAV) expressing ciliary neurotrophic factor (CNTF) (Hellstrom et al., 2011) or placental alkaline phosphatase (PLAP) was injected intravitreally in Opn4Cre/+;R26-tdTomatof/f and Opn4CreERT/+;R26-tdTomatof/f mice followed by optic nerve crush. Compared to other RGCs, ipRGCs labeled by the Cre recombinase lines have significantly higher survival (Figures S1A-S1F), consistent with previous work (Perez de Sevilla Muller et al., 2014). In AAV-CNTF treated animals, substantial regeneration of tdTomato+ axons is seen in both mouse lines (Figures 1E-1J). Of the CTB+ regenerating axons, ~50% are tdTomato+ in the Opn4Cre/+;R26-tdTomatof/f mice (Figure 1G). Remarkably, in the Opn4CreERT/+;R26-tdTomatof/f mice in which the tdTomato-labeled ipRGCs constitute only about 0.5% of total RGCs, about 15% of regenerating axons are positive for tdTomato (Figure 1J). Thus, considering that ipRGCs make up only a small portion of RGCs, yet their axons make up a significant portion of regenerating axons, these results demonstrate that ipRGCs are substantially more likely to regenerate an axon relative to other RGCs.
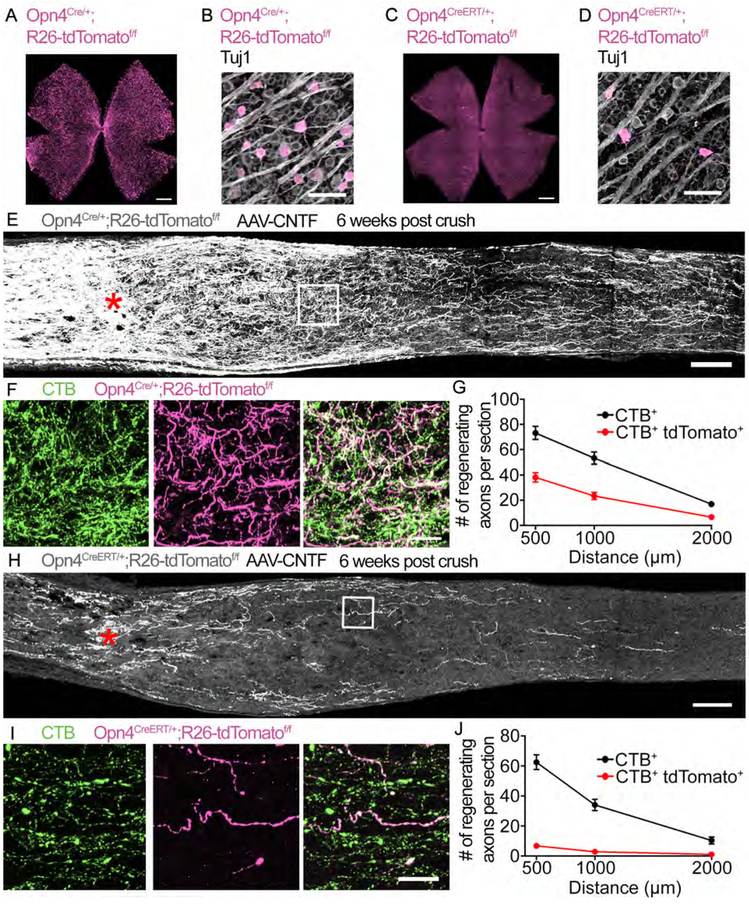
Figure 1: ipRGCs are capable of robust axon regeneration.
(A) Whole mount staining of an Opn4Cre/+;R26-tdTomatof/f retina for tdTomato (magenta).
(B) Higher magnification image showing colocalization of tdTomato+ RGCs with the pan neuronal marker Tuj1 (grey) in an Opn4Cre/+;R26-tdTomatof/f retina
(C) Whole mount staining of an Opn4CreERT/+;R26-tdTomatof/f retina for tdTomato.
(D) Higher magnification image showing colocalization of tdTomato+ RGCs with Tuj1 in an Opn4CreERT/+;R26-tdT omatof/f retina.
(E) Image of the optic nerve from Opn4Cre/+;R26-tdTomatof/f mice showing tdTomato (grey) labeled axons 6 weeks post crush. AAV-CNTF was injected intravitreally. Asterisks, lesion site.
(F) High-magnification images of the boxed area in (E), CTB (green), tdTomato (magenta), and merged image.
(G) Quantification of axon regeneration for (E). The average number of CTB+ and CTB+ tdTomato+ axons in Opn4Cre/+;R26-tdTomatof/f mice observed in each optic nerve section.
(H) Image of the optic nerve from Opn4CreERT/+;R26-tdTomatof/f mice showing tdTomato (grey) labeled axons 6 weeks post crush. AAV-CNTF was injected intravitreally.
(I) High-magnification images of the boxed area in (H), CTB (green), tdTomato (magenta), and merged image.
(J) Quantification axon regeneration for (H). The average number of CTB+ and CTB+ tdTomato+ axons in Opn4CreERT/+;R26-tdTomatof/f mice observed in each optic nerve section. n = 8 Opn4Cre/+;R26-tdTomatcff/f n = 7 Opn4CreERT/+;R26-tdTomatdf/f. wpc, weeks post crush. Error bars, SEM. Scale bars, 500 μm (A, C), 50 μm (B, D), 100 μm (E, H), 20 μm (F, I).
ipRGCs’ ability to regenerate axons under different conditions.
In the absence any regeneration-promoting treatment, a few RGC axons can nonetheless initiate spontaneous regeneration (Bray et al., 2017; de Lima et al., 2012). Thus, we examined whether ipRGCs also make up any of these spontaneously regenerating axons. Six weeks post-injury, we find a small but observable number of regenerating axons (Figures S1G-S2L). Remarkably, of the regenerating axons, ~35-50% are tdTomato+ in both ipRGC reporter lines (Figures S2I and S2L). No CTB+ axons are observed 2 mm and beyond, indicating that the crush was complete (Figures S1I and S1L). We next examined if ipRGC axon regeneration occurs under different gene treatment conditions. CNTF promotes regeneration primarily through activation of STAT3 (signal transducer and activator of transcription 3) (Leibinger et al., 2013). However, another form of regeneration is induced by PTEN (phosphatase and tensin homolog) deletion which relies on mTOR (mammalian target of rapamycin) activation (Park et al., 2008). To examine whether regeneration from ipRGCs is a common feature under the background of PTEN deletion, we generated triple transgenic mice, Opn4CreERT/+;Ptenf/f;R26-tdTomatof/f, in which PTEN is deleted conditionally in adult ipRGCs. In line with a previous study(Park et al., 2008), PTEN-deleted ipRGCs are protected from cell death (Figures 2A-2E). Moreover, significantly higher numbers of regenerating axons are found in the PTEN-deleted compared to the wild-type animals (Figures 2F-2H). The majority of these CTB+ regenerating axons are tdTomato+. These results demonstrate that ipRGCs’ high ability to regenerate axons is a feature not limited to CNTF treatment.
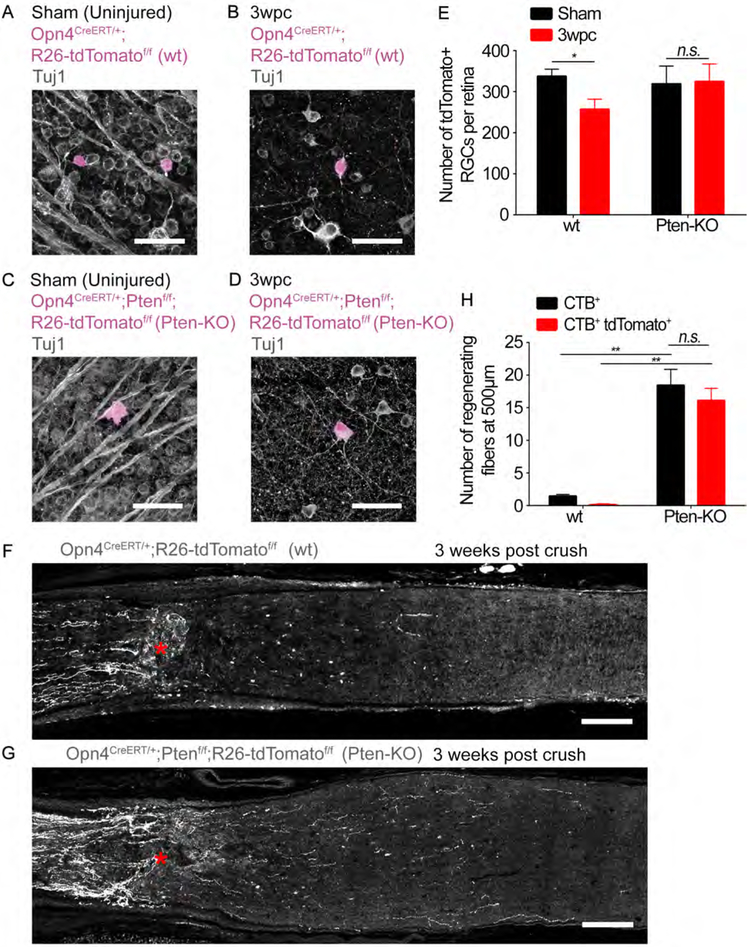
Figure 2: PTEN knockout promotes the regeneration of Opn4CreERT/+;R26-tdTomatof/f axons following optic nerve crush.
(A-D) Images of retinal whole mounts showing Tuj1-labeled RGCs (grey) and tdTomato-labeled ipRGCs (magenta). (A and B) Opn4CreERT/+;R26-tdTomatof/f (wt) or (C and D) Opn4CreERT/+;Ptenf/f;R26-tdTomatof/f (Pten-KO). (A and C) Sham uninjured (i.e. sham surgery only). (B and D) 3 weeks post crush. Scale bars, 50 μm.
(E) Quantification of RGC survival for (A-D). The number of tdTomato-labeled RGCs per retina in each condition. * p < 0.05, n.s. p ≥ 0.05, t-test 3wpc vs Sham for each group. Error bars, SEM. n =3 per group.
(G and H) Representative images of optic nerve sections showing tdTomato-labeled axons (grey) from (G) Opn4CreERT/+;R26-tdTomatof/f or (H) Opn4CreERT/+;Ptenf/f;R26-tdTomatof/f mice 3 weeks following crush. Lesion site marked by asterisks. Scale bars, 100 μm.
(F) Quantification of axon regeneration for (G and H). The number of CTB+ and CTB+ tdTomato+ axons at 500 μm distal to the lesion site. ** p<0.01, n.s. p>0.05 ANOVA with Tukey’s post-hoc. Error bars, SEM. n = 3 Opn4CreERT/+;Ptenf/f;R26-tdTomatdf/f, n = 4 Opn4CreERT/+;R26-tdTomatdf/f.
The effects of ablating known ipRGC signature genes Opn4, Igf1 and Tbr2 on ipRGC survival and axon regeneration.
We sought to examine what mechanisms contribute to the survival and regenerative abilities of ipRGCs. Melanopsin (Opn4) is a defining feature of ipRGCs. Overexpression of Opn4 was shown to promote axon regeneration in a light-dependent manner (Li et al., 2016). To determine if Opn4 is required for ipRGCs’ regenerative capacity, we used Opn4 knockout (KO) mice. AAV-CNTF was injected in Opn4Cre/+;R26-tdTomatof/f (Opn4-het) and Opn4Cre/Cre;R26-tdTomatof/f (Opn4-KO) mice. Knockout of Opn4 does not alter RGC survival or regeneration (Figures 3A-3C, Figures S2A-3E). Therefore, while ectopic overexpression of Opn4 can promote RGC axon regeneration (Li et al., 2016), these results demonstrate that Opn4 is dispensable for ipRGCs to initiate and sustain regeneration.
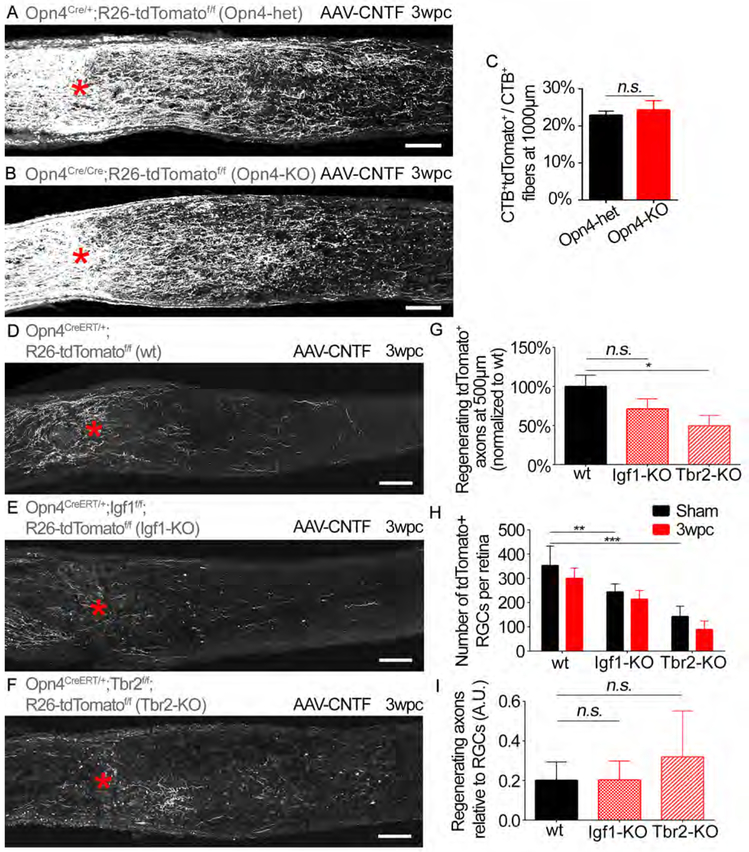
Figure 3: Assessing the effects of deleting known ipRGC signature genes on ipRGC survival and axon regeneration.
(A and B) Optic nerves from (A) Opn4Cre/+;R26-tdTomatof/f (Opn4-het) and (B) Opn4Cre/Cre;R26-tdTomatof/f (Opn4-KO) mice showing tdTomato (grey) labeled axons 3 weeks post crush. Asterisks, lesion site. Animals received AAV-CNTF injection. Scale bars, 100 μm.
(C) Quantification of axon regeneration for (A and B). Average number of CTB+ tdTomato+ axons per nerve section normalized to total CTB+ axons. t-test n.s. p>0.05. n=5 per condition.
(D-F) Optic nerves from (D) Opn4CreERT/+;R26-tdTomatof/f (wt), (E) Opn4CreERT/+;Igf1f/f;R26-tdTomatof/f (Igf1-KO), and (F) Opn4CreERT/+;Tbr2f/f;R26-tdTomatof/f (Tbr2-KO) mice showing tdTomato (grey) labeled axons. Scale bars, 100 μm.
(G) Quantification of axon regeneration for (D-F). Average number of CTB+ tdTomato+ axons per nerve section in each condition (i.e. wt, Igf1 KO and Tbr2 KO). ANOVA with Bonferroni’s post-hoc vs wt. * p≤0.05.
(H) Average number of tdTomato+ RGCs in the injured retina (“3wpc”) and uninjured retina contralateral to the injured side (i.e. “sham” surgery only) for each condition (i.e. wt, Igf1 KO and Tbr2 KO). ANOVA with Bonferroni’s post-hoc vs wt-sham. ** p≤0.01, *** p≤0.001.
(I) Axon regeneration relative to the number of surviving RGCs. ANOVA with Bonferroni’s post-hoc vs wt. n.s. p>0.05. (D-I) n=8 wt, n=6 Igf1-KO, n=7 Tbr2-KO. wpc, weeks post crush. A.U., arbitrary unit (axon/RGCs). Error bars, SEM.
Previously, two transcriptional profiling studies had identified mRNAs enriched in uninjured ipRGCs relative to other RGC types (Macosko et al., 2015; Siegert et al., 2012). Both studies identified 3 genes to be abundantly expressed in ipRGCs: Opn4, insulin like growth factor 1 (Igf1) and eomesodermin (Eomes/Tbr2). Igf1 has been shown to act in an autocrine manner, promotes neuronal survival and axon growth. Tbr2 plays a critical role in the development and maintenance of ipRGCs (Mao et al., 2014). We thus examined the requirement of Igf1 and Tbr2 in ipRGC survival and regeneration. Triple transgenic mice were generated, Opn4CreERT/+;Igf1f/f;tdTomatof/f (Igf1-KO) and Opn4CreERT/+;Tbr2f/f;tdTomatof/f (Tbr2-KO), in which Igf1 or Tbr2 are deleted conditionally in adult ipRGCs using tamoxifen injections. Knockout of Tbr2, but not Igf1 results in a relative decrease in ipRGC survival after optic nerve crush (Figures S2F-S2L), and a decrease in the number of regenerating CTB+ axons (Figures 3D-3G). We also observe that the number of ipRGCs without optic nerve crush is lower in both Igf1 and Tbr2 KO mice (Figure 3H), suggesting that Igf1, similar to Tbr2 (Mao et al., 2014) is required for the maintenance of ipRGCs. Since the baseline numbers of ipRGCs in the KO mice are lower than the wt mice, and this could have contributed to the reduction in ipRGC axon regeneration, we normalized the number of regenerating axons to the number of viable RGCs in each animal. In this case, the differences in regeneration in the KO mice compared to the wt mice are less apparent (Figure 3I). Overall, these results demonstrate that Igf1 and Tbr2 are important for the maintenance of ipRGCs, but deletion of either these genes does not eliminate ipRGC axon regeneration.
Transcriptional profiling identifies genes differentially expressed in the injured ipRGCs.
To explore the molecular basis of ipRGC survival and regeneration, we performed RNA-seq (GSE115661 and Data S1). We contrasted the profile of ipRGCs with an RGC subtype that cannot regenerate. Previously, ventrally-tuned ON-OFF Direction Selective RGCs (ooDSGCs) labeled by the HB9:GFP mouse were shown to die by two weeks following crush, and if any survived, they failed to regenerate an axon (Duan et al., 2015). Since the vast majority of ooDSGCs die, we generated HB9:GFP;Bax−/− mice to determine if ooDSGCs regenerate in the absence of cell death. We did not observe CTB+ GFP+ axons regenerating beyond the lesion (Figures S3A and S3B). As expected, GFP+ RGCs in the HB9:GFP;Bax−/− mice are completely prevented from cell death (Figures S3C-S3E), and these GFP+ RGCs are immunoreactive for cocaine and amphetamine regulated transcript (CART), a marker of DSGCs (Figures S3F and S3G). These findings confirm that that HB9:GFP RGCs (ooDSGCs) are incapable of axon regeneration, independent of their susceptibility to cell death.
Three days post-crush, retinas of Opn4CreERT/+;R26-tdTomatof/f and HB9:GFP mice were dissociated and individual fluorescently labeled RGCs were manually collected (Figure S4A). Thirty RGCs were pooled to create each sample. We chose a three day post-injury time point as spontaneous RGC axon regeneration is visible three days following injury (Bray et al., 2017). Pre-amplified polyA enriched RNA was sequenced to an average depth of 42 million reads. Transcripts for 13,406 genes were detected (i.e. expression ≥ 1 count per million in 3 of the 12 samples). Hierarchical clustering shows small intragroup differences and distinct separation of the Opn4 and HB9-RGC groups (Figure S4B). The molecular markers of ipRGCs (e.g. Opn4, Tbr2, Tbx20, Igf1 and Igfbp5) (Macosko et al., 2015) and ooDSGCs (e.g. Cartpt) show expected enrichment in the respective cell types, validating the experimental procedure (Figure S4C).
We used a series of comparisons to identify genes uniquely expressed by the Opn4-RGCs after injury in comparison to the HB9-RGC counterpart (i.e. putative novel neuroprotective and regeneration associated genes) (Figure 4A). Four genes were significantly upregulated with adjusted p-values ≤ 10−10: Jun, Atf3, Casp3, and Thbs1 (Figure 4B). 89 genes were upregulated in the Opn4-RGCs after injury, differential gene expression was defined as expression ±1 log2FC and adjusted (adj) p-value ≤ 0.05 (Figure 4C). Of those 89 genes, 17 genes were more highly expressed in injured Opn4 relative to HB9-RGCs (Figure 4D). Of the 17 genes, 6 increased with injury in HB9-RGCs, 10 did not significantly change, and 1 decreased in expression (Figure 4E). These 11 genes (10 + 1) are injury response genes distinctly expressed in Opn4-RGCs (Figure 4F). To validate the gene expression in vivo, we performed fluorescent in situ hybridization (FISH) for Thbs1. Indeed, we find that Thbs1 is expressed highly in the injured Opn4+ ipRGCs but undetectable in the Cartpt+ ooDSGCs (Figure S4D). Additionally, Cd24a is shown by our RNA-seq data to be also highly expressed in the Opn4-RGCs but expressed at a low level in the HB9-RGCs. FISH shows that Cd24a is indeed highly expressed in the injured ipRGCs but undetectable in the ooDSGCs (Figure S4E), further confirming the validity of RNA-seq results.
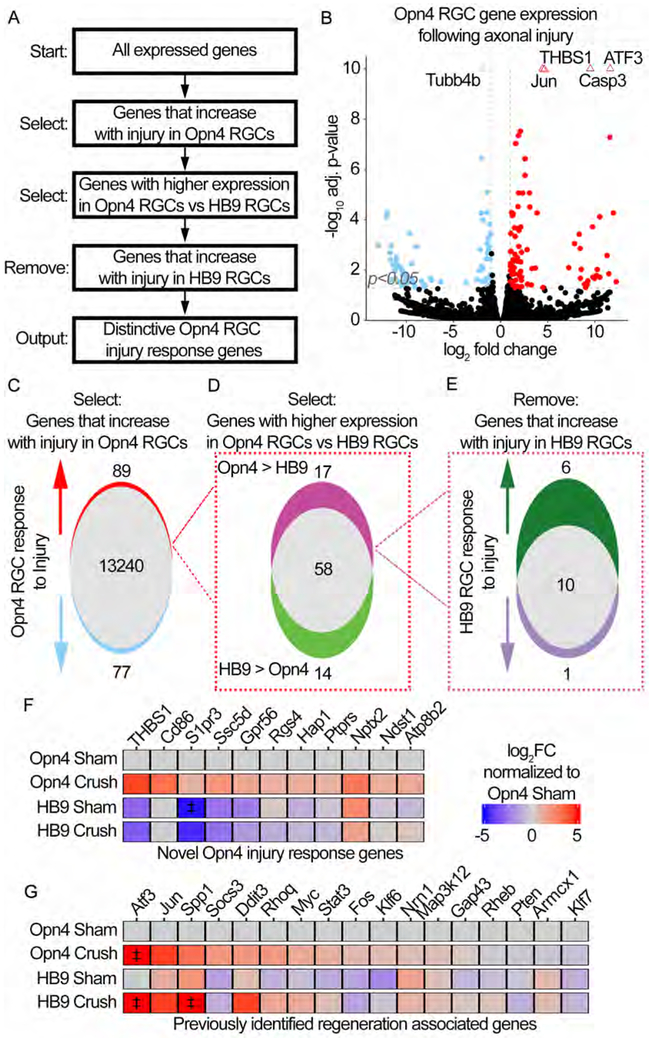
Figure 4: RNA-seq of regeneration competent ipRGCs reveals the expression of novel injury response genes.
(A) Multiple comparison methodology used to determine unique ipRGC injury response genes.
(B) Volcano plot showing differential gene expression in Opn4 RGCs following crush. Positive log2FC indicates an increase in Crush relative to Sham. Genes are considered significantly different if expression ±1 log2FC and adjusted (adj) p-value ≤0.05, vertical and horizontal reference lines at respective values. Triangles indicate genes with an adj p-value≤1*10−10, these values were fixed at 1*10−10.
(C) Venn-diagram representation of (B). 89 genes are upregulated following crush in Opn4 RGCs.
(D) Venn-diagram dividing the 89 genes from (C) based on the relative expression in injured Opn4 vs HB9 RGCs. 17 of the 89 genes are enriched in Opn4 RGCs.
(E) Venn-diagram dividing the 17 genes from (D) based on the expression change in HB9 RGCs following crush.
(F-G) Heatmaps showing the expression of indicated genes. Expression is presented as the log2FC relative to Sham Opn4 RGCs. ‡ indicates values that exceeded ±5 log2FC and were fixed at ±5 log2FC. (F) Heatmap of the 11 genes identified to be novel Opn4 injury response genes. (G) Heatmap of previously reported regeneration associated genes (RAGs).
Among previously described “regeneration associated genes” (RAGs), positive regulators of regeneration including Atf3, Jun, Spp1, cMyc and Stat3 (Chandran et al., 2016; Danzi et al., 2018) are upregulated in both Opn4 and HB9-RGCs and do not show distinct RGC type-specific expression patterns (Figure 4G). Of the KLF family members known to promote regeneration (Moore et al., 2009), Klf6, but not Klf7, is expressed at higher levels in the Opn4-RGCs relative to the HB9-RGCs (Figure 4G).
Gene set enrichment analysis shows that two of the top enriched biological processes in the injured Opn4-RGCs are “nervous system development” and “positive regulation of neuronal projection development” (Figure S5A), indicating reactivation of genes related to developmental axon growth.
Gene set enrichment analysis of injured HB9-RGCs shows that the majority of enriched GO biological processes relate to ER stress and apoptosis (Figure S5B).
Apoptosis-associated genes:
Many pro-apoptotic genes show similar levels of expression in the injured Opn4 and HB9-RGCs, except for Bad, Bax and Bnip3 which are expressed about 2-fold higher in the HB9-RGCs (Figure S5C). Of the anti-apoptotic genes, Igf1 is expressed higher in Opn4-RGCs (Figure S5D). Notably, of the genes known to ameliorate oxidative stress, glutathione peroxidase 3(Gpx3) is highly enriched in Opn4-RGCs (Figure S5E), suggesting a possible role of this gene in promoting RGC survival. Additionally, RNA-seq suggests that ipRGCs may activate ER pathways that protect them from cell death and allow axon regeneration. Differential activation of the unfolded protein response pathways has been shown to affect the fate of RGCs following axotomy; high levels of activated Xbp1 is associated with RGC survival whereas high Ddit3 and Atf4 expression is associated with RGC death (Hu et al., 2012. Injured HB9-RGCs express higher amounts of Atf4 and Ddit3; conversely, injured Opn4-RGCs express a higher level of Xbp1 (Figure S5F).
Ectopic expression of THBS1 in RGCs promotes axon regeneration.
Next, we examined whether any of the differentially expressed genes regulate RGC survival and regeneration. One possibility is that some genes highly enriched in the Opn4-RGCs might act as positive regulators, promoting cell survival and/or regeneration. In this regard, we reasoned that the 11 genes uniquely enriched in the ipRGCs derived from our stringent filtering (Figure 4F) should be good candidates. We examined 3 genes for functional analysis in vivo; Rgs4, Cd86 and Thbs1. RGS4 is involved in axonogenesis during embryogenesis (Cheng et al., 2013). CD86 is a membrane protein known to promote cell survival (Gavile et al., 2017). THBS1 is the most significantly different gene when comparing injured Opn4 and HB9-RGCs. THBS1 promotes neurite outgrowth in neurons in vitro (O'Shea et al., 1991; Osterhout et al., 1992), but the role of neuronally-expressed THBS1 is unknown.
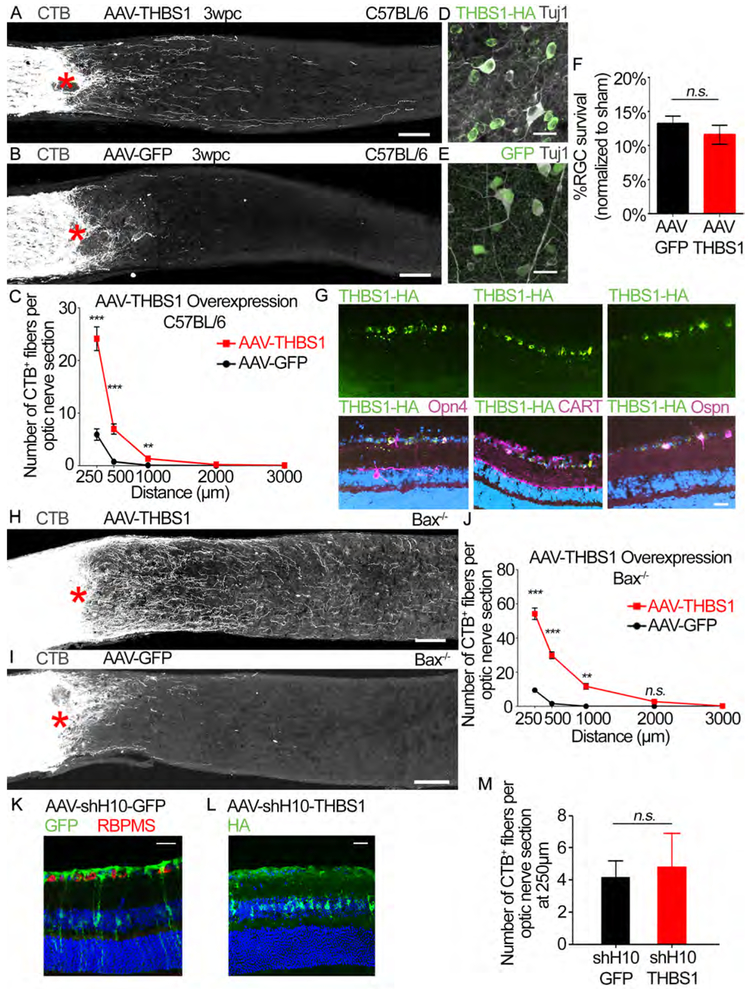
AAVs expressing the gene of interest were injected two weeks prior to optic nerve crush, leading to extensive transgene expression in RGCs (Figures S5G and S5H). AAV-RGS4 led to a small but statistically insignificant increase in regeneration. There was no increase in regeneration after AAV-CD86 injection (Figures S5I-S5K). We then evaluated if THBS1 overexpression induces axon regeneration. We observed a significant increase in regeneration in the AAV-THBS1 treated animals (Figures 5A-5C). We also examined RGC survival; THBS1 overexpression does not protect RGCs (Figures 5D-5F). Immunostaining of retinas shows that AAV-THBS1 used in this study causes THBS1 overexpression predominantly in RGCs among the retinal cell types (Figure 5G). We also observe that THBS1 is overexpressed in ipRGCs as well as other RGC types (Figure 5G), demonstrating that the ectopic THBS1 expression is not specific to one particular RGC type. To examine the degree of regeneration in the absence of RGC death, we injected AAV-THBS1 in the Bax−/− mice. In these animals, the regeneration is even more robust with many axons regenerating beyond 1 mm from the lesion site compared to the control animals (Figures 5H-5J). Since THBS1 normally acts as a secreted factor, we sought to examine whether overexpression of THBS1 in non-RGCs also promotes regeneration. To this end, we generated AAV serotype shH10 with GFAP promoter known to induce transgene expression predominantly in Müller cells. Consistent with previous studies (Klimczak et al., 2009), we confirmed that intravitreal injection of AAV-shH10-GFP and AAV-shH10-THBS1 induce transgene expression mostly in Muller cells and not in RGCs (Figures 5K and L). In line with the previous study (Klimczak et al., 2009), we estimate that approximately 20-40% of Muller cells express the transgene after AAV injection. AAV-shH10-THBS1 however, did not increase axon regeneration (Figure 5M). These results suggest that THBS1 promotes axon regeneration via intrinsic mechanisms within RGCs.
Figure 5: Ectopic THBS1 expression promotes RGC axon regeneration.
(A and B) Images of optic nerve sections showing CTB-labeled axons (grey) in C57BL/6J mice injected with either (A) AAV-THBS1 or (B) AAV-GFP. Asterisks, lesion site. Scale bars, 100 μm.
(C) Quantification of axon regeneration for (A and B). Average number of CTB+ fibers per optic nerve section at indicated distances from the lesion. n=10 per condition
(D and E) Representative retina whole-mount images showing Tuj1+ (grey) and (D) THBS1-HA (green) or (E) GFP labeled cells in C57BL/6J mice shown in (A and B). Scale bars, 25 μm.
(F) Quantification of RGC survival for (D-E). Average % survival of RGCs (Tuj1+ RGCs) in injured retina normalized to uninjured (sham) retina. n=10 per condition.
(G) Representative retinal sections from AAV-THBS1-HA injected mouse stained with antibodies against HA (green) and indicated RGC subtype markers (magenta; Opn4, CART and Ospn which are markers of ipRGCs, DSGCs and alpha RGCs, respectively). Dapi in blue. Scale bar, 25 μm.
(H and I) Images of optic nerve sections showing CTB-labeled axons (grey) in Bax−/− mice injected with either (H) AAV-THBS1 or (I) AAV-GFP. Asterisks, lesion site. Scale bars, 100 μm.
(J) Quantification of axon regeneration for (H and I). Average number of CTB+ fibers per optic nerve section at difference distances from lesion site. n=5 per condition.
(K) Representative retinal section from AAV-shHIO-GFP injected mouse stained with GFP (green) and RBPMS (RNA-Binding Protein With Multiple Splicing, a RGC marker; red) antibodies. Dapi in blue. Note that GFP expression is predominantly in non-RGCs. Scale bar, 25 μm.
(L) Representative retinal section from AAV-shH10-THBS1-HA injected mouse stained with HA antibody. Dapi in blue. Scale bar, 25 μm.
(M) Quantification of axon regeneration for the AAV-shH10 animals in (K and L). Average number of CTB+ fibers per optic nerve section at 250 μm from the lesion site. n=6 for AAV-shH10-GFP and 7 for AAV-shH10-THBS1.
Statistics, (C, and J) Unpaired t-test, 2 tailed, at each distance; (F and M) Unpaired t-test, 2 tailed, n.s. p>0.05, *p≤0.05, **p≤0.01, ***p≤0.001, Error bars, SEM.
RGC expression of THBS1 is indispensable for RGC axon regeneration.
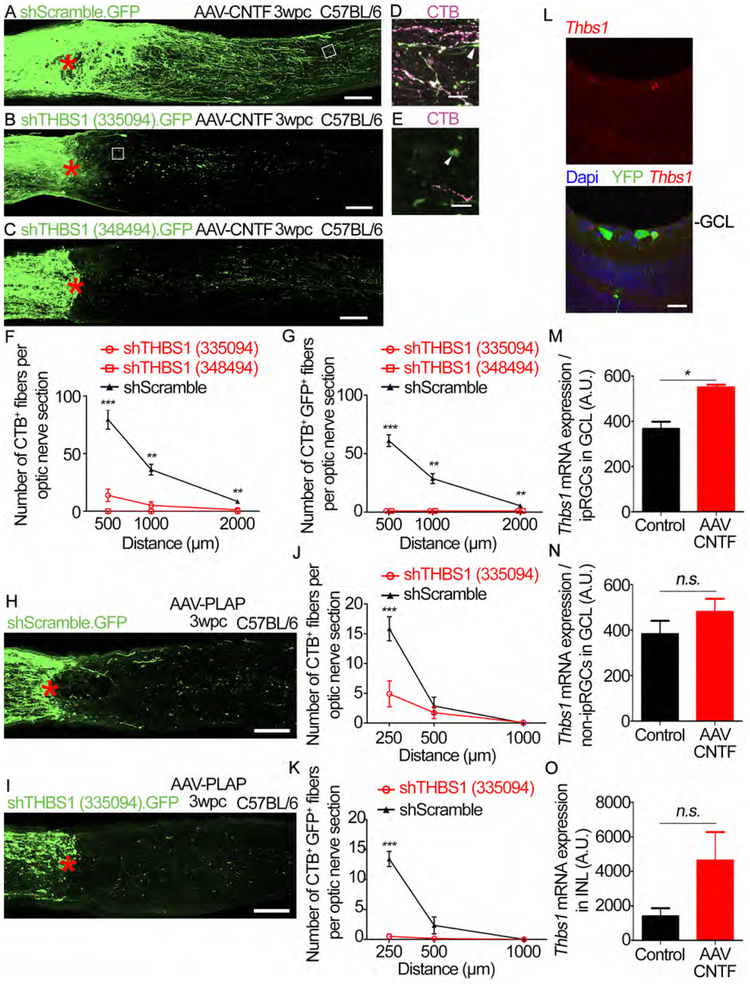
Next, we examined whether THBS1 in RGCs is necessary for regeneration. AAVs carrying dual expression cassettes for shRNAs and eGFP were given two weeks prior to injury, and AAV-CNTF or AAV-PLAP were injected prior to injury. We validated knockdown of THBS1 with two independent shRNAs (Figures S6A-S6D). These AAV-shRNAs block AAV-CNTF induced-axon regeneration (Figures 6A-6G). Furthermore, THBS1 knockdown blocks spontaneous regeneration that occurs without CNTF (Figures 6H-6K). THBS1 knockdown abolishes the pro-survival effects of AAV-CNTF (Figures S6E-SG and S6J). On the other hand, THBS1 knockdown does not alter RGC survival in the absence of CNTF (Figures S6H-S6J). We also examined whether AAV-CNTF induces Thbs1 expression. Using two color-FISH, we observe that CNTF increases Thbs1 expression in Opn4+ ipRGCs after injury (Figures 6L and 6M). There is a small but insignificant increase in the non-ipRGC cells in the ganglion cell layer after AAV-CNTF injection (Figure 6N). We observe that Thbs1 expression is scarce in the inner nuclear layer (INL) where multiple cell types are located including the Müller cells. There is a higher level of Thbs1 expression in the INL after AAV-CNTF (Figure 6O). These results demonstrate that CNTF increases Thbs1 expression in some RGCs including ipRGCs after axotomy. We further confirmed that THBS1 is required for axon regeneration independent of any effects on cell survival by using Bax−/− mice (Figures S6K-S6N). These results demonstrate that THBS1 expression in RGCs is required for RGC axon regeneration.
Figure 6: THBS1 expression is required for RGC axon regeneration.
(A) An image of optic nerve section showing GFP-labeled axons (green) in mice co-injected with AAV-CNTF and AAV-shScramble.GFP. Asterisk, lesion site.
(B) An image of optic nerve section showing GFP-labeled axons in mice co-injected with AAV-CNTF and AAV-shTHBS1 (335094). GFP.
(C) An image of optic nerve section showing GFP-labeled axons in mice co-injected with AAV-CNTF and AAV-shTHBS (348494). GFP.
(D) Higher magnification of the boxed region in (A). CTB (magenta), GFP (green).
(E) Higher magnification of the boxed region in (B). CTB (magenta). GFP (green).
(F) Quantification of axon regeneration for (A-E). Average number of CTB+ fibers per optic nerve section at various distances from the lesion site.
(G) Quantification of axon regeneration for (A-E). Average number of CTB+ GFP+ fibers per optic nerve section at various distances from the lesion site.
(H) An image of optic nerve section showing GFP-labeled axons (green) in mice co-injected with AAV-PLAP and AAV-shScramble.GFP.
(I) An image of optic nerve section showing GFP-labeled axons (green) in mice co-injected with AAV-PLAP and AAV-shTHBS1(335094).GFP.
(J) Quantification of axon regeneration for (H and I). Average number of CTB+ fibers per optic nerve section at various distances from the lesion site.
(K) Quantification of axon regeneration for (H and I). Average number of CTB+ GFP+ fibers per optic nerve section at various distances from the lesion site.
(L) Representative retinal section from an AAV-CNTF injected Opn4Cre/+;Rosa26-YFPf/f mouse. Fluorescent in situ hybridization (FISH) was performed on retina sections 3 days post-crush. Thbs1 mRNA (red), YFP protein expression, Dapi in blue. GCL, ganglion cell layer. Scale bar, 25 μm.
(M) Quantification of Thbs1 mRNA expression in ipRGCs (i.e. YFP+RGCs) in AAV-CNTF-treated or animals without AAV injection (“Control”) presented as fluorescent signal intensity. A.U., arbitrary unit.
(N) Quantification of Thbs1 mRNA expression in non-ipRGCs (i.e. YFP− RGCs) in the ganglion cell layer of AAV-CNTF-treated or animals without AAV injection (“Control”) presented as fluorescent signal intensity.
(O) Quantification of Thbs1 mRNA expression in the inner nuclear layer (INL) in AAV-CNTF-treated or animals without AAV injection (“Control”) presented as fluorescent signal intensity. Statistics, (F and G) ANOVA with Bonferroni’spost-hoc vs shScramble; (JandK) Unpaired t-test; n=5 per condition. (M-O) Unpairedt-test, n=2 per condition. * p≤0.05, ** p≤0.01, *** p≤0.001. Error bars, SEM. Scale bars, 100 μm (A-C, H and I), 25 μm (D, E and L).
THBS1’s regenerative effects require trimerization and C-terminal domain, but not the type 1 signature repeats.
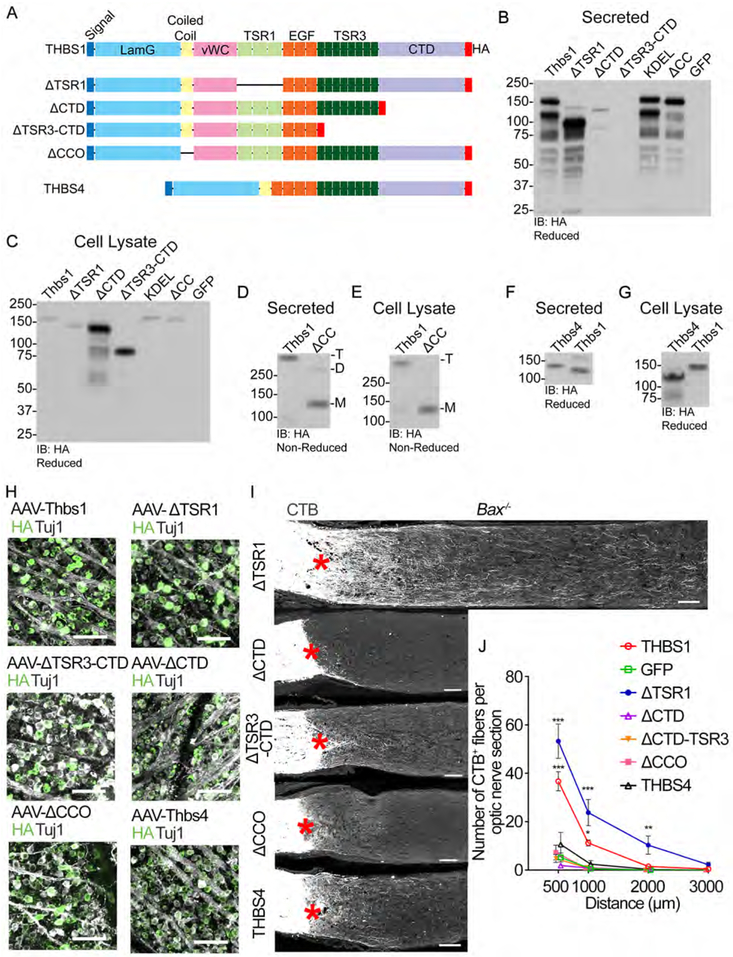
The neurite outgrowth-promoting effects of THBS1 in vitro has been attributed to the type 1 signature repeats (TSR1) domain (Osterhout et al., 1992). To further examine how THBS1 promotes axon regeneration, we examined the contribution of different domains within THBS1. To this end, we generated AAVs to express THBS1 with deletion of either the type 1 signature repeats (ΔTSR1), C-terminal domain (ΔCTD), CTD and type 3 signature repeats (ΔCTD-TSR3) or the coiled-coil oligomerization domain (ΔCCO) (Figure 7A). Using western blot analyses, we validated the size of the truncated proteins as well as the lack of oligomerization for the ΔCCO form (Figures 7B-7G). In retina, we confirmed that these mutant THBS1 proteins are indeed expressed in RGCs following AAV injection (Figure 7H), demonstrating that the mutant forms are translated despite the mutations. To examine the effects of various versions of THBS1, AAVs for each construct were given to Bax−/− mice. As described above, full length THBS1 promotes significant axon regeneration. AAV-ΔTSR1 also increases axon regeneration compared to the GFP controls, demonstrating that TSR1 domain is not required to promote axon regeneration (Figures 7I and 7J). In contrast, both AAV-ΔCTD and ΔAAV-CTD-TSR3 fail to promote regeneration. AAV-ΔCCO also fails to promote regeneration, suggesting that oligomerization is needed for THBS1 to promote regeneration (Figures 7I and 7J).
Figure 7: THBS1’s regenerative effects require trimerization and CTD, but not TSR1 domains.
(A) A schematic of THBS1 mutants investigated. All constructs contain the N-terminal signal peptide and have a C-terminal HA tag. Laminin G domain (LamG), oligomerization coiled coil (CC) domain, von Willebrand complex like domain (vWC), thrombospondin type 1 repeat domain (TSR1), epidermal growth factor-like repeat domains (EGF), type 3 repeat domain (TSR3), and the thrombospondin C-terminal domain (CTD). THBS4 is shown for comparison to THBS1.
(B-E) Validation of THBS1 constructs. Wild-type and mutant THBS1 vectors were expressed in HEK293T cells. All blots are immunoblotted (IB) for HA. (B and D) Secreted proteins from conditioned media. (C and E) Proteins from cell lysate. (B and C) Expression of THBS1, THBS1-ΔTSR1, THBS1-ΔCTD, THBS1-ΔTSR3-CTD, THBS1-KDEL, THBS1ΔCC, and GFP. Proteins reduced with DTT. THBS1-KDEL (nuclear exclusion sequence) was included to see if this form retains THBS1 expression inside the cells (refer to Discussion). (D and E) THBS1 and THBS1ΔCC were blotted under non-reducing conditions to validate oligomerization. Predicted oligomerization: M: monomeric, D: dimeric, and T: trimeric.
(F and G) Size validation of THBS4 construct. (F) Secreted proteins from conditioned media. (G) Proteins from cell lysate.
(H) Representative retinal whole mount images from mice injected with AAVs expressing various THBS forms stained for HA (green) and Tuj1 (grey). Scale bars, 100 μm.
(I) Images of optic nerve sections showing CTB-labeled axons (grey) in mice injected with AAV-THBS1ΔTSR1, AAV- THBS1ΔCTD, AAV- THBS1ΔTSR3-CTD, AAV- THBS1ΔCC or AAV-THBS4. Asterisks, lesion site. Scale bars, 100 μm.
(J) Quantification of axon regeneration for (I). Average number of CTB+ fibers per optic nerve section. ANOVA with Bonferroni’s post-hoc vs AAV-GFP, * p≤0.05, ** p≤0.01, *** p<0.001. n=5 for AAV-GFP, AAV-THBS1, AAV-THBS1ΔCTD, and AAV-THBS4, 4 for AAV-THBS1ΔTSR1, AAV-THBS1ΔTSR3-CTD, and AAV-THBS1ΔCC. Error bars, SEM.
We next investigated if THBS1 action is conserved in another thrombospondin family member. THBS4 lacks the vWC and TSR1 domains, has 4 EGF repeats, and forms a pentamer36. RNA-seq shows that Thbs4 is neither expressed in the ipRGCs nor ooDSGCs before and after injury. AAV-THBS4 does not promote axon regeneration (Figures 7I and 7J). Collectively, these results demonstrate that the effects of THBS1 on regeneration require at a minimum, the C-terminal and CCO domains, whereas the TSR1 domain shown previously to be important for THBS1 to promote neurite outgrowth in vitro is dispensable.
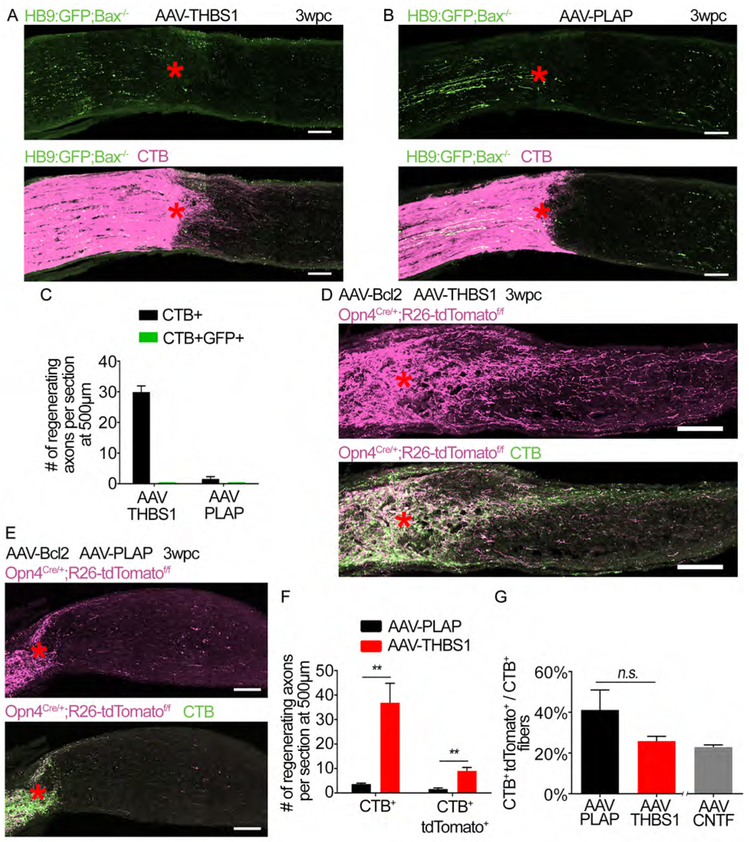
Ectopic overexpression of THBS1 promotes regeneration in ipRGCs as well as non-ipRGCs.
Is THBS1 expression capable of promoting axon regeneration only in ipRGCs? Next, we asked if THBS1 promotes the regeneration of specific RGC subtypes. To examine whether ooDSGCs is responsive to THBS1 overexpression, we injected AAV-THBS1 in the HB9:GFP;Bax−/− mice. We did not observe CTB+ GFP+ axons beyond the lesion site (Figures 8A-8C). Therefore, THBS1 is not sufficient to promote regeneration in ooDSGCs. These results indicate that ooDSGCs lack factors other than THBS1 needed to regenerate an axon. Next, we tested if regeneration by THBS1 overexpression is restricted to ipRGCs. To this end, we injected AAV-THBS1 in the Opn4Cre/+;R26-tdTomatof/f mice. AAV-Bcl2 was co-injected to prevent RGC death (i.e. as an alternative to generating a triple mutant, Opn4Cre/+;R26-tdTomatof/f;Bax−/−). At three weeks post crush, THBS1 induces significant regeneration of both CTB+ and CTB+ tdTomato+ axons (Figures 8D-8F). The percentage of regenerating axons that arise from ipRGCs is not significantly different between treatments (Figure 8G). These results demonstrate that THBS1 overexpression does not selectively promote regeneration in ipRGCs.
Figure 8: Ectopic overexpression of THBS1 promotes regeneration in ipRGCs as well as non-ipRGCs.
(A) Images of optic nerve section showing GFP-labeled axons (green) from HB9:GFP;Bax−/− mice and CTB (magenta) following injection with AAV-THBS1 and optic nerve crush. Asterisks, lesion site.
(B) Images of optic nerve section showing GFP-labeled axons (green) from HB9:GFP;Bax−/− mice and CTB (magenta) following injection AAV-PLAP. Asterisks, lesion site.
(C) Quantification of axon regeneration for (A and B). The number of CTB+ and CTB+ GFP+ axons at 500 μm distal to the lesion site. n=3 per condition. Error bars, SEM.
(D) Images of optic nerve section showing tdTomato (magenta) labeled axons from Opn4Cre/+;R26-tdTomatof/f mice and CTB in green. Mice were co-injected with AAV-Bcl2 and AAV-THBS1. Asterisks, lesion site.
(E) Images of optic nerve section showing tdTomato (magenta) labeled axons from Opn4Cre/+;R26-tdTomatof/f mice and CTB in green. Mice were co-injected with AAV-Bcl2 and AAV-PLAP. Asterisks, lesion site.
(F) Quantification of axon regeneration for (D and E). Average number of CTB+ and CTB+ tdTomato+ axons per optic nerve at 500 μm distal to the lesion site.
(G) Percentage of CTB+ tdTomato+ relative to CTB+ axons in each condition. An “AAV-CNTF” bar from Figure 3C (i.e. Opn4Cre/+;R26-tdTomatof/f mice treated with AAV-CNTF) is included as a comparison to AAV-THBS1 treatment.
Statistics in (F and G), t-test n.s. p>0.05, ** p≤0.01, n=3 for AAV-THBS1, 4 for AAV-PLAP. Scale bars, 100 μm. Error bars, SEM.
Examination of the involvement of ATF6, mTOR and STAT3 on THBS1-induced regeneration.
In the CNS, THBS1 is known widely as an astrocyte-derived factor during development that promotes synapse formation (Christopherson et al., 2005; Eroglu et al., 2009). How does neuronally expressed THBS1 promote axon regeneration? Like other extracellular matrix (ECM) molecules (e.g. laminin and vitronectin), THBS1 is an excellent substrate for neurite adhesion (Neugebauer et al., 1991; O'Shea et al., 1991). We sought to examine whether overexpressed THBS1 (i.e. THBS1-HA) is present in the axons close to the lesion site, perhaps acting locally to promote regeneration. Immunohistochemistry using HA antibody failed to detect THBS1-HA in the RGC axons of AAV-THBS1-HA injected, optic nerve crushed animals. On the other hand, using western blot as a more sensitive way to detect protein expression, we did observe THBS1-HA in the optic nerve tissues of AAV-THBS1-HA injected, optic nerve crushed animals (data not shown), indicating that THBS1-HA is transported to the axons. Using FISH, we also observe that Thbs1 expression is induced in the injured optic nerve, mostly likely in the macrophages (Figures S7A and S7B). Hence, although overexpressed THBS1 is likely present in the axons, the fact that THBS1 and many other ECM molecules are expressed in the injury site (i.e. and could support axon adhesion) (Chamak et al., 1994), it is questionable if RGC-derived THBS1 promotes regeneration by enhancing axon adhesion locally in the injured optic nerve. AAV expressing vitronectin (Neugebauer et al., 1991) in RGCs fails to promote regeneration (Figures S7C and S7D), indicating that the THBS1’s effects on axon regeneration is not a common feature shared by all adhesion-promoting ECM proteins. Notably, the results above showing that THBS1 overexpression in Müller cells does not enhance regeneration indicate that THBS1 might promote regeneration in a cell-autonomous manner. Previous studies have demonstrated intracellular roles of the thrombospondin members in regulating biological functions (Lynch et al., 2012); THBS1 and THBS4 localize to ER and promote activation of ER-resident ATF6, resulting in upregulation of protective genes against ER stress. Deletion of ATF6 blocked this effect, demonstrating a critical role of ATF6 in mediating THBS’ intracellular functions (Lynch et al., 2012). To examine the involvement of intracellular THBS1-ATF6 signaling in promoting regeneration, we deleted ATF6 in adult RGCs. AAV-Cre mediated knockout of ATF6 does not reduce regeneration, (Figures S7E-S7G and S7I-S7K). RGC survival is unchanged after ATF6 deletion (Figures S7H and S7L). We also observe that THBS1 overexpression neither increases the numbers of RGCs with high mTOR activity (i.e. indicated by phosphorylated S6 (pS6) immunoreactivity) (Park et al., 2008) nor expression of phosphorylated STAT3 (pSTAT3) after axonal injury (Sun et al., 2011) (Figures S7M-S7P), suggesting that THBS1-induced effect is unlikely to be due to enhancing these two known positive regulators of regeneration.
THBS1-induced axon regeneration requires syndecan-1.
Since many proteins can bind to THBS1, the multifaceted action of THBS1 depends on the availability of THBS1’s different ligands (Adams and Lawler, 2004; Resovi et al., 2014). We find that many of the genes known to bind to THBS1 and mediate THBS1’s functions (Resovi et al., 2014) are enriched in the Opn4-RGCs. These genes include syndecans, insulin-like growth factor binding proteins and integrins (Figure S8A) (Edwards and Hammarlund, 2014; Guo et al., 2018; Nieuwenhuis et al., 2018). Syndecans are transmembrane proteins known to mediate multiple biological functions including cell adhesion, growth and migration. While syndencan-1 and THBS1 are co-expressed in many tissues (Corless et al., 1992), their co-expression has not been investigated previously in neurons. Interestingly, we see that syndecan-1 is one of the most highly enriched THBS1 ligands in the ipRGCs (Figure S8A). Since previous studies have pointed to a role of syndecan-1 in mediating axon growth (Edwards and Hammarlund, 2014; Murakami et al., 2015), we examined whether syndecan-1 contributes to THBS1-induced regeneration. Strikingly, expression of shRNAs against syndecan-1 (Figure S8B; two different shRNAs tested) in RGCs completely eliminates THBS1-induced regeneration (Figures S8C and S8D). We also observe that syndecan-1 knockdown eliminates spontaneous regeneration that occurs in the absence of AAV-THBS1 (Figures S8C and S8D). RGC survival was examined, and we find that survival is higher in animals treated with one of the shRNAs tested compared to shRNA scramble controls (Figure S8E), indicating that loss of axon regeneration is unlikely to be a result of increase in RGC death. Taken together, these results demonstrate that syndecan-1 expression is specific to some RGC subtypes including ipRGCs, and that syndecan-1 is required for THBS1-induced axon regeneration.
Discussion:
ipRGCs’ regenerative capacity were examined previously with varying results. An Opn4 antibody approach indicated that ipRGC axons regenerate poorly into a PN graft (Robinson and Madison, 2004). Opn4-GFP mice showed that ipRGCs do not regenerate in the background of PTEN/SOCS3 deletion (Li et al., 2016). Interestingly, Lim et al., also used Opn4-GFP mice, but demonstrated that ipRGCs do regenerate axons (no quantitative data) when regeneration is stimulated by visual stimulation (Lim et al., 2016). It is also worth noting that Duan et al. used a transgenic mice that label all αRGCs (Duan et al., 2015), demonstrating that regenerating RGC axons after PTEN deletion are derived mostly from αRGCs. Since a subtype of ipRGCs (M4 ipRGCs) makes up a portion of αRGCs (i.e. ON αRGCs) (Sanes and Masland, 2015; Schmidt et al., 2011), the previous result suggests that at least some M4 ipRGCs may regenerate their axons. Taken together, the varying degrees of regeneration ability seen previously are likely due not only to the different approaches used to identify ipRGCs but perhaps also to the types of regeneration-inducing treatments used. In the case of Robinson et al. study, a PN graft which contains Schwann cells known to secrete a repertoire of growth factors and highly permissive for axon growth may have induced regeneration perhaps more robustly in several RGC types other than ipRGCs.
To systemically determine the ability of ipRGCs to regenerate an axon, we tested different growth promoting-strategies in two different Cre driver lines. Using a line that labels all ipRGCs, we determined that they make up a significant portion of the regenerating axons. We further corroborated this finding using an inducible CreERT line that labels predominantly the M1 and M2/3 ipRGCs. Since only a very few M4 ipRGCs are labelled in the CreERT line, it is likely that M1-M3 ipRGCs make up the regenerated axons seen in this mouse line. Thus, in addition to αRGCs already described previously (Duan et al., 2015; Watanabe et al., 1993), M1-M3 ipRGCs represent additional subsets that possess a high regenerative capacity. In support of this, it has been demonstrated that the M1 ipRGCs normally (and after injury) express a high level of mTOR activity (Li et al., 2016). We note that we were unable to determine the precise contribution of each ipRGC subtype making up the regenerating axons. Nonetheless, our results establish that ipRGCs collectively make up a large population of the spontaneously regenerating axons and those under CNTF stimulation.
Here we have demonstrated that neuronal expression of THBS1 is critical for RGC axon regeneration. Notably, among the five thrombospondin family members, RNA-seq shows that only Thbs1 is expressed, indicating that Thbs1 is under a unique transcriptional control and has an exclusive ability as a thrombospondin member to promote regeneration. How does THBS1 promote RGC axon regeneration? It seemed plausible that THBS1 acts extracellularly for subpopulations of RGCs. However, counter to this idea is that glial-expression of THBS1 failed to promote regeneration. These results are consistent with THBS1’s action being cell-autonomous in RGCs. We also note, in order to examine the role of intracellular vs. extracellular THBS1 in promoting regeneration, we have also tested overexpression studies using THBS1 with KDEL sequence (i.e. sequence which prevents the protein from being secreted from the ER) but this was insufficient to prevent the secretion of THBS1 when transfected in primary neurons, thus we were unable to test whether restricting THBS1 inside the cells affects its ability to promote regeneration. Since recent studies demonstrated that THBS1 and THBS4 function intracellularly through activating ATF6 (Lynch et al., 2012), we examined whether this intracellular pathway is involved. However, our results demonstrate that regeneration is not reduced after ATF6 deletion, arguing against a critical role of intracellular THBS1-ATF6 pathway in mediating RGC axon regeneration.
Syndecan-1 has been shown to promote axon regeneration in different model systems. In C. elegans, sdc1 mutant axons have defects in regeneration due to collapse of growth cones. It is thought that syndecan-1 stabilizes growth cones via modulating cytoskeletal proteins intracellularly (Edwards and Hammarlund, 2014). Others have shown that knocking down sdc1 in sensory neurons decreases neurite extension (Murakami et al., 2015). Notably, binding of THBS1 to syndecan-1 has been shown to stimulate cell migration, an effect that requires trimeric assembly of the THBS1’s C-terminal region (Adams et al., 2001; Anilkumar et al., 2002). This function of THBS1 and syndecan-1 is achieved by organization of actin and fascin bundles (Adams et al., 2001; Anilkumar et al., 2002), a process also critical for growth cone formation and axon growth. Thus, our results showing cell-type specific expression of Thbs1 and sdc1 in RGCs, together with the complete elimination of regeneration after sdc1 knockdown raise the possibility that this protein pair may participate in promoting RGC axon regeneration. One possibility is that THBS1 binding to syndecan-1 intracellularly (and possibly by interacting with other molecules whose expression is also enriched in ipRGCs) strengthens growth cone stabilization, leading to successful axon regeneration. Having said that, whether THBS1 directly interacts with syndecan-1 to promote axon regeneration remains to be determined. The site of THBS1’s actions and the downstream mechanisms that involve syndecan-1 also warrant further investigation.
It is worth commenting the differential effects of THBS1 on RGC survival. THBS1 overexpression did not increase overall RGC survival in wt mice after injury. On the other hand, THBS1 knockdown eliminated the increase in RGC survival seen after CNTF treatment. It is plausible that injured RGCs typically rely on multiple factors other than THBS1 for survival whereas THBS1 might play a more critical role in the case of CNTF treatment. It has been shown previously that the degree to which different intracellular signaling pathways contribute to RGC survival is different in RGCs depending whether or not they are treated with CNTF (Park et al., 2004). Thus, the mechanisms that confer RGC survival are complex, and might involve different factors for RGCs under different conditions.
RNA-seq in the two contrasting population of RGCs (i.e. high probability to regenerate vs. death- prone) in this study provides a valuable resource for probing the mechanisms that underlie the cell survival and regeneration following CNS injury. Although our initial screen led to characterization of THBS1, many of the unique genes seen in the ipRGCs (or in ooDSGCs) after injury are understudied in relation to regulating neuronal survival and regeneration. It remains to be seen whether these genes also play a role in these processes.
In summary, our study demonstrated a previously unappreciated regenerative ability of ipRGCs and revealed transcription of genes unique to these RGCs. Our results indicate that neuronal expression of THBS1 together with its ligands might represent an alternative strategy to improve axon regeneration.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kevin K. Park at kpark@miami.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal experimental procedures were performed in compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Miami. Animals used are C57BL/6J (Jackson Laboratory stock number: 000664), Opn4Cre and Opn4CreERT (a gift from Dr. Samer Hattar, Johns Hopkins University), Ptenf/f (Jackson Laboratory, stock number: 006440), R26 loxP-STOP-loxP-tdTomato (R26-tdTomato, a gift from Dr. Fan Wang, Duke University), HB9:GFP (Jackson Laboratory, stock number: 005029), Igf1f/f (Jackson Laboratory, stock number 016831), Tbr2f/f (Jackson Laboratory, stock number 017293), Bax−/− (Jackson Laboratory, stock number 002994), and Atf6f/f (Jackson Laboratory, stock number 028253). Glt1-eGFP (a gift from Dr. Jeffrey Rothstein, Johns Hopkins University). All animals were housed in a viral antigen free facility and kept under standard 12-hour light-dark conditions. For all surgical procedures, mice were anaesthetized with ketamine and xylazine. For analgesia buprenorphine (0.05 mg/kg) was administered post-operatively. Animals of both sexes were used.
Cell lines
HEK293T cells and Neuro-2a cells
Neuro-2a cells (ATCC; #CCL-131) were cultured in Eagle’s MEM medium (Sigma-Aldrich) supplemented with 10% FBS (Thermo Fisher Scientific). HEK293T cells (ATCC; #CRL-3216) were maintained in DMEM (Thermo Fisher Scientific) with 10% FBS (Thermo Fisher Scientific). The medium also contained 100 IU/mL penicillin (Lonza), and 100 μg/mL streptomycin (Lonza). Cells were maintained in a humidified incubator at 37°C with 5% CO2.
Mouse embryonic fibroblasts
Mouse embryonic fibroblasts (MEFs-Sufu null) were a gift from Dr. Anthony Capobianco and were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were maintained in a humidified incubator at 37°C with 5% CO2.
The cell lines used in this study were not further authenticated and not found to be on the list of commonly misidentified cell lines (International Cell Line Authentication Committee).
METHOD DETAILS
Injury and injections
Optic nerve crush
For the optic nerve crush procedure, the left optic nerve was exposed intra-orbitally by blunt dissection. The optic nerve was crushed with forceps (#5 Dumont, Fine Science Tools) for 10 seconds ~1 mm distal to the emergence from the globe. Animals received optic nerve crush at various time points after AAV injection as indicated for each experiment. For animals with quantification of RGC number, sham surgery was performed on the contralateral (right) side. Sham surgery involved exposure of the optic nerve without nerve crush. These contralateral eyes are referred in the main text as “Sham (uninjured)” retina. Following various time points after crush, animals were humanely euthanized and tissues processed for further analyses. Time points are indicated in each figure legend and described also in the main text.
Intravitreal injection
Female and male mice 6 to 8 weeks old underwent unilateral AAV injection. A fine glass micropipette was inserted into the posterior chamber taking care to avoid damaging the lens. Using a Hamilton syringe (Hamilton 80900) 2 μl of virus was slowly injected. Anterograde labeling of regenerating axons was performed by injecting 2 μl of 2 μg/μl Alexa-488 or Alexa-555 conjugated cholera toxin β subunit (CTB) (ThermoFisher, C22841 and C22843) 2-3 days before euthanasia. For in vivo experiment with intravitreal co-injection of two AAVs, 1 ul of each AAV was mixed together prior to injection.
Tamoxifen injection
Injections were started at about postnatal day 21 (for all experiments involving the Opn4CreERT mice), intravitreal injections and optic nerve crush experiments began at postnatal 6-8 weeks old. Procedure time-lines are outlined in the text and figure legends. The exact number of animals used for each group is indicated in the main text and figure legends. Opn4CreERT mice received intraperitoneal injection of tamoxifen (0.124 mg/g body weight) for 5 consecutive days. The mice were given a 2-week washout period prior to starting experimental procedures.
General considerations for experimental design
Replication
Unless stated otherwise, every in vivo experiment to quantitate axon regeneration and RGC survival was conducted once. Exceptions include AAV-THBS1 overexpression experiment to examine regeneration and survival with the experiment testing various THBS1 mutants as described in the Results.
Sample size estimation
We did not use statistical methods to pre-determine sample sizes, but our sample sizes are similar to those generally used in the field.
Animal Exclusion
Samples were excluded if lens shows signs of injury or if eyes have signs of infection, cataract, or degeneration.
Blinding and randomization
The evaluator of RGC, axon, immune-reactive cell counts was blind to the specific treatment in each eye for the following in vivo experiment; AAV-shSDC1 experiment to examine SDC1 knockdown effects on RGC survival and regeneration, ATF6 floxed mice experiment, and pS6 and pSTAT3+ RGC quantification.
Cell and tissue staining
Immunohistochemistry/Immunocytochemistry
Mice were euthanized by transcardial perfusion with ice cold PBS and 4% paraformaldehyde. The optic nerve was cut proximal to the optic chiasm. The globe with attached optic nerve were post-fixed in 4% paraformaldehyde overnight at 4°C. For retinal whole mount staining, the retina was carefully dissected out of the globe and derestricting cuts were placed in each quadrant. For cyrosectioning the optic nerve or globe was placed in 30% sucrose PBS for 48 hours at 40C. Tissue for sectioning was embeded (Tissue-Tek O.C.T.) and sectioned: 12 μm for the optic nerve and 20 μm for the globe. Tissue was washed with PBS, 5 × 5min (sections) or 5 × 30min (whole mount) at room temperature and blocked in 5% normal donkey serum in PBS + 0.3% Triton-X100 (PBST) for 1 hour at room temperature. Primary antibodies in blocking solution, 40C overnight: Rabbit anti-CART (55-102) 1:500 (Phoenix Pharmaceuticals H-003-62), Rabbit anti-pS6 1:500 (Cell Signaling #2211), Rabbit anti-pSTAT3(Y705) (Cell Signaling #9131), Rabbit anti-pSmad2 1:500 (Cell Signaling #8828), Mouse anti-Thrombospondin 1:50 (Santa Cruz, sc-59887, A6.1), Goat anti-GFP 1:1000 (Abcam ab6673), Rabbit anti-GFP 1:1000 (Millipore, ab3080), Chicken anti-Beta-3 tubulin 1:500 (Abcam ab107216), Rabbit anti-Beta-3 tubulin 1:500 (Tuj1, Sigma-Aldrich, T2200), Rabbit anti-RFP 1:1000 (Rockland, 600-401-379), and Rat anti-HA 1:500 (Roche, 11867423001). Following incubation with primary antibody, tissue was washed 3x PBST. Tissue was then incubated in 5% normal donkey serum containing secondary antibodies, 1 hour at room temperature for sections, overnight at 40C for whole mount. Secondary antibodies all at 1:500: Donkey anti-Rat Alexa-488 (Abcam ab150153), Donkey anti-Rat Alexa-568 (Abcam ab175475), Donkey anti-Mouse Lylight-649 (Jackson Immuno Research, 709-496-149), Donkey anti-Rabbit Alexa-488 (ThermoFisher, A-21206), Donkey anti-Rabbit Alexa-568 (ThermoFisher, A-100042), Donkey anti-Rabbit Alexa-647 (ThermoFisher, A-31573), Donkey anti-Goat Alexa-488 (ThermoFisher, A-11055), and Donkey anti-Goat Alexa-647 (ThermoFisher, A-21447). Following 3x washes with PBST, tissue was mounted with Prolong Diamond or Slowfade Diamond (ThermoFisher P36971, S36973).
Fluorescent In Situ Hybridization (FISH):
FISH was performed using an RNAscope kit (RNAscope® Multiplex Fluorescent Reagent Kit v2; Catalog No. 323100) according to the manufactures protocol (ACD-Bio). Target probes used are as follows: RNAscope® Probe- Mm-Thbs1-C3 (Cat No. 457891-C3), RNAscope® Probe- Mm-Cartpt-C2 (Cat No. 432001-C2), RNAscope® Probe- Mm-Opn4 (Cat No. 438061), RNAscope® Probe- Mm-Cd24a-C3 (Cat No. 432691-C3). TSA-based fluorophores were from Perkin Elmer (TSA Plus Fluorescein, PN NEL741001KT; TSA Plus Cyanine 3, PN NEL744001KT; TSA Plus Cyanine 5, NEL745001KT). Imaging was done on a Nikon Ti epifluorescence and Olympus FV1000 confocal microscopes.
FISH- Thbs1 mRNA expression quantification:
To quantify Thbs1 mRNA intensity signal in retinal sections, regions of interest (ROI) were drawn using ImageJ. The area, integrated density and background noise were measured and calculated by using the corrected total cell fluorescence (CTCF) formula (CTCF = integrated density – (area of selected cell × mean fluorescence of background readings)), and data presented as arbitrary unit (A.U.). For quantifying Thbs1 expression in ipRGCs, fluorescent signal intensity in individual YFP-positive cell in the ganglion cell layer of Opn4Cre;R26-YFPf/f mice was measured. A total of 70-80 YFP-positive cells in retinal sections from 2 animals per group was analyzed. For quantifying Thbs1 expression in non-ipRGC cells in the ganglion cell layer, fluorescent signal intensity in YFP-negative cells was measured. A total of 70-80 YFP-negative cells in retinal sections from 2 animals per group was analyzed. For quantifying Thbs1 expression in the inner nuclear layer, fluorescent signal intensity in the entire inner nuclear layer marked by the Dapi (nuclear) stain in each section was measured. A total of 25-30 sections (16 μm thickness) from 2 animals per group was analyzed.
RNA-sequencing in different RGC types
Manual cell sorting
Opn4CreERT/+;R26-tdTomatof/f (Opn4) and HB9:GFP (HB9) mice were used for the RNA sequencing experiment. HB9:GFP mice were also injected with tamoxifen as outlined above. This was done to control for any off-target effects of tamoxifen on translation. Animals underwent optic nerve crush (Crush) or sham surgery 3 days prior to tissue collection. Adult mouse RGCs were dissociated as follows; the retina was removed in HBSS (ThermoFisher, 14170161) and transferred to DMEM (ThermoFisher, 11995065) and 17 U/ml papain (Worthington Biochem, PAPL). The retina was enzymatically dissociated for 30 minutes at 37°C in a water bath. Following incubation with papain, the retina was washed 5 times with DMEM. The retina was then triturated in FluoroBrite DMEM (ThermoFisher, A1896701) with B27 supplement and 20 mM HEPES (ThermoFisher, 15630080). The cells were briefly spun down, resuspended in FluoroBrite DMEM + B27 + HEPES, and passed through a 100 μm filter. Cells were plated at low density and allowed to settle. Under epifluorescent illumination tdTomato+ or GFP+, RGCs were located. Phase contrast was used to visualize that the cell was isolated from other cells. Pipettes were pulled with a Sutter Instrument puller. Tips were trimmed to the desired size. A Sutter Instrument micromanipulator was loaded with a pipette with an opening slightly larger than the cell to be captured. Care was taken to prevent excess media from entering the pipette as it was positioned in the media. A cell was only collected if it could be picked up without any additional contaminating cells. The collected cell was promptly lysed in RNA lysis buffer. Cells were only collected for up to 2 hours after the animal was euthanized. 10 cells were collected from each animal. 3 pools of 10 cells were combined to make 1 biological sample.
RNA-purification for sequencing
The Absolutely RNA Nanoprep kit (Agilent, 400753) was used to purify RNA. RNA was purified according to the manufacturer’s protocol. 70% ethanol was substituted for 80% sulfolane. RNA was eluted in 11 μl of 1:1 elution buffer:water. RNA was reverse transcribed and pre-amplified for 11 cycles with the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech, 634888) according to the manufacturer’s protocol. An aliquot of the pre-amplified product was reserved for quality control qPCR.
RNA-sequencing and analysis
Ocean Ridge Biosciences (Deerfield Beach, FL) sequenced the RNA. Briefly, cDNA was assessed for quality by capillary electrophoresis on Agilent 2100 Bioanalyzer High Sensitivity DNA chips (Agilent Technologies, Santa Clara, CA). 100 pg of pre-amplified cDNA was used for library preparation. The Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA) was used to prepare cDNA libraries for sequencing. The libraries were pooled at equimolar concentrations and sequenced on an Illumina HiSeq Flow Cell v4 with 50-bp paired-end reads. All samples had a minimum of 30 million passed-filter 50nt paired-end reads. Reads were aligned using TopHat v2.0.11 to the mm10 Bowtie genome index. Reads were counted using the Bioconductor package easyRNASeq v1.6.0. edgeR v3.14.0 was used to calculate differential gene expression. Genes were included if they were expressed at ≥ 1 count per million in at least 3 of the 12 samples. Genes were considered differentially expressed if log2FC ≥ 1 and the FDR adjusted p-value ≤ 0.05. Pathway analysis was performed using Enrichr (http://amp.pharm.mssm.edu/Enrichr/).
RNA-purification and reverse transcription for PCR and cloning
Tissue was lysed in TRIzol (ThermoFisher, 15596026). RNA was purified using the Direct-zol Miniprep kit (Zymo, R2051) according to the manufacturer’s protocol. Isolated RNA was converted to cDNA using Superscript III reverse transcriptase (ThermoFisher, 18080-093) according to the manufacturer’s protocol. Oligo(dT)12-18 primers (ThermoFisher, 18418-012) were used to prime reverse transcription.
Viruses and Cloning
qPCR and end-point PCR
qPCR was performed using SYBR Green PCR master mix (ThermoFisher, 4309155) and a CFX Connect real-time PCR detection system (Bio-Rad). Primers were designed using NCBI primer blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primers used are listed in Table S1. Primers were designed to amplify all isoforms if applicable. All primers had a melt curve with a single peak and produced an amplicon of the predicted length. End-point PCR was performed using GoTaq G2 green master mix (Promega, M7828) and a S1000 Thermal Cycler (Bio-Rad).
Cloning:
Enzymes used: AgeI (NEB, R3552), BamHI (NEB, R3136), BglII (NEB, R0144), BlpI (NEB, R0585), BspEI (NEB, R0540) EcoRI (NEB, R3101), HindIII (NEB, R3104), KpnI (NEB, R3142), MfeI (NEB, R3589), MluI (NEB, R3198), NheI (NEB, R0131), NotI (R3189), and XhoI (NEB, R0146). DNA polymerase I, large (Klenow) fragment (NEB, M0210), T4 polynucleotide kinase (NEB, M0201), Calf intestinal alkaline phosphatase (NEB, M0491), and Q5 high-fidelity DNA polymerase (NEB, M0491).
Gel electrophoresis was used to analyze cut fragments. Fragments were recovered by gel extraction (Qiagen, 28704). Ligated plasmids were transformed into NEB Stable Competent E. coli (NEB, C3040H), and plated on LB agar (Millipore, 71752-5) with 100 μg/ml ampicillin (SigmaAldrich A9518). When applicable, PCR was used to analyze colonies. Picked colonies were grown overnight in LB broth (FisherScientific, 244620) with 100 μg/ml ampicillin. Plasmid DNA was column purified (Qiagen, 27104).
Primers used for PCR cloning are listed in Table S2. PCR based cloning was done using Q5 high-fidelity DNA polymerase (NEB, M0491S). Gel electrophoresis was used to confirm PCR amplification. PCR product was purified from gel or column purified (Qiagen, 28104). Sanger sequencing was used to confirm all PCR fragments.
For shRNAs: pAAV.U6.shRNA.CMV.eGFP.WPRE was cloned from pAAV2.CMV.eGFP.WPRE. First, the EcoRI and AgeI sites were removed (primers: CMV_mutant F, and R). Next, a U6 promoter with 5’ AgeI and 3’ EcoRI cloning sites was introduced. pKLO.shTHBS1 TCRN0000348494 (SigmaAldrich) served as the template to generate the U6.shRNA sequence (primers: MluI-U6 F, and R). The resulting plasmid (pAAV.U6.shTHBS1(348494).CMV.eGFP.WPRE) was digested with EcoRI and AgeI to accept oligonucleotides for other shRNAs (e.g. syndecan-1). shRNAs were picked from the Broad Institute’s shRNA database (https://portals.broadinstitute.org/gpp/public/) for THBS1. shRNAs for synedcan-1 were those that target UTR either at nucleotides 1420-1439 or nucleotides 1376-1396 of Mus musculus syndecan 1 (Sdc1) mRNA, GenBank accession number NM_011519 (see Table S2 for more details). Oligonucleotides were annealed by mixing equimolar amounts of the plus and minus strand oligonucleotide and NEB buffer 3.1. Oligonucleotides were heated to 95°C and allowed to slowly cool to room temperature over 1.5 hours. Sanger sequencing was used to confirm oligonucleotide integration.
The pAAV2.CMV.BG.mcs.hGH(polyA) vector has an insert capacity of ~2.7kb. The BG intron and hGH(polyA) were replaced with SV40 intron and polyA to increase insert capacity to ~3.7kb. The pscAAV2.CMV.SV40.eGFP.SV40(polyA) plasmid served as a template for the intra-ITR sequence56. First, the eGFP sequence was replaced with a mcs sequence and the NotI site was removed. The plasmid was cut with BamHI and NotI. A mcs sequence was introduced from annealed oligonucleotides (Oligo_SV40_mcs). Next, the intra-ITR sequence was PCR amplified (primers: NotI_SV40 F, and R). pAAV2.CMV.BG.mcs.hGH(polyA) was cut with NotI. These fragments were ligated to create pAAV2.CMV.SV40.mcs.SV40(polyA). All genes were cloned into this high capacity AAV2 plasmid.
pXY.THBS1 (BC6405710) was use as a template for the THBS1 CDS. The 3’ end of the CDS was modified to remove the STOP codon and introduce a C-terminal HA tag (primers: THBS1-TSS F, and THBS1-HA R). The resulting PCR fragment was cloned into the pAAV.CMV.SV40.mcs.SV40(polyA) expression plasmid.
To aid in the production of multiple C-terminal HA tagged constructs, a pAAV vector with a C-terminal HA tag was created. pAAV.CMV.mcs was digested with HindIII and BglII, and anneal oligos containing a C-terminal HA tag were incorporated (Oligo_C-HA). All cDNAs to be expressed as recombinant proteins with C-thermal HA-tag were PCR amplified and ligated into pAAV.CMV.C-HA digested with HindIII and blunted using DNA Polymerase I, large (Klenow) fragment (NEB, M0210).
Thbs4 was PCR amplified (primers: Thbs4-F and Thbs4-R). Cd86 was PCR amplified (primers: Cd86-F and Cd86-R). Vitronectin was PCR amplified (primers: Vtn-F and Vtn-R). Bcl2 was PCR amplified (primers: Bcl2-F and Bcl2-R). THBS1ΔCTD (AA 1-954) was PCR amplified (primers: THBS1-TSS F and THBS1-T3-8 R). THBS1-ΔTSR3-CTD (AA 1-690) was PCR amplified (primers: THBS1-TSS F and THBS1-EGFL1-3 R).
THBS1-ΔTSR1 was generated by bridge PCR. Two PCR fragments were amplified using pairs of oligos TSR1+TSR4 and TSR3+TSR2. Fragments purified in agarose gel and combined into one by bridge PCR with TSR1+TSR2 oligos. The r fragment was digested with BlpI and KpnI to obtain the 1835 bp mutant fragment and replaced the 2333 bp wild type fragment.
THBS1ΔCC was generated by bridge PCR. Two fragments with Coiled-Coil domain deletion were amplified using pairs of oligos CC1+CC4 and CC3+CC2. Fragments were purified in agarose gel and combined into one by bridge PCR with CC1 and CC2 oligos. The resulting DNA fragment was digested with SapI to obtain a 680 bp mutant fragment and to replace the 800 bp wild type fragment.
CD86 coding sequence (937 bp) was PCR amplified from mouse spleen total cDNA using Q5 DNA polymerase and the following oligos: CD86-F and CD86-R. Agarose gel purified DNA fragment was ligated into pAAV.CMV.C-HA plasmid as described earlier to generate CD86 expression construct with C-terminal HA-tag.
RGS4 coding sequence (614 bp) was PCR amplified fron mouse heart total cDNA using Q5 DNA polymerase and the following oligos: RGS4-F and RGS4-R. Agarose gel purified DNA fragment was ligated into pAAV.CMV.C-HA plasmid as described earlier to generate RGS4 expression construct with C-terminal HA-tag.
To generate the pAAV.GFAP.SV40.Thbs1-HA.SV40(polyA) plasmid, the 377 bp CMV promoter coding sequence was deleted from pAAV.CMV.SV40.THBS1-HA.SV40(polyA) plasmid by digestion with AvrII and BspEI restriction enzymes. pAAV.GFAP.EGFP plasmid (Addgene #50473) was used as template to PCR amplify short GFAP promoter sequence using the following oligonucleotides: GFAP-F and GFAP-R. The resulting 685bp DNA fragment was digested using NheI and BspEI enzymes, gel purified and ligated with the vector backbone.
To generate the pAAV.GFAP.SV40.eGFP.SV40(polyA) plasmid, the 3555 bp Thbs1 and HA-tag coding sequence was deleted by digestion with HindIII and XhoI restriction enzymes. EGFP coding sequence was PCR amplified from pAAV.GFAP.eGFP plasmid (Addgene #50473) using the following oligonucleotides: EGFP-F and EGFP-R. The resulting 728 bp DNA fragment was digested with HindIII and XhoI, gel purified and ligated with the vector backbone.
All plasmids were verified by restriction analysis and sequencing.
shRNA and mutant vector validation
shTHBS1 validation
HEK293T cells were cotransfected with shRNAs and the pAAV.CMV.THBS1-HA vector at a 6:1 ratio. 48 hours following transfection cells were lysed in RIPA buffer (Cell Signaling, 9806S) with protease inhibitors (SigmaAldrich, 4693124001) and the protein concentration was determined using the Pierce BCA kit (ThermoFisher 23225). Approximately 20μg of protein was mixed with NuPAGE LDS sample buffer (ThermoFisher NP0007) + 100mM DTT, loaded and separated in a 4%-20% gradient gel (EenScript, M01115). The protein was transferred onto a PVDF membrane overnight, blocked with 5% skim milk in 0.1% Tween-20 in tris buffered saline (TBST). Primary antibodies: Rat anti-HA 1:1000 and Mouse β-actin 1:10,000 (ThermoFisher, A1978), were diluted in blocking reagent and incubated overnight at 40C. Following extensive washing with TBST, membranes were incubated in HRP-conjugated secondary antibodies (Cell Signaling Technologies, 1:1000) for 1 hour at room temperature. After washing with TBST, membranes were developed using SuperSignal West Pico or Femto chemiluminescent substrate (ThermoFisher 34080, 34095). The intensity of each band was quantified using Image J. Bands were quantified as their ratio to β-actin, the mean was obtained from 4 biological replicates. For immunocytochemistry, mouse embryonic fibroblasts (MEFs) were nucleofected with either shScramble or shTHBS1 plasmid, and 72 hours later, immunostained with antibodies against GFP and THBS1, and stained for phalloidin and hoechst.
Thbs1 mutant analysis
HEK293T cells were transfected with Thbs1, Thbs1 mutants, Thbs4 or GFP. At 24 hours after transfection, cells were washed 2x with DPBS, and then cultured overnight in serum free DMEM. The conditioned medium containing secreted proteins was centrifuged at 11,000g for 5 minutes and an aliquot of the conditioned medium was mixed with NuPAGE LDS sample buffer + 100mM DTT. The cells were washed 2x with DPBS and lysed with NuPAGE LDS sample buffer + 100mM DTT. Protein samples were processed as stated above. Thbs1- ΔCC oligomerization was tested by omitting DTT from the sample buffer.
shSDC1 validation
shRNA-GFP plasmids encoding control (shScramble) or syndecan-1-targeting shRNAs (shSDC1) were validated in the mouse brain neuroblastoma Neuro-2a cells. The plasmids were transfected into Neuro-2a cells in triplicates in 24-well plate. Cells were harvested 3 days after transfection, sorted using FACS and co-expressing GFP as a marker, and cultured for additional 2 days. Subsequently, cells were harvested and the total RNA was isolated. SDC1 mRNA expression was analyzed using qPCR and normalized to PGK1. qPCR primer sequences used are listed in Table S1.
Quantification and Statistical Analysis
Quantification of RGC Survival
To estimate the number of surviving RGCs, the whole mount retina was stained with Tuj1 (beta III tubulin) antibody. Retinas with tdTomato- or GFP-labeled RGCs were co-stained with anti-RFP or GFP antibodies, respectively. Eight to ten 40x images were taken that tiled evenly across the retina and were used to determine the average RGC number per retina. RGC subtypes were quantified as the number of Tuj1+ RGCs that co-expressed the subtype specific reporter (e.g. tdTomato, GFP).
ipRGC subtype determination
To determine ipRGC subtypes labeled by the Opn4CreERT/+;R26-tdTomatof/f line, whole mount retinas were stained for tdTomato and a nuclear marker (DAPI). A 10x tiled image of the retina was first acquired. Then individual RGCs were visually assessed under 40x magnification. ipRGC subtype was determined by soma location (ganglion cell layer vs. inner nuclear layer), dendritic stratification, soma size, and dendritic field size. M1 was determined as having all dendrites in the OFF sublamina, Whereas M2 and M3 by having any dendrites in the ON sublamina with a dendritic field size equivalent to M1 ipRGCs. M4 were completely ON stratifying with a large soma and large dendritic field. M5 were ON stratifying with a small dendritic field. An ipRGC whose subtype could not be accurately determined was classified as Unidentified.
Quantification of Axon Regeneration
To quantify the number of regenerating RGC axons, the number of CTB+ axons was determined at multiple distances beyond the optic nerve crush site. 40x images were taken at each distance to be assessed. A line transecting the section was drawn. At least 4 sections per animal were analyzed. CTB+ axons crossing the transection line were quantified. Axon numbers were normalized to section width. Axons labeled by tdTomato or GFP were considered regenerating only if they were also CTB+.
Quantification of pS6+ RGCs
Retinal sections were immunostained with antibodies against pS6 and Tuj1. Data are presented as mean percentages of pS6+ Tuj1+ cells among total Tuj1+ cells in the ganglion cell layer of each retinal section examined. Only cells with bright pS6 fluorescent signal intensity above threshold level set by the evaluator were included in the count. Cell counts were performed on at least 5 non-consecutive sections, from three mice per group.
Statistical analysis
Statistical analysis and graph creation were performed using GraphPad Prism 6 (GraphPad Software, Inc.). Data were analyzed using ANOVA and the Bonferroni within-groups comparison with additional testing using Tukey’s test or Student’s t-test. Values of P < 0.05 were considered significant. No tests of normality were done, assuming normal distribution based on similar previous regeneration studies. All error bars indicate standard error of mean (SEM) unless indicated otherwise in the Figure Legends. Each Figure Legend lists the number of animals used each animal group and the statistical tests used.
Data and Software Availability
RNAseq data of RGCs are deposited to Gene Expression Omnibus (GEO: GSE115661). Full RNAseq dataset is also included Data S1 in the present study. The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Data S1. RNAseq entire gene list (Related to Figure 4). Triplicates for each group (Opn4 RGC Sham, Opn4 RGC Crush, HB9 RGC Sham and HB9 RGC Crush). RGCs were isolated 3 days after optic nerve crush for sequencing.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CART (55-102) | Phoenix Pharmaceuticals | H-003-62, RRID:AB_2313614 |
| Rabbit anti-pS6 (Ser235/236) | Cell Signaling | #2211, RRID:AB_331679 |
| Rabbit anti-pSTAT3(Y705) | Cell Signaling | #9131, RRID:AB_331586 |
| Mouse anti-Thrombospondin (A6.1) | Santa Cruz | Sc-59887, RRID:AB_793045 |
| Goat anti-GFP | Abcam | Ab6673, RRID:AB_305643 |
| Rabbit anti-GFP | Millipore | Ab3080, RRID:AB_91337 |
| Chicken anti-Beta-3 tubulin | Abcam | Ab107216, RRID:AB_10899689 |
| Rabbit anti-Beta-3 tubulin | Sigma-Aldrich | T2200, RRID:AB_262133 |
| Rabbit anti-RFP | Rockland | 600-401-379, RRID:AB_2209751 |
| Rat anti-HA | Roche | 11867423001, RRID:AB_390918 |
| Rabbit anti-RBPMS | PhosphoSolutions | 1830-RBPMS, RRID:AB_2492225 |
| Mouse anti-osteopontin | R & D System | AF808, RRID:AB_2194992 |
| Rabbit anti-melanopsin | Advanced Targeting System | AB-N38, RRID:AB_1608077 |
| Bacterial and Virus Strains | ||
| AAV2-Cre | Boston Childrens Hospital Viral Core | NA |
| AAV-shH10-GFAP-GFP | Park Lab | NA |
| AAV-ShH10-GFAP-THBS1 | Park Lab | NA |
| AAV2-THBS1 and THBS4 (various forms) | Park Lab | NA |
| AAV2-RGS4, VTN, CD86 and various shRNA | Park Lab | NA |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| KRFYVVMWKK | GenScript | NA |
| KRFYGGMWKK | GenScript | NA |
| Critical Commercial Assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit V2 | ACD-Bio | 323100 |
| SMART-Seq v4 Ultra Low Input RNA kit | Clontech | 634888 |
| Deposited Data | ||
| Raw and analyzed data | Present manuscript | GEO: GSE115661 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T cells | ATCC | CRL-3216, RRID:CVCL_0063 |
| Mouse: mouse embryonic fibroblasts | ATCC | A gift from Dr. A. Capobianco |
| Neuro-2a cells | ATCC | CCL-131 RRID:CVCL_0470 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Opn4Cre | Ecker et al. 2010 | NA |
| Mouse: Opn4CreERT | Chen et al. 2011 | NA |
| Mouse: C57BL/6J | Jackson Labs | 000664, RRID:IMSR_JAX:000664 |
| Mouse: B6.129S4-Ptentm1Hwu/J | Jackson Labs | 006440, RRID:IMSR_JAX:006440 |
| Mouse: R26 loP-STOP-loxP-tdTomato | Dr. Fan Wang, Duke University | NA |
| Mouse: B6.Cg-Tg(Hlxb9-GFP)1Tmj/J | Jackson Labs | 005029, RRID:IMSR_JAX:005029 |
| Mouse: B6.129(FVB)-Igf1tm1Dlr/J | Jackson Labs | 016831, RRID:IMSR_JAX:016831 |
| Mouse: B6.129S1(Cg)-Eomestm1.1Bflu/J | Jackson Labs | 017293, RRID:IMSR_JAX:017293 |
| Mouse: B6.129X1-Baxtm1Sjk/J | Jackson Labs | 002994, RRID:IMSR_JAX:002994 |
| Mouse: B6(Cg)-Atf6tm1Hota/J | Jackson Labs | 028253, RRID:IMSR_JAX:028253 |
| Mouse: Glt1-eGFP BAC promoter reporter | Dr. Jeffrey Rothstein, Johns Hopkins University | NA |
| Oligonucleotides | ||
| qPCR primers: Supplemental Table 1 | Present manuscript | NA |
| Cloning primers and oligonucleotides: Supplemental Table 2 | Present manuscript | NA |
| Recombinant DNA | ||
| pAAV2.CMV.mcs | Stratagen | 240071 |
| pscAAV2-GFP | Addgene Gray et al Methods Mol Biol. 2011;807:25-46 |
Addgene: 32396, RRID:Addgene_32396 |
| pAAV.GFAP.eGFP | Addgene gift from Bryan Roth |
Addgene: 50473, RRID:Addgene_50473 |
| shh10 | Addgene | Addgene: 64867, RRID:Addgene_64867 |
| Software and Algorithms | ||
| TopHat v2.0.11 | PMID: 23618408 | https://ccb.jhu.edu/software/tophat/index.shtml |
| easyRNASeq v1.6.0 | PMID: 22847932 | http://bioconductor.org/packages/release/bioc/html/easyRNASeq.html |
| edgeR v3.14.0 | PMID: 19910308 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| R v3.3.1 | https://cran.r-project.org) | |
| GraphPad Prism 6 | GraphPad Software, Inc | https://www.graphpad.com/scientific-software/prism/ |
| FIJI/ImageJ | PMID: 29187165 | https://fiji.sc/ |
| Adobe Photoshop CC | Adobe | https://www.adobe.com/products/photoshop.html |
Highlights:
ipRGCs survive and regenerate axons well after injury.
RNAseq reveals unique sets of genes enriched in injured ipRGCs including Thbs1 and Sdc1.
Neural THBS1 promotes axon regeneration in various RGC types.
Neural THBS1 promotes axon regeneration in syndecan1-dependent manner.
Acknowledgements:
This work was supported by grants from NEI 1R01EY022961-01 (K.K.P), NEI 1U01EY027257-01 (K.K.P and V.P.L), NIDCD R01 DC006308 (N.C), NIDCD R01 DC 014420 (N.C.), NICHD HD057632 (V.P.L) NEI 1F30EY025527-01 (ERB), DOD VRP W81XWH-12-1-0319 (K.K.P), Glaucoma Research Foundation (K.K.P), The Lois Pope LIFE Fellowship (E.R.B), The Miami Project to Cure Paralysis, and the Buoniconti Fund (K.K.P and V.P.L). V.P.L. holds the Walter G. Ross Distinguished Chair in Developmental Neuroscience. The authors thank Alan C. Rupp for insightful discussion, and Katherine Narvaez, Yadira Salgueiro and Shaffiat Karmally for assistance with the animals and histology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References:
- Adams JC, Kureishy N, and Taylor AL (2001). A role for syndecan-1 in coupling fascin spike formation by thrombospondin-1. J Cell Biol 152, 1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JC, and Lawler J (2004). The thrombospondins. Int J Biochem Cell Biol 36, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar N, Annis DS, Mosher DF, and Adams JC (2002). Trimeric assembly of the C-terminal region of thrombospondin-1 or thrombospondin-2 is necessary for cell spreading and fascin spike organisation. J Cell Sci 115, 2357–2366. [DOI] [PubMed] [Google Scholar]
- Bray ER, Noga M, Thakor K, Wang Y, Lemmon VP, Park KK, and Tsoulfas P (2017). 3D Visualization of Individual Regenerating Retinal Ganglion Cell Axons Reveals Surprisingly Complex Growth Paths. eNeuro 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafferty WB, McGee AW, and Strittmatter SM (2008). Axonal growth therapeutics: regeneration or sprouting or plasticity? Trends Neurosci 31, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamak B, Morandi V, and Mallat M (1994). Brain macrophages stimulate neurite growth and regeneration by secreting thrombospondin. J Neurosci Res 38, 221–233. [DOI] [PubMed] [Google Scholar]
- Chandran V, Coppola G, Nawabi H, Omura T, Versano R, Huebner EA, Zhang A, Costigan M, Yekkirala A, Barrett L, et al. (2016). A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron 89, 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, and Zheng B (2014). Axon plasticity in the mammalian central nervous system after injury. Trends Neurosci 37, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Badea TC, and Hattar S (2011). Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC, Scotting PJ, Hsu LS, Lin SJ, Shih HY, Hsieh FY, Wu HL, Tsao CL, and Shen CJ (2013). Zebrafish rgs4 is essential for motility and axonogenesis mediated by Akt signaling. Cell Mol Life Sci 70, 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, and Barres BA (2005). Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433. [DOI] [PubMed] [Google Scholar]
- Corless CL, Mendoza A, Collins T, and Lawler J (1992). Colocalization of thrombospondin and syndecan during murine development. Dev Dyn 193, 346–358. [DOI] [PubMed] [Google Scholar]
- Danzi MC, Mehta ST, Dulla K, Zunino G, Cooper DJ, Bixby JL, and Lemmon VP (2018). The effect of Jun dimerization on neurite outgrowth and motif binding. Mol Cell Neurosci 92, 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima S, Koriyama Y, Kurimoto T, Oliveira JT, Yin Y, Li Y, Gilbert HY, Fagiolini M, Martinez AM, and Benowitz L (2012). Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc Natl Acad Sci U S A 109, 9149–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Qiao M, Bei F, Kim IJ, He Z, and Sanes JR (2015). Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron 85, 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TJ, and Hammarlund M (2014). Syndecan promotes axon regeneration by stabilizing growth cone migration. Cell Rep 8, 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, Allen NJ, Susman MW, O'Rourke NA, Park CY, Ozkan E, Chakraborty C, Mulinyawe SB, Annis DS, Huberman AD, et al. (2009). Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell 139, 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Petkova V, Thanos S, and Benowitz LI (2004). Switching mature retinal ganglion cells to a robust growth state in vivo: gene expression and synergy with RhoA inactivation. J Neurosci 24, 8726–8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavile CM, Barwick BG, Newman S, Neri P, Nooka AK, Lonial S, Lee KP, and Boise LH (2017). CD86 regulates myeloma cell survival. Blood Adv 1, 2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Cho KS, Li Y, Tchedre K, Antolik C, Ma J, Chew J, Utheim TP, Huang XA, Yu H, et al. (2018). IGFBPL1 Regulates Axon Growth through IGF-1-mediated Signaling Cascades. Sci Rep 8, 2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal J (2006). Regulation of melanopsin expression. Chronobiol Int 23, 159–166. [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Pollett MA, and Harvey AR (2011). Post-injury delivery of rAAV2-CNTF combined with short-term pharmacotherapy is neuroprotective and promotes extensive axonal regeneration after optic nerve trauma. J Neurotrauma 28, 2475–2483. [DOI] [PubMed] [Google Scholar]
- Hu Y, Park KK, Yang L, Wei X, Yang Q, Cho KS, Thielen P, Lee AH, Cartoni R, Glimcher LH, et al. (2012). Differential effects of unfolded protein response pathways on axon injury-induced death of retinal ganglion cells. Neuron 73, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak RR, Koerber JT, Dalkara D, Flannery JG, and Schaffer DV (2009). A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Muller cells. PLoS One 4, e7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Morgia C, Ross-Cisneros FN, Sadun AA, Hannibal J, Munarini A, Mantovani V, Barboni P, Cantalupo G, Tozer KR, Sancisi E, et al. (2010). Melanopsin retinal ganglion cells are resistant to neurodegeneration in mitochondrial optic neuropathies. Brain 133, 2426–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibinger M, Andreadaki A, Diekmann H, and Fischer D (2013). Neuronal STAT3 activation is essential for CNTF- and inflammatory stimulation-induced CNS axon regeneration. Cell Death Dis 4, e805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RS, Chen BY, Tay DK, Chan HH, Pu ML, and So KF (2006). Melanopsin-expressing retinal ganglion cells are more injury-resistant in a chronic ocular hypertension model. Invest Ophthalmol Vis Sci 47, 2951–2958. [DOI] [PubMed] [Google Scholar]
- Li S, Yang C, Zhang L, Gao X, Wang X, Liu W, Wang Y, Jiang S, Wong YH, Zhang Y, and Liu K (2016). Promoting axon regeneration in the adult CNS by modulation of the melanopsin/GPCR signaling. Proc Natl Acad Sci U S A 113, 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Stafford BK, Nguyen PL, Lien BV, Wang C, Zukor K, He Z, and Huberman AD (2016). Neural activity promotes long-distance, target-specific regeneration of adult retinal axons. Nat Neurosci 19, 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, Lynch KA, Okada T, Aronow BJ, Osinska H, et al. (2012). A thrombospondin-dependent pathway for a protective ER stress response. Cell 149, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Li H, Zhang Z, Kiyama T, Panda S, Hattar S, Ribelayga CP, Mills SL, and Wang SW (2014). T-box transcription regulator Tbr2 is essential for the formation and maintenance of Opn4/melanopsin-expressing intrinsically photosensitive retinal ganglion cells. J Neurosci 34, 13083–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, and Goldberg JL (2009). KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Tanaka T, Bando Y, and Yoshida S (2015). Nerve injury induces the expression of syndecan-1 heparan sulfate proteoglycan in primary sensory neurons. Neuroscience 300, 338–350. [DOI] [PubMed] [Google Scholar]
- Nadal-Nicolas FM, Madeira MH, Salinas-Navarro M, Jimenez-Lopez M, Galindo-Romero C, Ortin-Martinez A, Santiago AR, Vidal-Sanz M, and Agudo-Barriuso M (2015). Transient Downregulation of Melanopsin Expression After Retrograde Tracing or Optic Nerve Injury in Adult Rats. Invest Ophthalmol Vis Sci 56, 4309–4323. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Emmett CJ, Venstrom KA, and Reichardt LF (1991). Vitronectin and thrombospondin promote retinal neurite outgrowth: developmental regulation and role of integrins. Neuron 6, 345–358. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis B, Haenzi B, Andrews MR, Verhaagen J, and Fawcett JW (2018). Integrins promote axonal regeneration after injury of the nervous system. Biol Rev Camb Philos Soc 93, 1339–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea KS, Liu LH, and Dixit VM (1991). Thrombospondin and a 140 kd fragment promote adhesion and neurite outgrowth from embryonic central and peripheral neurons and from PC12 cells. Neuron 7, 231–237. [DOI] [PubMed] [Google Scholar]
- Osterhout DJ, Frazier WA, and Higgins D (1992). Thrombospondin promotes process outgrowth in neurons from the peripheral and central nervous systems. Dev Biol 150, 256–265. [DOI] [PubMed] [Google Scholar]
- Park K, Luo JM, Hisheh S, Harvey AR, and Cui Q (2004). Cellular mechanisms associated with spontaneous and ciliary neurotrophic factor-cAMP-induced survival and axonal regeneration of adult retinal ganglion cells. J Neurosci 24, 10806–10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, and He Z (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Sevilla Muller L, Sargoy A, Rodriguez AR, and Brecha NC (2014). Melanopsin ganglion cells are the most resistant retinal ganglion cell type to axonal injury in the rat retina. PLoS One 9, e93274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard GE, and Sollars PJ (2012). Intrinsically photosensitive retinal ganglion cells. Rev Physiol Biochem Pharmacol 162, 59–90. [DOI] [PubMed] [Google Scholar]
- Resovi A, Pinessi D, Chiorino G, and Taraboletti G (2014). Current understanding of the thrombospondin-1 interactome. Matrix Biol 37, 83–91. [DOI] [PubMed] [Google Scholar]
- Robinson GA, and Madison RD (2004). Axotomized mouse retinal ganglion cells containing melanopsin show enhanced survival, but not enhanced axon regrowth into a peripheral nerve graft. Vision Res 44, 2667–2674. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Masland RH (2015). The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci 38, 221–246. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Do MT, Dacey D, Lucas R, Hattar S, and Matynia A (2011). Melanopsin-positive intrinsically photosensitive retinal ganglion cells: from form to function. J Neurosci 31, 16094–16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TM, Taniguchi K, and Kofuji P (2008). Intrinsic and extrinsic light responses in melanopsin-expressing ganglion cells during mouse development. J Neurophysiol 100, 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, and Roska B (2012). Transcriptional code and disease map for adult retinal cell types. Nat Neurosci 15, 487–495, S481-482. [DOI] [PubMed] [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, and He Z (2011). Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 480, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Sawai H, and Fukuda Y (1993). Number, distribution, and morphology of retinal ganglion cells with axons regenerated into peripheral nerve graft in adult cats. J Neurosci 13, 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. RNAseq entire gene list (Related to Figure 4). Triplicates for each group (Opn4 RGC Sham, Opn4 RGC Crush, HB9 RGC Sham and HB9 RGC Crush). RGCs were isolated 3 days after optic nerve crush for sequencing.
Data Availability Statement
RNAseq data of RGCs are deposited to Gene Expression Omnibus (GEO: GSE115661). Full RNAseq dataset is also included Data S1 in the present study. The data that support the findings of this study are available from the corresponding author upon request.