SUMMARY
Dorsal root ganglion (DRG) sensory neuron subtypes defined by their in vivo properties display distinct intrinsic electrical properties. We used bulk RNA-sequencing of genetically-labeled neurons and electrophysiological analyses to define ion channel contributions to the intrinsic electrical properties of DRG neuron subtypes. The transcriptome profiles of eight DRG neuron subtypes revealed differentially expressed and functionally relevant genes, including voltage-gated ion channels. Guided by these data, electrophysiological analyses using pharmacological and genetic manipulations as well as computational modeling of DRG neuron subtypes were undertaken to assess the functions of select voltage-gated potassium channels (Kv1, Kv2, Kv3, and Kv4) in shaping action potential (AP) waveforms and firing patterns. Our findings show that the transcriptome profiles have predictive value for defining ion channel contributions to sensory neuron subtype-specific intrinsic physiological properties. The distinct ensembles of voltage-gated ion channels predicted to underlie the unique intrinsic physiological properties of eight DRG neuron subtypes are presented.
ETOC
Zheng et al. defined distinct intrinsic physiological properties and transcriptome profiles of eight somatosensory neuron subtypes and further revealed subtype-specific contributions of differentially expressed potassium channels to the intrinsic membrane properties in these sensory neurons.
INTRODUCTION
Dorsal root ganglia (DRG) neurons are the first-order neurons in the somatosensory afferent pathway, transducing physical, thermal and chemical stimuli acting in the periphery into electrical signals and conveying these signals to the central nervous system for action and perception. DRG neurons are pseudo-unipolar, with one axonal branch that innervates peripheral tissues, including the skin, and another that projects to the spinal cord and, in some cases, the dorsal column nuclei of the brainstem (Abraira and Ginty, 2013). Decades of research has shown that DRG sensory neurons exhibit a wide range of peripheral ending morphologies, central projection patterns, and physiological properties, and that they are tuned to distinct modalities or features of sensory stimuli. DRG neurons are thus classified into distinct subtypes (Abraira and Ginty, 2013; Basbaum et al., 2009; Delmas et al., 2011; Willis and Coggeshall, 2004; Woolf and Ma, 2007; Zimmerman et al., 2014).
How do DRG neuron subtypes transduce sensory stimuli into patterns of APs that propagate to the CNS? Stimuli impinging on the skin and other organs activate transduction receptors or ion channels on peripheral terminals of DRG sensory neurons, resulting in the generation of receptor potentials. Transduction channels include thermosensitive and mechanosensitive cation channels, such as TrpV1, TrpM8 and Piezo2 (Julius, 2013; Ranade et al., 2015). Receptor potentials generated at axon terminals may be integrated over space and time and subject to modulation by locally expressed voltage-gated ion channels (Bai et al., 2015; François et al., 2015a; Grigg, 1986; Heidenreich et al., 2012; Wang and Lewin, 2011). Suprathreshold receptor potentials trigger activation of voltage-gated ion channels at initiation sites to generate APs that propagate along the length of the axon, into its central axonal branches and terminals. While the transduction machinery determines the general stimulus modality to which DRG sensory neurons respond, voltage-gated ion channels modulate generator currents and control AP thresholds, AP firing patterns, propagation, and synaptic transmission (Du et al., 2014; Heidenreich et al., 2012; Muqeem et al., 2018; Sundt et al., 2015; Tsantoulas and McMahon, 2014; Waxman and Zamponi, 2014) and are therefore determinants of DRG sensory neuron function.
It has long been established that a wide range of voltage-gated sodium (Nav), calcium (Cav), and potassium (Kv) channels are expressed in DRG neurons, and several of these ion channels are now appreciated as targets for developing drugs to treat pain (Dib-Hajj et al., 2010; Tsantoulas and McMahon, 2014; Waxman and Zamponi, 2014). Most electrophysiological studies have focused on small-diameter DRG neurons, most (but not all) of which correspond to unmyelinated nociceptors, with some studies distinguishing sub-populations of small-diameter neurons based on lectin binding or capsaicin sensitivity (Du and Gamper, 2013; Petruska et al., 2000; Rau et al., 2014; Vydyanathan et al., 2005). Much less is known about the complement of ion channels in physiologically-defined DRG neuron subtypes (for example, mechanoreceptors) or how they relate to the firing behaviors in these subtypes, especially medium-and large-diameter DRG neurons, which comprise a highly heterogenous population. Therefore, a major current challenge is determining how intrinsic electrical properties of the diverse, physiologically distinct mammalian somatosensory neuron subtypes are specified.
Recent advances in the development of mouse genetic tools to interrogate the major classes of DRG sensory neurons afford an opportunity to investigate the relationship between patterns of voltage-gated ion channel expression and the intrinsic physiological properties of DRG neuron subtypes. Genetic tools now exist for labeling five principal low-threshold mechanoreceptor subtypes (LTMRs, i.e., touch receptors) that innervate hairy skin: these are the C-LTMRs, Aδ-LTMRs, Aβ RA-LTMRs, Aβ SA1-LTMRs, and Aβ Field-LTMRs (Zimmerman et al., 2014; Bai et al., 2015). Here, we used genetic tools to identify intrinsic molecular determinants of the physiological properties of LTMR subtypes and, for comparison, nociceptor subtypes and proprioceptors (Hippenmeyer et al., 2005; Zylka et al., 2005). We employed genetic labeling to purify these DRG neuron subtypes and performed deep RNA-sequencing to identify candidate genes that underlie their subtype-specific intrinsic electrophysiological properties. Our analysis revealed many genes, including voltage-gated ion channels, that are differentially expressed across the five LTMR subtypes, peptidergic and polymodal nonpeptidergic nociceptors, and proprioceptors. Guided by these gene expression profiles, we performed electrophysiological experiments and computational modeling to define contributions of differentially expressed Kv channels to sensory neuron subtype-specific intrinsic electrical properties. Our findings reveal the distinct voltage-gated ion channel constellations that support unique intrinsic properties of the major DRG sensory neuron classes.
RESULTS
Somatosensory neurons exhibit subtype-specific intrinsic membrane properties and mechanosensitivity
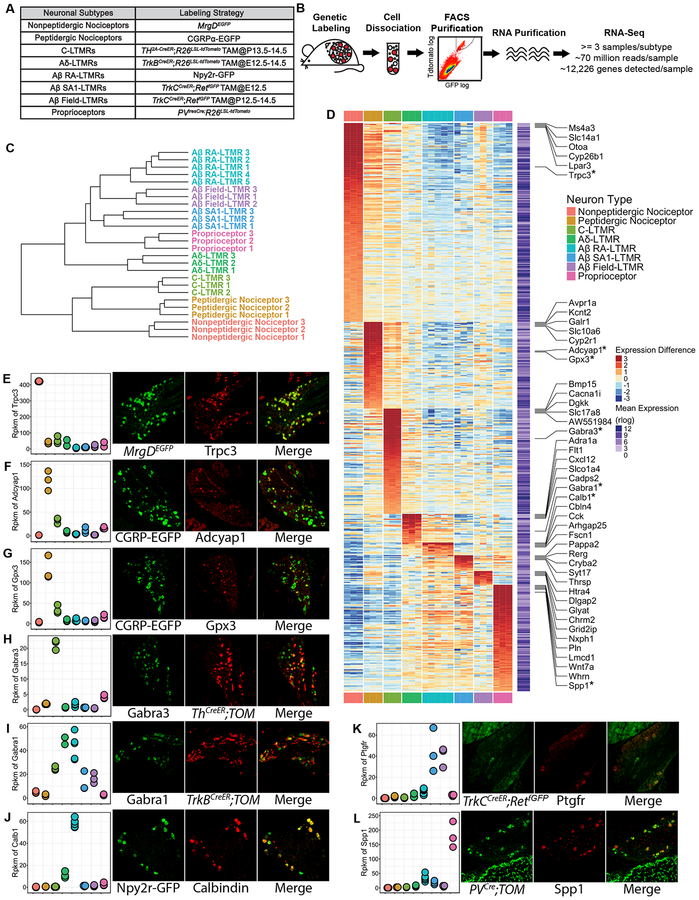
The availability of mouse genetic tools for selective labeling of functionally defined sensory neuron subtypes in vivo enables a comprehensive analysis of subtype-specific intrinsic physiological properties of these subtypes. We focused on eight major classes of DRG sensory neurons labeled in vivo using genetically modified mice. These are the MrgD+ polymodal nonpeptidergic nociceptors, the broad class of CGRP+ (peptidergic) nociceptors, five functionally defined LTMR subtypes (C-LTMRs, Aδ-LTMRs, Aβ RALTMRs, Aβ SA1-LTMRs, Aβ Field-LTMRs), and proprioceptors. The genetic tools used to label each of these neuronal populations are: MrgDEGFP mice to label MrgD+ nonpeptidergic nociceptors (Zylka et al., 2005); Calca(CGRP)-EGFP BAC transgenic mice for peptidergic nociceptors (Bai et al., 2015); Th2A-CreER; R26lsl-tdTomato (Ai14) mice (Tamoxifen 2mg/day at P13–14) for C-LTMRs (Abraira et al., 2017); TrkBCreER; Ai14 mice (Tamoxifen 2mg/day at E12.5–13.5) for Aδ-LTMRs (Rutlin et al., 2014); Npy2r-GFP BAC transgenic mice for Aβ RA-LTMRs (Li et al., 2011); TrkCCreER; RetfGFP mice (Tamoxifen 3mg at E12.5) for Aβ SA1-LTMRs (Bai et al., 2015; Jain et al., 2006); TrkCCreER; RetfGFP mice (Tamoxifen 2mg/day at P13–14) for Aβ Field-LTMRs (Bai et al., 2015); and PVIRES-Cre; Ai14 mice to label proprioceptors (Hippenmeyer et al., 2005) (Figure 2A). We estimate that these eight populations account for ~85% of all DRG neurons (Bai et al., 2015; Gorokhova et al., 2014; Li et al., 2011; Lu et al., 2017).
Figure 2. Transcriptome profiling of eight major DRG neuron subtypes.
(A) Genetic toolbox for labeling each of the eight DRG neuron subtypes.
(B) Schematic of the RNA-sequencing workflow.
(C) Hierarchical clustering of samples based on the expression (represented in rlog transformed count values) of the top 1000 genes that display the highest expression variance across samples using Euclidean distance.
(D) Heatmap depicting expression patterns of SUEGs. Expression (represented in rlog transformed count values) differences compared to the average expression levels for each gene are plotted in the main heatmap. Average expression level of a gene was calculated by averaging expression across all samples and is plotted in a second heatmap next to the main heatmap. Only highly expressed genes (average expression levels in the upper 75th percentile) were selected for this analysis. Genes are ordered based on the subtype in which expression is enriched, and their degree of enrichment. The top 5 most enriched genes for each of the neuronal subtypes are labeled. Genes whose expression were further tested with experiments as shown in (E-L) are labeled and marked with an asterisk.
(E-L) Left-most panels: Dotplots depicting expression levels of selected genes across subtypes in rpkm (reads per kilobase per million reads) values. Three right panels: Representative images of double immunostaining (E, H, I, J) or in situ hybridization (F, G, K, L) of fluorescent reporters and select genes using DRG sections from genetically labeled mice. Sensory neuron subtype reporters and tested genes are shown in separate channels, and the degree of overlap between the two is shown in the merged images. n=3 mice for (E-K), n=2 mice for L.
See STAR Methods for details.
Firing patterns and AP waveforms.
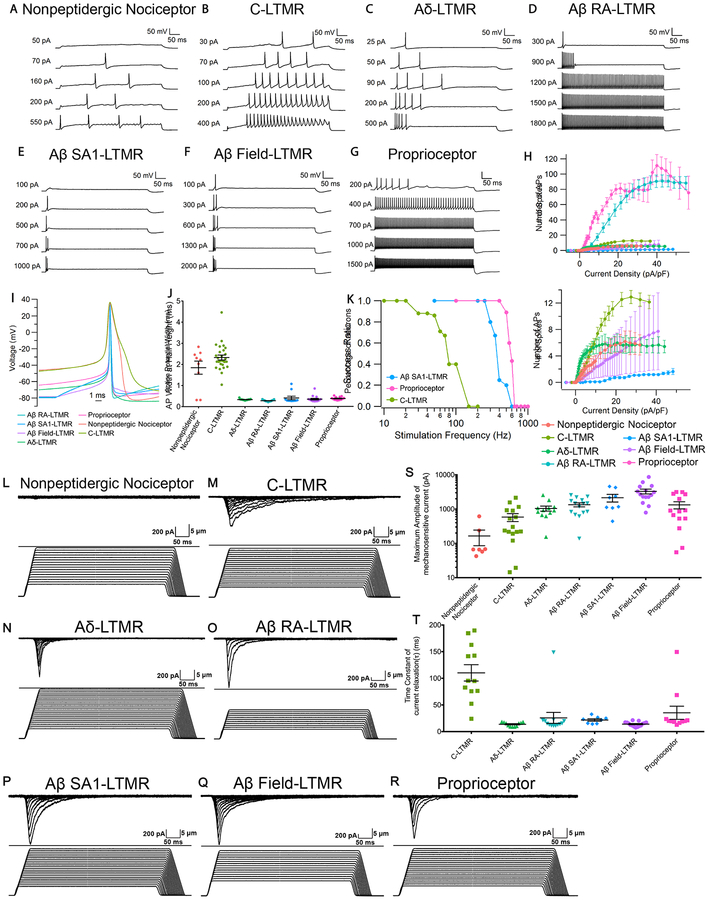
To define the intrinsic electrical properties of DRG neuron subtypes, neurons from each of the genetically labeled mouse lines were acutely dissociated and used for whole-cell patch-clamp recordings. The neuron subtypes displayed distinct electrophysiological signatures. MrgD+ nonpeptidergic nociceptors and C-LTMRs both showed repetitive low-frequency firing in response to current injection, reaching maximum frequencies of ~12 Hz and ~26 Hz, respectively (Figure 1A, 1B and 1H). C-LTMRs exhibited a distinctive delayed firing pattern with a long latency to the first AP followed by regular repetitive spiking (Figure 1B). Aδ-LTMRs showed strong adaptation, firing 5–6 spikes in the first few hundred ms and then becoming silent (Figure 1C and 1H). Aβ RA-LTMRs and proprioceptors both fired repetitively over a wide range of frequencies, with the frequency increasing with current amplitude to a maximum of ~180 and ~220 Hz, respectively (Figure 1D, 1G and 1H). Both Aβ Field-LTMRs and Aβ SA1-LTMRs typically displayed very strong adaptation, firing only in the first 50 ms of current injection (Figure 1E, 1F and 1H). Strikingly, Aβ SA1-LTMRs fired no more than two spikes regardless of the current intensity used for stimulation.
Figure 1. Distinct intrinsic physiological properties and mechanosensitivity of eight major DRG sensory neuron subtypes.
(A-G) Representative in vitro spiking patterns of DRG neuron subtypes during 500-ms injections of depolarizing current steps of increasing magnitude.
(H) Number of APs during 500-ms current injections for each subtype (mean ± standard error of the mean (SEM)) plotted against current density, with expanded y-axis shown in the bottom panel. N=9 for MrgD+ nonpeptidergic nociceptors; n=28 for C-LTMRs; n=19 for Aδ-LTMRs; n=25 for Aβ RA-LTMRs; n=11 for Aβ SA1-LTMRs; n=16 for Aβ Field-LTMRs; n=14 for proprioceptors.
(I) Representative AP waveforms of DRG neuron subtype (first AP evoked by 500-ms current injection at or near threshold).
(J) AP widths (measured at half-maximal spike amplitude) of DRG neuron subtypes (mean ± SEM, n’s as in H). One-way ANOVA with Tukey’s multiple comparisons test showed that MrgD+ nonpeptidergic nociceptors and C-LTMRs are different from each other (p=0.036) and from all other subtypes (p<0.001).
(K) Survival plot depicting the percentages of C-LTMRs (n=8), Aβ SA1-LTMRs (n=5) and proprioceptors (n=9) whose repetitive firing successfully followed 250 μs pulse stimulation at the indicated frequencies.
(L-R) Representative traces of Imech during 500-ms step displacements of increasing magnitudes for each DRG neuron subtype.
(S) Maximal amplitudes of Imech for each DRG neuron subtypes (mean ± SEM). The maximal amplitudes of Imech are significantly different (p<0.0001). In particular, maximal amplitudes of Imech measured in MrgD+ nonpeptidergic nociceptors are significantly smaller than those measured in Aβ RA-LTMRs (p=0.0072), Aβ SA1-LTMRs (p=0.0014) and Aβ Field-LTMRs (p<0.0001) and proprioceptors (p=0.0345). Kruskal-Wallis test with multiple comparisons of every subtype against MrgD+ nonpeptidergic nociceptors. Imech measured in MrgD+ nonpeptidergic nociceptors is not significantly different from 100pA, which is the noise level (p=0.4450), while Imech measured in five LTMRs and proprioceptors are (p<0.01). One-sample t-test. n=7 for MrgD+ nonpeptidergic nociceptors; n=16 for C-LTMRs; n=12 for Aδ-LTMRs; n=15 for Aβ RA-LTMRs; n=8 for Aβ SA1-LTMRs; n=15 for Aβ Field-LTMRs and n=14 for proprioceptors.
(T) Time constant (t) of Imech relaxation kinetics for each DRG neuron subtype. Group mean and SEM are shown using error bars. τ of Imech are significantly different among subtypes (p<0.0001). In particular, τ in C-LTMRs are significantly larger than those in Aδ-LTMRs (p<0.0001), Aβ RA-LTMRs (p=0.0006), and Aβ Field-LTMRs (p<0.0001), suggesting Imech in C-LTMRs decays slower. Kruskal-Wallis test with Dunn’s multiple comparisons test. n=12 for C-LTMRs; n=11 for Aδ-LTMRs; n=13 for Aβ RA-LTMRs; n=8 for Aβ SA1-LTMRs; n=15 for Aβ Field-LTMRs and n=11 for proprioceptors.
See also Figure S1.
The finding that Aβ SA1-LTMRs fire non-repetitively in response to sustained current injection in vitro is counterintuitive because these neurons fire repetitively and adapt slowly to sustained skin indentation in vivo (Wellnitz et al., 2010; Zimmerman et al., 2014). We therefore asked whether Aβ SA1-LTMRs could fire repetitively in vitro by applying a train of short current pulses (250 μs). Indeed, all Aβ SA1-LTMRs faithfully followed pulse stimulation up to ~300 Hz (Figure 1E). In vivo, Aβ SA1-LTMR cutaneous endings are associated with Merkel cells, which are themselves mechanosensitive and transmit excitatory signals to Aβ SA1-LTMRs (Chang et al., 2016; Ikeda et al., 2014; Maksimovic et al., 2013, 2014; Woo et al., 2014). These in vitro results are thus consistent with the notion that the repetitive firing of Aβ SA1-LTMRs in response to static indentation in vivo depends on their association with Merkel cells. Proprioceptors could follow repetitive stimulation at even higher frequencies (400–600 Hz) than Aβ SA1-LTMRs and maximal firing frequencies of both were far higher than that achieved by C-LTMRs (30–100 Hz).
The DRG neuron subtypes also display different AP waveforms. MrgD+ nonpeptidergic nociceptors and C-LTMRs have much wider APs (widths ~2 ms at half spike height) than other LTMR subtypes and proprioceptors, with AP widths < 0.5 ms (Figure 1B and 1D). The broad APs seen in MrgD+ nonpeptidergic nociceptors and C-LTMRs agree with previous results showing broad APs in cell bodies associated with unmyelinated C-fibers in rats (Harper and Lawson, 1985). Broad APs may facilitate AP propagation in small unmyelinated axons with short length constants, which are susceptible to spike failure, especially at the T-junction within the dorsal root ganglion (Gemes et al., 2013; Sundt et al., 2015).
Our electrophysiological analyses of MrgD+ nonpeptidergic nociceptors, the five LTMR subtypes, and proprioceptors indicate that these populations are largely homogeneous in terms of within-group firing patterns and AP waveforms. On the other hand, the CGRP+ peptidergic nociceptors displayed a range of firing patterns and AP waveforms (Figure S1), consistent with the idea that CGRP labels a large, functionally and morphologically heterogeneous group of neurons that can be further subdivided based on their intrinsic physiological properties (Arcourt et al., 2017; Bardoni et al., 2014; Han et al., 2013; Patil et al., 2018).
Mechanically activated currents.
To assess intrinsic mechanosensitivity of the major DRG sensory neuron subtypes, genetically labeled DRG neurons were used for in vitro whole-cell mechanoclamp experiments in which a controlled mechanical stimulus was applied to the cell body using a piezo-driven glass probe and resulting currents were recorded (Hao and Delmas, 2011; McCarter et al., 1999). All labeled neuronal types were used for this analysis, except for CGRP+ peptidergic nociceptors which were excluded because of their heterogeneous firing properties (Figure S1). With a series of 500-ms mechanical stimulations of increasing displacement, all five LTMR subtypes and proprioceptors displayed obvious mechanically-activated inward currents (Imech) (Figure 1M–1R and 1S), while most MrgD+ nonpeptidergic nociceptors displayed minimal Imech indistinguishable from noise (100pA) (Figure 1L and 1S). Imech in DRG neuron subtypes also displayed different decay kinetics. Imech of Aδ-LTMRs, Aβ-LTMRs, and proprioceptors have fast relaxation kinetics with a time constant mostly shorter than 30 ms (Figure 1T). In comparison, Imech of C-LTMRs decays slower with an average time constant around 100 ms.
Taken together, these in vitro electrophysiological recordings indicate that most functionally distinct DRG neuron subtypes are distinguished by unique combinations of firing patterns, AP waveforms, and mechanical sensitivities; Aβ Field-LTMRs and Aβ SA1-LTMRs displayed similar intrinsic properties despite major differences in their in vivo responses to mechanical stimuli acting on the skin, likely reflecting their unique peripheral terminal morphologies and the skin cell types with which they associate (Bai et al., 2015). Differences in intrinsic physiological properties among the subtypes are likely due to differences in ion channels and components of the mechanotransduction machinery that they express.
Deep RNA-sequencing to generate transcriptome profiles of major DRG neuron subtypes
To explore the molecular basis of the distinct intrinsic membrane properties and other distinguishing features of DRG neuron subtypes, we undertook an RNA-sequencing approach to generate transcriptome profiles for each of the eight major DRG neuron subtypes. We made use of the aforementioned genetic tools that selectively label each of these subtypes with fluorescent reporters, first purifying labeled neurons to homogeneity using flow cytometry (FACS) and then extracting RNA from these purified neuronal populations (Figure 2A and 2B). DRGs from all axial levels were used for FACS, except for proprioceptors which were purified from thoracic ganglia, because the PVIRES-Cre; Ai14 proprioceptor labeling strategy was found to label a subset of limb level cutaneous LTMRs as well. DRGs from multiple mice were combined for FACS, and neurons from multiple rounds of sorting were combined to obtain sufficient amounts of RNA for each sequencing reaction. At least three biological replicates were sequenced for each neuronal subtype (Figure 2B). RNA libraries were prepared and subsequently sequenced using an Illumina HiSeq2000 platform at an average depth of ~70 million mapped reads per sample. This depth translates to an average detection level of 12,226 genes per sample. By using well characterized mouse lines for specific labeling of neuronal subtypes, this analysis links gene expression patterns to sensory neuron subtypes defined by their distinct in vivo properties as well as their corresponding intrinsic properties, described above. This is particularly important for the three Aβ-LTMR subtypes, which account for low percentages of DRG neurons and have not been previously resolved by single cell sequencing approaches (Usoskin et al., 2015). Moreover, bulk RNA-sequencing at the high depth afforded by our analysis approaches or reaches saturation of gene detection, including the detection of low-expressed genes, and facilitates identification of differentially expressed genes (DEGs) (Li et al., 2016). Using this deep sequencing strategy, we generated a matrix of read counts of all genes for each of the eight major DRG neuron subtypes.
The count matrix was transformed using a regularized logarithm (rlog) implemented in DESeq2 and thus rendered homoscedastic (Love et al., 2014). Unsupervised hierarchical clustering and principal component analysis (PCA), using rlog values as measures of gene expression, showed that biological replicates for the same neuronal subtype cluster with each other (Figure 2C and S2A). This finding suggests that differences in gene expression across neuronal subtypes are not masked by variations across sequencing batches or noise introduced by experimental procedures.
We assessed the validity of the transcriptome profiles by examining expression patterns of several marker genes, including genes employed for the genetic tools and those whose expression patterns are well established in the DRG, as well as subtype-uniquely enriched genes (SUEGs) (Figure S2B and S2D) (See Star Methods for identification of SUEGs). For the former, expression patterns of several marker genes were found to be consistent with established knowledge (Figure S2B). For the latter, many SUEGs found in MrgD+ nonpeptidergic nociceptors, CGRP+ peptidergic nociceptors, C-LTMRs, and proprioceptors are in agreement with prior single-cell sequencing studies that identified these subtypes (data not shown) (Usoskin et al., 2015). Importantly, many SUEGs found in Aβ RA-LTMRs, Aβ SA1-LTMRs, and Aβ Field-LTMRs were identified here for the first time. Several SUEGs were also selected for validation by double immunostaining, double fluorescent in situ hybridization, and through characterization of a new mouse genetic tool (Calb1dgCre), which preferentially labels Aβ RA-LTMRs (Figure 2D–2L, Figure S2C–S2J). The deep transcriptome analysis of each of the eight major DRG neuron subtypes thus allows for the identification of gene candidates underlying subtype specific properties, including intrinsic physiological properties.
Functionally relevant genes are differentially expressed across DRG neuron subtypes
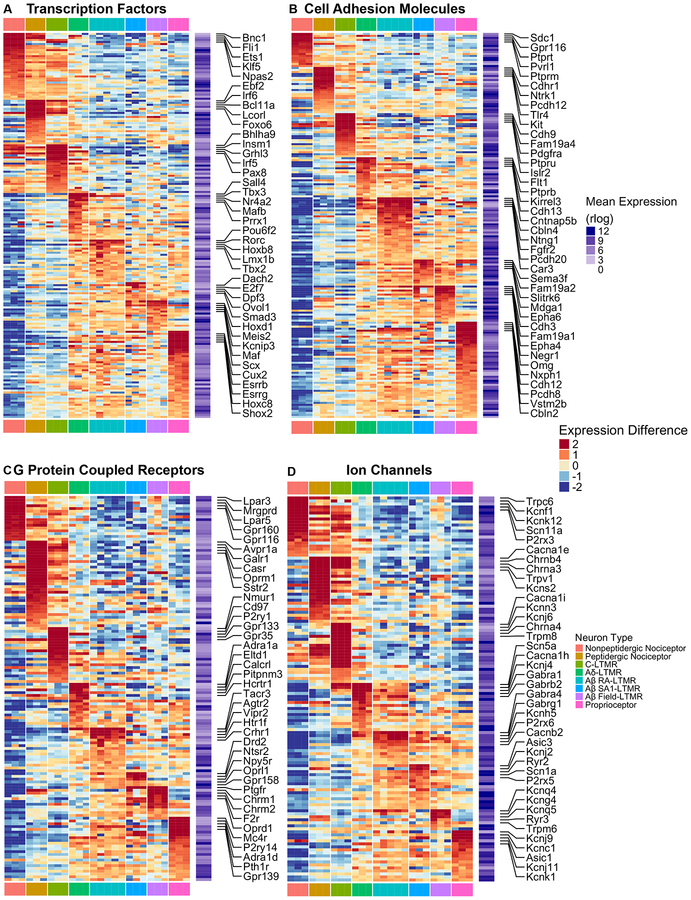
In addition to differences in intrinsic physiological properties, DRG neuron subtypes differ in their developmental trajectories, peripheral terminal morphologies, response properties, central projection patterns and postsynaptic partners, and modes of presynaptic modulation of their central terminals (Abraira and Ginty, 2013; Dubin and Patapoutian, 2010; Fleming and Luo, 2013; Marmigère and Ernfors, 2007; Rudomin, 1999; Zimmerman et al., 2014, 2019). To determine gene expression patterns that may underlie these and other subtype-specific properties, we next examined the expression patterns of genes that fall within six functionally relevant categories implicated in shaping neuronal phenotype. The gene categories explored are: transcription factors (TFs), cell adhesion molecules (CAMs), neuropeptides, synaptic exocytosis machinery, G protein-coupled receptors (GPCRs), and ion channels (Chawla et al., 2013; Földy et al., 2016; Paul et al., 2017; Südhof, 2012). Among the top 75% of expressed genes ranked by average rlog values, there are 511 transcription factors, 306 cell adhesion molecules, 183 GPCRs, 175 ion channel genes, 94 synaptic vesicle exocytosis genes and 23 neuropeptide genes. Genes from each of the categories are differentially expressed among the DRG neuron subtypes (Figure 3, S3A and S3B, See STAR Methods for selection of genes for plotting). The most dramatic differences in gene expression patterns were often observed between small-diameter neuron subtypes (MrgD+ nonpeptidergic nociceptors, CGRP+ peptidergic nociceptors, and C-LTMRs) and medium/large-diameter subtypes (Aδ-LTMRs, the three Aβ-LTMR subtypes, and proprioceptors), which is consistent with the expression structure revealed by hierarchical clustering and PCA analysis (Figure 2C and S2A). For example, most neuropeptide genes and several TRP channels, including TrpA, TrpM8, and TrpV1, are expressed at higher levels in the small-diameter neuron subtypes compared to large diameter subtypes (Figure S3A and S3C). In addition, most synaptic exocytosis machinery genes display elevated expression in either small-diameter or large-diameter neuron subtype clusters (Figure S3B). On the other hand, expression differences between small-diameter neuron subtypes or between medium/large-diameter neuron subtypes are less dramatic, although many notable differences exist (Figure 3, also see Figure 2C and S2A). These data also revealed many TFs, CAMs, receptors, and ion channels that are uniquely enriched in each of the three small-diameter neuron subtypes, Aδ-LTMRs and proprioceptors, however fewer genes in these families distinguish the three Aβ-LTMR subtypes, which likely contributed to the difficulty resolving Aβ-LTMR subtypes in prior single cell analyses.
Figure 3. Functionally relevant genes are differentially expressed across DRG sensory neuron subtypes.
(A-D) Heatmaps depicting expression patterns for genes encoding transcription factors and transcriptional regulators (A), cell adhesion molecules (B), G protein-coupled receptors (C), and ion channels (D). Plotting scheme, calculation for average expression and expression deviation from average are the same as in Figure 2D. Only genes that are both highly expressed (average expression in the upper 75th percentile) and highly variable (at least three samples have an expression deviation from average larger than 1) were included in the heatmaps.
See STAR Methods for details.
Expression patterns of ion channel genes in DRG neuron subtypes
Our transcriptome profile data resolve patterns and levels of expression of the genes encoding the principal or α subunits and auxiliary subunits of voltage-gated ion channels expressed in the DRG. This information provides a valuable guide for revealing the molecular identity of the ion channels that underlie subtype-specific intrinsic membrane properties and firing patterns, defined in experiments shown in Figure 1. Therefore, we focused on the patterns of expression of voltage-gated ion channels for each major DRG neuron subtype.
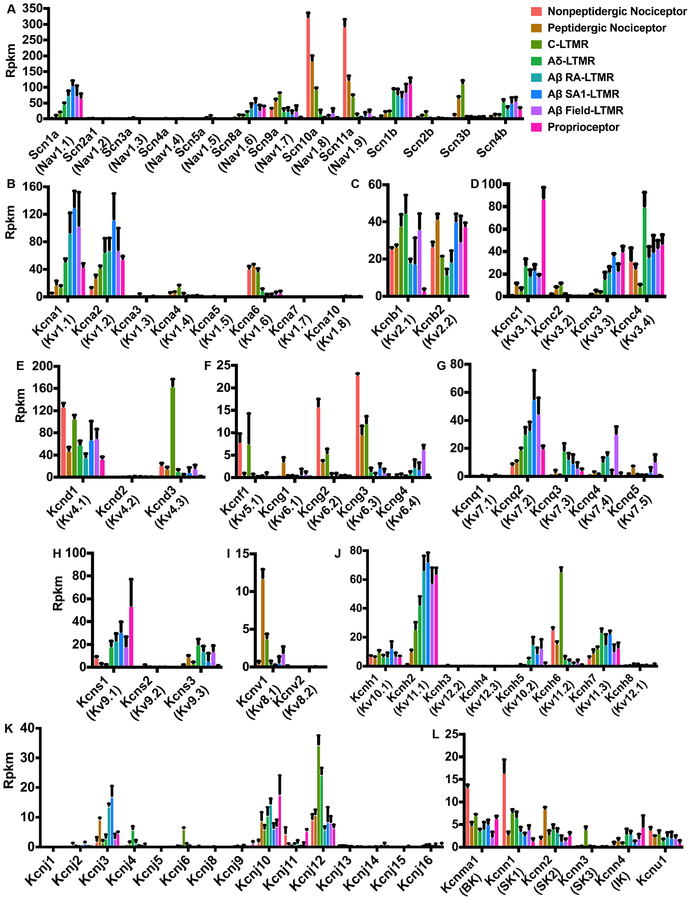
Nav channel α subunit genes show a clear distinction among the neuronal subtypes (Figure 4A). The most highly expressed Nav channels are Nav1.1, Nav1.6, Nav1.7, Nav1.8 and Nav1.9. Nav1.7 is expressed across all subtypes, with the notable exception of proprioceptors. On the other hand, Nav1.8 and Nav1.9 are most highly expressed in small-diameter neuron subtypes, whereas Nav1.1 and Nav1.6 are abundant in Aδ-LTMRs, the three Aβ-LTMR subtypes, and proprioceptors, but very low or undetectable in the small-diameter neuron types.
Figure 4. Expression patterns of genes encoding Nav and Kv channel subunits.
(A-L) Bar plots depicting expression patterns of genes encoding Nav channel α and β subunits (A), Kv1 subunits (B), Kv2 subunits (C), Kv3 subunits (D), Kv4 subunits (E), Kv5 and Kv6 subunits (F), Kv7 subunits (G), Kv8 subunits (I), Kv9 subunits (H), Kv10 subunits (J), inward-rectifier potassium channel subunits (Kir, IRK) (K) and calcium-activated potassium channel α subunits (L). Data are reported as mean ± standard deviation (SD).
K channel genes exhibit the greatest diversity among voltage-gated ion channels, with over ninety K channel subunit genes in the mouse genome (Coetzee et al., 1999; Vacher et al., 2008), and many show remarkably different patterns of expression among the DRG neuron subtypes (Figure 4B–4L, S4A–S4D and S3D). The expression of many different types of K channels is consistent with previous recordings showing multiple components of K current in DRG neurons, with considerable heterogeneity among neurons of different sizes (Everill et al., 1998; Gold et al., 1996). Within the Kv1 subfamily, Kv1.1 expression is elevated in large-diameter neuron subtypes compared to small-diameter neuron subtypes (Figure 4B). The pattern of Kv1.1 expression was confirmed by immunostaining experiments: most Kv1.1+ neurons are NF200+, but they do not express MrgD or TH, markers of MrgD+ nonpeptidergic nociceptors and C-LTMRs, respectively (Figure S5C and S5D). Kv1.2 expression is also relatively high in the large-diameter neuronal subtypes, especially compared to MrgD+ nonpeptidergic nociceptors, and, as with Kv1.1, this finding was supported by immunostaining: most Kv1.2+ neurons are NF200+, and Kv1.2+ neurons do not express MrgD (Figure S5E). Kv1.6 expression, on the other hand, is elevated in the small-diameter neuron subtypes and low or undetectable in large-diameter neuron subtypes. At least one Kv2 channel family member is expressed relatively abundantly (rpkm ≥15) in each of the neuronal subtypes (Figure 4C). Of the Kv3 channel family members, Kv3.1 and Kv3.3 are both enriched in the large-diameter neuron subtypes (Figure 4D); Kv3.1 is particularly highly enriched in proprioceptors where it may be critical for their fast spiking property. Both peptidergic and nonpeptidergic nociceptors express Kv3.4 channels, consistent with recent electrophysiology, single-cell qPCR, and immunocytochemistry (Muqeem et al., 2018; Ritter et al., 2012, 2015). Of the Kv4 family, Kv4.1 was expressed in all eight subtypes, consistent with widespread expression in DRGs seen by immunocytochemistry (Matsuyoshi et al., 2012; Phuket and Covarrubias, 2009), while Kv4.3 is uniquely and highly expressed in C-LTMRs (Figure 4E); this pattern was verified by immunostaining using Kv4.3-specific antibodies (Figure S5F). These results identify C-LTMRs as the population of small diameter DRG neurons previously associated with high Kv4.3 expression (Phuket and Covarrubias, 2009).
Among other differentially expressed K channel α and auxiliary subunit genes, BK and SK1 channels are elevated in MrgD+ nonpeptidergic nociceptors (consistent with previous work showing enrichment of BK current in a subpopulation of nociceptors (Zhang et al., 2010)), and Kv7.4 expression is elevated in Aδ-LTMRs, Aβ RA-LTMRs and Aβ Field-LTMRs (Figure 4G and 4L). The latter observation is consistent with the finding that Kv7.4 protein is detected in lanceolate endings and circumferential endings, but not in Merkel cell associated endings (Heidenreich et al., 2012). The highly diverse patterns of expression of Kv5, Kv6, Kv8 and Kv9 family members (Figure 4F, 4H and 4I) suggests that the complexity of K channel compositions among the DRG neuron subtypes is further increased by distinct heteromers formed between these electrically-silent (modulatory) channel subunits and Kv2 channels (Bocksteins and Snyders, 2012). Other voltage-gated ion channel genes, including Cav channels and HCN family members, are also differentially expressed among the eight major DRG neuron subtypes (Figure S4E–I). Cav2.1 (P-type) and Cav2.2 (N-type) calcium channels are relatively highly expressed in all subtypes, with much lower expression of Cav1.2 L-type calcium channels and very low expression of Cav1.3 channels. For T-type calcium channels, the expression of Cav3.2 is elevated in C-LTMRs and Aδ-LTMRs (Figure S4E), consistent with previous studies (François et al., 2015b; Wang and Lewin, 2011). Thus, each DRG neuronal subtype exhibits a unique combination of voltage-gated ion channels that together shape transduction of sensory information.
Major K channel families differentially contribute to outward potassium currents in a subtype-specific manner
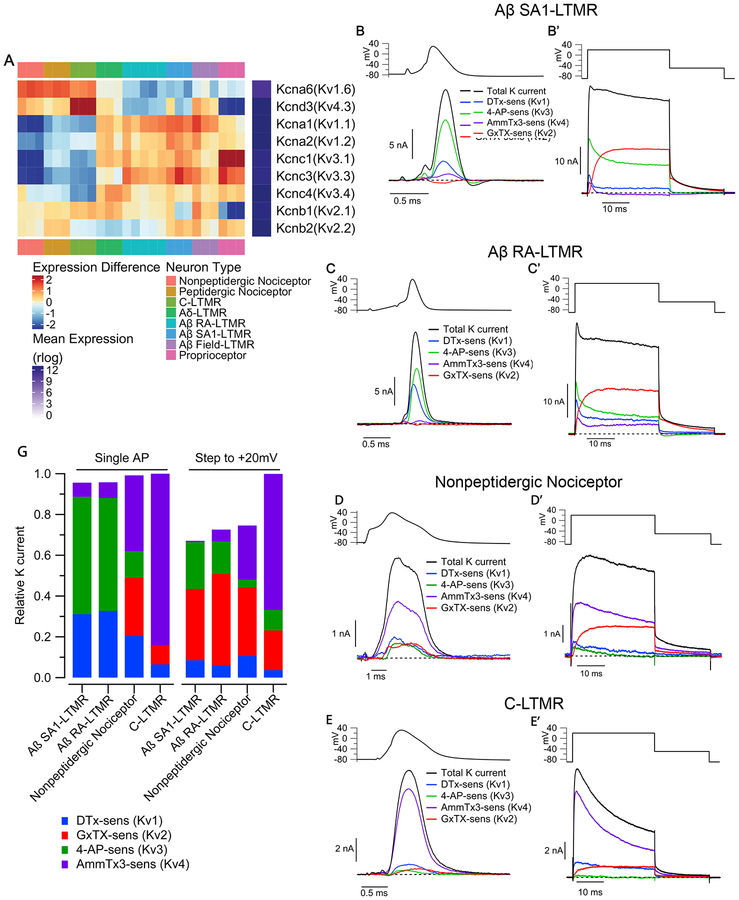
The unique combinations of Kv channel expression patterns among DRG sensory neuron subtypes suggests that differential expression of these particular ion channels is a key determinant of subtype-specific intrinsic physiological properties. The expression patterns of Kv1-Kv4 channels (Figure 5A, 4A–D) suggest that Kv1, Kv2, Kv3, but not Kv4.3, may mediate large outward potassium currents in Aδ-LTMR and Aβ-LTMR subtypes. Another prediction is that Kv4.3, but not Kv3 or Kv1, carries large outward potassium current in C-LTMRs. To test these and other possibilities raised by the sequencing analysis, we quantified the contributions of Kv1, Kv2, Kv3 and Kv4 channels to total potassium current in the genetically-defined subtypes using pharmacological dissection of the current.
Figure 5. Kv channel families differentially contribute to outward current in sensory neuron subtypes.
(A) Heatmap depicting expression patterns of the most abundantly expressed Kv1, Kv2, Kv3 and Kv4 subunits. Plotting scheme is as Figure 2D.
(B-E) Components of Kv current during the AP of sensory neuron subtypes. Currentwas evoked by AP waveforms (previously recorded in a different cell of each type), and components of current were isolated by sequential cumulative application of 100 nM α-Dendrotoxin (DTX), 100 μM 4-aminopyridine (4-AP), 3 μM AmmTx3, and 100 nM Guangxitoxin-1E (GxTX) to identify Kv1, Kv3, Kv4, and Kv2 currents, respectively. (B’-E’) Currents evoked by a step depolarization to +20 mV (30 ms), applied in parallel with the AP commands in the same cells as B-E.
(F) Stacked bar plots showing the average fraction of total outward K+ current carried by Kv1, Kv2, Kv3, Kv4 channels in Aβ SA1-LTMRs (AP waveform: 31 ± 3% Kv1, 0 ± 1% Kv2, 59 ± 3% Kv3, 7 ± 1% Kv4; Step to +20 mV: 9 ± 2% Kv1, 35 ± 3% Kv2, 22 ± 3% Kv3, 1 ± 1% Kv4; n=15), Aβ RA-LTMRs (AP waveform: 33 ± 4% Kv1, 0 ± 2% Kv2, 58 ± 4% Kv3, 8 ± 2% Kv4; Step to +20 mV: 6 ± 3% Kv1, 45 ± 8% Kv2, 16 ± 7% Kv3, 6 ± 3% Kv4; n=11), MrgD+ nonpeptidergic nociceptors (AP waveform: 20 ± 4% Kv1, 29 ± 6% Kv2, 13 ± 3% Kv3, 37 ± 6% Kv4; Step to +20 mV: 11 ± 3% Kv1, 34 Kv4; n=11; mean ± SEM). Current contributions were quantified by integrating the currents during the AP or the 30ms step depolarization.
We quantified the fraction of each current involved in repolarizing the AP in each cell type by using previously-recorded AP waveforms in each DRG neuron subtype (Figure 1) as a voltage command and then cumulatively applying a series of Kv1, Kv2, Kv3, and Kv4 inhibitors in an order designed to maximize selective inhibition at each step (Figure S5A, S5A’, and S5B). Total K current was defined at the end of the sequence using a high concentration (158.5 mM) of TEA together with all the Kv channel inhibitors.
Using this protocol, we found that in Aβ SA1-LTMRs and Aβ RA-LTMRs, Kv1 and Kv3 account for > 80% of total potassium current during their APs (Figure 5B, 5C, and 5G). These two Kv channel families also account for the majority of potassium current during the AP in proprioceptors (19 ± 5% from Kv1 and 40 ± 6% from Kv3, n=11). We compared the currents activated during the AP waveform with that activated by a step to +20 mV for 30 ms, long enough to achieve maximal activation for each channel type. Interestingly, in Aβ SA1-LTMRs and Aβ RA-LTMRs, Kv2 channels underlie a major component (~60–70%) of potassium current during 30 ms step depolarizations to +20 mV but contribute almost no current during the AP (Figure 5B’, 5C’ and 5G), apparently because of the slow opening kinetics of Kv2 channels and the relatively narrow AP waveforms of Aβ-LTMR subtypes. On the other hand, in MrgD+ nonpeptidergic nociceptors, which have wide AP waveforms, Kv2 channels contribute substantial current during both AP and step depolarization commands, along with Kv1 and Kv4 channels; Kv3 channels also carry significant current during the AP (13% of the total; Figure 5D, 5D’ and 5G). Strikingly, in C-LTMRs, Kv4 channels are the primary mediator (>80%) of potassium current during both step depolarization and the AP (Figure 5E, 5E’ and 5G), thus revealing a major difference compared to Aβ-LTMRs, where Kv4-mediated current is minor. Thus, overall potassium current is comprised of considerably different combinations of current from the various Kv channel families in the different DRG neuron subtypes.
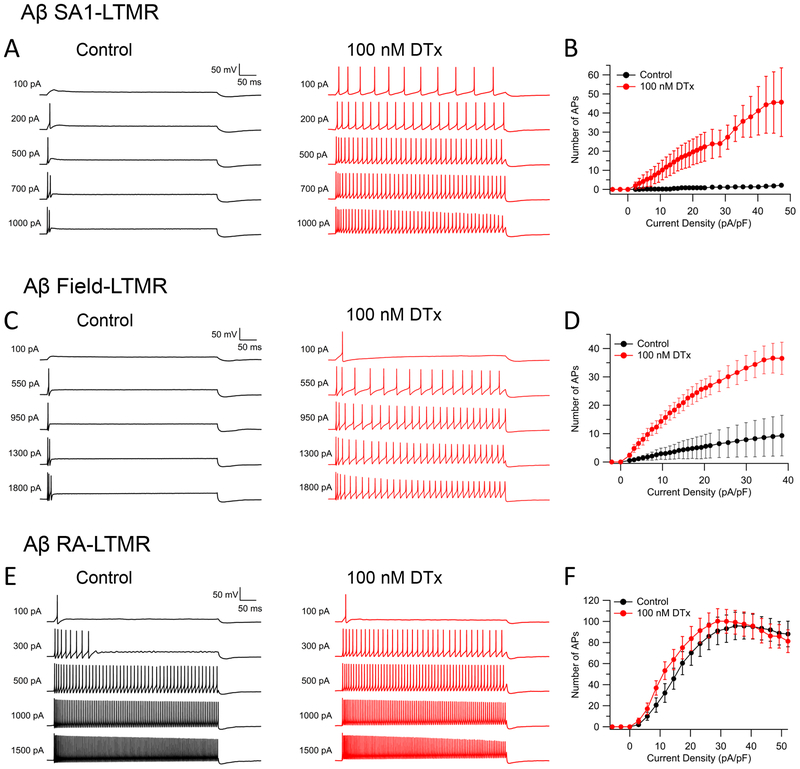
Kv1 channels govern firing patterns in a subtype-specific manner
We next explored the functional roles of key differentially-expressed Kv channels in shaping subtype-specific firing properties. We explored the effect of inhibiting Kv1 channels using the Kv1 blocker α-DTX. All the cell types expressed various Kv1 subunits, primarily Kv1.1, Kv1.2, and Kv1.6, with differing expression levels in the different cell types of each subunit, all of which are targeted by α-DTX. In Aβ SA1-LTMRs and Aβ Field-LTMRs, which both express high levels of Kv1.1 and Kv1.2, blocking Kv1 channels with α-DTX dramatically converted their characteristic strongly-adapting firing patterns to sustained repetitive firing lasting throughout a 500-ms current injection (Figure 6A–6D). In Aβ RA-LTMRs, which express substantial Kv1.1 and Kv1.2 subunits (but less than Aβ SA1-LTMRs), α-DTX enhanced firing for small depolarizations but had little effect on repetitive firing evoked by large current injections (Figure 6E–6F). In both MrgD+ nonpeptidergic nociceptors and C-LTMRs, Kv1.6 is enriched, compared to medium/large-diameter neuron subtypes. These two subtypes of small-diameter neurons both responded to α-DTX application but in different ways, with α-DTX substantially increasing the maximal firing rate of MrgD+ nonpeptidergic nociceptors which is relatively low (~10 Hz) in control conditions (Figure S6A–S6B), while having little effect on maximal firing rate of C-LTMRs (which in control conditions fire up to ~28 Hz) but inducing an early cessation of firing and thus a rapid drop in spike number during large current injections (Figure S6C–S6D). This effect may result from a more depolarized membrane potential during the trough between APs, facilitating inactivation of sodium channels. These findings show that inhibiting Kv1 currents alters firing of most DRG subtypes, in line with previous reports on both large-diameter (Glazebrook et al., 2002) and small-diameter (Chi and Nicol, 2007) primary sensory neurons, but with a wide range of effects on firing in different cell types. The differing effects likely reflect a combination of the levels of expression together with the different context of all the other channel types in the neurons as well as potentially different kinetic properties of Kv1 channel subunit compositions (including accessory subunits).
Figure 6. Blocking Kv1 channels dramatically increases repetitive spiking in Aβ SA1-LTMRs and Aβ Field-LTMRs but not in Aβ RA-LTMRs.
(A, C, E) Representative voltage traces showing the effect of 100 nM α-DTX on spike patterns of an Aβ SA1-LTMR (A), an Aβ Field-LTMR (C), and an Aβ RA-LTMR (E) during 500-ms current injections.
(B, D, F) Number of spikes during 500-ms current injections are plotted against injected current density before (black curve) and after (red curve) α-DTX for Aβ SA1-LTMRs (B, n=6), Aβ Field-LTMRs (D, n=13) and Aβ RA-LTMRs (F, n=15). Data are represented as mean ± SEM.
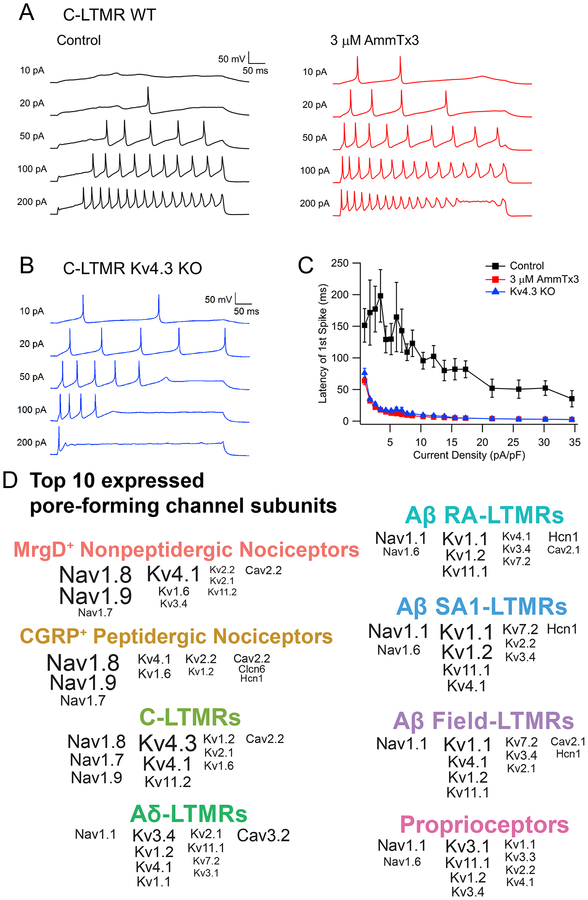
Kv4.3 uniquely regulates the delayed firing pattern of C-LTMRs
In contrast to Aβ-LTMRs, Kv4 channels are the major source of K current in C-LTMRs. In addition, in vivo immunostaining showed that Kv4.3 is present on peripheral endings and likely at central terminals of C-LTMRs, suggesting a unique function of Kv4.3 in these neurons (Figure S7). Therefore, we next recorded C-LTMR firing patterns in the presence and absence of the Kv4 channel blocker AmmTx3. Interestingly, AmmTx3 application greatly reduced the long latency to the first AP following current injection, normally a distinguishing feature of C-LTMRs (Figure 7A and 7C). Complementary experiments were performed using mice in which the Kcnd3 (Kv4.3) gene was ablated (Figure 7B and 7C). Consistent with the pharmacology experiments, C-LTMRs with a targeted deletion of Kv4.3 exhibited a markedly reduced latency to the first AP, confirming that Kv4.3 is indeed essential for the delayed firing of C-LTMRs. Thus, as with Kv1 family members and Aβ-LTMR subtypes, Kv4.3 is uniquely expressed in C-LTMRs and governs unique aspects of their intrinsic physiological properties.
Figure 7. Kv4.3 underlies the delayed firing pattern of C-LTMRs.
(A) Representative voltage traces showing the spike patterns of a C-LTMR during 500-ms sustained current injections before and after application of the Kv4 channel inhibitor AmmTx3.
(B) Representative spike pattern from a C-LTMR neuron from a Kv4.3D mouse.(C) Plot depicting latencies of the first spike as a function of injected current density for wild-type C-LTMRs, before (black squares) and after (red squares) AmmTx3 application (n=8), and for C-LTMRs from Kv4.3D mice (blue triangles) (n=13). Data are represented as mean ± SEM.
(D) Summary plot showing the ten most highly expressed voltage-gated ion channel α subunits in each sensory neuron subtype, ranked by rpkm value. The font size is proportional to the expression level in rpkm, but maxed out at rpkm=150.
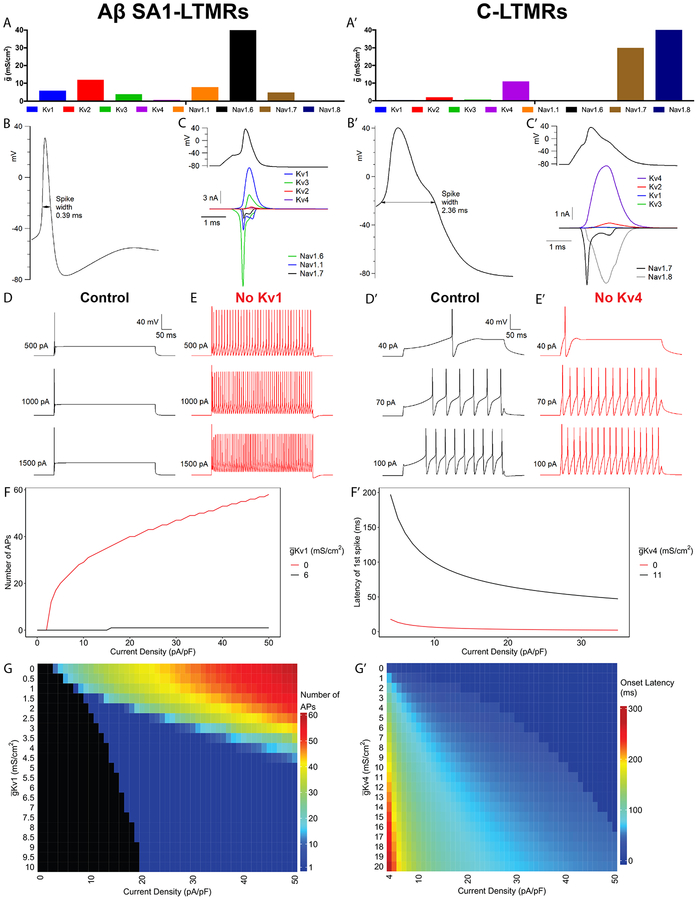
Computational modeling defines roles of Kv1 and Kv4.3 channels
These pharmacological and genetic manipulation experiments support the notion that expression of unique combinations of voltage-gated ion channels in the eight DRG sensory neuron subtypes (summarized in Figure 7D) are critical for subtype-specific intrinsic physiological properties. To further explore how expression levels of key channels control neuronal firing properties, we constructed computational models incorporating the major subtypes of Nav and Kv channels expressed in the sensory neuron subtypes (Figure 8). Setting the size of each conductance guided by the relative levels of RNA and the maximal conductances defined by voltage steps resulted in action potential shapes and firing patterns similar to experimental recordings (Figure 8A–D, A’–D’). The models recapitulated the essential role of Kv1 channels in producing non-repetitive firing in Aβ SA1-LTMRs (Figure 8D–F) and the key role of Kv4 channels in controlling the long onset latency of firing in C-LTMRs seen experimentally (Figure D’–F’). The model showed that the effect of Kv4 to produce delayed firing, also noted in some central neurons (Molineux, 2005; Shibata et al., 2000), occurs because Kv4 current activates rapidly at subthreshold voltages but then inactivates over ~100 ms, producing a slow approach to threshold. This mechanism is particularly effective in C-LTMRs because spike threshold is relatively depolarized (~−25 mV) compared to Aβ neurons (~−50 mV) as a result of lack of low-threshold Nav1.6 expression and the depolarized activation range of Nav1.8 channels.
Figure 8. Computational modeling suggests key roles of Kv1 and Kv4 in firing patterns of Aβ SA1-LTMRs and C-LTMRs.
(A-A’) Maximum conductances (ḡ) of each channel for Aβ SA1-LTMR (A) and C-LTMR (A’) models.
(B, B’) Waveforms of the first AP in response to near threshold 500-ms sustained current injection in Aβ SA1-LTMR (450pA) (B) and C-LTMR (40pA) (B’) models.
(C, C’) Current carried by each channel during the AP of Aβ SA1-LTMR (C) and C-LTMR (C’) models, using an AP waveform evoked by a 0.5-ms current injection.
(D-E) Firing patterns of Aβ SA1-LTMR model neuron evoked by 500-ms sustained current injections with Kv1 (ḡKv1 = 6 mS/cm2) (D) or with Kv1 removed from the model (E).
(F) Number of APs in the Aβ SA1-LTMR model as a function of injected current density (500-ms current injection) with Kv1 (ḡ Kv1 = 6 mS/cm2; black trace) or with Kv1 removed (red trace).
(G) Heatmap of the number of APs during 500 Kv1-ms current injections as a function of current density as well as ḡKv1.
(D’-E’) Firing patterns of C-LTMR model evoked by 500-ms current injections with Kv4 (ḡKv4 = 11 mS/cm2) (D’) or with Kv4 removed (E’).
(F’) Onset latency in current-clamp simulations of C-LTMR model as in (D’-E’) is plotted against varying levels of injected current density with Kv4 (ḡKv4 = 11 mS/cm2; black trace) or with Kv4 removed (red trace).
(G’) Heatmap of onset latency during 500-ms current injection as a function of current density as well as ḡ Kv4.
We also explored the impact of varying the levels of Kv1 and Kv4 in the Aβ SA1-LTMR and C-LTMR model neurons, respectively (Figure 8G, G’). In the Aβ SA1-LTMR model, the range of injected current amplitudes for which the model neuron fired a single spike displayed a near-linear change as ḡKv1 was varied between 1 to 5 mS/cm2. This suggests that differences in Kv1 levels observed between Aβ SA1-LTMRs and Aβ RALTMRs in the RNA-Seq data and also in voltage-clamp recordings may explain the respective nonrepetitive-firing and repetitive firing patterns of these two LTMR subtypes in the experimental recordings. For the C-LTMR computational model, increasing ḡ Kv4 increased the AP onset latency at all levels of current injection, as larger Kv4 current required more complete inactivation for spike threshold to be reached.
DISCUSSION
While it is well established that primary somatosensory neurons of the DRG are tunedto distinct stimuli and display unique firing patterns, the molecular machinery underlying their unique properties has remained largely elusive. Here, we used a combination of sensory neuron subtype-specific genetic labeling and deep RNA-sequencing to generate transcriptome profiles for each of eight major DRG neuron subtypes. The deep transcriptome analysis dataset has unique value for assessing the contributions of genes in a sensory neuron subtype-specific manner. While prior bulk sequencing or microarray studies using genetically labeled DRG neuron subtypes have been reported, these studies either used genetic tools targeting broad populations of DRG neurons rather than functionally or physiologically defined subsets or sequenced a limited number of subtypes (Chiu et al., 2014; Fleming and Luo, 2013). In addition, while previous unsupervised single cell RNA sequencing (scRNA-seq) studies of DRG neurons have provided valuable resources (Li et al., 2016; Usoskin et al., 2015), these prior studies did not resolve several key functionally distinct neuronal subtypes, notably the Aβ-LTMR subtypes. This is likely due to the relatively small numbers of Aβ-LTMRs, similarities in their gene expression patterns, especially for Aβ SA1-LTMRs and Aβ Field-LTMRs, the small numbers of neurons sequenced in prior studies, and the potential difficulty maintaining the integrity of large diameter DRG neurons during isolation procedures. For guiding functional analyses, the transcriptome profiles generated in the present study cover eight major DRG neuron subtypes, including three Aβ-LTMR subtypes, Aδ-LTMRs and C-LTMRs, with high sequencing depth. Indeed, the transcriptome profiles reported here reveal a large number of functionally relevant genes, including voltage gated Na+, K+ and Ca2+ channels, and many lowly expressed genes that are differentially expressed across DRG neuron subtypes, thus providing a resource for identifying molecular underpinnings of subtype-specific electrophysiological properties and functions. The transcriptome profiles also reveal genes useful for generating new genetic tools, as demonstrated here for Calb1 (Figure S2C–S2J). Thus, the present deep sequencing analysis of genetically defined DRG sensory neuron subtypes complements and extends previous RNA-seq studies. A notable disadvantage of our present bulk sequencing strategy is that we could not identify new neuronal subtypes within the broad CGRP+ peptidergic nociceptor population, which we found to be physiologically heterogeneous. Moreover, our genetic labeling strategies did not account for ~15% of DRG neurons, which likely include several small-diameter nonpeptidergic neuron populations and select subtypes of glabrous skin and deep tissue-innervating LTMRs. Future work using advanced scRNA-Seq approaches for larger scale analyses may be useful for defining transcriptional profiles of neuronal subtypes not covered in the present study.
The transcriptome profiles presented here reveal a great diversity of voltage-gated ion channels expressed among the neuronal subtypes. One of the most notable differences observed is for Nav channels, of which the expression patterns are largely consistent with previous work that assessed the distribution of Nav channels in DRG neurons with different sizes (Dib-Hajj et al., 2010). Although a recent study suggested that Nav1.1 uniquely functions in myelinated nociceptors (Osteen et al., 2016), our findings implicate this channel, along with Nav1.6, as the major determinants of Nav currents in most and possibly all large-diameter DRG sensory neuron subtypes. This is particularly interesting when considering approaches to block or reduce excitability of large diameter sensory neuron types, including Aβ-LTMR subtypes for the prevention of tactile hypersensitivity and mechanical allodynia in neuropathic pain states. Future work will be required to assess the relative contributions of Nav1.1 and Nav1.6 in Aδ-LTMRs, Aβ RA-LTMRs, Aβ SA1-LTMRs and Aβ Field-LTMRs under both normal and disease conditions.
Towards our goal of defining the molecular basis of sensory neuron subtype-specific intrinsic membrane properties, we used the transcriptome findings to guide an analysis of the contributions of several families of Kv channels. The results of both pharmacological block and computational modeling suggest that Kv1.1 and Kv1.2, which are highly expressed in Aβ SA1-LTMRs and Aβ Field-LTMRs, are responsible for the strong adaptation of these neurons. C-LTMRs are unusual in that the dominant potassium current is from A-type Kv4 channels, reflecting high expression of Kv4.3 subunits. Genetic deletion, pharmacological inhibition, and computational modeling all showed that Kv4.3-mediated current is critical for the delayed firing property of C-LTMRs, which is a unique electrophysiological signature of these neurons.
Our electrophysiological analysis focused on the firing patterns of DRG cell bodies. Of course, different cell regions almost certainly have different expression levels of the channels described herein (Trimmer, 2015). In future work it will be important to explore possible region-specific expression of the channels. In the case of Kv4.3 in C-LTMRs, immunohistochemical analysis showed that this channel is distributed throughout the entire cell surface of C-LTMRs, including its peripheral lanceolate endings around hair follicles and likely its central terminals in dorsal horn lamina IIiv. This subcellular localization pattern is in contrast to brain neurons, in which Kv4.3 is highly polarized, with localization limited to dendrites, and in certain cells to cell bodies, but not on axons in any cell types investigated (Trimmer, 2015; Vacher et al., 2008). Considering that Kv4.3 operates within the subthreshold voltage range, this channel may function in the cutaneous ending to mediate signal integration and govern AP initiation, which in turn may explain the observation that C-LTMRs are most highly sensitive to stroking across the skin at low speeds (Bessou et al., 1971; Löken et al., 2009).
In summary, deep sequencing of genetically labeled sensory neuron subtypes has afforded an opportunity to explore the molecular determinants of DRG sensory neuron subtype-specific intrinsic physiological properties. Our pharmacological and genetic dissection of a subset of K channels as well as computational modeling reveal unique roles of channel families in shaping firing properties. These analyses demonstrate the predictive value of the transcriptome data for revealing ion channels of somatosensory neuron subtype-specific intrinsic firing properties. The transcriptome profiles may also guide analyses of functions of other genes in somatosensation.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact David Ginty david_ginty@hms.harvard.edu.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mouse lines are listed in the Key Resources Table. Mice were group housed in a climate-controlled environment with ad libitum access of food and water, on a 12-hour light/dark cycle (7AM lights on and 7PM lights off). Mice were handled and housed in accordance with the Harvard Medical School and Johns Hopkins University IACUC guidelines. For all analyses, the morning after coitus (vaginal plug is seen) was designated as embryonic day 0.5 (E0.5) and the day of birth as postnatal day 0 (P0). Mice were ear tagged before postnatal day 21 or toe tagged at postnatal day 5–7 for determination of their genotypes. Chemical administration protocols for certain mouse lines are described in the METHOD DETAILS section. For all experiments, both male and female adult mice (over postnatal day 21 old) on a mixed background were used.
METHOD DETAILS
Tamoxifen treatment
Tamoxifen was dissolved in ethanol (20 mg/ml), which was stored at −20°C, then mixed with double volume of sun flower seed oil (Sigma), vortexed for 5–10 minutes and centrifuged under vacuum for 20–30 min to remove the ethanol. The working solution (10 mg/ml) was kept at −20°C and delivered via oral gavage. For labeling C-LTMRs, 2 mg/day for two days of tamoxifen were given at either P13–14 or P21–22. The latter was used for Th2A-CreER; R26lsl-tdTomato; Kcnd3−/− mice after weaning, so that parents avoided exposure to tamoxifen. Labeling specificity at both time windows is high. For labeling Aδ-LTMRs, tamoxifen was given to TrkBCreER; R26lsl-tdTomato (Ai14) mice at E12.5–14.5 to reduce glial cell labeling, which was critical for FACS sorting. For labeling Aβ SA1-LTMRs, tamoxifen was given to TrkBCreER; R26lsl-tdTomato (Ai14) mice at E12.5–14.5 to reduce glial cell labeling. For labeling Aβ SA1-LTMRs, tamoxifen was given to TrkCCreER; RetfGFP at E12.5 before 1PM; tamoxifen delivery at a later time point may increase offt-arget labeling.
Electrophysiology
Preparation of DRG neurons.
Acutely dissociated DRG neurons were prepared using enzymatic treatment as previously described (Liu et al., 2017). Briefly, DRG neurons were removed from mice (over P21), cut in half and treated for 20 minutes at 37°C with 20 U/ml papain (Worthington Biochemical, Lakewood, NJ) and 5 mM DL-cysteine in a calcium-and magnesium-free (CMF) Hank’s buffer containing 137 mM NaCl, 5.36 mM KCl, 0.33 mM Na2HP4, 0.44 mM KH2PO4, 5 mM HEPES, 5.55 mM glucose, 0.001% phenol red, pH 7.40 adjusted with NaOH; 300~310 mOsm. Ganglia were then treated for 20 minutes at 37°C with 3 mg/ml collagenase (ty pe I; Roche Diagnostics, Indianapolis, IN) and 4 mg/ml dispase II (Roche Diagnostics) in CMF Hank’s buffer. Cells were dispersed by trituration with a fire-polished glass Pasteur pipette in a solution composed of two media combined in a 1:1 ratio: Leibovitz’s L-15 medium (Invitrogen, Grand Island, NY) supplemented with 5 mM HEPES, and DMEM/F12 medium (Invitrogen); this solution also had added 100 ng/ml nerve growth factor (NGF) (Invitrogen). Cells were then plated on glass coverslips treated with 40 μg/ml poly-D-lysine and then 20 μg/ml laminin (Invitrogen). Then cells were incubated at 37°C (95% O2, 5% CO2) for 3 hours, after which Neurobasal medium (Invitrogen) containing B-27 supplement (Invitrogen), penicillin and streptomycin (Sigma, St. Louis, MO), and 100 ng/ml NGF was added to the petri dish. Cells were stored at 4°C and used within 48 hours. DRG neurons with specific molecular markers were chosen for following electrophysiological recordings.
Electrophysiology.
Recordings were performed at 37 °C using an Axon I nstruments Multiclamp 700B Amplifier (Molecular Devices). Voltage or current commands were delivered and signals were recorded using a Digidata 1321A data acquisition system (Molecular Devices) controlled by pCLAMP 9.2 software (Molecular Devices). Electrodes were pulled on a Sutter P-97 puller (Sutter Instruments) and shanks were wrapped with Parafilm (American National Can Company) to allow optimal series resistance compensation without oscillation. The resistances of the pipettes were 2–4 M. Seals were formed in Tyrode’s solution consisting of 155 mM NaCl, 3.5 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, pH 7.4 adjusted with NaOH. After establishing whole-cell recording, cell capacitance was nulled and series resistance was partially (70–85%) compensated. The cell was then lifted and placed in front of a series of quartz fiber flow pipes attached with cyanoacrylate glue to a rectangular aluminum rod (cross section 1.5 cm × 0.5 cm) whose temperature was controlled to 38°C using resistive heating elements and a feedback-controlled temperature controller (TC-344B, Warner Instruments). The end of the rod as well as the flow pipes (extending 1 mm from the end of the rod) were lowered just to the surface of the bulk chamber solution, which was locally quickly warmed by the rod; with the temperature of the aluminum rod set to 38°C, the so lution exiting from the flow pipes was measured at 37°C (Carter and Bean, 2009). All f low pipes are heated identically and continuously, allowing rapid solution changes without fluctuations of temperature. Solutions were changed (in ~1 second) by moving the cell from one pipe to another. Solutions. The standard recording solutions had quasi-physiological ionic composition, with an internal solution consisting of 140 mM K aspartate, 13.5 mM NaCl, 1.6 mM MgCl2, 0.09 mM EGTA, 9 mM HEPES, 14 mM creatine phosphate (Tris salt), 4 mM MgATP, 0.3 mM Tris-GTP, pH 7.2 adjusted with KOH and an external Tyrode’s solution consisting of 155 mM NaCl, 3.5 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, pH 7.4 adjusted with NaOH.
Sequence of inhibitors for dissecting K current components.
The inhibitors were applied cumulatively in a sequence designed to maximize the effective selectivity for identifying each component of K current by applying imperfectly-selective inhibitors after the main off-target channels had already been inhibited. The K channel inhibitors were applied on a background of TTX (1 μM) and A-803467 (1 μM) to inhibit Na channels. First, α-dendrotoxin (Alomone Labs), which is highly selective for Kv1.1, Kv1.2, and Kv1.6 channels (Harvey and Robertson, 2004), was applied at 100 nM. Next, 4-aminopyridine (4-AP) (Sigma) was applied at 100 μM, a concentration which should inhibit Kv3 channels almost completely while having minimal effects on other channels except Kv1, which was already blocked (Coetzee et al., 1999; Gutman, 2005). Next the Kv4 inhibitor AmmTx3 (Smartox Biotechnology) (Maffie et al., 2013; Pathak et al., 2016) was applied at 3 mM, followed by the Kv2 inhibitor Guangxitoxin-1E (Peptide Institute) at 100 nM (Liu and Bean, 2014). Finally, a high concentration of TEA was applied (in the continuing presence of the other inhibitors) to inhibit any remaining K current, using a solution in which TEA replaced both Na and K (158.5 mM TEACl, 1.5 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, pH 7.4 adjusted with TEAOH); block by this solution relative to control was considered to comprise the total K current.
In vitro mechanoclamp experiments
Cell culture.
Digestion solution, which contained 5mg/mL dispase (Gibco Invitrogen, 17105-041), 2mg/mL collagenase (Type I, Worthington, LS004196) and 0.1mg/mL DNase (Sigma, DN25) in HBSS, was aliquoted and stored in −80°C. DH10 solution was made with 1´Penicillin-Streptomycin (Pen-Strep) (Life Technologies, 15070–063), 10% SCG FBS (Life Technologies, 16000–044, thawed and heat-inactivated in 55°C water bath for 30 minutes, aliquoted and stored in −20°C) in DMEM (high glucose DMEM, Life Technologies, 11965–118), filtered through 0.2 m filters, and stored in 4°C for up to a month. Poly-D-lysine coated glass bottom petri dishes (MatTek, P35GC-1.5–10-C) were coated with 200 L laminin (10 g/mL) (Life Technologies, 23017–015), incubated at 37°C overnight. The petri dishes were rinsed with ddH2O three times and dried in a sterile fume hood. DRGs were freshly dissected out from mice (over P21) and placed into ice-cold DH10, briefly washed once in 1´HBSS, then digested in 1mL digestion solution at 37°C for 30–40 minutes under constant rotation, and then centrifuged at 200 g for 5 minutes. The supernatant was removed while the precipitant was carefully resuspended in 1mL DH10. Resuspended cells were gently triturated for 10–15 times using a pipette tip opening diameter no smaller than 0.5 mm, then allowed to recover for 1–2 minutes before the supernatant was collected and plated on glass areas in the dishes. Petri dishes were incubated in 37°C humidified atmosphere (5% CO2) for 20 minutes, then ~2 mL pre-warmed DH10 medium together with 25 ng/mL nerve growth factor (NGF), 25 ng/mL brain-derived neurotrophic factor (BDNF), 25 ng/mL neurotrophin (NT3) and 2 ng/mL glia-derived neurotrophic factor (GDNF) was added. Neurons were kept in the incubator overnight.
Voltage-clamp with controlled mechanical stimulations.
The method for in vitro mechanoclamp experiments was modified from that of Hao and Delmas, 2011. Mechanical probes and patch pipettes were fabricated from borosilicate glass tubing (Sutter, BF150-86-10). Pipettes were pulled on a P-97 pipette puller (Sutter) and fire-polished using a microforge (Narishige, MF-830). Mechanical probes were polished to obtain a blunt tip that had a diameter of 3–4 μm. Patch pipettes has a resistance of 2~4 MW. For voltage-clamp recordings, internal solutions contained 125 mM CsCl, 1 mM MgCl2, 4.8 mM CaCl2, 10 mM HEPES, 10 mM EGTA, 4 mM Mg-ATP and 0.4 mM Na-GTP with pH adjusted with CsOH to 7.4, osmolarity adjusted to 300 mOsm with CsCl. Extracellular solutions contained 132 mM NaCl, 3 mM KCl, 1 mM MgCl2, 2.5 mM CaCl2, 10 mM HEPES and 10 mM glucose with pH adjusted with CsOH to 7.4, osmolarity adjusted to 300 mOsm with CsCl. For mechanical stimulation, a piezoelectric actuator (P-601.1S, Physik Instrument) was fixed on a micromanipulator and a pipette holder was mounted on the actuator. The mechanical probe was positioned at an angle 45–60° from the horizontal plane. Movement of the mechanical probe was controlled and monitored using the pCLAMP program. A step protocol was used to deliver mechanical stimulations with increasing displacements at 1 μm increments. The probe moved at a speed of 500 m/s (measured between 10%−90% distance) and displacement lasted for 500 ms, followed by a 10s interval before the next stimulation. The zero position was found as follows: first, the probe tip was moved close to the cell under visual guidance; then a protocol with 4,5,6 μm stimulations was run. When the 4-μm stimulation did not yield any membrane deformation while 6-m movement yielded visible membrane deformation, the zero position was defined as 5-m movement of the probe.
Fluorescence-activated cell sorting (FACS)
For preparation, three solutions were made beforehand. Digestion solution and DH10 medium were prepared and stored as described for the cell culture and in vitro mechanoclamp procedures. FACS solution included 50 mg/mL bovine serum albumin (BSA) (Sigma, A6003), 1M HEPES (Sigma, H3375), 1´Pen-Strep, 0.5 mg/mL DNase in L-15 medium (Gibco, 11415). FACS solution was made fresh for every FACS experiment. For tissue collection, DRGs from all axial levels (except for proprioceptors, for which only non-limb level DRGs were used, because the genetic strategy used for labeling proprioceptors also labels limb level cutaneous LTMRs.) were freshly dissected from mice (3–12 weeks of age) and placed into ice-cold DH10 with roots carefully removed, briefly washed once in 1´HBSS, then digested in 1mL digestion solution at 37°C under constant rotation. DRGs from up to six mice were collected for each FACS experiment and the total dissection time was kept under three hours. The digestant was centrifuged at 200 g for 5 minutes. The supernatant was removed while the precipitant was carefully resuspended in 1 mL DH10. Resuspended solutions were gently triturated 10–15 times using a pipette tip opening diameter no smaller than 0.5 mm, rested for 1–2 minutes before supernatants were collected to a new tube and then the precipitants underwent the same process of trituration, rest and collection of supernatants for a second time. Collected supernatants were passed through a 100 μm cell strainer (pre-rinsed with DH10) (Fisher, 08-771-19) followed by 2–3 mL DH10, and all solutions were collected and centrifuged at 200–300 g for 5 minutes. Supernatants were removed, and precipitants were gently resuspended in 8 mL DH10, centrifuged again and the pellets were resuspended in 1 mL FACS solution. The samples were immediately transported to a flow-cytometry facility on ice. MoFlo (Johns Hopkins School of Medicine) and Avalon (Harvard Medical School) sorters with 100 μm or 150 μm nozzles were used to sort fluorescent cells under pressure lower than ~20 psi. Sorting criteria were selected monitoring distribution of GFP and tdTomato signals, forward scatter, side scatter and pulse widths. Propidium iodide (PI) staining was sometimes used to exclude dead cells. A small number of sorted cells was collected on a slide or culture medium and examined under fluorescent microscope during sorting for confirmation of fluorescence. Limited by the minimum amount of input RNA needed for sequencing, at least 2000 neurons were collected for each sample, except for one Aβ RA-LTMR sample that has ~1600 neurons. Sorted cells used for RNA extraction were collected in RNAlater solution (Life Technologies, AM7022), centrifuged, placed in 4°C overnight and sometimes stored in −80°C for several days. RNA was purified using the Absolutely RNA nanoprep kit (Agilent, 400753).
RNA sequencing
Purified RNA samples with a concentration of 1–10 ng/μL for 8–10 μL and RNA integrity number (RIN) above 7 (except for one sample for Aβ SA1-LTMRs that had a RIN ~6) were selected for library preparation and RNA-sequencing at the Johns Hopkins Deep Sequencing & Microarray Core. cDNA libraries were prepared using Nugen Ovation RNA-Seq and then Illumina TruSeq RNA Library Prep Kit. The cDNA libraries were then sequenced using the Illumina HiSeq 2000 platform with 50 bp single-end reads with 2´ multiplexing (two samples were combined and run for each lane).
Trimethoprim treatment and characterization of Calb1dgCre mice
Trimethoprim (TMP) (Sigma, T78883) was dissolved in DMSO (50 mg/ml) and prepared fresh for intraperitoneal injection of mice carrying Calb1dgCre. Mice were treated with two doses of TMP (100 μg per gram of body weight per day at P18,19) and then sacrificed at P21-P28 for tissue harvesting (Sando et al., 2013). Calb1dgCre; R26LSL-FSF-tdTomato or Calb1dgCre; R26LSL-FSF-tdTomato mice display low level of baseline recombination. TMP treatment induced ~10–20 fold increase in the number of tdTomato+ cells in DRGs (data not shown). Variable baseline Cre activity was observed when using Brn3aCKOAP (Badea et al., 2012) as the reporter line between different litters (data not shown), and it is recommended that littermate controls always be used as controls to monitor baseline reporter expression or Cre activity.
Immunohistochemistry of cryosections
Mice (3–6 weeks) were anesthetized with CO2 and perfused using 5–10 ml PBS followed by 10–20 ml of 4% paraformaldehyde (PFA) in PBS at room temperature (RT). Vertebral columns (including spinal cords and DRG) and skin were dissected from perfused mice. For immunostaining experiments other than those for Kv4.3, neuronal tissues were post-fixed in 4% PFA at 4°C for 2–4 hours, while skin was post-fixed in Zamboni’s fixation buffer at 4°C for 48 hours. For Kv4.3 immunostaining, no post-fixation was used for vertebral columns. For co-staining of Kv4.3 and tdTomato (related to Figure S7), the skin was post-fixed in Zamboni’s fixation buffer at 4°C for one hour. In general, the tdTomato signal decreased as fixation times shortened, while the staining background increased as fixation times increased. Using one-hour post-fixation, both signals could be observed, although the tdTomato signal was weak. Tissues were then washed 3×20 minutes with PBS at RT. Tissues were cryoprotected in 30% sucrose in PBS at 4°C overnight, embedded in OCT and frozen at −20°C, stored at −20°C for short term and-80°C for long term, and sectioned at 20–25 μm using a cryostat. Sections were collected on slides and outlined using an ImmEdge Hydrophobic Barrier Pen. Vertebral column sections were dried at RT for at least 30 minutes or overnight before staining. Skin sections were dried at RT overnight to 48 hours. Slides were washed 3×10min with PBS containing 0.1% Triton X-100 (0.1% PBST) and then blocked in 0.1% PBST containing 10% normal goat serum (Vector Labs, S-1000) or normal donkey serum (Jackson Immuno, 005-000-121) for 30 minutes at RT. Slides were incubated with primary antibodies (listed in Key Resources Table) diluted in 0.1% PBST containing 5% serum at 4°C overnight covered with parafilm (Fisher, PM996). The next day, Slides were washed 3×10 minutes with 0.1% PBST, and incubated with secondary antibodies diluted (species-specific Alexa Fluor 488, 546, and 647 conjugated IgGs, Invitrogen) 1:500 in 0.1% PBST containing 5% serum at room temperature for 2 hours, washed again 3×10 minutes with 0.1% PBST, and mounted with fluoromount-G (Southern Biotech).
Double fluorescent in situ hybridization (FISH) of cryosections
Digoxygenin (DIG) (DIG RNA labeling mix, Roche, 11277073910)-labeled sense and antisense probes were generated for Lpar3, Adcyap1, Gpx3, Ptgfr, Spp1, and Fluorescein (Fluorescein RNA labeling mix, Roche, 11685619910)-labeled sense and antisense probes were generated for tdTomato and GFP. Five probes for each target gene were generated and tested using single-color in situ hybridization, and only probes that displayed robust signals were selected for subsequent analyses. For experiments, multiple solutions were prepared beforehand. Diethyl pyrocarbonate (DEPC)-ddH2O and DEPC-PBS are ddH2O and PBS treated with DEPC (Sigma, D5758) at 1:1000 ratio, mixed at 37°C for at least 1 hour, then autoclaved. Washing buffer was 100 mM Tris-HCL, 150 mM NaCl, 0.05% Tween-20, pH 7.5 at 20°C. Detection buffer was 100 mM Tris-HCL, 100 mM NaCl, 10 mM MgCl2, and pH 8.0 at 20°C. All the solutions weremade in advance and stored in RT. Hybridization solution contained 50% formamide, 5ŚSC, 0.3 mg/ml yeast tRNA (Sigma, R6750), 100 μg/ml heparin (Sigma, H3393), 1´Denhardt’s (Sigma, D2532), 0.1% Tween 20, 5 mM EDTA in DEPC-ddH2O. Hybridization solution was made in advance and stored at −20°C. Vertebral columns were freshly dissected from mice (3–6 weeks) and frozen in OCT, then sectioned at 20–25 m using a cryostat. Sections from at least one pair of cervical, thoracic, and lumbar DRGs were sectioned and sections were collected on slides and warmed up at RT for 30 minutes. Sections were fixed in 4% PFA, then washed using freshly prepared DEPC in DEPC-PBS (1/1000) for 10 minutes and washed again with DEPC-PBS for 5 minutes. Slides were incubated in 50 g/ml proteinase K for 1 to 5 minutes, then washed in DEPC-PBS for 5 minutes and rinsed with DEPC-ddH2O (ddH2O pretreated with DEPC, and autoclaved). Slides were treated with freshly made 0.1 M TEA buffer (triethanolamine and acetic anhydride in DEPC-ddH2O) for 10 minutes at RT, then washed with DEPC-PBS for 5 minutes. Slides were then incubated in hybridization buffer and covered with parafilm and incubated at 65°C in a humidified chamber for 30 minutes, then in hybridization buffer with probes (1 DIG-and 1 fluorescein-labeled) diluted at 1:100 ratio at 65°C overnight. On the second day, slides were washed with 0.2ŚSC once for 15 minutes at 65°C, then twice for 30 minutes each at 65°C. Slides were then incubated with 20% normal goat serum in 0.1% PBST for 30 minutes to an hour at RT, then added with AP labeled antibody (anti-DIG-AP, Roche, 11093274910) diluted at 1:1000–1:2000 in 20% normal goat serum in 0.1% PBST and incubated at RT for 2 hours, then washed 3´10 minutes in PBS or washing buffer. HNPP/Fast Red TR mix was prepared fresh, diluting 10 μL HNPP and 10 μL Fast Red TR stock solution in 1 mL detection buffer (HNPP fluorescent detection kit, Roche, 11758888001), then filtered with 0.2 μm filter. Slides were then incubated in HNPP/Fast Red TR mix for 30 minutes at RT, and then washed in washing buffer. Next, slides were incubated in HNPP/Fast Red TR mix and washed repetitively for 3 times, then washed in PBS for 5 minutes, blocked with 0.5% blocking solution (blocking reagent (Roche, 11096176001) diluted in PBS) for 1 hour at RT, and then incubated in POD labeled antibody (anti-fluorescein-POD, Roche, 11426346910) (1:250 diluted in blocking solution). Slides were washed in PBS 3´10 minutes at RT, then incubated with TSA Plus Solution (TSA diluted in Amplification Plus Buffer at 1:100) (TSA plus fluorescein system, PekinElmer NEL741001KT) for 15–30 minutes at RT, then rinsed again with PBS. Sections were mounted with fluoromount-G before being examined using confocal microscopy.
QUANTIFICATION AND STATISTICAL ANALYSIS
In vitro electrophysiology analysis.
Current and voltage records were filtered at 10 kHz and digitized at 200 kHz. Analysis was performed with Igor Pro 6.12 (Wavemetrics, Lake Oswego, OR), using DataAccess (Bruxton Software) to import pClamp data. Reported membrane potentials are corrected for a liquid junction potential of −10 mV between the internal solution and the Tyrode’s solution in which current was zeroed before sealing onto the cell, measured using a flowing 3 M KCl reference electrode as described by Neher (1992). In current clamp experiments, resting potential was adjusted to −80 mV using a steady-holding current. In quantifying action potential firing, action potentials were defined by the criteria that peak voltage is above −20 mV and spike height is at least 30 mV.
Voltage-clamp current records were corrected for linear capacitative and leak current by subtracting scaled responses to 5 mV hyperpolarizations delivered from the holding potential. In AP clamp experiments, APs were evoked by current injection (just above threshold for 0.5 ms) in each cell type and then used as the command waveform in voltage clamp experiments with that cell type. To measure total net outward current during the repolarization phase of the AP, the current was integrated from the time that net current under control conditions became net outward (near the peak of the AP) to the time of the trough following the AP, when net current under control conditions shifted from outward to inward.
In vitro mechanoclamp.
Current and voltage records were filtered at 10 kHz and digitized at 20 kHz. Series resistance and junction potential were not compensated or corrected. Access resistance was assessed using membrane test with 5mV hyperpolarizations from the holding potential throughout recordings, and cells displaying significant access time constant change were eliminated from further analysis. Kinetics of mechanosensitive currents were analyzed using pClamp and determined by using nonlinear least-squares regression analysis applied to the decay phase of the currents. Traces were fit by one or two exponential components as follows: . When two component fitting was required, the faster time constant was used in Figure 1T. Only traces with a fitting correlation over 80% were selected for quantification.
RNA sequencing.
Reads were aligned and counted using STAR aligner and Htseq at Harvard Chan Bioinformatics Core. Both read count matrix and normalized data using reads per kilobase per million reads (rpkm) were obtained. Sequencing data quality was assessed by examining the number of mapped reads (average=73,506,450), genomic mapping rate (average=0.93), number of genes detected (19,601 genes that have a count number >=1, 12,226 genes that have a count per million (CPM) value>=1, 11,160 genes that have fpkm value>=1), exonic mapping rate (average=0.67), and rRNA mapping rate (average=1.86´10−6). Count matrix values were transformed using regularized log transformation implemented in DESeq2 R package and rendered homoscedastic (Love et al., 2014). Correlations between samples were assessed by clustering samples using all rlog transformed count data and multiple ways to calculate distances (Euclidean Distance, Pearson Correlation, Spearman Correlation). Samples of the same neuronal subtypes generally clustered with each other, and two major-group structures were observed regardless of the methods used (data not shown) (related to Figure 2C and Figure S2A). This also held true with performing analysis using kmeans clustering with a k value of 8 (data not shown). Differential expression tests were performed for every pair of neuronal subtypes, also using the DESeq2 package. Differentially expressed genes (DEGs) were defined using the criteria of a 2-fold change with a false discovery rate (FDR) <0.01. A differential expression test was also performed comparing small-diameter neuron subtypes to medium/large-diameter neuron subtypes. In the text, all genes that were described as differentially expressed comparing small-diameter to medium/large diameter neuron subtypes satisfy the aforementioned criteria. Subtype uniquely enriched genes (SUEGs) were selected for those that are enriched in the subtype in every pair comparison, thus the FDR criteria for SUEGs after correction for multiple comparisons is ~0.07 (related to Figure 2D). Genes whose expression patterns were tested in Figure 2E–L were selected from SUEGs, except for Ptgfr, which was found to be enriched in both Aβ SA1-LTMRs andAβ Field-LTMRs, compared to other subtypes. For the generation of heatmaps shown in Figure 2D, Figure S2B, Figure 3, Figure S3A–S3B and Figure 5A, rlog transformed count data was used as expression measurements and as inputs. Genes were first filtered using the criteria of average rlog value across all samples >0, which yielded 18,755 genes, and defined as expressed in Results sections related to individual figures. For the 18,755 genes detected, rlog averaged across 26 samples for every gene was calculated as average rlog values to represent average expression levels. Average rlog is 14.28 at 99th percentile, 12.59 at 95th percentile, 11.76 at 90th percentile, 10.14 at 75th percentile, 7.74 at 50th percentile and 3.60 at 25th percentile. For the 18,755 genes, rlog differences from the average rlog values across all samples for each gene were calculated to represent relative expression enrichment or reduction for each sample. A difference of 1 roughly translates to 2-fold change. Rlog differences werethen averaged across samples from the same neuronal subtype to represent subtype gene expression differences. Genes were then grouped based on the neuronal subtype their expression is enriched in, and then ordered based on the enrichment in a descending manner. Only genes that had average rlog values within the top 75% (average rlog>3.60) and displayed expression differences over 1 in three samples (the number of replicates for one neuronal subtype) were selected for displaying the expression pattern. Rlog difference from the average rlog value was plotted in the main heatmaps using a scale of −2 to 2 represented using blue to red color scale for the heatmaps. Average rlog values were plotted in a second heatmap to the right of the main heatmap. Figure S3C–S3D used the same plotting scheme, but without implementing any filters and all genes belonged to the categories were included in the heatmaps. Genes were ordered alphabetically. In bar plots displaying gene expression using rpkm values, error bars represent mean ± SD (related to Figure 4 and Figure S4). In dotplots diplaying gene expression using rpkm values, all data points were plotted (related to Figure 2E–2L). The bar plots were plotted using Prism 7, while heatmaps and dotplots were plotted using the ggplot2 package in R. All the n’s are as indicated in figure legends and corresponding methods section.
Computational Modeling
Computational models were implemented using the NEURON simulation environment (Hines and Carnevale, 1997, 2000). Single cylindrical compartment models were constructed with diameter and length = 19.55 μm to model C-LTMRs (total membrane capacitance Cm = 12 pF), and with diameter and length = 30.90 μm to model Aβ SA1-LTMRs (Cm = 30 pF). Specific membrane capacitance (cm) was 1 μF/cm and specific resistivity of the internal solution (Ri) was 100 Ω*cm. For ion channels, the voltage-gated ion channels Nav1.1, Nav1.6, Nav1.7, Nav1.8, Kv1, Kv2, Kv3, Kv4 and leak channels were included. The voltage-dependence and kinetics of current carried by each voltage-dependent channels was described by i=ḡmph(V-Vrev), where I is the current, ḡ is the maximal conductance with all channels open, V is the membrane voltage, and Vrev is the reversal potential of the channel. Vrev was +55 mV for sodium channels and −88 mV for potassium channels. Gating of channels was described by the Hodgkin-Huxley-style parameters m (governing activation) and h (governing inactivation), each varying between 0 and 1 in a voltage-and time-dependent manner, with p being an exponent varying from 1 to 4 for different channels. The steady-state voltage dependence of m was described by a Boltzmann function of the form , where Vh is the voltage at which m is 0.5 and k is the slope factor describing the steepness of the function. The steady-state voltage-dependence of h was described by a Boltzmann function with the opposite direction of voltage-dependence, . The kinetics of gating of m and h were expressed by defining a time-constant of gating at each voltage, allowing a more flexible definition of kinetics than is possible by separately defining backward and forward rate constants for each gating particle. The voltage-dependence of the m or h time-constants was described by a rising Boltzmann function at more negative voltages and a falling Boltzmann at more positive voltages, allowing more accurate description of gating in cases in which time constants measured by activation and deactivation kinetics fail to converge (e.g. Liu and Bean, 2015). The formula for the time-constant at more negative (“left-side”) voltages was , where aL corresponds to an asymptotic time-constant at strongly negative voltages and bL to a projected asymptote at positive voltages, with cL and dL governing the shape of the voltage-dependence. The formula for the time-constant at more positive (“right-side”) voltages was , where aR corresponds to an asymptotic time-constant at strongly positive voltages and bR to a projected asymptote at more negative voltages, with cR and dR governing the shape of the voltage-dependence. Time-constants were defined by the “left-side” formula at voltages more negative than a “break” voltage (denoted by “VBrk” in Table S1) and by the “right-side” formula at voltages positive to the break voltage (cf. (Korngreen and Sakmann, 2000)).
Parameters describing voltage dependence and kinetics were initially derived from previous studies (Nav1.1: (Rhodes et al., 2004); Nav1.6: (Carter and Bean, 2011; Royeck et al., 2008; Schwarz and Eikhof, 1987); Nav1.7: (Blair and Bean, 2002); Nav1.8: (Blair and Bean, 2002); Kv1: (Glazebrook et al., 2002; Guan et al., 2006); Kv2: (Liu and Bean, 2014); Kv3: (Lien and Jonas, 2003; Martina et al., 2007); Kv4: (Jackson and Bean, 2007)). Gating parameters along with current densities were then adjusted to better match the experimental data from DRG recordings at 37°C. The values for the gating parameters for each type of channel are listed in Table S1. The models for firing of Aβ SA1-LTMRs and C-LTMRs used identical parameters for gating of the voltage-dependent channels, with different combinations and magnitudes of each conductance. Aβ SA1-LTMRs contained Nav1.1 (8 mS/cm2), Nav1.6 (40 mS/cm2), Nav1.7 (5 mS/cm2), Kv1 (6 mS/cm2), Kv2 (12 mS/cm2), Kv3 (4 mS/cm2) and Kv4 (0.8 mS/cm2) conductances, as well as a non-voltage dependent leak conductance (0.1 mS/cm2) with a reversal potential of −80 mV. C-LTMRs contained Nav1.7 (30 mS/cm2), Nav1.8 (40 mS/cm2), Kv1 (0.06 mS/cm2), Kv2 (2 mS/cm2), Kv3 (0.05 mS/cm2) and Kv4 (11 mS/cm2) conductances, and a non-voltage dependent leak conductance (0.02 mS/cm2) with a reversal potential of −80 mV. For displaying the firing behavior in Figure 8, the model was implemented in the IgorPRO as well as in NEURON. Firing properties were analyzed and heatmaps were constructed using R.
Data and Software Availability
Computational models are available at ModelDB (https://senselab.med.yale.edu/modeldb/, Accession No. 256632). RNA-seq data is available on GEO (Accession No. GSE131230).
Supplementary Material
Highlights.
Eight DRG neuron subtypes display distinct intrinsic physiological properties.
Transcriptome profiles revealed gene expression patterns of these subtypes.
Differentially expressed K channels regulate firing in a subtype-specific manner.
Acknowledgements
We thank Ting Guo for advice during early stages of this project, and David Corey, Stephen Liberles, King-Wai Yau and the Ginty lab for comments on this manuscript. We thank Hao Zhang at the Johns Hopkins Bloomberg Flow Cytometry and Immunology Core for help with FACS, Haiping Hao, Jasmeet Sethi and Linda Orzolek at the Johns Hopkins Deep Sequencing & Microarray Core Facility for help with RNA-sequencing, Meeta Mistry at the Harvard School of Public Health Bioinformatics Core for help with Bioinformatics, Wenqin Hu for advice on computational modeling, Nat Blair, Alex Jackson, Tilia Kimm, and Brett Carter for channel kinetics data used for the computational model, Sarah Bahai and Sabrina Belozerova for help with mouse husbandry and histological experiments, and Gui-Lan Yao for help with histological experiments. This work was supported by NIH grants NS97344 (D.D.G.), NS036855 (B.P.B.), a Bertarelli Fund Collaborative Award (B.P.B. and D.D.G) and the Edward R. and Anne G. Lefler Center for Neurodegenerative Disorders. D.D.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no competing interests.
References
- Abraira VE, and Ginty DD (2013). The sensory neurons of touch. Neuron 79, 618–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, Orefice LL, Boyle KA, Bai L, Song BJ, et al. (2017). The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell 168, 295–310.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcourt A, Gorham L, Dhandapani R, Prato V, Taberner FJ, Wende H, Gangadharan V, Birchmeier C, Heppenstall PA, and Lechner SG (2017). Touch Receptor-Derived Sensory Information Alleviates Acute Pain Signaling and Fine-Tunes Nociceptive Reflex Coordination. Neuron 93, 179–193. [DOI] [PubMed] [Google Scholar]
- Badea TC, Williams J, Smallwood P, Shi M, Motajo O, and Nathans J (2012). Combinatorial Expression of Brn3 Transcription Factors in Somatosensory Neurons: Genetic and Morphologic Analysis. J. Neurosci 32, 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Lehnert BP, Liu J, Neubarth NL, Dickendesher TL, Nwe PH, Cassidy C, Woodbury CJ, and Ginty DD (2015). Genetic Identification of an Expansive Mechanoreceptor Sensitive to Skin Stroking. Cell 163, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni R, Tawfik VL, Wang D, François A, Solorzano C, Shuster SA, Choudhury P, Betelli C, Cassidy C, Smith K, et al. (2014). Delta opioid receptors presynaptically regulate cutaneous mechanosensory neuron input to the spinal cord dorsal horn. Neuron 81, 1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, and Julius D (2009). Cellular and Molecular Mechanisms of Pain. Cell 139, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessou P, Burgess PR, Perl ER, and Taylor CB (1971). Dynamic properties of mechanoreceptors with unmyelinated (C) fibers. J. Neurophysiol 34, 116–131. [DOI] [PubMed] [Google Scholar]
- Blair NT, and Bean BP (2002). Roles of tetrodotoxin (TTX)-sensitive Na+ current, TTX-resistant Na+ current, and Ca2+ current in the action potentials of nociceptive sensory neurons. J. Neurosci 22, 10277–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocksteins E, and Snyders DJ (2012). Electrically Silent Kv Subunits: Their Molecular and Functional Characteristics. Physiology 27, 73–84. [DOI] [PubMed] [Google Scholar]
- Carter BC, and Bean BP (2009). Sodium Entry during Action Potentials of Mammalian Neurons: Incomplete Inactivation and Reduced Metabolic Efficiency in Fast-Spiking Neurons. Neuron 64, 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BC, and Bean BP (2011). Incomplete Inactivation and Rapid Recovery of Voltage-Dependent Sodium Channels During High-Frequency Firing in Cerebellar Purkinje Neurons. J. Neurophysiol 105, 860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Kanda H, Ikeda R, Ling J, DeBerry JJ, and Gu JG (2016). Merkel disc is a serotonergic synapse in the epidermis for transmitting tactile signals in mammals. Proc. Natl. Acad. Sci. U. S. A 113, E5491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla K, Tripathi S, Thommesen L, Lægreid A, and Kuiper M (2013). TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics 29, 2519–2520. [DOI] [PubMed] [Google Scholar]
- Chi XX, and Nicol GD (2007). Manipulation of the potassium channel Kv1.1 and its effect on neuronal excitability in rat sensory neurons. J. Neurophysiol 98, 2683–2692. [DOI] [PubMed] [Google Scholar]
- Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman GS, Roberson DP, Ghasemlou N, et al. (2014). Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife 3, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee WA, Amarillo Y, Chiu J, Chow A, Lau D, McCormack T, Moreno H, Nadal MS, Ozaita A, Pountney D, et al. (1999). Molecular diversity of K+ channels. Ann. N. Y. Acad. Sci 868, 233–285. [DOI] [PubMed] [Google Scholar]
- Delmas P, Hao J, and Rodat-Despoix L (2011). Molecular mechanisms of mechanotransduction in mammalian sensory neurons. Nat. Rev. Neurosci 12, 139–153. [DOI] [PubMed] [Google Scholar]
- Dib-Hajj SD, Cummins TR, Black J. a, and Waxman SG (2010). Sodium channels in normal and pathological pain. Annu. Rev. Neurosci 33, 325–347. [DOI] [PubMed] [Google Scholar]
- Du X, and Gamper N (2013). Potassium channels in peripheral pain pathways: expression, function and therapeutic potential. Curr. Neuropharmacol 11, 621–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Hao H, Gigout S, Huang D, Yang Y, Li L, Wang C, Sundt D, Jaffe DB, Zhang H, et al. (2014). Control of somatic membrane potential in nociceptive neurons and its implications for peripheral nociceptive transmission. Pain 155, 2306–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, and Patapoutian A (2010). Nociceptors: the sensors of the pain pathway. J. Clin. Invest 120, 3760–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everill B, Rizzo MA, and Kocsis JD (1998). Morphologically identified cutaneous afferent DRG neurons express three different potassium currents in varying proportions. J. Neurophysiol 79, 1814–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MS, and Luo W (2013). The anatomy, function, and development of mammalian Aβ low-threshold mechanoreceptors. Front. Biol. (Beijing) 8, 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földy C, Darmanis S, Aoto J, Malenka RC, Quake SR, and Südhof TC (2016). Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. U. S. A 113, E5222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François A, Schüetter N, Laffray S, Sanguesa J, Pizzoccaro A, Dubel S, Mantilleri A, Nargeot J, Noël J, Wood JN, et al. (2015a). The Low-Threshold Calcium Channel Cav3.2 Determines Low-Threshold Mechanoreceptor Function. Cell Rep 10, 370–382. [DOI] [PubMed] [Google Scholar]
- François A, Schüetter N, Laffray S, Sanguesa J, Pizzoccaro A, Dubel S, Mantilleri A, Nargeot J, Noël J, Wood JN, et al. (2015b). The Low-Threshold Calcium Channel Cav3.2 Determines Low-Threshold Mechanoreceptor Function. Cell Rep 10, 370–382. [DOI] [PubMed] [Google Scholar]
- Gemes G, Koopmeiners A, Rigaud M, Lirk P, Sapunar D, Bangaru ML, Vilceanu D, Garrison SR, Ljubkovic M, Mueller SJ, et al. (2013). Failure of action potential propagation in sensory neurons: mechanisms and loss of afferent filtering in C-type units after painful nerve injury. J. Physiol 591, 1111–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook PA, Ramirez AN, Schild JH, Shieh C-C, Doan T, Wible BA, and Kunze DL (2002). Potassium channels Kv1.1, Kv1.2 and Kv1.6 influence excitability of rat visceral sensory neurons. J. Physiol 541, 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MS, Shuster MJ, and Levine JD (1996). Characterization of six voltage-gated K+ currents in adult rat sensory neurons. J. Neurophysiol 75, 2629–2646. [DOI] [PubMed] [Google Scholar]
- Gorokhova S, Gaillard S, Urien L, Malapert P, Legha W, Baronian G, Desvignes J-P, Alonso S, and Moqrich A (2014). Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor. PLoS Genet 10, e1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg P (1986). Biophysical studies of mechanoreceptors. J. Appl. Physiol 60, 1107–1115. [DOI] [PubMed] [Google Scholar]
- Guan D, Lee JCF, Tkatch T, Surmeier DJ, Armstrong WE, and Foehring RC (2006). Expression and biophysical properties of Kv1 channels in supragranular neocortical pyramidal neurones. J. Physiol 571, 371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman GA (2005). International Union of Pharmacology. LIII. Nomenclature and Molecular Relationships of Voltage-Gated Potassium Channels. Pharmacol. Rev 57, 473–508. [DOI] [PubMed] [Google Scholar]
- Han L, Ma C, Liu Q, Weng H-J, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, et al. (2013). A subpopulation of nociceptors specifically linked to itch. Nat. Neurosci 16, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, and Delmas P (2011). Recording of mechanosensitive currents using piezoelectrically driven mechanostimulator. Nat. Protoc 6, 979–990. [DOI] [PubMed] [Google Scholar]
- Harper AA, and Lawson SN (1985). Electrical properties of rat dorsal root ganglion neurones with different peripheral nerve conduction velocities. J. Physiol 359, 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AL, and Robertson B (2004). Dendrotoxins: structure-activity relationships and effects on potassium ion channels. Curr. Med. Chem 11, 3065–3072. [DOI] [PubMed] [Google Scholar]
- Heidenreich M, Lechner SG, Vardanyan V, Wetzel C, Cremers CW, De Leenheer EM, Aránguez G, Moreno-Pelayo MÁ, Jentsch TJ, and Lewin GR (2012). KCNQ4 K(+) channels tune mechanoreceptors for normal touch sensation in mouse and man. Nat. Neurosci 15, 138–145. [DOI] [PubMed] [Google Scholar]
- Hines ML, and Carnevale NT (1997). The NEURON simulation environment. Neural Comput 9, 1179–1209. [DOI] [PubMed] [Google Scholar]
- Hines ML, and Carnevale NT (2000). Expanding NEURON’s repertoire of mechanisms with NMODL. Neural Comput 12, 995–1007. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, and Arber S (2005). A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 3, 0878–0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Cha M, Ling J, Jia Z, Coyle D, and Gu JG (2014). Merkel cells transduce and encode tactile stimuli to drive aβ-afferent impulses. Cell 157, 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, and Bean BP (2007). State-Dependent Enhancement of Subthreshold A-Type Potassium Current by 4-Aminopyridine in Tuberomammillary Nucleus Neurons. J. Neurosci 27, 10785–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Golden JP, Wozniak D, Pehek E, Johnson EM, and Milbrandt J (2006). RET Is Dispensable for Maintenance of Midbrain Dopaminergic Neurons in Adult Mice. J. Neurosci 26, 11230–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D (2013). TRP channels and pain. Annu. Rev. Cell Dev. Biol 29, 355–384. [DOI] [PubMed] [Google Scholar]
- Korngreen A, and Sakmann B (2000). Voltage-gated K+ channels in layer 5 neocortical pyramidal neurones from young rats: subtypes and gradients. J. Physiol 525 Pt 3, 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-L, Li K-C, Wu D, Chen Y, Luo H, Zhao J-R, Wang S-S, Sun M-M, Lu Y-J, Zhong Y-Q, et al. (2016). Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 26, 83–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, et al. (2011). The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 147, 1615–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C-C, and Jonas P (2003). Kv3 potassium conductance is necessary and kinetically optimized for high-frequency action potential generation in hippocampal interneurons. J. Neurosci 23, 2058–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PW, and Bean BP (2014). Kv2 channel regulation of action potential repolarization and firing patterns in superior cervical ganglion neurons and hippocampal CA1 pyramidal neurons. J. Neurosci 34, 4991–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PW, Blair NT, and Bean BP (2017). Action Potential Broadening in Capsaicin-Sensitive DRG Neurons from Frequency-Dependent Reduction of Kv3 Current. J. Neurosci 37, 9705–9714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löken LS, Wessberg J, Morrison I, McGlone F, and Olausson H (2009). Codingof pleasant touch by unmyelinated afferents in humans. Nat. Neurosci 12, 547–548. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Flauaus C, Kennel L, Petersen J, Drees O, Kallenborn-Gerhardt W, Ruth P, Lukowski R, and Schmidtko A (2017). KCa3.1 channels modulate the processing of noxious chemical stimuli in mice. Neuropharmacology 125, 386–395. [DOI] [PubMed] [Google Scholar]
- Maffie JK, Dvoretskova E, Bougis PE, Martin-Eauclaire M-F, and Rudy B (2013). Dipeptidyl-peptidase-like-proteins confer high sensitivity to the scorpion toxin AmmTX3 to Kv4-mediated A-type K + channels. J. Physiol 591, 2419–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic S, Baba Y, and Lumpkin EA (2013). Neurotransmitters and synaptic components in the Merkel cell-neurite complex, a gentle-touch receptor. Ann. N. Y. Acad. Sci 1279, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic S, Nakatani M, Baba Y, Nelson AM, Marshall KL, Wellnitz S. a, Firozi P, Woo S-H, Ranade S, Patapoutian A, et al. (2014). Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature 509, 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigère F, and Ernfors P (2007). Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci 8, 114–127. [DOI] [PubMed] [Google Scholar]
- Martina M, Metz AE, and Bean BP (2007). Voltage-Dependent Potassium Currents During Fast Spikes of Rat Cerebellar Purkinje Neurons: Inhibition by BDS-I Toxin. J. Neurophysiol 97, 563–571. [DOI] [PubMed] [Google Scholar]
- Matsuyoshi H, Takimoto K, Yunoki T, Erickson VL, Tyagi P, Hirao Y, Wanaka A, and Yoshimura N (2012). Distinct cellular distributions of Kv4 pore-forming and auxiliary subunits in rat dorsal root ganglion neurons. Life Sci 91, 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter GC, Reichling DB, and Levine JD (1999). Mechanical transduction by rat dorsal root ganglion neurons in vitro. Neurosci. Lett 273, 179–182. [DOI] [PubMed] [Google Scholar]
- Molineux ML (2005). A-Type and T-Type Currents Interact to Produce a Novel Spike Latency-Voltage Relationship in Cerebellar Stellate Cells. J. Neurosci 25, 10863–10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqeem T, Ghosh B, Pinto V, Lepore AC, and Covarrubias M (2018). Regulation of Nociceptive Glutamatergic Signaling by Presynaptic Kv3.4 Channels in the Rat Spinal Dorsal Horn. J. Neurosci 38, 3729–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteen JD, Herzig V, Gilchrist J, Emrick JJ, Zhang C, Wang X, Castro J, Garcia-Caraballo S, Grundy L, Rychkov GY, et al. (2016). Selective spider toxins reveal a role for the Nav1.1 channel in mechanical pain. Nature 534, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak D, Guan D, and Foehring RC (2016). Roles of specific Kv channel types in repolarization of the action potential in genetically identified subclasses of pyramidal neurons in mouse neocortex. J. Neurophysiol 115, 2317–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil MJ, Hovhannisyan AH, and Akopian AN (2018). Characteristics of sensory neuronal groups in CGRP-cre-ER reporter mice: Comparison to Nav1.8-cre, TRPV1-cre and TRPV1-GFP mouse lines. PLoS One 13, e0198601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, and Huang ZJ (2017). Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell 171, 522–539.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska JC, Napaporn J, Johnson RD, Gu JG, and Cooper BY (2000). Subclassified acutely dissociated cells of rat DRG: histochemistry and patterns of capsaicin-, proton-, and ATP-activated currents. J. Neurophysiol 84, 2365–2379. [DOI] [PubMed] [Google Scholar]
- Phuket TRN, and Covarrubias M (2009). Kv4 Channels Underlie the Subthreshold-Operating A-type K+-current in Nociceptive Dorsal Root Ganglion Neurons. Front. Mol. Neurosci 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Syeda R, and Patapoutian A (2015). Mechanically Activated Ion Channels. Neuron 87, 1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau KK, Petruska JC, Cooper BY, and Johnson RD (2014). Distinct subclassification of DRG neurons innervating the distal colon and glans penis/distal urethra based on the electrophysiological current signature. J. Neurophysiol 112, 1392–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes TH, Lossin C, Vanoye CG, Wang DW, and George AL (2004). Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc. Natl. Acad. Sci 101, 11147–11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter DM, Ho C, O’Leary ME, and Covarrubias M (2012). Modulation of Kv3.4 channel N-type inactivation by protein kinase C shapes the action potential in dorsal root ganglion neurons. J. Physiol 590, 145–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter DM, Zemel BM, Hala TJ, O’Leary ME, Lepore AC, and Covarrubias M (2015). Dysregulation of Kv3.4 channels in dorsal root ganglia following spinal cord injury. J. Neurosci 35, 1260–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royeck M, Horstmann M-T, Remy S, Reitze M, Yaari Y, and Beck H (2008). Role of Axonal Na V 1.6 Sodium Channels in Action Potential Initiation of CA1 Pyramidal Neurons. J. Neurophysiol 100, 2361–2380. [DOI] [PubMed] [Google Scholar]
- Rudomin P (1999). Presynaptic selection of afferent inflow in the spinal cord. J. Physiol 93, 329–347. [DOI] [PubMed] [Google Scholar]
- Rutlin M, Ho C-Y, Abraira VE, Cassidy C, Bai L, Woodbury CJ, and Ginty DD (2014). The Cellular and Molecular Basis of Direction Selectivity of Aδ-LTMRs. Cell 159, 1640–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R, Baumgaertel K, Pieraut S, Torabi-Rander N, Wandless TJ, Mayford M, and Maximov A (2013). Inducible control of gene expression with destabilized Cre. Nat. Methods 10, 1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JR, and Eikhof G (1987). Na currents and action potentials in rat myelinated nerve fibres at 20 and 37 degrees C. Pflugers Arch 409, 569–577. [DOI] [PubMed] [Google Scholar]
- Shibata R, Nakahira K, Shibasaki K, Wakazono Y, Imoto K, and Ikenaka K (2000). A-type K+ current mediated by the Kv4 channel regulates the generation of action potential in developing cerebellar granule cells. J. Neurosci 20, 4145–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC (2012). The presynaptic active zone. Neuron 75, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundt D, Gamper N, and Jaffe DB (2015). Spike propagation through the dorsalroot ganglia in an unmyelinated sensory neuron: a modeling study. J. Neurophysiol 114, 3140–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS (2015). Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron 85, 238–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulas C, and McMahon SB (2014). Opening paths to novel analgesics: the role of potassium channels in chronic pain. Trends Neurosci 37, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci 18, 145–153. [DOI] [PubMed] [Google Scholar]
- Vacher H, Mohapatra DP, and Trimmer JS (2008). Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol. Rev 88, 1407–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vydyanathan A, Wu Z-Z, Chen S-R, and Pan H-L (2005). A-type voltage-gated K+ currents influence firing properties of isolectin B4-positive but not isolectin B4-negative primary sensory neurons. J. Neurophysiol 93, 3401–3409. [DOI] [PubMed] [Google Scholar]
- Wang R, and Lewin GR (2011). The Cav3.2 T-type calcium channel regulates temporal coding in mouse mechanoreceptors. J. Physiol 589, 2229–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman SG, and Zamponi GW (2014). Regulating excitability of peripheral afferents: emerging ion channel targets. Nat. Neurosci 17, 153–163. [DOI] [PubMed] [Google Scholar]
- Wellnitz S. a, Lesniak DR, Gerling GJ, and Lumpkin EA (2010). The regularity of sustained firing reveals two populations of slowly adapting touch receptors in mouse hairy skin. J. Neurophysiol 103, 3378–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis WDJ, and Coggeshall RE (2004). Sensory Mechanisms of the Spinal Cord-Volume 2-Back Matter In Sensory Mechanisms of the Spinal Cord, (Boston, MA: Springer US; ), pp. 2675–2690. [Google Scholar]
- Woo S-H, Ranade S, Weyer AD, Dubin AE, Baba Y, Qiu Z, Petrus M, Miyamoto T, Reddy K, Lumpkin E. a, et al. (2014). Piezo2 is required for Merkel-cell mechanotransduction. Nature 509, 622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf CJ, and Ma Q (2007). Nociceptors—Noxious Stimulus Detectors. Neuron 55, 353–364. [DOI] [PubMed] [Google Scholar]
- Zhang X-L, Mok L-P, Katz EJ, and Gold MS (2010). BK Ca currents are enriched in a subpopulation of adult rat cutaneous nociceptive dorsal root ganglion neurons. Eur. J. Neurosci 31, 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman A, Bai L, and Ginty DD (2014). The gentle touch receptors of mammalian skin. Science (80-.) 346, 950–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AL, Kovatsis EM, Pozsgai RY, Tasnim A, Zhang Q, and Ginty DD (2019). Distinct Modes of Presynaptic Inhibition of Cutaneous Afferents and Their Functions in Behavior. Neuron [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Rice FL, and Anderson DJ (2005). Topographically Distinct Epidermal Nociceptive Circuits Revealed by Axonal Tracers Targeted to Mrgprd. Neuron 45, 17–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Computational models are available at ModelDB (https://senselab.med.yale.edu/modeldb/, Accession No. 256632). RNA-seq data is available on GEO (Accession No. GSE131230).