Abstract
Mechanisms for making and breaking the heme b cofactor (heme) are more diverse than previously expected. Biosynthetic pathways have diverged at least twice along taxonomic lines, reflecting differences in membrane organization and O2 utilization among major groups of organisms. At least three families of heme degradases are now known, again differing in whether and how O2 is used by the organism and possibly the purpose for turning over the tetrapyrrole. Understanding these enzymes and pathways offers a handles for antimicrobial development and for monitoring heme use in organismal and ecological systems.
Introduction
Heme biosynthesis and degradation are important and highly regulated processes due to the significance of the cofactor and its breakdown products to cellular and organismal health. Over the past two decades, it has become clear that the biosynthetic steps for making heme are not universal [1]. The canonical pathway is limited to eukaryotic organisms and gram-negative bacteria, while gram-positive bacteria, archaea, and denitrifying and sulfate-reducing bacteria possess partly unique and more ancient ways to produce heme [2,3•] (Figure 1). By the same token, different groups of organisms break the heme tetrapyrrole under a variety of conditions using different catalytic strategies. Many discoveries along the heme synthesis and degradation pathways have been made fairly recently due to genome sequencing. Some significant ones are highlighted in this minireview.
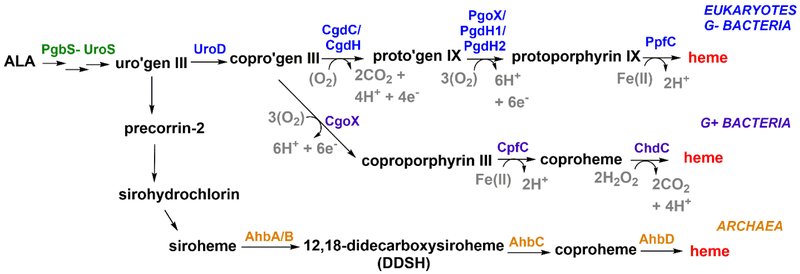
Figure 1. Multiple biosynthesis pathways leading to heme b.
Eukaryotes and gram-negative bacteria share the canonical pathway (blue). Gram-positive bacteria (purple) and denitrifying/sulfate reducing bacteria and archaea (orange) have distinct pathways that branch off following the coproporphyrinogen III (copro’gen) intermediate. The oxidation of porphyrinogens can use O2 as a 2e-/2H+ acceptor, generating H2O2 as a byproduct (i.e., steps catalyzed by CgdC, PgoX); alternatively, NAD(P)+ can serve as a 2e-/H+ acceptor in the presence of an active site acid (PgdH1, PgdH2). CgdH is a radical SAM-dependent enzyme. The electron acceptors for CgdH and CgoX have not been experimentally verified.
Multiple Routes to Heme b
All known pathways to heme commence with the irreversible synthesis of 5-aminolevulinic acid (ALA), are the same up to the intermediate uroporphyrinogen III, and then diverge along taxonomic lines. The “Alternate heme biosynthesis” (Ahb) pathway [3•], used by archaea/sulfate-reducing/denitrifying bacteria, was the first non-canonical pathway to be discovered (Figure 1). Uroporphyrinogen III is converted on this pathway to siroheme, the prosthetic group of sulfite and nitrite reductases. Siroheme is further processed into heme by AhbA, AhbB, AhbC, and AhbD (Figure 1) [3•]. The Ahb pathway offers a fully anaerobic route to heme and several of its intermediates for ancient organisms dwelling in anaerobic niches.
Coproporphyrinogen III, the product of the decarboxylation of uroporphyrinogen III by uroporphyrinogen decarboxylase (UroD), is the last common intermediate for the other two pathways. These begin with either a decarboxylation to yield protoporphyrinogen IX (protoporphyrin-dependent [(PPD) branch] or an oxidation step to yield coproporphyrin III (coproporphyrin-dependent [(CPD) branch]. The PPD branch is found in eukaryotes and gram negative bacteria, which make use of membrane-enclosed compartments to protect and steer toxic reactants (e.g., light-sensitive porphyrins, O2, H2O2, and Fe(II)). The CPD branch is used by gram positive bacteria, which lack either mitochondria or inner/outer membranes but must nonetheless exert control over similar substrates. Notably, the PPD branch has both O2-dependent and –independent catalysts for its two oxidative steps, suggesting that it can function under anaerobic conditions. One of the oxidative reactions of the CPD branch depends on H2O2 (ChdC); the electron acceptor for the other reaction (CgoX) has not been verified.
Decarboxylation of the Ring A- and B- Propionate Groups (CgdC, CgdH, and ChdC).
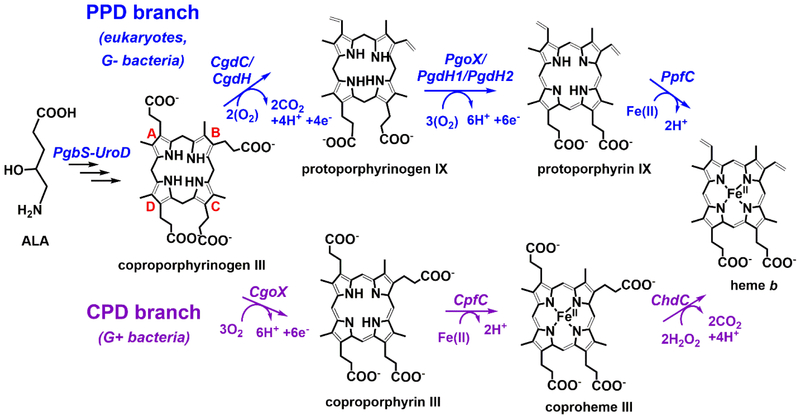
The first chemical transformation in the PPD branch is the oxidative decarboxylation of the ring A- and B- propionate groups to yield protoporphyrinogen IX (Figure 2). This reaction can be catalyzed by coproporphyrinogen decarboxylase (CgdC) or coproporphyrinogen dehydrogenase (CgdH) in an oxygen-dependent or -independent manner, respectively.
Figure 2. The PPD and CPD branches of heme b biosynthesis carry out similar chemical transformations in different orders.
Although some of the enzymes between the branches are homologous, a change in the order in which each is used creates different porphyrin intermediates. The terminal enzyme of the CPD pathway (ChdC) is completely unique. As knowledge about these pathways has evolved, new nomenclature for the enzymes that catalyze each individual step has been developed. For clarity, the most recently proposed nomenclature is used here3.
CgdC, found almost exclusively in mitochondria, is a dimeric membrane-bound enzyme [4] (Figure 3a). Its conserved active site base Asp is thought to facilitate direct, cofactor-independent reactions between the propionate side chains of coproporphyrinogen III and 2O2 [5••,6••·]. The reactions occur in a sequential and clockwise manner, yielding 2CO2 and 2H2O2. By contrast, CgdH is a soluble, monomeric, radical SAM (S-adenosylmethionine)/[4Fe-4S]-dependent enzyme that is exclusive to gram-negative bacteria and is structurally and mechanistically unrelated to CgdC (Figure 3) [7,8•, 9]. The 5’-deoxyadenosyl radical initiates the decarboxylation by stereospecific hydrogen-atom abstraction at the beta-position of the propionate side chains, producing a carbon radical [10]. This reaction requires an electron donor at the start of the reaction (to fill the iron-sulfur cluster electron hole) and an electron acceptor at the end (to quench the radical), neither of which has been identified [7]. As with CgdC, the decarboxylations are sequential and clockwise, generating 2CO2.
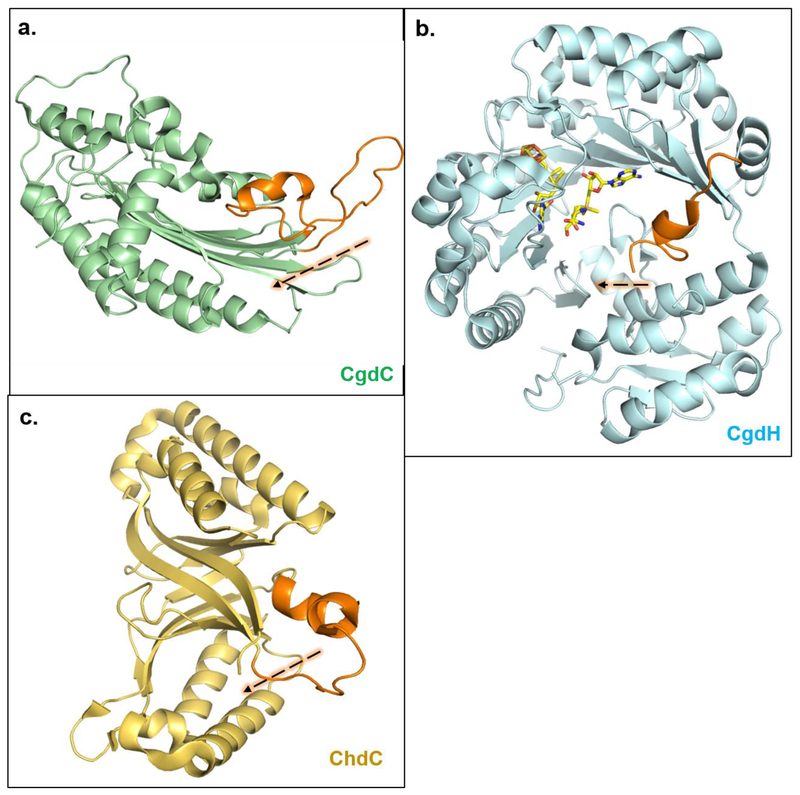
Figure 3. CgdC, CgdH, and ChdC all have an active site gate which closes upon substrate binding.
CgdC ((a); PDBID: 1TLB), CgdH ((b), PDBID: 1OLT) and ChdC ((c), PDBID: 1T0T) are isofunctional enzymes with no structural relationship and highly divergent reaction mechanisms. Interestingly, they all bind substrate at each monomeric site and structurally they share an “active site gate” (shown orange) made up of a partially disordered and mobile alpha helix, which closes in towards substrate upon its binding [4,9,11]. Note: Structures shown here are of individual monomers in their apo-/open- form. CgdC is a dimer, CgdH is a monomer, and ChdC is a pentamer in solution.
In the CPD branch, decarboxylation of the A- and B-ring propionate groups, by contrast, occurs at the end of the pathway. This reaction, catalyzed by coproheme decarboxylase (ChdC), occurs with ferric coproheme III acting as both substrate and cofactor, yielding heme as the final product (Figure 2) [2,12,13]. ChdC is evolutionarily and structurally unrelated to either CgdC or CgdH and is exclusive to gram-positive bacteria (Figure 3c). It is a member of the chlorite dismutase (Cld) family of proteins, which share a homopentameric structure [12]. Each ChdC monomer ChdC binds a coproheme III molecule. Oxidative decarboxylation is mediated by H2O2 and a conserved Tyr residue in the active site which forms a reactive radical, via a high-valent Fe(IV)=O(coproprophyrin▪+) intermediate [11,14,15]. Analogous to the 5’deoxyadenosyl radical in CgdH, the Tyr radical performs a hydrogen-atom abstraction at the beta-position of each reactive propionate, producing a radical in that position that leads to decarboxylation and formation of a vinyl group. Decarboxylation of the two propionates by ChdC also occurs in a clockwise fashion, generating 2CO2 and 4H2O. H2O2 is an unusual cellular substrate, particularly in catalase-rich organisms. Its source remains to be elucidated.
Oxidation of the Tetrapyrrole Rings (PgoX, PgdH1, PgdH2 and CgoX).
Diversity is most pronounced for the enzymes catalyzing oxidations of the tetrapyrrole ring. This penultimate step in the PPD branch is catalyzed by either PgoX, PgdH1, or PghH2, isofunctional enzymes that are non-homologous and structurally dissimilar [16].
Protoporphyrinogen oxidase (PgoX) catalyzes the six-electron oxidation of the protoporphyrinogen IX ring to protoporphyrin IX (PPIX) using O2. A total of 3O2/6H+ are consumed in three sequential rounds of oxidation, generating 3H2O2 and double bonds at all the meso-carbon positions [17,18•]. PgoX is a homodimeric, membrane-associated enzyme with a single FAD acting as a 2e- cofactor (Figure 4a). It is found almost exclusively in eukaryotes and a few gram-negative bacteria [19,20].
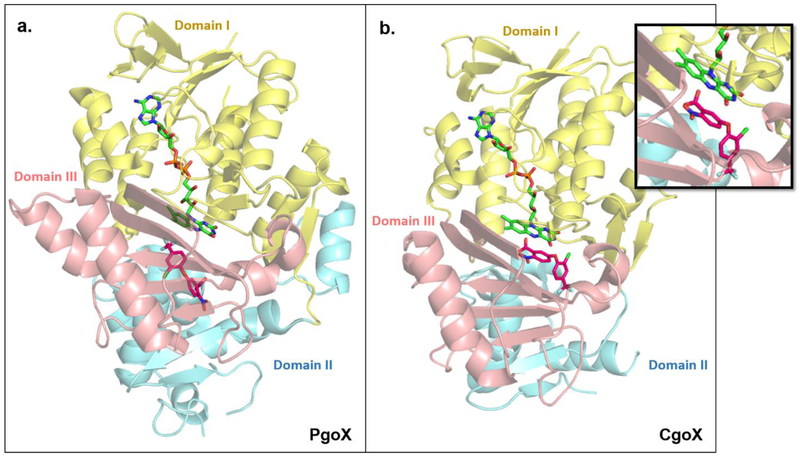
Figure 4. PgoX and CgoX share similar structural folds but show large differences in their active sites.
PgoX ((a); PDBID: 3NKS) and CgoX ((b); PDBID: 3I6D) share an overall folding pattern, consisting of three domains; (I) FAD-binding (yellow) domain, (II) membrane-binding domain (blue), and (III) substrate-binding domain (pink). Note: about 40% of amino acids in domain II of CgoX could not be crystallographically characterized [22]. PgoX and CgoX both non-covalently bind FAD (lime green) in the same location and orientation. The well-known PgoX inhibitor acifluoren (hot pink), also binds in both enzymes, but is mostly solvent-exposed in the CgoX active site, while it is buried deep inside the active site of PgoX. The different binding location and orientation of acifluoren in CgoX positions it to have a pi-stacking interaction with FAD (inset). This and the significantly larger active site in CgoX support the possibility that CgoX may use NAD(P) as an alternate electron acceptor to O2 to catalyze its reaction. Structures of PgdH1 and PgdH2 are not available to date.
Protoporphyrinogen can also be oxidized by the O2-independent enzymes protoporphyrinogen dehydrogenase 1 and 2 (PgdH1, PgdH2). PgdH1 is found mainly in γ-proteobacteria. It is a membrane-bound member of the long-chain flavodoxin family whose members use FMN to mediate electron transfer reactions [20]. PgdH1 is thought to use respiratory electron carriers such as menadione and ubiquinone for catalysis, in lieu of O2 [5••, 19]. PgdH2 is present in heme-synthesizing, non-enteric gram-negative bacteria such as Acinetobacter (γ-proteobacteria) and Synechocystis (cyanobacteria), which lack both pgoX and pgdH1 [5••,19,21]. Although PgdH2 is the most abundant of the three isofunctional enzymes, little is known about its mechanism. No cofactor has been identified for PgdH2 and it has been reported that PgdH2 is not able to complement a PgdH1-deficient E. coli strain, emphasizing its distinction from PgdH1 [5••,20].
Along the CPD branch, oxidative conversion of coproporphyrinogen III to coproporphyrin III by coproporphyrinogen oxidase (CgoX) is the first committed step. CgoX bears strong similarities in sequence and structure to PgoX, including a non-covalently bound FAD molecule [16,22•]. However, CgoX is monomeric instead of dimeric and is reported to be cytoplasmic instead of membrane-associated [2,22•,23]. The most widely studied CgoX is from Bacillus subtilis. Comparison of its crystal structure (PDBID: 3I6D) [24] with PgoX structures from Myxococcus xanthus (PDBID: 3NKS) [25] and Nicotana tabacum (PDBID: 1SEZ) [17] showed that the CgoX active site is 2-3 times larger (1173 Å vs 450-550 Å) and significantly more positively charged, allowing it to accommodate its larger and more negatively charged substrate [22•]. CgoX has been described as having the same reaction mechanism as PgoX. However, studies on CgoX have been conducted with a protoporphyrinogen IX substrate, and the requirement for O2 as the electron acceptor has not been experimentally validated [24,26]. This begs the question of whether CgoX may catalyze its reaction in an O2-independent manner, particularly in the facultative anaerobes in which it is sometimes found as the only coproporphyrinogen-converting enzyme. Interestingly, the PgoX in Plasmodium falciparium catalyzes its reaction using NAD(P) as an electron acceptor under anaerobic conditions [27]. It is possible that the much larger active site in CgoX can accommodate an electron acceptor such as NAD(P); pi-stacking between FAD and the inhibitor acifluoren in the B.subtilis CgoX alludes to this possibility (Figure 4b).
Over the past two decades, it has been reported that CgoX, unlike PgoX, is a cytosolic enzyme [2,5••,23,28]. However, studies with recombinant B. subtilis CgoX have shown that CgoX is found in the cell membrane fraction when expressed in E. coli [24,26], and its purification required detergent and denaturing conditions to maintain the enzyme in solution [24]. These observations, along with the fact that CgoX contains the membrane-binding “Domain II” that is also found in PgoX enzymes [22•], suggest that CgoX may be membrane-associated.
Tetrapyrrole Metalation (PpfC, CdfC).
Insertion of Fe(II) into the oxidized tetrapyrrole occurs as the final step in the PPD branch, catalyzed by protoporphyrin IX ferrochelatase (PpfC) (Figure 2). PpfC is homodimeric and thought to localize to the inner mitochondrial membrane of eukaryotes and the inner cellular membrane of gram-negative bacteria [29]. Each monomer contains a [2Fe-2S] cluster that does not participate in catalysis [6••,29,30]. Notably, the PpfC structure has a so-called “active site lip” that assumes an open state in the apo-protein and a closed state upon PPIX binding. In the absence of substrate or upon product formation, the active-site lip reopens, becoming solvent-exposed for substrate entry or product egress (Figure 5a).
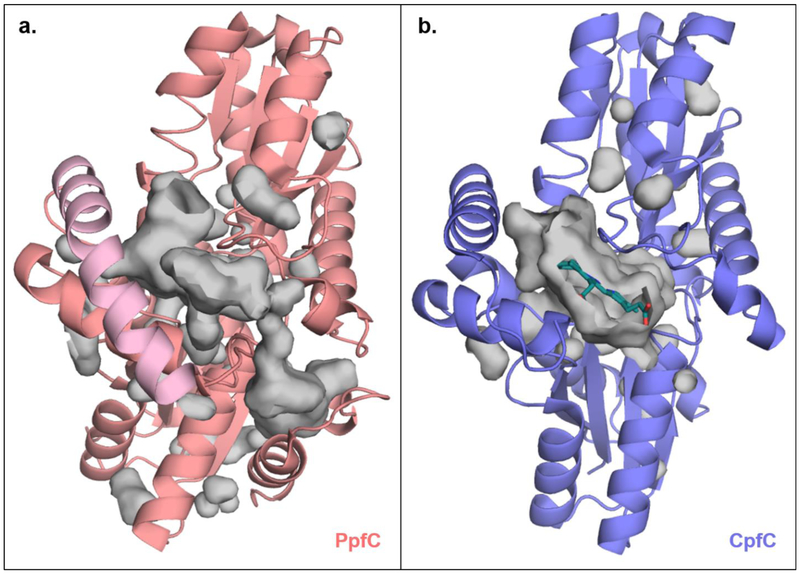
Figure 5. CpfC does not have the “active site lip” characteristic of canonical PpfC enzymes, leaving its active site largely solvent exposed.
Juxtaposed structures of (a) H. sapien PfpC bound to protoporphyrin IX (only one monomeric unit shown, pink, PDBID: 2QD1) and (b) B. subtilis CpfC bound to N-methylmesoporphyin (purple with porphyrin shown in teal, PDBID: 1C1H). PpfC has a characteristic “active site lip” (shown in light pink) that moves toward and completely occludes the active site upon substrate binding (surface of active site cavity shown in grey). CpfC enzymes lack this “active site lip” and have an active site that is largely solvent exposed even in the presence of substrate. Accessibility to the porphyrin substrate/product in CpfC may allow for ease in porphyrin transfer to the succeeding enzyme, ChdC.
The PpfC reaction mechanism requires Fe(II) and PPIX binding, deprotonation of two pyrrole ring nitrogens in PPIX, Fe(II)-insertion into the macrocycle, and product release [5••,29,31,32]. Fe(II) is directly delivered to PpfC by frataxin (Frda in human, Yfh1 in yeast), a homotrimeric membrane-bound enzyme involved in iron storage and delivery to [Fe-S]-cluster and heme biosynthetic enzymes [33]. Frataxin directly interacts with PpfC and transfers Fe(II) to conserved His and Glu residues in the active site (Figure 6) [31,32]. Upon binding PPIX, the active site lip closes in, causing rearrangement of an active site hydrogen-bonding network and distortion to the planarity of the PPIX ring [5••,29,34]. Both conformational changes trigger pyrrole deprotonation via a well-conserved His residue, and facilitating metal insertion into the porphyrin. Protonation of the His also reorients its side-chain and causes the final conformational change in the PpfC reaction cycle: opening of the active site lip for product release [5••,29]. The mechanism of PpfC is assumed to be the same in eukaryotes and gram-negative bacteria; however, further characterization of bacterial enzymes is required to address known differences in the ligation and metal content of their [Fe-S] clusters [5••,30].
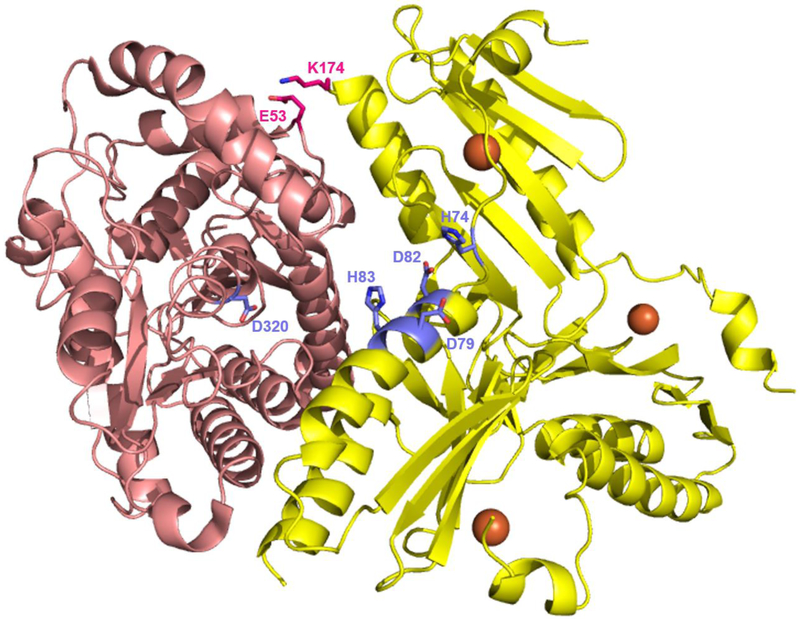
Figure 6. Interaction between yeast frataxin (Yfh1) and PpfC.
Complex formation between Yfh1 and PpfC in yeast involves one PfpC monomer (pink, PDBID: 1LBQ) and one Yfh1 homotrimer (yellow, PDBID: 4EC2). Each Yfh1 subunit binds two Fe(II) molecules (orange spheres, only one per monomer shown). Out of the three Yfh1 monomers, one subunit directly interacts with a PpfC monomer (interacting residues shown in hot pink), a second one is positioned for Fe(II) delivery (residues important for iron transfer shown in purple), and a third does not interact at all [33].
Metalation of the oxidized tetrapyrrole is the penultimate step of the CPD branch. Coproporphyrin ferrochelatase (CpfC) catalyzes insertion of iron into coproporphyrin III. CpfC has significantly lower binding affinities and reaction rates with PPIX, relative to PpfC [26,31]. Moreover, instead of a membrane-bound dimer, CpfC is a cytosolic monomer, and although similar in structure, CpfC shares less than 15% sequence identity with PpfC (Figure 5b) [2,26,29,31]. CpfC contains all of the conserved active site residues found in PpfC but lacks the active site lip, perhaps allowing for accommodation of its larger substrate, which binds in a different location than in PpfC (rotated ~100° and ~4.5Å further from the inside of the active site, leaving two propionate groups exposed to solvent). The porphyrin in CpfC still interacts with the conserved active site His, and CpfC binds Fe(II) in a similar manner with its His-Glu residues [31,32]. In the gram-positive B. subtilis, Fe(II) is directly delivered to CpfC by Fra, the B. subtilis frataxin homolog [33•,35] and metalation is thought to occur through a similar mechanism to PpfC, although further work with this enzyme and its substrate is needed.
Multiple ways to break heme
There are multiple reasons to degrade heme, and organisms from different taxonomic groups have evolved unique enzymes and pathways for doing so. Tetrapyrrole degradation allows for iron utilization or resorption in humans (during erythrocyte recycling) and in pathogenic bacteria (during infection through hemolysis and heme sequestration). In animals, heme turnover produces bilirubin and CO, which act systemically as signaling molecules. Heme is also a source of PPIX, used as a precursor for synthesis of photoactive compounds called phytochromes in cyanobacteria, red algae, and plants. Lastly, heme is a redox- and photo- active molecule which can cause oxidative stress. Complete degradation of heme and isolation of the iron is essential to not only pathogens, but also serves a self-protective function.
Heme as an iron source.
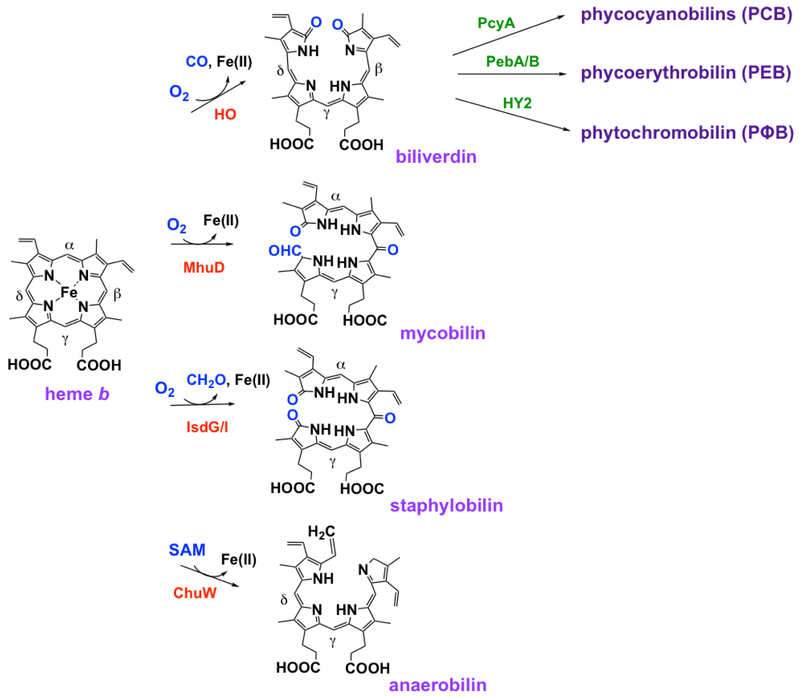
Heme can be non-enzymatically and non-stereospecifically degraded in the presence of either H2O2 or O2 with a reducing agent like ascorbate, releasing Fe(II), CO, and 4 isomers of the green product verdoheme [36,37]. By contrast, enzymes whose cellular role is to degrade heme (heme oxygenase, HO break the heme ring stereospecifically in an O2-dependent manner, usually producing α-verdoheme and CO [37,38•]. Reduction and addition of 2O2 yields α-biliverdin and Fe(II) (Figure 7). Mammalian isoform 1 of HO (HO1) is most abundant in the spleen, at the site of erythrocyte degradation and iron recycling. Soon after the discovery of HO1, prokaryotic homologs were found to catalyze the same reaction in pathogenic bacteria (e.g., HmuO in Corynebacterium and PigA/HemO in proteobacteria), this time as a way to harvest host-derived heme as a source of nutritional iron [37]. Termed canonical HOs, these enzymes have highly conserved α-helical structures, active sites with extensive hydrogen-bonding, and an α-helical loop that interacts with the distal side of the porphyrin and is responsible for stereospecific cleavage of heme at the α-meso carbon (Figure 8a) [39]. Because the heme ring is rotated ~100° in the PigA/HemO active site relative to the other HOs, this enzyme produces the β- and γ-products in a 3:7 ratio (Figure 8b) [36,37].
Figure 7. Different mechanisms and products of heme degradation.
Organisms have developed various mechanisms to break down heme in order to meet their specific needs. Each reaction is catalyzed by a different enzyme and generates unique products.
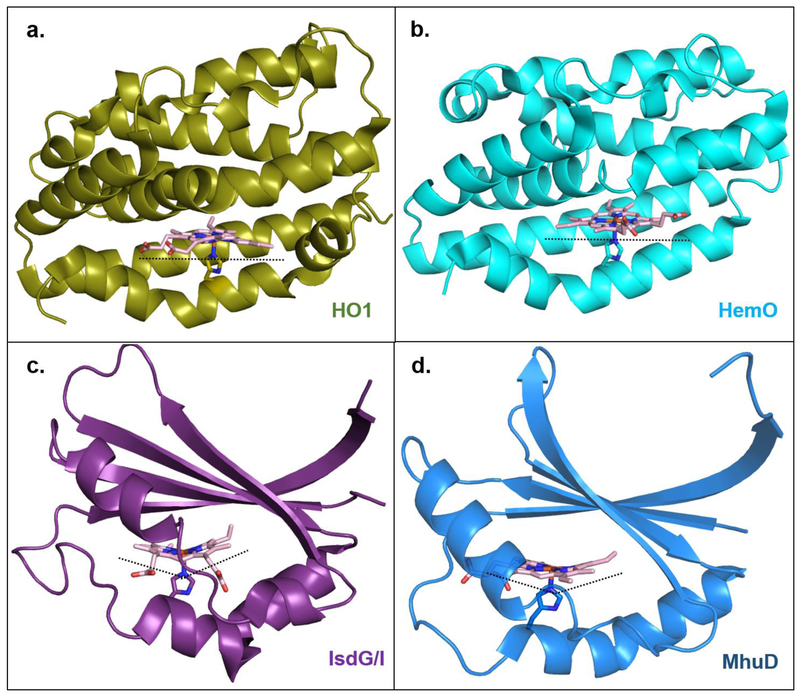
Figure 8. Multiple enzymes, multiple binding modes, multiple ways to degrade heme.
Canonical heme oxygenase enzymes HO1 and HemO ((a); PDBID: 1N45 and (b); PDBID: 1SK7), respectively) bind heme in a planar orientation. HO1 stereospecifically produces α-biliverdin, while HemO produces β- and γ-biliverdin due to the ~100° rotation of the heme ring in this active site relative to HO1. IsdG-like heme oxygenase enzymes IsdG/I and MhuD ((c); PDBID: 2ZDO and (d); PDBID: 4NL5, respectively) “ruffle” the heme porphyrin which results in cleavage of meso-carbons and products that are different than canonical HO’s (see figure 7).The heme ring in MhuD is rotated ~90° relative to the heme in IsdG/I, also contributing to the different bilin products that are produced.
Conceivably as a way to avoid production of CO, an immune system activator, some organisms such as Staphylococcus aureus and Mycobacterium tuberculosis have a different mechanism for degrading heme. Known as IsdG-like HOs, IsdG or IsdI and MhuD are dimeric enzymes that have ferredoxin-like α-/β-folds that form a β-barrel at the monomer interface (Figure 8C and 8D, respectively) [37,40••]. Each monomer binds heme, distorting the planarity of the ring. Porphyrin ruffling is believed to promote stereospecific cleavage at the β- and δ- positions or the α-position in IsdG/I or MhuD, respectively [41,42]. The products of IsdG/I are β- and δ-staphylobilin, Fe(II), and formaldehyde, while cleavage by MhuD (where the heme ring is rotated ~90° relative to the heme in IsdG/I) forms a different linear tetrapyrrole, α-mycobilin, Fe(II), and no 1-carbon product (Figure 7). Interestingly, the major chlorophyll breakdown pathway in green plants uses O2 and a Rieske-type monooxygenase, pheophorbide a oxygenase, to generate a structural analog of α-mycobilin called red chlorophyll catabolite, Mg(II), and no 1-carbon product [43,44]. This enzyme bears no sequence or structural similarity to MhuD.
The IsdG-like HOs exhibit further diversity among the species yet studied. The pathogen L. monocytogenes, in addition to its IsdG-like HO, encodes another IsdG-like protein called Isd- LmHde. This protein shares structural similarity to IsdG, and binds and degrades heme at both domains and folds as a monomer instead of a dimer [37]. By contrast, S. aureus expresses both IsdG and IsdI, paralogous proteins where the former is believed to carry out “housekeeping” heme degradation, while the latter serves a regulatory function.
Heme degradation in photosynthetic organisms.
Phytochromes are a class of photoreceptors in plants and photosynthetic bacteria. These molecules are synthesized via the conversion of heme to α-biliverdin by HO1 and the subsequent reduction of α-biliverdin to species specific phytochrome in a ferredoxin-dependent manner [45]. Cyanobacterial and plant HO1 is homologous to canonical HOs of non-photosynthetic organisms and similarly releases Fe(II) and CO during catalysis.
O2-independent heme degradation.
Organisms residing under anoxic conditions also have a mechanism to degrade heme. ChuW is a radical SAM methytransferase which degrades heme in an O2-independent manner. Using the 5’-deoxyadenosyl radical, a methyl group from a second molecule of SAM is transferred to heme, yielding the linear tetrapyrrole anaerobilin and Fe(II) [8•] (Figure 7). ChuW contains a [4Fe-4S] cluster and uses flavodoxin as an electron source. Heme degradation in the absence of O2 and without the concomitant production of immunogenic CO makes sense not only for obligate anaerobic pathogens, but also enteric commensal bacteria, where it may support nutritional iron absorption and/or relief from oxidative stress.
Conclusions
The diverse means by which organisms make and break heme speak to the importance of the tetrapyrrole scaffold and iron ion that is harbored inside it. Pathway diversity appears to have had both ecological and cell-biological drivers. It seems likely that, as genomes and ecosystems are further explored, tetrapyrrole biology will continue to be a locus for future discovery.
Highlights.
Pathways for heme biosynthesis and degradation are not universally conserved
Pathway diversification may have been both ecologically and biologically driven
Unique molecules from each pathway may serve as therapeutic targets or biomarkers
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review have been highlighted as:
• Denotes article of special interest
•• Denotes article of outstanding interest
- 1.Panek H, O'Brian MR: A whole genome view of prokaryotic haem biosynthesis. Microbiology 2002, 148:2273–2282. [DOI] [PubMed] [Google Scholar]
- 2.Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD: Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci U S A 2015, 112:2210–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, Howard MJ, Ferguson SJ, Warren MJ: Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc Natl Acad Sci U S A 2011, 108:18260–18265.• First article to describe a heme biosynthetic pathway for Archaea that differs from the canonical (PPD branch) heme biosynthesis pathway.
- 4.Phillips JD, Whitby FG, Warby CA, Labbe P, Yang C, Pflugrath JW, Ferrara JD, Robinson H, Kushner JP, Hill CP: Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae. Journal of Biological Chemistry 2004, 279:38960–38968. [DOI] [PubMed] [Google Scholar]
- 5.Dailey HA, Dailey TA, Gerdes S, Jahn D, Jahn M, O'Brian MR, Warren MJ: Prokaryotic Heme biosynthesis: Multiple pathways to a common essential product. Microbiol Mol Biol Rev 2017, 81.•• Extensive and thorough review article that describes and compares all heme biosynthesis pathways, and the involved enzymes, known to date; the PPD branch, the CPD branch, and the Ahb pathway branch.
- 6.Hamza I, Dailey HA: One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim Biophys Acta 2012, 1823:1617–1632.•• Review summarizes current knowledge about heme biosynthesis regulation and intermadiate trafficking in metazoans (PPD brach of heme biosynthesis). Review discusses the potential of multiezyme complex formation in order to effectively achive pathway regulation.
- 7.Layer G, Grage K, Teschner T, Schunemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein AX, Jahn D: Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines. J Biol Chem 2005, 280:29038–29046. [DOI] [PubMed] [Google Scholar]
- 8.LaMattina JW, Nix DB, Lanzilotta WN: Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proc Natl Acad Sci U S A 2016, 113:12138–12143.• Article provides evidence for a novel heme degration reaction that is O2-independent, exhibits a unique reaction mechanism, and produces distinct bilin products than described for other well-studied canonical and non-canonical heme oxygensases.
- 9.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD: Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes. The EMBO Journal 2003, 22:6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Layer G, Pierik AJ, Trost M, Rigby SE, Leech HK, Grage K, Breckau D, Astner I, Jansch L, Heathcote P, et al. : The substrate radical of Escherichia coli oxygen-independent coproporphyrinogen III oxidase HemN. The Journal of biological chemistry 2006, 281:15727–15734. [DOI] [PubMed] [Google Scholar]
- 11.AI Celis, Gauss GH Streit BR, Shisler K Moraski GC, Rodgers KR Lukat-Rodgers GS, Peters JW, DuBois JL: Structure-based mechanism for oxidative decarboxylation reactions mediated by amino acids and heme propionates in Coproheme Decarboxylase (HemQ). Journal of the American Chemical Society 2017, 139:1900–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.AI Celis, DuBois JL: Substrate, product, and cofactor: The extraordinarily flexible relationship between the CDE superfamily and heme. Arch Biochem Biophys 2015, 574:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayfield JA, Hammer ND, Kurker RC, Chen TK, Ojha S, Skaar EP, DuBois JL: The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J Biol Chem 2013, 288:23488–23504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AI Celis, Streit BR Moraski GC, Kant R, Lash TD, Lukat-Rodgers GS, Rodgers KR, DuBois JL: Unusual peroxide-dependent, heme-transforming reaction catalyzed by HemQ. Biochemistry 2015, 54:4022–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streit BR, Celis AI, Moraski GC, Shisler KA, Shepard EM, Rodgers KR, Lukat-Rodgers GS, DuBois JL: Decarboxylation involving a ferryl, propionate, and a tyrosyl group in a radical relay yields heme b. Journal of Biological Chemistry 2018, 293:3989–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi K, Masuda T, Tajima N, Wada H, Sato N: Molecular phylogeny and intricate evolutionary history of the three isofunctional enzymes involved in the oxidation of protoporphyrinogen IX. Genome Biol Evol 2014, 6:2141–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A: Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. The EMBO journal 2004, 23:1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira GC, Andrew TL, Karr SW, Dailey HA: Organization of the terminal two enzymes of the heme biosynthetic pathway. Orientation of protoporphyrinogen oxidase and evidence for a membrane complex. Journal of Biological Chemistry 1988, 263:3835–3839.• One of the first studies presenting evidence for mitochondrial membrane localization, complex formation, and substrate channeling by the terminal two enzymes in the PPD heme biosynthesis pathway.
- 19.Boynton TO, Daugherty LE, Dailey TA, Dailey HA: Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 2009, 48:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boynton TO, Gerdes S, Craven SH, Neidle EL, Phillips JD, Dailey HA: Discovery of a gene involved in a third bacterial protoporphyrinogen oxidase activity through comparative genomic analysis and functional complementation. Appl Environ Microbiol 2011, 77:4795–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato K, Tanaka R, Sano S, Tanaka A, Hosaka H: Identification of a gene essential for protoporphyrinogen IX oxidase activity in the cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A 2010, 107:16649–16654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin X, Sun L, Wen X, Yang X, Tan Y, Jin H, Cao Q, Zhou W, Xi Z, Shen Y: Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis. J Struct Biol 2010, 170:76–82.• Article throroughly compares the PgoX and CgoX enzyme structures, once thoughy to act on the same substrate, providing insight on how these enzymes are more different than what has been described.
- 23.Heinemann IU, Diekmann N, Masoumi A, Koch M, Messerschmidt A, Jahn M, Jahn D: Functional definition of the tobacco protoporphyrinogen IX oxidase substratebinding site. The Biochemical journal 2007, 402:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corrigal AV, Siziba KB, Maneli MH, Shepard EG, Ziman M, Dailey TA, Dailey HA, Kirsch RE, Meissner PN: Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus subtilis. Arch Biochem Biophys 1998, 358:251–256. [DOI] [PubMed] [Google Scholar]
- 25.Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED, Meissner PN, Acharya KR: Crystal Structure of Protoporphyrinogen Oxidase from Myxococcus xanthus and Its Complex with the Inhibitor Acifluorfen. Journal of Biological Chemistry 2006, 281:38625–38633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansson M, Hederstedt L: Bacillus subtilis HemY is a peripheral membrane protein essential for protoheme IX synthesis which can oxidize coproporphyrinogen III and protoporphyrinogen IX. Journal of Bacteriology 1994, 176:5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaraj VA, Arumugam R, Prasad D, Rangarajan PN, Padmanaban G: Protoporphyrinogen IX oxidase from Plasmodium falciparum is anaerobic and is localized to the mitochondrion. Mol Biochem Parasitol 2010, 174:44–52. [DOI] [PubMed] [Google Scholar]
- 28.Lobo SA, Scott A, Videira MA, Winpenny D, Gardner M, Palmer MJ, Schroeder S, Lawrence AD, Parkinson T, Warren MJ, et al. : Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol Microbiol 2015, 97:472–487. [DOI] [PubMed] [Google Scholar]
- 29.Medlock A, Swartz L, Dailey TA, Dailey HA, Lanzilotta WN: Substrate interactions with human ferrochelatase. Proc Natl Acad Sci U S A 2007, 104:1789–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepherd M, Dailey TA, Dailey HA: A new class of [2Fe-2S]-cluster-containing protoporphyrin (IX) ferrochelatases. Biochem J 2006, 397:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Henderstedt L: Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure 1997, 5:1501–1510. [DOI] [PubMed] [Google Scholar]
- 32.Lecerof D, Fodje MN, Alvarez Leon R, Olsson U, Hansson A, Sigfridsson E, Ryde U, Hansson M, Al-Karadaghi S: Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites. J Biol Inorg Chem 2003, 8:452–458. [DOI] [PubMed] [Google Scholar]
- 33.Soderberg C, Gillam ME, Ahlgren EC, Hunter GA, Gakh O, Isaya G, Ferreira GC, Al-Karadaghi S: The structure of the complex between yeast frataxin and ferrochelatase: Chracterization and pre-steady state reaction of ferrous iron delivery and heme synthesis.. J Biol Chem 2016, 291:11887–11898.• Article deliniates the site of interaction fetween frataxin and ferrochelatase (PpfC) in yeast by using protein docking, cross-linking-MS, an electron microscopy.
- 34.Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, Phillips JD, Dailey HA: Identification of the mitochondrial heme metabolism complex. PLoS One 2015, 10:e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mielcarek A, Blauenburg B, Miethke M, Marahiel MA: Molecular insights into frataxin-mediated iron supply for heme biosynthesis in Bacillus subtilis. PLoS One 2015, 10:e0122538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilks A, Heinzl G: Heme oxygenation and the widening paradigm of heme degradation. Arch Biochem Biophys 2014, 544:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyles KV, Eichenbaum Z: From host heme to iron: The expanding spectrum of heme degrading enzymes used by pathogenic bacteria. Frontiers in cellular and infection microbiology 2018, 8:198–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avila L, Huang H-w, Damaso CO, Lu S, Moënne-Loccoz P, Rivera M: Coupled oxidation vs heme oxygenation: Insights from axial ligand mutants of mitochondrial cytochrome b5. Journal of the American Chemical Society 2003, 125:4103–4110.• Article describes mechanistic difference between enzyme catalyzed and non-enzyme catalyzed heme degradation, emphasizing the need for extensive characterization of heme enzymes before ascribing a heme oxygenase role.
- 39.Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL: Crystal structure of human heme oxygenase-1. Nature Structural Biology 1999, 6:860. [DOI] [PubMed] [Google Scholar]
- 40.Wilks A, Ikeda-Saito M: Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc Chem Res 2014, 47:2291–2298.•• Review describes and summarizes the the O2-dependent heme degradation reactions, the unique enzymes that catlayze them, and the unique products that are produced.
- 41.Graves AB, Morse RP, Chao A, Iniguez A, Goulding CW, Liptak MD: Crystallographic and spectroscopic insights into heme degradation by Mycobacterium tuberculosis MhuD. Inorganic Chemistry 2014, 53:5931–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee WC, Reniere ML, Skaar EP, Murphy MEP: Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. Journal of Biological Chemistry 2008, 283:30957–30963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hörtensteiner S, Wüthrich KL, Matile P, Ongania K-H, Kräutler B: The key step in chlorophyll breakdown in higher plants: Clevage of pheophorbide a macrocyle by a monooxygenase. Journal of Biological Chemistry 1998, 273:15335–15339. [DOI] [PubMed] [Google Scholar]
- 44.Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S: Chlorophyll breakdown: Pheophorbide oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death gene. Proceedings of the National Academy of Sciences 2003, 100:15259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shekhawat GS, Verma K: Haem oxygenase (HO): an overlooked enzyme of plant metabolism and defence. Journal of Experimental Botany 2010, 61:2255–2270. [DOI] [PubMed] [Google Scholar]