Abstract
Biofilms are matrix-associated communities that enable bacteria to colonise environments unsuitable for free-living bacteria. The facultative intracellular pathogen Francisella tularensis can persist in water, amoebae, and arthropods, as well as within mammalian macrophages. F. tularensis Types A and B form poor biofilms, but F. tularensis mutants lacking lipopolysaccharide O-antigen, O-antigen capsule, and capsule-like complex formed up to 15-fold more biofilm than fully glycosylated cells. The Type B live vaccine strain was also 50% less capable of initiating surface attachment than mutants deficient in O-antigen and capsule-like complex. However, the growth medium of all strains tested also influenced the formation of biofilm, which contained a novel exopolysaccharide consisting of an amylose-like glucan. In addition, the surface polysaccharide composition of the bacterium affected the protein:DNA:polysaccharide composition of the biofilm matrix. In contrast, F. novicida attached to surfaces more efficiently and made a more robust biofilm than Type A or B strains, but loss of O-antigen or capsule-like complex did not significantly affect F. novicida biofilm formation. These results indicated that suppression of surface polysaccharides may promote biofilm formation by F. tularensis Types A and B. Whether biofilm formation enhances survival of F. tularensis in aquatic or other environmental niches has yet to be determined.
Subject terms: Bacteriology, Biofilms, Pathogens
Introduction
Francisella tularensis is a gram-negative, facultative intracellular bacterium that can infect numerous mammals, arthropods and other eukaryotes, and is the etiologic agent of tularemia or “rabbit fever”1–3. F. tularensis is categorised by the Centers for Disease Control and Prevention (CDC) as a Tier I select agent due to its capacity to cause fatal disease by as few as 10 cells and has the capability to be weaponised. Isolates that are most commonly responsible for disease belong to subsp. tularensis (Type A) and subsp. holarctica (Type B). Each year in the U.S. there are over 100 cases of naturally occurring tularemia, predominately by Type A strains, with the majority of transmissions occurring through tick bites4. F. tularensis subsp. holarctica is less virulent to humans than subsp. tularensis, and is more common in Europe where it is not uncommon for over one thousand human cases to occur in a given year5. The route of infection for the majority of cases in northern Europe is suspected to be mosquito-borne, as most patients are near water and report mosquito bites6. Most mammals are susceptible to infection, and typically die from the disease, although some animals are more resistant to disease and can clear the bacteria6; therefore, it is likely that F. tularensis can persist in nature in non-mammalian sites. Studies using molecular typing methods have shown that F. tularensis can persist in water, mud, and insects7–9, supporting persistence of F. tularensis in the environment, but also raising question as to how this fastidious bacterium does persist in such sites. Most of the current research on F. tularensis has focused on mammalian infection, growth in macrophages and other cells, host immune response, and vaccine development, but other facets of the biology of this bacterium require investigation to more thoroughly understand the life cycle and transmission of F. tularensis.
In order to persist in terrestrial and aquatic habitats, especially where nutrients are limited, bacteria commonly form a biofilm. A biofilm is an aggregated microbial community that forms within an extracellular polymeric substance, normally composed of exopolysaccharide (EPS), extracellular DNA (eDNA), and proteins10. F. novicida is a distinct species or subsp. of F. tularensis that is of low virulence for humans, but highly pathogenic for mice, and antigenically distinct from subsp. tularensis and holarctica. F. novicida readily forms biofilms on a variety of surfaces including plastics, glass, and crab shells11. In contrast, F. tularensis subsp. tularensis and subsp. holarctica generally do not form a substantial biofilm, although they are capable of developing some biofilm on polystyrene 96-well plates12.
Despite their genetic similarity, the less virulent F. novicida strains differ from Types A and B (A/B) strains in their polysaccharide surface components. The composition and antigenic reactivity of the lipopolysaccharide (LPS) O-antigen of Types A/B strains is distinct from that of F. novicida13. Type A strain SchuS4 and Type B live vaccine strain (LVS) have O-Ag structures that consist of a 4-glycose unit of two internal carbohydrate residues (α-D-GalNAcAN–α-D-GalNAcAN) and two peripheral residues (β-D-Qui4NFm and β-D-QuiNAc)14, while F. novicida O-Ag shares the same two internal residues, but has different terminal residues (α-D-GalNAcAN and α-D-QuiNAc4NAc)13. Genomic analysis of the O-Ag gene clusters (wbt locus) of the two species confirms that while genes encoding for proteins responsible for O-Ag synthesis in Types A/B strains are identical, the F. novicida O-Ag locus has fewer and some different genes, and a lower G + C ratio15,16. The LPS is an important virulence factor for all subspecies, as it protects the bacterium against innate host defenses, such as complement-mediated killing17, and mutants lacking O-Ag are highly attenuated in mice15,18. Subspecies tularensis and holarctica also produce an O-Ag capsule, but if only the O-Ag capsule is absent, the bacteria remain serum-resistant19. Types A/B strains and F. novicida also produce a novel capsule-like complex (CLC) containing glycosylated proteins20,21. Whether the differences in surface glycosylation between Types A/B strains and F. novicida contribute to their differences in capability to form biofilm or development of the biofilm matrix has yet to be determined.
In this investigation the capability of the more virulent Types A/B strains to form a biofilm in comparison to glycose-deficient mutants and F. novicida was examined. Whereas wildtype F. novicida made a robust biofilm within 2–3 days of incubation in vitro, Types A/B strains required at least 10 days to form a similar biofilm. However, loss of the LPS O-Ag and/or loss of the capability to glycosylate CLC proteins resulted in enhanced biofilm formation sooner by Types A/B strains, but not F. novicida. Furthermore, following F. tularensis biofilm formation the EPS from the matrix material was isolated and identified as glucan, making this the first report of glucan production by F. tularensis. We hypothesise that F. tularensis can phase vary its surface polysaccharides to successfully transition between a phenotype that forms a biofilm, which may enhance survival in the environment, and a planktonic phenotype that is a facultative intracellular pathogen in the mammalian host.
Results
Effect of incubation time and bacterial surface components on attachment and biofilm development
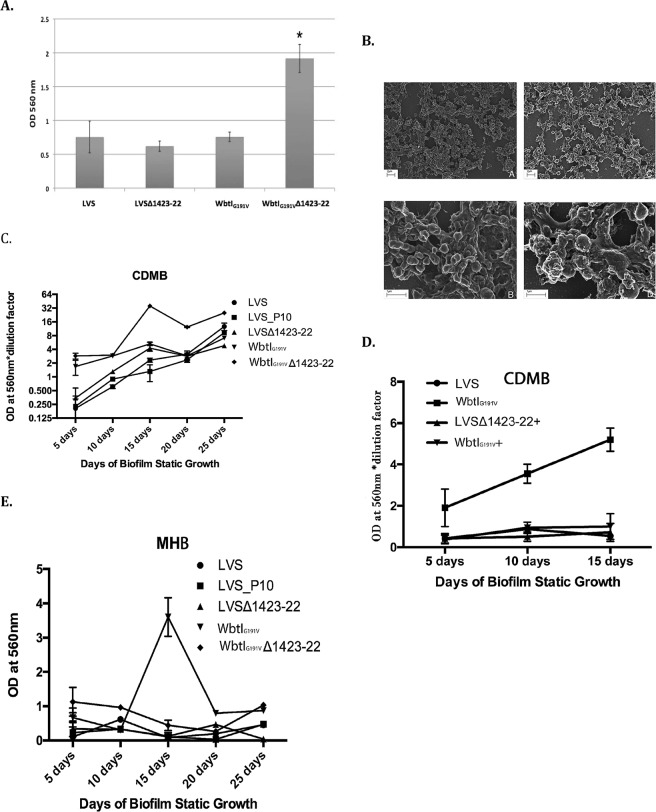
Static attachment of LVS to a polyvinyl surface occurred within 1 hour (Fig. 1A), and micro-colony and some multi-layering of cells occurred after 5 days incubation (Fig. 1B). However, there was no significant difference between the number of LVS cells and LVS mutants (Table 1) deficient in O-antigen (Ag) (WbtIG191V) or CLC (LVSΔ1423-22) that attached to polyvinyl 96-well plates (Fig. 1A), or the time required for attachment (1, 2, or 4 hours; data not shown). However, the LVS double mutant lacking O-Ag and CLC (WbtIG191VΔ1423-22) attached significantly better (p = 0.0101) (about 2-fold) than the parent LVS strain or mutants deficient in only O-Ag (p = 0.0025) or only CLC (p = 0.0019) (Fig. 1A).
Figure 1.
Biofilm development by LVS and surface antigen mutants: attachment and static growth. (A) CV attachment assay. Bacteria were grown to stationary phase, inoculated into 96-well plates in triplicate, incubated for 2 hours at 37 °C, washed, and stained with CV to detect cells that have attached. The absorbance was determined at 560 nm after the CV was solubilised with 95% ethanol. *p value < 0.005. (B) SEM of F. tularensis biofilms. Biofilms were grown on silicone disks for 5 days and carbon-coated (panels and strains: (A,B), LVS; (C,D), LVSΔ1423-22). After 5 days growth, no difference was seen in overall attachment or biofilm formation. At increased magnification the surface of LVSΔ1423-22 appeared more granular than the parent, which could be the result of CLC deficiency. (C,D,E) Effect of growth medium on biofilm formation by LVS, glycose-deficient mutants, and complemented strains. C, biofilm formation by parent and mutant strains grown in CDMB; D, biofilm formation by parent and complemented mutant strains (LVSΔ1423-22+ and WbtIG191V+) grown in CDMB with O-Ag mutant WbtIG191V included for comparison; E, biofilm formation by parent and mutant strains grown in MHB. A marked increase in biofilm formation by WbtIG191VΔ1423-22 occurred in MHB medium after 15 days incubation that was not observed following growth in CDMB.
Table 1.
Strains used in this study.
| Strain | Characteristics | LPS/O-Ag | O-Ag capsule | CLC | Attenuated | Source/Reference |
|---|---|---|---|---|---|---|
| F. tularensis LVS (Type B) | Attenuated Live Vaccine Strain | + | + | + | N | Dr. May Chu, CDC |
| LVS_P10 | LVS subcultured 10 consecutive days in broth CDMB | + | + | +++ | N | 20 |
| LVSΔ1423-22 | LVS deficient in CLC due to deletion of genes encoding for glycosyltransferases | + | + | − | Y | 20 |
| WbtIG191V | O-Ag mutant of LVS due to point mutation in wbtI | − | − | + | Y | 18 |
| WbtIG191V_P10 | WbtIG101V subcultured 10 consecutive days in CDMB | − | − | +++ | Y | 20 |
| WbtIG191VΔ1423-22 | O-Ag and CLC deficient double mutant. WbtIG191V with deletion of FTL_1423 and 1422 | − | − | − | Y | 20 |
| WbtIG191V+ | Complemented strain of respective mutant with O-Ag restored | + | + | + | NDb | 18 |
| LVSΔ1423-22+ | Complemented strain of respective mutant with CLC restored | + | + | + | ND | 20 |
| KY99-3387 | Wild Type B strain | + | + | + | N | BEI Resources |
| KY99-3387_P10 | KY99-3387 subcultured 10 consecutive days in broth CDMB | + | + | +++ | N | This work |
| F. novicida U112 | Parent F. novicida strain | + | ND | +/− | N | Dr. Karen Elkins, FDA |
| F. novicida_P10 | Strain U112 subcultured 10 consecutive days in CDMB | + | ND | +++ | N | 21 |
| F. novicidaΔ1212-18 | CLC-deficient strain of U112 due to loss of FTN_1212-1218 in CLC glycosylation locus | + | ND | − | Y | 21 |
| F. tularensis SchuS4 (Type A) | Virulent Type A strain parent | + | + | + | N | Dr. Mark Wolcott, USAMRIID |
| F. tularensis TI0902 (Type A) | Highly virulent Type A strain parent | + | + | + | N | 62 |
| F. tularensis TIGB03 | Chemical mutant of TI0902 lacking O-Ag | − | − | + | Y | 25 |
(+, present; +++, enhanced; −, not present).
aNot determined.
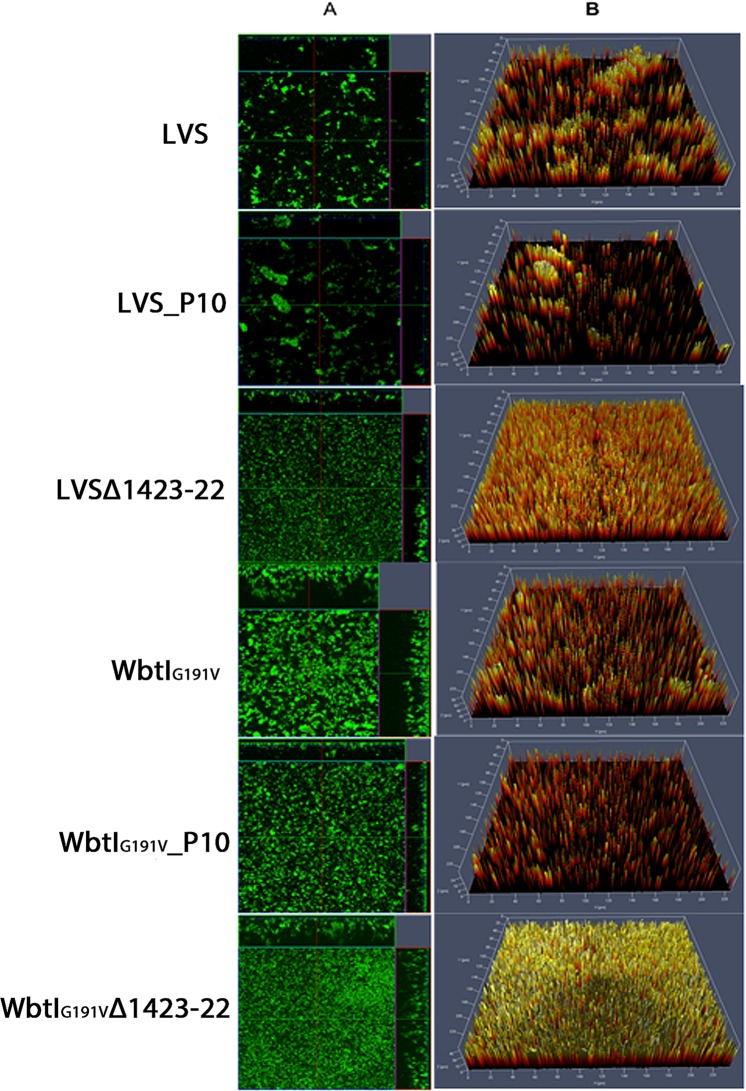
Previous studies of biofilm formation by F. novicida and F. tularensis strains measured biofilm formation after seven days or less11,22,23. However, we observed that at least 10–15 days incubation was required for optimum biofilm development by LVS, and to fully differentiate biofilm development between LVS and LVS mutants deficient in O-Ag or glycosylation of CLC surface structures (WbtIG191V, LVSΔ1423-22, and WbtIG191VΔ1423-22, respectively). However, production of a clear biofilm by the O-Ag mutants was evident within 5 days of incubation. Nonetheless, a mature biofilm by all strains tested required about 15 days incubation, at which time differences in thickness and overall mass between the parent and each mutant were maximised (Fig. 1C). Both the single mutant (LVSΔ1423-22) and double mutant (WbtIG191VΔ1423-22) with deletions in the polysaccharide locus that glycosylates CLC (CLC-deficient) formed 2 to 4-fold and 15-fold more biofilm, respectively, than their parent strain by day 15 (Fig. 1C). After 15 days incubation there was a reduction in the more mature biofilms, indicating that dispersal of planktonic cells occurred; after about 20 days the biofilm cycle began again with reattachment of cells and biofilm formation (Fig. 1C). Complementation of the O-Ag mutation or the CLC mutation in trans completely reversed the enhancement in biofilm formation (Fig. 1D). Furthermore, LVS formed 46% more biofilm than the CLC-enhanced strain LVS_P10 (LVS subcultured in Chamberlains Defined Medium Broth (CDMB) for 10 consecutive days) after 15 days incubation (Table 2). Therefore, cell attachment and the amount of biofilm formed was inversely proportional to the amount of cell-associated carbohydrate on the bacterial surface. These differences in biofilm formation by parent, mutants, and CLC-enhanced strains were also evident by confocal laser scanning microscopy (CLSM) (Fig. 2), particularly after 15 days of incubation. LVS, and to a greater degree LVS_P10, developed more sparse micro-colonies, whereas the mutants, particularly WbtIG191VΔ1423-22, developed more confluent, thicker biofilms. However, O-Ag-deficient variant WbtIG191V_P10, which is enhanced for CLC, also produces a less dense biofilm compared to the non-subcultured mutant (Fig. 2). The mutants that made a more robust biofilm did not increase the amount of substrata surface coverage as much as they increased the formation of dense, cellular aggregates that adhered to the surface, as well as floating pellicles.
Table 2.
Fold-difference in biofilm developmenta in CDM or MH compared to LVS.
| Day 10 | Day 15 | |||
|---|---|---|---|---|
| CDMB | MHB | CDMB | MHB | |
| LVS | 1 | 1 | 1 | 1 |
| LVS_P10 | 1.5 ± 0.20 | −2.5 ± 0.01* | −1.8 ± 0.4* | 1.3 ± 0.75 |
| LVSΔ1423-22 | 2.1 ± 0.16 | −2.5 ± 0.01* | 1.8 ± 0.1 | 1.56 ± 0.91 |
| LVSΔ1423-22+b | −1.05 ± 0.17 | NDc | −1.48 ± 0.44* | ND |
| WbtIG191V | 4.6 ± 0.17** | −2.7 ± 0.01 | 2.3 ± 0.1 | 45.2 ± 0.138** |
| WbtIG191V+ | −1.38 ± 0.20 | ND | −1.48 ± 0.16 | ND |
| WbtIG191V_Δ1423-22 | 4.9 ± 0.10** | 1.23 ± 0.1 | 15.3 ± 0.5** | 5.5 ± 0.34** |
aFold-difference calculated from OD560nm of CV assays.
b+-strain with mutation complemented.
cND-Not determined.
*Indicates P < 0.05; **Indicates P < 0.01.
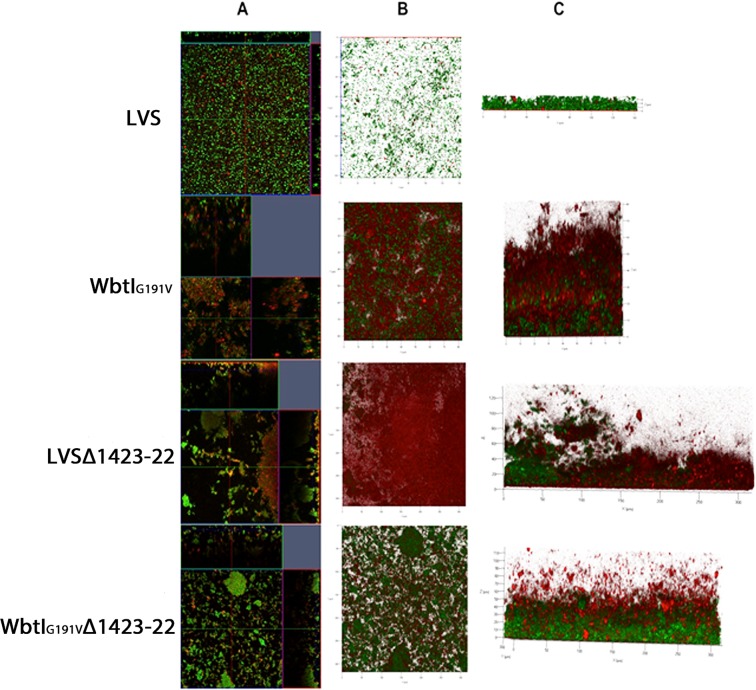
Figure 2.
Comparison of biofilm formation by F. tularensis LVS, WbtIG191V (LPS O-Ag mutant), LVSΔ1423-22 (CLC mutant), and O-Ag and CLC double mutant before and after passage in CDMB by confocal laser scanning microscopy. Panel (A) contains orthogonal sections showing horizontal (z) and side views (x and y) of reconstructed three-dimensional biofilm images at a magnification of 25x. Biofilms were stained with Syto9 showing both live and dead bacterial cells. Panel (B) shows topographical images of biofilm growth of the parent LVS and its surface structure mutant strains. Loss of surface carbohydrate was associated with denser biofilms, but subculture passage in CDMB resulted in thinner biofilms.
LVS is not a wildtype Type B strain, as it has been extensively subcultured and is known to contain multiple mutations within its genome24. Therefore, wildtype Type B strain KY99-3387 was subcultured in CDMB for 10 passages to obtain KY99-3387_P10 to enhance CLC expression (attempts to isolate a grey, O-antigen-deficient variant of strain KY99-3387 were unsuccessful). As for LVS, strain KY99-3387_P10 formed significantly less biofilm by 10 and 15 days incubation than non-subcultured strain KY99-3387 (Fig. 3), supporting the evidence that growth conditions that enhance surface carbohydrate expression reduce biofilm formation of type B strains.
Figure 3.
Biofilm formation by a F. tularensis subspecies holarctica wildtype strain following subculture passage in CDMB. A wildtype Type B strain was incubated in microtiter plates under biofilm forming conditions before and after 10 daily subculture passages in CDMB. After 5, 10, or 15 days incubation the wells were rinsed to remove planktonic cells, stained with CV, and the CV solubilized with ethanol and the absorbance determined at 560 nm. Five to six replicates of the parent and the passed variant were tested at each time period. *Indicates a statistically significant difference (P < 0.05).
Effects of growth medium on biofilm development by F. tularensis
Growth curves of all F. tularensis strains and mutants grown in either CDMB or Mueller-Hinton broth (MHB) were similar (data not shown). However, the amount of biofilm formed (as determined by crystal violet (CV) assay) was greatly enhanced when the bacteria were grown in CDMB (Fig. 1C) compared to MHB (Fig. 1E) for all strains by post-inoculation days 10 and 15 (Table 2) and beyond. The fold-difference in biofilm formed increased the most for the mutant lacking both O-Ag and CLC (WbtIG191VΔ1423-22) in CDMB, particularly 15 days post-inoculation when compared to the parent (15.3-fold in CDMB). Overall there was little biofilm formed by the parent, CLC-enhanced parent, or mutants over 25 days of growth in MHB. The exception was mutant WbtIG191VΔ1423-22, which spiked a 5.5-fold increase in biofilm formation by 15 days incubation in MHB, but was reduced again by 20 days incubation (Fig. 1E). Any increase in biofilm formation by the mutants was eliminated when the mutation was complemented (Table 2). These differences indicated that both the growth medium, and the presence or absence of bacterial surface polysaccharides (O-Ag and CLC), influenced biofilm development in F. tularensis.
Extraction of the CLC from biofilm cells
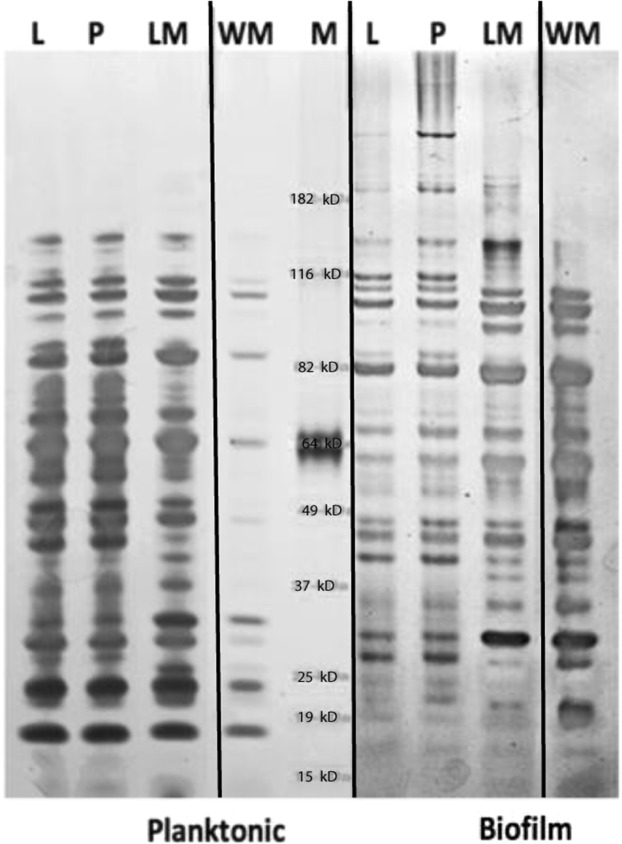
LVS and LVS_P10 cells grown as biofilms both produced high molecular size (HMS) components that have been associated with the CLC20 (Fig. 4, Biofilm plate; L [LVS] and P [LVS_P10], respectively). These HMS components were clearly more prominent following repeated subculture of LVS in CDMB, which greatly enhances CLC formation20. However, the HMS bands were diminished in extracts from the biofilm of LVSΔ1423-22 (LM) and absent from the biofilm of WbtIG191VΔ1423-22 (WM), from which biofilms were more prominent, but CLC is not produced20. HMS bands were also not present in any of the bacteria or mutants in planktonic phase, from which CLC is not formed20. There were also clear differences in the overall protein profiles of planktonic bacteria and biofilm bacteria, as well as in O-Ag and CLC mutants (Fig. 4). Therefore, growth as a biofilm and loss of surface carbohydrates also affected the protein profile of F. tularensis.
Figure 4.
Electrophoretic protein profiles of LVS variants or mutants grown as a biofilm or are planktonic cells from the biofilm broth supernatant. Bacterial strains were grown as a biofilm in CDMB for 25 days, the medium above the biofilm was removed, and the planktonic cells harvested by centrifugation. Planktonic cells in the medium and cells in the biofilm were extracted with 1 M urea. The protein profiles were resolved by SDS-PAGE on 4-12% gels and stained with silver. Strains: L, LVS; P, LVS_P10; LM, LVSΔ1423-22; WM, WbtIG191VΔ1423-22; M, Molecular size standards. The figure is a compilation of four separate gels containing samples prepared during the same experiment (left two are from planktonic phase cells and right two are from biofilm cells), which have been cropped to present the most relevant information (original shown in Supplementary Fig. S3). Precipitates of the broth medium are not shown. Lanes of WbtIG191V (W) have been deleted from the planktonic cells (far left) and from the biofilm cells (far right) due to lack of differences with WbtIG191VΔ1423-22 (WM). Repeated subculture in CDMB had little effect on protein composition, but loss of surface glycoses, particularly O-Ag, or growth as a biofilm substantially altered the protein profile.
Composition and analysis of the extracellular matrix exopolysaccharide (EPS)
EPS was extracted from WbtIG191VΔ1423-22, which made the most biofilm. The extracted EPS was visualised following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Pro-Q Emerald 300 fluorescent carbohydrate stain. The EPS appeared as a diffuse smear, but included large molecular size material >200-kDa that was not visible with silver stain alone (data not shown). Based on gas chromatography-mass spectrometry (GC-MS), the composition of the purified EPS was determined to be 99.9 mole% glucose and 0.1 mole% mannose. Subsequent linkage analysis indicated that 54% of the glucose residues were terminal and that 33% were 1,4-linked and that 1.6% of the glucose was 1,6-linked, indicating the composition of the EPS was consistent with amylose (Supplementary Fig. S1). An additional 4.4% of the EPS contained 1,4-linked mannose residues. The presence of 1,6-linked poly-N-acetylglucosamine (PNAG) was not identified in the biofilm based on GC-MS, lack of reactivity by immunoblotting with antibodies to PNAG (Sigma-Aldrich, St. Louis, MO), or by fluorescence microscopy with wheat germ agglutinin (lectin to PNAG) (data not shown). Texas Red-conjugated Concanavalin A (ConA), which binds to α-D-mannosyl and α-D-glucosyl groups, reacted strongly with LVS, WbtIG191V, and WbtIG191VΔ1423-22 biofilms (Fig. 5C). An increase in the amount of EPS/matrix (red) was evident in the biofilms of both mutants. However, the biofilm matrix could not be clearly resolved by CLSM in conjunction with ConA due to the thickness of the matrix. A BLASTP search of proteins responsible for glycogen expression and regulation in Escherichia coli (GlgXBACP) identified genes with substantial identity in F. tularensis SchuS4 (Table 3). The most closely related proteins were from Clostridium spp., but also from Massilia spp., Lachnospiraceae spp., Burkholderia spp., and Lachnospiraceae spp. Of interest was that glgC is described as a pseudo gene in Type A F. tularensis, but is a functional glucose-1-phosphate adenylyltransferase in Type B LVS.
Figure 5.
Comparison of biofilm matrix development by F. tularensis LVS and its surface structure mutants by CLSM. Panel (A) shows orthogonal sections of horizontal (z) and side views (x and y) of reconstructed three-dimensional biofilm images at a magnification of 25x. Nucleic acids were stained with Syto9 (green) and the matrix EPS stained with Texas Red-ConA (red). Panel (B) is an overview (x and y) view of the reconstructed three-dimensional z-stack of each biofilm growth on the coverslip. Panel C is the side view of the three-dimensional z-stack. Each picture was obtained from biofilms grown for 15 days.
Table 3.
Francisella tularensis Type A strain SchuS4 genes with amino acid identity to glycogen synthesis proteins in heterologous bacteria.
| F. tularensis | Gene symbol | Putative gene product | Percent amino acid | Gene Size (bp) |
|---|---|---|---|---|
| Locus taga | identity to:b | |||
| FTT_0412 | pulB | type I pullulanase | Clostridium spp.-50% 3212 | 3212 |
| FTL_RS02530 | F. tularensis LVS-97% | |||
| FTT_0413 | glgB | 1,4-α-glucanotransferase | Clostridium perfringens-63% | 1,922 |
| FTL_RS02535 | glycogen branching enzyme | F. tularensis LVS-99% | ||
| FTT_0414 | pgm | α-D-glucose phosphoglucomutase | Massilia spp.-68% | 1,634 |
| FTL_RS02540 | F. tularensis LVS-99% | |||
| FTT_0415 | glgC | Pseudo | 1,269 | |
| FTLRS02545 | glucose-1-phosphate adenylyltransferase | Lachnospiraceae spp.-63% | 1,271 | |
| F. tularensis LVS-99% | ||||
| FTT_0416 | glgA | glycogen synthase | Burkholderia ubonensis-63% | 1,469 |
| FTL_RS02550 | F. tularensis LVS-96% | |||
| FTT_0417 | malP | maltodextrin phosphorylase | Lachnospiraceae bacterium-63% | 2,273 |
| FTL_RS02555 | F. tularensis LVS-99% | |||
| FTT_0418 | malA | 4-α-glucanotransferase (amylomaltase) | Clostridium spp.-53% | 1,463 |
| FTL_RS02560 | F. tularensisLVS-97% |
aType A SCHUS4 locus tag, followed by the Type B LVS locus tag listed underneath.
bGenus other than Francisella with the closest amino acid identity.
Enzymatic treatment of the biofilms using sodium m-periodate reduced biofilm mass for all strains, but there was a significant reduction in biofilm for only strains WbtIG191V (p = 0.0246) and WbtIG191VΔ1423-22 (p = 0.0055) compared to treatment of the biofilms with phosphate buffered saline, pH 7.4 (PBS) (Fig. 6). Detachment of the biofilm by sodium m-periodate supported that these mutant strains, though producing less surface polysaccharide, may contain relatively more EPS in their biofilms. Only the WbtIG191V biofilm was significantly diminished after treatment with DNase (p = 0.0190), and WbtIG191VΔ1423-22 was the only strain whose biofilm was significantly decreased after treatment with Proteinase K (p = 0.0162). These differences indicated that changes in the surface antigen structures on Francisella impact the proportional composition of the biofilm matrix, in addition to the amount of biofilm produced.
Figure 6.
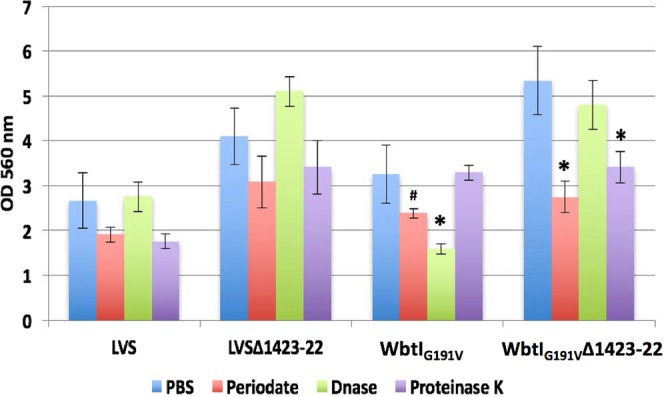
Effect of periodate, DNAse, or Proteinase K on biofilm integrity. Ten-day old biofilms were treated with sodium m-periodate, DNase, Proteinase K, or PBS for two hours at 37 °C, washed with distilled water and left to dry overnight before CV staining. Only WbtIG191V and WbtIG191VΔ1423-22 contained significantly less biofilm (#p = 0.0246, *p = 0.0055) after periodate treatment when compared to untreated biofilm from the same strain, indicating these strains may produce more of the glucan EPS in their biofilms compared with the LVS parent strain. The WbtIG191V biofilm only was significantly diminished after treatment with DNase (*p = 0.0190), whereas only the WbtIG191VΔ1423-22 biofilm was significantly affected by treatment with Proteinase K (*p = 0.0162).
Biofilm formation by Type A F. tularensis
Overall, Type A strains developed less biofilm than LVS or KY99-3387. By 15 days incubation, the Type A strains appeared to have detached and new/continued growth of biofilm was not detected even with the addition of fresh medium, indicating the biofilm lifecycle of Type A strains is shorter than for LVS. The Type A mutant TIGB03 is deficient in O-Ag25 and like its LVS counterpart, developed more biofilm compared to parent strain TI0902. After 5 days growth in CDMB, TIGB03 produced significantly more biofilm (~2-fold) than parent strain TI0902 or SCHUS4 in CDMB or in MHB (p = 0.0321) (Supplementary Fig. S2). However, by day 10 parent strains SCHUS4 and TI0902 developed prominent biofilms when grown in CDMB that were not significantly different from the biofilm formed by TIGB03 (p = 0.78) (Fig. S2). The wildtype strains never developed a prominent biofilm in MHB, but O-Ag mutant TIGB03 did.
F. novicida biofilm
In contrast to the differences in biofilm development observed between Type A strains, the Type B LVS strain, and their surface antigen mutants when grown in CDMB versus MHB, there was not a significant difference in biofilm formation by F. novicida grown in CDMB or MHB. However, F. novicida did produce significantly more (p < 0.0001) biofilm when the bacteria were grown in BHI or tryptic soy broth (TSB) (OD560 1.5 ± 0.2 and 1.3 ± 0.1, respectively) than in MHB (0.7 ± 0.1). Similar results were obtained for attachment of the cells to microtiter wells (data not shown). As for Type B strains LVS and KY99-3387, increased CLC production (by F. novicida_P10) negatively affected biofilm formation (Fig. 7A). However, in contrast to parent strain U112, the CLC-deficient mutant F. novicidaΔ1212-1218 did not produce more biofilm than the parent when grown in either medium, but there was a significant increase (~2-fold) in biofilm development when the cells were grown in CDMB compared to MHB (p = 0.0050) by days 10 and 15 post-inoculation (Fig. 7B). Surprisingly, most of the F. novicida O-Ag mutants tested (Supplementary Table S1) made less biofilm than the parent, except for those with mutations in wbtN and wbtQ (Fig. 7C). F. novicida also attached more uniformly to the glass coverslip than Type A or LVS strains, including mutants (data not shown). Therefore, there were stark differences in biofilm formation, and the factors that influence biofilm formation, by F. novicida compared to Types A and B strains, although there were also some similarities.
Figure 7.
Comparison of biofilm development by F. novicida and mutant strains. (A) CLSM: Orthogonal sections of horizontal (z) and side views (x and y) of reconstructed three-dimensional biofilm images at a magnification of 25x. Nucleic acids were stained with Syto9 (green) and the matrix EPS was stained with Texas Red-Con A (red). Passage of F. novicida in CDMB to increase CLC production resulted in a less robust biofilm by day 10, but initial attachment did not appear to be affected. Large, dense aggregates of cells were evident by day 10. (B) Comparison of biofilm development by F. novicida and its CLC-deficient mutant grown in CDMB and MHB. No significant differences were seen between parent and mutant biofilms formed in either medium. Like Type B strains, biofilm development peaked at ~15 days. (C) Biofilm development of O-Ag mutants of F. novicida. F. novicida biofilms were grown for 5 days in CDMB and stained with CV. Data are shown as percent comparison to the parent F. novicida strain. All O-Ag mutants produced less biofilm than the parent except mutants in the wbtN and wbtQ genes.
Discussion
The capability of Francisella species to form biofilms has been confirmed in subspecies tularensis, holarctica, and novicida12, but the biofilm made by subspecies novicida forms earlier and is denser and more developed than biofilms formed by the other two subspecies. However, details regarding the factors that influence biofilm formation, particularly in Type A and B strains, have not been described. Francisella is a unique intracellular pathogen with an extremely low infectious dose, but the role of biofilm formation in the life cycle of this bacterium is unknown. One hypothesis is that biofilm may contribute to survival of Francisella spp. outside of the mammalian host12,26,27. As a pathogen that can kill most of its hosts, F. tularensis must survive in a wide range of environmental conditions (hot and arid, cold waterways, and even intracellularly in amoebae)7,27,28. The shorter biofilm cycle and overall less biofilm formed by Type A strains, even compared to Type B strains, could be an indicator of the differences in environmental niches that these two biotypes inhabit. Type A strains are found predominately only in drier environments while Type B strains are commonly found in waterways, particularly in Europe, but also in the U.S.29–31. The initial attachment and structural stability of biofilms from F. novicida and Type B strains may need to be thicker and more developed than from Type A strains in order to survive the sheer force of flowing water and the natural predation that occurs in waterways from amoebae and vectors, such as mosquito larvae2,7,28. Formation of a biofilm by Francisella spp. may also aid survival while inside arthropod vectors26. Yersinia pestis survives inside the flea gut in biofilm form, and F. novicida can form biofilms on chitin surfaces11,32. Chitin is found in many arthropod vector exoskeletons, and we have found that F. tularensis Types A/B strains will form biofilms, albeit less prominent (data not shown), at room temperature (~22 °C), which is similar to the internal temperature of a tick11,33. Further exploration of this observation is needed.
Besides the subspecies of Francisella, the amount of cell surface carbohydrate was found to be the major factor influencing biofilm formation by Types A and B strains. The glycosylated CLC on the cell surface is greatly enhanced when F. tularensis is serially passed in and grown on medium that simulates in vivo growth conditions, such as CDMB34. Although LVS mutants that lack CLC on the cell surface are attenuated20, biofilm formation by these mutants was also enhanced. When CLC expression was enhanced (by subculture in CDMB) in both LVS and virulent Type B strain KY99-3387, whether O-Ag was present or not, biofilm formation was diminished after 10-15 days incubation. This interference in biofilm formation also occurs in encapsulated bacteria such as Porphyromonas gingivalis35 and Pasteurella multocida36. In these species enhanced biofilm formation is associated with loss of capsular polysaccharide. The CLC is composed of multiple glycoproteins, and its expression on the bacterial surface is negatively affected by deletion of two glycosyltransferases in a proposed O-linked protein glycosylation locus (FTT_0789-0800)20,37. Similar results have been reported in Acinetobacter baumannii lacking PglC, the initiating glycosyltransferases (iGT) in a glycan synthesis locus38. The proposed iGT of the F. tularensis CLC locus, FTT_0790, and the iGT of the O-Ag locus wbtB, are homologous to pglC in A. baumannii and could indicate en bloc glycan synthesis is involved in CLC O-glycosylation as well39.
O-Ag deficient strains of F. tularensis Types A and B produced 2-5-fold more well-developed biofilms than their more virulent parent strains, yet are attenuated in the highly susceptible mouse model, and compromised in intracellular growth18,25,40. F. tularensis has the ability to phase vary from smooth (blue colonies, O-Ag positive) to rough (grey colonies, O-Ag negative) colony phenotypes41, and one potential benefit to this phase variation would be to increase viability in environmental niches through enhanced biofilm formation if O-Ag is lost. Therefore, these results support the hypothesis that phase-variable loss of surface carbohydrates enhance formation of a biofilm, which may contribute to survival of Francisella species in the environment, rather than as a virulence mechanism in the host. In other bacterial species truncation of the LPS O-Ag also affects biofilm formation. The loss of O-Ag is thought to result in a change in the cell surface charge, altering cell surface hydrophobicity42,43. In Escherichia coli, hld mutants that express a rough LPS demonstrate an increase in biofilm formation and expression of several surface bacterial factors affecting virulence44. Alteration of the cell surface charge has also been shown in Pseudomonas aeruginosa and Bradyrhizobium japonicum when these bacteria lack O-Ag polysaccharide45,46. Differences in biofilm formation by F. novicida have also been attributed to changes in surface hydrophobicity26. However, we were unable to establish any significant differences between the hydrophobicity of F. tularensis LVS with any of its O-Ag mutants, as all LVS-derived strains and the parent were not incorporated into a hydrophobic hexadecane layer (data not shown). F. tularensis has a unique O-Ag consisting of two proximal carbohydrate residues (α-D-GalNAcAN–α-D-GalNAcAN) and two distal residues (β-D-Qui4NFm and β-D-QuiNAc), while the F. novicida O-Ag shares the same two proximal residues, but has different distal glycoses (α-D-GalNAcAN and β-DQui2NAc4NAc)17. The difference in O-Ag surface glycoses between the two species could explain, in part, the differences in biofilm formation between the parent strains of F. novicida and F. tularensis. While F. novicida formed a uniform film across the coverslip surface (data not shown), F. tularensis attached in non-uniform clumps. Removal of the dideoxy O-Ag sugars in F. tularensis (WbtIG191V) allowed for more uniform attachment across the surface, although the biofilm coverage on the coverslip was still not comparable to that of F. novicida. However, most O-Ag mutants of F. novicida did not produce more biofilm, as Types A and B O-Ag mutants did, but enhancement or loss of CLC by F. novicida grown in CDMB did reduce or enhance biofilm formation, respectively, as it did for Type A and B strains.
A double mutant lacking both O-Ag and CLC20 formed 7-fold or more biofilm than either the O-Ag mutant or CLC mutant, further supporting an inverse relationship between cell surface-associated glycosylation and biofilm formation. Furthermore, adherence is the first step in biofilm formation, and double mutant WbtIG191VΔ1423-22 lacking O-Ag and CLC adhered significantly better to surfaces than cells whose surfaces were glycosylated. Therefore, the presence of glycosylated CLC, O-Ag LPS, and an O-Ag capsule may sterically interfere with protein adhesins47–49, thereby reducing adherence and initiation or delay of biofilm formation.
In addition to the O-Ag and CLC, the growth medium also affected biofilm development. Zarrella et al.50 reported that F. tularensis undergoes host-adaptation that includes changes in multiple surface polysaccharides, and that these changes can be reproduced by growing F. tularensis in some, but not all, growth media. Zarrella et al. demonstrated that F. tularensis grown in brain hear infusion broth (BHIB) expresses the same surface polysaccharides when growing in the host, whereas the bacteria did not express these components after growth in MHB. We have determined that F. tularensis produces the HMS material when grown in CDMB, as well as when it is grown in BHIB, and that this component is associated with the CLC20,50. Biofilm formation was also affected when the bacteria were grown in CDMB versus MHB, and an increase in biofilm formation occurred in all surface antigen mutants when grown in CDMB. A significant increase in biofilm formation in WbtIG191VΔ1423-22 occurred only when the bacteria were grown in BHIB and CDMB. Overall less biofilm was present when the bacteria were grown in MHB, and the differences observed in the CLC-deficient mutant grown in BHIB were not found in the double mutant WbtIG191VΔ1423-22 grown in MHB. Therefore, there are likely factor(s) available in BHIB or CDMB that stimulate biofilm formation, possibly by upregulating adherence factors. Of interest was that growth of F. novicida in CDMB did not enhance biofilm formation, but growth in BHIB and TSB did. These differences on factors that influence biofilm formation between F. novicida and Types A and B strains may be related to genetic regulators that are governed by environmental adaptations required by the different species.
Mutations in the only two polysaccharide loci thus far identified in Francisella positively enhanced biofilm production, questioning what the composition is of the EPS forming the biofilm matrix. The F. tularensis biofilm is susceptible to enzymes that degrade DNA, proteins, and/or polysaccharide, and therefore is likely to contain each of these components at some stage in development. Cywes-Bentley et al. have reported that a monoclonal antibody to β-(1,6)-linked poly-N-acetyl-D-glucosamine (PNAG), which is a common EPS in the biofilms of many bacterial species, reacts with F. tularensis51. However, our attempts to detect PNAG in the F. tularensis biofilm using a fluorescent-tagged wheat germ agglutinin lectin (WGA) (specific to PNAG), or antibodies to PNAG were unsuccessful. Extracts from F. tularensis biofilm as well as CLC were also incubated with fluorescein-tagged WGA, but no fluorescence was detected (data not shown). Phenol extraction of the EPS from the most prominent biofilm-forming strain, WbtIG191VΔ1423-22, and subsequent compositional and linkage analysis indicated the EPS consisted predominately of end-linked and 1,4-linked glucose, suggesting that the EPS was similar to amylose. Glycogen expression and regulation in E. coli is controlled by the genes within the operon glgBXCAP52. Proteins with low identity or similarity to GLGBXCAP were identified in F. tularensis. In addition, F. tularensis gene FTT_1629c, which annotated as a hypothetical gene, had 25.8% identity to the Streptococcus pneumoniae tts gene, which is solely responsible for producing the type 37 capsule that is comprised of (1,3)-Β-glucan53. FTT_1629c also includes a DXD motif (glycose binding domain), which indicates it is likely a glycosyltransferase. Alternatively, there may be more than one F. tularensis EPS, and the predominant EPS of the WbtIG191VΔ1423-22 mutant may differ from that of Type A strains. One of the glycosyl transferases deleted in WbtIG191VΔ1423-22, FTL_1423, has some identity to the Y. pestis hmr gene, which is similar to icaA, an N-glycosyltransferase used for PNAG production54.
In conclusion, lack of the surface antigens CLC, LPS O-Ag, and O-Ag capsule resulted in poorer biofilm formation by F. tularensis Types A and B in vitro. Strains lacking one or all surface carbohydrate antigens formed a more robust biofilm than the parent strain, and such loss could occur naturally. Biofilm formation was also affected by incubation time and the specific growth medium. For the first time, the extended biofilm development cycle of F. tularensis Types A and B strains has been described, a previously unreported glucan polysaccharide that makes up the biofilm matrix has been identified, and cell-associated surface polysaccharides of F. tularensis Types A and B were shown to interfere with biofilm development, which is not required for bacterial virulence.
Methods
Bacterial strains and growth conditions
The strains used in this study are described in Table 1. Generation of mutants WbtIG191V and LVSΔ1423-22 and their complementation to obtain WbtIG191V+ and LVSΔ1423-22+ has been previously reported18,20. Strains were cultured on either CDMB with 1.5% glucose, BHIB, TSB, or MHB (BD, Franklin Lakes, NJ) (all supplemented with 0.1% cysteine) at 37 °C in 6% CO234. To enhance CLC expression the bacteria were subcultured in CDMB for 10 subsequent passages, then grown on CDMB agar at 32 °C in 6% CO2 for 5 days, as previously described20. All subcultured strains, parents, and mutants were stored at −80 °C in sterile 10% skim milk until needed and were not subcultured unintentionally. Recombinant strains were grown in CDMB with 15 μg/ml kanamycin (Kan). All experiments with LVS, F. novicida, and mutants were carried out in biosafety level (BSL)-2 facilities in an approved biosafety cabinet. All Type A strains were grown in BSL-3 facilities, and removed from the facility only when loss of viability was confirmed by subculture and 5 days incubation.
Static biofilm formation
A modified version of the method by Stepanovic et al.55 was used to grow biofilms in round-bottomed, poly-L-lysine-coated, polystyrene microtiter plates (Nunc, Rochester, NY). Briefly, overnight cultures of F. tularensis strains were grown at 37 °C and stationary phase bacteria were diluted 1:4 in either CDMB or MHB. The samples were incubated for 5, 10, 15, 20, or 25 days at 37 °C, and the medium replaced every 5 days. The attached biofilms were quantified by solubilising the CV with 95% ethanol and determining the absorbance at 560 nm. For plates with sample absorbance values greater than 4.0, a 1:3-1:10 dilution was made of all samples on the plate. All results involving the CV biofilm assay with mutant strains were calculated and compared to the parent strain and blank control in the same 96-well plate to eliminate technical bias.
Attachment assay
Triplicate cultures of all strains were grown with shaking (180 rpm) overnight at 37 °C to stationary phase. Cultures were transferred (200 μl) to 96-well polystyrene plates that had been coated with poly-L-lysine (Lab-Tek II/Thermo Fisher Scientific, Waltham, MA) and allowed to adhere for 1, 2, or 4 hours statically at 37 °C. The medium was removed and the wells stained with CV as described above. All assays were repeated in triplicate.
Confocal laser scanning microscopy and image analysis
Biofilms were statically grown for 5, 10, or 15 days on 8-well chambered glass coverslips coated with poly-L-lysine (Lab-Tek II/Thermo Fisher Scientific). Medium and non-adherent cells were replaced with fresh medium every 5 days (flow cells were not used due to restrictions in the BSL-3 laboratory). At each time point, the medium was removed from the well, the coverslip was washed with sterile saline twice, and 40 μM/ml Syto9 in sterile saline from the BacLight stain (Molecular Probes, Inc., Eugene, OR) or 50 μg/ml Texas Red-labeled ConA in saline was added. After 15 min incubation at room temperature in the dark, the coverslip wells were washed in saline and visualised by CLSM using a ZEISS LSM 510 laser-scanning microscope, mounted on an inverted Axio Observer Z1 (Carl Zeiss, Jena, Germany). Laser scanning image browser software was used for analysis of the biofilm images, including collection of z-stacks, three-dimensional reconstruction, and data analysis. Images were acquired from the center of the coverslip in 1.1-μm sections throughout the biofilm depth, with the number of sections varying depending on the thickness of the biofilm.
Electron microscopy
Each strain was grown as a biofilm on 10-mm diameter silicone disks in CDMB for 5 days under static conditions at 32 °C for examination by scanning electron microscopy (SEM). After removal of the medium, biofilms were fixed in 2% glutaraldehyde, followed by 1% osmium tetraoxide for 30 minutes each. The samples were dehydrated in a sequential series of ethanol solutions (increasing from 10% to 100%). The disks were critical point-dried, sputter-coated with carbon, and examined using a Zeiss EVO40 scanning electron microscope.
Exopolysaccharide isolation and analysis
EPS was isolated from LVS mutant WbtIG191VΔ1423-22, which lacked both O-Ag and CLC glycosylation, and formed the most robust biofilm. The strain was grown statically in 200-ml volumes of BHIB or CDMB supplemented with 1% glucose, in large tissue culture flasks coated with poly-L-lysine at 32 °C or 37 °C for 10-15 days. The supernatant was carefully removed, the sediment extracted with 45% aqueous phenol, and the EPS purified as previously described56.
Glycose composition analysis was performed by GC-MS of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis, as described57. Briefly, 300 μg of sample was added to a tube containing 20 µg of inositol as an internal standard. Methyl glycosides were prepared from the dry sample following methanolysis in 1 M HCl in methanol at 80 °C (16 hours), followed by re-N-acetylation with pyridine and acetic anhydride in methanol (for detection of amino sugars). The sample was then per-O-trimethylsilylated by treatment with Tri-Sil (Pierce) at 80 °C for 30 min. Gas chromatography-mass spectrometry (GC/MS) analysis of the TMS methyl glycosides was performed on an Agilent 7890 A GC interfaced to a 5975 C mass selective detector using Agilent DB-1 fused silica capillary column (30 m × 0.25 mm ID). For glycose linkage analysis, approximately 1 mg of the sample was suspended in 200 μl of dimethyl sulfoxide, and stirred for 1 day. The sample was permethylated as described by Ciucanu and Kerek58 with the base prepared according to Anumula and Taylor59. Briefly, the sample was subjected to the NaOH base for 15 min, followed by addition of methyl iodide and left for 45 minutes. This procedure was repeated one more time to ensure complete methylation of the polymer. The permethylated material was hydrolysed using 2 M trifluoroacetic acid (2 h in sealed tube at 121 °C), reduced with NaBD4, and acetylated using acetic anhydride/TFA. The resulting partially methylated alditol acetates were analysed on the same equipment described above, except separation was performed on a 30 m Supelco SP-2331 bonded phase fused silica capillary column, as described by York et al.60. Identification of genes that may be involved in expression and regulation of glycogen synthesis was determined by BLASTP61.
Enzymatic detachment assays
Biofilms were grown in CDMB for 10 days in 96-well plates as described above. Wells were washed once with PBS and the biofilm treated with Proteinase K (100 μg/ml in 10 mM Tris-HCl pH 7.5, Sigma-Aldrich), 40 mM sodium m-periodate, or DNase I (100 μg/ml in 150 mM NaCl/1 mM CaCl2) (Sigma-Aldrich). Enzymes were added to designated wells in a volume of 200 μl and incubated at room temperature for 2 hours. Enzyme solutions were removed and the wells washed once with PBS and then stained with CV, as described above. All assays were performed in triplicate unless otherwise stated.
Analysis of large molecular size components associated with CLC
F. tularensis strains were grown with shaking at 37 °C overnight in either CDMB or MHB and 500 μl was used to inoculate 10 ml of new MHB or CDMB in 50-ml conical tubes (Corning, Oneonta, NY). The tubes were held stationary at 37 °C for 10 days to allow bacteria to adhere and form biofilms. The medium above the biofilm was then removed (broth supernatant) and non-adherent, planktonic cells were sedimented by centrifugation at 6,000 × g for 15 min. Proteins in the supernatant were precipitated by addition of 3 volumes of 95% ethanol, sedimented by centrifugation (as above), resuspended in a small volume of distilled water, and the protein concentration determined by BCA assay (Thermo Fisher Scientific). Surface components of the biofilm and planktonic cells were extracted by suspending each bacterial phenotype in 1–5 ml of 1 M urea and holding the bacteria without shaking at room temperature for 15 min. The cells were removed by centrifugation (6,000 × g for 15 minutes) and the urea extracts stored at −20 °C until analysed. Solubilisation buffer (2×) was added to an equal volume of the urea extract and 15 μl was loaded onto 4–12% NuPAGE gels (Invitrogen/Thermo Fisher Scientific). Following electrophoresis the gels were stained for proteins with silver stain (Pierce, Rockford, IL), for carbohydrates with Pro-Q Emerald 300 (Molecular Probes, Grand Island, NY), or for CLC with StainsAll/silver stain (Pierce/Sigma-Aldrich)20.
Statistical analyses
Statistical analyses were performed using Microsoft Excel software (Redmond, WA) and Prism GraphPad 6 software (San Diego, CA). Data obtained with LVS strains were analysed using the student t-test and expressed as the mean ± standard deviation. Results with a p value of less than 0.05 were considered statistically significant. One-way ANOVA was used to evaluate significant differences in CV absorbance of F. novicida and the F. novicida O-Ag mutants. Dunnet’s multiple comparison test was performed after completion of the one-way ANOVA to determine significance between the CV absorbance of F. novicida with the absorbance of a specific F. novicida O-Ag mutant. Adjusted p-values were determined for each comparison.
Supplementary information
Acknowledgements
We thank Kristi DeCourcy at the Fralin Life Science Institute for her technical expertise performing and training for the CLSM. We also thank Parastoo Azadi and Ian Black at The Complex Carbohydrate Research Center, University of Georgia, Athens, GA, for compositional analysis of the EPS. This work was supported, in part, by funds from the Virginia-Maryland College of Veterinary Medicine and foundation funds from the Tyler J. and Frances F. Young endowment. The EPS compositional work was supported by grant DE-FG02-93ER20097 to Parastoo Azadi from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, U.S. Department of Energy.
Author Contributions
A.E.C. and T.J.I. conceived and designed the experiments. A.E.C., K.C.F.-C. and A.B.B. carried out the experimental work. A.E.C. and T.J.I. drafted the manuscript. All authors analysed the data, and reviewed and revised the manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available in the U.S. National Library of Medicine National Center for Biotechnology Information repository (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). Other data generated or analysed during this study are included in this published article (and its Supplementary Information files), or available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48697-x.
References
- 1.McLendon MK, Apicella MA, Allen LA. Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Ann. Rev. Microbiol. 2006;60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morner T. The ecology of tularaemia. Rev. Scientif. Techniq. 1992;11:1123–1130. [PubMed] [Google Scholar]
- 3.Nigrovic LE, Wingerter SL. Tularemia. Infect. Dis. Clinics North Amer. 2008;22:489–504. doi: 10.1016/j.idc.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Pedati, C. et al. Vol. 64 Morbid. Mortal. Weekly Report 1317–1318 (Centers for Disease Control and Prevention, 2015).
- 5.Hestvik G, et al. The status of tularemia in Europe in a one-health context: a review. Epidemiol. Infect. 2015;143:2137–2160. doi: 10.1017/S0950268814002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjostedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. New York Acad. Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 7.Abd H, Johansson T, Golovliov I, Sandstrom G, Forsman M. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl.Environmen. Microbiol. 2003;69:600–606. doi: 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eliasson H, Back E. Tularaemia in an emergent area in Sweden: an analysis of 234 cases in five years. Scand. J. Infect. Dis. 2007;39:880–889. doi: 10.1080/00365540701402970. [DOI] [PubMed] [Google Scholar]
- 9.Petersen JM, et al. Methods for enhanced culture recovery of Francisella tularensis. Appl. Environmen. Microbiol. 2004;70:3733–3735. doi: 10.1128/AEM.70.6.3733-3735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial Biofilms. Ann. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 11.Margolis JJ, et al. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl. Environ. Microbiol. 2010;76:596–608. doi: 10.1128/AEM.02037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Hoek ML. Biofilms: an advancement in our understanding of Francisella species. Virulence. 2013;4:833–846. doi: 10.4161/viru.27023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Europ. J. Biochem. / FEBS. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- 14.Vinogradov EV, et al. Structure of the O-antigen of Francisella tularensis strain 15. Carbohydr. Res. 1991;214:289–297. doi: 10.1016/0008-6215(91)80036-M. [DOI] [PubMed] [Google Scholar]
- 15.Thomas RM, et al. The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect. Immun. 2007;75:371–378. doi: 10.1128/IAI.01241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raynaud C, et al. Role of the wbt locus of Francisella tularensis in lipopolysaccharide O-antigen biogenesis and pathogenicity. Infect. Immun. 2007;75:536–541. doi: 10.1128/IAI.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn JS, Ernst RK. The structure and function of Francisella lipopolysaccharide. Ann. New York Acad. Sci. 2007;1105:202–218. doi: 10.1196/annals.1409.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, et al. Attenuation and protective efficacy of an O-antigen-deficient mutant of Francisella tularensis LVS. Microbiol. 2007;153:3141–3153. doi: 10.1099/mic.0.2007/006460-0. [DOI] [PubMed] [Google Scholar]
- 19.Apicella MA, et al. Identification, characterization and immunogenicity of an O-antigen capsular polysaccharide of Francisella tularensis. PloS One. 2010;5:e11060. doi: 10.1371/journal.pone.0011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandara AB, et al. Isolation and mutagenesis of a capsule-like complex (CLC) from Francisella tularensis, and contribution of the CLC to F. tularensis virulence in mice. PloS One. 2011;6:e19003. doi: 10.1371/journal.pone.0019003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freudenberger Catanzaro KC, Champion AE, Mohapatra N, Cecere T, Inzana TJ. Glycosylation of a capsule-like complex (CLC) by Francisella novicida is required for virulence and partial protective immunity in mice. Front. Microbiol. 2017;8:935. doi: 10.3389/fmicb.2017.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durham-Colleran MW, Verhoeven AB, van Hoek ML. Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb. Ecol. 2010;59:457–465. doi: 10.1007/s00248-009-9586-9. [DOI] [PubMed] [Google Scholar]
- 23.Zogaj X, Wyatt GC, Klose KE. Cyclic di-GMP stimulates biofilm formation and inhibits virulence of Francisella novicida. Infect. Immun. 2012;80:4239–4247. doi: 10.1128/IAI.00702-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomonsson E, et al. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect. Immun. 2009;77:3424–3431. doi: 10.1128/IAI.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modise T, et al. Genomic comparison between a virulent type A1 strain of Francisella tularensis and its attenuated O-antigen mutant. J. Bacteriol. 2012;194:2775–2776. doi: 10.1128/JB.00152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung MC, Dean S, Marakasova ES, Nwabueze AO, van Hoek ML. Chitinases are negative regulators of Francisella novicida biofilms. PLoS One. 2014;9:e93119. doi: 10.1371/journal.pone.0093119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhoeven AB, Durham-Colleran MW, Pierson T, Boswell WT, Van Hoek ML. Francisella philomiragia biofilm formation and interaction with the aquatic protist Acanthamoeba castellanii. Biolog. Bull. 2010;219:178–188. doi: 10.1086/BBLv219n2p178. [DOI] [PubMed] [Google Scholar]
- 28.El-Etr SH, et al. Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl. Environ. Microbiol. 2009;75:7488–7500. doi: 10.1128/AEM.01829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopla CE. The ecology of tularemia. Adv. Vet. Sci. Comp. Med. 1974;18:25–53. [PubMed] [Google Scholar]
- 30.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann. New York Acad. Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 31.Farlow J, et al. Francisella tularensis in the United States. Emerg. Infect. Dis. 2005;11:1835–1841. doi: 10.3201/eid1112.050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarrett CO, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 33.Hopla CE, Durden LA, Keirans JE. Ectoparasites and classification. Rev. Scientif. Techniq. 1994;13:985–1017. doi: 10.20506/rst.13.4.815. [DOI] [PubMed] [Google Scholar]
- 34.Cherwonogrodzky JW, Knodel MH, Spence MR. Increased encapsulation and virulence of Francisella tularensis live vaccine strain (LVS) by subculturing on synthetic medium. Vaccine. 1994;12:773–775. doi: 10.1016/0264-410X(94)90284-4. [DOI] [PubMed] [Google Scholar]
- 35.Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 2006;188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petruzzi, B. et al. Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida serogroup A. MBio8 (2017). [DOI] [PMC free article] [PubMed]
- 37.Thomas RM, et al. Glycosylation of DsbA in Francisella tularensis subsp. tularensis. J. Bacteriol. 2011;193:5498–5509. doi: 10.1128/JB.00438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lees-Miller RG, et al. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 2013;89:816–830. doi: 10.1111/mmi.12300. [DOI] [PubMed] [Google Scholar]
- 39.Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiol. 2011;21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen JA, et al. Francisella tularensis Schu S4 lipopolysaccharide core sugar and O-antigen mutants are attenuated in a mouse model of tularemia. Infect. Immun. 2014;82:1523–1539. doi: 10.1128/IAI.01640-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soni S, et al. Francisella tularensis blue-gray phase variation involves structural modifications of lipopolysaccharide o-antigen, core and lipid a and affects intramacrophage survival and vaccine efficacy. Front. Microbiol. 2010;1:129. doi: 10.3389/fmicb.2010.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park KM, So JS. Altered cell surface hydrophobicity of lipopolysaccharide-deficient mutant of Bradyrhizobium japonicum. J. Microbiol. Methods. 2000;41:219–226. doi: 10.1016/S0167-7012(00)00155-X. [DOI] [PubMed] [Google Scholar]
- 43.Kempf M, et al. Cell surface properties of two differently virulent strains of Acinetobacter baumannii isolated from a patient. Can. J. Microbiol. 2012;58:311–317. doi: 10.1139/w11-131. [DOI] [PubMed] [Google Scholar]
- 44.Nakao R, Ramstedt M, Wai SN, Uhlin BE. Enhanced biofilm formation by Escherichia coli LPS mutants defective in Hep biosynthesis. PloS One. 2012;7:e51241. doi: 10.1371/journal.pone.0051241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee YW, et al. Lack of O-polysaccharide enhances biofilm formation by Bradyrhizobium japonicum. Lett. Appl. Microbiol. 2010;50:452–456. doi: 10.1111/j.1472-765X.2010.02813.x. [DOI] [PubMed] [Google Scholar]
- 46.Murphy K, et al. Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2014;196:1306–1317. doi: 10.1128/JB.01463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakraborty S, et al. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect. Immun. 2008;76:2852–2861. doi: 10.1128/IAI.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall JD, Craven RR, Fuller JR, Pickles RJ, Kawula TH. Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect. Immun. 2007;75:1034–1039. doi: 10.1128/IAI.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melillo A, et al. Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 2006;263:102–108. doi: 10.1111/j.1574-6968.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- 50.Zarrella TM, et al. Host-adaptation of Francisella tularensis alters the bacterium’s surface-carbohydrates to hinder effectors of innate and adaptive immunity. PloS One. 2011;6:e22335. doi: 10.1371/journal.pone.0022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cywes-Bentley C, et al. Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc. Natl. Acad. Sci. USA. 2013;110:E2209–2218. doi: 10.1073/pnas.1303573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montero M, et al. Escherichia coli glycogen genes are organized in a single glgBXCAP transcriptional unit possessing an alternative suboperonic promoter within glgC that directs glgAP expression. Biochem. J. 2011;433:107–117. doi: 10.1042/BJ20101186. [DOI] [PubMed] [Google Scholar]
- 53.Llull D, Munoz R, Lopez R, Garcia E. A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide. Type 37 pneumococci are natural, genetically binary strains. J. Exp. Med. 1999;190:241–251. doi: 10.1084/jem.190.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 55.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods. 2000;40:175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 56.Sandal I, et al. Identification, structure, and characterization of an exopolysaccharide produced by Histophilus somni during biofilm formation. BMC Microbiol. 2011;11:186. doi: 10.1186/1471-2180-11-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merkle RK, Poppe I. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- 58.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984;131:209–217. doi: 10.1016/0008-6215(84)85242-8. [DOI] [Google Scholar]
- 59.Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 1992;203:101–108. doi: 10.1016/0003-2697(92)90048-C. [DOI] [PubMed] [Google Scholar]
- 60.York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1986;118:3–40. doi: 10.1016/0076-6879(86)18062-1. [DOI] [Google Scholar]
- 61.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inzana TJ, et al. Characterization of a wildtype strain of Francisella tularensis isolated from a cat. J. Vet. Diagn. Invest. 2004;16:374–381. doi: 10.1177/104063870401600502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the U.S. National Library of Medicine National Center for Biotechnology Information repository (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). Other data generated or analysed during this study are included in this published article (and its Supplementary Information files), or available from the corresponding author on reasonable request.