Abstract
The lymphatic vasculature mediates essential physiological functions including fluid homeostasis, lipid and hormone transport, and immune cell trafficking. Recent studies have suggested that promoting lymphangiogenesis enhances cardiac repair following injury, but it is unknown whether lymphangiogenesis is required for cardiac regeneration. Here, we describe the anatomical distribution, regulation, and function of the cardiac lymphatic network in a highly regenerative zebrafish model system using transgenic reporter lines and loss-of-function approaches. We show that zebrafish lacking functional vegfc and vegfd signaling are devoid of a cardiac lymphatic network and display cardiac hypertrophy in the absence of injury, suggesting a role for these vessels in cardiac tissue homeostasis. Using two different cardiac injury models, we report a robust lymphangiogenic response following cryoinjury, but not following apical resection injury. Although the majority of mutants lacking functional vegfc and vegfd signaling were able to mount a full regenerative response even in the complete absence of a cardiac lymphatic vasculature, cardiac regeneration was severely impaired in a subset of mutants, which was associated with heightened pro-inflammatory cytokine signaling. These findings reveal a context-dependent requirement for the lymphatic vasculature during cardiac growth and regeneration.
Subject terms: Angiogenesis, Cardiac regeneration
Introduction
In adults, lymphangiogenesis frequently occurs at sites of tissue injury and inflammation.1–4 This rapid lymphangiogenic response to injury is thought to contribute to the resolution of edema and clearance of immune cells at the wound site.1,5–7 Promoting lymphangiogenesis has therefore emerged as an attractive therapeutic strategy to facilitate tissue repair.8–11 However, whether the lymphangiogenic response influences endogenous tissue repair in highly regenerative organisms such as zebrafish is currently unclear. Identifying the cellular mechanisms and physiological processes that govern cardiac regeneration has the potential to inform new therapeutic approaches for ischemic heart disease and heart failure.
The heart relies on lymphatic vessels to maintain fluid balance, which must be tightly controlled to ensure normal cardiac output. The heart comprises an extensive lymphatic vascular plexus, which was originally thought to be entirely derived from the venous circuitry during development.12,13 However, a recent study has suggested that cardiac lymphatics may have a developmentally heterogeneous origin, whereby formation of part of the cardiac lymphatic network occurs independent of sprouting from the veins.11 The lymphatic vasculature has also emerged as a potential therapeutic target for ischemic heart disease with two independent studies reporting beneficial effects of recombinant vascular endothelial growth factor type C (VEGFC) delivery on cardiac function following experimental myocardial infarction (MI) in mice and rats.10,11 While these studies suggest that VEGFC delivery following MI is cardioprotective, the underlying cellular and molecular mechanisms are still poorly understood. Given that VEGFC has immunomodulatory functions that extend beyond its role on lymphatic endothelial cells (LECs),14 it is currently unclear whether the cardioprotective actions of VEGFC treatment following MI are due to promotion of VEGFC-dependent lymphangiogenesis.
VEGF family members are key mediators of blood vessel growth (angiogenesis) and lymphatic vessel growth (lymphangiogenesis). The VEGF family includes VEGFA, VEGFB, VEGFC, VEGFD, VEGFE, PLGF-1, and PLGF-2. VEFC and VEGFD are the most potent regulators of lymphangiogenesis. In mice and zebrafish, lymphatic vessel network assembly is dependent on VEGFR3 signaling through its ligand VEGFC.6,15–17 Heterozygous deletion of Vegfr3 in mice causes severe lymphatic defects18 and mice lacking Vegfc fail to develop a lymphatic vasculature.16 In humans, patients harboring loss-of-function mutations in either VEGFC or VEGFR3 develop lymphedema.19 Moreover, application of recombinant VEGFC provides a powerful stimulus for lymphangiogenesis in adults, which could be exploited in a number of therapeutic contexts including management of lymphedema and tissue repair.20 The other ligand for VEGFR3 is VEGFD, which is not required for development of the lymphatic vasculature in mice,21 but can compensate for loss of VEGFC in some contexts.22 VEGFD also functions at postnatal stages during the maturation of the lymphatic vascular network in the lung and dermal tissues.23 In zebrafish, vegfd is required for the development of facial lymphatics24,25 and is expressed throughout the embryonic trunk.26 Furthermore, genetic disruption of vegfd in a hypomorphic vegfc mutant background gives rise to severe lymphatic vascular defects in the trunk suggesting a redundant role for these two growth factors during lymphangiogenesis.25 In addition, vegfd is also required for artery hyperbranching during primary angiogenesis in zebrafish.25 Further, the genetic interaction of this growth factor with the transcription factor SOX18 has been shown to control artery caliber and angiogenic remodeling during mouse embryogenesis.26 Although VEGFA primarily drives angiogenesis via VEGFR2-dependent signaling, there is some evidence that VEGFA can also regulate lymphangiogenesis following inflammation.27,28 Due to their central roles as key mediators of blood vessel growth, VEGF family members are being pursued as therapeutic targets for a variety of disorders including cardiovascular disease.
To investigate the potential function of the lymphatic vasculature in cardiac homeostasis and tissue repair, we used vegfc/d loss-of-function in vivo to prevent lymphatic vascular remodeling and determine its impact on cardiac growth and endogenous regeneration following injury in zebrafish. Using a series of lymphatic reporter lines, we describe the major cardiac lymphatic vessels in adult zebrafish and use this system to analyze lymphangiogenesis following cardiac injury. Genetic deletion of vegfc and vegfd established a requirement for these signaling pathways for development of the cardiac lymphatic vasculature and for maintenance of heart size. Using two independent models of cardiac regeneration, we found a robust lymphangiogenic response occurred following cryoinjury, but not following apical resection injury. The regenerative response following cryoinjury was severely impaired in a subset of mutants lacking functional vegfc/d signaling, which was associated with an exacerbated inflammatory response. These findings reveal a surprising context-dependent requirement for the lymphatic vasculature during physiological cardiac growth and regeneration.
Results
Anatomical organization of the cardiac lymphatic network in adult zebrafish
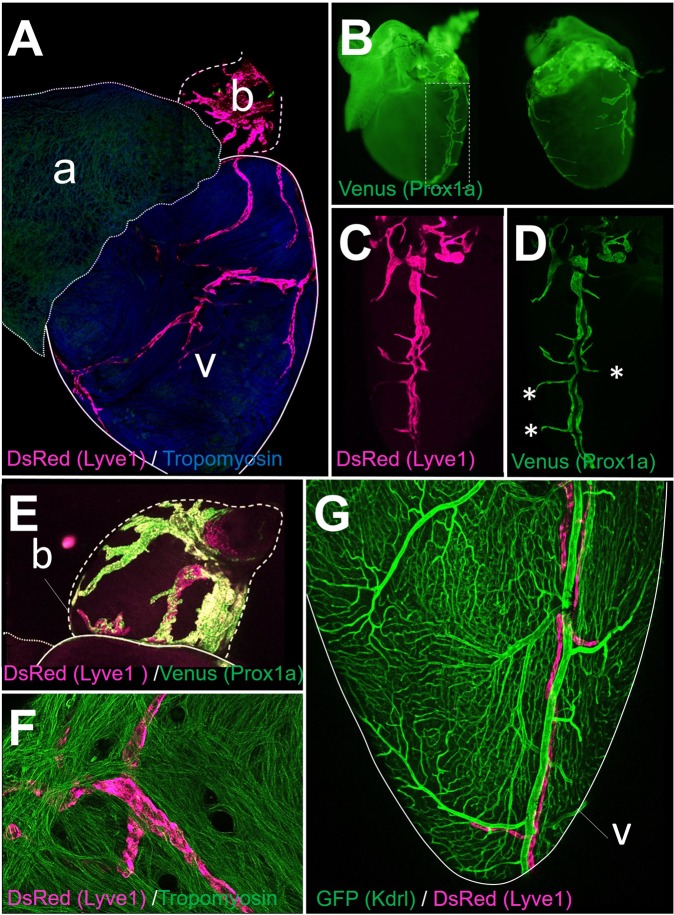
We firstly characterized the anatomical organization of the cardiac lymphatic vasculature in size-matched adult zebrafish using well established lymphatic reporter lines: Tg(−5.2lyve1b:DsRed)nz101 29 and Tg(prox1a:KalTA4)uq3bh.30 Consistent with a LECs molecular identity, we observed a vessel network co-expressing both Prox1a and Lyve1 reporters in the adult zebrafish heart (Fig. 1). A highly-branched network of large lymphatic vessels was present on the bulbus arteriosus, while a less complex network of lymphatics harboring short ramifications was present on the cardiac ventricle (Fig. 1a–e). Ventricular lymphatics were primarily detected at the epicardial surface of the ventricle (Fig. 1a–c), but no vessels were detected on the atrium (Fig. 1a). To determine the proximity of the lymphatics and blood vessels of the heart, we used the Tg(kdrl:EGFP)s834:Tg(−5.2lyve1b:DsRed)nz101 reporter line,29,31 which allows for visualization of both blood vessels and lymphatics. As expected, the cardiac lymphatic network was clearly distinguishable from the coronary vasculature based on the visualization of this dual transgenic reporter line (Fig. 1g). The main lymphatic vessel in the cardiac ventricle ran parallel to the major coronary arteries in close proximity (Fig. 1g), but not all coronary vessels were surrounded by lymphatics (Fig. 1g). Together, these findings demonstrate that, in the absence of injury, the adult zebrafish heart contains a dense network of lymphatic vessels on the bulbous arteriosus and a prominent lymphatic vessel on the epicardial surface of the ventricle with few lymphatics penetrating deeper into the ventricular myocardium and no lymphatics within the atrium.
Fig. 1.
Anatomical organization of cardiac lymphatic vessels in adult zebrafish. a DsRed and Tropomyosin staining was performed on Tg(−5.2lyve1b:DsRed)nz101 adult zebrafish hearts. A dense lymphatic plexus was present in the bulbous (b) and a prominent lymphatic vascular network was detected on the ventricle (v). No lymphatics were detected on the atrium (a). b DsRed and Venus staining was performed on Tg(−5.2lyve1b:DsRed)nz101:Tg(prox1a:KalTA4)uq3bh adult zebrafish hearts. Venus expression (Prox1a reporter) shows the same anatomical distribution of lymphatic vessels in the adult zebrafish heart compared with the Lyve1 reporter (a). High magnification images of the major lymphatic vessel and associated ramifications in the heart of Prox1 (c) and Lyve1 (d) reporter fish. There is a strong overlap in Lyve1 and Prox1 reporter expression, with the exception of a few small ramifications that were positive for Prox1 and negative for Lyve1 (*). e A broad network of large cardiac lymphatic vessels (DsRed and Venus stained) were detected on the bulbus arteriosus (b). f Ventricular lymphatics were predominantly located at the epicardial surface of the ventricle (confocal image taken from epicardial surface of the heart following whole-mount immunostaining for DsRed and Tropomyosin staining on Tg(−5.2lyve1b:DsRed)nz101 adult zebrafish heart). g Co-GFP and DsRed staining of Tg(kdrl:EGFP)s834:Tg(−5.2lyve1b:DsRed)nz101 adult zebrafish hearts. Lymphatic vessels (DsRed stained) are clearly distinct from blood vessels expressing Kdrl (GFP stained)
Impairment of both vegfc and vegfd function prevents the formation of lymphatic vessels but does not block coronary vasculature assembly in the heart
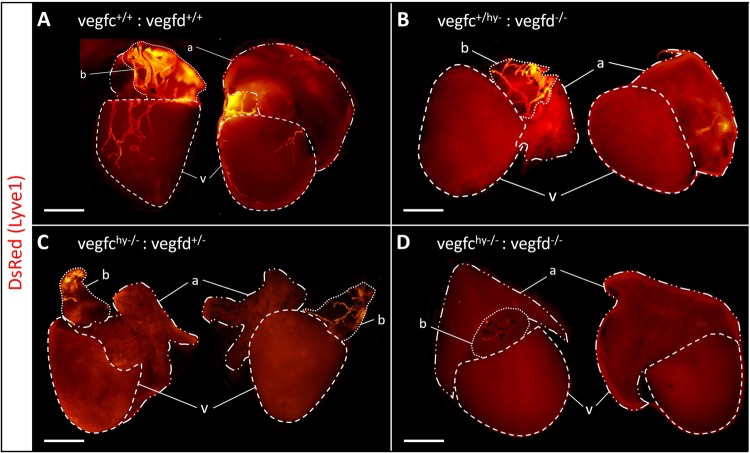
The formation of lymphatic vessels in zebrafish is highly dependent on the Vegfc/d-Vegfr3 signaling axis.24,25,32,33 Due to the early lethality of vegfc null mutants at day 7 postfertilization,33 we used hypomorphic vegfc mutants crossed to vegfd null mutants (vegfduq9bh:vegfchu5055 double mutants hereto referred to as vegfchy−/−;vegfd−/−), which survive to adulthood but have severely impaired lymphangiogenesis.25 In addition, vegfc and vegfd mutants were backcrossed into a Lyve1 transgenic reporter background (Tg(−5.2lyve1b:DsRed)nz101) to enable the visualization of the lymphatic vasculature. As expected, in control vegfc+/+;vegfd+/+:Tg(−5.2lyve1b:DsRed) fish DsRed expression was detected in lymphatic vessels within the bulbus arteriosus and within the cardiac ventricle, but was once again absent within the atrium (Fig. 2a). In vegfc+/hy−:vegfd−/−;Tg(−5.2lyve1b:DsRed) or vegfchy−/−;vegfd+/−:Tg(−5.2lyve1b:DsRed) fish, lymphatics were absent around the ventricle, but were still detected in the bulbous arteriosus (Fig. 2b, c). This indicates that one functional allele of vegfc or vegfd alone is sufficient to establish and maintain the lymphatic vasculature in the bulbus arteriosus, which contains the highest density of lymphatic vessels in the zebrafish heart. In contrast, double mutants lacking both vegfc and vegfd alleles were almost completely devoid of lymphatic vessels on the bulbus arteriosus and did not comprise any lymphatics on the cardiac ventricle (Fig. 2d). There was no overt lethality in fish lacking functional vegfc and vegfd, indicating cardiac function at homeostasis was sufficient for survival without any requirement for a lymphatic vasculature in the heart. Importantly, cardiac blood vessels were detectable in the epicardium and myocardium of both vegfc+/+:vegfd+/+control and vegfchy−/−;vegfd−/−mutant fish (Supplementary Fig. 1) indicating that loss of vegfc/d function did not overtly impact the development of the coronary vasculature in zebrafish.
Fig. 2.
Vegfc and Vegfd are required for maintenance of the adult cardiac lymphatic vasculature. a–d Lymphatic reporter gene expression (Tg(−5.2lyve1b:Dred) in vegfc and vegfd mutant lines. There is no ventricular lymphatic network in vegfchy−/−;vegfd+/− and vegfc+/hy−:vegfd−/− hearts (b, c). Also, there is an absence of lymphatic vascular network in both the ventricle and bulbus arteriosus of vegfchy−/−;vegfd−/− double mutants (d). Scale bar length: 1 mm
Dual loss-of-function of vegfc and vegfd causes cardiac hypertrophy in adult zebrafish
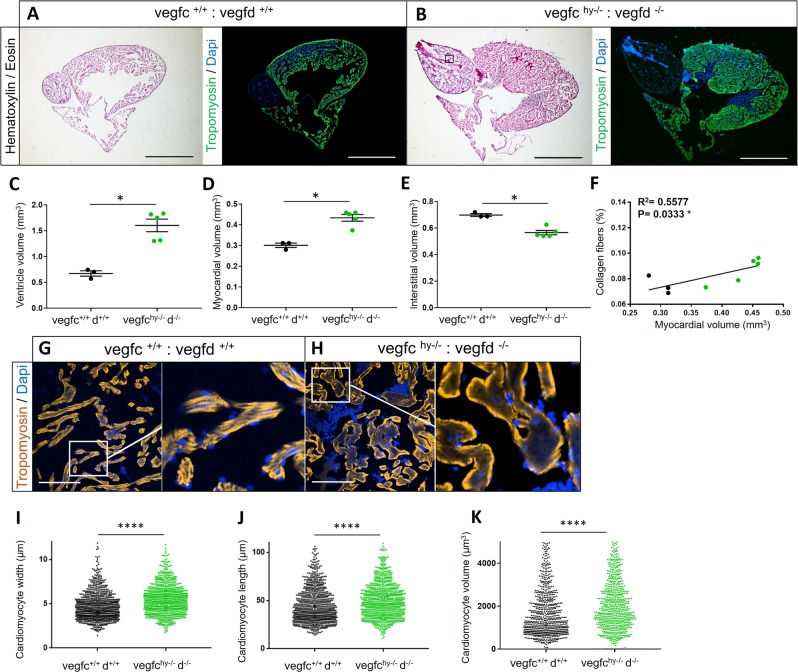
We next examined the hearts of vegfchy−/−;vegfd−/−;Tg(−5.2lyve1b:DsRed) fish to identify any potential phenotypic outcome. Ventricular and myocardial volume were significantly increased by ~2.5-fold and ~1.5-fold, respectively, in vegfchy−/−;vegfd−/−;Tg(−5.2lyve1b:DsRed) mutant hearts compared with controls, suggestive of a cardiac hypertrophic response (Fig. 3a–d). The hypertrophic phenotype was highly penetrant with 100% of the mutants having a lager ventricle volume (Fig. 3c) and increased myocardial volume (Fig. 3d) when compared with the controls. Interstitial volume was significantly reduced in vegfchy−/−;vegfd−/− hearts (Fig. 3e) due to an increased deposition of interstitial nonmuscular, non-collagenous tissue (Fig. 3a, b and Supplementary Fig. 2). There was no statistically significant difference in fibrosis between control and mutant hearts (Supplementary Fig. 2), although there was a highly significant correlation between fibrosis and the hypertrophic phenotype in the absence of injury (Fig. 3f). To determine whether the cardiac hypertrophic phenotype was due to an increase in cardiomyocyte size, we isolated cardiomyocytes by enzymatic digestion of control and mutant ventricles (Supplementary Fig. 2). Cardiomyocyte width, length, and volume were increased (Fig. 3g–k, Supplementary Fig. 2 and Supplementary Data Set 1) in vegfchy−/−;vegfd−/− mutant hearts consistent with hypertrophy of cardiomyocytes. These findings suggest that loss of vegfc/d function, which is required for the formation of lymphatic vessels in the heart, results in a highly penetrant cardiac hypertrophy phenotype.
Fig. 3.
Loss of Vegfc/d signaling causes cardiac hypertrophy. Hematoxylin and Eosin or Tropomyosin and Dapi staining were performed on vegfc+/+:vegfd+/+ (a) and vegfc−/−:vegfdhy−/− hearts (b). Scale bar length: 0.8 mm. Quantification of ventricle volume (c), myocardial volume (d), and interstitial volume (e) in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts. There is marked cardiac hypertrophy with a reduced interstitial volume in vegfchy−/−:vegfd−/− double mutants. n = 3 vegfc+/+:vegfd+/+ control hearts and 4 vegfchy−/−:vegfd−/− mutant hearts. Statistical analysis: unpaired, nonparametric, Mann–Whitney test was used to compare two means. Data are presented as the mean ± standard error of the mean (sem). f Correlation analysis between myocardial volume and collagen content in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts. Black plots represent vegfc+/+:vegfd+/+ samples, green plots represent vegfchy−/−:vegfd−/− samples. There is a highly significant correlation between fibrosis and myocardial volume. g, h Tropomyosin and Dapi staining were performed on vegfc+/+:vegfd+/+ and vegfc−/−:vegfdhy−/− hearts. Scale bar length: 100 μm. i–k Enzymatic digestion was performed on vegfc+/+:vegfd+/+ or vegfchy−/−:vegfd−/− mutant ventricles and isolated cardiomyocytes were measured. The black dots represents the vegfc+/+:vegfd+/+cardiomyocytes width (i), length (j), or volume (k) and the green dots represent vegfchy−/−:vegfd−/− mutant cardiomyocytes width (i), length (j), or volume (k). There are statistically significant differences in cardiomyocyte size between control and mutant hearts. Frequency distribution calculated from a total of ~200 cardiomyocytes per samples (~800 cardiomyocytes per group) comprising n = 4 vegfchy−/−:vegfd−/− ventricles and n = 4 vegfc+/+:vegfd+/+ ventricles, respectively. Statistical analysis: unpaired, nonparametric, Mann–Whitney test was used to compare the two groups. Significant differences are marked by an asterisk ****p < 0.0001
Transcriptomic profiling of vegfchy−/−;vegfd−/− mutant hearts reveals an upregulation of genes involved in cell proliferation and lipid metabolism
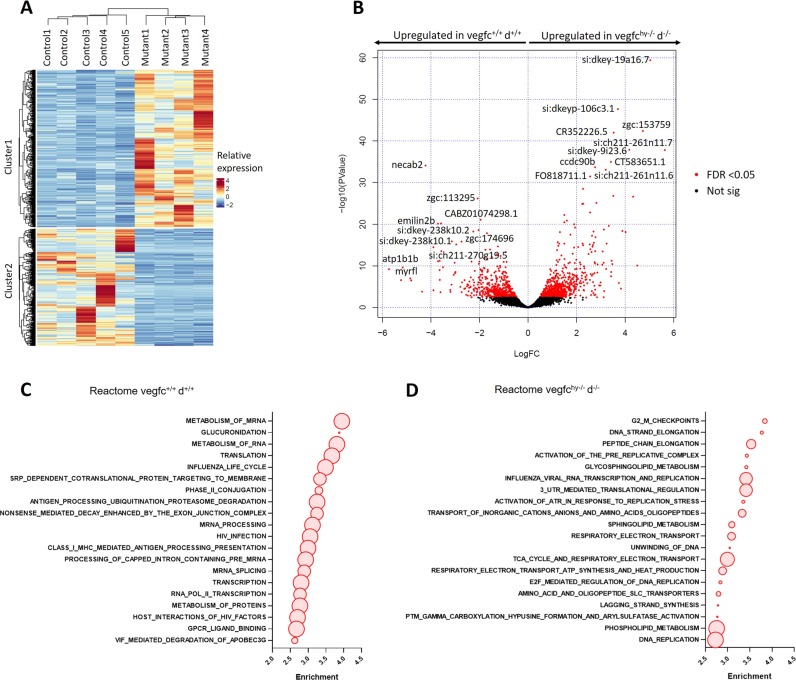
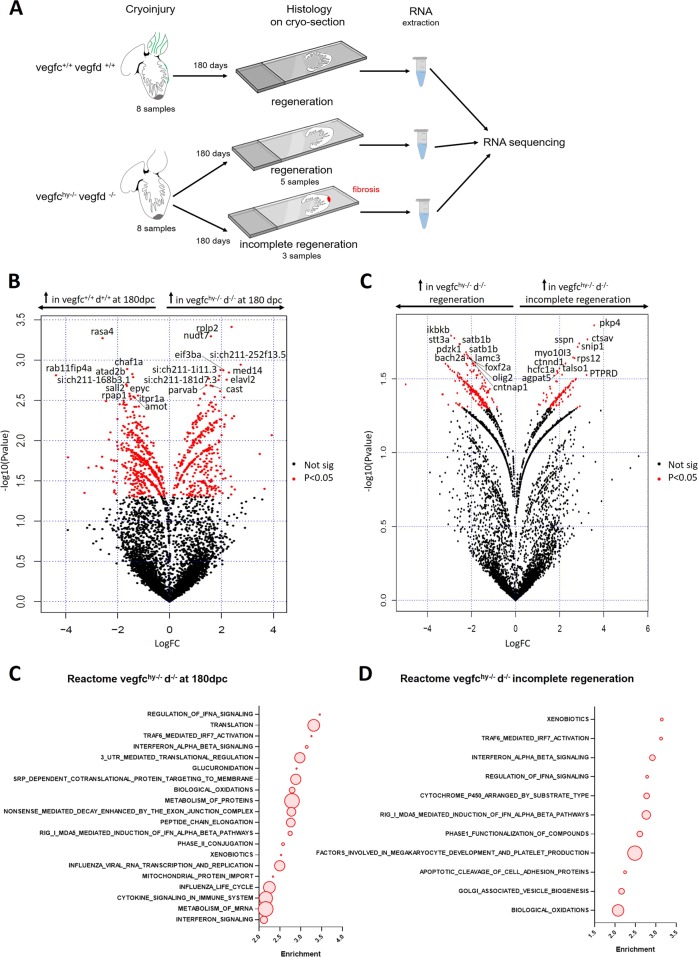
To profile molecular pathways contributing to cardiac hypertrophy in vegfchy−/−;vegfd−/− mutants, we performed RNA-seq on cardiac ventricles from control and mutant zebrafish at 5 months of age (Fig. 4 and Supplementary Data Set 2). Unsupervised hierarchical clustering and principal component analysis (PCA) revealed distinct transcriptional signatures for control and mutant ventricles (Fig. 4a). In total, 616 genes with FDR < 0.05 were upregulated in control hearts and 828 genes were upregulated in mutant hearts (Fig. 4b). Gene set enrichment analysis (GSEA) revealed an enrichment for pathways related to mRNA processing and transcription in control ventricles (Fig. 4c), whereas mutant ventricles were characterized by an upregulation of pathways related to cell proliferation and lipid metabolism, in particular sphingolipid and phospholipid metabolism (Fig. 4d). Interestingly, vegfchy−/−;vegfd−/− mutant ventricles were not characterized by an upregulation of classic stress markers associated with pathological hypertrophy or hypertrophic cardiomyopathy in mammals (e.g., nppa, nppb, myh7 or acta1a)34 (Supplementary Fig. 3). As cardiomyocyte hypertrophy is an uncommon phenotype in zebrafish and there are no established experimental models of cardiac hypertrophy in fish, we sought to determine the extent to which the transcriptional profile of vegfchy−/−;vegfd−/− mutant ventricles overlapped with the known molecular phenotype of established cardiac hypertrophy models in mice. To identify transcriptional networks that were common to the hypertrophic vegfchy−/−;vegfd−/− mutant zebrafish and established cardiac hypertrophy models, we crossed our data set with published data sets for both physiological and pathological cardiac hypertrophy induced by swim training and trans-thoracic aortic constriction (TAC) in mice, respectively (Supplementary Figs 3 and 4).35 GSEA revealed a number of transcriptional pathways that were shared between vegfchy−/−;vegfd−/− mutants and both physiological and pathological cardiac hypertrophy models35 (Supplementary Figs 3 and 4). Of note, cell cycle and lipid metabolism pathways were upregulated in both vegfchy−/−;vegfd−/− mutant zebrafish hearts and adult mouse hearts subjected to pressure overload following TAC (Supplementary Fig. 3). Conversely, genes involved in oxidative metabolism and the tricarboxylic acid (TCA) cycle were downregulated in both vegfchy−/−;vegfd−/− mutant zebrafish hearts and adult mouse hearts subjected to pressure overload following TAC. In contrast, pathways related to protein and RNA metabolism were shared between vegfchy−/−;vegfd−/− mutant zebrafish hearts and adult mouse hearts subjected to swim training (i.e., physiological hypertrophy) (Supplementary Fig. 4). Thus, some common features of the molecular cardiac hypertrophy program are shared between vegfchy−/−;vegfd−/− mutant zebrafish and established models of pathological and physiological cardiac hypertrophy in mice.
Fig. 4.
Transcriptomic profiling of vegfchy−/−;vegfd−/− mutant hearts reveals a disruption in cell proliferation and lipid metabolism. a Unsupervised hierarchical clustering showing distinct transcriptional profiles for vegfc+/+:vegfd+/+ control ventricles and in vegfchy−/−:vegfd−/− mutant ventricles. b Volcano plot of vegfc+/+:vegfd+/+ control ventricles and vegfchy−/−:vegfd−/− mutant ventricles. The top 10 upregulated genes for each comparison are indicated. Genes highlighted in red have a FDR < 0.05. Gene set enrichment analysis (GSEA) of vegfc+/+:vegfd+/+ control (c) and vegfchy−/−;vegfd−/− mutant ventricles (d). Representative pathways are highlighted with circles and gene set sizes are represented by circle sizes. GSEA indicates that the vegfchy−/−;vegfd−/− mutant ventricles are characterized by an upregulation of pathways related to cell proliferation and lipid metabolism compared with vegfc+/+:vegfd+/+ control hearts
Different types of injury cause distinct lymphangiogenic responses
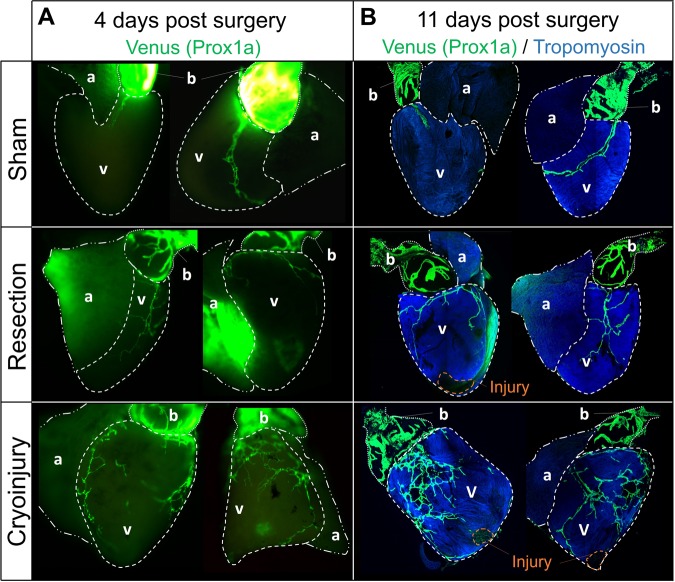
To test whether the cardiac lymphatic network is altered following cardiac injury, we subjected Tg(prox1a:KalTA4)uq3bh lymphatic reporter fish to either apical resection or cryoinjury, both of which stimulate a robust cardiac regenerative response in zebrafish.36–40 To control for the impact of chest incision without myocardial damage on the cardiac lymphatic network, we performed sham surgeries and assessed the cardiac lymphatic response in the Tg(prox1a:KalTA4)uq3bh reporter fish line. Following sham surgery, lymphatic vessels were detected on the bulbus arteriosus and the cardiac ventricle (Fig. 5), with a similar network coverage to wild-type control zebrafish (see Fig. 1), indicating that thoracic incision alone was insufficient to promote cardiac neo-lymphangiogenesis. We next assessed the cardiac lymphatics at 4 days or 11 days following apical resection injury. Apical resection injury did not stimulate overt cardiac lymphangiogenesis in adult zebrafish (Fig. 5). In contrast, global cardiac neo-lymphangiogenesis was clearly observed at 4 days and 11 days after cryoinjury, with the entire cardiac ventricle covered by neo-formed lymphatic vessels displaying an intricate network of ramifications (Fig. 5). Given this variable phenotypic response, we next wanted to assess whether this injury-specific response was also conserved in the regeneration of other organs. For these experiments we chose the fin, as both resection and cryoinjury also result in regeneration of this tissue.41,42 Consistent with the response of the heart, cryoinjury of the caudal fin stimulated an organ-wide activation of the lymphatic reporter transgene whereas amputation of this appendage failed to elicit any lymphatic vascular outgrowth (Supplementary Fig. 5). Overall, these results reveal that the lymphangiogenic response to tissue injury is highly model-dependent with cryoinjury providing a particularly powerful stimulus for neo-lymphangiogenesis.
Fig. 5.
Cryoinjury stimulates a lymphangiogenic response in the adult zebrafish heart. a Whole-mount immunofluorescence staining of Tg(prox1a:KalTA4)uq3bh adult zebrafish hearts at day 4 following sham surgery, apical resection, or cryoinjury. b Co-Venus and Tropomyosin staining was performed on Tg(prox1a:KalTA4)uq3bh adult zebrafish hearts at day 11 following sham surgery, apical resection or cryoinjury. Note robust global lymphangiogenic response to cryoinjury compared with either apical resection or sham surgery
Context-dependent requirement for lymphangiogenesis for heart regeneration following cryoinjury
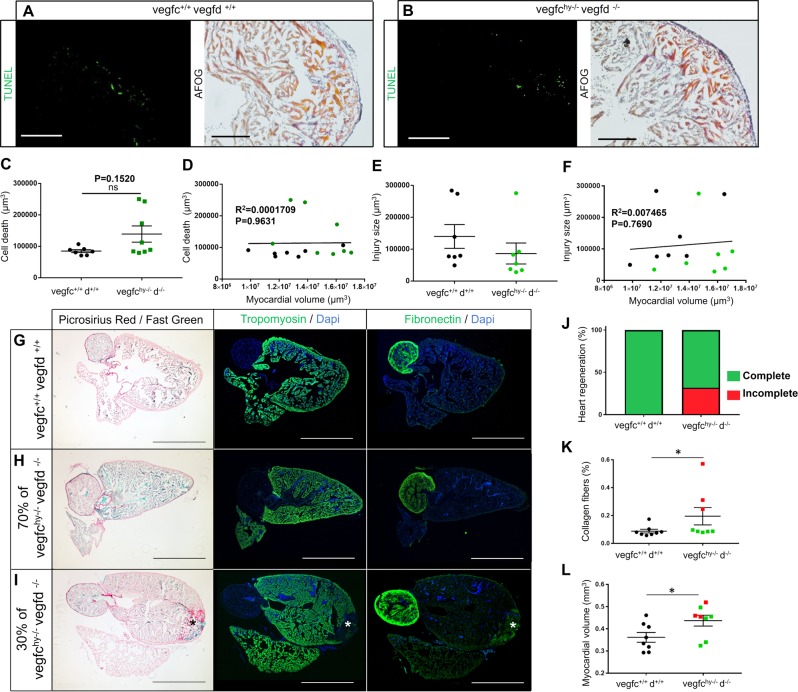
Compared with apex resection, cryoinjury is a more relevant pathophysiological model of infarction because the damaged tissue requires clearance and subsequent repair instead of a regrowth-based mechanism. To test whether lymphangiogenesis is required for cardiac regeneration, we subjected adult fish devoid of cardiac lymphatics (vegfchy/−;vegfd−/− double mutants) to cryoinjury, as this infarction model stimulated a reproducible and quantifiable lymphangiogenic response (Fig. 5). Hearts of both vegfc+/+:vegfd+/+ control fish or vegfchy−/−;vegfd−/− mutant fish were collected and analyzed at 1 day after cryoinjury to monitor the degree of cardiac damage (Fig. 6a–f). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and Acid Fucshin Orange G (AFOG) staining revealed comparable infarct sizes between control and mutant hearts following cryoinjury (Fig. 6a–f). However, we noted that a subset of mutant hearts (3/8) displayed an accumulation of TUNEL-positive apoptotic/necrotic cells despite having similar infarct sizes (Fig. 6c and Supplementary Fig. 6), suggesting that a subset of vegfchy−/−;vegfd−/− mutants have an impaired capacity to clear dead cells following cryoinjury. Cell death and injury size were not correlated with myocardial volume (Fig. 6d, f), indicating that the degree of cardiac tissue damage was not influenced by heart size.
Fig. 6.
Vegfc/d-dependent lymphangiogenesis is dispensable for cardiac regeneration in adult zebrafish. TUNEL staining and AFOG staining were performed on vegfc+/+:vegfd+/+ (a) and vegfchy−/−:vegfd−/− hearts (b) at 1 day after cryoinjury. Scale bar length: 200 μm (TUNEL) or 100 μm (AFOG). c Quantification of cell death in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts 1 day after cryoinjury. d Correlation analysis between myocardial volume and cell death in vegfc+/+:vegfd+/+ control and vegfchy−/−:vegfd−/− mutant hearts at 1 day post cryoinjury. Black plots represent vegfc+/+:vegfd+/+ samples, green plots represent vegfchy−/−:vegfd−/− samples. e Quantification of injury size in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts at 1 day after cryoinjury. f Correlation analysis between myocardial volume and injury size in vegfc+/+:vegfd+/+ control and vegfchy−/−:vegfd−/− mutant hearts at 1 day post cryoinjury. Black plots represent vegfc+/+:vegfd+/+ samples, green plots represent vegfchy−/−:vegfd−/− samples. Statistical analysis: unpaired, nonparametric, Mann–Whitney test was used to compare two means. Data are presented as the mean ± standard error of the mean (sem), n = 8 vegfchy−/−:vegfd−/−, n = 7 vegfc+/+:vegfd+/+. g–i First column: picrosirius red and fast green staining was performed on vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− hearts 6 months (180 days) after cryoinjury. Second column: tropomyosin staining was performed on vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− hearts 6 months after cryoinjury. Third column: fibronectin staining was performed on vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− hearts 6 months after cryoinjury. 30% of vegfchy−/−:vegfd−/− hearts display an impaired regenerative response with collagen deposition and fibrosis evident at the injury site (*). Scale bar length: 0.8 mm. j Percentage of hearts showing complete or incomplete cardiac regeneration at 180 days post cryoinjury. Quantification of collagen content (k) or myocardial volume (l) in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts 6 months after cryoinjury. Black plots represent vegfc+/+:vegfd+/+ hearts, green plots represent vegfchy−/−:vegfd−/− hearts that showed complete heart regeneration, red plots represent vegfchy−/−:vegfd−/− hearts that had impaired cardiac regenerative capacity. Statistical analysis: unpaired, nonparametric, Mann–Whitney tests was used to compare two means. Data are presented as the mean ± standard error of the mean (sem), n = 8 vegfchy−/−:vegfd−/−, n = 8 vegfc+/+:vegfd+/+. Significant differences are marked by an asterisk (*p < 0.05)
To determine whether lymphangiogenesis is required for cardiac regeneration, vegfchy/−;vegfd−/− double mutants and control hearts were collected and analyzed at 180 days after cryoinjury, which corresponds to a timepoint when the regenerative response is normally completed in wild-type zebrafish.38–40,43 Surprisingly, the vast majority (~70%) of vegfchy−/−;vegfd−/− mutants, which completely lack cardiac lymphatics, were able to mount a complete regenerative response without any signs of fibrosis (Fig. 6g–k and Supplementary Fig. 8). We found that the cardiac regenerative response was impaired only in a subset (3/8) of vegfchy−/−;vegfd−/− mutants, which were characterized by large deposits of fibrotic scar tissue present at the injury site (Fig. 6i-I”). Given that cardiomyocyte proliferation is required for zebrafish heart regeneration, we assessed vegfchy−/−;vegfd−/− mutant hearts at both early (day 19) and late (day 180) timepoints following cryoinjury to determine whether there was a defect in proliferation. Cardiomyocyte proliferation rates were unaffected at either timepoint in vegfchy−/−;vegfd−/− mutant hearts (Supplementary Figs. 8 and 9), including the 30% of mutants that had an impaired regenerative response (Supplementary Fig. 8). Conversely, in the subset of mutant hearts that displayed an impaired regenerative response, large infarcts and severe hypertrophy were evident at early and late timepoints following injury (Fig. 6k, l, and Supplementary Figs 8 and 9). Thus, vegfc/d signaling and lymphangiogenesis are not required for cardiomyocyte proliferation but appear to be required for efficient cellular clearance and scar resolution in a subset of zebrafish following cryoinjury.
Transcriptomic profiling of non-regenerative vegfchy−/−;vegfd−/− mutant hearts reveals a marked inflammatory response
To profile vegfc/d-dependent genetic pathways contributing to lymphangiogenesis in cardiac regeneration, we performed RNA-seq on cryosections from control and mutant ventricles at 180 days following cryoinjury. This analysis enabled us to directly compare transcriptional profiles with regenerative outcomes for the samples presented in Fig. 6g–l (Fig. 7a). Overall, vegfchy−/−;vegfd−/− mutant ventricles were characterized by an upregulation of pathways related to RNA and protein metabolism, cytokine signaling, and inflammation (Fig. 7c and Supplementary Data Set 3). In order to identify which pathways were specifically associated with injury in the mutant hearts, we cross-referenced these data to the baseline RNA-seq data set obtained from vegfchy−/−;vegfd−/− mutant hearts without injury (presented in Fig. 4) and then repeated the GSEA (Supplementary Fig. 10). Pathways related to cytokine signaling, innate immune system and inflammation were uniquely enriched after injury in the vegfchy−/−;vegfd−/− mutant hearts (Supplementary Fig. 10).
Fig. 7.
Transcriptomic profiling of vegfchy−/−;vegfd−/− mutant hearts at 180 days following cryoinjury. a Schematic representation of experimental design for RNA sequencing on cryosections. RNA-seq was performed on cryosections from control and mutant ventricles at 180 days following cryoinjury to compare transcriptional profiles to regenerative outcomes for the samples presented in Fig. 6g–l. b Volcano plot showing genes enriched in vegfc+/+:vegfd+/+ control ventricles at 180 days post cryoinjury and in vegfchy−/−:vegfd−/− mutant ventricles at 180 days after cryoinjury. The top 10 upregulated genes are indicated for each comparison. Genes highlighted in red have a p-value < 0.05. c Volcano plot showing genes enriched in vegfchy−/−:vegfd−/− mutant ventricles that did not regenerate compared with vegfchy−/−:vegfd−/− mutant ventricles that did regenerate at 180 days after cryoinjury. The top 10 upregulated genes are indicated for each comparison. Genes highlighted in red have a p-value < 0.05. d GSEA for vegfchy−/−:vegfd−/− mutant ventricles at 180 days after cryoinjury compared with vegfc+/+:vegfd+/+ control ventricles at 180 days post cryoinjury. Representative pathways are highlighted with circles and gene set sizes are represented by circle sizes. e GSEA for vegfchy−/−:vegfd−/− mutant ventricles that did not regenerate compared with vegfchy−/−:vegfd−/− mutant ventricles that did regenerate at 180 days post cryoinjury. Representative pathways are highlighted with circles and gene set sizes are represented by circle sizes. These data indicate that vegfchy−/−;vegfd−/− mutant ventricles are characterized by an upregulation of pathways related to inflammation, cytokine signaling and immune response compared with vegfc+/+:vegfd+/+ control hearts at 180 days post cryoinjury
To determine the molecular basis of the regenerative capacity within the mutant fish population, we next compared the transcriptional profiles of vegfchy−/−;vegfd−/− mutant hearts that did not regenerate (n = 3) with the profiles of mutant hearts that recovered completely (n = 5) at 180 days post cryoinjury (presented in Fig. 6g–l). The subset of vegfchy−/−;vegfd−/− mutant hearts that failed to regenerate were characterized by an enrichment of inflammatory pathways, in particular TRAF6-mediated IRF7 activation, interferon alpha beta signaling and regulation of IFNA signaling (Fig. 7d, and Supplementary Data Set 4). These findings indicate that loss of the vegfc/d signaling axis is associated with a sustained and pronounced inflammatory response following injury. This change in the cardiac micro-environment seems to favor adverse conditions, which impair the regenerative process.
Discussion
Here, we characterize the lymphatic vasculature of the adult zebrafish heart and assess its potential role in cardiac regeneration. Analysis of the lymphatic vasculature revealed a prominent cardiac lymphatic vessel network on the epicardial surface of the heart, which included a major lymphatic vessel located in close proximity to the main coronary artery and numerous branching vessels. In contrast, lymphatics were not detected in the atrium. The anatomical organization of the cardiac lymphatic vasculature in zebrafish is similar to the mammalian heart, which comprises a greater density of lymphatic vessels on the ventricles compared with the atria.44 Interestingly, the fish heart is also characterized by a dense lymphatic network in the bulbus arteriosus, which could reflect the specialized physiological function of this anatomical structure in the fish circulatory system. The bulbus arteriosus has a thick fibro-elastic wall and functions as a capacitor to normalize the ventricular pulse pressure as blood exits the ventricle and is pumped out to the gills.45 Thus, the presence of lymphatic vessels in the zebrafish heart is correlated with blood pressure in the different chambers, where the chambers under the highest pressure have the greatest density of lymphatic vessels. This likely reflects the central role of the lymphatic system in fluid homeostasis.
Our findings indicate that the development of the cardiac lymphatic vasculature is dependent on VEGFC and VEGFD signaling. Genetic loss-of-function of either vegfc or vegfd was sufficient to abolish the development of the ventricular lymphatic network in a gene dosage dependent manner. In contrast, a single copy of either vegfc or vegfd was sufficient to maintain the lymphatic vascular network on the bulbus arteriosus, which was only abolished when both vegfc and vegfd function were compromised. However, it should be noted that a few lymphatic vessels could still be detected on the bulbous arteriosus of vegfchy−/−;vegfd−/− double mutants. The presence of a few lyve1-expressing cells on the bulbus arteriosus of double mutant fish might reflect the existence of an alternative Vegfc/d-independent pathway for lymphangiogenesis in the heart or, alternatively, could be indicative of residual Vegfc signaling in this hypomorphic mutant.32 Nevertheless, vegfchy−/−:vegfd−/− double mutants completely lacked a ventricular lymphatic network, which did not affect organismal viability. Importantly, there was also no effect on the development of the coronary vasculature, which enabled the specificity of the lymphatic network to be studied.
A robust lymphangiogenic response occurred following cryoinjury, but not following resection injury in the heart and caudal fin. It is important to note that a regenerative response is observed in the heart and caudal fin following either resection or cryoinjury despite the apparent discrepancy in lymphangiogenic responses to different types of tissue damage.36–38,40–43 Surgical amputation (resection) and cryoinjury models differ with respect to the size of the injury, presence of cellular debris following tissue damage, and the degree of inflammation induced after injury, which are all more pronounced following cryoinjury.36–38,40,43,46 Interestingly, macrophages can secrete VEGF-C to promote lymphangiogenesis, which could provide a potential link between inflammation and lymphangiogenesis depending on injury context. While the precise mechanisms accounting for differences in the lymphatic response to apical resection versus cryoinjury require further investigation, it is likely that the degree of inflammation is a key regulator that dictates the nature of the lymphangiogenic response.
We were surprised to find that the majority of adult zebrafish were still capable of mounting a complete regenerative response following cryoinjury even in the absence of a functional cardiac lymphatic network. Therefore, the cardiac lymphatic vasculature appears to be dispensable for cardiomyocyte proliferation and clearance of fibrotic scar tissue in the majority of regenerating hearts. However, cardiac regeneration was severely impaired in a subset of mutants, which were characterized by fibrotic scars, severe cardiac hypertrophy, and a prominent inflammatory signature up to 180 days following cryoinjury. This variable response to cryoinjury in a subset of mutants was apparent as early as 1 or 19 days following cryoinjury, where some mutants were characterized by an accumulation of apoptotic/necrotic cells at day 1 and larger infarcts at day 19. This phenotype is consistent with the known role of lymphatics in the clearance of cellular debris following injury.47 In addition, non-regenerative mutant hearts displayed an enrichment in inflammatory pathways associated with TRAF6-mediated IRF7 activation and interferon alpha/beta signaling, which are involved in immune cell activation, recruitment, and clearance.48,49 In the absence of a functional lymphatic network, vegfchy−/−:vegfd−/− mutant hearts may have an impaired capacity to resolve inflammation following injury, which could impair the regenerative process when inflammation is heightened under certain conditions. Intrinsic differences in the susceptibility to myocardial injury and inflammation could be related to differences in genetic background, which is known to influence infarct size in mice.50 Together, these findings suggest that Vegfc/d-dependent lymphangiogenesis may facilitate cardiac regeneration under certain circumstances, for example, when there is a heightened inflammatory response to injury.
The most striking cardiac phenotype observed in zebrafish lacking Vegfc and Vegfd was a marked increase in myocardial wall volume, ventricular enlargement, and hypertrophy of cardiomyocytes. Several potential mechanisms could contribute to cardiac hypertrophy in vegfchy−/−:vegfd−/− mutants. Hypertrophy of mutant cardiomyocytes is consistent with increased myocardial wall stress due to pressure overload,51 which would be expected if there is interstitial edema in mutant hearts lacking a functional lymphatic system (Fig. 8). Interestingly, classic molecular markers of pathological cardiac hypertrophy such as nppa, nppb, myh7, and acta1a were not upregulated in mutant zebrafish hearts.34 However there was some overlap between transcriptional pathways in the vegfchy−/−;vegfd−/− mutant ventricles and the molecular programs associated with established experimental models of physiological and pathological cardiac hypertrophy in mice.35 In particular, pathways related to cell proliferation and lipid metabolism were enriched in mutant zebrafish hearts and mouse hearts subjected to pressure overload. The enrichment of transcripts related to cell proliferation likely reflects an infiltration of proliferating non-myocytes rather than cardiomyocyte proliferation, as cardiomyocyte proliferation indices were not different between mutant and control ventricles and cell cycle-associated transcripts are typically enriched in non-myocyte populations in the adult mouse heart following injury.52 Although the precise mechanism driving cardiac hypertrophy in vegfchy−/−:vegfd−/− mutants is unclear, a striking enrichment of genes implicated in sphingolipid and phospholipid metabolism was observed. The lymphatic vasculature is known to play an important role in lipid transport.53 It is currently unclear how dysregulated lipid metabolism might contribute to cardiac hypertrophy, but a recent study reported that physiological and pathological cardiac hypertrophy are characterized by distinct lipid profiles including enrichment of several sphingolipid and phospholipid species.54 Sphingolipid metabolites are also important regulators of inflammation and immune cell trafficking,55 which could be linked to the heightened expression of pathways related to inflammation and cytokine signaling in vegfchy−/−;vegfd−/− mutant zebrafish following cryoinjury. Further studies are required to dissect the role of sphingolipid and phospholipid metabolism in cardiac hypertrophy and inflammation.
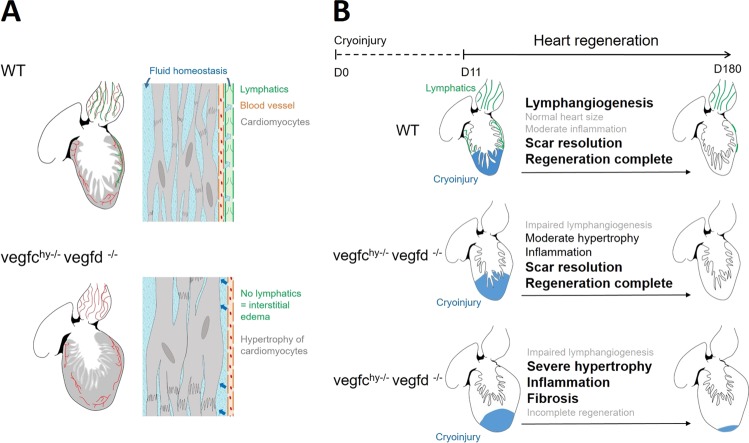
Fig. 8.
Proposed model showing relationship between lymphangiogenesis, cardiac hypertrophy and cardiac regeneration. a In wild-type (WT) zebrafish, the cardiac lymphatic network (shown in green) is present on the bulbus arteriosus and on the ventricle but is not detected on the atrium. Genetic deletion of either vegfc or vegfd is sufficient to abolish the development of the cardiac lymphatic network and induces a marked increase in myocardial wall volume, ventricular enlargement, hypertrophy of cardiomyocytes and a decrease in myocardial interstitial volume. The absence of lymphatics can cause interstitial edema because of the impaired regulation of fluid balance. Blood vessels (shown in red) are not affected by vegfc/vegfd and are present in both vegfc+/+:vegfd+/+ control hearts and vegfchy−/−:vegfd−/− mutants hearts. b A robust lymphangiogenic response is induced by cryoinjury in WT fish. The majority of vegfchy−/−:vegfd−/− mutants display moderate hypertrophy and are able to fully regenerate their heart even in the absence of a functional cardiac lymphatic vascular network and moderate inflammation. However, a subset of mutants display an incomplete cardiac regenerative response following cryoinjury. A heightened and persistent inflammatory response to injury could in part explain why a subset of mutants have an impaired regenerative response and severe cardiac hypertrophy following cryoinjury
A recent study also reported that vegfaa over-expression results in changes in cardiac energy utilization in zebrafish, suggesting that VEGF family members are important regulators of myocardial metabolism.28 Similarly, genes related to oxidative metabolism and TCA cycle were downregulated in vegfchy−/−:vegfd−/− mutants, which also occurs following pressure overload in adult mice. There is a strong interplay between myocardial metabolism, activation of inflammatory signaling pathways, expression of cytokines, and immune cell infiltration in heart failure and many of these processes are characteristic of maladaptive hypertrophy of the heart in mammals.28,56,57 Mechanisms linking these processes are beginning to be unraveled, but given the strong cross talk between these processes it has been difficult to precisely deconvolute the molecular interplay.57 The current study reveals an association between cardiac hypertrophy, metabolic disruption, and inflammation in organisms lacking a functional lymphatic vascular system. Given that VEGF-C/VEGFR3 pathway inhibitors are being developed for cancer, it will be important to consider the impact of these treatments on the cardiovascular system before translation to the clinic.
Our findings reveal a previously unappreciated, context-dependent requirement for the lymphatic vasculature in cardiac growth and regeneration. Cardiac hypertrophy, metabolic disruption, and inflammation are likely to influence the outcome of therapeutic approaches designed to target lymphangiogenesis in ischemic heart disease.
Methods
Zebrafish strains
Ethical approval for zebrafish procedures was obtained from The University of Queensland Animal Ethics Committee (AE21513). All zebrafish strains were maintained at The University of Queensland under standard husbandry conditions with a 14 h light, 10 h dark cycles. Transgenic zebrafish lines used in this study included Tg(−5.2lyve1b:DsRed)nz101,29 Tg(prox1a:KalTA4)uq3bh:Tg(10xUAS: Venus),30 Tg(kdrl:EGFP)s834, vegfduq9bh and vegfchu5055.25 In addition, to label the lymphatic vasculature in vegf mutants, vegfduq9bh;vegfchu5055 double mutants25 were outcrossed to Tg(−5.2lyve1b:DsRed).29 For mutant genotyping, DNA extraction from fin clips was performed as previously described and genotyping was performed using KASP assays (LGC Genomics). Fish body length was manually measured with a ruler, from the tip of the mouth to the body/caudal fin juncture. All experiments were performed on size-matched fish between 28 and 31 mm in length.
Cardiac injury in adult zebrafish
Adult fish were anesthetized in aquarium water containing 0.2 mg/ml of Tricaine and secured ventral side up in a slotted sponge. Watchmaker forceps (Dumont #5 91150-20) were used to remove the surface scales and penetrate the skin, muscle, and pericardial sac. Ventricular resection was performed as previously described.36,37,58 The ventricle was gently pulled at the apex and 20% of the ventricular apex was cut with Vannas scissors (8 cm, Angled 5 mm, 0.1 mm Tips). Cryoinjury was performed as previously described.38–40,43 The silvery epithelial layer of the hypodermis was gently torn with the tip of the scissors to directly access the beating ventricle. The cryoprobe was made with stainless steel, the tip of the applicator was 6 mm long and 0.8 mm in diameter. The cryoprobe was taken out of liquid nitrogen for 10 s before being placed on the ventricle for 24 s, as previously described.38 After surgeries, fish were immediately returned to system water. For EdU labeling, fish were anesthetized in water containing 0.2 mg/ml of Tricaine and 10 μl of EdU (5-ethynyl-2′-deoxyuridine from Thermo Fisher) solution (1 μg/μl in PBS) was injected into the abdominal cavity at specified timepoints.
Fin injury in adult zebrafish
Adult fish were anesthetized in aquarium water containing 0.2 mg/ml of Tricaine. For resection injury, an excision of a small portion of the caudal fin was performed using a scalpel as previously described.41 For cryoinjury, the cryoprobe was made with stainless steel, the tip of the applicator was 6 mm long and 0.8 mm in diameter. The cryoprobe was taken out of liquid nitrogen for 10 s before being placed on the caudal fin for 1 min. After surgery, fish were immediately returned to system water.
Sample preparation for histology
Zebrafish were anesthetized in aquarium water containing 0.6 mg/ml of Tricaine and then euthanized by rapid decapitation. Zebrafish were collected intact and paraformaldehyde-fixed (4% in phosphate buffered saline (PBS; Sigma)) overnight at 4 °C. Hearts were subsequently excised and dissected and the pericardial membrane removed before being placed in paraformaldehyde (4% in PBS) for 1 h at room temperature. Prior to further processing, hearts were washed three times in PBS and DsRed2 and/or Venus reporter expression was monitored in whole hearts before further analyses. For histology, hearts were equilibrated overnight in 15% sucrose in PBS, mounted in Tissue-Tek® O.C.T. compound (ProSciTech), and frozen for cryosectioning.
Immunohistochemistry
For whole-mount immunostaining, hearts were permeabilized with PBST (PBS + 0.3% Triton X-100 (AMRESCO) for 30 min and incubated with 1% BSA in PBST overnight at 4 °C to block nonspecific binding of the antibodies. Hearts were incubated with the diluted primary antibody in blocking solution (1% BSA in PBST) in a humidified chamber for 1 h at room temperature and overnight at 4 °C. The following primary antibodies were used: chicken anti-GFP (1:200; ABCAM ab13970), rabbit anti-RFP (1:100; ABCAM ab62341), mouse anti-Tropomyosin (1:200; Sigma T9283), and rabbit anti-Fibronectin (1:50; F3648 Sigma). Hearts were washed six times with PBST and incubated with the appropriate secondary fluorescent antibodies (1:400: Goat anti-rabbit/mouse/or chicken Alexa Fluor Thermo Fisher Scientific) in blocking solution in a humidified chamber overnight at 4 °C. Samples were then washed six times in PBST and analyzed. Sagittal sections were obtained (8-μm thickness) at −24 °C using a cryostat. For immunodetection on heart sections, cryosections were dried for 20 min at room temperature, permeabilized with PBSTw (PBS + 0.1% Tween-20 (Sigma)) for 15 min and incubated with 10% Goat Serum (PCN5000, Life Technologies) in PBSTw for 1 h at room temperature to block nonspecific binding of the antibodies. Sections were then incubated with the diluted primary antibody in 5% Goat Serum/PBSTw solution in a humidified chamber overnight at 4 °C. Rabbit anti-fibronectin (1:50; Sigma F3648), rabbit anti-RFP (1:100; ABCAM ab62341) and mouse anti-Tropomyosin (1:200; Sigma T9283) primary antibodies were used. Sections were washed three times with PBSTw and incubated with the appropriate secondary fluorescent antibodies (1:400: Goat anti-rabbit or mouse Alexa Fluor) in 5% Normal Goat Serum (Life technologies, PCN5000)/PBSTw solution in a humidified chamber overnight at 4 °C. Sections were then washed three times in PBSTw and analyzed. EdU labeling was carried out using Click-iT EdU Imaging kit C10338 (Invitrogen), according to the manufacturer’s instructions. For cell death detection, cryosections were dried for 20 min at room temperature and then processed using the In Situ Cell Death Detection Kit (TUNEL kit, 11684795910, Roche), according to the manufacturer’s instructions
Picrosirius Red/Fast Green, hematoxylin/eosin, and acid fushin orange G (AFOG) staining
For Picrosirius Red/Fast Green staining, sections were postfixed for 30 min in 10% formalin (Sigma), followed by overnight fixation in Bouin’s solution (Sigma), and then extensively washed with tap water. Sections were incubated for 1 h in Picrosirius Red and Fast Green solution: 0.1% direct red 80 (Sigma) and 0.1% fast green FCF (Sigma) in saturated aqueous picric acid (1.2% picric acid in water; Sigma). Sections were quickly washed ten times in distilled water, twice in 70% ethanol, twice in 90% ethanol, then in 100% ethanol (2 × 15 min), and finally in Safesolv solution (Q Path®, VWR International; 2 × 15 min), before being mounted in safemount medium (Q Path®, VWR International). For Hematoxylin and Eosin staining, sections were incubated for 5 min in Hematoxylin solution, washed ten times for 1 min in running tap water, and incubated for 3 min in Eosin solution. Sections were washed three times in distilled water, twice in 70% ethanol, twice in 90% ethanol, then in 100% ethanol (2 × 15 min), and finally in Safesolv solution (Q Path®, VWR International; 2 × 15 min), before being mounted in safemount medium (Q Path®, VWR International). For AFOG staining, an AFOG kit (100 T, Clinisciences) was used, according to the manufacturer’s instructions, before being mounted in safemount medium (Q Path®, VWR International).
In situ hybridization
The flt4 probe was prepared as previously described.59 Cryosections were dried for 20 min at room temperature, permeabilized with PBSTw for 20 min, and incubated for 5 min with proteinase K (5 μg/ml, Sigma 3115887001), sections were then postfixed for 10 min in 4% PFA at room temperature. Sections were pre-hybridized with hybridization buffer (50% formamide, 5× SSC, ARNt 1 mg/ml, Heparin 100 ug/ml, Denhart’s 1×, 0.1% Tween 20, 10% CHAPS, 10 mnol/l EDTA) for 24 h in a humidified chamber at 70 °C. Sections were then hybridized with flt4 probe in hybridization buffer (90 ng/ml) for 24 h in a humidified chamber at 70 °C. Sections were then washed in a humidified chamber at 70 °C with hybridization buffer (100%, 75% in 2× SSC, 50% in 2× SSC and 25% in 2× SSC), followed by 2× SSC and twice with 0.2× SSC. Sections were incubated with Anti-Digoxigenin-AP, Fab fragments (1:200: Sigma 11093274910) in 2% Sheep Serum/BSA (2 mg/ml)/PBSTw solution in a humidified chamber overnight at 4 °C. Sections were washed three times with PBSTw and incubated with BM-Purple (1:50, Sigma11442074001) in staining buffer (100 mM Tris-HCl pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20) at room temperature for 6 h. Sections were then washed twice in PBS and analyzed.
RNA extraction from fresh tissue or from cryosections and cDNA library preparation for RNA-seq
For the transcriptomic analyses presented in Fig. 4 and Supplementary Figs 3 and 4, RNA extraction was performed from fresh tissue using Direct-zol™ RNA Miniprep kit (Zymo/ Integrated Science, R2051), according to the manufacturer’s instructions. For the transcriptomic analyses presented in Fig. 7 and Supplementary Fig. 10, RNA extraction was performed from PFA-fixed (4% PFA) and Tissue-Tek® O.C.T. compound (ProSciTech)-embedded heart cryosections (8 μm). A scalpel was used to collect 12 sections of the ventricle per sample. Sections were transferred to a tube on ice containing 100 μl of TRIzol® Reagent (15596018, Invitrogen) and then processed with Direct-zol™ RNA Microprep kit (Zymo/ Integrated Science, R2061), according to the manufacturer’s instructions. RNA quality was examined with the High Sensitivity RNA ScreenTape on the TapeStation (Agilent) before library preparation. For RNA-seq using fresh tissue, libraries were generated using TruSeq Stranded mRNA Library Prep Kit (#20020595, Illumina) according to the manufacturer’s instructions. For RNA-seq using fixed tissue, ribosomal RNA was depleted from total RNA using NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (E6310, New England Biolabs) followed by library generation using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (E7760, New England Biolabs) to select library fragment sizes of 200–400 bp, according to the manufacturer’s protocol. Libraries were validated on the TapeStation and sequenced for 150 cycles, 25 million reads, paired-end sequencing on HiSeq4000 for RNA-seq using fixed tissue, or NextSeq550 for RNA-seq using fresh tissue (Illumina).
RNA-seq and bioinformatic analyses
Paired sequence reads underwent quality trimming using the Skewer (version 0.2.2) with default setting.60 For RNA-seq using fresh tissue, sequencing reads were aligned to the zebrafish genome (GRCz11.95) using STAR (version 2.5.2a) with default settings. For RNA-seq using fixed tissue, reads were aligned to the reference genome using Burrows Wheeler Aligner (version 0.7.17) with default settings.61 Reads aligning to Ensembl-annotated whole genes on the correct strand with a strict mapping quality (Q ≥ 20) were counted with feature Counts and used to construct a data matrix comprising genes with an average of 10 reads or more per sample across the experiment. Heatmaps were generated from normalized and scaled gene expression values using Complex Heatmap package (version 1.18.1) with R in RStudio.62 Differential gene expression analysis was performed using edgeR (v3.22.4) with R in RStudio.63 GSEA was performed on these data as previously described using publicly available gene sets (www.broadinstitute.org/gsea/msigdb) to determine the enrichment of Reactome pathways.
Imaging
Imaging of heart slices was performed using a Dragonfly Spinning Disk confocal microscope (ANDOR Oxford Instruments Company). For whole-mount imaging, the heart was mounted in 0.5% low-melting-point agarose and imaged using a Zeiss LSM 710 FCS confocal microscope. Images were processed using ImageJ 1.47 software (National Institute of Health).
Enzymatic dissociation of cardiomyocytes, staining, and quantification of cardiomyocyte size
Zebrafish ventricles were excised and dissected and the pericardial membrane was removed before being placed in paraformaldehyde (4% in PBS) for 1 h at room temperature, followed by three times washes in PBS. Ventricles were transferred in collagenase solution at 0.5 U/mL in 0.2% NaN3/PBS (Collagenase B 11088807001, Roche) in a tube shaker with oscillation at 1000 rpm at 37 °C. Collection of supernatant was performed every 4 h and the supernatant was transferred to the same volume of 0.2% NaN3/FBS. Digested material was stored at 4 °C between collections. Collection of supernatant was performed every 4 h until the entire ventricle was dissociated. Once dissociation was completed, cardiomyocytes were centrifuged at 1000 × g for 3 min. For immunostaining, cardiomyocytes were incubated with 4% BSA, 0.2% Triton X-100, 1 mM EDTA, 0.02% NaN3 in PBS (referred to as blocking buffer) for 10 min at room temperature, then cardiomyocytes were incubated with the anti-Tropomyosin (1:200; Sigma T9283) antibody in blocking buffer for 1 h at room temperature. Cardiomyocytes were washed with blocking buffer and incubated with the appropriate secondary fluorescent antibody (1:400: Goat anti-mouse Alexa Fluor Thermo Fisher Scientific) in blocking buffer for 1 h at room temperature. Cardiomyocytes were then washed in blocking buffer before being mounted in Fluoromount-G (Thermo Fisher Scientific) and analyzed. For quantification of cardiomyocyte volume, width, and length, Z-stack imaging was performed at ×60 magnification for a total of ~1600 cardiomyocytes (~200 per sample, n = 8 samples) using a Dragonfly Spinning Disk confocal microscope (ANDOR Oxford Instruments Company). All images were similarly processed using ImageJ 1.47 software (National Institute of Health). Automated quantification of cardiomyocyte volume was performed using ImageJ Trainable Weka Segmentation macro.64 Cardiomyocyte width and length were quantified manually using ImageJ measurement tools. The code can be accessed via GitHub using the following link (https://github.com/Jacob-Smith-MCRI/Cardiomyocyte-Volume-Calculator).
Quantification of ventricle size, myocardial volume, collagen deposition, interstitial volume, cell death, and injury size
Heart sections labeled with Picrosirius Red/Fast Green were used to analyze ventricle volume, myocardium volume and the presence of collagen and non-collagenous proteins. Whole hearts were sectioned (sagittal sections of 8 μm) and Picrosirius Red/Fast Green labeling was performed at 56 μm intervals. Images were captured for each labeled section. All images were similarly processed using ImageJ 1.47 software (National Institute of Health). For ventricle size and myocardium volume measurements, selections were carefully traced around the entire ventricle and around the myocardial wall. Selected surface areas were then quantified using ImageJ software. Surface areas were then summed giving the total ventricle volume and myocardial volume for each sample. Myocardial volume was then normalized to ventricle volume. For interstitial volume measurements, color thresholds were adjusted to select the area corresponding to the interstitial space and interstitial volumes were measured and normalized to ventricular volumes. For quantification of collagen fibers, manual selections were captured within the ventricle on RGB images and color thresholds were adjusted to only select the area corresponding to collagen fiber staining (dark pink); four sections were processed for each sample. Collagen surface areas were measured and normalized to ventricular surface areas. Transverse heart sections of 8 μm labeled with Acid Fushin Orange G were used to quantify infarct size. Color thresholds were adjusted to dark orange to select the area corresponding to the damaged tissue and damaged tissue volumes were measured (eight sections were processed for each sample). Selected surface areas of transverse heart sections labeled with TUNEL (11684795910, Roche) were quantified using ImageJ software. Surface areas were then summed giving the total cell death volume for each sample (six sections were processed for each sample).
Quantification of cardiomyocyte number and proliferation
Heart sections co-labeled with Tropomyosin, EdU, and Dapi were used to analyze cardiomyocyte proliferation or number. Whole hearts were sectioned (sagittal sections of 8 μm) and EdU, Dapi, and Tropomyosin co-labeling were performed at 56 μm intervals. For quantification of cardiomyocyte number, five sections were processed for each sample. Dapi positive/Tropomyosin positive nuclei were manually counted in a surface selection of the ventricle and normalized to total surface area (1000 μm2). For cardiomyocyte proliferation analysis, three sections corresponding to the injured area were processed for each sample. EdU positive/Tropomyosin positive nuclei were manually counted in a surface selection at the apex of the ventricle and normalized to the tropomyosin positive surface selection area.
Statistical analysis
The nonparametric Mann–Whitney test was used for comparison of two means and linear regression analysis was performed to analyze correlations using Prism software (GraphPad). Kolmogorov-Smirnov tests was used for comparison of two frequency distributions using Prism software (GraphPad). Comparison of means are presented as the mean ± standard error of the mean (sem). Correlation analyses are presented as XY dot plots with the slope, R square, and P-value. For all analyses P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We would like to thank Dr. David Elliott (Murdoch Children’s Research Institute) for helpful discussions and critical appraisal of our manuscript. This work was supported by fellowships and project grants from the National Health and Medical Research Council of Australia (E.R.P., J.E.H, M.F., B.M.H. and C.P.), Heart Foundation of Australia (E.R.P, J.E.H., B.M.H.) and The University of Queensland (E.R.P., J.E.H., M.F., B.M.H.). The Murdoch Children’s Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program.
Author contributions
E.R.P., J.E.H. and M.F. conceptualized the study; C.V. and C.P.T. are co-first authors and contributed equally to this work; E.R.P., M.F., J.E.H., C.V. and C.P.T. designed the experiments; C.V., C.P.T. and J.S. conducted the experiments and performed data analysis; C.B.S. conducted the RNA-sequencing analysis; N.I.B. and B.M.H. generated the vegfc/vegfd zebrafish transgenic lines; E.R.P. and C.V. wrote the paper; E.R.P, J.E.H. and M.F. managed and funded the project; All authors contributed to data interpretation and editing of the manuscript.
Data availability
All data sets associated with the current study are available from the corresponding author on reasonable request. RNA-seq data have been deposited at the Gene Expression Omnibus under the accession number GSE133131.65
Code availability
Automated quantification of cardiomyocyte volume was performed using ImageJ 1.47 software (National Institute of Health) with ImageJ Trainable Weka Segmentation macro.64 The code can be accessed via GitHub using the following link (https://github.com/Jacob-Smith-MCRI/Cardiomyocyte-Volume-Calculator).
Competing interest
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Céline J. Vivien, Cathy Pichol-Thievend
Contributor Information
James E. Hudson, Phone: +61 7 3362 0104, Email: james.hudson@qimrberghofer.edu.au
Mathias Francois, Phone: +61 7 3346 2494, Email: m.francois@imb.uq.edu.au.
Enzo R. Porrello, Phone: +61 3 9936 6140, Email: enzo.porrello@mcri.edu.au
Supplementary information
Supplementary Information accompanies the paper on the npj Regenerative Medicine website (10.1038/s41536-019-0079-2).
References
- 1.Abouelkheir GR, Upchurch BD, Rutkowski JM. Lymphangiogenesis: fuel, smoke, or extinguisher of inflammation’s fire? Exp. Biol. Med (Maywood) 2017;242:884–895. doi: 10.1177/1535370217697385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cimpean AM, Raica M. Lymphangiogenesis and Inflammation-Looking for the “Missing Pieces” of the Puzzle. Arch. Immunol. Ther. Exp. (Warsz.) 2015;63:415–426. doi: 10.1007/s00005-015-0349-7. [DOI] [PubMed] [Google Scholar]
- 3.Corliss BA, Azimi MS, Munson JM, Peirce SM, Murfee WL. Macrophages: an inflammatory link between angiogenesis and lymphangiogenesis. Microcirculation. 2016;23:95–121. doi: 10.1111/micc.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuda KS, et al. A zebrafish model of inflammatory lymphangiogenesis. Biol. Open. 2015;4:1270–1280. doi: 10.1242/bio.013540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji RC. Lymphatic endothelial cells, lymphedematous lymphangiogenesis, and molecular control of edema formation. Lymphat. Res. Biol. 2008;6:123–137. doi: 10.1089/lrb.2008.1005. [DOI] [PubMed] [Google Scholar]
- 6.Alitalo K, Tammela T, Petrova TV. Lymphangiogenesis in development and human disease. Nature. 2005;438:946–953. doi: 10.1038/nature04480. [DOI] [PubMed] [Google Scholar]
- 7.Zampell JC, et al. HIF-1alpha coordinates lymphangiogenesis during wound healing and in response to inflammation. FASEB J. 2012;26:1027–1039. doi: 10.1096/fj.11-195321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang LH, Lavine KJ, Randolph GJ. Cardiac lymphatic vessels, transport, and healing of the infarcted heart. JACC Basic Transl. Sci. 2017;2:477–483. doi: 10.1016/j.jacbts.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angeli V, Harvey NL. Lymphatic vessels at the heart of the matter. Cell Metab. 2015;22:56–58. doi: 10.1016/j.cmet.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Henri O, et al. Selective stimulation of cardiac lymphangiogenesis reduces myocardial edema and fibrosis leading to improved cardiac function following myocardial infarction. Circulation. 2016;133:1484–1497. doi: 10.1161/CIRCULATIONAHA.115.020143. [DOI] [PubMed] [Google Scholar]
- 11.Klotz L, et al. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature. 2015;522:62–67. doi: 10.1038/nature14483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabin F. On the origin of the lymphatic system from the veins and the development of the lymph hearts and thoracic duct in the pig. Am. J. Anat. 1902;1:367. doi: 10.1002/aja.1000010310. [DOI] [Google Scholar]
- 13.Norman S, Riley PR. Anatomy and development of the cardiac lymphatic vasculature: its role in injury and disease. Clin. Anat. 2016;29:305–315. doi: 10.1002/ca.22638. [DOI] [PubMed] [Google Scholar]
- 14.Li YL, Zhao H, Ren XB. Relationship of VEGF/VEGFR with immune and cancer cells: staggering or forward? Cancer Biol. Med. 2016;13:206–214. doi: 10.20892/j.issn.2095-3941.2015.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin M, et al. Correction: Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Development. 2017;144:531. doi: 10.1242/dev.148569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karkkainen MJ, et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat. Immunol. 2004;5:74–80. doi: 10.1038/ni1013. [DOI] [PubMed] [Google Scholar]
- 17.Bahram F, Claesson-Welsh L. VEGF-mediated signal transduction in lymphatic endothelial cells. Pathophysiology. 2010;17:253–261. doi: 10.1016/j.pathophys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Karkkainen MJ, et al. A model for gene therapy of human hereditary lymphedema. Proc. Natl Acad. Sci. USA. 2001;98:12677–12682. doi: 10.1073/pnas.221449198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon K, et al. Mutation in vascular endothelial growth factor-C, a ligand for vascular endothelial growth factor receptor-3, is associated with autosomal dominant milroy-like primary lymphedema. Circ. Res. 2013;112:956–960. doi: 10.1161/CIRCRESAHA.113.300350. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Nakagami H, Kaneda Y, Morishita R. Lymphedema and therapeutic lymphangiogenesis. Biomed. Res. Int. 2013;2013:804675. doi: 10.1155/2013/804675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldwin ME, et al. Vascular endothelial growth factor D is dispensable for development of the lymphatic system. Mol. Cell Biol. 2005;25:2441–2449. doi: 10.1128/MCB.25.6.2441-2449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiko P, et al. Deletion of vascular endothelial growth factor C (VEGF-C) and VEGF-D is not equivalent to VEGF receptor 3 deletion in mouse embryos. Mol. Cell Biol. 2008;28:4843–4850. doi: 10.1128/MCB.02214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paquet-Fifield S, et al. Vascular endothelial growth factor-d modulates caliber and function of initial lymphatics in the dermis. J. Invest. Dermatol. 2013;133:2074–2084. doi: 10.1038/jid.2013.83. [DOI] [PubMed] [Google Scholar]
- 24.Astin JW, et al. Vegfd can compensate for loss of Vegfc in zebrafish facial lymphatic sprouting. Development. 2014;141:2680–2690. doi: 10.1242/dev.106591. [DOI] [PubMed] [Google Scholar]
- 25.Bower NI, et al. Vegfd modulates both angiogenesis and lymphangiogenesis during zebrafish embryonic development. Development. 2017;144:507–518. doi: 10.1242/dev.146969. [DOI] [PubMed] [Google Scholar]
- 26.Duong T, et al. VEGFD regulates blood vascular development by modulating SOX18 activity. Blood. 2014;123:1102–1112. doi: 10.1182/blood-2013-04-495432. [DOI] [PubMed] [Google Scholar]
- 27.Kataru RP, et al. Critical role of CD11b+ macrophages and VEGF in inflammatory lymphangiogenesis, antigen clearance, and inflammation resolution. Blood. 2009;113:5650–5659. doi: 10.1182/blood-2008-09-176776. [DOI] [PubMed] [Google Scholar]
- 28.Cursiefen C, et al. VEGF-A stimulates lymphangiogenesis and hemangiogenesis in inflammatory neovascularization via macrophage recruitment. J. Clin. Invest. 2004;113:1040–1050. doi: 10.1172/JCI20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuda KS, et al. lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish. Development. 2012;139:2381–2391. doi: 10.1242/dev.077701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koltowska K, et al. Vegfc regulates bipotential precursor division and Prox1 expression to promote lymphatic identity in zebrafish. Cell Rep. 2015;13:1828–1841. doi: 10.1016/j.celrep.2015.10.055. [DOI] [PubMed] [Google Scholar]
- 31.Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- 32.Le Guen L, et al. Ccbe1 regulates Vegfc-mediated induction of Vegfr3 signaling during embryonic lymphangiogenesis. Development. 2014;141:1239–1249. doi: 10.1242/dev.100495. [DOI] [PubMed] [Google Scholar]
- 33.Villefranc JA, et al. A truncation allele in vascular endothelial growth factor c reveals distinct modes of signaling during lymphatic and vascular development. Development. 2013;140:1497–1506. doi: 10.1242/dev.084152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernardo BC, Weeks KL, Pretorius L, McMullen JR. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharm. Ther. 2010;128:191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Song HK, Hong SE, Kim T, Kim DH, Deep RNA. sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS ONE. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 38.Chablais F, Jazwinska A. Induction of myocardial infarction in adult zebrafish using cryoinjury. J. Vis. Exp. 2012;18:3666. doi: 10.3791/3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 40.Schnabel K, Wu CC, Kurth T, Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfefferli C, Jazwinska A. The art of fin regeneration in zebrafish. Regen. (Oxf.) 2015;2:72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chassot B, Pury D, Jazwinska A. Zebrafish fin regeneration after cryoinjury-induced tissue damage. Biol. Open. 2016;5:819–828. doi: 10.1242/bio.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hein SJ, et al. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS ONE. 2015;10:e0122665. doi: 10.1371/journal.pone.0122665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patek Paul R. The morphology of the lymphactics of the mammalian heart. American Journal of Anatomy. 1939;64(2):203–249. doi: 10.1002/aja.1000640202. [DOI] [Google Scholar]
- 45.Hu N, Yost HJ, Clark EB. Cardiac morphology and blood pressure in the adult zebrafish. Anat. Rec. 2001;264:1–12. doi: 10.1002/ar.1111. [DOI] [PubMed] [Google Scholar]
- 46.Vivien CJ, Hudson JE, Porrello ER. Evolution, comparative biology and ontogeny of vertebrate heart regeneration. NPJ Regen. Med. 2016;1:16012. doi: 10.1038/npjregenmed.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bower NI, Hogan BM. Brain drains: new insights into brain clearance pathways from lymphatic biology. J. Mol. Med. (Berl.) 2018;96:383–390. doi: 10.1007/s00109-018-1634-9. [DOI] [PubMed] [Google Scholar]
- 48.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makris S, Paulsen M, Johansson C, Type I. Interferons as regulators of lung inflammation. Front Immunol. 2017;8:259. doi: 10.3389/fimmu.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Y, et al. Genetic background, gender, age, body temperature, and arterial blood pH have a major impact on myocardial infarct size in the mouse and need to be carefully measured and/or taken into account: results of a comprehensive analysis of determinants of infarct size in 1,074 mice. Basic Res. Cardiol. 2012;107:288. doi: 10.1007/s00395-012-0288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grossman W, Paulus WJ. Myocardial stress and hypertrophy: a complex interface between biophysics and cardiac remodeling. J. Clin. Invest. 2013;123:3701–3703. doi: 10.1172/JCI69830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quaife-Ryan GA, et al. Multicellular transcriptional analysis of mammalian heart regeneration. Circulation. 2017;136:1123–1139. doi: 10.1161/CIRCULATIONAHA.117.028252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon JB. Lymphatic lipid transport: sewer or subway? Trends Endocrinol. Metab. 2010;21:480–487. doi: 10.1016/j.tem.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tham YK, et al. Lipidomic profiles of the heart and circulation in response to exercise versus cardiac pathology: a resource of potential biomarkers and drug targets. Cell Rep. 2018;24:2757–2772. doi: 10.1016/j.celrep.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 55.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samak M, et al. Cardiac hypertrophy: an introduction to molecular and cellular basis. Med. Sci. Monit. Basic Res. 2016;22:75–79. doi: 10.12659/MSMBR.900437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Linthout S, Tschope C. Inflammation—cause or consequence of heart failure or both? Curr. Heart Fail Rep. 2017;14:251–265. doi: 10.1007/s11897-017-0337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hogan BM, et al. Vegfc/Flt4 signalling is suppressed by Dll4 in developing zebrafish intersegmental arteries. Development. 2009;136:4001–4009. doi: 10.1242/dev.039990. [DOI] [PubMed] [Google Scholar]
- 60.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 63.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arganda-Carreras I, et al. Trainable Weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 65.Clough E, Barrett T. The gene expression omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets associated with the current study are available from the corresponding author on reasonable request. RNA-seq data have been deposited at the Gene Expression Omnibus under the accession number GSE133131.65
Automated quantification of cardiomyocyte volume was performed using ImageJ 1.47 software (National Institute of Health) with ImageJ Trainable Weka Segmentation macro.64 The code can be accessed via GitHub using the following link (https://github.com/Jacob-Smith-MCRI/Cardiomyocyte-Volume-Calculator).