Abstract
Obesity increases the risk of cancer. Increased levels of hormones (such as oestrogen, insulin, insulin-like growth factor, and leptin), free fatty acid-induced production of reactive oxygen species, an altered intestinal microbiome and chronic inflammation are known to be associated with an increased cancer risk in obese subjects. However, the mechanism underlying the connection between obesity and cancer development remains elusive. Here, we show that a high-fat diet (HFD) promotes tumour initiation/progression and induces a phenotypic switch from PD-1− CD8+ non-exhausted T cells to PD-1+ CD8+ exhausted T cells in a murine breast cancer model. While PD-1− CD8+ non-exhausted T cells predominated in the mammary glands of normal diet (ND)-fed mice, PD-1+ CD8+ exhausted T cells accumulated in the developing tumours of HFD-fed mice. Gene expression profiles indicated that PD-1+ CD8+ T cells expressed higher levels of the tumour-trophic gene Opn and lower levels of the cytotoxic genes Ifng and Gzmb than did PD-1− CD8+ T cells. Our study provides a possible mechanistic linkage between obesity and cancer.
Subject terms: Obesity, Cancer microenvironment
Introduction
The obesity epidemic is a worldwide problem. Obesity is linked to a number of complications, such as diabetes, dyslipidaemia, cardiovascular disease, kidney disease, nonalcoholic steatohepatitis, obstructive sleep apnoea and cancer1. Obesity increases the risk of several types of cancers, including breast cancer, colorectal cancer, liver cancer, and pancreatic cancer2–6. Previous studies have shown that obesity-associated cancer is linked to high levels of hormones (oestrogen, insulin, insulin-like growth factor and leptin), the induction of reactive oxygen species production by free fatty acids (which are derived from adipocytes), an altered intestinal microbiome and chronic inflammation in the tumour microenvironment2,7. However, the mechanism underlying the connection between obesity and cancer development remains elusive.
Chronic inflammation is associated with cancer development through tumour-promoting cytokines, including IL-1β, IL-6 and osteopontin (OPN)2,8. Although immune cells play a major role in chronic inflammation, the understanding of how obesity directly affects the tumour immune microenvironment is not complete. In adipose tissue, obesity induces a phenotypic switch in adipose tissue macrophages from M2-like macrophages (anti-inflammatory phenotype) to M1-like macrophages (proinflammatory phenotype)9. Macrophages have phenotypic diversity. Although the M1/M2 polarization model seems to be oversimplified, M1-like macrophages are considered to be anti-tumourigenic, while M2-like macrophages are considered to be pro-tumourigenic10. Immunomodulatory cells such as M2-like macrophages modify the tumour microenvironment by secreting immunosuppressive factors, angiogenic factors, and proinflammatory cytokines11–13. In chronic infectious diseases and cancers, CD8+ effector T cells gradually lose their ability to secrete IFNγ, IL-2, and TNFα due to continuous T-cell antigen receptor (TCR) stimulation by viral and tumour antigens14,15. These T cells are referred to as exhausted T cells, and they lack proliferative activity and cytotoxic functions16,17. Programmed death-1 (PD-1) is expressed by exhausted T cells16,17, and the presence of PD-1+ tumour-infiltrating lymphocytes is associated with a poor prognosis in human breast cancer18. An increase in the number of PD-1+ CD8+ T cells in tumours was shown to be associated with tumour progression in a murine MMTV-PyMT (PyMT) breast cancer model19. We therefore hypothesized that T cell exhaustion is involved in the mechanism underlying high-fat diet (HFD)-induced tumourigenesis, and we conducted this study to test our hypothesis.
Results
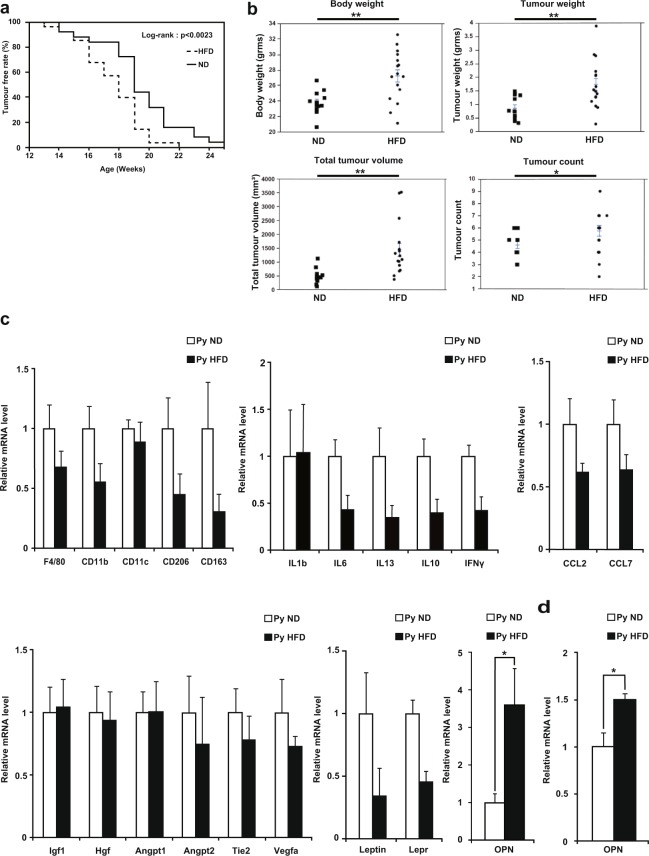
HFD promotes tumour initiation and progression
We used the murine PyMT breast cancer model to investigate the mechanism by which a HFD induces or promotes cancer development20,21. PyMT mice were fed either a normal diet (ND) or HFD, and the results showed that the HFD accelerated tumour initiation and was associated with increased tumour weights, tumour volumes, and tumour counts as well as elevated body weights (Fig. 1a,b). To clarify the mechanisms, we compared the gene expression profiles of the tumours of the ND-fed PyMT mice and those of the HFD-fed PyMT mice by performing quantitative RT-PCR (qPCR), but the results showed no significant differences in the expression of macrophage markers (F4/80, CD11b, CD11c, CD206, and CD163), proinflammatory cytokines (Il1b, Il6, Il10, and Ifng), angiogenic markers (Igf1, Hgf, Angpt1, Angpt2, Tie2, and Vegfa), myeloid chemoattractants (Ccl2 and Ccl7), Leptin, or Lepr between the two groups (Fig. 1c). Interestingly, the expression of Opn, which contributes to tumour growth via angiogenesis promotion22–26, apoptosis inhibition22,26–28, epithelial-mesenchymal transition (EMT) induction22,29,30, bone marrow-derived cell recruitment22,25,31, and cytotoxic CD8+ T cell suppression32, was significantly upregulated in the tumours of the HFD-fed PyMT mice (Fig. 1c). We then analysed Opn expression in fractions that had been depleted of adipocytes by centrifugation, leaving tumour cells and stromal vascular cells, and confirmed that Opn expression was upregulated in the tumours of the HFD-fed PyMT mice compared with the tumours of the ND-fed PyMT mice (Fig. 1d).
Figure 1.
High-fat diet promotes tumour initiation and progression. (a) Survival curves of tumour-free ND-fed PyMT mice (n = 25) and HFD-fed PyMT mice (n = 28). (b) Body weights, tumour weights, total tumour volumes, and tumour counts of ND-fed PyMT mice (n = 13) and HFD-fed PyMT mice (n = 16) at 20 weeks of age. (c) Quantitative RT-PCR analysis of the mRNA levels of macrophage markers, proinflammatory cytokines, angiogenic markers, myeloid chemoattractants, Opn, Leptin, and Lepr in tumours harvested from ND-fed PyMT mice and HFD-fed PyMT mice at 20 weeks of age (n = 4 per group). (d) Quantitative RT-PCR analysis of Opn mRNA expression in fractions of tumours from ND-fed PyMT mice and HFD-fed PyMT mice after depleting adipocytes by centrifugation (n = 3 per group). Error bars indicate the s.e.m. *P < 0.05 and **P < 0.01.
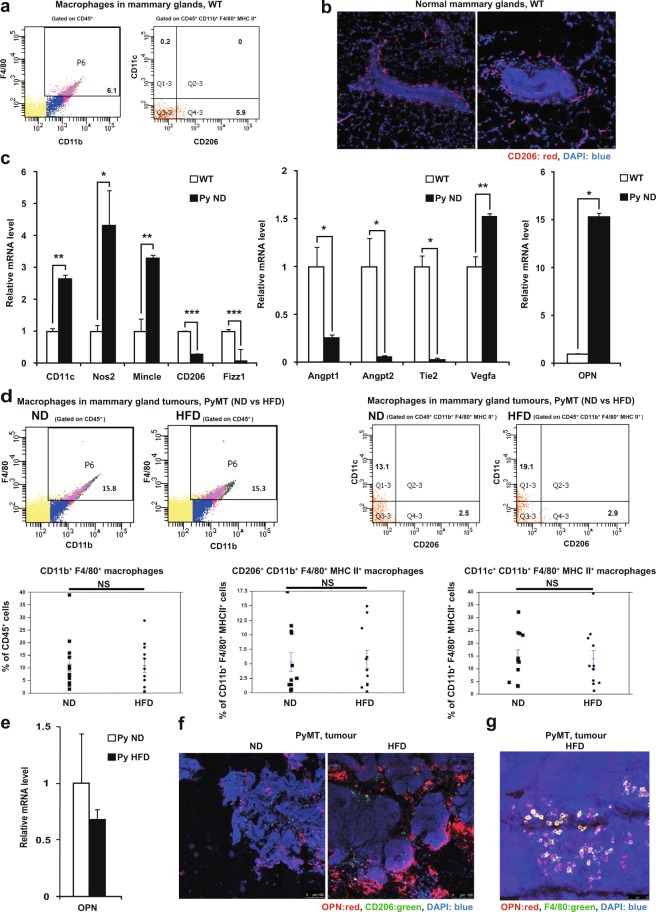
This finding was confirmed by an immunohistochemical analysis showing higher OPN expression in the tumours of the HFD-fed PyMT mice than in the tumours of the ND-fed PyMT mice (Fig. 2f). Since macrophages and T cells have been reported to be the major sources of OPN22,29, we focused our analyses on these two cell types.
Figure 2.
CD11c+ M1-like macrophages are the major macrophage population in tumours, while CD206+ M2-like macrophages are the major resident macrophage population in the mammary gland. (a) Flow cytometric analysis of macrophage markers in mammary glands from WT mice at 20 weeks of age (n = 4). (b) Representative CD206 (red) immunostaining with DAPI (blue) staining of mammary glands from WT mice. (c) Quantitative RT-PCR analysis of the mRNA levels of macrophage markers and angiogenic markers in F4/80+ macrophages from the mammary glands of WT mice and from the tumour tissue of ND-fed PyMT mice (n = 3 per group). (d) Flow cytometric analysis of macrophage marker expression in tumours from ND-fed PyMT mice (n = 12) and HFD-fed PyMT mice (n = 11) at 20 weeks of age. (e) Quantitative RT-PCR analysis of Opn mRNA expression in F4/80+ macrophages from the tumours of ND-fed PyMT mice and HFD-fed PyMT mice (n = 3 per group). (f) Representative CD206 (green) and OPN (red) immunostaining with DAPI (blue) staining of tumours from ND-fed PyMT mice and HFD-fed PyMT mice. (g) Representative F4/80 (green) and OPN (red) immunostaining with DAPI (blue) staining of tumours from HFD-fed PyMT mice. Error bars indicate the s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001; NS, not significant.
Macrophages in HFD-fed PyMT mice
First, we determined the phenotypes of macrophages in PyMT mouse tumours by comparing the phenotypes of these macrophages with those of macrophages in the normal mammary glands of wild-type (WT) mice. Flow cytometric analysis indicated that CD206+ M2-like macrophages were the major resident macrophage population in the normal mammary glands of the WT mice (Fig. 2a), whereas CD206− macrophages were the major macrophage population in the tumours of the PyMT mice (Fig. 2d). Immunohistochemical staining demonstrated that the CD206+ M2-like macrophages in the normal mammary glands were mainly localized around the mammary ducts (Fig. 2b).
We then examined the gene expression profiles of F4/80+ macrophages that had been isolated from the normal mammary glands of WT mice or the tumours of PyMT mice fed an ND by using magnetic beads. The F4/80+ macrophages isolated from the normal mammary glands of the WT mice showed high levels of angiogenic marker genes (Angpt1, Angpt2, and Tie2) and M2 marker genes (CD206 and Fizz1), whereas the F4/80+ macrophages from the tumours of the PyMT mice fed the ND showed high levels of Opn, Vegfa, and M1 marker genes (CD11c, Nos2, and Mincle) (Fig. 2c).
Next, we examined the effect of a HFD on changes in macrophage subtypes in tumours. Flow cytometric analysis revealed no significant difference in the number of F4/80+ macrophages between tumours in ND-fed PyMT mice and those in HFD-fed PyMT mice, and CD11c+ M1-like macrophages, not CD206+ M2-like macrophages, were the major macrophage subtype in the tumours in both the ND-fed PyMT mice and the HFD-fed PyMT mice (Fig. 2d). There were no significant differences in the proportions of CD11c+ M1-like and CD206+ M2-like macrophages between the tumours of the ND-fed PyMT mice and those of the HFD-fed PyMT mice (Fig. 2d).
We then compared the Opn expression levels in tumour macrophages from HFD-fed PyMT mice and ND-fed PyMT mice. F4/80+ macrophages were isolated by using magnetic beads, and an assessment of the Opn gene expression levels by qPCR did not show a significant difference between the groups (Fig. 2e).
When we examined OPN protein expression by immunohistochemical staining, tumours in HFD-fed PyMT mice showed higher OPN expression than those in ND-fed PyMT mice (Fig. 2f). Moreover, the majority of F4/80+ macrophages in the tumours of the HFD-fed PyMT mice expressed the OPN protein (Fig. 2g). Because the results showed that the HFD affected neither the Opn expression levels in F4/80+ macrophages nor the proportions of CD11c+ M1-like macrophages and CD206+ M2-like macrophages in the tumours, the marked induction of Opn expression in the HFD-fed PyMT mouse tumours (Fig. 1c) must be attributed to other cells, not macrophages.
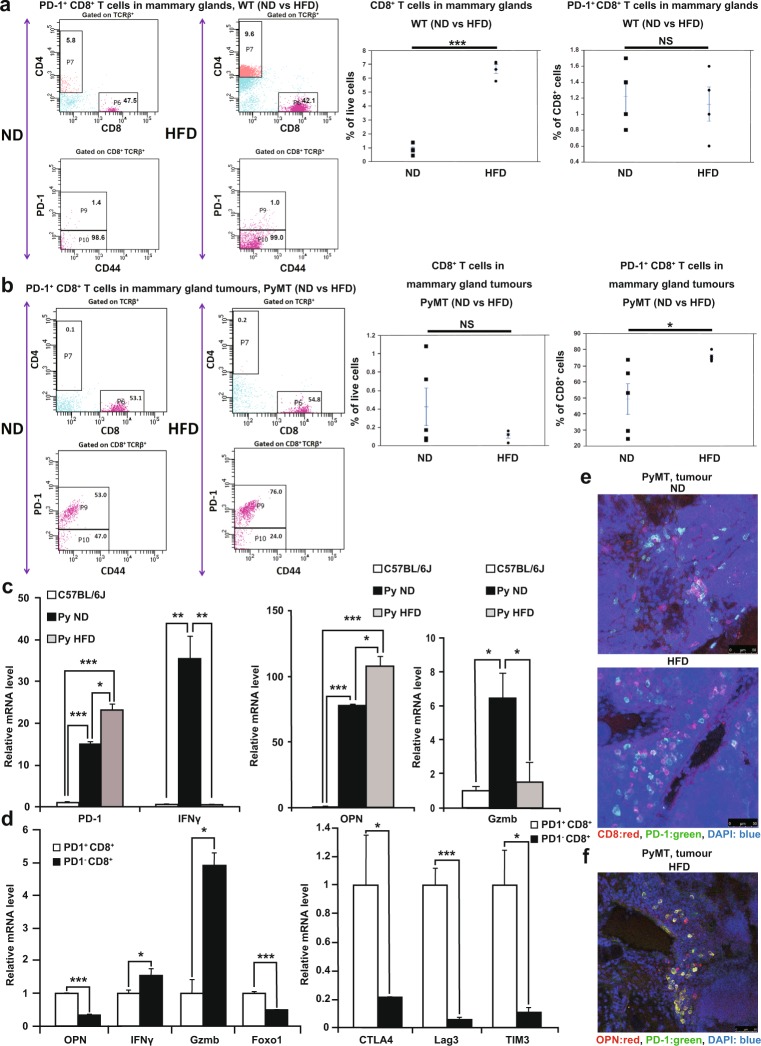
CD8+ T cells in HFD-fed PyMT mice
Next, we investigated the effect of a HFD on CD8+ T cells in the normal mammary glands of WT mice. We examined the expression of PD-1, a marker of exhaustion. Flow cytometric analysis showed that the majority of the CD8+ T cells in the normal mammary glands of ND-fed WT mice were PD-1− CD8+ T cells (Fig. 3a). The results showed that although the HFD induced a higher number of CD8+ T cells than did the ND in WT mice (Fig. 3a), the HFD did not affect the proportion of PD-1+ CD8+ T cells in the total CD8+ T cell population in the mammary glands (Fig. 3a).
Figure 3.
High-fat diet promotes the accumulation of PD-1+ CD8+ exhausted T cells. (a) Flow cytometric analysis of PD-1+ CD8+ T cells in mammary glands harvested from ND-fed WT mice and HFD-fed WT mice at 20 weeks of age (n = 4 per group). (b) Flow cytometric analysis of PD-1+ CD8+ T cells in tumours harvested from ND-fed PyMT mice and HFD-fed PyMT mice at 20 weeks of age (n = 5 per group). (c) Quantitative RT-PCR analysis of PD-1, Ifng, Opn and Gzmb mRNA expression in CD8+ T cells from the mammary glands of WT mice and from the tumour tissue of ND-fed PyMT mice and HFD-fed PyMT mice (n = 3 per group). (d) Quantitative RT-PCR analysis of Opn, Ifng, Gzmb, Foxo1, CTLA4, LAG3 and TIM3 mRNA expression in PD-1+ CD8+ T cells and PD1- CD8+ T cells from the tumour tissue of HFD-fed PyMT mice (n = 3 per group). (e) Representative PD-1 (green) and CD8 (red) immunostaining with DAPI (blue) staining of tumours from ND-fed PyMT mice and HFD-fed PyMT mice. (f) Representative PD-1 (green) and OPN (red) immunostaining with DAPI (blue) staining of tumours from HFD-fed PyMT mice. Error bars indicate the s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001; NS, not significant.
We then investigated Opn expression in CD8+ T cells in tumours. Flow cytometric analysis revealed no significant difference in the number of CD8+ T cells between ND-fed PyMT mice and HFD-fed PyMT mice (Fig. 3b). Interestingly, the tumours of the HFD-fed PyMT mice contained higher percentages of PD-1+ CD8+ T cells than the tumours of the ND-fed PyMT mice (Fig. 3b). We further analysed the phenotypes of CD8+ T cells isolated from tumours of ND-fed PyMT mice and HFD-fed PyMT mice by using magnetic beads. The results showed that the CD8+ T cells obtained from the tumours of the HFD-fed PyMT mice expressed the highest levels of PD-1 and Opn, the lowest levels of Ifng, and low levels of Gzmb (Fig. 3c). We then examined the gene expression profiles of PD-1+ CD8+ T cells and PD-1− CD8+ T cells that were isolated with magnetic beads. Thus, we were able to determine that PD-1+ CD8+ T cells exhibited the characteristics of “exhausted T cells”11,16,33, which express high levels of Opn, forkhead box protein O1 (Foxo1), and inhibitory receptor genes (CTLA4, LAG3 and TIM3) and low levels of Ifng and Gzmb (Fig. 3d and Extended data Fig. 1). Immunohistochemical staining confirmed that higher numbers of PD-1+ CD8+ exhausted T cells accumulated in the tumours of HFD-fed PyMT mice than in the tumours of ND-fed PyMT mice (Fig. 3e). Immunohistochemical staining also showed that PD-1+ cells in HFD-fed PyMT mice exhibited high levels of OPN expression (Fig. 3f). Thus, these findings suggested that the accumulation of PD-1+ CD8+ exhausted T cells contributed to HFD-induced tumourigenesis.
Discussion
In this study, we showed that CD206+ M2-like macrophages were the major resident macrophage population in the normal mammary gland and that CD11c+ M1-like macrophages, not CD206+ M2-like macrophages, were the major macrophage subtype in tumour tissue. These findings are consistent with those of a previous report that used the same model19. In addition, we found that obesity did not induce a phenotypic switch from M2-like macrophages to M1-like macrophages in tumours, but this switch has been shown to occur in adipose tissue macrophages9. Moreover, we showed that the levels of M2 markers (CD206 and Fizz1) were higher in normal mammary gland-resident macrophages than in tumour macrophages. Jäppinen et al. also reported CD206 as a useful marker of tissue-resident macrophages in the normal mammary gland34. The mechanism underlying why M2 markers are expressed at a high level in normal mammary gland-resident macrophages is uncertain. This is an important topic to be investigated.
Interestingly, we showed that a HFD promoted tumour initiation/progression and induced a phenotypic switch from PD-1− CD8+ T cells to PD-1+ CD8+ T cells in a murine breast cancer model. While PD-1− CD8+ non-exhausted T cells predominated in the mammary glands of ND-fed mice, PD-1+ CD8+ exhausted T cells accumulated in developing tumours in HFD-fed mice. Gene expression profiles indicated that PD-1+ CD8+ T cells expressed higher levels of Opn, a tumour-trophic gene, and lower levels of Ifng and Gzmb, cytotoxic genes, than PD-1− CD8+ T cells. Franklin et al. also reported that the increase in PD-1+ CD8+ T cell population and the decrease in PD-1− CD8+ T cell population were associated with tumour progression in the PyMT model19. Thus, the HFD-induced increase in PD-1+ CD8+ T cell population and decrease in PD-1− CD8+ T cell population can explain the HFD-induced tumour progression.
Chronic inflammation in the mammary glands may induce T cell exhaustion via cytokines that upregulate PD-1 expression (e.g., IL-2, IL-6, IL-7, IL-12, IL-15, IL-21, and VEGFa)17,35–37, and most of these cytokines are known to be associated with obesity-induced inflammation. Although further studies are needed, the cytokines that induce T cell exhaustion may be produced by multiple cell types, including adipocytes, fibroblasts, and immune cells. Moreover, we found that PD-1+ CD8+ T cells were a major source of OPN. OPN has a role in mediating tumour progression by regulating various pathways, such as angiogenesis promotion22–26, apoptosis inhibition22,26–28, EMT induction22,29,30, bone marrow-derived cell recruitment22,25,31, and cytotoxic CD8+ T cell suppression32. Although the mechanism involving OPN in tumour growth was unclear in the present study, a previous report using a PyMT-derived tumour cell line transplantation model indicated that OPN facilitated tumour growth29. Our study showed that OPN expression was elevated in tumours in HFD-fed PyMT mice, but this increased expression was not associated with angiogenesis promotion or bone marrow-derived cell recruitment. This discrepancy with previous studies may be caused by experimental differences, including the models and methods used. Although further research is needed, the expression of OPN by PD-1+ CD8+ T cells may be a therapeutic target in obesity-associated cancer.
Discrepancies between mouse and human immune systems are a problem in translating mouse data into the clinic. Therefore, we assessed the gene expression profiles of human breast carcinomas by searching a web-based cancer database (https://www.oncomine.org/, August 2015, Thermo Fisher Scientific, Ann Arbor, MI and http://kmplot.com/analysis/). Although the levels of CD11c (ITGAX), OPN (SPP1), and PD-1 (PDCD1) in human invasive breast carcinomas were significantly higher than those in normal human mammary glands (Extended Data Fig. 2a), there was no significant difference in CD206 (MRC1) expression between these two types of tissue (Extended Data Fig. 2a). There was a correlation between OPN expression levels in breast cancers and patient outcomes; high levels of OPN but not PD-1 correlated with poor outcomes (Extended Data Fig. 2b,c). These findings suggest that CD11c+ M1-like macrophages predominate over CD206+ M2-like macrophages in human breast cancers, that the number of PD-1+ CD8+ exhausted T cells increases in human breast cancers and that OPN is a prognostic marker in human breast cancer. In fact, previous studies have already shown that OPN is a prognostic marker in human breast cancer38–40.
In conclusion, our findings show that obesity promotes the accumulation of PD-1+ CD8+ exhausted T cells in tumours and that PD-1+ CD8+ T cells are a major source of OPN. Our study provides a possible mechanistic linkage between obesity and cancer.
Materials and Methods
Mice
MMTV-PyMT mice were purchased from The Jackson Laboratory (USA) and backcrossed onto the C57BL/6 J background. They were fed a standard diet containing 10% of calories from fat (Nosan Corporation, Yokohama, Japan) or a HFD containing 60% of calories from fat (D12492; Research Diets, NJ, USA) for 14–20 weeks beginning at the age of 6 weeks. Littermate controls were used in all experiments when possible. All the experiments were performed in accordance with the relevant guidelines and regulations and were approved by the Committee for Institutional Animal Care and Use at the University of Toyama (Toyama, Japan).
Tumour measurement
Tumours were measured with a calliper. Tumour volume was calculated by using the following formula: m1 × m1 × m2 × 0.5236, where m1 was the length of the shorter axis, and m2 was the length of the longer axis. Individual tumour volumes were added together to calculate the total tumour volume19,41.
Quantitative RT-PCR
Total RNA was extracted from tissue samples or cells by using an RNeasy Mini kit (Qiagen). cDNA was synthesized with multiscribe reverse transcriptase, and qPCR was performed using a Takara RNA PCR kit (Takara Bio) and the QuantiTect SYBR Green PCR Kit (Qiagen) as previously described42,43.
Flow cytometry
Tissues were excised from mice, minced and digested in Collagenase for 30 min. The samples were filtered through a 40-μm filter. Erythrocytes were removed with Lysing buffer (BD Biosciences). The samples were then incubated for 10 min with anti-CD16/CD32 blocking antibodies (BD Biosciences). The cells were stained with the following antibodies: anti-CD206 (C068C2, BioLegend), anti-CD11c (HL3, BD Biosciences), anti-CD11b (M1/70, BioLegend), anti-CD45 (30-F11, eBioscience), anti-F4/80 (CI: A3-1, BD Bioscience), anti-I-A/I-E (M5/114.15.2, BioLegend), anti-CD8a (53-6.7, BioLegend), anti-CD4 (GK1.5, BioLegend), anti-TCRβ (H57-597, BioLegend), anti-PD-1 (29F.1A12, BioLegend), and anti-CD44 (IM7, BioLegend). The samples were washed, incubated with 7-amino-actinomycin D (BD Biosciences) and then analysed on a FACSAria II (BD Biosciences).
Immunofluorescence
Immunostaining was performed with 10- to 15-μm-thick OCT frozen sections. The sections were permeabilized in a 0.3% Triton X-100/PBS solution and then incubated with primary antibodies diluted in 0.03% Triton X-100/PBS with 10% normal goat serum or 5% bovine serum albumin. The sections were washed, incubated with secondary antibodies and counterstained with DAPI. Imaging was performed using a Leica TCS SP5 confocal system (Leica Microsystems, Wetzlar, Germany).
Isolation of macrophages and CD8+ T cells by bead separation
Macrophages were purified by using an anti-F4/80 FITC-conjugated antibody and anti-FITC microbeads (Miltenyi Biotec) according to the manufacturer’s protocol. CD8+ T cells were purified by using an anti-CD8 APC-conjugated antibody and anti-APC microbeads (Miltenyi Biotec). PD1+ cells were purified by using an anti-PD-1 FITC-conjugated antibody and anti-FITC microbeads (Miltenyi Biotec).
Statistical analysis
Statistical analysis was performed with JMP11 software. The statistical significance of between-group differences was evaluated by using unpaired two-tailed Student’s t-tests. Differences among more than two groups were evaluated for statistical significance by ANOVA with the Tukey-Kramer HSD post hoc test for multiple comparisons. The results of survival studies were evaluated by Kaplan-Meier curves and related tests (log-rank and Wilcoxon tests). Data are expressed as the mean ± s.e.m., and P < 0.05 was considered evidence of statistical significance.
Supplementary information
Acknowledgements
We are grateful to H. Mori and S. Yamamoto for their technical support. We thanks J. Piao and K. Sugihara for their assistance with breeding and maintaining the mice used in this study. This work was supported by the Uehara Memorial Foundation, the Naito Foundation, and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21591126 to 26461327 K.T.).
Author Contributions
T.K. and K.T. designed and performed the experiments, and they analysed the data. N.A., A.T. and I.U. assisted in acquiring and analysing the data. T.K. and K.T. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48678-0.
References
- 1.Heymsfield SB, Wadden TA. Mechanisms, Pathophysiology, and Management of Obesity. N. Engl. J. Med. 2017;376:254–266. doi: 10.1056/NEJMc1701944. [DOI] [PubMed] [Google Scholar]
- 2.Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nat. Immunol. 2017;18:843–850. doi: 10.1038/ni.3754. [DOI] [PubMed] [Google Scholar]
- 3.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 4.Font-Burgada J, Sun B, Karin M. Obesity and Cancer: The Oil that Feeds the Flame. Cell Metab. 2016;23:48–62. doi: 10.1016/j.cmet.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer. 2011;11:886–895. doi: 10.1038/nrc3174. [DOI] [PubMed] [Google Scholar]
- 6.Park J, Morley TS, Kim M, Clegg DJ, Scherer PE. Obesity and cancer–mechanisms underlying tumour progression and recurrence. Nat Rev Endocrinol. 2014;10:455–465. doi: 10.1038/nrendo.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avgerinos KI, Spyrou N, Mantzoros CS, Dalamaga M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism. 2019;92:121–135. doi: 10.1016/j.metabol.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Deng T, Hsueh WA. Obesity, Inflammation, and Cancer. Annu Rev Pathol. 2016;11:421–449. doi: 10.1146/annurev-pathol-012615-044359. [DOI] [PubMed] [Google Scholar]
- 9.McNelis JC, Olefsky JM. Macrophages, Immunity, and Metabolic Disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Aras S, Zaidi MR. TAMeless traitors: macrophages in cancer progression and metastasis. Br. J. Cancer. 2017;117:1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahmar Q, et al. Tissue-resident versus monocyte-derived macrophages in the tumor microenvironment. Biochimica Et Biophysica Acta Bba - Rev Cancer. 2016;1865:23–34. doi: 10.1016/j.bbcan.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Eikawa S, et al. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proceedings of the National Academy of Sciences. 2015;112:1809–1814. doi: 10.1073/pnas.1417636112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wherry EJ. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 17.Chinai JM, et al. New immunotherapies targeting the PD-1 pathway. Trends Pharmacol. Sci. 2015;36:587–595. doi: 10.1016/j.tips.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muenst S, et al. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2013;139:667–676. doi: 10.1007/s10549-013-2581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/MCB.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castello LM, et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Mediators Inflamm. 2017;2017:4049098. doi: 10.1155/2017/4049098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty G, Jain S, Kundu GC. Osteopontin promotes vascular endothelial growth factor-dependent breast tumor growth and angiogenesis via autocrine and paracrine mechanisms. Cancer Res. 2008;68:152–161. doi: 10.1158/0008-5472.CAN-07-2126. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi F, et al. Osteopontin induces angiogenesis of murine neuroblastoma cells in mice. Int. J. Cancer. 2002;98:707–712. doi: 10.1002/ijc.10261. [DOI] [PubMed] [Google Scholar]
- 25.Anborgh PH, Mutrie JC, Tuck AB, Chambers AF. Role of the metastasis-promoting protein osteopontin in the tumour microenvironment. J. Cell. Mol. Med. 2010;14:2037–2044. doi: 10.1111/j.1582-4934.2010.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, et al. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 2018;9:356. doi: 10.1038/s41419-018-0391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S-H, et al. Suppression of osteopontin inhibits chemically induced hepatic carcinogenesis by induction of apoptosis in mice. Oncotarget. 2016;7:87219–87231. doi: 10.18632/oncotarget.13529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh YH, et al. Papilloma development is delayed in osteopontin-null mice: Implicating an antiapoptosis role for osteopontin. Cancer Res. 2006;66:7119–7127. doi: 10.1158/0008-5472.CAN-06-1002. [DOI] [PubMed] [Google Scholar]
- 29.Sharon Y, et al. Tumor-derived osteopontin reprograms normal mammary fibroblasts to promote inflammation and tumor growth in breast cancer. Cancer Res. 2015;75:963–973. doi: 10.1158/0008-5472.CAN-14-1990. [DOI] [PubMed] [Google Scholar]
- 30.Li NY, et al. Osteopontin up-regulates critical epithelial-mesenchymal transition transcription factors to induce an aggressive breast cancer phenotype. J. Am. Coll. Surg. 2013;217(discussion 26):17–26. doi: 10.1016/j.jamcollsurg.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 31.Atai NA, et al. Osteopontin is up-regulated and associated with neutrophil and macrophage infiltration in glioblastoma. Immunology. 2011;132:39–48. doi: 10.1111/j.1365-2567.2010.03335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klement JD, et al. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Invest. 2018;128:5549–5560. doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimatani K, Nakashima Y, Hattori M, Hamazaki Y, Minato N. PD-1+ memory phenotype CD4+ T cells expressing C/EBPα underlie T cell immunodepression in senescence and leukemia. Proceedings of the National Academy of Sciences. 2009;106:15807–15812. doi: 10.1073/pnas.0908805106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jäppinen, N. et al. Fetal-derived macrophages dominate in adult mammary glands. Nat. Commun. 10, 10.1038/s41467-018-08065-1 (2019). [DOI] [PMC free article] [PubMed]
- 35.Aguilar EG, Murphy WJ. Obesity induced T cell dysfunction and implications for cancer immunotherapy. Curr. Opin. Immunol. 2018;51:181–186. doi: 10.1016/j.coi.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palucka AK, Coussens LM. The Basis of Oncoimmunology. Cell. 2016;164:1233–1247. doi: 10.1016/j.cell.2016.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bengsch B, et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8(+) T Cell Exhaustion. Immunity. 2016;45:358–373. doi: 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y-Y, Zhang Y-Y, Lu W-F, Mi Y-J, Chen Y-Q. Prognostic value of osteopontin expression in breast cancer: A meta-analysis. Mol. Clin. Oncol. 2014;3:357–362. doi: 10.3892/mco.2014.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei R, Wong JPC, Kwok HF. Osteopontin - A promising biomarker for cancer therapy. J. Cancer. 2017;8:2173–2183. doi: 10.7150/jca.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patani N, Jouhra F, Jiang W, Mokbel K. Osteopontin expression profiles predict pathological and clinical outcome in breast cancer. Anticancer Res. 2008;28:4105–4110. [PubMed] [Google Scholar]
- 41.Mazzieri R, et al. Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells. Cancer Cell. 2011;19:512–526. doi: 10.1016/j.ccr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Kambara K, et al. In vivo depletion of CD206+ M2 macrophages exaggerates lung injury in endotoxemic mice. Am. J. Pathol. 2015;185:162–171. doi: 10.1016/j.ajpath.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Nawaz A, et al. CD206+ M2-like macrophages regulate systemic glucose metabolism by inhibiting proliferation of adipocyte progenitors. Nat Commun. 2017;8:286. doi: 10.1038/s41467-017-00231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.