Abstract
Selective gene delivery into subtypes of interneurons remains an important challenge in vector development. Adeno-associated virus (AAV) vector particles are especially promising for intracerebral injections. For cell entry, AAV2 particles are supposed to attach to heparan-sulfate proteoglycans (HSPGs) followed by endocytosis via the AAV receptor (AAVR). Here, we assessed engineered AAV particles deficient in HSPG attachment but competent in recognizing the glutamate receptor 4 (GluA4, also known as GluRD or GRIA4) through a displayed GluA4-specific DARPin (designed ankyrin repeat protein). When injected into the mouse brain, histological evaluation revealed that in various regions, more than 90% of the transduced cells were interneurons, mainly of the parvalbumin-positive subtype. Although part of the selectivity was mediated by the DARPin, the chosen spleen focus-forming virus (SFFV) promoter had contributed as well. Further analysis revealed that the DARPin mediated selective attachment to GluA4-positive cells, whereas gene delivery required expression of AAVR. Our data suggest that cell selectivity of AAV particles can be modified rationally and efficiently through DARPins, but expression of the AAV entry receptor remains essential.
Keywords: GluA4, AAV, interneuron, PV, parvalbumin, targeting, AAVR
Introduction
Interneurons form a diverse class of neurons that establish inhibitory circuits in the brain. They have been suggested as crucial players in a number of brain disorders including schizophrenia, epilepsy, autism, and intellectual disabilities.1, 2 Compared with excitatory neurons, they are less abundant (about 5-fold) and form synapses preferentially with local neurons, i.e., neurons of the network that they themselves reside in. Interneurons are often differentiated into four types based on the expression of particular proteins: parvalbumin (PV)-expressing interneurons, a heterogeneous population of somatostatin (SST)-expressing interneurons, the population of the ionotropic serotonin receptor 5HT3a (5HT3aR)-expressing interneurons, and neuron-derived neurotrophic factor-expressing neurons; together, these subtypes make up the mouse neocortical GABAergic interneuronal population.3 Beyond that, interneurons can be further divided into several subgroups.4, 5 The selective genetic manipulation of particular interneuronal subtypes is key to developing novel treatment strategies and to improving our understanding about fundamental processes in brain function. In mice, this task has been tackled by the generation of transgenic animals harboring specific promoter and enhancer elements that had been found active in distinct interneuronal subtypes. However, other vertebrate species are less amenable for this approach. Especially in non-human primates, high maintenance costs, long generation cycles, and ethical concerns are limiting factors.6, 7 Further, the selectivity mediated by these elements is often impaired or even lost when their size is reduced to become compatible with the packaging capacity of viral vectors. This holds especially true for adeno-associated virus (AAV) vector particles, which are preferred for intracerebral administrations because of their efficient spreading from the injection site.
Although some progress has been made in targeting neurons in a region-specific fashion with viral vectors, restricted gene delivery to defined neuronal subtypes is still scarce. A single publication described unmodified AAV particles derived from the AAV1 serotype that showed some preference for murine interneurons. This, however, was only the case when the particles were injected at a low dose.8 Although this approach has not been pursued by other scientists, it suggested that at least in principle interneuron targeting with AAVs may be possible. Important progress has then been made by inserting genetic elements into the AAV transfer vector. Restricted gene expression in GABAergic interneurons in various brain regions and different species was achieved with the enhancer elements of the distal-less homeobox family Dlx1/2 or Dlx5/6.9, 10 In a detargeting approach, the insertion of a series of microRNA (miRNA) target sites mediated the selective gene expression in GABAergic interneurons of the mouse cortex.11
An alternative strategy in targeted gene delivery is to modify the surface of viral vectors such that they attach selectively to the target cell population.12 The rational approach relies on displaying single-chain antibodies or designed ankyrin repeat proteins (DARPins) on the particle surface, thereby achieving attachment to a cell surface marker of choice with high affinity.13 This approach was initially described for lentiviral vectors.14 These were pseudotyped with paramyxoviral glycoproteins engineered such that usage of the natural entry receptors was replaced by the targeted surface protein.15 In an attempt to transfer this strategy to AAV vector particles, which are most commonly used for gene therapies and optogenetic studies, the AAV2 capsid proteins were mutated in residues R585A and R588A to destroy binding to heparan-sulfate proteoglycan (HSPG), the primary AAV2 attachment site,16, 17, 18 and fused to DARPins recognizing tumor surface antigens.19, 20 These AAV particles were highly selective for tumor cells expressing the targeted antigens even upon systemic injection into tumor-bearing mice.
In order to apply this approach for a targeted gene delivery into interneurons with AAV, we started out by targeting the glutamate receptor 4 (GluA4), which we had previously found to be expressed preferentially in PV-positive interneurons.21, 22 To generate GluA4-specific DARPins, we expressed and purified the extracellular amino-terminal domain (ATD) of murine GluA4 and used it as bait for several rounds of selection of a library covering more than 1013 variants of DARPins by ribosomal display. The ATD of GluA4 differs in only two amino acid positions between mouse, rat, marmoset, and human but exhibits only up to 62% sequence identity to the other glutamate receptor family members GluA1–3. The subsequent analysis of binders revealed DARPin 2K19 to be most efficient in mediating gene delivery by AAV into GluA4-expressing Chinese hamster ovary (CHO) cells. Parental CHO or CHO cells expressing GluA1 or GluA2 were not or were only minimally transduced, thus confirming the selectivity of the resulting GluA4-AAV.23
Here, we investigated the gene delivery activity of GluA4-AAV in vivo upon intracerebral injections into various brain regions. Our analysis revealed a high preference for interneurons, especially those being PV-positive. Besides attachment to GluA4, also promoter choice contributed to this selectivity. Gene transfer studies with cell lines being deficient for the recently identified AAV receptor (AAVR; also named KIAA0319L)24, 25 demonstrated the display of 2K19-mediated cell attachment, whereas AAVR expression was required for functional gene delivery.
Results
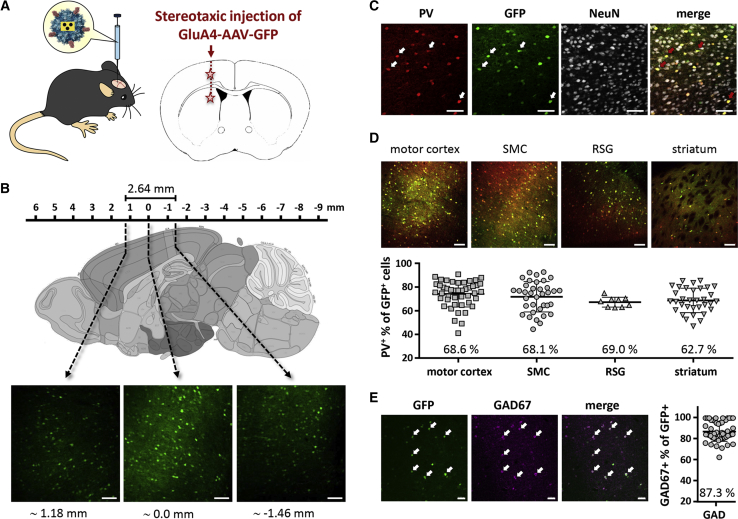
To evaluate the cell-type selectivity of GluA4-AAV in vivo, we packaged the reporter gene for the GFP under the control of the spleen focus-forming virus (SFFV) promoter into the particles, which were then injected stereotactically into two brain regions, allowing analysis of gene delivery into the motor cortex, the sensorimotor cortex (SMC), the retrosplenial granular (RSG) cortex, and the striatum (Figure 1A). Three weeks after injection, the analysis of consecutive brain slices revealed that the injected particles had spread from the injection site covering a distance of at least 2.6 mm (Figure 1B). To determine whether PV+ interneurons had been hit by the particles, the sections were co-stained for PV and the neuronal marker NeuN. Strikingly, almost every PV+ cell was GFP+, whereas only a small fraction of the NeuN+ cells was GFP+ (Figure 1C). This was valid for all four brain regions that were evaluated (Figure 1D). For quantification of the fraction of PV+ interneurons among the GFP+ cells, we chose a conservative approach taking only those events into account that were double positive without doubt. Accordingly, cells that gave signals only marginally above background or did not show up on at least two consecutive slices were not taken into account (Figure S1). The obtained average numbers for the different brain regions ranging between 63% and 69% thus represented minimum values rather underestimating the fraction of PV+/GFP+ interneurons (Figure 1D). To identify the source of the other GFP+ cells, we stained against the general interneuron marker GAD67. Almost 90% of the cells were on average also positive for GAD67, thus confirming the high selectivity of GluA4-AAV for interneurons (Figure 1E).
Figure 1.
Specific In Vivo Transduction of Interneurons with GluA4-AAV
(A) Schematic representation of GluA4-AAV particles and their stereotactic injection into the striatum and motor cortex of 4-week-old C57BL/6 mice. (B–E) Immunofluorescent staining of coronal brain sections from different brain regions 3 weeks after injection of GluA4-AAV-encoding GFP. (B) Representative sections stained with a GFP-specific antibody from three different cortex regions as depicted in the schematic sagittal plane (lateral 0.60 mm). Reference to bregma is given below the images. Scale bar, 100 μm. (C) Representative section of motor cortex stained with a GFP-specific antibody in combination with an antibody to PV and NeuN. Single-staining and merge images are shown. Arrows indicate exemplary cells positive for GFP and PV. Scale bar, 50 μm. (D) Selectivity of GluA4-AAV for PV-positive neurons in different brain regions. Representative merge images of sections from motor cortex, somatosensory cortex (SMC), retrosplenial granular cortex (RSG), and striatum stained with a GFP-specific antibody in combination with an antibody to PV (top panel). Scale bar, 100 μm. Amount in percent of GFP and PV double-positive cells from six different mice in the indicated brain region (bottom panel). For each region and mice, one to five different coronal sections were analyzed. From each section, up to four individual images were counted per region. One data point represents one image. Mean value and SD are shown. (E) Selectivity of GluA4-AAV for GAD67-positive neurons. Representative section of motor cortex stained with a GFP-specific antibody in combination with an antibody to GAD67. Single-staining and merge images are shown. Arrows indicate exemplary cells positive for GFP and GAD67 (left panel). Scale bar, 50 μm. Amount in percent of GFP and GAD67 double-positive cells from six different mice in combined cortex regions (motor cortex, SMC, and RSG; right panel). For each region and mice, one to five different coronal sections were analyzed. From each section, up to four individual images were counted per region. One data point represents one image. Mean value and SD are shown.
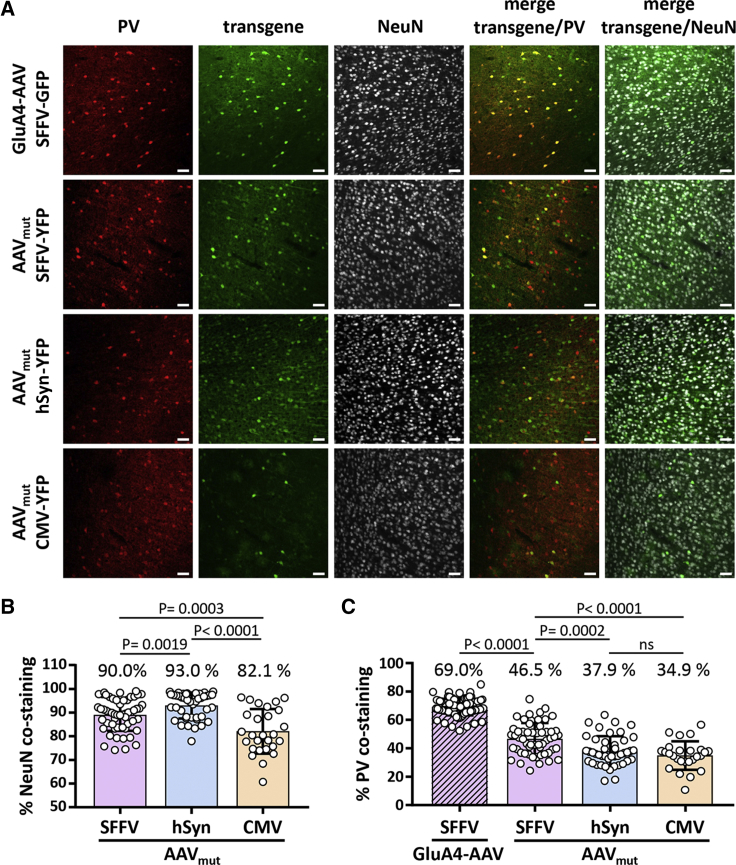
In order to assess the contribution of the GluA4-specific DARPin to the selectivity for PV+ interneurons, we generated AAV particles carrying the HSPG mutations but displaying no targeting ligand (AAVmut). Interestingly, these particles also exhibited some selectivity for interneurons upon injection into the cortex, indicating that the observed transduction profile was not solely due to the displayed GluA4-targeting ligand, but possibly the SFFV promoter used to drive the yellow fluorescent protein (YFP) reporter as well. To assess this systematically and in a quantitative way, we put the reporter gene under control of the human synapsin (hSyn) or the CMV promoter and packaged these constructs into AAVmut particles (Figure S2). These particles were then injected into the cortex along with GluA4-AAV. Brain slices were stained for transgene expression, as well as for PV and the neuronal cell marker NeuN (Figure 2A). Quantification of co-staining with NeuN revealed that the SFFV promoter was equally neuron specific as the hSyn promoter (Figure 2B). Both were significantly more selective for neuronal cells than the CMV promoter (Figure 2B). When quantifying PV+ co-staining, the SFFV promoter mediated indeed a significantly more selective YFP expression in these cells than the hSyn or CMV promoter (Figure 2C). This selectivity was substantially enhanced when the particles were displaying the GluA4-specific DARPin (Figure 2C). This analysis revealed that the SFFV promoter in GluA4-AAV had contributed to its selectivity for PV+ interneurons, whereas the main effect was mediated by the DARPin.
Figure 2.
Contribution of Promoter Usage to Interneuron Targeting
(A) Immunofluorescent staining of coronal brain sections from different brain regions 3 weeks after intracerebral injection of targeted (GluA4-AAV) or HSPG-blinded AAV (AAVmut) vector particles harboring an SFFV, hSyn, or CMV promoter (GluA4-AAV-SFFV-GFP, AAVmut-SFFV-YFP, AAVmut-hSyn-YFP, AAVmut-CMV-YFP), respectively. Single-staining and merge images of representative sections of cortex regions stained with a GFP-specific antibody in combination with an antibody to PV and NeuN are shown. Scale bar, 50 μm. Schematic drawings of the transgene cassettes used are provided in Figure S2. (B and C) Specificity of the indicated AAV vectors toward NeuN-positive (B) and PV-positive (C) neurons in the cortex. Percentage of GFP+/PV+ or GFP+/NeuN+ double-positive cells from at least two different mice, respectively. For each mouse, two to five different coronal sections were analyzed. From each section, up to eight individual images were counted. One data point represents one image. Mean value and SD are shown. Unpaired t test was performed to determine significance.
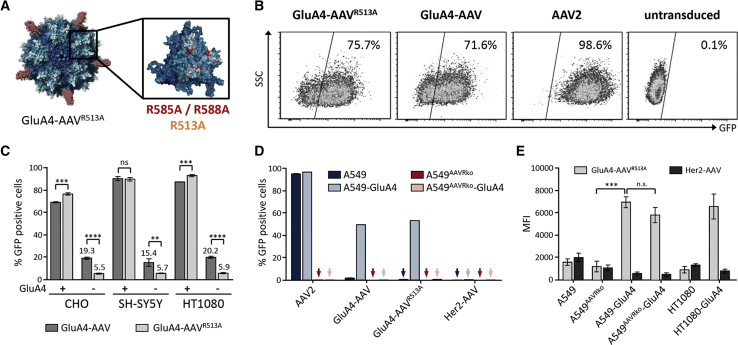
We next addressed whether the AAV secondary receptors α5β1 integrin and AAVR were involved in cell entry of GluA4-AAV and thus gene delivery. Because the capsid residue R513 mediates binding to α5β1,26 we exchanged this residue against alanine and generated the corresponding vector particles GluA4-AAVR513A (Figure 3A). These particles transduced GluA4-positive cells at least as efficiently as GluA4-AAV (Figure 3B), but exhibited an about 4-fold reduced off-target activity on GluA4-negative cells (Figure 3C; Figure S3). Notably, these cell lines were proven to express the integrin α5β1 on their surface (Figure S4A). Capsid mutations can impact assembly and/or stability of AAV particles. To address this issue, we analyzed biophysical properties of GluA4-AAVR513A and compared them with GluA4-AAV and AAV2 particles. Although particle numbers were similar for the three vector types, both DARPin-displaying particles contained about two times more empty particles (Figures S5A and S5B). This is well in line with previous reports about AAVs displaying DARPins with other specificity.19 Thus, mutating R513 to alanine did not impact capsid assembly. Although the thermostability of GluA4-AAVR513A was slightly reduced compared with its counterparts (Figure S5C), temperatures as high as 55°C were still tolerated, indicating that gene delivery with this vector under physiological conditions should proceed unimpaired.
Figure 3.
AAVR Is Essential for Entry of GluA4-AAV
(A) Model of a GluA4-AAVR513A vector particle is shown. DARPins (red) are extruding from pores at the 5-fold symmetry axis of the AAV capsid (left). Point mutations inserted in the capsid are shown in red and orange on the blow-up of the 3-fold axis (right). The illustration was created using Pymol. Adapted from Münch and colleagues.19 (B) GluA4-targeted vectors harboring two (R585A and R588A; GluA4-AAV) or three point mutations (R585A, R588A, and R513A; GluA4-AAVR513A) transduce GluA4-positive CHO-GluA4 cells. Cells were incubated with the indicated AAV vectors encoding GFP at a GOI of 1.25 × 106 (GluA4-AAV, GluA4-AAVR531A) or 1 × 104 (AAV2). Cells were analyzed 72 h after transduction by flow cytometry. Untransduced cells were used as control. (C) Reduced off-target transduction with GluA4-AAVR513A. GluA4-positive or -negative cells of the indicated cell lines were incubated with GluA4-AAVmut or GluA4-AAVmutR513A vectors encoding GFP at a GOI of 7.9 × 105 (CHO) or 2.6 × 105 (SH-SY5Y and HT1080). Cells were analyzed 72 h after transduction by flow cytometry. Each transduction experiment was performed in technical triplicates. The mean and SD are shown. ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05. ns, not significant by unpaired t test. Representative dot plots are shown in Figure S3. (D) AAV vectors transduce AAVR-positive cell lines only. AAVR-positive and -negative cell lines were incubated with the indicated vector stocks encoding GFP at a GOI of 4.9 × 105 (GluA4-AAV, GluA4-AAVR531A, Her2-AAV) or 1 × 104 (AAV2). Cells were analyzed 72 h after transduction by flow cytometry. Each transduction experiment was performed in technical triplicates. Mean value and SD are shown. (E) GluA4-AAV vector particles bind to AAVR-positive and -negative GluA4-expressing cells. The indicated cell lines were incubated with GluA4-AAVR513A at a GOI of 3 × 104 for 1 h at 4°C. Bound particles were stained with an AAV capsid-specific antibody in combination with a PE-conjugated secondary antibody. Mean PE fluorescence intensity (MFI) was measured via flow cytometry. An unrelated vector targeted to Her2 (Her2-AAV; GOI 8 × 104) was used as control. Each binding experiment was performed in technical triplicates. Mean value and SD are shown. ***p ≤ 0.001. ns, not significant by unpaired t test.
To assess the role of AAVR in the entry of GluA4-AAV, we made use of the cell line A549, which had been genetically engineered to destroy AAVR expression.24 We equipped the parental and the knockout (ko) line with GluA4 and verified the presence of GluA4 and absence of AAVR by flow cytometry or western blot analysis (Figures S4B and S4C). The absence of AAVR in knockout cell lines rendered these cells resistant to transduction, not only by AAV2, but also by GluA4-AAV. Expression of GluA4 did not change this picture (Figure 3D). However, the attachment of GluA4-AAV to GluA4-positive cells was equally efficient independently of whether these cells expressed AAVR or not (Figure 3E). Control AAV particles displaying a Her2/neu-specific DARPin did not bind to these cells (Figure 3E). Thus, GluA4-AAV used GluA4 as its primary attachment site on these cells. This was confirmed in experiments employing HT1080 and HT1080-GluA4 cells (Figure 3E), implying that AAVR was not involved in binding but rather mediated internalization and trafficking of GluA4-AAV.
Discussion
Here we provide proof-of-principle that surface engineering and attachment of vector particles to GluA4 result in selective gene delivery to interneurons. This is to our knowledge the first description of applying a rational design-based surface engineering strategy for interneuron targeting with AAV vectors. Especially PV+ interneurons were hit by GluA4-AAV, which is well in agreement with several independent studies confirming that GluA4 protein and mRNA are selectively upregulated in PV+ interneurons.22, 27, 28, 29 The selectivity of GluA4-AAV for interneurons appears to be well in the range of that observed with AAVs targeted to interneurons via enhancer9 or miRNA11 targeting strategies. Notably, both can be easily combined with GluA4-AAV by inserting these elements into the transfer vector plasmid. PV+ interneurons are central to the maintenance of an excitation-inhibition balance and to the generation of gamma-band rhythms, thereby playing an important role in several psychiatric diseases.4, 30, 31 Thus, a dysfunction of PV+ interneurons leads to de-synchronization of neuronal circuits in the brain and ultimately to behavioral deficits.32 Accordingly, GluA4-AAV can be regarded as a novel instrumental tool for studies into the pathogenicity and therapy of these disorders.
To characterize the entry properties of GluA4-AAV, we made use of information and tools available for the AAV secondary receptors α5β1 integrin and AAVR. Our data are in line with the view that α5β1 integrin serves as an attachment, but not as an entry receptor, because GluA4 particles, in which the contact residue R513 was mutated, were unaffected in gene delivery to GluA4-positive cells, but exhibited reduced off-target activity on GluA4-negative cells. Much stronger effects were caused by the presence or absence of AAVR on target cells. Although GluA4-AAV bound efficiently to AAVR-deficient cells via the displayed DARPin, gene transfer, and thus functional particle entry, was not detectable.
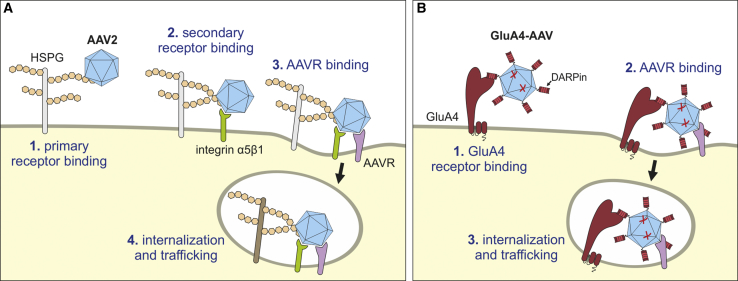
This strict dependency of GluA4-AAV on the presence of AAVR has multiple consequences and raises important questions for the cell entry mode of capsid engineered AAVs in general. First, GluA4-AAV can deliver genes to AAVR+ cells only. Although the high affinity of the DARPin attaches the particles preferentially to GluA4+ cells, only those cells that are also AAVR+ can express the transferred gene. Thus, GluA4 serves as attachment receptor, but not as entry receptor for GluA4-AAV. GluA4 thus replaces HSPG but cannot overcome the dependency on AAVR (Figure 4). Our data suggest that binding of AAV to AAVR has essential roles in AAV entry beyond cell attachment. Analogous to other well-characterized functional virus receptors, these roles could be triggering of endocytosis, facilitating intracellular trafficking, or inducing a conformational change in the viral particle to enable crossing of the endosomal membrane.24, 33 Second, AAV particles deficient in both HSPG and GluA4 binding still can deliver genes upon intracerebral injection. From our data, it cannot be distinguished whether these particles used another attachment receptor first or contacted AAVR immediately. Especially in a tissue as densely packed with cells as brain, the latter scenario may be a possibility. Histological data about the AAVR expression pattern in mouse and human brain will be instrumental to better predict and understand the biodistribution of these particles and of GluA4-AAV upon intracerebral injection.
Figure 4.
Model for the Surface Proteins Involved in Entry of GluA4-AAV
(A) Cell entry of AAV2. Cooperative binding of AAV particles to the primary attachment receptor HSPG and possibly also integrin α5β1 mediates contact to AAVR, which then leads to internalization and trafficking through the cytoplasm. (B) GluA4-AAV does not bind to HSPG because of the introduced capsid mutations (R585A and R588A; depicted as red crosses). Primary cell binding is mediated by the GluA4-receptor via the displayed DARPin. Afterward, cooperative binding to AAVR takes place leading to internalization and trafficking through the cytoplasm.
Receptor-targeted AAVs thus differ principally from their lentiviral vector-based counterparts, because the latter can be targeted to their actual entry receptor.34 Although the current study focuses on GluA4-AAV, it is likely that also other receptor-targeted AAVs displaying DARPins, like Her2-AAV, epithelial cell adhesion molecule (EpCAM)-AAV, and CD4-AAV, rely on AAVR expression in their entry modes. Beyond that, it will be important to address this issue also for capsid-engineered AAVs selected from AAV libraries. With the AAV2 capsid residues being in contact with AAVR having recently become available,35 it will now be possible to clarify whether AAVs can be engineered such that AAVR usage can be replaced by cell surface receptors of choice.
Materials and Methods
Plasmid Constructions
For generation of AAV transfer vector plasmids encoding the YFP under the control of either the promoter from the SFFV, the promoter of hSyn or the promoter of the cytomegalovirus (CMV) (Figure S2), we first exchanged the mouse APPsα coding sequence under the control of the hSyn promoter in pssAAV-hSyn-APPsa36 via BamHI and HindIII restriction cloning resulting in pssAAV-hSyn-eYFP (enhanced YFP). Afterward, the hSyn promoter was replaced by the SFFV promoter via PCR amplification (primers 2298 and 2723, template pSEW23) and subsequent restriction cloning with MulI and BamHI generating pssAAV-SFFV-eYFP. The CMV promoter was directly derived from pcDNA3.1(+) and inserted into pssAAV-hSyn-eYFP using the restriction sites MulI and BamHI, resulting in pssAAV-CMV-eYFP.
The AAV2 capsid residue R513 was mutated to alanine in the plasmid pRCVP2koA, encoding the rep and the mutated cap proteins of AAV-2 deficient for HSPG binding (R585A and R588A) and carrying a mutated VP2 start codon,19 and in pGluA4.2K19-VP2 encoding the GluA4 DARPin 2K19 in fusion with HSPG-blinded VP2 protein.23 In brief, a part of the coding sequences rep (primers 2195, 2198, 2199, and 2202) was amplified by overlapping size extension PCR introducing point mutation R513A. The generated fragment was inserted into pRCVP2koA and pGluA4.2K19-VP2 by sticky end ligation via the restriction sites BsiWI/XcmI, respectively, resulting in pRCVP2koA-R513A and pGluA4.2K19-VP2-R513A. All primer sequences for plasmid construction are provided in Table S1.
Cultivation and Generation of Cell Lines
HEK293T (ATCC CRL-11268), CHO-K1 (ATCC CCL-61), HT1080 (ATCC CCL-121), and A549 (ATCC CCL-185) cells were cultivated in DMEM (Sigma-Aldrich, Munich, Germany) supplemented with 10% fetal calf serum (FCS; Biochrom, Berlin, Germany) and 2 mM L-glutamine (Sigma-Aldrich, Munich, Germany). SH-SY5Y (ATCC CRL-2266) cells were cultivated in DMEM/F-12 Nutrient Mixture (Ratio 1:1; Thermo Fisher Scientific, Darmstadt, Germany) supplemented with 10% FCS and 2 mM L-glutamine. The cell lines CHO-GluA4 and HT1080-GluA4 were derived from CHO-K1 and HT1080 cells by lentiviral transduction, respectively,15, 23 and cultivated in the same medium in presence of 10 μg/mL puromycin (Thermo Fisher Scientific, Darmstadt, Germany). A549AAVRko cells were derived from A549 cells by CRISPR-Cas9 technology24 and cultivated in the same medium. SH-SY5Y-GluA4 cells, A549-GluA4 cells, and A549AAVRko-GluA4 were established by lentiviral transduction of the respective parental cell line with lentiviral particles having packaged the coding sequence for GluA4 with an N-terminal myc tag.15 Transduced cells were selected and cultured in media containing 2.5 μg/mL (A549-GluA4, A549AAVRko-GluA4) or 10 μg/mL (SH-SY5Y-GluA4) puromycin. Transgene expression was verified via flow cytometry analysis.
AAV Vector Production
GluA4-AAV and AAV2 vector particles were generated using the adenovirus-helper-free AAV-packaging strategy as described before using polyethylenimine (PEI).20, 37 In brief, 24 h prior to transfection, 2 × 107 HEK293T cells were seeded per T175 flask. On the day of transfection, the cell culture medium was replaced by 10 mL DMEM with 15% FCS and 2 mM L-glutamine (DMEM+FCS). For the transfection mix, 35 μg of total DNA was mixed with 2.3 mL of DMEM without additives and added to 2.2 mL DMEM supplemented with 140 μL of 18 mM PEI solution. After incubation for 20 min at room temperature, the transfection mix was added to the HEK293T cells. Forty-eight hours after transfection, cells were scraped off, pelleted (400 × g, 10 min, 4°C), and lysed using Tris-HCl NaCl (pH 8.5). After three freeze and thaw cycles in liquid nitrogen, cells were treated with Benzonase (50 U/mL cell lysate; Sigma-Aldrich, Hamburg, Germany) for 30 min at 37°C. The cleared lysates (3,700 × g, 20 min, 4°C) were purified by iodixanol gradient purification for 2 h at 290,000 × g in a Beckman 70Ti rotor, and AAV particles were harvested from the 40% iodixanol layer. For production of AAV2, HEK293T cells were transfected with plasmids pXX6-80,37 pRC,38 and the self-complementary transfer vector pscAAV-SFFV-GFP.20 For production of targeted AAVmut or AAVmutR531A vectors, the plasmid pRC was substituted by plasmid pRCVP2koA and pDARPin-VP2 (AAVmut) or pRCVP2koA-R513A and pDARPin-VP2-R531A (AAVmutR531A). For the production of non-targeted AAVmut vectors encoding YFP under the control of the SFFV, hSyn, or CMV promoter, the plasmids pXX6-80 and pRCVP2koA and one of the following single-stranded transfer vector plasmids were used: pssAAV-SFFV-eYFP, pssAAV-hSyn-eYFP, or pssAAV-CMV-eYFP. Plasmid ratios for the generation of AAV vectors are provided in Table S2. Genomic AAV vector titers were determined via free inverted terminal repeat (ITR)-specific qPCR as described before.39
Transduction of Cell Lines and Flow Cytometry Analysis
For transduction, 0.8–2.0 × 104 cells of indicated cell lines were seeded into a single well of a 96-well plate. Twenty-four hours later, the cell culture medium was exchanged by 50 μL AAV vector stock per well at a given genomic particle of infection (GOI). After 2 h, 150 μL cell culture medium was added per well. At day 3 post transduction, transduced cells were determined by flow cytometry analysis based on the indicated percentage of green fluorescent cells. Flow cytometry analysis was performed on the MACSQuant Analyzer 10 (Miltenyi Biotec, Bergisch Gladbach, Germany). Data were analyzed using FCS Express version 4.0 (DeNovo Software).
AAV Cell Binding Assay
A total of 5 × 104 cells were incubated with 1.5 × 109 particles of GluA4-AAVmutR513A or Her2-AAVmut for 1 h at 4°C. Afterwards, cells were washed twice with washing buffer (PBS, 2% FCS, 0.1% NaN3). Bound particles were subsequently stained by incubation of cells with an anti-AAV2 intact particle antibody (clone A20, diluted 1:20; Progen, Heidelberg, Germany) and a PE-conjugated anti-mouse antibody (diluted 1:50; Miltenyi Biotec, Bergisch-Gladbach, Germany). After each antibody incubation step, cells were washed twice with washing buffer. Bound vector particles were measured by flow cytometry analysis (MACSQuant Analyzer 10; Miltenyi Biotec, Bergisch Gladbach, Germany) detecting mean PE fluorescence intensity. Data were analyzed using FCS Express version 4.0 (DeNovo Software).
Intracerebral Injection of AAV Vectors
Stereotactic delivery of AAV vectors into the brain of mice has been approved by the responsible authorities. Injections into the cortex (coordinates: 0.5 mm anterior, 1.8 mm lateral, and 1.0–2.0 mm ventral to bregma) and striatum (coordinates: 0.5 mm anterior, 1.7 mm lateral, and 3.0–4.0 mm ventral to bregma) were performed in 4-week-old C57BL/6 mice as described previously.40, 41 Brains of mice were analyzed 3 weeks after intracerebral injection of AAV vectors.
Immunostainings and Microscopy
For immunohistochemistry after intracerebral injections, mice were transcardially perfused with 4% PFA in PBS. Brains were embedded in 4% agar in PBS and cut into 50- to 70-μm-thick coronal sections (Leica VT1200S vibratome). The sections were blocked and permeabilized either in 0.4% Triton X-100, 5% donkey serum, PBS (PV staining) or 0.05% Triton X-100, 5% donkey serum, and PBS (GAD67 staining) for 30–60 min at room temperature under slight agitation. Brain slices were stained with a primary antibody cocktail consisting of a chicken antibody to EGFP (1:1,000; Abcam), a guinea pig antibody to NeuN (1:1,000; Millipore), and a rabbit antibody to parvalbumin (1:1,000; Sigma) or a mouse antibody to GAD67 (1:500; Millipore) overnight at 4°C under slight agitation. On the next day, slices were washed three times with PBS, before being incubated with a secondary antibody cocktail consisting of an Alexa 488-conjugated anti-chicken antibody (1:250; Invitrogen), an Alexa 647-conjugated anti-guinea pig antibody (1:1,000; Invitrogen), and an Alexa 594-conjugated anti-rabbit antibody (1:250; Jackson) or an Alexa 594-conjugated anti-mouse antibody (1:1,000; Invitrogen) for 2–3 h at room temperature and slight agitation. Antibody cocktails were either prepared in 0.2% Triton X-100, PBS (PV staining), or PBS only (GAD67 staining). Brain slices were washed twice with PBS and mounted on coverslips. Slices were analyzed using a laser scanning fluorescence microscope (Zeiss). Images were acquired with an Optronics Magnafire Camera using Zeiss Zen software and a 16× (Uplan apo) objective lens. Image analysis was performed using Zeiss Zen software. Cell counting was performed using Cell Profiler software.
Statistical Analysis
Statistical analyses were performed with Prism 7 software (GraphPad). Tests for statistical significance used the unpaired two-tailed Student’s t test as indicated; p values less than 0.05 were considered significant.
Author Contributions
J.H., F.B.T., F.H., and T.K designed and performed experiments. J.H., F.B.T., and C.J.B. evaluated data. K.K., D.G.-G., H.M., H.B., J.E.C., I.D., and P.F. contributed protocols and reagents and to writing of the manuscript. C.J.B., I.D., H.M., and P.F. conceived and designed the study. C.J.B., F.B.T., and J.H. supervised work, acquired grants, and wrote the manuscript.
Conflicts of Interest
J.E.C. is a co-inventor on a Stanford-owned patent about the use of AAVR for modulating AAV infection. All other authors declare no competing interests.
Acknowledgments
This work was supported by grants from the LOEWE Center for Cell and Gene Therapy Frankfurt funded by the Hessische Ministerium für Wissenschaft und Kunst (HMWK; funding reference number III L 5-518/17.004 [2013] to J.H.) and the Deutsche Forschungsgemeinschaft SPP 1665 (BU1301/4-1 to C.J.B., FR2557/1-1 to P.F., and DI 1908/3-1 to I.D.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.07.004.
Supplemental Information
References
- 1.Marín O. Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 2.Pelkey K.A., Chittajallu R., Craig M.T., Tricoire L., Wester J.C., McBain C.J. Hippocampal GABAergic Inhibitory Interneurons. Physiol. Rev. 2017;97:1619–1747. doi: 10.1152/physrev.00007.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelsom C., Lu W. Development and specification of GABAergic cortical interneurons. Cell Biosci. 2013;3:19. doi: 10.1186/2045-3701-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudy B., Fishell G., Lee S., Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepecs A., Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park J.E., Silva A.C. Generation of genetically engineered non-human primate models of brain function and neurological disorders. Am. J. Primatol. 2019;81:e22931. doi: 10.1002/ajp.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deubner J., Coulon P., Diester I. Optogenetic approaches to study the mammalian brain. Curr. Opin. Struct. Biol. 2019;57:157–163. doi: 10.1016/j.sbi.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Nathanson J.L., Yanagawa Y., Obata K., Callaway E.M. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.-A., Guo L., Xu Q., Liu R., Lu C., Chu J. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee A.T., Vogt D., Rubenstein J.L., Sohal V.S. A class of GABAergic neurons in the prefrontal cortex sends long-range projections to the nucleus accumbens and elicits acute avoidance behavior. J. Neurosci. 2014;34:11519–11525. doi: 10.1523/JNEUROSCI.1157-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keaveney M.K., Tseng H.-A., Ta T.L., Gritton H.J., Man H.-Y., Han X. A MicroRNA-Based Gene-Targeting Tool for Virally Labeling Interneurons in the Rodent Cortex. Cell Rep. 2018;24:294–303. doi: 10.1016/j.celrep.2018.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm D., Büning H. Small But Increasingly Mighty: Latest Advances in AAV Vector Research, Design, and Evolution. Hum. Gene Ther. 2017;28:1075–1086. doi: 10.1089/hum.2017.172. [DOI] [PubMed] [Google Scholar]
- 13.Buchholz C.J., Friedel T., Büning H. Surface-Engineered Viral Vectors for Selective and Cell Type-Specific Gene Delivery. Trends Biotechnol. 2015;33:777–790. doi: 10.1016/j.tibtech.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Anliker B., Abel T., Kneissl S., Hlavaty J., Caputi A., Brynza J., Schneider I.C., Münch R.C., Petznek H., Kontermann R.E. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat. Methods. 2010;7:929–935. doi: 10.1038/nmeth.1514. [DOI] [PubMed] [Google Scholar]
- 15.Bender R.R., Muth A., Schneider I.C., Friedel T., Hartmann J., Plückthun A., Maisner A., Buchholz C.J. Receptor-Targeted Nipah Virus Glycoproteins Improve Cell-Type Selective Gene Delivery and Reveal a Preference for Membrane-Proximal Cell Attachment. PLoS Pathog. 2016;12:e1005641. doi: 10.1371/journal.ppat.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opie S.R., Warrington K.H., Jr., Agbandje-McKenna M., Zolotukhin S., Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kern A., Schmidt K., Leder C., Müller O.J., Wobus C.E., Bettinger K., Von der Lieth C.W., King J.A., Kleinschmidt J.A. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillay S., Carette J.E. Host determinants of adeno-associated viral vector entry. Curr. Opin. Virol. 2017;24:124–131. doi: 10.1016/j.coviro.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Münch R.C., Janicki H., Völker I., Rasbach A., Hallek M., Büning H., Buchholz C.J. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol. Ther. 2013;21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Münch R.C., Muth A., Muik A., Friedel T., Schmatz J., Dreier B., Trkola A., Plückthun A., Büning H., Buchholz C.J. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun. 2015;6:6246. doi: 10.1038/ncomms7246. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs E.C., Zivkovic A.R., Cunningham M.O., Middleton S., Lebeau F.E.N., Bannerman D.M., Rozov A., Whittington M.A., Traub R.D., Rawlins J.N., Monyer H. Recruitment of parvalbumin-positive interneurons determines hippocampal function and associated behavior. Neuron. 2007;53:591–604. doi: 10.1016/j.neuron.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 22.Geiger J.R., Melcher T., Koh D.S., Sakmann B., Seeburg P.H., Jonas P., Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 23.Hartmann J., Münch R.C., Freiling R.-T., Schneider I.C., Dreier B., Samukange W., Koch J., Seeger M.A., Plückthun A., Buchholz C.J. A Library-Based Screening Strategy for the Identification of DARPins as Ligands for Receptor-Targeted AAV and Lentiviral Vectors. Mol. Ther. Methods Clin. Dev. 2018;10:128–143. doi: 10.1016/j.omtm.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillay S., Meyer N.L., Puschnik A.S., Davulcu O., Diep J., Ishikawa Y., Jae L.T., Wosen J.E., Nagamine C.M., Chapman M.S., Carette J.E. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillay S., Zou W., Cheng F., Puschnik A.S., Meyer N.L., Ganaie S.S., Deng X., Wosen J.E., Davulcu O., Yan Z. Adeno-associated virus (AAV) serotypes have distinctive interactions with domains of the cellular AAV receptor. J. Virol. 2017;91 doi: 10.1128/JVI.00391-17. e00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asokan A., Hamra J.B., Govindasamy L., Agbandje-McKenna M., Samulski R.J. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J. Virol. 2006;80:8961–8969. doi: 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamasaki M., Fukaya M., Yamazaki M., Azechi H., Natsume R., Abe M., Sakimura K., Watanabe M. TARP γ-2 and γ-8 Differentially Control AMPAR Density Across Schaffer Collateral/Commissural Synapses in the Hippocampal CA1 Area. J. Neurosci. 2016;36:4296–4312. doi: 10.1523/JNEUROSCI.4178-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambolez B., Ropert N., Perrais D., Rossier J., Hestrin S. Correlation between kinetics and RNA splicing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors in neocortical neurons. Proc. Natl. Acad. Sci. USA. 1996;93:1797–1802. doi: 10.1073/pnas.93.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelkey K.A., Barksdale E., Craig M.T., Yuan X., Sukumaran M., Vargish G.A., Mitchell R.M., Wyeth M.S., Petralia R.S., Chittajallu R. Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron. 2015;85:1257–1272. doi: 10.1016/j.neuron.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferguson B.R., Gao W.-J. PV interneurons: critical regulators of E/I balance for prefrontal cortex-dependent behavior and psychiatric disorders. Front. Neural Circuits. 2018;12:37. doi: 10.3389/fncir.2018.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Magueresse C., Monyer H. GABAergic interneurons shape the functional maturation of the cortex. Neuron. 2013;77:388–405. doi: 10.1016/j.neuron.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Hamm J.P., Peterka D.S., Gogos J.A., Yuste R. Altered Cortical Ensembles in Mouse Models of Schizophrenia. Neuron. 2017;94:153–167.e8. doi: 10.1016/j.neuron.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wien M.W., Chow M., Hogle J.M. Poliovirus: new insights from an old paradigm. Structure. 1996;4:763–767. doi: 10.1016/s0969-2126(96)00082-2. [DOI] [PubMed] [Google Scholar]
- 34.Frank A.M., Buchholz C.J. Surface-engineered lentiviral vectors for selective gene transfer into subtypes of lymphocytes. Mol. Ther. Methods Clin. Dev. 2018;12:19–31. doi: 10.1016/j.omtm.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R., Cao L., Cui M., Sun Z., Hu M., Zhang R., Stuart W., Zhao X., Yang Z., Li X. Adeno-associated virus 2 bound to its cellular receptor AAVR. Nat. Microbiol. 2019;4:675–682. doi: 10.1038/s41564-018-0356-7. [DOI] [PubMed] [Google Scholar]
- 36.Fol R., Braudeau J., Ludewig S., Abel T., Weyer S.W., Roederer J.-P., Brod F., Audrain M., Bemelmans A.P., Buchholz C.J. Viral gene transfer of APPsα rescues synaptic failure in an Alzheimer’s disease mouse model. Acta Neuropathol. 2016;131:247–266. doi: 10.1007/s00401-015-1498-9. [DOI] [PubMed] [Google Scholar]
- 37.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girod A., Ried M., Wobus C., Lahm H., Leike K., Kleinschmidt J., Deléage G., Hallek M. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 1999;5:1052–1056. doi: 10.1038/12491. [DOI] [PubMed] [Google Scholar]
- 39.Muik A., Reul J., Friedel T., Muth A., Hartmann K.P., Schneider I.C., Münch R.C., Buchholz C.J. Covalent coupling of high-affinity ligands to the surface of viral vector particles by protein trans-splicing mediates cell type-specific gene transfer. Biomaterials. 2017;144:84–94. doi: 10.1016/j.biomaterials.2017.07.032. [DOI] [PubMed] [Google Scholar]
- 40.Liehl B., Hlavaty J., Moldzio R., Tonar Z., Unger H., Salmons B., Günzburg W.H., Renner M. Simian immunodeficiency virus vector pseudotypes differ in transduction efficiency and target cell specificity in brain. Gene Ther. 2007;14:1330–1343. doi: 10.1038/sj.gt.3302988. [DOI] [PubMed] [Google Scholar]
- 41.Cetin A., Komai S., Eliava M., Seeburg P.H., Osten P. Stereotaxic gene delivery in the rodent brain. Nat. Protoc. 2006;1:3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.