Abstract
Down-regulation or loss of MHC class I expression is a major mechanism used by cancer cells to evade immunosurveillance and increase their oncogenic potential. MHC I mediated antigen presentation is a complex regulatory process, controlled by antigen processing machinery (APM) dictating immune response. Transcriptional regulation of the APM that can modulate gene expression profile and their correlation to MHC I mediated antigen presentation in cancer cells remain enigmatic. Here, we reveal that Scaffold/Matrix-Associated Region 1- binding protein (SMAR1), positively regulates MHC I surface expression by down-regulating calnexin, an important component of antigen processing machinery (APM) in cancer cells. SMAR1, a bonafide MAR binding protein acts as a transcriptional repressor of several oncogenes. It is down-regulated in higher grades of cancers either through proteasomal degradation or through loss of heterozygosity (LOH) at the Chr.16q24.3 locus where the human homolog of SMAR1 (BANP) has been mapped. It binds to a short MAR region of the calnexin promoter forming a repressor complex in association with GATA2 and HDAC1. A reverse correlation between SMAR1 and calnexin was thus observed in SMAR1-LOH cells and also in tissues from breast cancer patients. To further extrapolate our findings, influenza A (H1N1) virus infection assay was performed. Upon viral infection, the levels of SMAR1 significantly increased resulting in reduced calnexin expression and increased MHC I presentation. Taken together, our observations establish that increased expression of SMAR1 in cancers can positively regulate MHC I surface expression thereby leading to higher chances of tumor regression and elimination of cancer cells.
Introduction
Oncogenic transformations occur by either activation of oncogenes or down-regulation of tumor suppressor genes. However, not all such incidences result in appearance of tumor mass. This is because of the ability of immune system to recognize and clear-off tumor antigens via MHC I mediated presentation to cytotoxic T-lymphocytes (CTLs) [1]. Cancer cells are known to deploy escape strategies which bypass the host immunosurveillance. Loss or down-regulation of MHC I expression associated with malignant transformation is a key feature of immune escape mechanism [2]. This decreased MHC I expression on cancer cell surface results in inefficient recognition by CTLs thereby favoring tumor progression [3].
Antigen processing and presentation by MHC I is a fine interplay of several components including the protein breakdown molecules, peptide transport machinery, chaperones like calreticulin and calnexin, protein trimming machinery and the structural components of MHC I molecule (HLA-B and β2M) forming the antigen processing machinery (APM) [4]. Proper functioning of all these components is necessary for antigen presentation and any alterations in these factors are directly associated with diminished or inefficient antigen presentation [5]. Several cancers both solid and hematological have been linked to APM dysfunction leading to down-regulation of MHC I expression and poor prognosis [6]. Regulation of the genes of APM and their effects on elimination of tumor cells is poorly understood. Our lab is working on a MAR binding protein SMAR1, established to have both tumor suppressor as well as immuno-modulatory functions [7], [8], [9], [10]. We speculated that apart from its tumor suppressor function, SMAR1 might also be involved in immunosurveillance of cancer cells by regulating MHC I.
SMAR1 gene was mapped at 16q24.3 loci of chromosome 16 of mice and this region also codes for many other tumor suppressors [11]. LOH of this locus has already been reported in hepatocellular, prostate, breast, head and neck cancers [12]. SMAR1 has been shown to be down regulated in higher grades of cancer either through Cdc20 mediated proteasomal degradation or through LOH at the Chr.16q24.3 locus where the human homolog of SMAR1 (BANP) has been mapped [13], [14]. It is known to coordinate with p53 for modulating expression of various genes that decide cell fate under various pathophysiological conditions [9]. It acts as tumor suppressor by repressing cyclinD1 expression and arresting cells in G1 phase [15]. SMAR1 is also known to stabilize p53 by preventing its MDM2 mediated degradation [16]. Reports have further implicated its role as a stress responsive protein as evident from regulation of Bax and Puma under genotoxic conditions [9]. Owing to its ability to regulate diverse set of proteins and modulate various functions, a high throughput proteomic profiling was carried out in colorectal carcinoma cells after knocking down SMAR1. Interestingly, calnexin, a component of the antigen processing machinery was observed to be one of the up-regulated proteins in SMAR1 knockdown condition.
Calnexin is an ER resident protein with calcium binding ability. It has known functions in glycoprotein folding and maturation [17], [18], [19]. Cumulative evidences indicate the implication of calnexin in apoptosis induced by ER stress. Calnexin gene silencing in lung cancer cell line was shown to decrease cancer cell survival leading to effective chemotherapy [20]. In addition, serum calnexin was earlier reported as early diagnostic marker in lung cancer and as prognostic marker for colorectal cancer [20], [21]. Calnexin is also known to induce impairment of proliferation and effector functions of CD4+ and CD8+ T cells, thereby promoting tumor growth [22]. It is therefore evident that higher calnexin expression can lead to altered antigen presentation and cancer immune response. However, there is a lacunae in the understanding of regulation of calnexin gene expression, though some evidences support that hyperactivity of EGFR pathway and ER stress up-regulates calnexin expression in cancer cells [23], [24].
Herein, we establish that SMAR1 regulates MHC I processing and antigen presentation by suppressing calnexin gene expression in cancer cells. SMAR1 along with GATA2 and HDAC1 forms a repressor complex which binds to the MAR site on the calnexin promoter thereby repressing its transcription. Breast cancer cell lines as well as cancer patient samples revealed an inverse correlation between SMAR1 and calnexin. Higher calnexin expression is directly related to lower MHC I expression and cancer progression. Our in-vivo results also indicate that SMAR1 and MHC I mediated antigen presentation has positive correlation leading to tumor regression. Apart from repression of calnexin, SMAR1 also acts through p53 axis by up-regulating ERAP1 expression which is a known p53 target protein and positively regulate MHC I. This study highlights the role of tumor suppressor SMAR1 in regulation of calnexin and MHC I expression and ultimately antigen presentation by cancer cells.
Experimental Procedures
Cell Lines and Cell Culture
HCT116 p53+/+ and HCT116 p53−/− were kind gifts from Dr. Bert Vogelstein, John Hopkins University. MO4 cells were kind gift from Dr. Kenneth Rock, UMass Medical School. HEK293T, MCF7, NIH3T3, B16F10, A549, MDA-MB231 and T47D were obtained from NCCS cell repository. HEK293T, HCT116 p53+/+, HCT116 p53−/−, MO4, NIH3T3, B16F10, A549, cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 1x Pen Strep antibiotic (1X, Gibco) at 37 °C with 5% CO2. T47D was cultured in RPMI (Invitrogen) and MDA-MB231 was cultured in L15 (Invitrogen) under similar conditions. These cultured cells were used for further experiments.
Plasmids, Transfections and Lenti/Adenoviral Transductions
Qiagen Midiprep kit was used to prepare the plasmids for transfection experiments. Lipofectamine-2000 in OptiMEM without FBS was used for carrying out transfection in cells. Plasmid constructs used for overexpression were GFP-SMAR1, flag-SMAR1, flag-HDAC1, myc-GATA1, myc-GATA2. GFP-vector and Flag vector were used as control. Sh1077 and lentiviral shRNA were used for knockdown of SMAR1. Lentiviral shRNA for BANP was purchased from Thermo Scientific (USA) and packaged and transduced as described by Nakka et al. [30] Replication deficient recombinant SMAR1 adenovirus Ad-SMAR1 was generated and target cells were transduced for 72 hours to achieve maximum SMAR1 overexpression.
Influenza A Virus Experiment
The pandemic influenza A(H1N1) pdm09 virus (A/India/Jln_NIV9436/2009) used in this study was isolated at NIV, Pune [46] (GenBank accession nos: HM204573, HM241701–07). The stock of the virus was prepared by propagating the virus in embryonated chicken eggs in Biosafety level-2 (BSL-2) laboratory [48]. In brief, the allantoic cavity of 10-day-old chicken embryos were inoculated with the virus and the allantoic fluid was harvested 72 h post inoculation. The virus titer was determined by hemagglutination (HA) assay. Pooled samples with HA titers of ≥64 were used to make the virus stock and stored at −80 °C. The same stock of viruses was used for all the experiments. The infectious virus titer was determined with the help of plaque assay [65].
RNA Extraction and quantitative Real-Time PCR
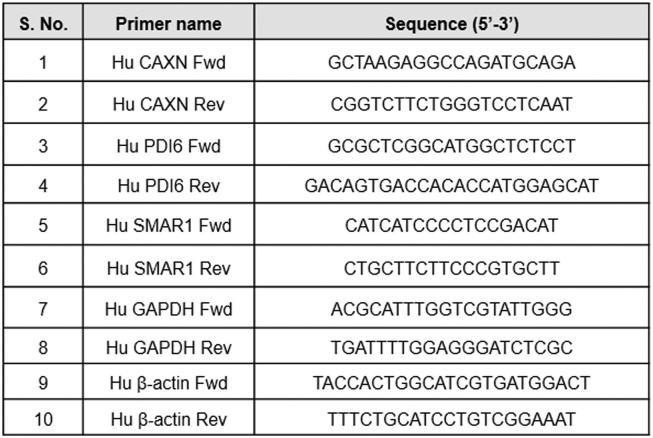
Total RNA was extracted from cells or tissues using TRIzol reagent according to manufacturer's (Invitrogen, USA) protocol. 2 μg of this RNA was used for cDNA synthesis. For quantitative analysis of expression of genes in this study, Real Time PCR was performed in Realplex (Ependorf) and StepOne™ (Applied Biosystems) Real-Time PCR Systems. In a 10 μl PCR reaction, cDNA was amplified using Power SYBR® Green PCR Master Mix. List of primers and their sequences are given in Supplementary table 2.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was done using HCT116 p53+/+ cells as described earlier (9) using a chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology) following the manufacturer's instructions. PCR was performed using promoter specific primers. Chromatin immunoprecipitation (ChIP) was carried out using anti-SMAR1 (Bethyl Laboratories), anti-HDAC1 (Cell Signaling), anti-GATA2, anti-p300 (Santacruz), anti-H4k20m (Cell Signaling), anti-h3k9 acetyl (Cell Signaling), anti-H3k27me3(Cell Signaling) and normal IgG control antibody (Santacruz). MAR-Wiz (http://genomecluster.secs.oakland.edu/MarWiz/) was used for determining MAR potential of the sequence of interest. The following parameters were used: Window width- 1000, Slide distance- 100, Cut-off threshold- 0.60 and Run length- 3. Primers used for amplifying potential matrix attachment (mar) region in calnexin promoter are forward 5′ACAGAGCAAGACTTCGTCTC3′, reverse ′5CAGCGAATGTATCACTGATCT3′.
Luciferase Reporter Assay
A 1.5 kb region of calnexin promoter was cloned in pGL3 enhancer vector. This segment was PCR amplified from genomic DNA isolated from HCT116 p53+/+ cells using promoter specific primers. Forward primer used was ‘5CTGGGTACCCATCAAGTAAGTCTATAAGC3’ and Reverse was ‘5GTCAAGCTTATGGCTCGGCCCAGGAGCCT-3’. It was then cloned in pGL3 enhancer vector using Kpn I and Hind III restriction sites and referred as calnexin promoter construct in this study. Co-transfection of cells was carried out by Flag vector, Flag SMAR1 and HDAC1 along with this Luc construct and luciferase activities were measured after 48 hours of transfection. Luciferase assay kit (Promega) was used to quantitate the luciferase activity on Fluoroskan Ascent Luminometer (Lab Systems).
Electrophoretic Mobility Shift Assay (EMSA)
A 100 bp long probe complementary to Calnexin promoter MAR region was used for studying protein-DNA interaction. Purified recombinant SMAR1 protein in increasing concentrations with poly (dI-dC) (Poly(deoxyinosinic-deoxycytidylic)) from (Sigma) at 1 mg/ml was incubated with the probe at room temperature for 1 hour in 50 mM Tris buffer (pH 7.5); 8% native-PAGE was pre-run at 100 V constant voltage at 40 °C for 1 hour. Samples were then run on this gel. The gel was stained in 0.5 μg/ml ethidium bromide solution, rinsed with water and visualized on Versadoc (Bio-Rad).
Western Blot Analysis
Levels of desired proteins were determined by immunoblotting. Total protein was extracted using protein extraction buffer (20 mM Tris–HCl pH- 7.8, 1 mM EDTA, 1 mM PMSF, 0.1% (v/v) Triton X 100, PI cocktail). It was then quantified using Bradford's reagent (BioRad) and equal amount was loaded on 8% or 10% SDS polyacrylamide gels. Proteins were electro transferred to PVDF membrane (Amersham) at 100 V constant voltage; blots were saturated with 5% (w/v) BSA, followed by incubation with specific primary antibodies, and further incubated with respective secondary antibodies tagged with horseradish peroxidase. Signals were detected by chemiluminescence using ECL Chemiluminescence substrate (Thermo Scientific). Immunoblotting was used to determine levels of SMAR1 (Bethyl Laboratories), Calnexin (BD), β-actin (Sigma), GATA2 (Abcam), myc-tag (Sigma), p53 (Santacruz), HLA ABC (Abcam) and HDAC1 (Cell signaling).
Coimmunoprecipitation and Immunoblot Analysis
Protein–protein interaction between target proteins was checked using coimmunoprecipitation studies. Cells were lysed in TNN buffer (50 mM Tris-Cl pH 7.5, 1% NP-40, 150 mM NaCl, and 1 mM DTT) supplemented with protease-inhibitor cocktail (Roche). The lysate obtained was diluted in 1XPBS containing 0.3–0.5% NP40 IP buffer. Preclearing was done with control IgG and 15 μl of protein A/G beads for 1 h at 4 °C. The precleared lysates were further incubated with specific antibodies overnight at 4 °C with constant rotation. Antibody bound complexes were then pulled down with protein A/G beads. Non-specific binding was removed by washing them five times in IP buffer. Elution of bound proteins from A/G beads was done using 2X SDS sample buffer and detection was done by immunoblotting (as described above) with specific antibody. The antibodies used were SMAR1, HDAC1 and GATA2.
Immunofluorescence Analysis
1*104 cells were seeded on sterilized glass coverslips. Twenty-four hours post seeding, the cells were washed with cold 1X PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. This was followed by quenching with 0.1% (W/V) glycine in 1X PBS. 0.1% Triton-X 100 was used for permeabilization and 10% FCS in PBS for blocking. After blocking, cells were incubated with specific primary antibodies. Primary antibody (1:150 in blocking solution) was added, followed by specific fluorophore-tagged secondary antibodies (1:1000 in blocking solution). The cells were then washed, and coverslips were mounted in Fluoroshield with DAPI (Sigma) and examined under confocal laser microscope (LSM 510 version 2.01; Zeiss). Antibodies used were SMAR1, calnexin, HLA ABC. Secondary antibodies used are Cy3 and Alexa 488 labeled. Influenza A (H1N1) pdm09 virus infection in A549 cells was confirmed by immunofluorescence assay (IFA) against the nucleoprotein (NP) of influenza (MAB8257, Millipore, USA) as described by Thube et al., 2018 [65]. Blocks of tumor and normal breast tissue samples were archival materials provided by department of pathology, Netaji Subhas Chandra Bose Cancer Hospital and Research Institute, Kolkata, India. Tissues were fixed with 10% formalin, paraffin embedded and cut into 5-micron sections. Sections were deparaffinized and antigen retrieval was done by boiling slides in sodium citrate buffer, pH 6. Sections were stained with hematoxylin and eosin (H&E stain) or with SMAR1, MHC I antibody or calnexin antibody. Next day slides were washed thrice with 1XPBST and incubated with anti-rabbit Alexa 488 and anti-mouse Alexa 647 conjugated secondary antibodies for 2 hr. at room temperature. Nuclei were visualized by Hoechst 33258. Slides were mounted with Prolong Gold antifade. Sections were observed at 20X magnification using Leica TCS-SP8 Confocal microscope (Leica Microsystems, Wetzlar, Germany).
Flow Cytometry
Cells were trypsinized using 0.05% trypsin–EDTA (Life technologies), washed with PBS and resuspended in labeling buffer (PBS supplemented with 2% FBS) to a final concentration of 1*106 cells per mL. They were stained with HLA ABC monoclonal antibody (BD Biosciences, USA), H-2Kb antibody (BioLegend, USA) and OVA257–264-H-2Kb antibody (BioLegend, USA) at 4 °C for 1 hour. Cells were washed after each incubation and resuspended in 0.5 mL of labeling buffer for flow cytometry using Calibur flow cytometer (Becton Dickinson). The results were analyzed and MFI was calculated by Cell Quest software.
Peritoneal Macrophage Isolation
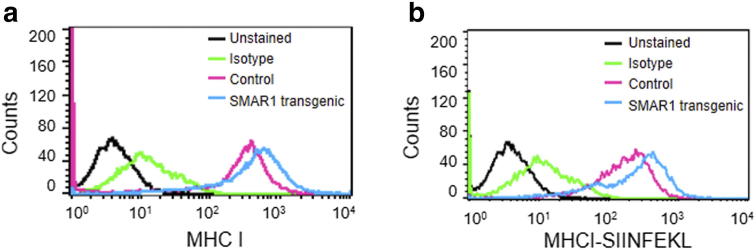
Peritoneal macrophages were isolated from SMAR1 transgenic mice and their control littermates and treated with MHC I peptide SIINFEKL. Antigen presentation of SIINFEKL peptide on MHC I was quantitated by FACS using an antibody that recognizes SIINFEKL bound to H-2Kb MHC I.
Two-Dimension Gel Electrophoresis and MALDI
Protein samples were mixed with rehydration buffer (7 M Urea, 2 M thiourea, 2% CHAPS, 0.8% ampholyte, 0.02% bromophenol blue, and 20 mM diththiothreitol [DTT]), loaded onto 24 cm immobilized pH 4–7 gradient (IPG) strips and focused with Ettan IPGphor 3 isoelectric focusing unit (GE). The IPG strips were then equilibrated in equilibration buffer I [0.38 M Tris-base, pH 8.8, 6 M urea, 2% (w/v) SDS and 20% (v/v) glycerol] and equilibration II [2.5% (w/v) iodoacetamide with above mentioned buffer] for 10 min each and 2% (w/v) DTT in 5 mL/strip. After equilibration, each IPG strips were loaded onto the 12% polyacrylamide gel and sealed with 1% agarose in tris-glycine buffer containing bromophenol blue. The gels were electrophoresed at 200 V for 6 to 8 h, using ETTAN DALT6 (GE Healthcare) in a buffer containing 25 mM Tris, 190 mM glycine and 0.1% SDS. After electrophoresis, proteins were stained and visualized using Coomassie brilliant blue dye. Stained gels were scanned with Image Scanner (GE Healthcare) and IMP7 (GE Healthcare) was used for spot analysis and the proteins of interest were marked for excision. The gel plugs were transferred directly to 1 ml Eppendorf tubes. In-gel digestion with trypsin (Promega) was done to extract the peptides from the cut spots. Tryptic peptides were further purified by C-18 ZipTip. After ZipTip purification, the tryptic peptides were eluted from the ZipTip with 50% ACN/0.1% formic acid into 500 μl eppendorf tubes. Eluted proteins were further spotted onto a wax-coated matrix-assisted laser desorption/ionization (MALDI) plates for protein identification (Applied Biosystems, Framingham, MA).
In-Vivo Tumorigenicity and Imaging
All mice used in the animal experiments were bred at animal resource facility of National Centre for Cell Science (NCCS), Pune, India. Standard protocols approved and monitored by institutional animal ethical committee of NCCS were followed for all experiments. Control, Lenti-SMAR1 and Adeno-SMAR1 transduced MO4/luc/ova cells (1*106) in sterile 1× PBS were injected subcutaneously in the right flank of C57 BL/6 mice and tumor progression was studied by weekly measuring its size for 3 weeks post injection. Localization and/or metastasis of these injected cells were observed by in vivo imaging after anesthetizing the mice and injecting them with luciferin (PerkinElmer) substrate. In-vivo imaging system (Xenogen) was used for imaging animals.
ELISA
Blood was collected from the retro-orbital sinus at day 10 and day 21 post tumor cell injection. All sera were collected and stored at -80°C for ELISA analysis of the cytokines IFN-γ, IL-4, IL-2 and IL-6. Serum concentrations were assayed by sandwich ELISA. IL-4 and IL-2 were measured using (DuoSet kit, R&D Systems, Minneapolis, MN, USA) while IL-6 and IFNγ were assayed using BD ELISA kit according to the respective manufacturer's protocol.
Modeling and docking studies
To study the atomic minutiae of interaction pattern of SMAR1-GATA2-HDAC1 complex, structure of SMAR1 and GATA2 were build and further all were docked together to get an understanding of their interaction pattern. Full length SMAR1 (NP_001167010.1) and GATA2 (NP_001139133.1) structure was built with I-TASSER web-server [66] and the predicted structure was further prepared and minimized using Schrodinger PRIME module [67]. They were further simulated in Desmond module [68] for 100 ns. The X-Ray Crystallographic structure of HDAC1 protein (PDB ID – 4BKX) was already predicted, so the coordinate file was downloaded from RCSB PDB site and was further processed in Protein Preparation Wizard [69]. All the missing hydrogen along with missing side chain was added and then subjected to energy minimization by OPLS-2005 force field. Now to analyze the stable non-covalent interaction between SMAR1, GATA2 and HDAC1, we performed docking through ZDOCK (v–3.0.2) server [70]. ZDOCK is a Fast Fourier Transform based protein docking platform which searches all possible binding modes in the translational and rotational space among the two proteins and assesses each pose using an energy-based scoring function. First, SMAR1 and GATA2 were docked and then the complex was further docked with HDAC1. The final predicted docked structure was viewed and analyzed using PyMOL [71].
Survival Curve Analysis
Correlation of relapse free survival of breast cancer patients (n = 1660) with calnexin gene expression was analyzed using Kaplan–Meier Plotter (http://kmplot.com/analysis/).
Statistical Analysis
All the experiments were performed at least three times and the data are expressed as mean ± standard deviation. Statistical analysis was performed by the Student's t-test to determine significant difference. Differences at P ≤ .05/0.005 level were considered statistically significant.
Results
Proteomic Profiling of SMAR1 Regulated Gene by 2D Gel Electrophoresis
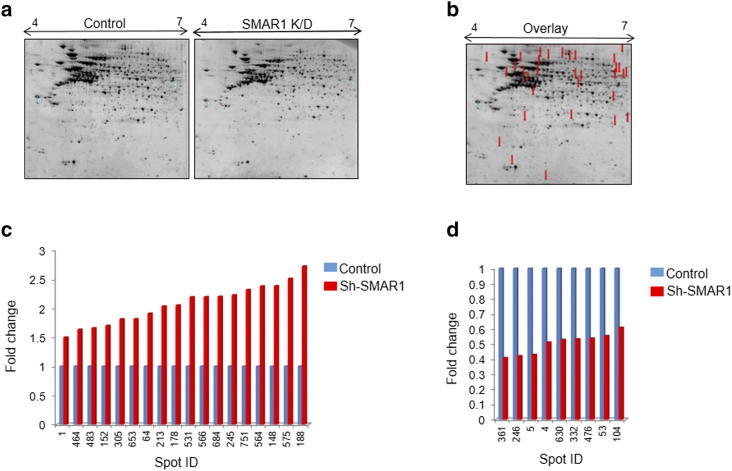
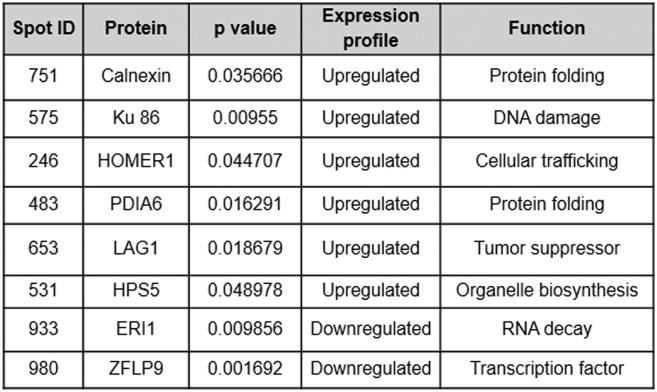
SMAR1 is a MAR binding protein and a vital component of the nuclear matrix which controls cellular homeostasis. SMAR1 is known to be down-regulated in various grades of breast cancers [25] and its expression is directly correlated with the survival of patients and disease outcome [26]. Loss of heterozygosity at the SMAR1 loci and Cdc20 mediated proteasomal degradation are main reason behind its dampened expression in cancer cells [12], [13], [14]. To identify the SMAR1 target genes involved in immune evasion of cancer cells, proteomic profiling was carried out in SMAR1 knockdown scenario. For this, total protein from control HCT116 p53+/+ cells and sh-SMAR1 transfected HCT116 p53+/+ cells were subjected to 2D gel electrophoresis (2DE) (Supplementary Figure 1A). In the three match sets (pH 4–7), up to 1200 protein spots were separated and detected by IMP7 software (GE Healthcare) analysis (data not shown). Of these, we found 41 protein spots with statistically significant differences in expression levels (P ≤ .05) (Supplementary Figure 1B). Statistical significance was defined as a minimum 1.5-fold increase or decrease in expression level. SMAR1 knockdown resulted in both up-regulation as well as down-regulation of proteins and interestingly some protein spots were present only in control cells. These results suggest that SMAR1 can act as a transcriptional repressor for some proteins and it may be required for the stable expression of others. Among these differentially expressed protein spots under SMAR1 knockdown condition, 18 spots were found to be up regulated (Supplementary Figure 1C), 9 spots were down regulated (Supplementary Figure 1D) and 16 spots were present only in control cells (data not shown). Identified protein spots are given in the Table (Supplementary Table 1). The notable ones of these are calnexin and PDIA6 which have well-known functions in protein folding. Other significant proteins include Ku 86, HOMER1, LAG1, HPS5, ERI1 and ZFLP9.
SMAR1 Regulates Calnexin Gene Expression
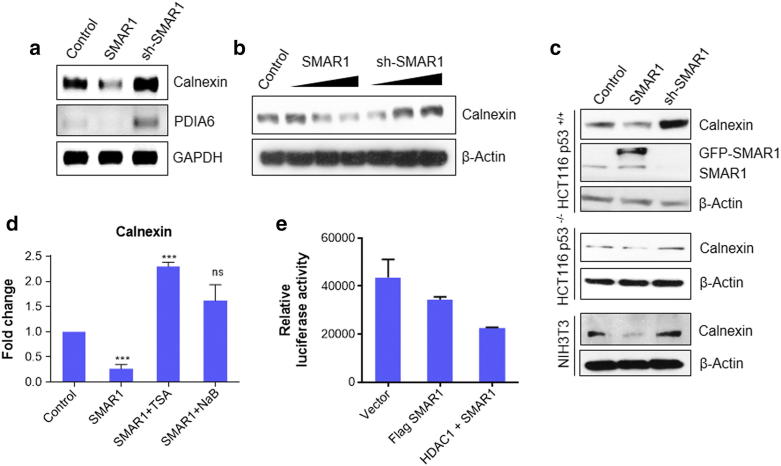
Out of all the differentially regulated proteins identified, calnexin showed highest up-regulation. It is known to have significant role in MHC I processing and presentation [27]. This led us to speculate SMAR1 may be directly involved in immune evasion mechanisms by regulating calnexin expression. To verify the 2DE results, semi-quantitative PCR analysis of calnexin and PDIA6 in HCT116 p53+/+ cells was performed. The results were mostly in agreement with the 2DE data. Both calnexin and PDIA6 were found to be prominently up regulated after SMAR1 knockdown whereas SMAR1 over-expression resulted in their down-regulation (Figure 1a). Further, to check the effect of SMAR1 on calnexin protein expression, we overexpressed and down-regulated SMAR1 in a dose dependent manner in HEK293T cells and performed western blot. This experiment confirmed our findings and SMAR1 levels were found to be inversely correlated with calnexin levels (Figure 1b). Next, we sought to check whether this phenomenon is p53 dependent. For this purpose, we used HCT116 p53+/+ and HCT116 p53−/− cells and interestingly found that SMAR1 can regulate calnexin in a p53 independent manner (Figure 1c). We further confirmed these results in NIH3T3 cell lines to confirm that our findings are not cell line specific. Similar trend was observed in NIH3T3 cells (Figure 1c). SMAR1 is known to form a repressor complex with HDAC1. In order to understand if HDAC1 is involved in calnexin gene regulation, we used trichostatin A (TSA) and sodium butyrate (NaB) which are general inhibitors of HDAC. SMAR1 overexpression was found to repress calnexin gene expression but overexpression of SMAR1 along with TSA or NaB treatment did not show any effect. (Figure 1d). Further to understand the transcriptional regulation of calnexin by SMAR1 and HDAC1, luciferase assay was performed on calnexin promoter. It was observed that there is a decrease in luciferase activity when SMAR1 was overexpressed and this repression was further enhanced when both SMAR1 and HDAC1 were overexpressed together (Figure 1e). This result confirms that HDAC1 along with SMAR1, plays an important role in calnexin gene regulation.
Figure 1.
SMAR1 regulates calnexin gene expression. (a) Semi-quantitative PCR to check the expression of calnexin, PDIA6 in HCT116 p53+/+ cells transfected with SMAR1 overexpression and sh-SMAR1 constructs. GAPDH was used as the loading control. (b) HEK293T cells were transfected with GFP-SMAR1 and sh-SMAR1 in a dose dependent manner. 2 μg, 4 μg and 6 μg of DNA was used for transfection. β-Actin was used as loading control (c) Western blotting of different cell lines transfected with GFP-vector, GFP-SMAR1 and sh-SMAR1 to check the expression profile of calnexin. Cell lines used were HCT116 p53+/+, HCT116 p53−/− and NIH3T3 cells. (d) Real-time PCR to check the gene regulation of calnexin by SMAR1. HCT116 p53+/+ cells were transfected with GFP-SMAR1 and treated with either 100 nM trichostatin A (TSA) or 1 mM sodium butyrate (NaB), the general inhibitors of histone deacytylase enzyme. (e) Luciferase assay was done in HEK293T cells transfected with pGL3-enhancer-calnexin promoter, Flag-SMAR1, and Flag-HDAC1. Cells were harvested after 48 h. Protein was extracted and luciferase assay was performed using the standard protocol from promega. (Bars represent standard deviation from three independent experiments. */** indicate statistically significant difference at P ≤ .05/0.005, respectively).
Although some evidences indicate that calnexin expression is modulated under various physiological conditions, but there is a dearth of literature showing direct calnexin gene regulation. Some reports link increased calnexin expression to ER stress and propose it as one of the ER stress responsive protein [28]. One report shows that tunicamycin, one of the ER stress inducer increases the expression of calnexin in MCF7 cells [29].
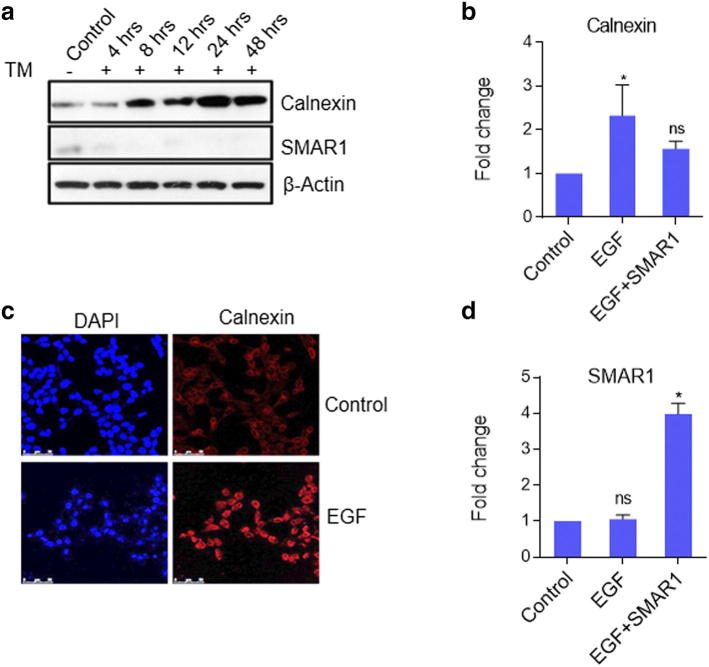
To check whether SMAR1 and calnexin expression have any correlation, 5 mM tunicamycin treatment was given to MCF7 cells in a time dependent manner and their expression kinetics was checked till 24 h. It was observed that calnexin expression increased at 4 h time point and kept on increasing with time whereas SMAR1 expression decreased significantly and gradually became negligible with time (Supplementary Figure 2A). This result implicated that under ER stress condition also SMAR1 and calnexin show negative correlation, supporting our earlier observation that SMAR1 negatively regulates calnexin expression. Further literature review revealed that Heregulin-β1 (HRG) regulates calnexin expression. Heregulin-β1 (HRG), a secretory polypeptide is known to up regulate calnexin gene expression by activating HER3 and HER4 signaling [23]. There are multiple EGF family-specific ligands which include EGF that can bind to HER1 and regulate downstream signaling of EGFR pathway. One report from our lab has shown that upon EGF treatment, SMAR1 gets phosphorylated and translocates from the nucleus to cytoplasm [30]. We therefore predicted that calnexin expression might be regulated by EGFR signaling following the translocation of SMAR1 to cytoplasm. EGF treatment leads to increased expression of calnexin both at RNA and protein levels (Supplementary Figure 2, B and C) whereas when SMAR1 is overexpressed along with EGF treatment, calnexin expression was not increased. Also, we checked SMAR1 expression upon EGF treatment as a control experiment. SMAR1 expression remained unperturbed upon EGF treatment while its expression increased upon SMAR1 transfection and EGF treatment (Supplementary Figure 2D). This suggests that EGF does not play any role in SMAR1 transcription and so increased calnexin expression is a result of SMAR1 cytoplasmic translocation as previously discussed.
SMAR1, HDAC1, and GATA2 Form Repressor Complex to Suppress Calnexin Expression
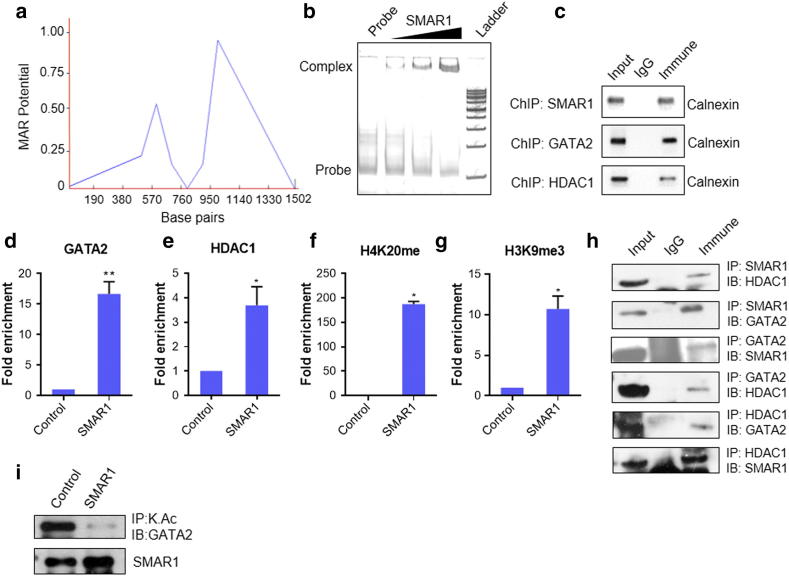
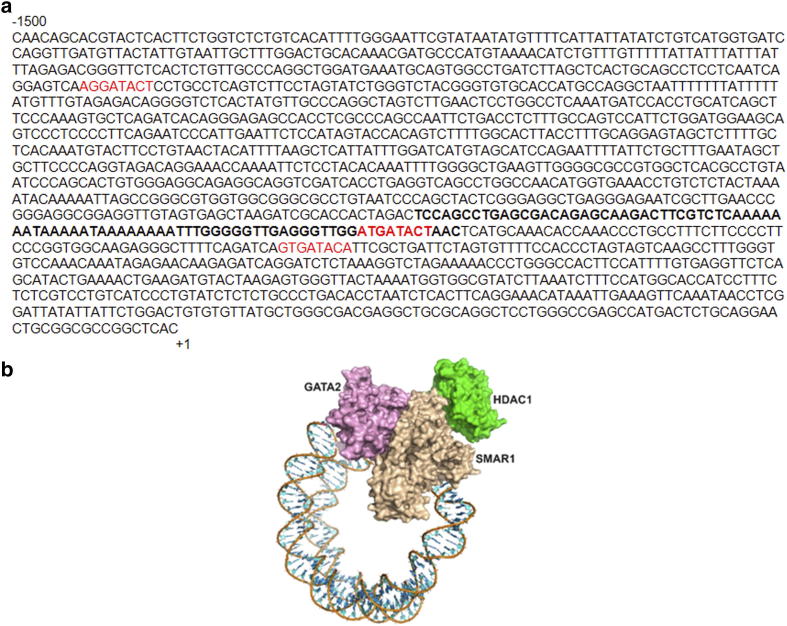
SMAR1 is a matrix attachment region binding protein (MARBP) and it binds to MAR sites throughout the genome. MAR region in calnexin promoter was analyzed using MAR-wiz software. It marked the presence of a potential MAR site at 550 bp upstream of the TSS on calnexin promoter (Figure 2A). SMAR1 is expected to bind to this region and suppresses the expression of calnexin under various physiological conditions. We know that SMAR1 binds to promoters of many genes and represses their transcription along with other factors. In-silico analysis was carried out to analyze other transcription factors binding to calnexin promoter. We found a GATA1 binding site proximal to SMAR1 binding site on calnexin promoter (Supplementary Figure 3a). However, non-hematopoietic cells do not express GATA1 [31]. Literature review suggested that GATA2 is dysregulated in cancer and associated with poor prognosis [32], [33]. Interestingly GATA2 competes with GATA1 for similar sequence binding [34]. So, we further studied SMAR1 and GATA2 binding for calnexin promoter regulation. EMSA and chromatin immunoprecipitation confirms the binding of SMAR1 and GATA2 on calnexin promoter (Figure 2, b and c). Previous study from our lab has proved that SMAR1 interacts with HDAC1 and forms a repressor complex [15]. So, we checked the binding of HDAC1 on calnexin promoter and found that HDAC1 also binds to calnexin promoter (Figure 2c). It was observed that SMAR1 enhanced the recruitment of GATA2 and HDAC1 at the MAR region of calnexin promoter (Figure 2, D and E). Simultaneously, there was increased enrichment of repression marks such as H3K9me3 and H4K20me at these regions confirming the role of SMAR1 in recruiting these factors for repressing calnexin gene expression (Figure 2, F and G).
Figure 2.
SMAR1, GATA2 and HDAC1 binds to calnexin promoter. (a) MAR-WIZ software analysis to predict MAR site in calnexin promoter (b) Electrophoretic mobility shift assay (EMSA): 500 ng, 1 μg and 2 μg of SMAR1 DNA binding domain was used. MAR region spanning 100 bp was used as probe. (c) Chromatin immunoprecipitation (ChIP) assay was done for the binding of SMAR1, GATA2 and HDAC1 on calnexin promoter MAR region. Chromatin from HCT116 p53+/+ cells was immunoprecipitated with α-SMAR1, α-GATA2 and α-HDAC1 and quantitative real time PCR was performed and the relative fold enrichment of (d) GATA2 (e) HDAC1 (f) H4k20me and (g) H3k9me3 on calnexin gene promoters was determined. Parallel ChIP with α-mouse and α-rabbit IgG was used as control depending upon the isotype of antibody used for the immunoprecipitation. (h) Immunoprecipitation (IP), cell lysate from HCT116 p53+/+ cells was immunoprecipitated with α-SMAR1, α-GATA2 and α-HDAC1 antibodies (lane 3). Parallel immunoprecipitation with control rabbit IgG antibody is shown in lane 2. Lane 1 denotes input control. (i) Immunoprecipitation of adeno-SMAR1 transduced HCT116 p53+/+cells with acetyl lysine antibody and probed with GATA2 antibody. (Bars represent standard deviation from three independent experiments. */** indicate statistically significant difference at P ≤ .05/0.005, respectively).
Since earlier results depicted that SMAR1 increases the enrichment of both GATA2 and HDAC1 at calnexin promoter, it gave us a clue that they might interact with each other and form a repressor complex. Also, there are reports which show that upon HDAC3 and HDAC5 mediated deacetylation, GATA2 acts as a repressor [35]. However, no reports are available showing its interaction with HDAC1. SMAR1 is known to interact with HDAC1, but its interaction with GATA2 was unknown. We hypothesized that SMAR1 might form a triple complex with GATA2 and HDAC1 resulting in deacetylation of GATA2. We then checked the interaction between SMAR1, GATA2 and HDAC1 and found them to be interacting with each other (Figure 2h). Acetylation status of GATA2 was also checked upon SMAR1 overexpression and it was found to be reduced. (Figure 2i). This result gave an insight into how an activator like GATA2 can become repressor in a context dependent manner.
GATA2 Acts as an Activator of Calnexin in the Absence of SMAR1
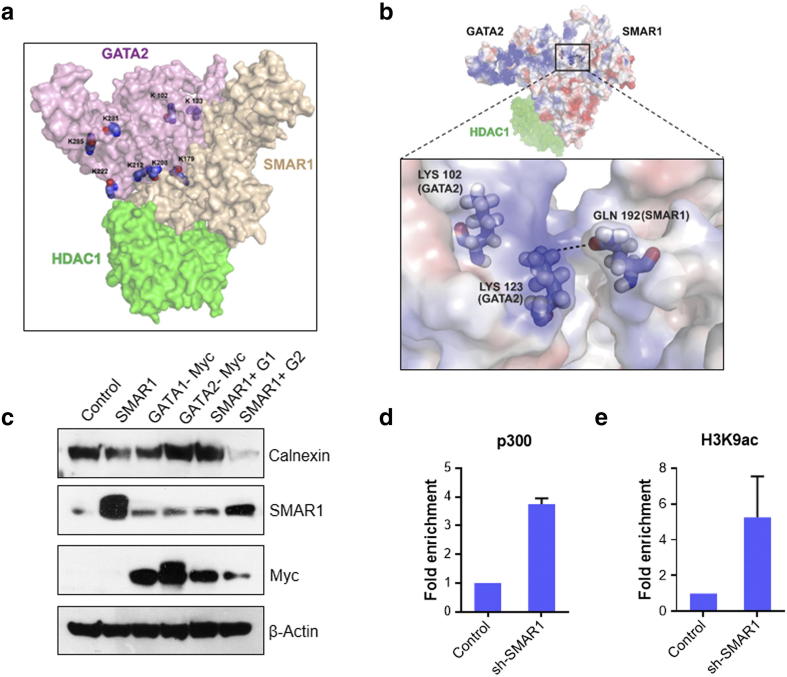
GATA2 is known to act as an activator under acetylated condition, this acetylation is generally carried out by p300, an important member of HAT family of proteins [36]. We established that SMAR1 forms a triple complex with GATA2 and HDAC1. In the presence of SMAR1, there is reduction in acetylation of GATA2. So, we further checked the coordinated regulation of calnexin gene expression by SMAR1 and GATA2. In-silico docking of the SMAR1, HDAC1 and GATA2 was carried out to understand the repressor complex (Supplementary Figure 3B). It revealed that residue 192–351 of SMAR1 is binding to GATA2. The N-terminal zinc finger domain of GATA2 contains many Lysine at positions 102, 123, 179, 208, 212, 222, 281 and 285. Amongst them the major acetylation sites are Lys 102, 123, 281 & 285 [36]. From the molecular docking studies, it was observed that on interacting with SMAR1, the Lys 102, 123 and 179 becomes unavailable for acetylation (Figure 3A). Lys 102 gets buried inside, Lys 123 forms a salt bridge with Gln 192 of SMAR1 and Lys 179 positions itself inside a cleft interacting with SMAR1 (Figure 3B). Further on docking HDAC1 with SMAR1-GATA2 complex, it is seen that Lys 222 of GATA2 is in close proximity to Gln 26 of HDAC1. That leaves only Lys 281 and Lys 285 on surface available for acetylation. It is evident that on interacting with SMAR1 and HDAC1, not enough Lysine of GATA2 are present to be acetylated. Thus, sufficient acetylation fails to occur and GATA2 in presence of SMAR1 and HDAC1 gets weakly acetylated. As per above findings, we over expressed GATA1 or GATA2 along with SMAR1 and checked calnexin expression levels. It revealed that GATA2 along with SMAR1 can repress calnexin gene expression to a higher extent as compared to SMAR1 alone (Figure 3C). SMAR1 overexpression along with GATA1 does not repress calnexin expression indicating it as an exclusive mechanism for GATA2. Further, we predict that upon SMAR1 knockdown, there will be no recruitment of HDAC1, letting GATA2 remain in acetylated condition. Interaction of p300 with GATA2 is known to keep it in acetylated condition which is very crucial for transactivation of different promoters. In our experiments, we observe a 4 fold increase in enrichment of p300 at calnexin promoter upon SMAR1 knockdown (Figure 3D). We also checked fold enrichment of H3K9 acetylation which is hallmark of most of the activated genes. Upon SMAR1 knockdown, enrichment of H3K9 acetylation was 6 folds higher than that in control condition (Figure 3E). This result establishes that in the absence of SMAR1, GATA2 is acetylated by p300 which then acts as an activator of calnexin gene expression.
Figure 3.
GATA2 act as an activator of calnexin in absence of SMAR1. (a) Model representing a triple complex of SMAR1, GATA2 and HDAC1 through docking studies highlighting the lysine residues which are getting masked upon this interaction. (b) Representative figure to show salt bridge between Gln 192 to SMAR1 and Lys 123 of GATA2. (c) HCT116 p53+/+ cells were transfected with flag-SMAR1, SMAR1 shRNA, myc-GATA1 and myc-GATA2. Western blot analysis was done to check the regulation calnexin by SMAR1, GATA2 and GATA1. Blots were probed with α-calnexin, α-flag, and α-myc tag antibodies. β-Actin was used as loading control. HCT116 p53+/+ cells were transfected with shRNA for SMAR1. Chromatin immunoprecipitation (ChIP) assay was done for the enrichment study of (d) p300 and (e) promoter activation mark H3K9 acetylation on MAR region of calnexin promoter. The relative promoter occupancy of all the above mentioned proteins were determined by comparing it with control IgG values.
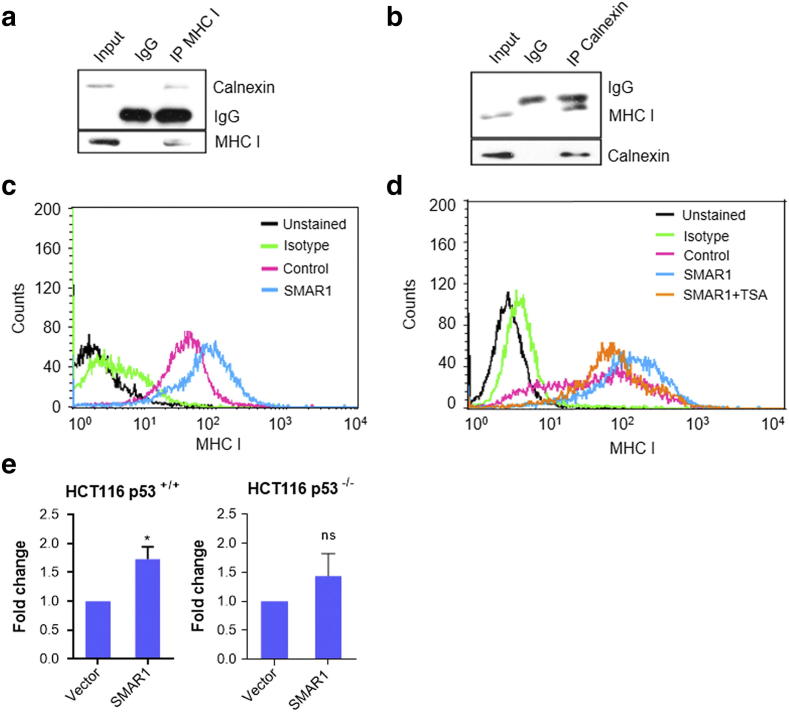
SMAR1 Positively Regulates MHC I Surface Expression on Cancer Cell
To delineate the biological significance of SMAR1 mediated calnexin gene regulation, antigen processing and presentation was studied. Calnexin is known to regulate MHC I heavy chain processing and antigen peptide loading. But the interaction between MHC I and calnexin is not reported in HCT116 p53+/+ cancer cells. So, we carried out an immunoprecipitation to check the same and found them to be interacting (Fig. 4, a and b). Further, to check whether regulation of calnexin has any effect on MHC I expression in cancer cells, SMAR1 was over expressed in HCT116 p53+/+ cells and MHC I surface staining was done with HLA ABC PE labeled antibody. Flow cytometry analysis revealed that upon SMAR1 over expression, MHC I surface expression also increased (Figure 4c). To understand whether SMAR1 mediated regulation of MHC I is HDAC1 dependent, trichostatin A, an HDAC inhibitor was used along with SMAR1 overexpression. It was observed that TSA largely reduced the SMAR1 mediated induction of MHC I (Figure 4d), clearly demonstrating the involvement of HDACs. We already have shown that SMAR1 suppresses calnexin expression through HDAC1.
Figure 4.
SMAR1 increases MHC I expression on cancer cells. (a and b) HCT116 p53+/+ cells were lysed and immunoprecipitation studies were carried out to check the interaction between calnexin and MHC I. IP was performed with α-calnexin and α-HLA ABC antibodies. Western blot analysis was done and blots were probed with α-calnexin and α-HLA ABC antibodies. (c) HCT116 p53+/+ cells were transfected with flag-vector and flag-SMAR1 constructs. Cells were harvested after 48 h and stained with PE-HLA ABC (MHC I) antibody. Flow cytometry acquisition was done with FACS calibur. (d) Flow cytometry was done with HCT116 p53+/+ cells transfected with flag-vector or flag-SMAR1 constructs, treated with 100 nM trichostatin A and stained with PE-HLA ABC (MHC I) antibody. Real-time PCR was done to check the relative gene expression of ERAP1 in (e) HCT116 p53+/+ and HCT116 p53−/− cells after SMAR1 overexpression. GAPDH was used as endogenous control.
Wang et al (2013) reported for the first time that p53 regulates MHC I in cancer and infection by up regulating ERAP1 (ER aminopeptidase1) [37]. Findings from our lab prove that SMAR1 stabilizes p53, so we checked the correlation between SMAR1 and ERAP1 to further strengthen our hypothesis. For this purpose, isogenic cell lines differing in the p53 status were taken. Overexpression of SMAR1 in HCT 116 p53+/+ cells resulted in increased expression of ERAP1 whereas SMAR1 overexpression in HCT 116 p53−/− cells does not affect ERAP1 expression (Figure 4, e). We conclude that SMAR1 stabilizes p53 which in turn increases ERAP1 expression. All these results led us to predict two tier regulation of MHC I by SMAR1 in both p53 dependent and independent manner.
Deregulation of MHC I Expression in Higher Grades of Breast Cancer or Cells Having LOH at Chr.16q24.3 Locus
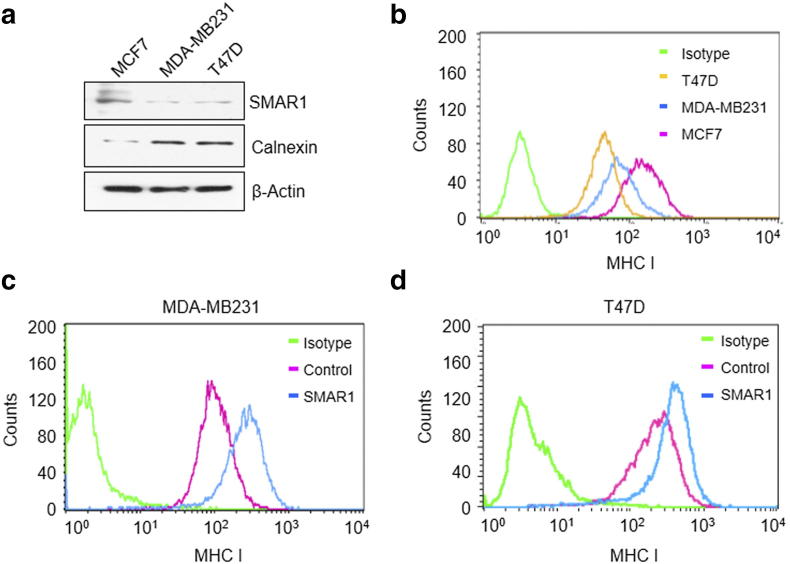
To investigate the role of SMAR1 in MHC I presentation pathway, different grades of breast cancer cell lines like MCF7, T47D and MDA-MB231 were used. MCF7 is non-metastatic and there is no LOH at Chr.16q24.3 locus. MDA-MB231 is highly metastatic whereas T47D reportedly have LOH at Chr.16q24.3 locus where SMAR1 homolog BANP is mapped [12], [38]. Hence, MCF7 cells have more SMAR1 expression compared to T47D and MDA-MB 231. Western blot analysis of these three cell lines revealed that MCF7 has low Calnexin expression whereas MDA-MB231 and T47D showed higher calnexin expression (Figure 5a). These results confirm the inverse correlation between calnexin and SMAR1 expression. Further, MHC I surface expression was studied in these three cell lines. MCF7 showed high MHC I expression compared to MDA-MB231 and T47D cell lines (Figure 5b). To establish that SMAR1 and calnexin expression have direct effect on MHC I surface expression, SMAR1 was overexpressed in MDA-MB231 and T47D. As per our hypothesis, MHC I surface expression was found to be rescued (Figure 5, c and d). Our results clearly suggest that SMAR1 down-regulates calnexin gene expression resulting in increased MHC I presentation and cell surface expression.
Figure 5.
SMAR1 rescues MHC I expression in higher grades of breast cancer cells. (a) Western blot showing calnexin and SMAR1 expression in different grades of breast cancer cell lines MCF7, MDA-MB231 and T47D. (b) FACS showing MHC I expression on different grades of breast cancer cell lines MCF7, MDA-MB231 and T47D. Cells were stained with PE-HLA ABC (MHC I) antibody. Flow cytometry acquisition was done in (c) MDA-MB 231 and (d) T47D cells after overexpressing SMAR1 and staining with PE-HLA ABC (MHC I) antibody.
Breast Cancer Patients Have High Calnexin and Decreased SMAR1, MHC I Expression
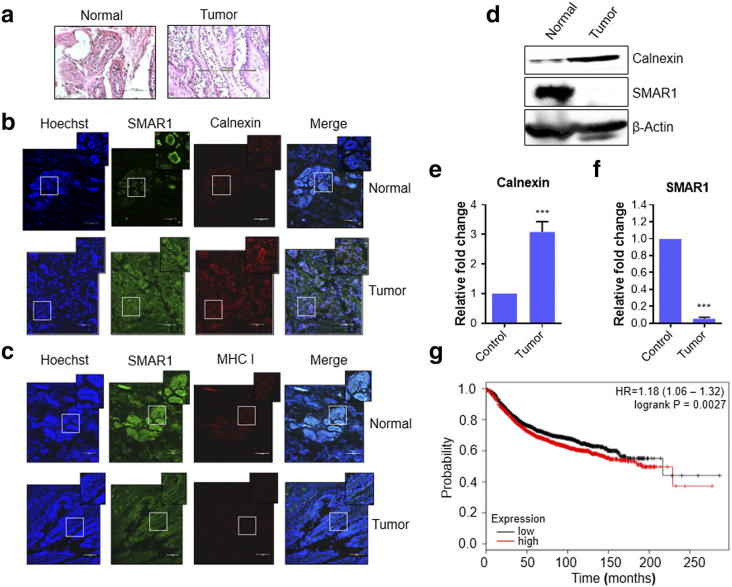
We explored the correlation of SMAR1 with either calnexin or MHC I in human breast normal and higher-grade tumor tissue samples (Figure 6a). It is evident that the tissue architecture is lost in cancer with a simultaneous decrease in nuclear localization of SMAR1 compared to adjacent normal tissue. The staining pattern of SMAR1 appears to be diffused. There is a significant increase in calnexin expression in cancer tissue (Figure 6b). Our data additionally implicates a role of SMAR1 in regulating calnexin as there is a negative correlation between the two proteins in tumor tissue. We find a parallel down-regulation of both SMAR1 and MHC I in tumor tissues of same patients in which there is up-regulation of calnexin as compared to adjacent normal tissues (Figure 6c). It is long known that class I MHC undergoes down-regulation in cancers aiding in the process of immune evasion [39]. Reduced expression of class I MHC has been associated with nodal metastasis in breast cancer [40]. This data therefore supports our in-vitro studies implicating a role of SMAR1 in down-regulating calnexin thereby allowing for proper class I MHC expression. We further performed western blot analysis of normal and tumor tissue samples and observe down-regulation of SMAR1 protein levels in tumor tissues whereas calnexin is up-regulated in the same samples (Figure 6d). Thus, the inverse correlation is evident in cancerous conditions also. Lower protein levels of SMAR1 could be a result of down-regulation at the transcript level, proteasomal degradation or it might be a combination of these mechanisms. Thus, we checked the levels of both SMAR1 and calnexin transcripts in these patient samples. We found this down-regulation of SMAR1 and up-regulation of calnexin in the transcript levels also (Figure 6, e and f). LOH at SMAR1 locus is also reported in different cancers including breast and colorectal cancer. This can also contribute to drastically low levels of SMAR1. Thus, different mechanisms operate in parallel to decrease SMAR1 causing up-regulation of calnexin in cancer that in turn results in lower class I MHC expression on tumor cells. The Kaplan–Meier plot was utilized to further extrapolate our findings to breast cancer patients by checking the correlation between calnexin expression profile and patient survival. This tool is capable of evaluating the effect of 22,277 genes on patient survival using 10,188 cancer patient samples. Of these samples, 4142 are breast cancer samples having a mean follow-up of 69 months [41]. We performed the survival analysis on 1975 distant metastasis free survival (DMFS) breast cancer patient data with respect to calnexin expression levels. The KM curve for calnexin established that higher expression levels of calnexin correlated with poor prognosis and survival (Figure 6g). This observation is in concordance with the fact that breast cancer patients with higher SMAR1 levels show better survival [26], establishing inverse correlation between SMAR1 and calnexin. Therefore, we conclude that SMAR1 expression positively correlates with disease free survival while calnexin is a predictor for poor prognosis and disease outcome.
Figure 6.
Breast cancer patients have high calnexin expression and decreased SMAR1, MHC I expression. (a) Control and tumor section are stained with Hematoxylin and Eosin (H&E) stain to visualize the cellular structure and morphology. (b) and (c) Immunofluorescence assay was done to visualize the expression of SMAR1, Calnexin and MHC I in normal and breast cancer tissue section. (d, e and f) Immunoblotting and quantitative real-time PCR to check the expression of SMAR1 and calnexin in normal and breast cancer tissue samples. β-Actin was used as loading control. (g) Kaplan–Meier distant metastasis free survival analysis for calnexin gene in 1975 breast cancer patients. Higher expression levels of calnexin are correlated with poor survival (P = .0027).
SMAR1 Causes Tumor Regression by Modulation of MHC I Antigen Presentation
Antigen presenting cells (APC) include dendritic cells, B-cells and macrophages which have the capacity to present antigen through MHC I as well as MHC II. MHC I surface expression has direct correlation with SMAR1 expression. To understand whether SMAR1 has any effect on MHC I antigen presentation by the APCs, we isolated peritoneal macrophages from control littermate and SMAR1 transgenic mice. Here we observe that macrophages isolated from SMAR1 transgenic mice show increased MHC I expression as compared to control macrophages (Supplementary Figure 4A). Further, these macrophages were treated with OVA 257–264 class 1-restricted peptide epitope of ovalbumin which is presented by MHC I molecule (SIINFEKL). The antigen presentation by macrophages isolated from SMAR1 transgenic mice was found to be higher than the control macrophages (Supplementary Figure 4B). This result indicates that SMAR1 modulation can alter antigen presentation not only in cancer cells but also in antigen presenting cells.
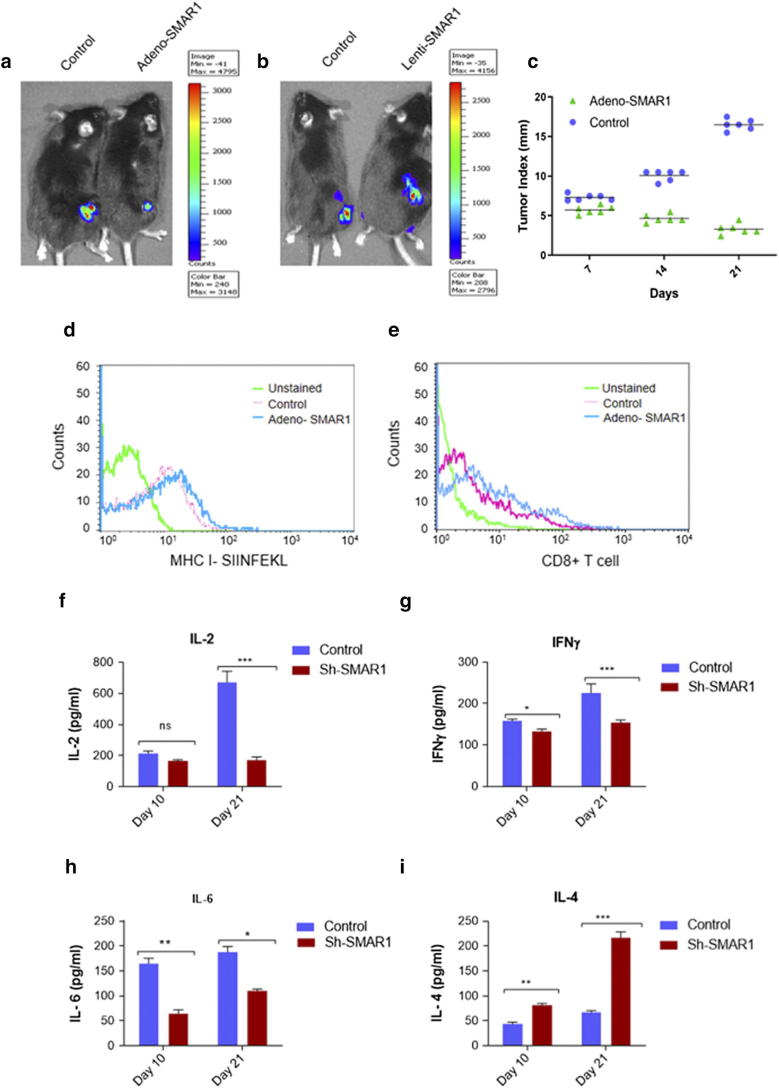
Role of SMAR1 as tumor suppressor is well established, but its function in immune response mediated cancer regression was unknown. Our findings from the present study led us to understand the role that SMAR1 plays in MHC class I mediated antigen processing and presentation. To understand its implication in immunity against cancer, tumor model was established in immunocompetent mice using a melanoma cell line MO4 cells expressing ovalbumin and luciferase proteins. The dual function of this cell line was exploited to study the exogenous ovalbumin antigen presentation as well as in vivo imaging in the mice tumor. To activate the immune system of mice, SIINFEKL peptide which is generally presented by MHC class I pathway was used. Six days after immunization, MO4 cells transduced with adeno-SMAR1 for overexpression and sh-lenti-SMAR1 for knockdown were injected subcutaneously in the flank region of C57BL/6 mice. After 4 weeks of injection, in vivo imaging was done and it was observed that tumors which overexpressed SMAR1 show less bioluminescence as compared to the control counterparts whereas inverse result was observed in Sh-lenti-SMAR1 tumors. Intensity of bioluminescence is directly proportional to the size of tumor. (Figure 7, a and b). Tumor progression studies also confirmed the regression in SMAR1 overexpressed tumors compared to the control (Figure 7c). Further, total MHC I antigen presentation by the tumor cells was studied by flow cytometry. We used an antibody which can bind to MHC-Ip (MHC I- SIINFEKL). There was a marked increase in MHC class I antigen presentation by SMAR1 transduced tumor cells (Figure 7d). Subsequently, immune infiltration of CD8+ T cells inside the tumors was also studied. Tumors were excised and single cell suspension was prepared by enzymatic digestion and flow cytometry analysis was done. SMAR1 transduced tumors show increased CD8+ T cell infiltration inside the tumor compared to the control (Figure 7e). CD8+ T cells infiltration has direct correlation with tumor regression and disease outcome. Immune cells are attracted to the tumor site by the signal of various chemoattractants [42]. Cytokines are the major chemoattractant produced by the tumor cells as well as infiltrating immune cells. IL-6, IL-2 and IFNγ are some of the proinflammatory cytokines produced by the infiltrating immune cells. IL-10, IL-4 and TGF-β are anti-inflammatory in nature that helps in tumor progression [43]. To understand the early and late cytokine response with respect to MHC I mediated antigen presentation and immune response, cytokine response was analyzed in normal and SMAR1 knockdown MO4 tumor models at day 10 and day 21 post injection. There is no significant change in IL-2 serum levels at day 10 in SMAR1 knockdown MO4 tumor mice compared to the control but at day 21, a 3-fold decrease in serum IL-2 was observed (Figure 7f). Decreased IL-2 expression can be directly correlated with decreased T cell activation and antigen presentation. Similar trend was observed with IFNγ serum levels, but IL-6 levels drastically decreased by 3 folds at day 10 and at day 21, a 2-fold decrease was observed contrary to other proinflammatory cytokines which showed delayed response (Figure 7, g and h). This suggests that absence of SMAR1 results in decreased proinflammatory serum cytokines levels and antitumor immune response. Moreover, IL-4 serum levels were found to be increased by 1.5-fold at day 10 and 4-fold at day 21 (Figure 7i). Increase in serum IL-4 levels indicates a bias towards Th2 response.
Figure 7.
Tumor regression by SMAR1 via increased antigen presentation by tumor cells. (a, b and c) Ova- Ip (SIINFEKL) immunized C57BL/6 mice were injected with MO4 cells for tumor generation followed by transduction with control, adeno-SMAR1 and lenti-SMAR1 and tumor progression was studied. In-vivo imaging was done to visualize the tumor growth. (d) FACS for Ova- Ip (SIINFEKL) antigen presented by MHC I on MO4 tumor cells transduced with control and Adeno-SMAR1 virus. (e) FACS for CD8+ T-cells infiltration inside the control and Ad-SMAR1 transduced MO4 tumors. (f-i) ELISA was performed to check the serum levels of IL-2, IFNγ, IL-6 and IL-4 in lenti-SMAR1 transduced MO4 tumor containing mice compared to control at day 10 and 21 of tumor progression.
All these observations provide experimental evidence for survival advantage to the cancer cells expressing low SMAR1 levels leading to their immune evasion. Transformed cells having higher SMAR1 expression levels were eliminated by the immune system leading to tumor regression.
Influenza A Virus Infection Increases SMAR1 Expression and Decreases Calnexin Expression
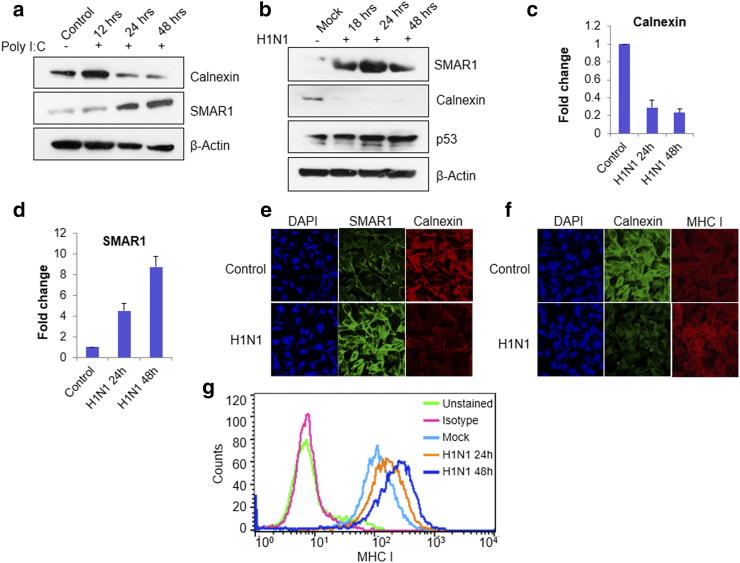
It has been reported that influenza A virus infection induces p53 expression leading to increased MHC I expression [44]. p53 target genes such as TAP1 and TAP2 which are a part of the APM and have direct role in MHC I processing and presentation [45]. A study by Wang et al. suggests that p53 regulates MHC I during influenza A virus infection by up regulating ERAP1 (ER aminopeptidase1) [37]. Our lab reported that SMAR1 stabilizes the p53 protein expression by down-regulating MDM2 [16]. Previous experiments in this study have already established that SMAR1 regulates MHC class I expression by down-regulating calnexin and also by increasing ERAP1 expression. To establish the role of SMAR1 in the context of influenza virus infection, we used Poly I:C, a double stranded RNA homolog which mimics viral infection and works through TLR3 receptors. When A549 cells were treated with poly I:C, SMAR1 expression increased whereas calnexin expression was down-regulated by 24 h (Figure 8A). This result suggests that TLR3 signaling can up-regulate SMAR1 expression and it has inverse correlation with calnexin expression. In order to correlate the data with viral infection, the pandemic influenza virus [influenza A(H1N1) pdm09] was used for the infection [46], [47], [48]. Infection was confirmed by immunofluorescence using antibody against the nucleoprotein (NP) of influenza A virus (Supplementary Figure 5, A and B). After infecting the cells, protein expression of SMAR1, calnexin and p53 was evaluated (Figure 8b), which revealed that SMAR1 and p53 expressions were up-regulated whereas calnexin expression was down-regulated. Higher SMAR1 protein expression was detected in the H1N1 pdm09 virus infected cells as early as 18 h post infection and continued to increase up to 48 h whereas calnexin expression was found to be down-regulated as early as 18 h post infection. To correlate the reduced calnexin expression to SMAR1, mRNA expression was measured by quantitative real-time PCR. It was found that upon H1N1 virus infection, mRNA levels of SMAR1 increased whereas it decreased the expression of calnexin (Figure 8c and d). An inverse correlation between SMAR1 and calnexin is evident during the influenza virus infection also. Immunostaining carried out on the virus infected A549 cells. Calnexin, SMAR1 and MHC I expression was visualized by fluorescence microscopy. Intense SMAR1 labelling was seen in the H1N1 virus infected cells which correlated with decreased calnexin protein expression and elevated MHC class I expression (Figure 8 e and f). Flow cytometry also suggested that around at 24 h post infection, up-regulation was marked in MHC I expression which kept increasing till 48 h (Figure 8G). These results are in concordance with our findings that within 18 h of infection, SMAR1 and p53 levels increase and calnexin expression shows a marked decrease. SMAR1 increases MHC I expression in a bilateral fashion: by down-regulating calnexin and/or by stabilizing p53, leading to increase in MHC I expression. These results are consistent in the influenza virus infection also where host immune system itself clears the viral load except for the genetically susceptible individuals with compromised immune system. Present study highlights the importance of the nuclear matrix protein SMAR1 in antigen processing and presentation during the influenza virus infection as well as in cancer conditions.
Figure 8.
Influenza A virus infection increases SMAR1 expression and decreases calnexin expression. (a) Western blot analysis of A549 cells transfected with Poly I:C, a double stranded synthetic RNA homolog. Blots were probed with α-SMAR1 and α-calnexin antibodies. (b) Western blot analysis of A549 cells upon influenza A virus (H1N1) infection at 0.5 MOI. Blots were probed with α-SMAR1, α-p53 and α-calnexin antibodies. (c) and (d) Real-time PCR analysis of A549 cells infected with H1N1 virus at 0.5 MOI for 24 h and 48 h. (e and f). Immunostaining was done in A549 cell infected with 0.5 MOI H1N1 influenza A virus and α-SMAR1, α-MHC I and α-calnexin antibodies were used. Cy3 and FITC labeled secondary antibodies were used against the primary antibody. (g) Flow cytometry analysis of A549 cells treated with H1N1 pandemic strain of influenza virus.
Discussion
SMAR1 is a MAR binding protein and it is an important component of the nuclear matrix which controls cellular homeostasis. SMAR1 expression is ubiquitous throughout various cell types; the redundancy in its expression makes it an indispensable component for cellular control and survival. During cancer conditions, the tight control of SMAR1 expression gets altered which can provide these transformed cells, the survival advantage. SMAR1 is known to be down-regulated in various grades of breast cancers [25] and its expression is directly correlated with the survival of patients and disease outcome [26]. It is known that SMAR1 gets down-regulated in higher grades of cancer through Cdc20 mediated proteasomal degradation [13], [14]. Apart from the above-mentioned studies, there are reports which demonstrate loss of heterozygosity at the SMAR1 loci leading to its dampened expression in cancer cells [12]. Non-canonical role of SMAR1, other than tumor suppressor functions in the context of immune system remained unanswered. Immune editing is a phenomenon which explains the elimination or survival of cancer cells other than tumor suppression theory. Present study highlights the importance of SMAR1 in immune editing mechanism for tumor suppression where we show its function in repressing calnexin expression and positively regulating MHC I surface expression.
MHC I mediated antigen presentation is a complex process and requires proper functioning of various components of antigen processing machinery. Once a foreign or mutated protein is recognized, a cascade of events starts to correctly present it and elicit immune response. Antigenic peptides are transported into the endoplasmic reticulum by Transporter Associated with Antigen Processing (TAP) and loaded onto newly synthesized MHC I molecule with the help of chaperone proteins calnexin, calreticulin, tapasin and ERp57 [49], [50]. These chaperone proteins play varied role during peptide loading. Calnexin has a vital role in MHC I heavy chain folding. When newly synthesized MHC I heavy chain enters ER lumen it acquires Glc1Man9GlcNAc2 glycan moiety that is recognized by calnexin [19]. The interaction between calnexin and MHC I heavy chain does not allow it's assembly with β2-microglobulin inhibiting the formation of complete MHC I molecule [51]. Binding of calreticulin triggers an open confirmation to heavy chain resulting in interaction with β2 microglobulin [52]. Once the final confirmation is attained, antigenic peptides are loaded on MHC I molecule with the assistance of TAP, ERAP1, ERAP2 and tapasin and transported to the surface though Golgi network [53]. Now these antigenic peptides complexed with MHC I can be recognized by the cognate TCRs present on CD8+ T cells and facilitates their interaction. CD8+ T cells elicit immune response on the basis of the origin of peptides; generally unmutated or self-proteins are ignored whereas pathogenic or mutated proteins are triggering factors. MHC I mediated intracellular surveillance by CD8+ T cells performs vital role in eradicating intracellular pathogens and malignancies. Cancer is one of the most evolved disease having an escape mechanism from the immune system for their survival. Down-regulation of MHC I surface expression is identified in breast cancer [54], colon cancer [1], melanoma [55], renal cancer [56] and cervical cancer [57] as one of the most common escape mechanisms. In fact, alteration in expression of APM components is a foremost reason for MHC I down-regulation and is observed in several cancers. Although most of the APM such as TAP, tapasin, ERAP1, LMPs, were down-regulated in different cancers, calnexin show inverse correlation with cancer progression [58]. Calnexin expression was found to be higher in plasma cells from multiple myeloma patients than in either premalignant plasma cells from monoclonal gammopathy of undetermined significance (MGUS) patients or normal plasma cells from healthy donors [59]. Also, in lung, colon and breast cancers, calnexin appears to have an inverse correlation with disease outcome [20], [21], [22].
The present study demonstrates that SMAR1 negatively regulates calnexin gene expression by forming a repressor complex with GATA2 and HDAC1. The functional relevance of calnexin gene regulation by SMAR1 in context of MHC I expression and antigen presentation were investigated. Since, calnexin expression was found to be drastically increased in cancer patients whereas SMAR1 and MHC I expression were found to be decreased, the involvement of SMAR1 in regulating antigen presentation by MHC I via regulation of calnexin expression was studied. To understand this further, we used different grades of cancer cells and compared MHC I expression levels in Ch16 q 24.3 LOH cells. Less MHC I surface expression was observed in the higher grade cancers and cells having LOH as compared to those non-metastatic cells. Also, similar observation was found in breast cancer patient samples. SMAR1 and MHC I were drastically reduced but calnexin expression increased with tumor progression. All these observations led us to understand that higher SMAR1 expression increases MHC I surface expression whereas calnexin shows an inhibitory effect. As the cancer progresses, SMAR1 gets down-regulated by various mechanisms leading to higher calnexin expression which ultimately results in decreased MHC I surface expression. This can be directly implicated in antigen presentation and immune response against the cancer cells. For this, we studied antigen presentation in SMAR1 transgenic mice. The peritoneal macrophages isolated from transgenic mice showed an increased MHC I expression as well as antigen presentation by MHC I. Remarkably, SMAR1 shows direct involvement in antigen presentation by the macrophages. Increased antigen presentation of cancer cells will lead to elimination of transformed cells. During oncogenic transformations, SMAR1 can augment cancer antigen presentation to CD8+ T cells and other immune cells. A tumor model was established in immunocompetent mice with the syngeneic MO4 cells expressing exogenous ovalbumin antigen. Tumors expressing higher SMAR1 levels show a progressive tumor regression and an increased exogenous ovalbumin antigen presentation. Increased antigen presentation will prime immune cells against the transformed tumor cells leading to their infiltration inside the tumor. Accordingly, SMAR1 overexpressing tumors showed increased CD8+ T cell infiltration inside the tumor microenvironment. It has been well established that cancer patients with higher CD8+ T cell tumor infiltration have better disease outcome and survival [60]. Cytokine profile also marks the onset of immune response against tumor cells leading to their elimination. Pro-inflammatory cytokines such as IL-2, IL-6 and IFNγ can elicit anti-tumor immune response [43]. Higher serum levels of IL2 and IFNγ are implicated in robust Th1 and CD8+ immune response against cancer cells [61]. Higher IL6 level is still contentious and shown to dampen Th1 mediated anti-tumor immune response in old mice compared to young mice [62]. Anti-inflammatory cytokines such as TGF-β, IL-10 and IL-4 marks reduced anti-tumor response and can be correlated with either tumor dormancy or escape phase [63]. Similar results were obtained in our study where IL-2, IL-6 and IFNγ serum levels decreased while IL-4 levels increased in MO4 tumors having reduced SMAR1 expression. This strengthens the observation that lower expression of SMAR1 in tumor can dampen immune response leading to increased tumor progression.
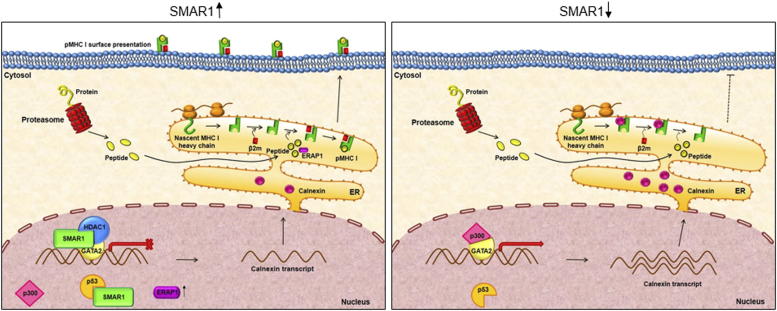
In the year 2013, a study by Wang et al. established that during influenza virus infection, p53 increases the MHC I expression through modulating ERAP1 expression, a component of MHC I antigen processing machinery [37]. SMAR1 is also known to stabilize p53 by bringing about phosphorylation at serine 15 residue [16]. This opened up another dimension of MHC I regulation, where SMAR1 stabilizes p53 leading to increased ERAP1 and MHC I surface expression during influenza virus infection. ERAP1 modulation by SMAR1 in a p53 dependent manner led us to speculate a two-tier regulation of MHC I; one via calnexin modulation and the other through the p53 mediated regulation of ERAP1. Similar results were obtained when influenza A virus infection model was used, showing increase in SMAR1 and MHC I expression concomitant with decrease in calnexin expression. SMAR1 induction during influenza virus infection most probably occurs due to TLR3 signaling which gets activated by double stranded RNA intermediate during viral replication [64]. Double stranded RNA analogue poly I:C experiment rationalized our hypothesis that TLR3 signaling can increase SMAR1 expression, implicating the significance of SMAR1 as host defense protein and can be associated to MHC I mediated clearance of influenza A virus. All these observations suggest that non-canonical tumor suppressor function of SMAR1 occurs by up-regulating MHC I and increasing antigen presentation in a context dependent manner. As indicated in Figure 9, SMAR1 forms repressor complex with HDAC1 and GATA2 thereby repressing calnexin expression. Low calnexin expression leads to higher MHC I surface expression. However, under lower levels of SMAR1, calnexin is up-regulated resulting in sequestration of MHC I heavy chain and therefore β2−microglobin cannot bind. This leads to reduction in MHC I molecule surface expression.
Figure 9.
Working model showing MHC I regulation by SMAR1. (Left) Endogenous SMAR1 interacts with HDAC1 and GATA2 thereby forming a repressor complex resulting in decreased calnexin expression. This decrease in calnexin expression as well as increased ERAP1 expression leads to increased MHC I expression. (Right) During low SMAR1 condition, GATA2 remains in acetylated condition and increases calnexin expression resulting in sequestration of MHC I heavy chain. This leads to low surface expression and antigen presentation.
This study gives a better insight into the intricate mechanism of MHC I regulation and antigen presentation by SMAR1 during cancer as well as infection. Clear understanding of the immune pathways that keep cancer under check, can be very effective in designing therapy against cancer. Apart from MHC I deregulation, SMAR1 down-regulation appears to be major hallmark of cancer progression. Stabilizing SMAR1 expression through some small compounds has enormous therapeutic potential to treat of cancer patients and increase their overall survival. We are screening some of the small compounds that can stabilize SMAR1 by inhibiting its proteasomal degradation. In future, we might have a drug that can boost host immune system to fight cancer.
Acknowledgments
Acknowledgement
We are thankful to National Centre for Cell Science, Pune and CSIR-Indian Institute of Chemical Biology for providing infrastructure facility. We also acknowledged DBT, DBT-SyMeC and Council of Scientific and Industrial Research, Government of India for funding the research work. We are thankful to Dr. Sanjeeva Srivastava, IIT Bombay and Dr. Srikant Rapole, NCCS, Pune, for their kind help in performing proteomics study. We are very much thankful to Dr. D.T. Mourya, Director, ICMR-NIV, Pune and Dr. M.S. Chadha for allowing the work to be conducted in NIV, Pune. We are thankful to UGC and DBT for fellowship. We are also thankful to Shounak Bhattacharya, senior technical officer CSIR-IICB for confocal microscopy. We are also thankful to Dr. Kenneth L. Rock, University of Massachusetts Medical School, Worcester, USA, and Hirotake Tsukamoto Awai, Kumamoto University, Japan for providing MO4 cell line.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions
AA and SC* conceptualized and designed the experiment. AA, NT, SP, VKS, RP performed experiments. AA, MT, JM performed viral infection experiments. TR, RB, RR, AM performed patient sample experiments. AA, DM did the animal experiments. NB, SC executed docking studies. AA, SP, SC wrote manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.07.002.
Appendix A. Supplementary Data
Supplementary figure 1.
Proteomic profiling of SMAR1 regulated genes by 2DE: (a) Comparative 2DE gels for control and SMAR1 knockdown HCT116 p53+/+ cells. Gels were run in triplicate for each sample. (b) Differentially expressed proteins were marked on 2DE gels. Bar graph showing (c) up-regulated and (d) down-regulated proteins upon SMAR1 knockdown in HCT116 p53+/+ cells compared to the control HCT116 p53+/+ cells.
Supplementary figure 2.
SMAR1 and calnexin show inverse correlation. (a) MCF7 cells were treated with 5mM Tunicamycin in a time-dependant manner. Western blot analysis was done to check the expression of calnexin and SMAR1. β-Actin was used as loading control. (b) HCT116 p53+/+ cells were treated with 100 ng/ml EGF and real-time PCR was done to check the relative gene expression of calnexin. (c) 100 ng/ml Epidermal Growth Factor (EGF) treatment was given to HCT116 p53 +/+ cells and stained with calnexin antibody. DAPI was used for staining nucleus. (d) HCT116 p53+/+ cells were transfected with FLAG-SMAR1 followed by treatment with 100 ng/ml EGF and real-time PCR was done to check the relative gene expression of SMAR1. GAPDH was used as internal control for quantitative PCRs.
Supplementary figure 3.
Calnexin promoter analysis and docking studies (a) Calnexin promoter spanning from -1500 to +1 region. MAR site is depicted in bold, GATA sites are marked in red. (b) Computational docking model depicting interaction of SMAR1, GATA2 and HDAC1 along with double stranded DNA.
Supplementary figure 4.
SMAR1 increases antigen presentation in peritoneal macrophages. (a) Flow cytometry analysis for MHC I expression on peritoneal macrophages isolated from SMAR1 transgenic mice and control littermate mice. (b) Ovalbumin (SIINFEKL) peptide (antigen) presentation study by flow cytometry. Peritoneal macrophages isolated from SMAR1 transgenic mice and control littermate were treated with 1 μg/ml SIINFEKL peptide followed by flow cytometry analysis using an antibody that stain MHC I-SIINFEKL complex.
Supplementary figure 5.
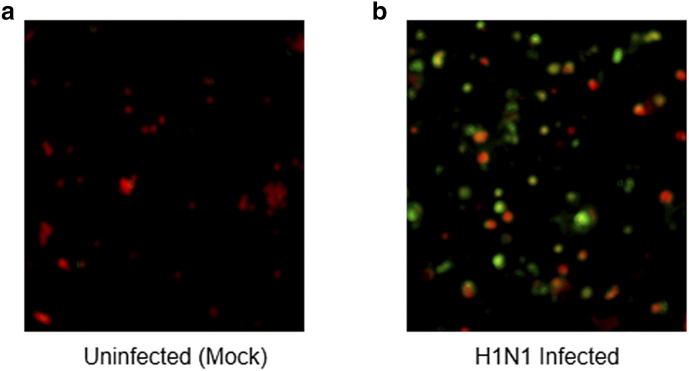
Immunofluorescence assay. Influenza A(H1N1) pdm09 virus infection in A549 cells was confirmed by immunofluorescence assay against the nucleoprotein (NP) of influenza post 24h of infection. (a) Control or uninfected A549 cells (red). (b) Influenza A(H1N1) pdm09 virus infected A549 cells (green).
Supplementary Table 1.
List of differentially regulated proteins upon knocking down SMAR1. Table depicts the list of up-regulated and down-regulated genes in HCT116 p53+/+ SMAR1 knockdown cells. Statistical analysis for up-regulation and down-regulation of differentially expressed spots was done with IMP7 software.
Supplementary Table 2.
List of primers and their sequences used in quantitative real time PCR for measuring relative expression levels of various genes.
References
- 1.Watson NFS, Ramage JM, Madjd Z. Immunosurveillance is active in colorectal cancer as downregulation but not complete loss of MHC class I expression correlates with a poor prognosis. Int J Cancer. 2006;118:6–10. doi: 10.1002/ijc.21303. [DOI] [PubMed] [Google Scholar]
- 2.Garrido F, Algarra I. MHC antigens and tumor escape from immune surveillance. Adv Cancer Res. 2001;83:117–158. doi: 10.1016/s0065-230x(01)83005-0. [DOI] [PubMed] [Google Scholar]
- 3.Lampen MH, van Hall T. Strategies to counteract MHC-I defects in tumors. Curr Opin Immunol. 2011;23:293–298. doi: 10.1016/j.coi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Pamer E, Cresswell P. Mechanisms of MHC class I–restricted antigen processing. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 5.Seliger B, Wollscheid U, Momburg F. Characterization of the major histocompatibility complex class I deficiencies in B16 melanoma cells. Cancer Res. 2001;61:1095–1099. [PubMed] [Google Scholar]
- 6.Reeves E, James E. Antigen processing and immune regulation in the response to tumours. Immunology. 2017;150:16–24. doi: 10.1111/imm.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay S, Kaul R, Charest A. SMAR1, a novel, alternatively spliced gene product, binds the scaffold/matrix-associated region at the T cell receptor β locus. Genomics. 2000;68:93–96. doi: 10.1006/geno.2000.6279. [DOI] [PubMed] [Google Scholar]
- 8.Chemmannur SV, Badhwar AJ, Mirlekar B. Nuclear matrix binding protein SMAR1 regulates T-cell differentiation and allergic airway disease. Mucosal Immunol. 2015;8:1201–1211. doi: 10.1038/mi.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha S, Malonia SK, Mittal SPKK. Coordinated regulation of p53 apoptotic targets BAX and PUMA by SMAR1 through an identical MAR element. EMBO J. 2010;29:830–842. doi: 10.1038/emboj.2009.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirlekar B, Ghorai S, Khetmalas M. Nuclear matrix protein SMAR1 control regulatory T-cell fate during inflammatory bowel disease (IBD) Mucosal Immunol. 2015;8:1184–1200. doi: 10.1038/mi.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birot A, Duret L, Bartholin L. Identification and molecular analysis of BANP. Gene. 2000;253:189–196. doi: 10.1016/s0378-1119(00)00244-4. [DOI] [PubMed] [Google Scholar]
- 12.Powell JA, Gardner AE, Bais AJ. Sequencing, transcript identification, and quantitative gene expression profiling in the breast cancer loss of heterozygosity region 16q24.3 reveal three potential tumor-suppressor genes. Genomics. 2002;80:303–310. doi: 10.1006/geno.2002.6828. [DOI] [PubMed] [Google Scholar]
- 13.Paul D, Ghorai S, Dinesh US. Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taye N, Alam A, Ghorai S. SMAR1 inhibits Wnt/β-catenin signaling and prevents colorectal cancer progression. Oncotarget. 2018;9:21322–21336. doi: 10.18632/oncotarget.25093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rampalli S, Pavithra L, Bhatt A, Kundu TK. Tumor Suppressor SMAR1 mediates Cyclin D1 repression by recruitment of the SIN3 / Histone deacetylase 1 complex. Mol Cell Biol. 2005;25:8415–8429. doi: 10.1128/MCB.25.19.8415-8429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jalota A, Singh K, Pavithra L. Tumor suppressor SMAR1 activates and stabilizes p53 through its arginine-serine-rich motif. J Biol Chem. 2005;280:16019–16029. doi: 10.1074/jbc.M413200200. [DOI] [PubMed] [Google Scholar]
- 17.Arunachalam B, Cresswell P. Molecular requirements for the interaction of Class II major histocompatibilty complex molecules and invariant chain with calnexin. J Biol Chem. 1995;270:2784–2790. doi: 10.1074/jbc.270.6.2784. [DOI] [PubMed] [Google Scholar]
- 18.Fewell SW, Travers KJ, Jonathan S, Brodsky JL. The action of moleclar chapreones in the early secretory pathway. Annu Rev Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 19.Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi M, Nagashio R, Jiang SX. Calnexin is a novel sero-diagnostic marker for lung cancer. Lung Cancer. 2015;90:342–345. doi: 10.1016/j.lungcan.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Ryan D, Carberry S, Murphy ÁC. Calnexin, an ER-induced protein, is a prognostic marker and potential therapeutic target in colorectal cancer. J Transl Med. 2016;14:1–10. doi: 10.1186/s12967-016-0948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Ma D, Wang X, et al (2018) Calnexin impairs the antitumor immunity of CD4+ and CD8+ T cells. Cancer Immunol Res canimm.0124.2018 . doi: 10.1158/2326-6066.CIR-18-0124 [DOI] [PubMed]
- 23.Li F, Mandal M, Barnes CJ. Growth factor regulation of the molecular chaperone calnexin. Biochem Biophys Res Commun. 2001;289:725–732. doi: 10.1006/bbrc.2001.6001. [DOI] [PubMed] [Google Scholar]
- 24.Boden G, Merali S. Measurement of the increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Methods Enzymol. 2011;489:67–82. doi: 10.1016/B978-0-12-385116-1.00004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh K, Mogare D, Giridharagopalan RO. p53 target gene SMAR1 is dysregulated in breast cancer: Its role in cancer cell migration and invasion. PLoS One. 2007;2:e660–e675. doi: 10.1371/journal.pone.0000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathai J, Mittal SPK, Alam A. SMAR1 binds to T(C/G) repeat and inhibits tumor progression by regulating miR-371-373 cluster. Sci Rep. 2016;6 doi: 10.1038/srep33779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris MR, Yu YY, Kindle CS. Calreticulin and calnexin interact with different protein and glycan determinants during the assembly of MHC class I. J Immunol. 1998;160:5404–5409. [PubMed] [Google Scholar]
- 28.Coe H, Bedard K, Groenendyk J. Endoplasmic reticulum stress in the absence of calnexin. Cell Stress Chaperones. 2008;13:497–507. doi: 10.1007/s12192-008-0049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delom F, Emadali A, Cocolakis E. Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ. 2007;14:586–596. doi: 10.1038/sj.cdd.4402012. [DOI] [PubMed] [Google Scholar]
- 30.Nakka KK, Chaudhary N, Joshi S. Nuclear matrix-associated protein SMAR1 regulates alternative splicing via HDAC6-mediated deacetylation of Sam68. Proc Natl Acad Sci. 2015;112:E3374–E3383. doi: 10.1073/pnas.1418603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function , a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu K, Wang J, Gao J. GATA binding protein 2 overexpression is associated with poor prognosis in KRAS mutant colorectal cancer. Oncol Rep. 2016;36:1672–1678. doi: 10.3892/or.2016.4961. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Jiang B, Wang Z. Expression and prognostic significance of GATA-binding protein 2 in colorectal cancer. Med Oncol. 2013;30 doi: 10.1007/s12032-013-0498-7. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki M, Kobayashi-Osaki M, Tsutsumi S. GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells. 2013;18:921–933. doi: 10.1111/gtc.12086. [DOI] [PubMed] [Google Scholar]
- 35.Ozawa Y, Towatari M, Tsuzuki S. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood. 2001;98:2116–2123. doi: 10.1182/blood.v98.7.2116. [DOI] [PubMed] [Google Scholar]
- 36.Hayakawa F, Towatari M, Ozawa Y. Functional regulation of GATA-2 by acetylation. J Leukoc Biol. 2003;75:529–540. doi: 10.1189/jlb.0603389. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Niu D, Lai L, Ren EC. P53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat Commun. 2013;4:1–11. doi: 10.1038/ncomms3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callen DF, Crawford J, Derwas C. Defining regions of loss of heterozygosity of 16q in breast cancer cell lines. Cancer Genet Cytogenet. 2002;133:76–82. doi: 10.1016/s0165-4608(01)00565-9. [DOI] [PubMed] [Google Scholar]
- 39.Atkins D, Ferrone S, Schmahl GE. Down-regulation of HLA class I antigen processing molecules: An immune escape mechanism of renal cell carcinoma? J Urol. 2004;171:885–889. doi: 10.1097/01.ju.0000094807.95420.fe. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko K, Ishigami S, Kijima Y. Clinical implication of HLA class I expression in breast cancer. BMC Cancer. 2011;11 doi: 10.1186/1471-2407-11-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gyorffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta - Mol Cell Res. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Zhu K, Wang J, Zhu J. p53 induces TAP1 and enhances the transport of MHC class I peptides. Oncogene. 1999;18:7740–7747. doi: 10.1038/sj.onc.1203235. [DOI] [PubMed] [Google Scholar]
- 45.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullick J, Cherian SS, Potdar VA. Evolutionary dynamics of the influenza A pandemic (H1N1) 2009 virus with emphasis on Indian isolates: Evidence for adaptive evolution in the HA gene. Infect Genet Evol. 2011;11:997–1005. doi: 10.1016/j.meegid.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Potdar VA, Chadha MS, Jadhav SM. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rattan A, Pawar SD, Nawadkar R. Synergy between the classical and alternative pathways of complement is essential for conferring effective protection against the pandemic influenza A(H1N1) 2009 virus infection. PLoS Pathog. 2017;13:1–26. doi: 10.1371/journal.ppat.1006248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tector M, Salter RD. Calnexin influences folding of human class I histocompatibility proteins but not their assembly with β2-microglobulin. J Biol Chem. 1995;270:19638–19642. doi: 10.1074/jbc.270.33.19638. [DOI] [PubMed] [Google Scholar]
- 50.Wright CA, Kozik P, Zacharias M, Springer S. Tapasin and other chaperones: Models of the MHC class I loading complex. Biol Chem. 2004;385:763–778. doi: 10.1515/BC.2004.100. [DOI] [PubMed] [Google Scholar]
- 51.Jackson MR, Cohen-Doyle MF, Peterson PA, Williams DB. Regulation of MHC class I transport by the molecular chaperone, calnexin (p88, IP90) Science. 1994;263:384–387. doi: 10.1126/science.8278813. [DOI] [PubMed] [Google Scholar]
- 52.Gao B, Adhikari R, Howarth M. Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity. 2002;16:99–109. doi: 10.1016/s1074-7613(01)00260-6. [DOI] [PubMed] [Google Scholar]
- 53.Elliott T, Neefjes J. The complex route to MHC class I-peptide complexes. Cell. 2006;127:249–251. doi: 10.1016/j.cell.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Inoue M, Mimura K, Izawa S. Expression of MHC Class I on breast cancer cells correlates inversely with HER2 expression. Oncoimmunology. 2012;1:1104–1110. doi: 10.4161/onci.21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kageshita T, Hirai S, Ono T. Down-regulation of HLA Class I antigen-processing molecules in malignant melanoma. Association with disease progression Am J Pathol. 1999;154:745–754. doi: 10.1016/S0002-9440(10)65321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seliger B, Atkins D, Bock M. Characterization of human lymphocyte antigen class I antigen-processing machinery defects in renal cell carcinoma lesions with special emphasis on transporter-associated with antigen-processing characterization of human lymphocyte antigen class I antigen. Clin Cancer Res. 2003;9:1721–1727. [PubMed] [Google Scholar]
- 57.Mehta AM, Jordanova ES, Corver WE. Single nucleotide polymorphisms in antigen processing machinery component ERAP1 significantly associate with clinical outcome in cervical carcinoma. Genes Chromosomes Cancer. 2009;48:410–418. doi: 10.1002/gcc.20648. [DOI] [PubMed] [Google Scholar]
- 58.Leone P, Shin EC, Perosa F. MHC class I antigen processing and presenting machinery: Organization, function, and defects in tumor cells. J Natl Cancer Inst. 2013;105:1172–1187. doi: 10.1093/jnci/djt184. [DOI] [PubMed] [Google Scholar]
- 59.Racanelli V, Leone P, Frassanito MA. Alterations in the antigen processing-presenting machinery of transformed plasma cells are associated with reduced recognition by CD8+ T cells and characterize the progression of MGUS to multiple myeloma. Blood. 2010;115:1185–1193. doi: 10.1182/blood-2009-06-228676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noble F, Mellows T, McCormick Matthews LH. Tumour infiltrating lymphocytes correlate with improved survival in patients with oesophageal adenocarcinoma. Cancer Immunol Immunother. 2016;65:651–662. doi: 10.1007/s00262-016-1826-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S, Margolin K. Cytokines in cancer immunotherapy. Cancers (Basel) 2011;3:3856–3893. doi: 10.3390/cancers3043856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsukamoto H, Senju S, Matsumura K. IL-6-mediated environmental conditioning of defective Th1 differentiation dampens antitumour immune responses in old age. Nat Commun. 2015;6:1–15. doi: 10.1038/ncomms7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jarnicki AG, Lysaght J, Todryk S, Mills KHG. Suppression of antitumor immunity by IL-10 and TGF-producing T cells infiltrating the growing tumor: influence of tumor environment on the induction of CD4+ and CD8+ regulatory T cells. J Immunol. 2006;177:896–904. doi: 10.4049/jimmunol.177.2.896. [DOI] [PubMed] [Google Scholar]
- 64.Le Goffic R, Pothlichet J, Vitour D. Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 65.Thube MM, Shil P, Kasbe R. Differences in Type I interferon response in human lung epithelial cells infected by highly pathogenic H5N1 and low pathogenic H11N1 avian influenza viruses. Virus Genes. 2018;54:414–423. doi: 10.1007/s11262-018-1556-1. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 68.Bowers KJ et al (2006) Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In: Proceedings of the ACM/IEEE Conference on Supercomputing (SC06). Tampa, Florida
- 69.Madhavi Sastry G, Adzhigirey M, Day T. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 70.Pierce BG, Wiehe K, Hwang H. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeLano WL. PyMOL: An open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr. 2002;40:82–92. [Google Scholar]