Abstract
The serum lipid profile and clinical outcomes of cancer patients are commonly correlated in a wide range of carcinomas. However, few studies have investigated the serum lipid profile of patients with thyroid cancer (TC). The present study therefore aimed to analyze the lipid profiles of patients with TC. The serum proteomes of 31 participants with stage I–IV TC were screened using Orbitrap Q Exactive Plus. Analytical data collected between November 1, 2013 and November 11, 2018 from the laboratory information system included the total cholesterol (CHO), triglyceride (TG), high-density lipoprotein-cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), apolipoprotein A1 (ApoA1), lipoprotein (a) and apolipoprotein B (ApoB) levels that were used to validate the screening results. A total of 3875 outpatients were enrolled in this study. A number of 17 differentially expressed proteins were identified. An Ingenuity pathway analysis identified activation of the liver X receptor/retinoid X receptor (LXR/RXR) activation, which is a crucial pathway involved in lipid metabolism. The results demonstrated that the total CHO levels were significantly different between patients with TC and control groups, both in men and women. In women, the levels of TG, HDL-C, Apo A1 and LDL-C/HDL-C were significantly different between patients with TC and control groups (all P<0.05). Higher concentrations of TG and LDL-C/HDL-C were observed in the cancer group compared with the control group. However, lower levels of Apo A1 and HDL-C were observed in women from the cancer group compared with the control group. The results from the present study revealed the presence of a disordered lipid profile in patients with TC. The molecular mechanism underlying the association between lipid metabolism and cancer requires further investigation and may be used to develop novel diagnostic biomarkers and therapeutic targets in human cancers.
Keywords: thyroid cancer, lipid profile, Q-Exactive-plus, serum
Introduction
Thyroid cancer (TC) is the most prevalent type of endocrine carcinoma worldwide (1). The incidence of this disease peaks in the third and fourth decades of life (2). TC originates from the follicular epithelium in >95% of all TCs, including papillary thyroid carcinoma (PTC; 85%), follicular thyroid carcinoma (FTC, 11%) and anaplastic thyroid carcinoma (ATC; 4%). Furthermore, medullary thyroid cancer, which originates from the parafollicular cells of the thyroid, accounts for <5% of all TCs (3,4).
According to the 1975–2016 Surveillance, Epidemiology, and End Results reports (5), the incidence of PTC increased from 4.8 to 14.9 per 100,000 people. The ratio of male to female papillary thyroid cancer is usually about 1:2.5. The highest proportion of women in 30–50 years old patients. This sex-based disparity is also specific to the histologic subtype of TC. Specifically, the more aggressive subtypes TC, ATC and MTC, the more similar rates of incidence in men and women have similar incidence rates in men and women (6).
At present, proteomics studies mainly use the techniques of matrix-assisted laser desorption ionization time-of-flight mass spectrometry and surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (7). These simple methods are used to identify spectral fingerprints for prognosis and diagnosis. However, these methodologies offer poor sensitivity and an inability to convincingly identify proteins (8,9). In addition these techniques require a labor-intensive sample preparation protocol when used for the analysis of large sample sets (10). Alternatively, Orbitrap Q Exactive plus is a method that can be used to screen differentially expressed proteins; this technology combines quadrupole ion selection with Orbitrap high-resolution scanning to provide high-quality full-scan and tandem mass spectrometry (MS/MS) data, yielding therefore a superior performance and operability compared with other types of hybrid MS/MS (11).
The study of abnormal tumor lipid metabolism represents a new field of research that has recently received increasing interest. The crosstalk between tumor cells and tumor-associated stromal cells can modulate the tumor high metabolic demands. Notably, tumor cells require a high fatty acid turnover rate to provide the energetic and synthetic requirements for the growing tumor (12). In addition, previous studies reported that patients' lipid profile, including lipid molecules [cholesterol (CHO), triglycerides (TG), high-density lipoprotein-cholesterol (HDL-C), low-density lipoprotein-cholesterol (LDL-C), apolipoprotein A1 (ApoA1), and apolipoprotein B (ApoB)] and their derivative indexes (LDL-C/HDL-C ratio and ApoB/ApoA1 ratio) are associated with various types of carcinoma (13,14).
To the best of our knowledge, only a few studies have investigated the use of a serum lipid profile as a diagnostic tool for TC. The present study aimed to identify a diagnostic serum lipid profile for TC by using a proteomics-based approach with the Orbitrap Q Exactive plus, and to validate this differential lipid profile in a large population.
Materials and methods
Training study and participants
A total of 61 participants were recruited from the Beijing Peking Union Medical College Hospital (PUMCH) and the Zhejiang Cancer Hospital between February 2017 and January 2018. Patients were divided into three groups as follows: i) 15 healthy controls (HC); ii) 15 patients with benign thyroid nodules (TN); and iii) 31 patients with TC. Patients with TC were further divided into three subgroups according to tumor histological analysis and morphology as follows: i) 15 patients with PTC; ii) 10 patients with medullary thyroid cancer (MTC); and iii) 6 patients with FTC.
Validation study design and participants
Data from patients with TC admitted between November 1, 2013 and November 11, 2018 were collected from the outpatient department of the PUMCH. The lipid metabolism data included the CHO, TG, HDL-C, LDL-C, ApoA1, lipoprotein (a) and ApoB levels. Additional information was obtained from the laboratory information system (LIS) A total of 3875 outpatients were enrolled in the study. Lipid profiles were divided into two groups, the cancer (PTC, FTC and MTC) and control (TN and HC) groups. This study enrolled 462 male patients with cancer, 889 male controls, 1255 female patients with cancer and 1269 female controls.
Each cancer group, according to whether there exists the cervical lymph node metastasis, was divided into the metastasis and non-metastasis groups. The information collected from the laboratory information system included the identification number, name, sex, age, hospital department, diagnosis and the results of thyroid function and lipid metabolism tests of patients with TC. The data were analyzed after removing all personal identification information. A statistical method was used to exclude outliers according to the exclusion criteria. Outlying values were identified using Tukey's method, which involves the computation of the 25th (Q1) and 75th (Q3) percentiles and the interquartile range (IQR=Q3-Q1). Outliers were excluded based on the following formula: Q1-3IQR and Q3+3IQR (15).
Inclusion criteria and exclusion criteria for the training and validation studies
The inclusion criteria for the training and validation studies were as follows: i) Patients with TC aged ≥18 years old; ii) patients diagnosed with PTC, MTC and FTC following postoperative histopathological analysis; and iii) patients with thyroid nodules measuring ≥1 cm in diameter. Furthermore, patients with TN were included according to the following features: i) They were aged ≥18 years old; ii) they had thyroid nodules ≥1 cm in diameter as confirmed via ultrasonography; and iii) they had been diagnosed with TN by a physician. HC group comprised healthy volunteers who underwent a health checkup at the PUMCH and who had clear thyroid ultrasonography results. Healthy volunteers were excluded according to the following criteria: i) They presented with systemic diseases, including diabetes mellitus, hypertension, cancer or other cardiovascular, renal, gastrointestinal or pulmonary diseases; ii) they had recurrent TC; iii) they had surgery within 6 months preceding the study; and/or (4) they were taking treatment for thyroid diseases prior to the analysis. No differences in diet were observed among all participants. A total of 1 ml serum was obtained from the eligible candidates. In the cancer group (PTC, MTC, FTC) and TN group, the serum was taken prior to surgery and stored at −80°C. The histopathological assessment and classification were conducted according to the criteria of the World Health Organization (16). Informed consent was obtained from each patient included in this study. The study and procedures were approved by the Institutional Research Ethics Committee of the PUMCH.
Sample preparation for the training study
Serum samples were depleted from immunoglobulin (Ig) G and albumin using the ProteoPrep Blue Albumin and IgG Depletion Kit (PROTBA; Sigma-Aldrich; Merck KGaA) according to the manufacturer s instructions. The protein concentration after depletion and vacuum concentration was evaluated by the Bradford method. A total of 40 µg protein lysate per sample was reduced with 25 mM DTT at 60°C for 30 min and alkylated with 50 mM iodoacetamide in the dark for 10 min. After alkylation, the sample was loaded onto an ultrafiltration filter (10 kDa cut-off; Sartorius AG) for FASP digestion. Trypsin was added at a ratio of 1:100 (enzyme: Protein) at 37°C for 14–16 h. The samples were spun at 20,000 g at 4°C for 10 min. Peptides were then desalted using Ziptip pipette tips containing C18 media (Merck KGaA) according to the manufacturer's instructions.
High pH reversed phase fractionation
The digests from sample preparation were further fractionated using high pH reversed phase chromatography. A reverse chromatography column (XBridge® peptide BEH C18; Waters Corp) combined with the RIGOL L-3000 system (Rigol Technologies, Inc.) were used to separate the mixed peptides in the sample. The peptide mixtures were dissolved in 100 µl mobile phase A [2% (v/v) acetonitrile, 98% (v/v) ddH2O; pH 10] and centrifuged at 14,000 × g at 4°C for 20 min. The supernatant loaded onto the column and eluted stepwise by injecting mobile B [98% (v/v) acetonitrile, 2% (v/v) ddH2O; pH 10]. The flow rate was set at 700 µl/min. The fractions were eluted (1.5 min each) and collected using step gradients of mobile phase B (5.0, 5.0, 8.5, 20.5, 31.0, 90.0, 95.0, 5.0 and 5.0%).
Mass spectrometric data analysis
Date from data-independent acquisition were analyzed using the Spectronaut Pulsar X software (Biognosys AG). Data-dependent acquisition (DDA) spectra were analyzed using the Proteome Discoverer analysis software (version 2.1; Thermo Fisher Scientific, Inc.) with the default settings ‘trypsin/P’ and ‘two missed cleavages’. The DDA files were searched against the human Swiss-Prot FASTA database (www.uniprot.org/uniprot/?query=proteome: UP000005640%20reviewed:yes; state 15.03.2018; 20,240 entries) and the Biognosys iRT peptides FASTA database (biognosys.com/shop/irt-kit#SupportMaterials; uploaded to the public repository).
Laboratory measurements for the validation study
The CHO, TG, HDL-C, LDL-C, ApoA1, ApoB, lipoprotein (a) and free fatty acid (FFA) levels were measured using a Beckman AU Series Automatic Biochemical Analyzer (Beckman Coulter, Inc.) and reagents from Sekisui Medical Co. Ltd. Biochemical laboratories that participated in this survey followed a common internal quality control program that had been standardized by the PUMCH.
Statistical analysis
The Kolmogorov-Smirnov test was used to estimate the distribution of data. Normally distributed continuous variables were presented as the means ± standard deviations. A one-way ANOVA followed by the Student-Newman-Keuls post hoc test was used for statistical analysis. Variables with a skewed distribution were presented as medians (interquartile ranges). The Student's t-test was used to determine the significance between two groups, the Mann-Whitney U test was used to compare differences between groups and the Kruskal-Wallis test was used to compare differences among groups. The quoted P-values were two-sided, and a P-value <0.05 was considered statistically significant. All statistical analyses were performed using SPSS 20.0 (IBM Corp.).
Results
Clinical characteristics of study participants
The demographic data of the participants are provided in Table I. A total of 61 samples were included in the five groups. Among the 15 patients with HC, 15 patients with TN and 15 patients with PTC, the male-to-female ratio was 1:3. No significant differences in age were observed among these three groups. However, age comparisons of participants with FTC and MTC vs. patients with TN, HC and PTC were all significant (P<0.05).
Table I.
Summary of the serum samples collected from the patients in the training study.
| Group | Number (n) | Age (mean ± SD) | P-value | Sex (male: female) |
|---|---|---|---|---|
| HC | 15 | 43.8±9.13 | 0.102a | 4:11 |
| TN | 15 | 49.6±10.38 | 4:11 | |
| PTC | 15 | 43±7.37 | 4:11 | |
| FTC | 6 | 60.2±17.66 | <0.05b | 1:5 |
| MTC | 10 | 54.1±13.2 | <0.05c | 2:3 |
Comparison of age among the three groups.
Age of individuals with FTC compared with those with TN, HC and PTC.
Age of individuals with MTC compared with those with TN, HC and PTC. FTC, follicular thyroid carcinoma; HC, healthy controls; MTC, medullary thyroid cancer; PTC, papillary thyroid carcinoma; SD, standard deviation; TN, thyroid nodules.
Detection and identification of total proteins and differentially expressed proteins
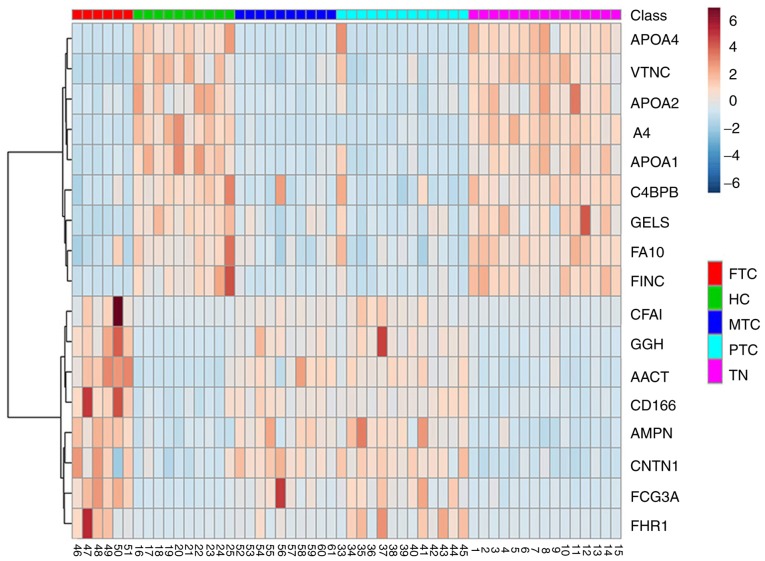
Total proteins were analyzed by Student's t-tests to determine the significance between two groups (FTC_HC, PTC_HC, MTC_HC, FTC_TN, PTC_TN, MTC_TN) with the Perseus software (version 1.5.0.31; Max-Planck Institute of Biochemistry). The q-values were calculated on the basis of permutation-based false-discovery rate (FDR) default setting in Perseus. The significance threshold of P-values was 0.05. Ratio cut-off values were set as 2 or 0.5. Differentially expressed proteins were designated when meeting the two following requirements simultaneously: i) P<0.05; and ii) ratios ≥2 (upregulated) or ≤0.5 (downregulated). The FDR values of the differentially expressed proteins are shown in Data S1. A total of 29 proteins expressed in patients with PTC, FC, MC and HC were identified. Furthermore, 39 proteins were expressed in patients with PTC, FC, MC and TN. In addition, the intersections between these two sets of proteins were assessed. After considering the P-values and ratio, 17 differentially expressed proteins were identified. The results are presented in Figs. 1 and 2. Eight and nine proteins were upregulated (ratio >2) and downregulated (ratio <0.5), respectively, in the TC group compared with the non-TC groups (Table II). These 17 proteins included ApoA1, ApoA2 and ApoA4, which are involved in lipid metabolism.
Figure 1.
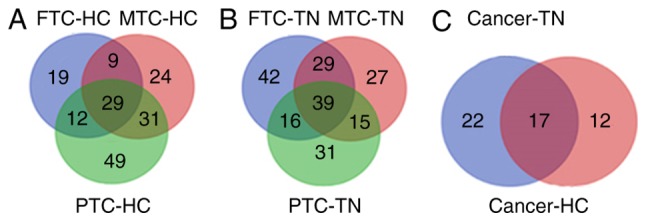
Venn diagrams used to identify 17 differentially expressed proteins. (A) Proteins expressed in the PTC, FTC, MTC and HC groups (n=29). (B) Proteins expressed in the PTC, FTC, MTC and TN groups (n=6). (C) Differentially expressed proteins (n=17). P<0.05 and ratio ≥2 or ≤0.5. FTC, follicular thyroid carcinoma; HC, healthy controls; MTC, medullary thyroid cancer; PTC, papillary thyroid carcinoma; TN, thyroid nodules.
Figure 2.
Heatmap of the expression of these 17 differentially expressed proteins. FTC, follicular thyroid carcinoma; HC, healthy controls; MTC, medullary thyroid cancer; PTC, papillary thyroid carcinoma; TN, thyroid nodules.
Table II.
Expression of the 17 differential proteins.
| Protein groups | Protein description | Up or downregulation | Fold change_(cancer/control) |
|---|---|---|---|
| A4_HUMAN | Amyloid beta A4 protein | Down | 0.0433 |
| APOA4_HUMAN | Apolipoprotein A-IV | Down | 0.134 |
| APOA1_HUMAN | Apolipoprotein A-I | Down | 0.258 |
| FINC_HUMAN | Fibronectin | Down | 0.259 |
| VTNC_HUMAN | Vitronectin | Down | 0.298 |
| APOA2_HUMAN | Apolipoprotein A-II | Down | 0.337 |
| GELS_HUMAN | Gelsolin | Down | 0.356 |
| C4BPB_HUMAN | C4b-binding protein beta chain | Down | 0.403 |
| FA10_HUMAN | Coagulation factor X | Down | 0.435 |
| CNTN1_HUMAN | Contactin-1 | Up | 2.373 |
| AMPN_HUMAN | Aminopeptidase N | Up | 2.566 |
| AACT_HUMAN | Alpha-1-antichymotrypsin | Up | 3.188 |
| FCG3A_HUMAN | Low affinity immunoglobulin gamma Fc region receptor III-A | Up | 3.194 |
| CD166_HUMAN | CD166 antigen | Up | 3.62 |
| CFAI_HUMAN | Complement factor I | Up | 3.814 |
| GGH_HUMAN | Gamma-glutamyl hydrolase | Up | 4.66 |
| FHR1_HUMAN | Complement factor H-related protein 1 | Up | 7.799 |
Ingenuity pathway analysis (IPA) network and pathway analysis
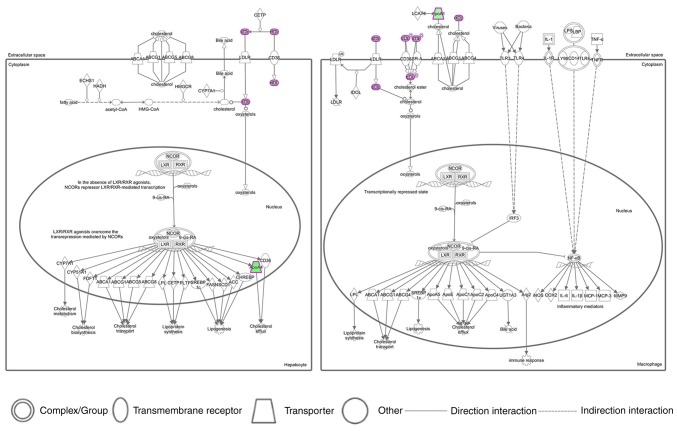
IPA software (Winter 2018 release; Qiagen, Inc.) was used to conduct network analyses of the 17 differentially expressed proteins in the serum samples of individuals with and without TC. The IPA software calculated the P-values using Fisher's exact test and the p score (p score=-lgP-value) in order to assess the probability that the submitted proteins in a protein-protein interaction would be matched by random chance. To do so, the 17 differentially expressed proteins were imported into IPA software, and the top six generated pathways are presented in Table III. The LXR/RXR activation networks are described in Fig. 3. The results demonstrated that the lipid profile of HDL, LDL, ApoA1 and ApoA4 existed in the LXR/RXR activation pathway, which suggested that these lipids may serve important roles in lipid metabolism.
Table III.
Top six canonical pathways enriched by IPA.
| Ingenuity canonical pathways | Log (P-value) | Ratio | Molecules |
|---|---|---|---|
| LXR/RXR activation | 5.70 | 0.0331 | APOA1, APOA4, APOA2, VTN |
| FXR/RXR activation | 5.63 | 0.0317 | APOA1, APOA4, APOA2, VTN |
| Atherosclerosis signaling | 3.92 | 0.0236 | APOA1, APOA4, APOA2 |
| Phagosome formation | 3.88 | 0.0229 | FN1, VTN, FCGR3A/FCGR3B |
| IL-12 signaling and production in macrophages | 3.74 | 0.0205 | APOA1, APOA4, APOA2 |
| Glutathione-mediated detoxification | 3.59 | 0.0645 | GGH, ANPEP |
FXR/RXR, farnesoid X receptor/retinoid X receptor; IL, interleukin; LXR/RXR, liver X receptor/retinoid X receptor.
Figure 3.
Top canonical pathways assigned by the ingenuity pathway analysis for the 17 differentially expressed proteins expressed in individuals with and without TC. The canonical pathway of LXR/RXR activation is presented. Each line and arrow indicates a known functional or physical interaction. Green colored shapes indicate the downregulated expression of a pathway-associated protein. Solid lines indicate direct interactions and dashed lines indicate indirect interactions. LXR/RXR, liver X receptor/retinoid X receptor.
Large data validation of the lipid profiles in the cancer and control groups
The lipid profiles in the cancer (PTC, FTC and MTC) and control (TN and HC) groups are presented in Table IV. The present study enrolled 462 male patients with cancer and 889 male controls, and 1255 female patients with cancer and 1269 female controls. Significant differences in the CHO level were observed between men and women in both groups. Among women, the TG, HDL-C, Apo A1 and LDL-C/HDL-C levels differed significantly between the cancer and control groups (all P<0.05). In particular, TG and LDL-C/HDL-C levels were higher in women from the cancer group compared with women from the control group. Conversely, Apo A1 and HDL-C levels were lower in women from the cancer group compared with women from the control group. Results from analysis of the impact of lymph node metastasis on lipid profile in patients with TC are presented in Table V. Among patients with FTC and MTC, no significant differences were observed between the metastasis and non-metastasis groups. However, among patients with PTC, the CHO, HDL-C, LDLC, Apo A1, Apo B and lipoprotein (a) levels were higher in the non-metastasis group compared with the metastasis group.
Table IV.
Level of lipid profile between the cancer and control groups.
| Men (n=1351) | Women (n=2524) | P-valuea | P-valueb | |||
|---|---|---|---|---|---|---|
| Variable | Cancer (n=462) | Control (n=889) | Cancer (n=1255) | Control (n=1269) | − | − |
| Age (years) | 47 (41–56) | 52 (47–57) | 46 (39–54) | 49 (41–58) | <0.001 | <0.001 |
| CHO (mmol/l) | 4.49 (4.00–5.11) | 4.63 (4.04–5.22) | 4.65 (4.07–5.36) | 4.78 (4.24–5.37) | 0.040 | 0.002 |
| TG (mmol/l) | 1.51 (1.04–2.23) | 1.52 (1.02–2.18) | 1.10 (0.74–1.65) | 0.99 (0.70–1.47) | 0.632 | <0.001 |
| HDL-C (mmol/l) | 1.02 (0.88–1.17) | 1.02 (0.89–1.18) | 1.22 (1.06–1.43) | 1.32 (1.11–1.53) | 0.472 | <0.001 |
| LDL-C (mmol/l) | 2.66 (2.21–3.22) | 2.74 (2.28–3.23) | 2.67 (2.22–3.23) | 2.75 (2.32–3.28) | 0.119 | <0.025 |
| ApoA1 (g/l) | 1.25 (1.13–1.35) | 1.24 (1.13–1.35) | 1.38 (1.26–1.51) | 1.42 (1.28–1.55) | 0.475 | 0.01 |
| ApoB (g/l) | 0.93 (0.80–1.08) | 0.96 (0.83–1.10) | 0.89 (0.75–1.06) | 0.89 (0.77–1.06) | 0.021 | 0.527 |
| Lipoprotein (a) (mg/l) | 77.5 (33–162.25) | 65.5 (35–158) | 85 (42–179) | 97 (47–192) | 0.416 | 0.004 |
| FFA (µmol/l) | 456.5 (340–575.25) | 442 (337–574) | 498 (344.5–665.0) | 471 (351.5–628.5) | 0.828 | 0.214 |
| LDL-C/HDL-C | 2.69 (2.13–3.22) | 2.65 (2.12–3.27) | 2.22 (1.76–2.73) | 2.11 (1.66–2.63) | 0.671 | 0.036 |
| ApoB/ApoA1 | 0.76 (0.62–0.88) | 0.78 (0.64–0.90) | 0.65 (0.53–0.78) | 0.64 (0.53–0.77) | 0.001 | 0.242 |
Male patients with TC vs. male healthy controls
Female patients with TC vs. female healthy controls. ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; CHO, total cholesterol; FFa, free fatty acid; HDL-C, high-density lipoprotein-cholesterol; TG, triglyceride.
Table V.
Association between lipid profile level in the different types of TC and lymph node metastasis among the participants.
| PTC | FTC | MTC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Metastasis | Non-metastasis | P-value | Metastasis | Non-metastasis | P-value | Metastasis | Non-metastasis | P-value |
| Age (years) | 45.00 (38.0053.00) | 50.00 (41.00–58.00) | 0.050 | 49.50 (44.25–54.25) | 51.00 (38.50–56.75) | 0.909 | 47.50 (44.00–53.25) | 48.00 (40.00–60.00) | 0.869 |
| CHO (mmol/l) | 4.41 (3.80–5.07) | 4.65 (4.07–5.31) | 0.001 | 4.05 (1.08–4.69) | 4.65 (3.91–5.47) | 0.148 | 4.44 (3.93–5.43) | 4.80 (4.18–5.26) | 0.803 |
| TG (mmol/l) | 1.15 (0.79–1.80) | 1.18 (0.78–1.78) | 0.654 | 3.69 (1.24–4.78) | 1.77 (1.08–2.34) | 0074 | 1.45 (1.12–2.29) | 1.40 (0.95–2.48) | 0.914 |
| HDL-C (mmol/l) | 1.12 (0.96–1.30) | 1.16 (1.00–1.37) | 0.024 | 1.32 (0.98–1.52) | 1.17 (0.95–1.38) | 0.481 | 1.29 (1.02–1.73) | 1.27 (1.04–1.51) | 0.266 |
| LDL-C (mmol/l) | 2.50 (2.11–3.14) | 2.69 (2.25–3.22) | 0.003 | 2.47 (2.05–3.43) | 2.86 (2.26–3.51) | 0.392 | 2.66 (2.24–3.08) | 2.83 (2.30–3.28) | 0.521 |
| Apo A1 (g/l) | 1.32 (1.20–1.43) | 1.35 (1.23–1.48) | 0.023 | 1.22 (1.13–1.44) | 1.35 (1.20–1.43) | 0.493 | 1.52 (1.22–1.74) | 1.36 (1.18–1.54) | 0.114 |
| Apo B(g/l) | 0.85 (0.72–1.03) | 0.91 (0.77–1.07) | 0.006 | 0.9350 (0.7775–1.0825) | 1.03 (0.77–1.15) | 0.879 | 0.83 (0.78–1.04) | 0.94 (0.80–1.07) | 0.353 |
| Lipoprotein (a) | 75.00 | 85.00 | 0.043 | 89.50 | 124.00 | 0.581 | 35.00 | 71.00 | 0.105 |
| (mg/l) | (33.75–149.75) | (42.00–177.00) | (57.25–158.50) | (47.75–379.25) | (18.50–119.20) | (29.75–192.00) | |||
| FFA (µmol/l) | 466.00 | 487.00 | 0.416 | 462.00 | 374.00 | 0.502 | 586.00 | 478.00 | 0.862 |
| (347.50–621.00) | (344.00–638.50) | (286.00–713.00) | (309.00–584.00) | (315.25–678.75) | (351.50–673.50) | ||||
| LDLC/HDL-C | 2.28 (1.80–2.88) | 2.33 (1.84–2.90) | 0.542 | 2.04 (1.75–2.51) | 2.77 (2.21–3.10) | 0.057 | 2.37 (1.41–2.71) | 2.26 (1.76–2.93) | 0.317 |
| ApoB/ApoA1 | 0.66 (0.64–0.80) | 0.67 (0.56–0.81) | 0.322 | 0.73 (0.60–0.94) | 0.75 (0.65–0.86) | 0.939 | 0.65 (0.43–0.79) | 0.67 (0.52–0.86) | 0.205 |
ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; CHO, total cholesterol; FFa, free fatty acid; FTC, follicular thyroid carcinoma; HC, healthy controls; HDL-C, high-density lipoprotein-cholesterol; MTC, medullary thyroid cancer; PTC, papillary thyroid carcinoma; TG, triglyceride.
Discussion
Recent studies revealed that lipid metabolism disorders are important in carcinogenesis and cancer development, as they induce abnormalities in the expression of various genes and proteins, and in the regulation of cytokines and signaling pathways (17,18). The present study demonstrated that lipid metabolism disorders occurred in patients with TC. To the best of our knowledge, this study was the first to validate the differential lipid profile in a large population of patients with TC.
As the incidence of TC varies by sex (6), the present study screened the differential lipid profiles in age- and sex-matching samples. A total of 17 proteins were designated as differentially expressed proteins. These proteins were therefore subjected to bioinformatics analyses. The IPA output identified the LXR/RXR activation pathway as the significant canonical pathway. In addition, significantly lower expression levels of ApoA1, ApoA2 and ApoA4 were observed in patients with TC, compared with healthy controls. Furthermore, after observing the significant modulation of expression in TC pathogenesis and etiology, this differential lipid profile was validated in a larger population.
A previous cohort study reported that the serum TG concentration is negatively and positively correlated with the risk of prostate and renal cancer, respectively, in men. Furthermore, serum TG level is positively correlated with the risk of gynecological cancer in women (19). Consistent with that study, the present study demonstrated that TG concentrations were higher in women with TC compared with women in the control group. In men however, no significant difference was observed in the TG level between cancer and control groups. A previous study demonstrated that TG levels are not associated with the risk of cancer-associated mortality (20). Further investigation is therefore needed to validate the importance of TG in tumor development.
ApoA-I (28 kDa), which is the major protein constituent of high-density lipoprotein, serves a crucial role in reverse cholesterol transport. This protein can transfer cholesterol and phospholipids from peripheral cells to the liver for excretion (21). A previous study identified HPX, POTEE and ApoA1 as the most significant genes associated with the proteomic profile. In addition, these genes are significantly correlated with the risk of breast cancer and could be used for the detection of disease progression (22). In the present study, women in the cancer group expressed lower levels of Apo A1 compared with women in the control group.
Numerous studies have validated the prognostic values of lipid molecules and their derivative indexes in various types of carcinoma (23,24). In the present study, a new derivative index was developed for the ApoB/ApoA1 and LDL-C/HDL-C ratios. Furthermore, the LDL-C/HDL-C and ApoB/ApoA1 ratios have significant diagnostic values in several diseases (25,26). Ma et al (14) reported that, among the various lipid molecules and derivative indexes, the ApoB/ApoA1 ratio could serve as an independent prognostic marker of gastric cancer. In the present study, the LDL-C/HDL-C level was significantly higher in women with TC compared with women in the control group, mainly because the former group had a decreased level of HDL-C.
Hong et al (23) reported that the preoperative serum lipid profile is correlated with the outcome of patients with non-metastatic colorectal cancer. In the present study, no significant differences between the metastasis and non-metastasis groups were observed in patients with FTC and MTC. The effect of lymph node metastasis on lipid metabolism requires therefore further investigation.
Preliminary exploration of lipid metabolism in patients with TC revealed the existence of lipid metabolism disorders (27). The present study demonstrated that ApoA1 and HDL-C levels, which have positive effects in human metabolism, were decreased Conversely, TG level and LDL-C/HDL-C ratio, which have negative effects on human metabolism, were increased. Identification of a lipid metabolism disorder in patients may therefore provide additional evidences leading to the diagnosis of TC. Subsequently, lipid profile analysis is essential and may yield valuable biomarkers in the early diagnosis of TC.
The present study had some major strengths. Firstly, it included a variety of patients with various types of TC. Secondly, the screening targeted differentially expressed proteins. Thirdly, the differentially expressed proteins involved in lipid mechanism between the groups were compared and validated in large samples. However, this study had certain limitations. Firstly, the screening and validation experiments applied age- and sex-matching to HC and patients with TN and PTC; however, as the incidence rates of MTC and FTC were extremely low, the sample sizes for these tumors were too small to allow appropriate matching. Secondly, the exact moment of tumor recurrence/progression could not be determined due to information bias. This study did therefore not evaluate the effects of lipid metabolism on the prognosis of TC. Thirdly, the underlying mechanism of lipid metabolism on the carcinogenesis and development of TC requires further investigation.
In conclusion, the present study identified lipid metabolism disorders in patients with TC. Notably, ApoA1, ApoA2 and ApoA4 levels were significantly lower in patients with TC compared with healthy controls. Following further validation, significantly lower levels of Apo A1 and HDL-C were identified in women with TC compared with female controls. The underlying molecular mechanism of lipid metabolism and cancer require further investigation in order to develop potential novel diagnostic biomarkers and therapeutic targets for human cancers.
Supplementary Material
Acknowledgements
The authors would like to thank Mr. Jun Wang (Laboratory Information System, Beijing Peking Union Medical College Hospital) for providing technical support.
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81702060; www.nsfc.gov.cn).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DL, JW and LQ designed the study. WC, YMZ and LZ collected the samples. DL and CM analyzed the data. SY, DW and YTZ made substantial contributions to conception and design and the acquisition of data and contributed reagents, materials and analysis tools. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Institutional Research Ethics Committee of PUMCH. Informed consent was obtained from each patient included in this study.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Scopa CD. Histopathology of thyroid tumors. An overview. Hormones (Athens) 2004;3:100–110. doi: 10.14310/horm.2002.11118. [DOI] [PubMed] [Google Scholar]
- 2.Holmes L, Jr, Hossain J, Opara F. Pediatric thyroid carcinoma incidence and temporal trends in the USA (1973–2007): Race or shifting diagnostic paradigm? ISRN Oncol. 2012;2012:906197. doi: 10.5402/2012/906197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faquin WC. The thyroid gland: Recurring problems in histologic and cytologic evaluation. Arch Pathol Lab Med. 2008;132:622–632. doi: 10.5858/2008-132-622-TTGRPI. [DOI] [PubMed] [Google Scholar]
- 4.Navas-Carrillo D, Rios A, Rodriguez JM, Parrilla P, Orenes-Piñero E. Familial nonmedullary thyroid cancer: Screening, clinical, molecular and genetic findings. Biochim Biophys Acta. 2014;1846:468–476. doi: 10.1016/j.bbcan.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al., editors. National Cancer Institute; Bethesda, MD: Apr, 2019. SEER Cancer Statistics Review, 1975–2016. based on November 2018 SEER data submission, posted to the SEER web site. [Google Scholar]
- 6.Rahbari R, Zhang L, Kebebew E. Thyroid cancer gender disparity. Future Oncol. 2010;6:1771–1779. doi: 10.2217/fon.10.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paricharttanakul NM, Saharat K, Chokchaichamnankit D, Punyarit P, Srisomsap C, Svasti J. Unveiling a novel biomarker panel for diagnosis and classification of well-differentiated thyroid carcinomas. Oncol Rep. 2016;35:2286–2296. doi: 10.3892/or.2016.4567. [DOI] [PubMed] [Google Scholar]
- 8.Villanueva J, Martorella AJ, Lawlor K, Philip J, Fleisher M, Robbins RJ, Tempst P. Serum peptidome patterns that distinguish metastatic thyroid carcinoma from cancer-free controls are unbiased by gender and age. Mol Cell Proteomics. 2006;5:1840–1852. doi: 10.1074/mcp.M600229-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Wang JX, Yu JK, Wang L, Liu QL, Zhang J, Zheng S. Application of serum protein fingerprint in diagnosis of papillary thyroid carcinoma. Proteomics. 2006;6:5344–5349. doi: 10.1002/pmic.200500833. [DOI] [PubMed] [Google Scholar]
- 10.Cohen A, Wang E, Chisholm KA, Kostyleva R, O'Connor-McCourt M, Pinto DM. A mass spectrometry-based plasma protein panel targeting the tumor microenvironment in patients with breast cancer. J Proteomics. 2013;81:135–147. doi: 10.1016/j.jprot.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, et al. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maan M, Peters JM, Dutta M, Patterson AD. Lipid metabolism and lipophagy in cancer. Biochem Biophys Res Commun. 2018;504:582–589. doi: 10.1016/j.bbrc.2018.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Horikawa M, Sato S, Miyake H, Setou M. Discovery of lipid biomarkers correlated with disease progression in clear cell renal cell carcinoma using desorption electrospray ionization imaging mass spectrometry. Oncotarget. 2019;10:1688–1703. doi: 10.18632/oncotarget.26706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma MZ, Yuan SQ, Chen YM, Zhou ZW. Preoperative apolipoprotein B/apolipoprotein A1 ratio: A novel prognostic factor for gastric cancer. Onco Targets Ther. 2018;11:2169–2176. doi: 10.2147/OTT.S156690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Cheng X, Yu S, Qiu L, Lian X, Guo X, Hu Y, Lu S, Yang G, Liu H. Data mining: Seasonal and temperature fluctuations in thyroid-stimulating hormone. Clin Biochem. 2018;60:59–63. doi: 10.1016/j.clinbiochem.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Thompson L. World Health Organization classification of tumours: Pathology and genetics of head and neck tumours. Ear Nose Throat J. 2006;85:74. doi: 10.1177/014556130608500201. [DOI] [PubMed] [Google Scholar]
- 17.Long J, Zhang CJ, Zhu N, Du K, Yin YF, Tan X, Liao DF, Qin L. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8:778–791. [PMC free article] [PubMed] [Google Scholar]
- 18.Nikitin PV, Potapov AA, Ryzhova MV, Shurkhay VA, Kulikov EE, Zhvanskiy ES, Popov IA, Nikolaev EN. The role of lipid metabolism disorders, atypical isoforms of protein kinase C, and mutational status of cytosolic and mitochondrial forms of isocitrate dehydrogenase in carcinogenesis of glial tumors. Zh Vopr Neirokhir Im N N Burdenko. 2018;82:112–120. doi: 10.17116/neiro2018823112. (In Russian) [DOI] [PubMed] [Google Scholar]
- 19.Ulmer H, Borena W, Rapp K, Klenk J, Strasak A, Diem G, Concin H, Nagel G. Serum triglyceride concentrations and cancer risk in a large cohort study in Austria. Br J Cancer. 2009;101:1202–1206. doi: 10.1038/sj.bjc.6605264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin RM, Vatten L, Gunnell D, Romundstad P, Nilsen TI. Components of the metabolic syndrome and risk of prostate cancer: The HUNT 2 cohort, Norway. Cancer Causes Control. 2009;20:1181–1192. doi: 10.1007/s10552-009-9319-x. [DOI] [PubMed] [Google Scholar]
- 21.Breslow JL, Ross D, McPherson J, Williams H, Kurnit D, Nussbaum AL, Karathanasis SK, Zannis VI. Isolation and characterization of cDNA clones for human apolipoprotein A-I. Proc Natl Acad Sci USA. 1982;79:6861–6865. doi: 10.1073/pnas.79.22.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cine N, Baykal AT, Sunnetci D, Canturk Z, Serhatli M, Savli H. Identification of ApoA1, HPX and POTEE genes by omic analysis in breast cancer. Oncol Rep. 2014;32:1078–1086. doi: 10.3892/or.2014.3277. [DOI] [PubMed] [Google Scholar]
- 23.Hong TT, Shen D, Chen XP, Wu XH, Hua D. Preoperative serum lipid profile and outcome in nonmetastatic colorectal cancer. Chronic Dis Transl Med. 2016;2:241–249. doi: 10.1016/j.cdtm.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao R, Cheng G, Wang B, Qin C, Liu Y, Pan Y, Wang J, Hua L, Zhu W, Wang Z. BMI and serum lipid parameters predict increasing risk and aggressive prostate cancer in Chinese people. Oncotarget. 2017;8:66051–66060. doi: 10.18632/oncotarget.19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu BZ, Tao L, Chen YZ, Li XZ, Dong YL, Ma YJ, Li SG, Li F, Zhang WJ. Preoperative body mass index, blood albumin and triglycerides predict survival for patients with gastric cancer. PLoS One. 2016;11:e0157401. doi: 10.1371/journal.pone.0157401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You S, Zhong C, Xu J, Han Q, Zhang X, Liu H, Zhang Y, Shi J, Huang Z, Xiao G, et al. LDL-C/HDL-C ratio and risk of all-cause mortality in patients with intracerebral hemorrhage. Neurol Res. 2016;38:903–908. doi: 10.1080/01616412.2016.1204797. [DOI] [PubMed] [Google Scholar]
- 27.Liao T, Wang YJ, Hu JQ, Wang Y, Han LT, Ma B, Shi RL, Qu N, Wei WJ, Guan Q, et al. Histone methyltransferase KMT5A gene modulates oncogenesis and lipid metabolism of papillary thyroid cancer in vitro. Oncol Rep. 2018;39:2185–2192. doi: 10.3892/or.2018.6295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.