Abstract
The transcriptional coactivator Amplified in Breast Cancer 1 (AIB1) plays a major role in the progression of hormone and HER2-dependent breast cancers but its role in triple negative breast cancer (TNBC) is undefined. Here, we report that established TNBC cell lines, as well as cells from a TNBC patient-derived xenograft (PDX) that survive chemotherapy treatment in vitro express lower levels of AIB1 protein. The surviving cell population has an impaired tube-formation phenotype when cultured onto basement membrane, a property shared with TNBC cells that survive shRNA-mediated depletion of AIB1 (AIB1LOW cells). DNA analysis by exome sequencing revealed that AIB1LOW cells represent a distinct subpopulation. Consistent with their in vitro phenotype AIB1LOW cells implanted orthotopically generated slower growing tumors with less capacity for pulmonary metastases. Gene expression analysis of cultured cells and tumors revealed that AIB1LOW cells display a distinct expression signature of genes in pro-inflammatory pathways, cell adhesion, proteolysis and tissue remodeling. Interestingly, the presence of this AIB1LOW expression signature in breast cancer specimens is associated with shorter disease free survival of chemotherapy treated patients. We concluded that TNBC cell lines contain heterogeneous populations with differential dependence on AIB1 and that the gene expression pattern of AIB1LOW cells may represent a signature indicative of poor response to chemotherapy in TNBC patients.
Introduction
Triple negative breast cancer (TNBC) is a breast cancer subtype that lacks expression of hormone receptors (ER, PR) and HER2 amplification [1], [2]. It represents 15–20% of all breast cancer cases in the United States. Gene expression profiling broadly classifies breast cancers into luminal A and B, HER2, and basal intrinsic molecular subtypes [3], [4]. Most TNBC tumors overlap with the basal intrinsic subtype, characterized by expression of basal keratins 5, 6, 14, and 17 [5], [6]. More recently, further classification of TNBC by gene expression has resulted in four major subtypes of TNBC [7], [8], including basal-like (BL) 1 and 2, mesenchymal (M), and luminal androgen-receptor (LAR). Despite the refinement of TNBC classification, it is not clear whether different subtypes of TNBC are driven by diverse signaling pathways during malignant initiation, progression or metastasis. Similarly, it is not yet clear whether patients assigned to these novel subtypes of TNBC present different therapeutic opportunities or whether each subtype has different levels of resistance to therapy, although results using small cohorts are consistent with this notion [9], [10].
Patients diagnosed with TNBC have significantly worse clinical outcomes than patients diagnosed with luminal disease [11], [12]. Furthermore, epidemiological studies in the US have reported an increased prevalence and higher mortality rate of TNBC in young African American women compared to other groups [13], [14], [15]. Targeted therapy for TNBC using EGFR [16], Src [17], and MEK [18] inhibitors have been tested in TNBC patients, but have not significantly improved the outcomes although PARP inhibitors have promising efficacy in patients whose tumors harbor BRCA mutations [19]. The current standard of care for TNBC consists of anthracycline and taxane-based chemotherapy regimens [20] in the neoadjuvant, adjuvant, and metastatic setting [21], [22]. Despite a high response rate of TNBC to chemotherapy, fewer than 30%, of those that progress to metastatic TNBC, survive 5 years after diagnosis [23], [24]. Currently the relationship between the different subtypes of TNBC and their response to treatment or their resistance to therapy is beginning to be elucidated [25], [26]. Furthermore it has been postulated that resistance to chemotherapy can occur in TNBC and other cancers because a subpopulation of cancer stem (CSC) cells are relatively resistant to chemotherapy (reviewed in [27]).
The oncogene AIB1 (AIB1/SRC3/NCOA3) is a member of the nuclear receptor coactivator family and interacts with nuclear receptors as well as a host of transcription factors, including NF-κB [28], E2F1 [29], STAT6 [30] to influence gene transcription (reviewed in [31], [32]). Clinical correlative data has shown that AIB1 expression is associated with worse outcomes in estrogen receptor (ER) positive luminal breast cancer [33] and contributes to anti-estrogen tamoxifen resistance [34], [35]. AIB1 also plays a role in the signaling and in the progression of HER2 amplified breast cancers [36], [37]. However, a role for AIB1 in TNBC is not well defined, although there is a reported association between higher mRNA levels of AIB1 and decreased overall survival of TNBC patients [38]. In the present study, we sought to determine the role of AIB1 in TNBC using established cell lines from African American women [39], [40], [41] and from a patient derived xenograft.
Results
TNBC Cells That Survive Chemotherapy Have Reduced Protein Levels of AIB1
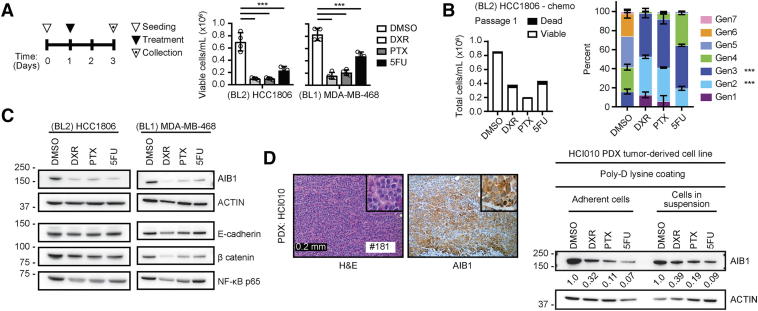
Chemotherapy treatment can result in the enrichment of slow-proliferating, stem-like, tumor initiating cells (TIC) that may lead to therapy resistance [42], [43], [44], [45]. We have previously reported that AIB1 is involved in the maintenance of TIC in a ductal carcinoma in situ (DCIS) cell line [46]. Thus, we sought to determine if cytotoxic chemotherapy could modulate the expression of AIB1 in BL1 (HCC1806) and BL2 (MDA-MB-468) TNBC lines. Single-agent, IC50, treatment (Figure 1A) with either Doxorubicin (DXR, 90 nM), Paclitaxel (PTX, 2 nM), or 5-Fluorouracil (5FU, 4.9 μM) resulted in significant cell death and reduced cell proliferation as measured by TrypanBlue exclusion or cell trace analysis (Figure 1, A and B). Interestingly, the chemotherapy surviving population of HCC1806 and MDA-MB-468 cells had reduced protein levels of AIB1 (Figure 1C). We thus evaluated the protein expression of known AIB1 downstream targets such as E-cadherin, β-catenin and NF-κB that have also been reported to contribute to TNBC chemotherapy resistance ([28], [47], [48], respectively). Indeed, we observed down-regulation of AIB1 downstream targets at the protein level in the chemotherapy surviving HCC1806 cells and MDA-MB-468 (Figure 1C and Supplementary Figure 1A). Of note is that flow cytometry analysis of AIB1 in untreated TNBC cell lines demonstrated that nearly all cells (>97%) expressed high and homogeneous levels of AIB1 (Supplementary Figure 1B). To extend these observations, we obtained a TNBC patient derived xenograft (PDX-HCI010) developed by DeRose and colleagues [49]. PDX models represent with high fidelity human tumor progression in immunocompromised mice [50]. We determined that the PDX tumor expressed AIB1 by IHC (Figure 1D, left). Subsequently, we established a cell line from the PDX tumor and treated this with DXR [0.25 μM], PTX [0.79 μM], or 5FU [32.8 μM] and also observed significant reduction of AIB1 protein levels in surviving cells under both adherent and in-suspension conditions (Figure 1D, right) as seen in the established TNBC cell lines.
Figure 1.
Chemotherapy downregulates AIB1 expression in TNBC cell lines. (A) HCC1806 and MDA-MB-468 cells were treated as shown (left panel) with DXR, PTX and 5FU. Viable cells were determined by Trypan blue exclusion (n = 4) (right panel) (B) Total count of HCC1806 cells labeled with Cell Trace Violet dye (left) following chemotherapy treatment (n = 2) and percent distribution of dividing cells by doubling generations (right). (C) Representative Western blot images for AIB1, E-cadherin, β catenin, and NF-kB from chemotherapy-treated surviving HCC1806 and MDA-MB-468 cells (n = 2) (D) H&E and AIB1 IHC staining of HCI010 PDX tumor grafts (left) and Western blot images (right) of HCI010 PDX-derived cell lines treated as in A. Graphs are representative of three independent experiments. Technical repeats shown. Mean ± SEM. Scale bar: 200 μm. One-way ANOVA followed by Sidak's (A and C) or Dunnett's (B) multiple comparisons test. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
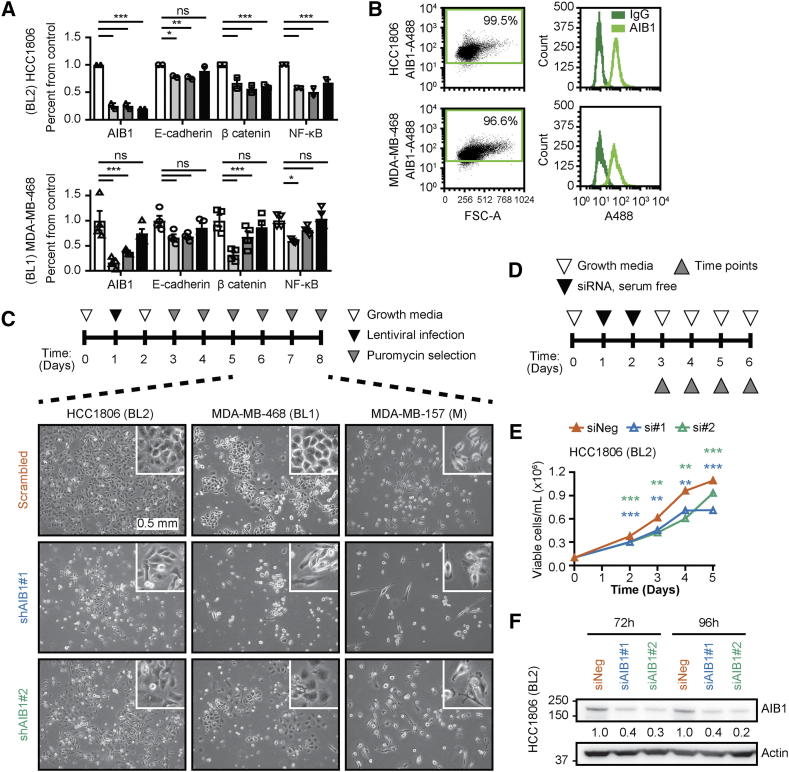
Supplementary Figure 1.
Molecular characterization of AIB1 expressing TNBC cell lines following chemotherapy and RNAi treatment. (A) Quantitation of band intensity normalized to Actin for listed proteins from Western blots in chemotherapy-treated surviving basal like subtypes of TNBC cell lines. (B) Intracellular AIB1 protein expression of parental HCC1806 and MDA-MB-468 cells analyzed by flow cytometry. (C) Schematic timeline for shRNA lentiviral infection and puromycin selection. Representative images of shRNA-treated TNBC cell lines after puromycin selection. (D) Schematic timeline for siRNA treatment of HCC1806 cell lines. (E) Proliferation of siRNA treated HCC1806 cells. (E) Representative Western blot image for AIB1 protein in siRNA-treated HCC1806 cells at 72 and 96 hours. Two-way ANOVA followed by Dunnett's multiple comparisons test in A. Linear regression (slope coefficient) in D. *P ≤ 0.05, **P ≤ .01, ***P ≤ .001.
Depletion of AIB1 From TNBC Cell Lines Selects for a Surviving Subpopulation Expressing Low Levels of AIB1
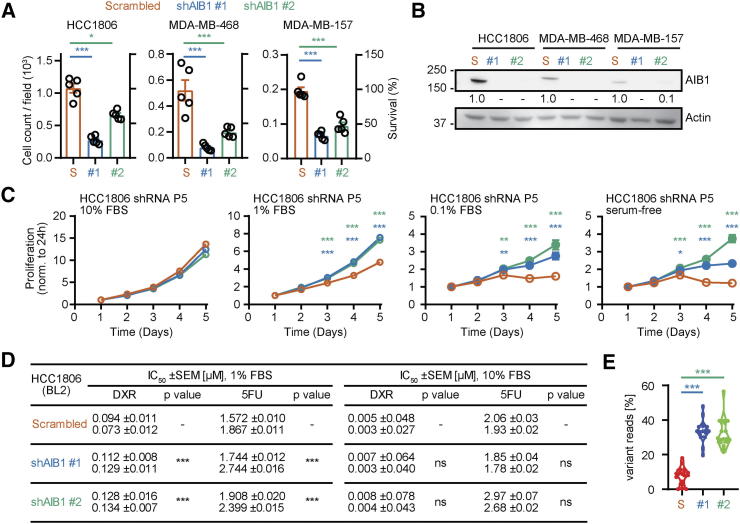
Our lab has previously shown that reduced expression of AIB1 affects the proliferation rate and the phenotype of MDA-MB-231 cells, a mesenchymal subtype of TNBC, in vitro [51]. To further investigate the functional significance of AIB1, we reduced AIB1 expression using lentiviral transduction with two distinct shRNAs (shAIB1#1 and shAIB1#2) and compared that to a scrambled control shRNA in cell lines representing three different subtypes of TNBC (Supplementary Figure 1C). Following puromycin selection, there was a significant reduction in cell survival (25%–60%) in the AIB1 shRNA transduced cells relative to their controls across the three subtypes of TNBC cell lines (Figure 2A) that correlated with reduced AIB1 protein levels (Figure 2B). Reduced cell viability after shRNA transduction was not an off-target effect of the lentiviral infection or puromycin selection because transient knock-down with AIB1 siRNA also reduced cell numbers that correlated with reduced levels of AIB1 protein (Supplementary Figure 1, D-F). Surviving cells after AIB1 shRNA maintained lower levels of AIB1 in serial passage (AIB1LOW) (Supplementary Figure 2A) and further experiments were conducted on HCC1806 AIB1LOW cells. The phenotype of the HCC1806 AIB1LOW cells differs from control shRNA cells showing increased proliferation in reduced serum containing media but not after 10%-serum supplement (Figure 2C and Supplementary Figure 2B). We also observed a small but significant increase in IC50 for both DXR and 5FU under low serum conditions in HCC1806 AIB1LOW compared to shRNA control cells (Figure 2D). Gene expression analysis of HCC1806 AIB1LOW cells showed a number of differentially regulated genes compared to control shRNA cells that are shown in Supplementary Figure 2C. Taken together, the reduction in AIB1 protein levels in TNBC cell lines appears to select for a cell subpopulation with a distinct gene expression profile.
Figure 2.
Phenotype of AIB1 shRNA on BL2-HCC1806 cells in vitro. (A) Cell count per field (n = 5) (left axis) and percent survival (right axis) of TNBC cell lines following AIB1 shRNA infection and selection relative to their respective control shRNA. (B) Representative Western images for AIB1 in cells from A. (C) Proliferation of serial passaged HCC1806 AIB1LOW relative to control shRNA in 10%, 1%, 0.1% serum-supplemented RPMI 1640 growth media or serum-free basal media. (D) IC50 from dose response curves of 72-hours chemotherapy treated HCC1806 AIB1LOW cells and control shRNA in 1% or 10% serum-supplemented culture conditions. Graphs are representative of three independent experiments. Mean ± SEM. Linear regression (slope coefficient) and Non-linear regression (least-squares) for each cell type. (E) Genomic variant analysis of HCC1806 AIB1LOW relative to control shRNA (passage 5). Quantification of variant reads (16 variants on 6 chromosomes, n = 2) is shown as a percentage of total reads. One-way ANOVA followed by Dunnett's multiple comparisons test. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
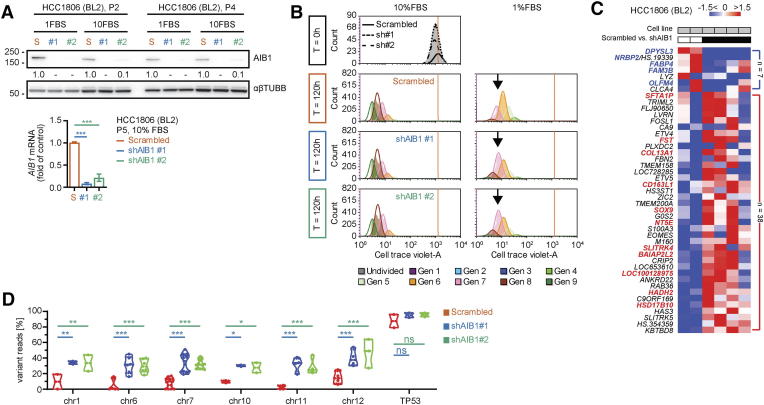
Supplementary Figure 2.
Molecular characterization of serial passaged HCC1806 AIB1LOWcells in vitro. (A) Representative Western blot images for AIB1 protein and PCR gene expression of AIB1 in serial passaged HCC1806 AIB1LOW relative to control shRNA. (B) Proliferation analysis by CellTrace Violet of serial passaged HCC1806 AIB1LOW relative to control shRNA in 10% or 1% serum-supplemented growth media after 72 hours. (C) Heatmap showing differentially expressed genes in HCC1806 AIB1LOW cells relative to control shRNA from cell lines (2D culture). (D) Quantification of genomic variant reads as percentage of total reads for each genomic locus in HCC1806 AIB1LOW cells relative to control shRNA. TP53 intron 6 is shown as a control (n = 2). One-way and two-way ANOVA followed by Dunnett's multiple comparisons test in A (lower panel) and D, respectively. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
AIB1LOW Cells Result From Clonal Expansion of a Subpopulation With a Distinct Genetic Profile
Cultured cell lines adapt to in vitro culture conditions although they maintain genetic, molecular, and phenotypic heterogeneity [52], [53] similar to intratumoral heterogeneity observed in human cancers [54]. As shown above the analysis of the parental TNBC cells for expression of the AIB1 protein by flow cytometry showed a relatively homogenous distribution of protein expression levels in HCC1806 or MDA468 cells (Supplemental Figure 1B). Therefore we speculated that the parental TNBC cell lines contain subpopulations with different dependence on AIB1 for their survival and that the depletion of endogenous AIB1 would result in the selection of cells that are less dependent on the expression of AIB1. To further assess the cell population heterogeneity we performed whole exome sequence analysis on cells that survived after control shRNA or AIB1 shRNA transduction and selection (passage 5). Homozygous mutations did not show a difference between the control and AIB1 shRNA cell lines. However, heterozygous alterations revealed differences: The comparison of the two HCC1806 AIB1low lines showed an indistinguishable pattern between them. A high portion of AIB1low cells (20% - 50%) carry a heterozygous mutation pattern that is also present in the control shRNA transduced cells yet diluted by other populations resulting in a significantly lower abundance (<20%) (Figure 2E). It is noteworthy that the higher portion of heterozygous mutations in the AIB1 depleted cell populations was evenly distributed across all chromosomes (Supplemental Figure 2D). This shows that depletion of AIB1 reveals a pre-existing subpopulation in the parental line that is less dependent on AIB1 expression for their survival.
TNBC Cells With Reduced Levels of AIB1 Have Reduced Tube Forming Capacity
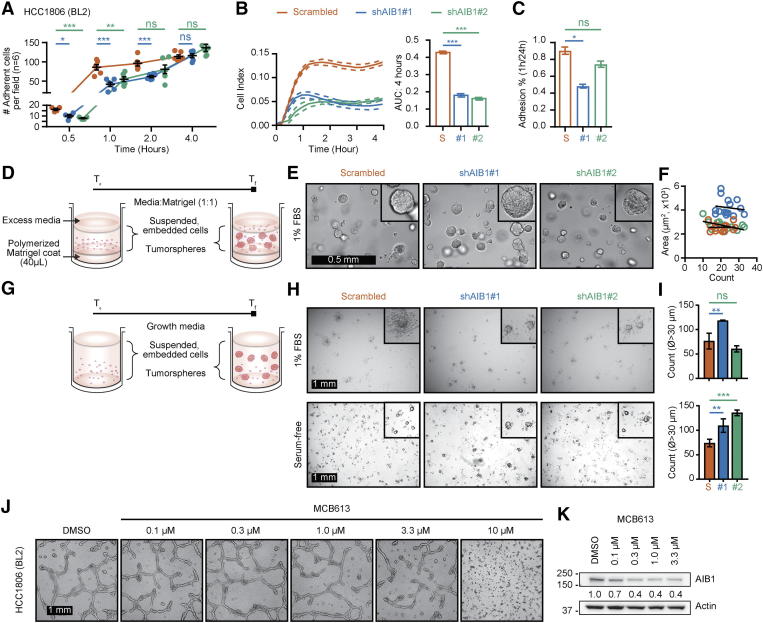
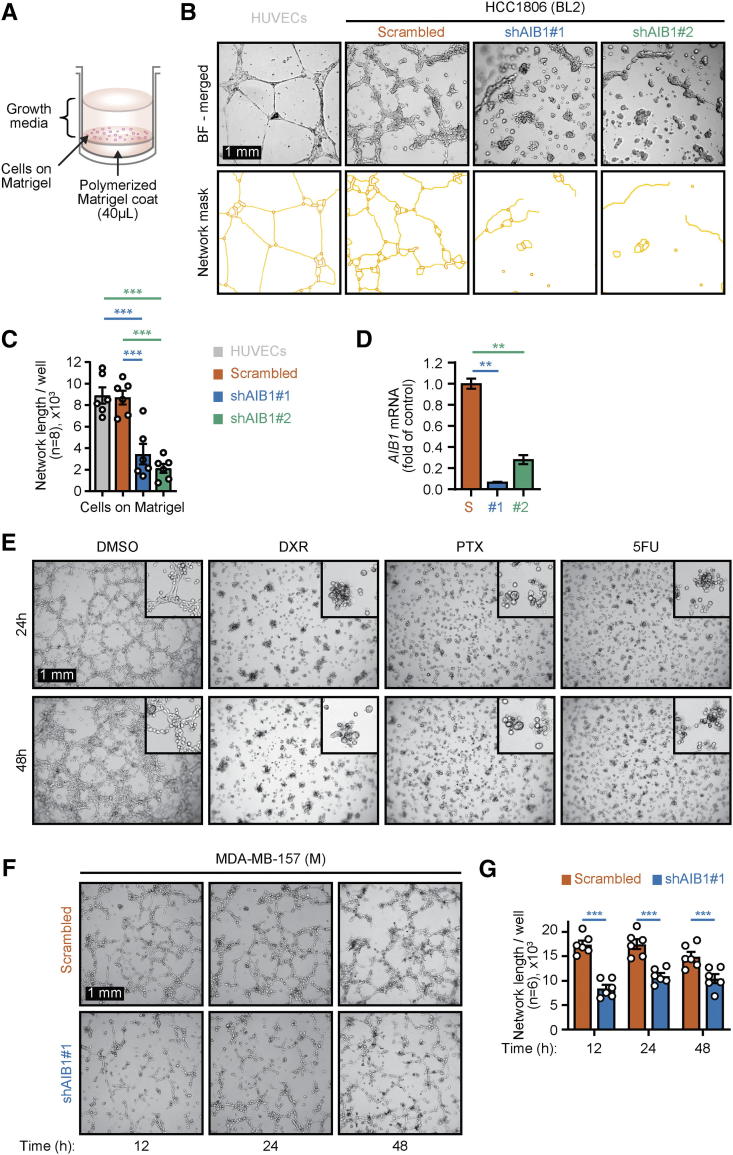
One property of cancer stem cells (CSC) in vitro is their ability to form spheres in non-adherent conditions [55]. Because we observed differences in attachment kinetics with HCC1806 AIB1LOW cells relative to control shRNA cells when seeded onto uncoated culture dishes (Supplementary Figure 3, A–C), we conjectured that AIB1LOW cells might be related to a cancer stem-like population. Tumorspheres embedded in Matrigel-or in ultra-low attachment (ULA) conditions [56], showed no differences in size or count between groups in low serum (1%) growth media (Supplementary Figures 3, D–I, top). However, we observed an increased number of tumorspheres with >30 μm diameter HCC1806 AIB1LOW cells relative to control shRNA when cultured in serum-free RPMI basal growth media (Supplementary Figure 3, H and I, bottom). An additional property of CSC is related to tube-formation, a phenomenon related to vascular mimicry in where cancer cells form channels that connect to blood vessels for nutrients and oxygen [57]. Tube-formation assays were initially described using human umbilical vein endothelial cells (HUVEC) [58] (Figure 3A). We have previously reported AIB1 depleted mouse endothelial cells have reduced tube formation capacity [59]. Control shRNA HCC1806 cells also have an intrinsic tube formation capacity (Figure 3B) but this phenotype is significantly reduced in HCC1806 AIB1LOW cells that maintain low expression of AIB1 mRNA following culture on a substrate matrix (Figure 3, B–D). Similarly, reducing AIB1 protein levels using a small molecule (MCB613) [60] also inhibited HCC1806 cell tube formation (Supplementary Figure 3 J and K). HCC1806 cells that survived DXR, PTX, or 5FU treatment with reduced levels of AIB1 protein (Figure 1C) have a similar decreased tube formation phenotype (Figure 3E). This effect is not restricted to basal like subtypes of TNBC because MDA-MB-157 AIB1LOW cells, a mesenchymal subtype of TNBC, also have a reduced tube formation phenotype relative to their respective control shRNA (Figures 3, F and G). Thus, AIB1 expression is important for tube formation in a number of TNBC cell lines, as well as in chemotherapy-treated surviving cells.
Supplementary Figure 3.
Characterization of HCC1806 AIB1LOW cells. (A) Quantification of adherent HCC1806 AIB1LOW on plastic compared to control shRNA cells seeded at similar densities and measured over time. Adherent cells were counted per well (n = 6) following decanting of the supernatant after seeding and stained with Crystal Violet. (B) Adhesion properties of HCC1806 AIB1LOW compared to control shRNA cells represented as cell index (left) measured by an electrical-cell impedance substrate array and the area under the curve (right). (C) Percentage of adhered HCC1806 AIB1LOW compared to control shRNA cells 1-hour post seeding and normalized to percentage of adhered cells 24-hours post seeding. (D) Schematic describing tumorsphere culture embedded in Matrigel TM. (E) Representative images of HCC1806 AIB1LOW compared to control shRNA cells forming tumorspheres. (F) Correlation analysis for tumorsphere size and count from HCC1806 AIB1LOW compared to control shRNA cells. (G) Schematic describing tumorsphere culture in suspension using ultra-low attachment plates. (H) Representative micrographs of serial passaged HCC1806 AIB1LOW compared to control shRNA cells cultured in 1% serum-supplemented (top) and serum-free conditions (bottom). (I) Tumorsphere count using diameter (ᴓ) of the longest axis. (J) Tube-formation assay of parental HCC1806 treated with increasing concentrations of MCB613 and DMSO for 12-hours, trypsinized, then seeded onto Matrigel for 72-hours. (K) Western blot analysis of AIB1 protein after MCB613 treatment of HCC1806 cells cultured onto Matrigel after 72h. Graphs are representative of at least two independent experiments. Mean ±SEM. Scale bar = 1 mm (J). Two-tailed t-test. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Figure 3.
AIB1LOWTNBC cells have reduced tube formation phenotype. (A) Schematic of tube formation assay on Matrigel™. (B) Representative images of 48-hour tube-formation assays showing HCC1806 AIB1LOW compared to control shRNA cells and (top) and network mask (bottom) (C) Bar graphs showing average of tube network length per well (n = 6). (D) AIB1 mRNA expression of cells from B. (E) Representative micrographs of 48-hours tube-formation assay of HCC1806 cells that survived chemotherapy vs DMSO control (F) Representative micrographs of tube-formation assays showing AIB1LOW compared to control shRNA MDA-MB-157 cells at indicated time points. (G) Average tube network length measured per image (n = 6) of tube-formation from F. Graphs are representative of three independent experiments. Mean ± SEM. Scale bar = 1 mm. Two-tailed t-test. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Differential Gene Expression in HCC1806 AIB1LOW Cells Grown on Matrigel™
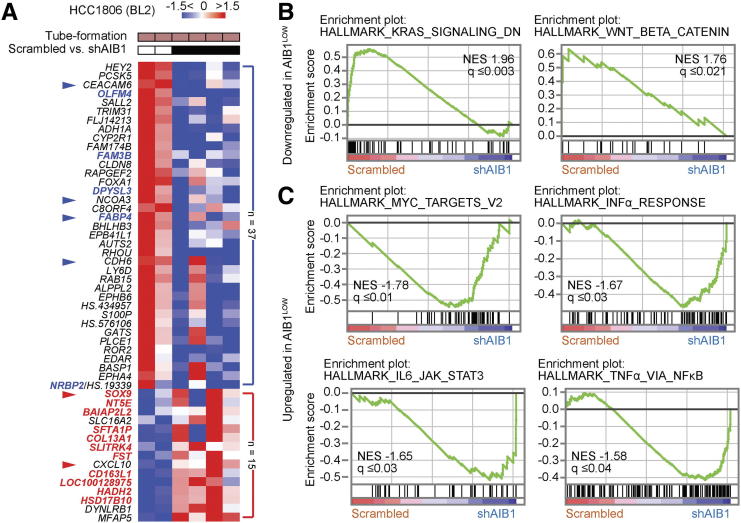
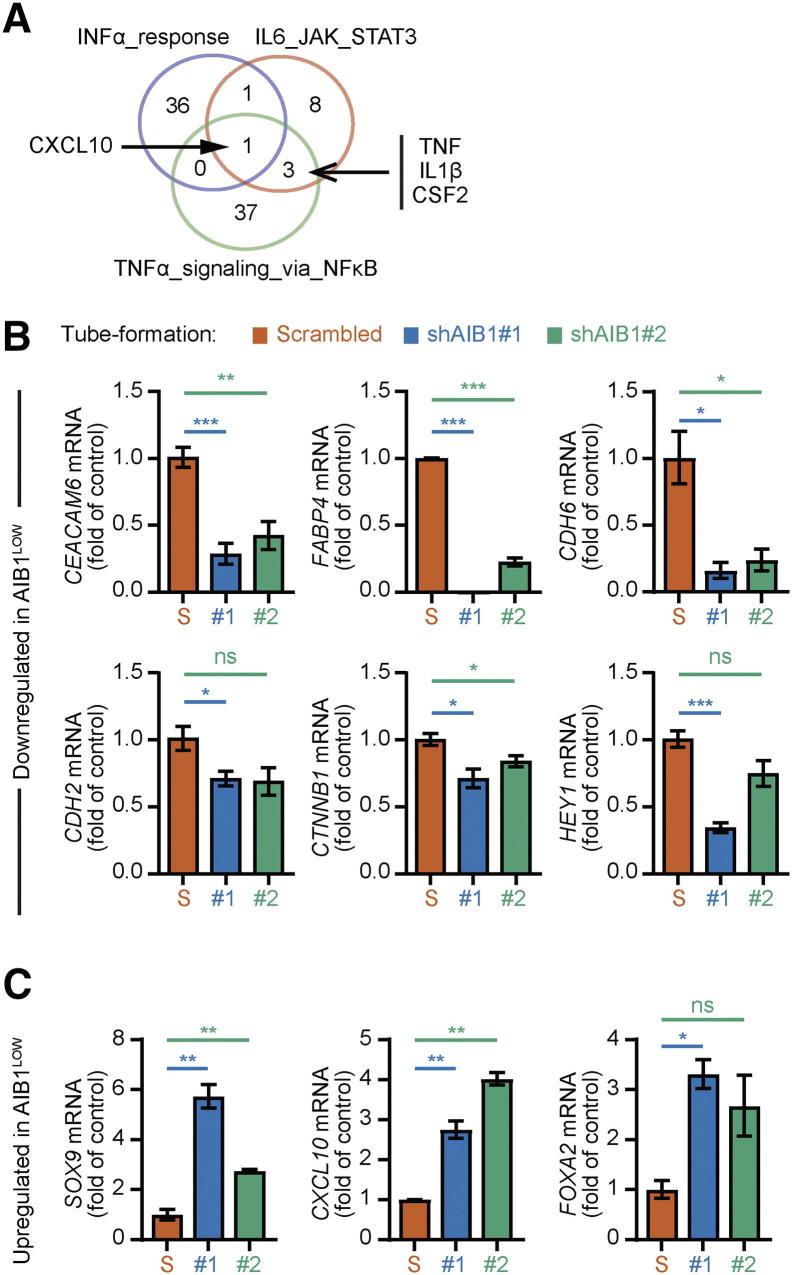
To define signaling pathways that could be involved in the impaired tube formation phenotype of HCC1806 AIB1LOW cells relative to control shRNA cells, we performed microarray analysis on cells grown on Matrigel™. The top 50 differentially expressed genes (DEGs) with >1.5-fold change and P < .05 are depicted in the heatmap in Figure 4A. The results from gene expression analysis revealed a dynamic shift of DEGs from cell lines grown on plastic compared to cells grown on Matrigel. For instance, overall upregulated genes in AIB1LOW cells from tube formation assays represented 29% (n = 15) compared to 84% (n = 38) in AIB1LOW cells grown in uncoated cell culture dishes (Figure 4A and Supplementary Figure 2C) highlighting cell differentiation states influenced by the culture conditions. However, the transcription factor SRY-Box9 (SOX9), a regulator of differentiation, was upregulated in (BL2) HCC1806 AIB1LOW cells independent of culture conditions (Figure 4A and Supplementary Figure 2C). Conversely, downregulated genes in AIB1LOW cells harvested from tube formation assays represented 71% (n = 37) compared to 16% (n = 7) in AIB1LOW cells grown in uncoated cell culture dishes (Figure 4A and Supplementary Figure 2C). Of most relevance to the tube formation phenotype, six of the top ten down-regulated genes (HEY2, PCSK5, CEACAM6, OLFM4, TRIM31, and PRR5L) in HCC1806 AIB1LOW cells are associated with integrin activity and/or cell adhesion consistent with our in vitro assays (Figure 3B and Supplementary Figure 3A). Gene set enrichment analysis (GSEA) using the detectable nearly 10,000 genes revealed KRAS as well as WNT-β-Catenin related pathways to be downregulated in tube-forming HCC1806 AIB1LOW cells relative to control shRNA cells (Figure 4B). Conversely, enrichment of MYC targets and inflammatory pathways was observed in the HCC1806 AIB1LOW cells (Figure 4C) and examination of overlapping genes within the leading-edge showed that CXCL10 was common to all inflammatory pathways. TNF, IL1β, and CSF2 were common to both IL6- and TNFα-related signaling pathways (Supplementary Figure 4A). We independently confirmed differential and leading-edge gene expression changes (Supplementary Figures 4, B and C). Thus, when TNBC cells are in contact with extracellular matrix (ECM), AIB1 regulates a number of adhesion and inflammatory signaling pathways associated with tube formation and this regulation is inhibited in AIB1LOW cells.
Figure 4.
Gene expression patterns of HCC1806 AIB1LOWcells with reduced tube-formation phenotype. (A) Heatmap showing differentially expressed genes in HCC1806 AIB1LOW cells relative to control shRNA from tube formation assays. Bold, overlapping genes with 2D cultures. Arrowheads, RT-qPCR validation performed. Gene enrichment plots showing (B) down-regulated and (C) up-regulated pathways in HCC1806 AIB1LOW cells relative to control shRNA (n = 2). Gene expression fold change and NES FDR as indicated.
Supplementary Figure 4.
Gene expression analysis of HCC1806 AIB1LOW cell lines on Matrigel. 4. (A) Venn Diagram showing overlap of leading-edge lists from inflammatory pathways in 4C. (B) Independent gene expression validation of down-regulated genes during tube-formation in HCC1806 control shRNA cells (Figure 4). Data analysis and normalization as described in Figure (C) Independent gene expression validation of upregulated genes during tube-formation in HCC1806-BL2 AIB1LOW cells. Two biological samples from each condition were submitted for microarray analysis and each sample was validated. Graphs are representative of at least two independent experiments. Mean ±SEM. Two-tailed t-test. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
HCC1806 AIB1LOW Cells Have Reduced Growth and Metastatic Capacity In Vivo
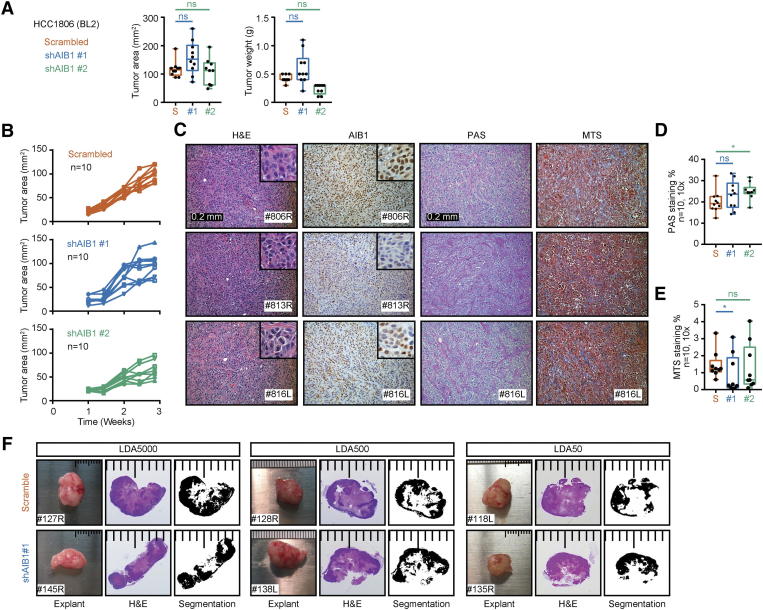
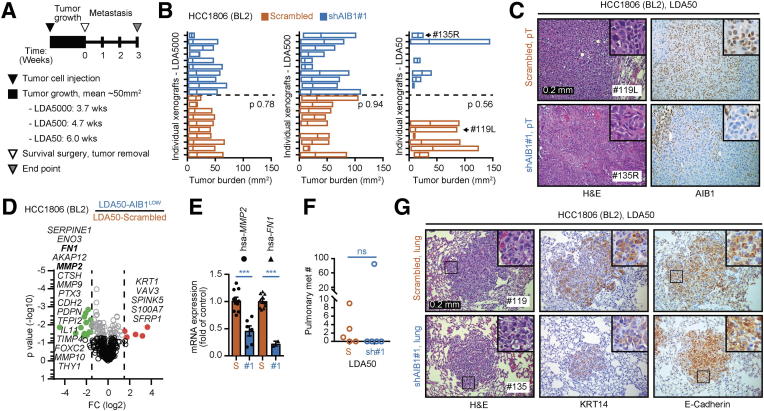
To further characterize HCC1806 AIBLOW cells, we examined their tumorigenicity and metastatic potential in vivo. Orthotopic injections of 1×106 AIB1LOW or control shRNA HCC1806 cells resulted in comparable tumor sizes by 3-weeks post injection (Supplementary Figure 5, A and B). Histological analysis showed similar highly invasive and aggressive tumors with high degree of ischemic necrosis comparable in both groups (Supplementary Figure 5C). HCC1806 AIB1LOW tumor cells showed reduced staining for AIB1 protein by IHC compared to control shRNA tumors (Supplementary Figure 5C) but stromal accumulation of collagen and glycoproteins showed no significant differences (Supplementary Figure 5D). HCC1806 cells are highly aggressive and grow rapidly in vivo. Therefore, to compare the tumorigenicity of HCC1806 AIB1LOW cells to control shRNA cells we set up limiting dilution assays (LDA) in vivo by injecting either 50, 500 or 5000 cells into the clear mammary fat pad of immunocompromised mice. After tumor size reached a group average of ~50 mm2, we performed survival surgery and monitored the mice for three additional weeks to detect metastasis (Figure 5A). Target tumor size in LDA5000 or LDA500 groups was reached by 3.7 and 4.7 weeks, respectively, with little tumor size variability (Figure 5B, left and middle panels). On the other hand, greater tumor size variability was observed in the LDA50 groups which took 6-weeks to reach the target size. Most of the LDA50-AIB1LOW xenografts were significantly smaller than the control shRNA xenografts (Figure 5B, right panel). We also detected negative to low reactivity for AIB1 protein in all LDA50-AIB1LOW xenografts relative to controls by IHC (Figure 5C). Thus, rapid growth of the outlier, LDA50-AIB1LOW #135R, xenograft is not caused by re-expression of AIB1. The overall histology for both LDA50-AIB1LOW and LDA50-control shRNA xenografts was comparable to injections using higher densities of tumor cells; although a trend showing reduced necrosis in LDA50-AIB1LOW xenografts may be associated with their reduced tumorigenicity (Supplementary Figure 5F). Analysis of human gene expression of the LDA50 tumors using the NanoString platform indicated that the AIB1LOW xenografts showed >1.5-fold down-regulation of several genes associated with tissue remodeling, including matrix metalloproteases (MMP) 2, 9 & 10, pentraxin 3 (PTX3), metallo-protease inhibitor TIMP4 and cathepsin (CTSH) [61], genes that predict chemotherapeutic response, such as SERPINE1 and A-kinase anchoring protein 12 (AKAP12) [62], as well as genes associated with inflammatory pathways, such as interleukin 11 (IL11) and podoplanin (PDPN) [63], [64], and the cellular adhesion gene Cadherin 2 (CDH2) [65]. Additionally, we found several genes involved in the blood clotting cascade and regulation of the vascular system, such as SERPINE1, Fibronectin (FN1), FOXC2, and PTX3 to be downregulated in LDA50-AIB1LOW xenografts relative to control xenografts (Figure 5D). We validated expression of FN1 and MMP2 by qRT-PCR (Figure 5E). Taken together, our results demonstrate that the reduced tumor size in LDA50 HCC1806 AIB1LOW correlates with gene expression related to tumor-stromal crosstalk. Notably pulmonary metastases were found in the majority of the LDA50 control shRNA mice whereas metastases were only detected in the outlier (#135) in the LDA-AIB1LOW mice; although, these were smaller relative to the pulmonary metastases of controls by H&E and Keratin 14 or E-cadherin IHC staining (Figure 5, F and G).
Supplementary Figure 5.
Analysis of HCC1806AIB1LOW orthotopic xenografts. (A) Average end-point tumor size and weight of 1×106 HCC1806AIB1LOW compared to control shRNA cells injected into the cleared mammary fat pad of 4-week old nude mice; n = 10. (B) Line graphs of HCC1806AIB1LOW cells compared to control shRNA cells showing tumor development over time. (C) Representative images of H&E and IHC staining from primary tumor graft sections of the 1×106 HCC1806 AIB1LOW compared to control shRNA xenografts. (D)Quantification of stromal glycoprotein stained with periodic-acid shift (PAS) stromal collagen or (E) stained with Masson’s trichome staining (MTS). (F) Representative H&E stains from LDA50-HCC1806AIB1LOW versus control shRNA showing levels of tissue necrosis.
Figure 5.
Limiting dilution HCC1806 AIB1LOWcells have reduced growth and metastatic capacity in vivo. (A) Schematic timeline for in vivo limiting dilution analysis. (B) Individual tumor size of HCC1806 AIB1LOW and control shRNA per LDA group (n = 10). (C) Representative images of H&E and IHC staining of primary tumor grafts for HCC1806 AIB1LOW and control shRNA. (D) Volcano plot showing differential gene expression of LDA50-AIB1LOW xenografts (n = 2) relative to control shRNA xenografts (n = 4). (E) PCR gene expression for MMP2 and FN1 in LDA50-AIB1LOW xenografts (n = 2) relative to control shRNA xenografts. (F) Number of mice with confirmed pulmonary metastasis by H&E in the LDA50 group. (G) Representative images of H&E, KRT14, and E-cadherin IHC staining in lung tissues for HCC1806 AIB1LOW and control shRNA. Mean ± SEM. Two-tailed t-test. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
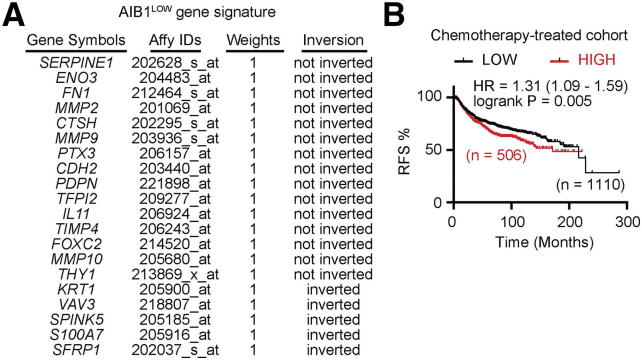
Finally, we explored the relevance of the gene expression changes seen in xenografts from AIB1LOW cells. We analyzed whether the combined signature of 20 differentially expressed genes (DEGs) from the PanCancer Progression NanoString array correlates with outcomes in patients following chemotherapy. For this, we used the Kaplan Meier plotter (www.kmplot.com), an interactive website that analyses the effect of gene expression on survival [66]. We restricted our analysis to breast cancer patients that received chemotherapy (n = 1616) as their only mode of treatment and excluded those that were in the untreated control group or that underwent endocrine therapy. Breast cancer patients that received chemotherapy treatment and that showed the 20 gene AIB1LOW expression signature in their tumors had an almost 4 years (45.3 months) shorter median relapse free survival (171.4 months) than patients whose tumors lacked the AIB1LOW expression signature (216.7 months; P < .005; Supplemental Figure 6).
Supplementary Figure 6.
Recurrence free survival (RFS) of chemotherapy treated patients with breast cancer relative to the AIB1LOW 20 gene expression signature. (A) List of signature genes derived from a comparison of HCC1806 AIB1LOW relative to control shRNA xenograft tumors. (B) Kaplan-Meier curve showing significantly longer RFS of patients which tumors lacking the AIB1LOW 20 gene expression signature. Log-rank p values and hazard ratios (HRs; 95% CI) are shown. Note: For signature genes whose expression was downregulated in AIB1LOW tumors the respective expression levels in breast cancer specimens were inverted for the combined analysis to take the directionality of expression of the signature genes into account.
Discussion
Here, we show that the major portion of basal like TNBC cells depend on AIB1 expression for survival. However, cells that survive depletion of AIB1 by shRNA-mediated knockdown (AIB1LOW) or after chemotherapy treatment, can be maintained in serial passage with low-to-undetectable AIB1 mRNA expression. These AIB1LOW cells are a distinct subpopulation that have reduced dependence on AIB1 signaling. The whole exome sequence analysis of the AIB1LOW population identifies these cells as a subset of the bulk population that survives selection after AIB1 depletion. Furthermore, our data suggest that the presence of an AIB1LOW cell gene expression signature in TNBC patient samples may be useful as a prognostic indicator of poor survival outcome following therapy, i.e., relative resistance to cytotoxic treatment. Thus AIB1LOW cells could well be useful as a model to study therapeutic targeting of metastasis and for understanding chemotherapy resistance in TNBC.
Despite their increased proliferation and reduced sensitivity to chemotherapy in reduced serum conditions, HCC1806 AIB1LOW cells did not show differences in classical CSC indicators in vitro or in vivo. Enrichment of CSC populations has been observed after chemotherapy treatment of TNBC [44] and CSC enrichment is hypothesized to contribute to resistance to treatment, disease recurrence, and metastasis [45]. Phenotypic markers to identify these rare populations continue to be an area of active research. However, the in vivo LDA results indicate that HCC1806 AIB1LOW have different properties compared to control shRNA cells. Although, we cannot rule out the presence of CSC in the AIB1LOW population, if present, they do not appear to influence the overall phenotype that we observe. The inability of AIB1LOW cells to form tubes on Matrigel and their slower tumor growth and metastasis in vivo along with reduced adhesion gene expression and changes in inflammatory signaling suggest that cross talk of the HCC1806 AIB1LOW cells with each other and/or their surrounding stroma environment is impeded, and this influences their tumorigenicity and metastasis potential.
The question then arises as to the downstream targets that play into differential AIB1 sensitivity in TNBC cells. Certainly, the regulation of adhesion and proteolytic genes, both in vitro and in vivo, is expected to influence the phenotype of AIB1LOW TNBC cells and their interactions with components in the tumor microenvironment including fibroblasts, adipocytes, endothelial, bone marrow derived and immune cells. In addition, our findings show that type 2 inflammatory response genes are regulated by AIB1 expression and could contribute to tumorigenicity. Reduced AIB1 in TNBC cells activates the IFNɑ signaling pathway upregulates the angiostatic cytokine CXCL10 and this could contribute to the impaired tube formation phenotype of AIB1LOW cell on Matrigel. It has been previously reported that CXCL10, and its cognate receptor CXCR3, have anti-angiogenic effects through the trafficking of immune cells or by activating endothelial cells; therefore, reducing vascular perfusion to sites of inflammation (reviewed in [67]). However, we did not observe significant differences in the vascularity of the tumors between groups suggesting that HCC1806 AIB1LOW cells are not altering vascular mimicry or endothelial cell function.
The reduced tumorigenicity of HCC1806 AIB1LOW cells in vivo became apparent only at low cell implant numbers suggesting that the pro-inflammatory effect exerted by too few tumor epithelial cells fails to overcome an anti-tumorigenic response from the stroma. Chemotherapy can reduce tumor size and our results show that a relatively indolent population of cells that survives chemotherapy have reduced levels of AIB1 protein indicative of tumor cells with the potential for recurrence or metastasis should cell density increase above a threshold. Thus, the AIB1LOW cells have some of the hallmarks of dormant cells although gene expression comparisons of AIB1LOW versus control shRNA cells on Matrigel™ did not show major changes in known dormancy gene signatures [68], [69].
The differences in the activated transcriptional pathways in HCC1806 AIB1LOW cells compared to control cells suggests that there may be significant differences in the engagement of this nuclear coactivator at the chromatin level. AIB1 is known to interact with multiple nuclear receptors and other transcription factors [31] and AIB1 genomic engagement is altered in different breast cancer cell types and under different conditions. We conjecture that patterns of epigenetic modifications could determine whether AIB1 is activating pro-survival gene pathways in a subset of parental cell lines, whereas in AIB1LOW cells these pathways are no longer controlled by the endogenous AIB1. In this regard it would be also interesting to compare the epigenetic profiles of the parental vs AIB1LOW cells and compare the pattern of chromatin engagement of AIB1. The fact that an AIB1LOW expression signature is correlated with worse outcome suggests that for future studies it will be important to determine the sensitivity of AIB1LOW cells to other therapies and to design robust gene expression signatures that can predict the presence of AIB1LOW cells in TNBC subtypes.
Materials and Methods
Cell Lines and Reagents
HCC1806 cell lines were purchased from ATCC® (#CRL-233, Manassas, VA, USA). MDA-MB-468 (#HTB-132, ATCC), MDA-MB-157 (#HTB-24, ATCC), and HEK293T (#CRL-3216) cells were obtained from the Georgetown University (GU) Tissue Culture Share Resource (TCSR) in Lombardi Comprehensive Cancer Center (LCCC). HUVEC cell lines were purchased from Lonza (#CC-2517A, Walkersville, MD, USA). Cell lines were authenticated using short tandem repeat (STR) analysis by the TCSR prior to use. Cell lines were maintained under sub-confluent conditions (70–80%) and media was replenished every three days. PDX-HCI010 grafts were obtained from Huntsman Cancer Center and expanded in immunocompromised mice as previously described [70]. Cell lines and PDX tissues were mycoplasma negative by RADIL-IMPACT testing results and throughout this project by TCSR periodic screening. For additional details of media recipes, cell culture and maintenance, see Supplementary Materials and Methods.
AIB1 mRNA Interference
RNAi targeting sequences in AIB1 exon 6 and exon 14 were described previously [51], [71]. Oligo nucleotides were purchased from Bioneer, Inc. (Alameda, CA, USA). Scramble (#1864) [72] and AIB1 short-hairpin RNAs (shRNAs) were purchased from Addgene (Cambridge, MA, USA). AIB1 shRNAs and Scramble plasmids were inserted into pLKO.1 puro (#8453; Addgene) lentiviral vectors [73]. For viral production and shRNA infection, see Supplementary Material and Methods. Cell lines were transfected with 200 nmol/L of siRNAs diluted in RPMI-1640 or DMEM with Lipofectamine 2000 (#11668027; Invitrogen, Carlsbad, CA, USA).
Cellular Phenotype
Cell proliferation, serum-free cell survival, viability assays, and cell trace assays were performed and analyzed as previously described [74], [75]. Indirect intracellular staining of AIB1 was performed using the Nuclear Factor Fixation and Permeabilization kit (#422601; BioLegend, San Diego, USA) following manufacturer's recommendations. Briefly, 1×106 cells/ml densities were fixed, permeabilized, then incubated with 1.2 μg/ml of anti-rabbit human AIB1 (#2126; Cell Signaling Technology, Danvers, MA, USA) primary antibody in suspension for 30–60 minutes. Upon incubation cells were rinse then incubated with 2.5 μg/ml of secondary isotype-specific fluorescent antibody, AlexaFluor488 goat anti–rabbit (#A11008; Invitrogen) for 30 minutes prior to submission for analysis. Samples were analyzed using the LSRFortessa™ cell analyzer (BD Biosciences, San Jose, CA, USA). Cells were labeled using the CellTrace Violet (CTV, #C34557, Invitrogen) dye. Briefly, cells (1×106) were incubated with 1–2 μM dye in 1 ml PBS for 20-minutes at 37 °C. Excess dye was rinsed off by adding 9 ml of serum-supplemented media, centrifuged at 1,000 rpm for 5-minutes, after removal of the supernatant the cell pellet was resuspended in fresh media. Resuspended cells (1×105) were seeded in 60×15 mm dishes to monitor mitotic index for 3 or 5 days (T3, T5) after seeding. Cell lines were trypsinized, counted, and fixed in 1%PFA prior to flow cytometry analysis. An aliquot of unlabeled and labeled cells at time zero (T0) were used as reference for the highest signal of the dye. Tube formation assays on Matrigel™ (#354230; BD Biosciences) and tumorsphere assays were conducted and analyzed as previously described [46], [59]. Separating cells from basement matrix was adapted from Lee and colleagues [56]. Briefly, media was removing from each well followed by a rinse with 200 μl of PBS per well. A volume of 200 μl of cold 5 mM EDTA-PBS was added to each well and the basement matrix with attached cells was scratched using a 1000 low-retention tip and transferred to a 15 ml conical tube. This was repeated for each well in each condition. The conical tube was then vortexed briefly and the volume of cold EDTA-PBS was doubled. The mix was incubated in ice for 5 minutes follow by a quick vortex prior to centrifugation at 1000 rpm at 4 °C for 10 minutes. Digestion of Matrigel required an approximate 1:10 ratio (i.e., 450 μl of EDTA-PBS to 50 μl of 100% Matrigel) at the time of incubation but this reference is dependent on the integrity of the Matrigel (length of culture and accumulation of proteases by cells) and the type of cells utilized. If Matrigel is not completely digested, removed the aqueous part and add 1:1 cold EDTA-PBS to the remaining polymerized Matrigel. Incubate in ice for 5 minutes, vortex briefly, then centrifuge. The resulting cell pellet was lysed for RNA or protein extraction. For additional details, see Supplementary Material and Methods.
RNA Extraction and Gene Expression Analysis
Gene expression of target genes was normalized (ΔCt) to the average expression of three housekeeping genes; ACTB, B2M, and GAPDH. Gene expression relative to control shRNA was calculated using 2-ΔΔCT, as previously described [76]. For the list of primers and their sequences, see Supplementary Material and Methods. For genome-wide gene expression analysis, total RNA samples with RNA integrity number (RIN) >8.0 were submitted to the UCLA Neuroscience Genomics Core (UNGC). The HumanHT-12 v4.0 Expression BeadChip (GRCh38/hg38) (Illumina, San Diego, CA, USA) covering more than 47,000 transcripts was utilized and direct hybridization was performed following manufacturer's recommendations. Average signal intensity was normalized and log-transformed using GenePattern v3.9.10 suite (Broad Institute of MIT and Harvard, Cambridge, MA, USA) as described previously [77]. Gene set enrichment analysis (GSEA) was carried out to identify common pathways affected by AIB1 silencing. For NanoString (Seattle, WA, USA) gene expression of HCC1806-BL2 AIB1LOW (n = 2) and control shRNA (n = 4) xenografts, total RNA and protein were extracted from snap frozen xenograft tissues (LDA50 experimental group) following manufacturer's recommendations [78]. nCounter® PanCancer Progression Panel (XT-CSO-PROG1–12; NanoString Technologies) was acquired through the GU Genomics and Epigenomics Shared Resources (GESR) in LCCC. Samples were hybridized and processed using the nCounter® SPRINT Profiler. nSolver v4.0 software (NanoString Technologies) was used for data analysis and for the generation of expression tables.
Next Generation Sequencing (NGS) and Exome Sequencing
Cells from two independent shRNA infections: control shRNA, shAIB1#1, shAIB1#2 (x2), were carried out for five passages. DNA was isolated using the DNeasy Blood&Tissue kit (Qiagen). Whole Exome sequencing was carried out by the UCLA Neuroscience Genomics Core. 65 Exome sequencing hybridized with probes from the Nimblegen SeqCap EZ Exome v2/3 Kapa/IVTL kit (Roche, Basel, Switzerland) and samples were run on an Illumina Hiseq4000 and an average of 61 million paired reads were acquired for each sample. Exome sequencing data were analyzed and mapped to HG38 using BWA [79] and the variant calling analysis was performed by Samtools and Bcftools [80], [81]. Reads were filtered by quality (QUAL >20) and number of reads (DP <100).
Protein Detection and Immuno-Blot (Western Blot)
Total protein lysates were extracted from end-point cell cultures following a rinse with cold PBS or from snap frozen cell pellets stored briefly at −80 °C as previously described [71]. Antibodies expression were verified with manufacture's control cell lines. For list of antibodies and additional information, see Supplementary Material and Methods.
Tumor Transplantation Experiments: Xenografts
Experiments involving animals were approved by the GU-IACUC and were conducted according to the NIH guidelines for the care and use of laboratory animals. Immunocompromised 3-weeks old female mice were purchased from Envigo (Athymic Nude-Foxn1nu, #6901F, Indianapolis, IN, USA) and from Jackson Laboratory (NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ), #005557, Bar Harbor, ME, USA). A week after acclimation, female mice underwent bilateral removal of the endogenous epithelium from the inguinal mammary fat pad (MFP) prior to injection of tumor cells in Matrigel™ suspension (20 μl) or tumor fragments as described previously [49], [82], [83]. Orthotopic injections or tumor fragments into the cleared MFP were monitored three times a week. Hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining were performed on 5-μm sections formalin-fixed, paraffin embedded (FFPE) tissue sections by GU Histopathology and Tissue Shared Resource (HTSR). Histopathology analysis was conducted by a board-certified pathologist at GU.
Statistics and Image Analysis
Statistical differences and linear regression analysis were performed using the GraphPad Prism software v8.0 (Graph-Pad Software Inc., San Diego, CA, USA). Phenotypic and gene expression differences were measured by unpaired student t-Test, One-way and Two-way ANOVA as indicated. Non-linear regression determined results in viability assays and data from population-based registries. Median overall survival (OS) was calculated using Kaplan–Meier estimates and compared using log-rank tests. The significance of change reflects P < .05, P < .01, and P < .001 and were considered statistically significant, unless stated otherwise. For information on image processing and analysis, see Supplementary Material and Methods.
The following are the supplementary data related to this article.
Author contributions
Conceived and designed the experiments: FRS, AW, ATR. Performed the experiments: FRS. Analyzed the data: FRS, MS, VO, AW, ATR. Contributed reagents/materials/analysis tools: BK, SCM, PAF. Contributed to the writing of the manuscript: FRS, VO, MS, BK, SCM, PAF, AW, ATR.
Acknowledgements
We thank Drs. Ivana Peran and Gai Yan for their valuable discussions towards the completion of this project. We also thank Maria Idalia Cruz for her technical assistance with in vivo experiments. This work was funded in part by R01 CA205632,R21 CA226542 (ATR). Funding for FRS was provided by the Susan G Komen Graduate Training in Breast Cancer Disparities at LCCC (GTDR15330383), VO was funded by the NCI T32 Training Grant in Tumor Biology (CA009686). The project described used the Tissue Culture & Biobanking, Flow Cytometry & Cell Sorting, Microscopy & Imaging, Animal Model, Histopathology & Tissue, and Genomics & Epigenomics Shared Resources which are partially supported by Award Number P30CA051008 (PI: Weiner) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Hammond MEH, Hayes DF, Wolff AC, Mangu PB, Temin S. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract. 2010;6(4):195–197. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolff AC. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2006;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sørlie T. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen TO Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 6.Charafe-Jauffret E. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann BD. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann BD. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda H. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19(19):5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abramson VG, Lehmann BD, Ballinger TJ, Pietenpol JA. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer. 2015;121(1):8–16. doi: 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakha EA. Basal phenotype identifies a poor prognostic subgroup of breast cancer of clinical importance. Eur J Cancer. 2006;42(18):3149–3156. doi: 10.1016/j.ejca.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Dent R. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15):4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 13.Carey LA. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295(21):2492. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 14.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 15.Stead LA. Triple-negative breast cancers are increased in black women regardless of age or body mass index. Breast Cancer Res. 2009;11(2):1–10. doi: 10.1186/bcr2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey LA. TBCRC 001: Randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30(21):2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RS. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/“triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105(3):319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 18.Duncan JS. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149(2):307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y. BMN673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 2013;19(18):5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carey LA. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 21.Albain K. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100 000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortazar P. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 23.Rouzier R. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 24.Liedtke C. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 25.Balko JM. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto JA et al. A prognostic signature based on three-genes expression in triple-negative breast tumours with residual disease. npj Genomic Med. 2016;1(1):15015. [DOI] [PMC free article] [PubMed]
- 27.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werbajh S, Nojek I, Lanz R, Costas MA. RAC-3 is a NF-κB coactivator. FEBS Lett. 2000;485(2–3):195–199. doi: 10.1016/s0014-5793(00)02223-7. [DOI] [PubMed] [Google Scholar]
- 29.Louie MC, Zou JX, Rabinovich A, Chen H-W. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24(12):5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arimura A, Van Peer M, Schröder AJ, Rothman PB. The transcriptional co-activator p/CIP (NCoA-3) is up-regulated by STAT6 and serves as a positive regulator of transcriptional activation by STAT6. J Biol Chem. 2004;279(30):31105–31112. doi: 10.1074/jbc.M404428200. [DOI] [PubMed] [Google Scholar]
- 31.Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116(2):225–237. doi: 10.1007/s10549-009-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9(9):615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao C et al. Elevated expression levels of NCOA3,TOP1, and TFAP2C in breast tumors as predictors of poor prognosis. Cancer 2003;98(1):18–23. [DOI] [PubMed]
- 34.Osborne CK. Role of the Estrogen Receptor Coactivator AIB1 (SRC-3) and HER-2/neu in Tamoxifen Resistance in Breast Cancer. JNCI J Natl Cancer Inst. 2003;95(5):353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 35.Dihge L. Epidermal growth factor receptor (EGFR) and the estrogen receptor modulator amplified in breast cancer (AIB1) for predicting clinical outcome after adjuvant tamoxifen in breast cancer. Breast Cancer Res Treat. 2008;109(2):255–262. doi: 10.1007/s10549-007-9645-1. [DOI] [PubMed] [Google Scholar]
- 36.Hurtado A. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456(7222):663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fereshteh MP. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res. 2008;68(10):3697–3706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song X. Steroid receptor coactivator-3 (SRC-3/AIB1) as a novel therapeutic target in triple negative breast cancer and its inhibition with a phospho-bufalin prodrug. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0140011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gazdar AF. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78(6):766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 40.Cailleau R, Olivé M, Cruciger QVJ. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro. 1978;14(11):911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 41.Young RK, Cailleau RM, Mackay B, Reeves WJ. Establishment of epithelial cell line MDA-MB-157 from metastatic pleural effusion of human breast carcinoma. In Vitro. 1974;9(4):239–245. doi: 10.1007/BF02616069. [DOI] [PubMed] [Google Scholar]
- 42.Creighton CJ. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 44.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 46.Ory V. The nuclear coactivator amplified in breast cancer 1 maintains tumor-initiating cells during development of ductal carcinoma in situ. Oncogene. 2014;33(23):3033–3042. doi: 10.1038/onc.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin L. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28(19):5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Creighton CJ. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeRose YS. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobrolecki LE. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016;35(4):547–573. doi: 10.1007/s10555-016-9653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lahusen T, Fereshteh M, Oh A, Wellstein A, Riegel AT. Epidermal growth factor receptor tyrosine phosphorylation and signaling controlled by a nuclear receptor coactivator, amplified in breast cancer 1. Cancer Res. 2007;67(15):7256–7265. doi: 10.1158/0008-5472.CAN-07-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navin N. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koboldt DC. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Weiswald L, Bellet D, Dangles-marie V. Spherical Cancer Models in Tumor. NEO. 2015;17(1):1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee GY, Kenny P. a, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrell JC. Endothelial-like properties of claudin-low breast cancer cells promote tumor vascular permeability and metastasis. Clin Exp Metastasis. 2014;31(1):33–45. doi: 10.1007/s10585-013-9607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubota Y, Kleinman HK, Marin GR. and Lawley TJ, Kubota Y, Kleinman HK, Martin GR, Lawley TJ. Role of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary like structures. J. Cell Biol. 1988;107(October):1589–1598. doi: 10.1083/jcb.107.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Otaiby M. Role of the nuclear receptor coactivator AIB1/SRC-3 in angiogenesis and wound healing. Am J Pathol. 2012;180(4):1474–1484. doi: 10.1016/j.ajpath.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L. Characterization of a steroid receptor coactivator small molecule stimulator that overstimulates cancer cells and leads to cell stress and death. Cancer Cell. 2015;28(2):240–252. doi: 10.1016/j.ccell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gautam J. Down-regulation of cathepsin S and matrix metalloproteinase-9 via Src, a non-receptor tyrosine kinase, suppresses triple-negative breast cancer growth and metastasis. Exp Mol Med. 2018;50(9):118. doi: 10.1038/s12276-018-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhat-Nakshatri P, Goswami CP, Badve S, Sledge GW, Nakshatri H. Identification of FDA-approved drugs targeting breast cancer stem cells along with biomarkers of sensitivity. Sci. Rep. 2013;3(Ml). doi: 10.1038/srep02530 [DOI] [PMC free article] [PubMed]
- 63.Roberti MP. Protein expression changes during human triple negative breast cancer cell line progression to lymph node metastasis in a xenografted model in nude mice. Cancer Biol Ther. 2012;13(11):1123–1140. doi: 10.4161/cbt.21187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cazet AS. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun. 2018;9(1):2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azimi I, Petersen RM, Thompson EW, Roberts-Thomson SJ, Monteith GR. Hypoxia-induced reactive oxygen species mediate N-cadherin and SERPINE1 expression, EGFR signalling and motility in MDA-MB-468 breast cancer cells. Sci Rep. 2017;7(1):15140. doi: 10.1038/s41598-017-15474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Györffy B. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 67.Liu M, Guo S, Stiles JK. The emerging role of CXCL10 in cancer. Oncol Lett. 2011;2(4):583–589. doi: 10.3892/ol.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim RS. Dormancy signatures and metastasis in estrogen receptor positive and negative breast cancer. PLoS One. 2012;7(4):1–8. doi: 10.1371/journal.pone.0035569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghajar CM. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15(7):807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeRose YS et al. Patient-Derived Models of Human Breast Cancer: Protocols for In Vitro and In Vivo Applications in Tumor Biology and Translational Medicine. In: Current Protocols in Pharmacology. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2013:1–43 [DOI] [PMC free article] [PubMed]
- 71.Oh A. The nuclear receptor coactivator AIB1 mediates insulin-like growth factor I-induced phenotypic changes in human breast cancer cells. Cancer Res. 2004;64(22):8299–8308. doi: 10.1158/0008-5472.CAN-04-0354. [DOI] [PubMed] [Google Scholar]
- 72.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway2005;596–603. [DOI] [PubMed]
- 73.Stewart SA. Lentivirus-delivered stable gene silencing by RNAi in primary cells Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gjoerup OV. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc Natl Acad Sci. 2007;104(20):8334–8339. doi: 10.1073/pnas.0703164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alley MC, Scudiero DA, Monks PA, Hursey ML. C, M.J., Fine, D.L., Abbott, B.J., Mayo, J.G., Shoemaker RH& B, M.R. Feasibility of drug screening with panels of human tumor cell lines using a mocroculture microculture tetrazolium assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 76.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 77.Reich M. GenePattern 2.0 [2]. Nat. Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 78.Geiss GK. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 79.Heng Li, Richard Durbin. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics [published online ahead of print: 2009]; doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed]
- 80.Narasimhan V. BCFtools/RoH: A hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics. 2016;32(11):1749–1751. doi: 10.1093/bioinformatics/btw044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deome KB, Bern HA, Blair PBFLJJ. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19(5):515–520. [PubMed] [Google Scholar]
- 83.Lawson DA, Werb Z, Zong Y, Goldstein AS. The cleared mammary fat pad transplantation assay for mammary epithelial organogenesis. Cold Spring Harb Protoc. 2015;2015(12):1064–1068. doi: 10.1101/pdb.prot078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.