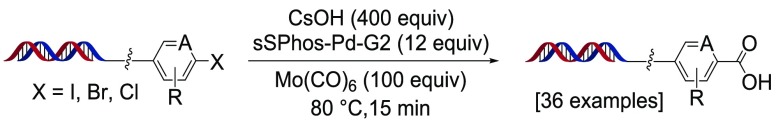Abstract
A strategy for DNA-compatible, palladium-catalyzed hydroxycarbonylation of (hetero)aryl halides on DNA–chemical conjugates has been developed. This method generally provided the corresponding carboxylic acids in moderate to very good conversions for (hetero)aryl iodides and bromides, and in poor to moderate conversions for (hetero)aryl chlorides. These conditions were further validated by application within a DNA-encoded chemical library synthesis and subsequent discovery of enriched features from the library in selection experiments against two protein targets.
Introduction
DNA-encoded chemical libraries (DECLs) are collections of small molecules connected to a unique DNA sequence that serves as a barcode for the small molecule structure.1−3 Due to the high fidelity of DNA amplification and high-throughput sequencing, multibillion member DECL pools may be used in affinity-based protein target screening campaigns to enrich small molecule binders whose identifying DNA tags may be amplified/decoded at subfemtomole scales.4−6 Although many types of DNA-compatible chemical transformations for solution-phase small molecule synthesis have been disclosed,7−15 a survey of reported DECLs show the majority are constructed with at least one amidation reaction,16 likely due to the inherent stability of amide bonds to aqueous conditions,17 the broad substrate scope of amidations on oligonucleotide–chemical conjugates,18,19 and the wide commercial availability of amines and carboxylic acids.20 Amidation substrates in DECL synthesis are typically prepared through attachment of a building block already containing the required functional group, such as a bifunctional protected amine (e.g., N-Fmoc, N-Boc, nitro21), bifunctional protected acid (e.g., ester), or by addition of an amino acid.22,23 Although many of such bifunctional building blocks are commercially available or known, an alternative paradigm would be to install the amino or carboxyl group through conversion of a DNA-attached functional group. This offers many benefits including access to an additional segments of novel chemical matter, extended commercial building block availability, clear orthogonality in the presence of another amino or carboxyl group, and potential use as an ultimately traceless handle to modulate building block reactivity for other transformations.
Palladium-catalyzed cross coupling is a widely used reaction class in drug discovery,24−26 and several aqueous adaptations of these transformations have been reported within various DECL platforms, including Suzuki,8,14,27,28 Heck,29,30 and Buchwald–Hartwig coupling.31,32 A powerful variant of this class is carbonylative cross-coupling, in which carbon monoxide migrates into a palladium-inserted electrophile forming an acylpalladium complex and ultimately a carbonylated cross-coupled product (Scheme 1a).33,34 Although this reaction is often conducted with carbon nucleophiles,35 inclusion of water/hydroxide is known to result in carboxylic acid products.36−38 Typically, carbonylative cross-coupling is conducted under a saturated or high-pressure carbon monoxide atmosphere to ensure efficient migration of carbon monoxide into the palladium–electrophile complex.33,34 However, the use of atmospheric or pressurized systems is inconvenient for plate-based DECL reaction screening, synthesis, or heating,39 and poor control over the carbon monoxide atmosphere may lead to inconsistent product distributions from competing noncarbonylative coupling,40 dehalogenation pathways,41 or carbon monoxide mediated suppression of oxidative addition. Alternatively, carbon monoxide may be generated in situ through decomposition of an added carbon monoxide source, which would be amenable to sealed plate formats and accurately measured on small scale. Several methods to generate carbon monoxide in situ from reagent decomposition in the reaction medium have been reported, including the use of N-formyl saccharin,42 molybdenum hexacarbonyl,43 and formyl acetates.36−38 Based upon our work27 and that of others8,14,28 on DNA-compatible Suzuki coupling using hydroxide base, we hypothesized that similar conditions utilizing an internal carbon monoxide source could yield (hetero)aromatic carboxylic acids from the corresponding (hetero)aromatic halides (Scheme 1b).
Scheme 1. Palladium-Catalyzed Carbonylative Cross-Coupling.
(a) General carbonylative cross-coupling; (b) On-DNA hydroxycarbonylation of (hetero)aryl halides.
Results and Discussion
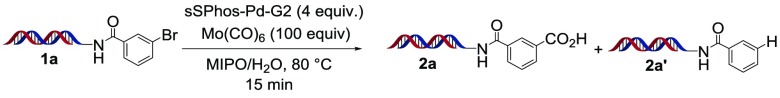
We initiated our studies with simple aryl bromide 1a using water-soluble palladium precatalyst sSPhos-Pd-G2,44,451-methoxy-2-propanol as organic cosolvent, and CsOH as base. Preliminary studies with N-formyl saccharin were not successful; however, upon switching to Mo(CO)6, we were pleased to observe moderate conversion to the desired benzoic acid 2a (Table 1, entry 1). A catalyst screen revealed sSPhos-Pd-G2 to be superior to all other catalysts tried, and as shown in Table 1, further studies focused on optimizing this catalyst system. Increasing the amount of hydroxide base, increasing the amount of Mo(CO)6, or changing the hydroxide counterion had little effect on the product distribution (Table 1, entries 2–5). Not surprisingly, replacing hydroxide with weaker carbonate or borate bases resulted in significantly lowered conversion to the benzoic acid (Table 1, entries 6–8). Substitution of 1-methoxy-2-propanol cosolvent with CH3CN, 1,4-dioxane, or dimethylacetamide appeared to suppress initial oxidative addition (Table 1, entries 9–11).46 Increasing the temperature to 90 or 95 °C lowered conversion to 2a and enhanced the formation of reduced side product 2a′ (Table 1, entries 12, 13). Doubling the catalyst loading led to enhanced conversion to the benzoic acid, albeit with a concomitant increase in formation of 2a′ (Table 1, entry 14). Ultimately, the use of 12 equiv of sSPhos-Pd-G2 and 400 equiv of CsOH with 1-methoxy-2-propanol as cosolvent at 80 °C was found to be optimal (Table 1, entry 15).
Table 1. Optimization of Reaction Conditions for On-DNA Hydroxycarbonylation.
| Entry | Base (equiv) | 1a | 2a | 2a′ |
|---|---|---|---|---|
| 1 | CsOH (400) | 58% | 41% | 1% |
| 2 | CsOH (800) | 62% | 34% | 4% |
| 3a | CsOH (400) | 62% | 36% | 2% |
| 4b | CsOH (400) | 66% | 33% | 1% |
| 5 | NaOH (400) | 68% | 31% | 1% |
| 6 | K2CO3 (400) | 91% | 9% | 0% |
| 7 | Cs2CO3 (400) | 89% | 11% | 0% |
| 8 | pH 9.5 borate (400) | 95% | 5% | 0% |
| 9c | CsOH (400) | 100% | 0% | 0% |
| 10d | CsOH (400) | 96% | 4% | 0% |
| 11e | CsOH (400) | 72% | 27% | 1% |
| 12f | CsOH (400) | 67% | 29% | 4% |
| 13g | CsOH (400) | 64% | 25% | 11% |
| 14h | CsOH (400) | 29% | 65% | 6% |
| 15i | CsOH (400) | 0% | 84% | 16% |
150 equiv of Mo(CO)6.
200 equiv of Mo(CO)6.
CH3CN as cosolvent.
1,4-dioxane as cosolvent.
DMA as cosolvent.
Temperature at 90 °C.
Temperature at 95 °C.
8 equiv of [Pd].
12 equiv of [Pd]
With this optimized condition in hand, we turned toward exploring the potential substrate scope of this transformation on a series of (hetero)aryl iodides, bromides, and chlorides (Scheme 2). Differentially substituted aryl iodides 1b–d, iodoquinoline 1e, and azole iodide 1j were transformed to the carboxylic products in very good conversions, and the presence of a nearby basic amine, such as in 1f and 1g, did not appreciably affect the conversion. However, hydroxycarbonylation with iodide at the labile 2-position of pyridine resulted in significant formation of the dehalogenation side product (2h vs 2i). Continuing to the hydroxycarbonylation of (hetero)aryl bromides, familial aryl bromides 1a and 1k both furnished the carboxylic acid product, and importantly, similar results were observed for 1a when performed on the 150 nmol scale.47 Although ortho-acyl bromide 1l mostly provided dehalogenation,48 the presence of other mild metal-coordinating groups (in 2m–p) did not fully suppress product formation. Electron-rich bromides 1q–t underwent effective hydroxycarbonylation, and while no reduction of the aldehyde was observed in 2t, the nitro group was fully reduced in 2u. This reaction was also effective on a variety of heterocyclic bromides, including pyridines 1v and 1x, quinazoline 1y, imidazolopyridine 1z, pyrimidine 1bb and thiazole 1cc. However, highly coordinating ortho-acyl pyridine 1w, pyrimidine 1aa, and pyrazole 1dd suffered from significant formation of byproducts. Finally, we attempted to extend this reaction to a series of electronically diverse aryl and heteroaryl chlorides (selected examples, 1ee–jj). Although the desired products were observed, lowered conversion to the carboxylic acid was realized, presumably from carbon monoxide suppression of oxidative addition.
Scheme 2. On-DNA Hydroxycarbonylation of (Hetero)aryl Halides.
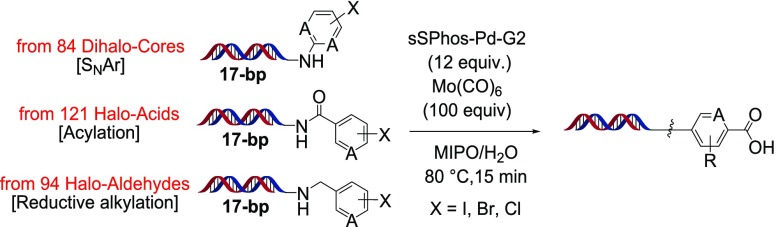
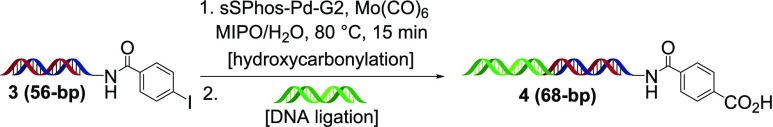
To further study this reaction for DECL synthesis, we prepared an expanded set of iodide, bromide, and chloride substrates using amino-terminated 17-bp dsDNA (Scheme 3). This substrate set was prepared in plate format through the nucleophilic substitution of 84 dihalo-cores, the acylation of 121 halo-acids, and the reductive alkylation of 94 halo-aldehydes. After ethanol precipitation, application of our optimized conditions to this set led to product distributions similar to those observed previously, with a variety of iodide and bromide substrates undergoing hydroxycarbonylation in moderate to very good conversions, and chlorides, in general, very poor to moderate conversions (see Supporting Information for full structural and product distribution details). To simulate a late-stage application of this transformation within a DECL, we prepared para-iodobenzamide 3 with an elongated 56-bp dsDNA tag (Scheme 4). This substrate underwent hydroxycarbonylation in comparable conversion to other iodide substrates, and after ethanol precipitation, could be fully ligated with a dsDNA tag to carboxylic acid 4 (see Supporting Information for full details).
Scheme 3. Hydroxycarbonylation on an Expanded Set of Substrates.
Scheme 4. Long DNA and Ligation Test.
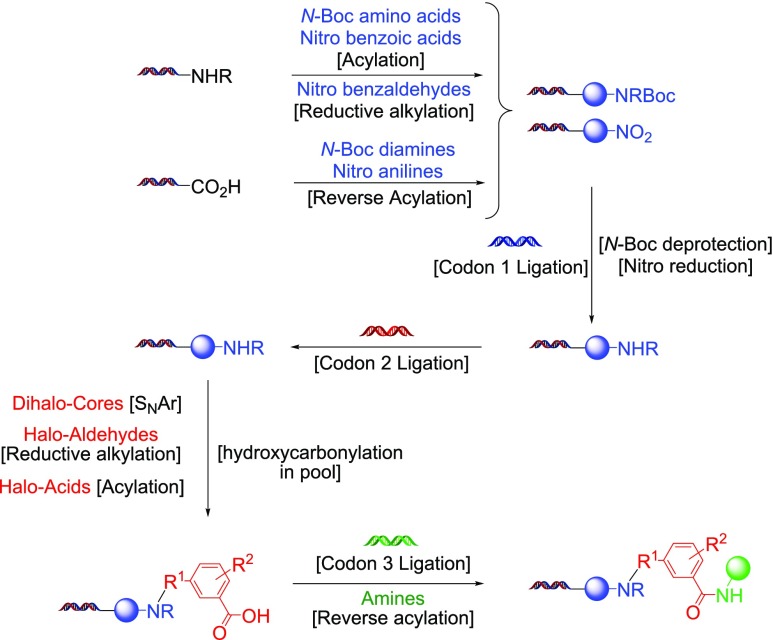
Although these tests suggested that we had developed a DNA-compatible process, application of the transformation within a pilot DECL synthesis, subsequent analysis of DECL DNA amplification efficiency and sequence distributions, and the discovery of hits are the definitive tests to determine a chemical transformation’s suitability for large-scale DECL production. To accomplish this, these conditions were used on a portion of the pooled cycle 2 material from a three-cycle, “split-and-pool” type DECL synthesis we described previously (Scheme 5).21 In this “split-and-pool” type process, each cycle consists of an initial split of DNA materials into many wells, a specific ligation of a unique encoding DNA tag, a specific chemical transformation and/or building block attachment, and a final pooling. In this DECL, hundreds of thousands of DNA-encoded, two-cycle (hetero)aryl halide substrates were prepared through an initial cycle 1 linkage of a bifunctional protected amine and, after N-deprotection, a subsequent attachment of a dihalo, halo-aldehyde or halo-acid core in cycle 2. To avoid unknown scale-induced effects,47 the two-cycle material was split into 150 nanomole portions in individual wells to undergo hydroxycarbonylation and then repooled. For the third cycle, the collection of generated carboxylic acids was split into wells, each well ligated with an encoding DNA-tag and the carboxylic acids were amidated through a “reverse” acylation with a unique amine to ultimately provide a DECL of ∼81 million compounds (DECL synthesis details are in the Supporting Information). After the ligation of a library-encoding DNA tag, a “naive” sample of this DECL (i.e., not subjected to protein binding experiments) was sequenced after elaboration with additional DNA segments that enable amplification, illumina sequencing, and bioinformatic analysis. Overall, DNA recovery was close to levels observed within similar productions, the amplification efficiency was consistent to other DECLs of similar DNA architecture, no base-specific DNA effects were detected, and normalized codon populations had narrow Gaussian-like distributions (Figure 1).
Scheme 5. Synthesis of the Hydroxycarbonylation DECL.
Figure 1.
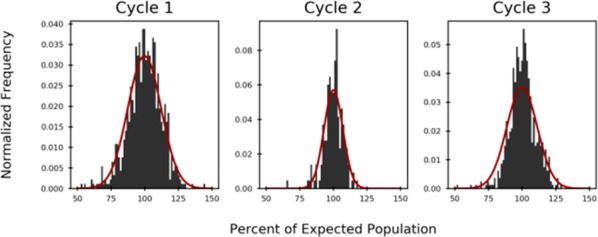
Codon distributions within the DECL.
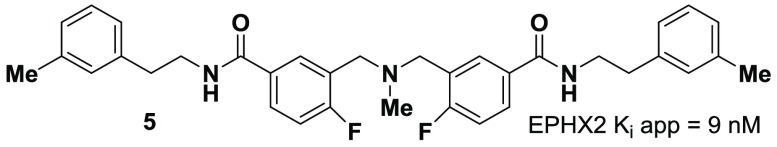
Finally, we sought to further validate the successful application of hydroxycarbonylation within this DECL through the rediscovery of known inhibitors or binding motifs through test “selections”49 against several previously studied protein targets. After a three-round “selection” against soluble epoxide hydrolase (sEH, EPHX2), a cardiovascular target that typically exhibits high DECL success rates due to its affinity for amide and urea pharmacophores,50 bio- and cheminformatics analysis51 revealed significant enrichment within a disynthon52 series. This disynthon hit series was produced through the cycle 2 reductive amination of 2-fluoro-5-iodobenzaldehyde and the cycle 3 “reverse” acylation of 3-methylphenylethylamine on the hydroxycarbonylation-generated carboxylic acid, a series which bears structural similarity to previously disclosed EPHX2 inhibitors.53 Synthesis of the dimeric54 compound truncated with a methyl group within the cycle 1 region provided 5 with an apparent Ki = 9 ± 2 nM derived from a fluorescent kinetic assay (Figure 2).55 An additional test set of “selections” was performed against L3MBTL1, a member of the malignant brain tumor (MBT) family of methylated lysine readers.56 After a four-round “selection”, bio- and cheminformatic analysis revealed high enrichment within cycle 3 monosynthons structurally similar to other known MBT-binders featuring benzamide-connected pyrrolidinyl amines (see Supporting Information for monosynthon analysis).57
Figure 2.
EPHX2 inhibitor identified from the DECL.
Conclusions
In summary, we have demonstrated that sSPhos-Pd-G2 is an efficient catalyst for DNA-compatible hydroxycarbonylation on a variety of DNA-linked (hetero)aryl halides, and that Mo(CO)6 may safely be used as a source of carbon monoxide within this system. This reaction was utilized within a DECL production and further validated through the discovery of hits. These results should be useful to future full-scale DECL builds and the development of other DNA-compatible carbonylative cross-coupling methods.
Experimental Procedures
Materials and Instrumentation
DTSU (“DEC-Tec Starting Unit”) (Figure S1) and 5′-phosphorylated oligonucleotides were obtained from LGC Biosearch Technologies. All DNA samples were assessed for purity through the general analytical procedure for DNA oligonucleotides before use. Sequences of the oligonucleotides were designed to maximize sequence-reads and sequencing data quality (by avoiding close similarities) and each duplexed DNA pair (“codon”) was designed to have divergent masses for efficient quality control analysis. A 10-mer DNA oligomer functionalized with a primary amine and cholesterol tag (“spike-in”) was obtained from Sigma to monitor chemical steps during postpooling manipulations (as the greasy oligo chromatographically separates from the main library peak). T4 DNA ligase was purchased from Enzymatics (Qiagen) and the activity (i.e., minimal amount that can provide full ligation under our standardized ligation conditions) was determined through test DNA ligations. Chemical building blocks and reagents were sourced from a variety of vendors and aliquots of building blocks were stored in pure or partially aqueous acetonitrile, dimethylacetamide, 1-methoxy-2-propanol, or DMSO solutions. Building block solutions were stored in either Tracetraq (Biosero) or Matrix (Fisher) barcoded tubes with either screw- or septa-caps. Barcoded tubes were read using a SampleScan 96 scanner (BiomicroLab) and decoded using Vortex software (Dotmatics) and Excel (Microsoft). All listed buffers, including HEPES 10X ligation buffer (300 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid, 100 mM MgCl2, 100 mM dithiothreitol, 10 mM adenosine triphosphate, pH 7.8), concentrated hydroxide solutions, and basic borate buffer (250 mM sodium borate/boric acid, pH 9.5), were prepared in-house. DECL working solutions were prepared using DNase free ultrapure water (Invitrogen), HPLC-grade acetonitrile (Fisher), HPLC-grade dimethylacetamide, HPLC-grade DMSO, or high-purity absolute ethanol (Koptec). LC/MS running solvents were made from Optima LC/MS grade water (Fisher), Optima LC/MS grade methanol (Fisher), 99+% purity hexafluoroisopropanol (Sigma), and HPLC-grade triethylamine (Fisher). Solution transfers were performed using Biotix or Fisher brand pipet tips and reservoirs (various sizes), reactions were generally performed in polypropylene, 96-well, deep-well plates (USA Scientific, various sizes), or 96-well PCR plates (Fisher), plates were sealed for incubation with AlumaSeal II foil seals (Excel Scientific) and large volume DNA precipitations were performed in polypropylene 250 mL screw-cap bottles (from various vendors). Heated reactions were either performed in ep384 Mastercyclers (Eppendorf) or in laboratory ovens (Fisher). Solutions were centrifuged in either Avanti J-30I or Allegra X-15R centrifuges (Beckman-Coulter). Optical density measurements were made using a Biophotometer (Eppendorf).
Data Analysis
Samples were analyzed on a Thermo Vanquish UHPLC system coupled to an electrospray LTQ ion trap mass spectrometer. An ion pairing mobile phase comprising 15 mM TEA/100 mM HFIP in a water/methanol solvent system was used in conjunction with an oligonucleotide column Thermo DNAPac RP (2.1 × 50 mm, 4 μm) for all the separations. All mass spectra were acquired in the full scan negative-ion mode over the mass range 500–2000 m/z. The data analysis was performed by exporting the raw instrument data (.RAW) to an automated biomolecule deconvolution and reporting software (ProMass) which uses a novel algorithm known as ZNova to produce artifact-free mass spectra. The following deconvolution parameters were applied: peak width 3.0, merge width 0.2, minimum and normalized scores of 2.0 and 1.0, respectively. The noise threshold was set at S/N 2.0. The processed data was directly exported to Microsoft Excel worksheets for further data comparisons.
Ethanol Precipitation and DNA Reconstitution
Based on the theoretical solution volume n, n/20 – n/10 volume of a 5 M NaCl stock solution was added and the solution was gently mixed. Then, absolute ethanol (3n volume, 75% v/v final ethanol concentration) was added, and the solution was thoroughly mixed and then stored at −20 °C overnight to precipitate the DNA. The resulting slurry was centrifuged (>10,000g for 1 h), the supernatant decanted, an addition 2n–3n of chilled 75% ethanol (v/v) was added, and the pellet was centrifuged again (>10,000g for 30 min). After removal of the supernatant, the pellet was dried (in open air or under gentle vacuum within a speed-vac) and then reconstituted in neutral water or buffer (to a concentration of ∼1 mM; assessed by optical density measurements).
General Procedure for DNA Compatible Hydroxycarbonylation Reaction
To the reconstituted DNA-linked halides (10 nmol, 10 μL, 1.0 mM in H2O, 1 equiv) was added CsOH (4000 nmol, 10 μL, 400 mM in H2O, 400 equiv) and Mo(CO)6 (1000 nmol, 5 μL, 200 mM in 1-methoxy-2-propanol (MIPO), 100 equiv), followed by adding sSPhos-Pd-G2 (120 nmol, 12 μL, 20 mM in MIPO, 12 equiv). The reaction mixture was heated at 80 °C for 15 min and was cooled to room temperature prior to EtOH precipitation.
Acknowledgments
This work was supported by the Welch Foundation (Grant Q-0042), a Core Facility Support Award (RP160805) from the Cancer Prevention Research Institute of Texas (CPRIT), grant OPP1160866 from The Bill and Melinda Gates Foundation, and National Institutes of Health grant P01HD087157 from The Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.9b00447.
Details of experimental procedures, DNA structures, DECL construction, and selection experiments (PDF)
Author Contributions
J.-Y.L. optimized reaction conditions, prepared substrates, explored substrate scope, and cobuilt the DECL. G.M. performed the L3MBTL1 selection. R.K.M. synthesized the EPHX2 hit. K.M.B. performed the EPHX2 assay. Z.F. performed the EPHX2 selection. M.P. conducted naive sequencing experiments. J.C.F. provided DECL cheminformatic analysis. K.R. provided DECL bioinformatics analysis. M.M.M. advised experiments. N.S. advised experiments and cobuilt the DECL.
The authors declare no competing financial interest.
Supplementary Material
References
- Neri D.; Lerner R. A. (2018) DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu. Rev. Biochem. 87, 479–502. 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satz A. L. (2018) What Do You Get from DNA-Encoded Libraries?. ACS Med. Chem. Lett. 9 (5), 408–410. 10.1021/acsmedchemlett.8b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow R. A.; Dumelin C. E. Jr; Keefe A. D. (2017) DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat. Rev. Drug Discovery 16, 131–147. 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- Decurtins W.; Wichert M.; Franzini R. M.; Buller F.; Stravs M. A.; Zhang Y.; Neri D.; Scheuermann J. (2016) Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat. Protoc. 11 (4), 764–780. 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B.; Zhou Y.; Huang Y.; Zhang J.; Li X. (2017) Recent advances on the encoding and selection methods of DNA-encoded chemical library. Bioorg. Med. Chem. Lett. 27 (3), 361–369. 10.1016/j.bmcl.2016.12.025. [DOI] [PubMed] [Google Scholar]
- Chan A. I.; McGregor L. M.; Liu D. R. (2015) Novel selection methods for DNA-encoded chemical libraries. Curr. Opin. Chem. Biol. 26, 55–61. 10.1016/j.cbpa.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.; Davie C. P. (2017) Zirconium(IV)-Catalyzed Ring Opening of on-DNA Epoxides in Water. ChemBioChem 18 (9), 843–847. 10.1002/cbic.201600563. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Franklin G. J.; DeLorey J. L.; Centrella P. A.; Mataruse S.; Clark M. A.; Skinner S. R.; Belyanskaya S. (2016) Design and Synthesis of Biaryl DNA-Encoded Libraries. ACS Comb. Sci. 18 (10), 625–629. 10.1021/acscombsci.6b00078. [DOI] [PubMed] [Google Scholar]
- Satz A. L.; Cai J.; Chen Y.; Goodnow R.; Gruber F.; Kowalczyk A.; Petersen A.; Naderi-Oboodi G.; Orzechowski L.; Strebel Q. (2015) DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjugate Chem. 26 (8), 1623–1632. 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- Kölmel D. K.; Loach R. P.; Knauber T.; Flanagan M. E. (2018) Employing Photoredox Catalysis for DNA-Encoded Chemistry: Decarboxylative Alkylation of a-Amino Acids. ChemMedChem 13 (20), 2159–2165. 10.1002/cmdc.201800492. [DOI] [PubMed] [Google Scholar]
- Du H.-C.; Huang H. (2017) DNA-Compatible Nitro Reduction and Synthesis of Benzimidazoles. Bioconjugate Chem. 28 (10), 2575–2580. 10.1021/acs.bioconjchem.7b00416. [DOI] [PubMed] [Google Scholar]
- Ruff Y.; Berst F. (2018) Efficient copper-catalyzed amination of DNA-conjugated aryl iodides under mild aqueous conditions. MedChemComm 9, 1188–1193. 10.1039/C8MD00185E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Lundberg H.; Asai S.; Martín-Acosta P.; Chen J. S.; Brown S.; Farrell W.; Dushin R. G.; O’Donnell C. J.; Ratnayake A. S.; et al. (2018) Kinetically guided radical-based synthesis of C(sp3)-C(sp3) linkages on DNA. Proc. Natl. Acad. Sci. U. S. A. 115 (28), E6404–E6410. 10.1073/pnas.1806900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Clark M. A. (2015) Robust Suzuki-Miyaura Cross-Coupling on DNA-Linked Substrates. ACS Comb. Sci. 17 (1), 1–4. 10.1021/co5001037. [DOI] [PubMed] [Google Scholar]
- Li H.; Sun Z.; Wu W.; Wang X.; Zhang M.; Lu X.; Zhong W.; Dai D. (2018) Inverse-Electron-Demand Diels-Alder Reactions for the Synthesis of Pyridazines on DNA. Org. Lett. 20 (22), 7186–7191. 10.1021/acs.orglett.8b03114. [DOI] [PubMed] [Google Scholar]
- Franzini R. M.; Randolph C. (2016) Chemical Space of DNA-Encoded Libraries. J. Med. Chem. 59 (14), 6629–6644. 10.1021/acs.jmedchem.5b01874. [DOI] [PubMed] [Google Scholar]
- Szostak M.; Yao L.; Aubé J. (2009) Stability of Medium-Bridged Twisted Amides in Aqueous Solutions. J. Org. Chem. 74 (5), 1869–1875. and references therein. 10.1021/jo802192v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Gabriele E.; Samain F.; Favalli N.; Sladojevich F.; Scheuermann J.; Neri D. (2016) Optimized Reaction Conditions for Amide Bond Formation in DNA-Encoded Combinatorial Libraries. ACS Comb. Sci. 18 (8), 438–443. 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone M.; Paegel B. M. (2016) What is a “DNA-Compatible” Reaction?. ACS Comb. Sci. 18 (4), 182–187. 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliokoski T. (2015) Price-Focused Analysis of Commercially Available Building Blocks for Combinatorial Library Synthesis. ACS Comb. Sci. 17 (10), 600–607. 10.1021/acscombsci.5b00063. [DOI] [PubMed] [Google Scholar]
- We recently explored the use of nitroarenes in DECL synthesis as a masked aniline, see:; Du H.-C.; Simmons N.; Faver J. C.; Yu Z.; Palanaippan M.; Riehle K.; Matzuk M. (2019) A mild, DNA-compatible nitro reduction using B2(OH)4. Org. Lett. 21 (7), 2194–2199. 10.1021/acs.orglett.9b00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. A.; Acharya R. A.; Arico-Muendel C. C.; Belyanskaya S. L.; Benjamin D. R.; Carlson N. R.; Centrella P. A.; Chiu C. H.; Creaser S. P.; Cuozzo J. W.; et al. (2009) Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 5, 647–654. 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- Deng H.; Zhou J.; Sundersingh F. S.; Summerfield J.; Somers D.; Messer J. A.; Satz A. L.; Ancellin N.; Arico-Muendel C. C.; Bedard K. L.; et al. (2015) Discovery, SAR, and X-ray Binding Mode Study of BCATm Inhibitors from a Novel DNA-Encoded Library. ACS Med. Chem. Lett. 6 (8), 919–924. 10.1021/acsmedchemlett.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Castillo P.; Buchwald S. L. (2016) Applications of Palladium-Catalyzed C-N Cross-Coupling Reactions. Chem. Rev. 116 (19), 12564–12649. 10.1021/acs.chemrev.6b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaura N.; Suzuki A. (1995) Palladium-Catalyzed Cross-Coupling Reactions of Organoboron Compounds. Chem. Rev. 95 (7), 2457–2483. 10.1021/cr00039a007. [DOI] [Google Scholar]
- Seechurn C. C. C. J.; Kitching M. O.; Colacot T. J.; Snieckus V. (2012) Palladium-catalyzed cross-coupling: a historical contextual perspective of the 2010 Nobel Prize. Angew. Chem., Int. Ed. 51 (21), 5062–5085. 10.1002/anie.201107017. [DOI] [PubMed] [Google Scholar]
- Li J.-Y.; Huang H. (2018) Development of DNA-Compatible Suzuki-Miyaura Reactions in Aqueous Media. Bioconjugate Chem. 29 (11), 3841–3846. 10.1021/acs.bioconjchem.8b00676. [DOI] [PubMed] [Google Scholar]
- Ding Y.; DeLorey J. L.; Clark M. A. (2016) Novel Catalyst System for Suzuki-Miyaura Coupling of Challenging DNA-Linked Aryl Chlorides. Bioconjugate Chem. 27 (11), 2597–2600. 10.1021/acs.bioconjchem.6b00541. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Kanan M. W.; Liu D. R. (2002) Expanding the reaction scope of DNA-templated synthesis. Angew. Chem., Int. Ed. 41 (10), 1796–1800. . [DOI] [PubMed] [Google Scholar]
- Wang X.; Sun H.; Liu J.; Zhong W.; Zhang M.; Zhou H.; Dai D.; Lu X. (2019) Palladium-Promoted DNA-Compatible Heck Reaction. Org. Lett. 21 (3), 719–723. 10.1021/acs.orglett.8b03926. [DOI] [PubMed] [Google Scholar]
- Lu X.; Roberts S. E.; Franklin G. J.; Davie C. P. (2017) On-DNA Pd and Cu promoted C-N cross-coupling reactions. MedChemComm 8, 1614–1617. 10.1039/C7MD00289K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro Beato E.; Priego J.; Gironda-Martinez A.; Gonzalez F.; Benavides J.; Blas J.; Martin-Ortega M. D.; Toledo M. A.; Ezquerra J.; Torrado A. (2019) Mild and Efficient Palladium-Mediated C-N Cross-Coupling Reaction between DNA-Conjugated Aryl Bromides and Aromatic Amines. ACS Comb. Sci. 21 (2), 69–74. 10.1021/acscombsci.8b00142. [DOI] [PubMed] [Google Scholar]
- Sergeev A. G.; Spannenberg A.; Beller M. (2008) Palladium-Catalyzed Formylation of Aryl Bromides: Elucidation of the Catalytic Cycle of an Industrially Applied Coupling Reaction. J. Am. Chem. Soc. 130 (46), 15549–15563. 10.1021/ja804997z. [DOI] [PubMed] [Google Scholar]
- Wu X.-F.; Neumann H.; Spannenberg A.; Schulz T.; Jiao H.; Beller M. (2010) Development of a General Palladium-Catalyzed Carbonylative Heck Reaction of Aryl Halides. J. Am. Chem. Soc. 132 (41), 14596–14602. 10.1021/ja1059922. [DOI] [PubMed] [Google Scholar]
- Wu X.-F.; Neumann H.; Beller M. (2011) Palladium-catalyzed carobonylative coupling reactions between Ar-X and carbon nucleophiles. Chem. Soc. Rev. 40 (10), 4986–5009. 10.1039/c1cs15109f. [DOI] [PubMed] [Google Scholar]
- Li C.-L.; Qi X.; Wu X.-F. (2016) Palladium-Catalyzed Hydroxycarbonylation of Aryl Halides with the in-situ Generation of CO and H2O. Chemistry Select 1 (8), 1702–1704. 10.1002/slct.201600514. [DOI] [Google Scholar]
- Gadakh A. V.; Chikanna D.; Rindhe S. S.; Karale B. K. (2012) Heteroaryl Hydroxycarbonylation: An Efficient, Robust, Practically Scalable Approach Using Formyl Acetate as the CO Source. Synth. Commun. 42 (5), 658–666. 10.1080/00397911.2010.528851. [DOI] [Google Scholar]
- Cacchi S.; Fabrizi G.; Goggiamani A. (2003) Palladium-Catalyzed Hydroxycarbonylation of Aryl and Vinyl Halides or Triflates by Acetic Anhydride and Formate Anions. Org. Lett. 5 (23), 4269–4272. 10.1021/ol0354371. [DOI] [PubMed] [Google Scholar]
- Commonly, on-DNA reaction screening is conducted at low nanomole scales in sealed PCR plates with thermocyler heating.
- For an example in which the noncarbonylative cross-coupling is competitive in the presence of CO, see:; O’Keefe B. M.; Simmons N.; Martin S. F. (2008) Carbonylative Cross-Coupling of ortho-Disubstituted Aryl Iodides. Convenient Synthesis of Sterically Hindered Aryl Ketones. Org. Lett. 10 (22), 5301–5304. 10.1021/ol802202j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcadi A.; Cerichelli G.; Chiarini M.; Correa M.; Zorzan D. (2003) A Mild and Versatile Method for Palladium-Catalyzed Cross-Coupling of Aryl Halides in Water and Surfactants. Eur. J. Org. Chem. 2003 (20), 4080–4086. 10.1002/ejoc.200300356. [DOI] [Google Scholar]
- Ueda T.; Konishi H.; Manabe K. (2013) Palladium-Catalyzed Reductive Carbonylation of Aryl Halides with N-Formylsaccharin as a CO Source. Angew. Chem., Int. Ed. 52 (33), 8611–8615. 10.1002/anie.201303926. [DOI] [PubMed] [Google Scholar]
- Jafarpour F.; Rashidi-Ranjbar P.; Kashani A. O. (2011) Easy-to-Execute Carbonylative Arylation of Aryl Halides using Molybdenum Hexacarbonyl: Efficient Synthesis of Unsymmetrical Diaryl Ketones. Eur. J. Org. Chem. 2011 (11), 2128–2132. 10.1002/ejoc.201001733. [DOI] [Google Scholar]
- Anderson K. W.; Buchwald S. L. (2005) General Catalysts for the Suzuki-Miyaura and Sonogashira Coupling Reactions of Aryl Chlorides and for the Coupling of Challenging Substrate Combinations in Water. Angew. Chem., Int. Ed. 44 (38), 6173–6177. 10.1002/anie.200502017. [DOI] [PubMed] [Google Scholar]
- Bruno N. C.; Tudge M. T.; Buchwald S. L. (2013) Design and Preparation of New Palladium Precatalysts for C-C and C-N Cross-Coupling Reactions. Chem. Sci. 4 (3), 916–920. 10.1039/C2SC20903A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- We have often observed suppressed rates of insertion of palladium catalysts for other on-DNA palladium-catalyzed cross couplings when these solvents are used. However, 1-methoxy-2-propanol is a hydride donor solvent that can promote undesired reduction side products (e.g., dehalogenation) in some transformations/conditions.
- As carbon monoxide is a gas and Mo(CO)6 is poorly soluble within this system, different product distributions due to scale effects associated with the rate of initial heating, solution volume of the reaction, headspace within the well, and the method of plate sealing could occur. Thus, the hydroxycarbonylation should only be used on previously studied scales and validated heating methods.
- The failure of these type of substrates in other palladium-catalyzed on-DNA reactions has been previously reported, see ref 32.
- “Selections” are experiments intended to find compounds within a DECL that bind to a protein target. This may be performed through multiple rounds of incubation of a DECL with a target, pull down, and subsequent elution of compounds off the protein to provide an enriched pool of small-molecule binders relative to the original DECL population. “Selection” parameters are often designed to find binders of micromolar to nanomolar affinities.
- Shen H. C. (2010) Soluble epoxide hydrolase inhibitors: a patent review. Expert Opin. Ther. Pat. 20 (7), 941–956. 10.1517/13543776.2010.484804. [DOI] [PubMed] [Google Scholar]
- Faver J. C.; Riehle K.; Lancia D. R.; Milbank J. B. J.; Kollmann C. S.; Simmons N.; Yu Z.; Matzuk M. M. (2019) Quantitative Comparison of Enrichment from DNA-Encoded Chemical Library Selections. ACS Comb. Sci. 21 (2), 75–82. 10.1021/acscombsci.8b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The term “di-synthon” refers to any two cycle building block/DNA encoding combination within a DECL. “Mono-synthon” and “tri-synthon” refers to the one and three cycle combinations, respectively.
- Kim I.-H.; Heirtzler F. R.; Morisseau C.; Nishi K.; Tsai H.-J.; Hammock B. D. (2005) Optimization of Amide-Based Inhibitors of Soluble Epoxide Hydrolase with Improved Water Solubility. J. Med. Chem. 48 (10), 3621–3629. 10.1021/jm0500929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Within this DECL, methods to produce a mono- versus a dialkylated product in the reductive amination of an aldehyde on a primary amine were encoded separately.
- Wolf N. M.; Morisseau C.; Jones P. D.; Hock B.; Hammock B. D. (2006) Development of a high-throughput screen for soluble epoxide hydrolase inhibition. Anal. Biochem. 355 (1), 71–80. 10.1016/j.ab.2006.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P.; Li G.; Sims R. J.; Vaquero A.; Kalakonda N.; Boccuni P.; Lee D.; Erdjument-Bromage H.; Tempst P.; Nimer S. D.; et al. (2007) L3MBTL1, a Histone-Methylation-Dependent Chromatin Lock. Cell 129 (5), 915–928. 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- James L. I.; Korboukh V. K.; Krichevsky L.; Baughman B. M.; Herold J. M.; Norris J. L.; Jin J.; Kireev D. B.; Janzen W. P.; Arrowshith C. H.; et al. (2013) Small-molecule Ligands of Methyl-Lysine Binding Proteins: Optimization of Selectivity for L3MBTL3. J. Med. Chem. 56 (18), 7358–7371. 10.1021/jm400919p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.