This cross-sectional study investigates if a genetic risk score that comprised 12 primary open-angle glaucoma genetic risk variants is associated with age at disease diagnosis.
Key Points
Question
Is a genetic risk score (GRS) using 12 primary open-angle glaucoma genetic risk variants associated with the age at diagnosis?
Findings
In this cross-sectional study of 3108 individuals with primary open-angle glaucoma, each higher GRS unit was associated with an earlier age at diagnosis, and affected individuals in the top 5% of the GRS had an earlier age at diagnosis compared with those in the bottom 5% GRS.
Meaning
These findings suggest aggregate primary open-angle glaucoma genetic risk variants can influence clinically relevant disease features such as age at diagnosis.
Abstract
Importance
Genetic variants associated with primary open-angle glaucoma (POAG) are known to influence disease risk. However, the clinical effect of associated variants individually or in aggregate is not known. Genetic risk scores (GRS) examine the cumulative genetic load by combining individual genetic variants into a single measure, which is assumed to have a larger effect and increased power to detect relevant disease-related associations.
Objective
To investigate if a GRS that comprised 12 POAG genetic risk variants is associated with age at disease diagnosis.
Design, Setting, and Participants
A cross-sectional study included individuals with POAG and controls from the Glaucoma Genes and Environment (GLAUGEN) study and the National Eye Institute Glaucoma Human Genetics Collaboration (NEIGHBOR) study. A GRS was formulated using 12 variants known to be associated with POAG, and the alleles associated with increasing risk of POAG were aligned in the case-control sets. In case-only analyses, the association of the GRS with age at diagnosis was analyzed as an estimate of disease onset. Results from cohort-specific analyses were combined with meta-analysis. Data collection started in August 2012 for the NEIGHBOR cohort and in July 2008 for the GLAUGEN cohort and were analyzed starting in March 2018.
Main Outcomes and Measures
Association of a 12 single-nucleotide polymorphism POAG GRS with age at diagnosis in individuals with POAG using linear regression.
Results
The GLAUGEN study included 976 individuals with POAG and 1140 controls. The NEIGHBOR study included 2132 individuals with POAG and 2290 controls. For individuals with POAG, the mean (SD) age at diagnosis was 63.6 (9.8) years in the GLAUGEN cohort and 66.0 (13.7) years in the NEIGHBOR cohort. For controls, the mean (SD) age at enrollment was 65.5 (9.2) years in the GLAUGEN cohort and 68.9 (11.4) years in the NEIGHBOR cohort. All study participants were European white. The GRS was strongly associated with POAG risk in case-control analysis (odds ratio per 1-point increase in score = 1.24; 95% CI, 1.21-1.27; P = 3.4 × 10−66). In case-only analyses, each higher GRS unit was associated with a 0.36-year earlier age at diagnosis (β = −0.36; 95% CI, −0.56 to −0.16; P = 4.0 × 10−4). Individuals in the top 5% of the GRS had a mean (SD) age at diagnosis of 5.2 (12.8) years earlier than those in the bottom 5% GRS (61.4 [12.7] vs 66.6 [12.9] years; P = 5.0 × 10−4).
Conclusions and Relevance
A higher dose of POAG risk alleles was associated with an earlier age at glaucoma diagnosis. On average, individuals with POAG with the highest GRS had 5.2-year earlier age at diagnosis of disease. These results suggest that a GRS that comprised genetic variants associated with POAG could help identify patients with risk of earlier disease onset impacting screening and therapeutic strategies.
Introduction
Primary open-angle glaucoma (POAG), a leading cause of blindness worldwide, is a genetically complex disease associated with multiple genetic and environmental risk factors. Genome-wide association studies have now identified over 20 loci associated with POAG in European white, Asian, and multiethnic populations.1 Although genetic variants located within these associated regions are known to influence disease risk, the clinical effect of associated variants individually or in aggregate is currently not known. A genetic risk score (GRS) examines the cumulative genetic load by combining individual genetic variants into a single measure, which is assumed to have a larger effect and increased power to detect relevant disease-related associations.2 While the heritability of POAG is substantial overall, the genetic effect may be largest in patients with earlier onset of disease.3 Identifying risk variants associated with earlier disease onset could influence monitoring and therapeutic strategies. In this study, we performed a retrospective cross-sectional study to evaluate the association of a cumulative dose of 12 established POAG genetic risk variants with age at diagnosis.
Methods
This study has been approved by the institutional review boards of Massachusetts Eye and Ear, Harvard School of Public Health, Brigham and Women’s Hospital, University of Pittsburgh, Johns Hopkins University, Duke University, University of West Virginia, University of Miami, University of Michigan, Stanford University, Marshfield Clinic, and the University of California, San Diego. Written informed consent was obtained from all participants. Data collection started in August 2012 for the NEIGHBOR cohort and in July 2008 for the GLAUGEN cohort and were analyzed starting in March 2018.
The Glaucoma Genes and Environment Study (GLAUGEN) genome-wide association study is part of the Genes Environment Association (GENEVA) studies,4 and the National Eye Institute Glaucoma Human Genetics Collaboration (NEIGHBOR) genome-wide association study is a collaboration of 12 groups in the United States.5 Detailed information on these data sets has been described previously.4,5,6 Both the GLAUGEN and NEIGHBOR data sets include individuals with POAG and controls.
A harmonized definition of POAG used the following criteria: (1) open anterior segment angles; (2) reproducible glaucomatous visual field loss on reliable tests; or (3) an eye with cup-disc ratio of at least 0.7 with 1 visual field showing glaucomatous loss; and (4) no identifiable secondary cause for optic nerve disease. Elevated intraocular pressure (IOP) was not a criterion for POAG definition, but if present, there had to be no secondary causes on anterior segment examination.5,6
The GRS was constructed based on the lead single-nucleotide polymorphisms (SNPs) for loci previously associated with POAG in European white populations (Table 16,7,8,9,10,11).1 For this study, the SNPs we selected for the GRS were the lead SNPs from the loci with statistical evidence for association with POAG in European white populations. The GRS corresponds with the unweighted sum of all POAG risk alleles (ie, dosage) under the assumption that all risk alleles have the same effect on the measure selected for analysis (age at diagnosis). To create the GRS, the risk alleles at each SNP were aligned in each data set.
Table 1. Association of Lead SNPs With Primary Open-Angle Glaucoma in the Combined NEIGHBOR and GLAUGEN Data Sets.
| SNP | Chromosome | Nearest Gene | Risk Allele | OR (95% CI) | P Value | Reported Effect | Source |
|---|---|---|---|---|---|---|---|
| rs4656461 | 1 | TMCO1 | G | 1.33 (1.20-1.47) | 6.8 × 10−8 | 1.51 | Burdon et al7 |
| rs6445055 | 3 | FNDC3B | G | 1.17 (1.06-1.29) | .002 | 1.09 | Hysi et al8 |
| rs4619890 | 4 | AFAP1 | G | 1.17 (1.09-1.25) | 2.2 × 10−5 | 1.20 | Gharahkhani et al9 |
| rs2745572 | 6 | FOXC1 | A | 1.26 (1.17-1.37) | 7.0 × 10−9 | 1.17 | Bailey et al10 |
| rs11969985 | 6 | GMDS | G | 1.17 (1.05-1.30) | .004 | 1.31 | Gharahkhani et al9 |
| rs4236601 | 7 | CAV1 | A | 1.21 (1.12-1.31) | 2.2 × 10−6 | 1.27 | Thorleifsson et al11 |
| rs2472493 | 9 | ABCA1 | G | 1.24 (1.16-1.34) | 1.9 × 10−9 | 1.31 | Gharahkhani et al9 |
| rs4977756 | 9 | CDKN2B-AS1 | A | 1.41 (1.30-1.52) | 5.5 × 10−19 | 1.39 | Burdon et al7 |
| rs7137828 | 12 | ATXN2 | T | 1.17 (1.09-1.26) | 1.4 × 10−5 | 1.17 | Bailey et al10 |
| rs10483727 | 14 | SIX1/SIX6 | A | 1.28 (1.19-1.37) | 1.7 × 10−11 | 1.32 | Wiggs et al6 |
| rs9897123 | 17 | GAS7 | C | 1.16 (1.08-1.25) | 3.7 × 10−5 | 1.20 | Bailey et al10 |
| rs35934224 | 22 | TXNRD2 | C | 1.36 (1.23-1.51) | 3.3 × 10−9 | 1.28 | Bailey et al10 |
| GRSa | NA | NA | NA | 1.24 (1.21-1.27) | 3.4 × 10−66 | NA | NA |
Abbreviations: GLAUGEN, Glaucoma Genes and Environment study; GRS, genetic risk score; NA, not applicable; NEIGHBOR, National Eye Institute Glaucoma Human Genetics Collaboration study; OR, odds ratio; SNP, single-nucleotide polymorphism.
Genetic risk score corresponds with the sum of all risk alleles.
To test for association, regression analyses were performed separately for the GLAUGEN and NEIGHBOR data sets using SAS, version 9.4 (SAS Institute). Logistic regression was performed for each SNP and for the cumulative GRS with POAG. Linear regression was performed for each SNP and for the cumulative GRS with age at diagnosis. Potential confounding factors such as sex, DNA source, and population structure (eigenvector 1, 2 and 6 for GLAUGEN and eigenvector 1 and 2 for NEIGHBOR) were included as covariates in the regression models.4,6 Age at diagnosis between individuals with POAG in the bottom 5% of the GRS and those in the top 5% of the GRS was compared using the t test. Histograms with kernel density curves and boxplots for age at diagnosis in the bottom 5% GRS and in the top 5% GRS were generated using the R package ggplot2 (R Foundation for Statistical Computing).
Meta-analysis was conducted to combine the results of the GLAUGEN and NEIGHBOR data sets. The inverse-variance weighting method was applied based on the regression coefficients and standard errors estimated from each study as implemented in the METAL software.12
Results
The GLAUGEN data set included 2116 individuals (976 individuals with POAG [46.1%] and 1140 controls [53.9%]), and the NEIGHBOR data set included 4422 individuals (2132 individuals with POAG [48.2%] and 2290 controls [51.8%]). The mean (SD) age at diagnosis was 63.6 (9.8) years for individuals with POAG in the GLAUGEN cohort and 66.0 (13.7) years for individuals with POAG in the NEIGHBOR cohort. The mean (SD) age at enrollment was 65.5 (9.2) years for GLAUGEN controls and 68.9 (11.4) years for NEIGHBOR controls. In the GLAUGEN group, of 976 individuals with POAG, 570 (58.4%) were women, and of 1140 controls, 682 (59.8%) were women. In the NEIGHBOR group, of 2132 individuals with POAG, 1153 (54.1%) were women, and of 2290 controls, 1297 (56.5%) were women.
All 12 lead SNPs were associated with POAG in case-control analyses in the meta-analysis of NEIGHBOR and GLAUGEN data sets (Table 16,7,8,9,10,11), and the GRS created using the 12 SNPs selected for this study was significantly associated with POAG risk (odds ratio, 1.24; 95% CI, 1.21-1.27; P = 3 × 10−66). For individuals with POAG, the mean (SD) number of risk alleles was 14.0 (2.2), ranging from 6 to 21.
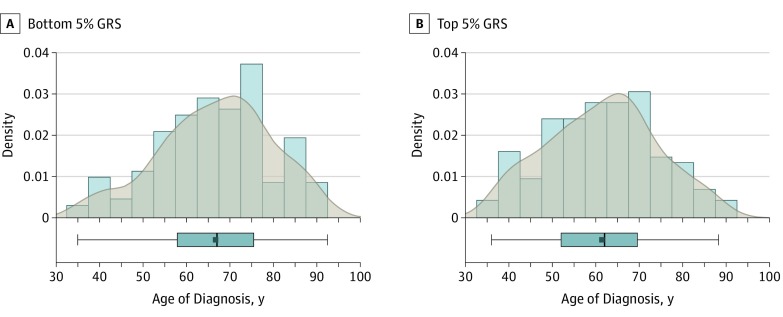
In case-only analyses, each higher GRS unit was associated with a 0.36-year earlier age at diagnosis (β = −0.36; 95% CI, −0.56 to −0.16; P = 4.0 × 10−4; Table 2). Four SNPs were individually nominally associated with earlier age at diagnosis: AFAP1 (β = −0.71; 95% CI, −1.31 to −0.11; P = .02), FOXC1 (β = −1.23; 95% CI, −1.93 to −0.53; P = 6.0 × 10−4), CDKN2B-AS1 (β = −0.73; 95% CI, −1.39 to −0.08; P = .03), and GAS7 (β = −0.88; 95% CI, −1.50 to −0.27; P = .005). Individuals in the top 5% of the GRS (equivalent to a mean of 18.3 risk allele dosage; range, 17.3-21.0) had a mean (SD) age at diagnosis of 5.2 (12.8) years earlier than those in the bottom 5% of the GRS (equivalent to a mean of 9.4 risk allele dosage; range, 6.0-10.1); mean [SD] ages in groups, 61.4 [12.7] vs 66.6 [12.9] years; P = 5.0 × 10−4; Figure).
Table 2. Association of Lead SNPs With Age at Diagnosis in GLAUGEN and NEIGHBOR Cases.
| SNP | Chromosome | Nearest Gene | Risk Allele | GLAUGEN (n = 973) | NEIGHBOR (n = 1974) | Meta-analysis (n = 2947) | |||
|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P Value | β (SE) | P Value | β (SE) | P Value | ||||
| rs4656461 | 1 | TMCO1 | G | 0.012 (0.629) | .98 | −1.549 (0.596) | .009 | −0.811 (0.433) | .06 |
| rs6445055 | 3 | FNDC3B | G | −0.044 (0.652) | .95 | 0.731 (0.628) | .24 | 0.358 (0.452) | .43 |
| rs4619890 | 4 | AFAP1 | G | 0.107 (0.447) | .81 | −1.435 (0.422) | 7 × 10−4 | −0.708 (0.307) | .02 |
| rs2745572 | 6 | FOXC1 | A | −1.105 (0.521) | .03 | −1.336 (0.487) | .006 | −1.228 (0.356) | 6 × 10−4 |
| rs11969985 | 6 | GMDS | G | −0.289 (0.705) | .68 | 0.466 (0.661) | .48 | 0.113 (0.482) | .82 |
| rs4236601 | 7 | CAV1 | A | −0.056 (0.477) | .91 | −0.639 (0.473) | .18 | −0.350 (0.336) | .30 |
| rs2472493 | 9 | ABCA1 | G | −0.243 (0.437) | .58 | 0.480 (0.439) | .27 | 0.117 (0.310) | .71 |
| rs4977756 | 9 | CDKN2B-AS1 | A | −0.445 (0.473) | .35 | −1.017 (0.470) | .03 | −0.733 (0.333) | .03 |
| rs7137828 | 12 | ATXN2 | T | 0.555 (0.439) | .21 | −0.074 (0.439) | .87 | 0.241 (0.310) | .44 |
| rs10483727 | 14 | SIX1/SIX6 | A | 0.234 (0.434) | .59 | −0.377 (0.438) | .39 | −0.069 (0.308) | .82 |
| rs9897123 | 17 | GAS7 | C | −0.558 (0.456) | .22 | −1.170 (0.432) | .007 | −0.881 (0.314) | .005 |
| rs35934224 | 22 | TXNRD2 | C | 0.059 (0.699) | .93 | 0.041 (0.647) | .95 | 0.049 (0.475) | .92 |
| GRSa | NA | NA | NA | −0.13 (0.14) | .38 | −0.58 (0.14) | 4 × 10−5 | −0.36 (0.10) | 4 × 10−4 |
Abbreviations: GLAUGEN, Glaucoma Genes and Environment study; GRS, genetic risk score; NA, not applicable; NEIGHBOR, National Eye Institute Glaucoma Human Genetics Collaboration study; SNP, single-nucleotide polymorphism.
Genetic risk score corresponds with the sum of all risk alleles.
Figure. Distribution of Age at Diagnosis Among Cases in the Bottom and Top 5% of the Genetic Risk Score (GRS) .
Histogram with kernel density curves and the boxplot for the age at diagnosis in cases (mean [SD] age, 66.6 [12.9] years) in the bottom 5% GRS (9.4 risk alleles; range, 6-10.1) (A) and for the age at diagnosis in cases (mean [SD] age, 61.4 [12.7] years) in the top 5% GRS (18.3 risk alleles; range, 17.3-21) (B); P = 5.0 × 10−4.
Discussion
In this study, we created a cumulative GRS for 12 SNPs previously shown to be significantly associated with POAG in European white individuals. We investigated the effect of this GRS on age at diagnosis, a disease feature with clinical relevance. Our results show that individuals with the largest number of risk variants (top fifth percentile) for the GRS have, on average, a 5.2-year earlier age at diagnosis compared with patients in the bottom fifth percentile (Figure). Patients with earlier disease onset will require more years of treatment and may be more likely to become blind from glaucoma.13Up to 50% of patients with an earlier diagnosis have affected first-degree relatives, suggesting that individuals in the top GRS percentiles could have affected family members who could benefit from clinical screening.14
Ten of the 12 POAG risk variants selected for the GRS are also known to be associated with IOP in population studies,15 and 2 of the 4 SNPs individually associated with earlier age at onset are also associated with IOP (FOXC1 and GAS7; Table 2). These results suggest that higher IOP is one factor that could contribute to the age at diagnosis effect.
Limitations
There are several limitations to this study. First, the retrospective study design can bias the results, and future prospective studies are necessary prior to the development of clinical tests using these or other genetic risk variants. Second, because many patients are unaware of glaucoma onset, the age at diagnosis may not accurately reflect the true age at disease onset. However, because onset is always before diagnosis, this difference would bias the results toward the null.
Conclusions
In summary, using a 12-SNP GRS, we have shown that an increased load of genetic risk variants was associated with an earlier age at disease diagnosis. These findings could lead to the development of screening tests that identify patients at risk for early disease, allowing for timely initiation of preventive treatment and surveillance strategies.
References
- 1.Wiggs JL, Pasquale LR. Genetics of glaucoma. Hum Mol Genet. 2017;26(r1):r21-r27. doi: 10.1093/hmg/ddx184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nannini DR, Kim H, Fan F, Gao X. Genetic risk score is associated with vertical cup-to-disc ratio and improves prediction of primary open-angle glaucoma in Latinos. Ophthalmology. 2018;125(6):815-821. doi: 10.1016/j.ophtha.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou T, Souzeau E, Siggs OM, et al. Contribution of mutations in known Mendelian glaucoma genes to advanced early-onset primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2017;58(3):1537-1544. doi: 10.1167/iovs.16-21049 [DOI] [PubMed] [Google Scholar]
- 4.Wiggs JL, Kang JH, Yaspan BL, et al. ; GENEVA Consortium . Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20(23):4707-4713. doi: 10.1093/hmg/ddr382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiggs JL, Hauser MA, Abdrabou W, et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013;22(7):517-525. doi: 10.1097/IJG.0b013e31824d4fd8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiggs JL, Yaspan BL, Hauser MA, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8(4):e1002654. doi: 10.1371/journal.pgen.1002654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43(6):574-578. doi: 10.1038/ng.824 [DOI] [PubMed] [Google Scholar]
- 8.Hysi PG, Cheng CY, Springelkamp H, et al. ; BMES GWAS Group; NEIGHBORHOOD Consortium; Wellcome Trust Case Control Consortium 2 . Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46(10):1126-1130. doi: 10.1038/ng.3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gharahkhani P, Burdon KP, Fogarty R, et al. ; Wellcome Trust Case Control Consortium 2, NEIGHBORHOOD consortium . Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46(10):1120-1125. doi: 10.1038/ng.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey JN, Loomis SJ, Kang JH, et al. ; ANZRAG Consortium . Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016;48(2):189-194. doi: 10.1038/ng.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42(10):906-909. doi: 10.1038/ng.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters D, Bengtsson B, Heijl A. Factors associated with lifetime risk of open-angle glaucoma blindness. Acta Ophthalmol. 2014;92(5):421-425. doi: 10.1111/aos.12203 [DOI] [PubMed] [Google Scholar]
- 14.Souzeau E, Goldberg I, Healey PR, et al. Australian and New Zealand Registry of Advanced Glaucoma: methodology and recruitment. Clin Exp Ophthalmol. 2012;40(6):569-575. doi: 10.1111/j.1442-9071.2011.02742.x [DOI] [PubMed] [Google Scholar]
- 15.Khawaja AP, Cooke Bailey JN, Wareham NJ, et al. ; UK Biobank Eye and Vision Consortium; NEIGHBORHOOD Consortium . Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018;50(6):778-782. doi: 10.1038/s41588-018-0126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]