Key Points
Question
What are the most important variables that predict the response to therapy to inhibit programmed cell death 1 and its ligand across different cancer types?
Findings
This analysis of multiomics data from the Cancer Genome Atlas cohort and objective response rates to therapy data across 21 cancer types found that estimated CD8+ T-cell abundance is the most predictive, followed by tumor mutational burden and the fraction of samples with high programmed cell death 1 gene expression.
Meaning
Immune, neoantigen, and checkpoint target variables are required in combination for accurately predicting Fthe response to therapy to inhibit programmed cell death 1 and its ligand across multiple cancers.
This cross-validation analysis of multi-omics data seeks to identify key neoantigen-, checkpoint-, and immune response–related variables associated with response to inhibition of programmed cell death 1 and its ligand across cancer types.
Abstract
Importance
Therapies to inhibit programmed cell death 1 and its ligand (anti–PD-1/PD-L1) provide significant survival benefits in many cancers, but the efficacy of these treatments varies considerably across different cancer types. Identifying the underlying variables associated with this cancer type–specific response remains an important open research challenge.
Objective
To evaluate systematically a multitude of neoantigen-, checkpoint-, and immune response–related variables to determine the key variables that accurately predict the response to anti–PD-1/PD-L1 therapy across different cancer types.
Design, Setting, and Participants
This analysis of a broad range of data used whole-exome and RNA sequencing of 7187 patients from the publicly available Cancer Genome Atlas and the objective response rate (ORR) data of 21 cancer types obtained from a collection of clinical trials. Thirty-six variables of 3 distinct classes considered were associated with (1) tumor neoantigens, (2) tumor microenvironment and inflammation, and (3) the checkpoint targets. The performance of each class of variables and their combinations in predicting the ORR to anti–PD-1/PD-L1 therapy was evaluated. Accuracy of predictions was quantified with Spearman correlation measured using a standard leave-one-out cross-validation, a statistical method of evaluating a statistical model by dividing data into 2 segments: one to train the model and the other to validate the model. Data were collected from October 19 through 31, 2018, and were analyzed from November 1 through December 14, 2018.
Main Outcomes and Measures
Response to anti-PD-1/PD-1 therapy.
Results
Among the 36 variables, estimated CD8+ T-cell abundance was the most predictive of the response to anti–PD-1/PD-L1 therapy across cancer types (Spearman R = 0.72; P < 2.3 × 10−4), followed by the tumor mutational burden (Spearman R = 0.68; P < 6.2 × 10−4), and the fraction of samples with high PD1 gene expression (Spearman R = 0.68; P < 6.9 × 10−4). Notably these top 3 variables cover the 3 classes considered, and their combination is highly correlated with response (Spearman R = 0.90; P < 4.1 × 10−8), explaining more than 80% of the ORR variance observed across different tumor types.
Conclusions and Relevance
That we know of, this is the first systematic evaluation of the different variables associated with anti–PD-1/PD-L1 therapy response across different tumor types. The findings suggest that the 3 key variables can explain most of the observed cross-cancer response variability, but their relative explanatory roles may vary in specific cancer types.
Introduction
Targeting programmed death 1 (PD-1) or its ligand (PD-L1) offers significant clinical benefits in many cancers. One emerging biomarker of response to anti–PD-1/PD-L1 therapy is the tumor mutational burden,1,2 which quantifies the number of somatic mutations in the tumor. The mutational burden has recently been shown by Yarchoan et al3 to strongly correlate with the response rate for anti–PD-1/PD-L1 therapies across multiple cancer types. However, tumors containing comparably high mutational burden may exhibit variable responses,4,5,6 suggesting that additional factors may contribute to anti–PD-1/PD-L1 response. Herein we systematically study this hypothesis, aiming to identify the most important additional factor(s) that contribute to response prediction. To this end we analyze whole-exome sequencing and RNA sequencing data of pretreated samples from 21 different cancer types in the Cancer Genome Atlas (TCGA). We find that a trivariate model composed of tumor mutational burden, estimated CD8+ T-cell abundance (eCD8T), and fraction of high PD-1 messenger RNA expression samples (fPD1) markedly enhances the prediction of the response to anti–PD-1/PD-L1 therapy across different cancer types.
Methods
Data were collected from October 19 through 31, 2018, and analyzed from November 1 through December 14, 2018. We considered 36 different variables that have been previously reported to be associated with anti–PD-1/PD-L1 response within a given cancer type, of 3 distinct classes: (1) descriptors associated with tumor neoantigens, including the tumor mutational burden, intratumor heterogeneity, neoantigen burden, and hydrophobicity7; (2) descriptors of the tumor microenvironment, including the estimated abundance of different immune cells, the cytolytic score,8 T-cell exhaustion signatures, interferon-γ signature, and estimated T-cell receptor diversity; and (3) checkpoint target–related variables, including PD-L1 protein expression, the combined positive score,9 and fPD110 (eMethods and eTable in the Supplement). All our predictions were performed using a standard leave-one-out cross-validation. This study does not require additional approval from an institutional review board because its human data is gathered from publicly available datasets (eg, TCGA) that already received such approvals.
Statistical Analysis
For this study, we performed a standard regression analysis with leave-one-out cross-validation in predicting objective response rate (ORR) across cancer types using the R package caret (R Foundation for Statistical Computing). The performance of the prediction was evaluated based on Spearman rank correlation (R) and unexplained variance (1-R2). For comparing goodness of fit between different models, a log-likelihood ratio test was performed. The association between tumor mutational burden, eCD8T, and their combination vs the patient survival after anti-PD-1/PD-L1 therapy in those with melanoma was evaluated using log-rank test. Statistical significance was set at 2-sided P < .05.
Results
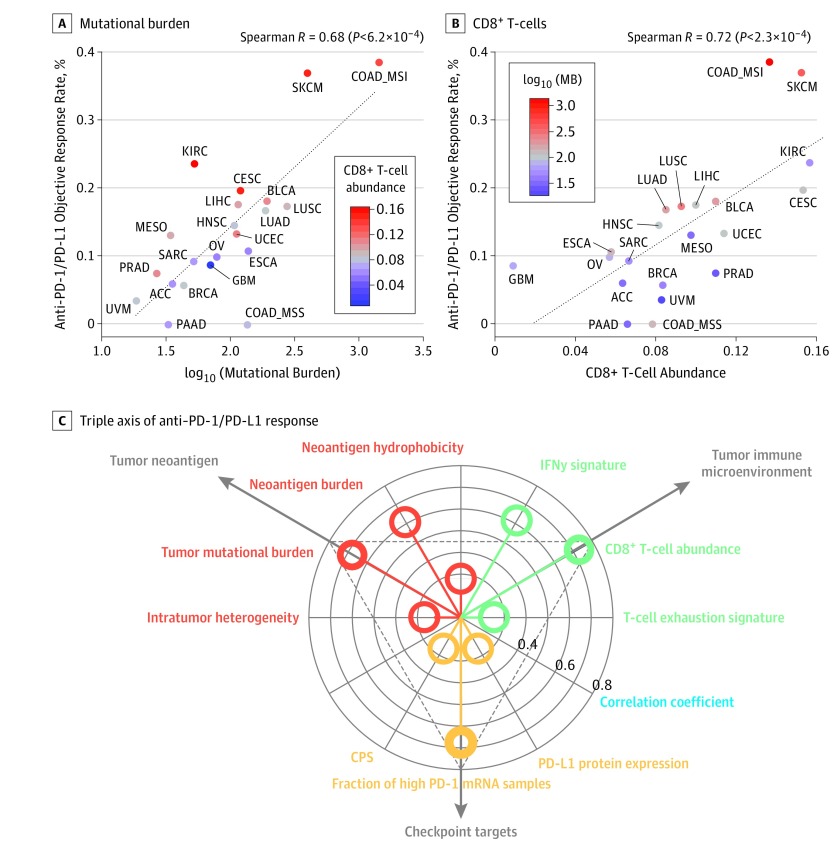
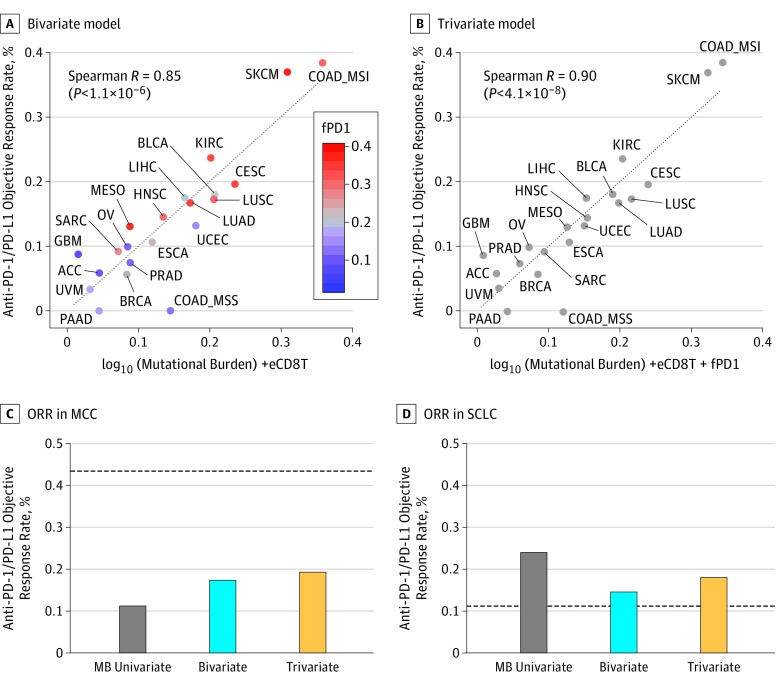
We studied 21 cancer types for which molecular TCGA data from 7187 patients and anti–PD-1/PD-L1 ORR from Yarchoan et al3 are available. First, we observed a high and significant correlation between mutational burden and ORR (Spearman R = 0.68; P < 6.2 × 10−4) (Figure 1A). The slightly higher correlation (R = 0.74; P < .001) Yarchoan et al3 originally reported may result from the consideration of a larger number of cancer types (n = 27) available from Chalmers et al.11 Second, we found that eCD8T was the strongest positive correlate of ORR (R = 0.72; P < 2.3 × 10−4) (Figure 1B). The estimated abundance of all other immune cell types was not significantly associated with PD-1/PD-L1 response, except for M1-macrophage (R = 0.51; P < .02) and CD4+ T-cell abundance (R = 0.44; P < .05) (eFigure 1A and B in the Supplement). The associations found for other neoantigen- or immune system–related variables were weaker (Figure 1C and eFigure 1C-I in the Supplement). Notably, most of the cancer types that showed higher response rates than predicted by the mutational burden regression model had higher eCD8T levels; those showing lower than mutational burden–based predicted response rates had lower eCD8T levels (Figure 1A). Correspondingly, most of the cancer types that showed a higher response rate than predicted by an eCD8T regression model had higher mutational burden and vice versa (Figure 1B). Accordingly, the combined mutational burden and eCD8T model markedly enhanced the response prediction (R = 0.85; P < 1.1 × 10−6) (Figure 2A) with a significant log-likelihood model improvement compared with each of the univariate models (P < .001). The resulting bivariate linear regression model assigned approximately equal weights to the mutational burden and eCD8T (when z scores of both variables were considered), which were not significantly correlated each other (R = 0.32; P = .16).
Figure 1. Systematic Evaluation of the Correlates of Therapy to Inhibit Programmed Cell Death 1 and Its Ligand (PD-1/PD-L1) Across Different Cancer Types.
A, Correlation of log10 (mutational burden) with the objective response rate to anti–PD-1/PD-L1 therapy across cancer types. The estimated CD8+ T-cell abundance (eCD8T) of each cancer type is color coded where red denotes high abundance and blue, low abundance. B, Correlation of eCD8T with the objective response rate to anti–PD-1/PD-L1 therapy across cancer types. The mutational burden of each cancer type is color coded where red denotes high mutational burden and blue, low mutational burden. C, Triple axis of anti–PD-1/PD-L1 response shows the distribution of Spearman correlation coefficients (blue) (radial axis) of the 3 classes of variables (polar axis) associated with tumor neoantigen (red), tumor immune microenvironment (green), and checkpoint targets (yellow). The dots inside the innermost circle denote the cases where the absolute value of the correlation coefficients are less than 0.30. ACC indicates adrenocortical carcinoma; BLCA, bladder carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD MSI, microsatellite-unstable colon adenocarcinoma; COAD MSS, microsatellite-stable colon adenocarcinoma; CPS, combined positive score; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; INF, interferon; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MB, mutational burden; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; and UVM, uveal melanoma.
Figure 2. Combined Regression Models for Predicting Therapy to Inhibit Programmed Cell Death 1 and Its Ligand (Anti–PD-1/PD-L1) Across Cancer Types.
A, Combined effect of the mutational burden and estimated CD8+ T-cell abundance (eCD8T) bivariate model (Spearman R = 0.85; P < 1.1 × 10−6). The regression formula for the objective response rate (ORR) is 0.13 × log10 (mutational burden) + 1.3 × eCD8T − 0.21. The fraction of high PD-1 messenger RNA expression samples (fPD1) of each cancer type is color coded where red denotes a high fraction and blue, a low fraction. B, Combined effect of the mutational burden–eCD8T-fPD1 trivariate model (Spearman R = 0.90; P < 4.1 × 10−8). The regression formula for the ORR is 0.12 × log10 (mutational burden) + 0.96 × eCD8T + 0.21 × fPD1 − 0.19. C and D, The estimated ORR to anti–PD-1/PD-L1 therapy in polyomavirus-positive Merkel cell carcinoma (MCC) (C) and small cell lung cancer (SCLC) (D) using a univariate mutational burden model (gray), the mutational burden–eCD8T bivariate model (blue), and the mutational burden–eCD8T-fPD1 trivariate model (yellow). The observed ORRs are depicted with dotted lines.
As exhibited in Figure 2A, several outlier cancer types had actual response rates that still significantly deviated from the predicted values of the combined bivariate regression model. We asked whether the third class of factors, which were associated with checkpoint targets, could possibly enhance the prediction accuracy. We focused on the fPD1 (>80-percentile across cancer types in TCGA) reported to correlate with anti–PD-1 ORR across a smaller set of cancer types (n = 17).10 We found that the performances of an fPD1-based model (R = 0.70) and the bivariate mutational burden–eCD8T model (R = 0.71) were comparable. In the larger data set of Yarchoan et al,3 however, the correlation of the fPD1 model (R = 0.68) was much weaker than that of the bivariate model (eFigure 1J in the Supplement). Notably, however, most cancer types that showed higher response rate than predicted by the mutational burden–eCD8T model had higher fPD1 levels (Figure 2A) and those showing lower than predicted response rates had lower fPD1 levels. Indeed, adding fPD1 to the bivariate mutational burden–eCD8T model led to a trivariate regression model with a significantly improved accuracy (R = 0.90; P < 4.1 × 10−8; log-likelihood ratio test P < .02) in the Yarchoan et al data set3 (Figure 2B). There were 2 clear outliers whose actual response rate was far from that predicted by the trivariate regression model, glioblastoma multiforme and microsatellite-stable colon cancer. The discrepancy observed for glioblastoma multiforme could be explained by its strong cytolytic score (eFigure 1F in the Supplement). Other factors in this class associated with checkpoint targets, including PD-L1 protein expression and combined positive score, were not predictive of the response (Figure 1C and eFigure 1K-L in the Supplement).
Some tumors are known to exhibit high response rates to anti–PD-1/PD-L1 therapy despite having low mutational burden, such as Merkel cell carcinoma, kidney renal cell carcinoma, and mesothelioma. On the other hand, some tumors such as small cell lung cancer have low response rates, despite their high mutational burden. The high response rate of kidney renal cell carcinoma and mesothelioma can be explained by the high eCD8T (Figure 1A) and fPD1 levels (eFigure 2A in the Supplement) observed in these cancer types. Because Merkel cell carcinoma and small cell lung cancer samples were not available in TCGA, we analyzed independent cohorts of these cancer types.12,13 Although incorporating eCD8T and fPD1 levels does not fully explain the observed ORRs, it significantly improved on the mutational burden–based predictions (Figure 2C-D). For Merkel cell carcinoma, the increase in predicted ORR was more prominent in polyomavirus-positive cases in which the mutational burden was low (Figure 2C) than in the polyomavirus-negative cases with a high mutational burden12 (eFigure 2B in the Supplement); however, better descriptors remain to be discovered in this case. As noted by Yarchoan et al,3 high CD8+ T-cell infiltration induced by presentation of viral antigens may confer an increased response rate to anti–PD-1/PD-L1 therapy in Merkel cell carcinoma.
Finally, we evaluated the performance of our models in predicting the PD-1/PD-L1 response of individual patients with melanoma by analyzing a large data set providing whole-exome and RNA sequencing data.14 Because fPD1 is a cancer type–specific property that is difficult to translate to individual patients in a straightforward manner, we resorted to testing the combined association of mutational burden and eCD8T and compared its performance to those of the respective univariate models. Although both individual variables were not predictive, their combination was (eFigure 2C in the Supplement).
Discussion
This systematic evaluation of the different variables associated with anti–PD-1/PD-L1 therapy response across different tumor types is, to our knowledge, the first such study. Going beyond a single variate mutational burden model to include immune- and checkpoint target-related variables has enabled us to markedly reduce the unexplained variance in predicting anti–PD-1/PD-L1 response rate across cancer types from 0.45 (1 – 0.742) to 0.19 (1 – 0.92). Both analyses were based on public data of the patients who were not given anti–PD-1/PD-L1 therapy. A potential limitation is that our analysis is based on bulk tumor data, which obviously only reflects the mean expression of different cell populations in the tumor’s microenvironment. Nonetheless, it is quite remarkable and nontrivial that such accuracy can be obtained from the analysis of bulk tumor data, considering the complex interplay between different cell types in the tumor microenvironment, which modulates the clinical response to checkpoint inhibitors. With the accumulation of tumor-specific single cell data and the advancement of computational deconvolution methods, we expect that our results could be further improved in a significant manner, possibly leading to a better understanding of the mechanisms underlying the variability of immune response observed across different cancer types.
Conclusions
Our findings show that variables from the 3 different classes all play an important and quite independent role in mediating anti–PD-1/PD-L1 therapy response across multiple cancers. Their relative explanatory roles may vary in specific cancer types.
eMethods. Tumor Neoantigen, Tumor Immune Microenvironment, Checkpoint Targets, and Statistics
eTable. ORR and Correlate Variables of anti–PD-1/PD-L1 Therapy Across Cancer Types
eFigure 1. Systematic Evaluation of Correlates of Response to Anti–PD-1/PD-L1 Therapy Across Different Cancer Types
eFigure 2. Combination of MB, eCD8T and fPD1 to Predict Anti–PD-1/PD-L1 Response in Individual Cancer Types
eReferences.
References
- 1.Rizvi NA, Hellmann MD, Snyder A, et al. . Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348(6230):124-128. doi: 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riaz N, Havel JJ, Makarov V, et al. . Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934-949.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500-2501. doi: 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275-287. doi: 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542-e551. doi: 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao D, Van Allen EM. Genomic determinants of cancer immunotherapy. Curr Opin Immunol. 2016;41:32-38. doi: 10.1016/j.coi.2016.05.010 [DOI] [PubMed] [Google Scholar]
- 7.Chowell D, Krishna S, Becker PD, et al. . TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci U S A. 2015;112(14):E1754-E1762. doi: 10.1073/pnas.1500973112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48-61. doi: 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow LQM, Haddad R, Gupta S, et al. . Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 Expansion Cohort. J Clin Oncol. 2016;34(32):3838-3845. doi: 10.1200/JCO.2016.68.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paré L, Pascual T, Seguí E, et al. . Association between PD1 mRNA and response to anti-PD1 monotherapy across multiple cancer types. Ann Oncol. 2018;29(10):2121-2128. [DOI] [PubMed] [Google Scholar]
- 11.Chalmers ZR, Connelly CF, Fabrizio D, et al. . Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms PW, Vats P, Verhaegen ME, et al. . The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75(18):3720-3727. doi: 10.1158/0008-5472.CAN-15-0702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George J, Lim JS, Jang SJ, et al. . Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47-53. doi: 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugo W, Zaretsky JM, Sun L, et al. . Genomic and transcriptomic features of response to anti–PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35-44. doi: 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Tumor Neoantigen, Tumor Immune Microenvironment, Checkpoint Targets, and Statistics
eTable. ORR and Correlate Variables of anti–PD-1/PD-L1 Therapy Across Cancer Types
eFigure 1. Systematic Evaluation of Correlates of Response to Anti–PD-1/PD-L1 Therapy Across Different Cancer Types
eFigure 2. Combination of MB, eCD8T and fPD1 to Predict Anti–PD-1/PD-L1 Response in Individual Cancer Types
eReferences.