Abstract
The genus Avena (oats) contains diploid, tetraploid and hexaploid species that evolved through hybridization and polyploidization. Four genome types (named A through D) are generally recognized. We used GBS markers to construct linkage maps of A genome diploid (Avena strigosa x A. wiestii, 2n = 14), and AB genome tetraploid (A. barbata 2n = 28) oats. These maps greatly improve coverage from older marker systems. Seven linkage groups in the tetraploid showed much stronger homology and synteny with the A genome diploids than did the other seven, implying an allopolyploid hybrid origin of A. barbata from distinct A and B genome diploid ancestors. Inferred homeologies within A. barbata revealed that the A and B genomes are differentiated by several translocations between chromosomes within each subgenome. However, no translocation exchanges were observed between A and B genomes. Comparison to a consensus map of ACD hexaploid A. sativa (2n = 42) revealed that the A and D genomes of A. sativa show parallel rearrangements when compared to the A genomes of the diploids and tetraploids. While intergenomic translocations are well known in polyploid Avena, our results are most parsimoniously explained if translocations also occurred in the A, B and D genome diploid ancestors of polyploid Avena.
Subject terms: Agricultural genetics, Evolutionary biology
Introduction
Among its many evolutionary consequences1–3, polyploidy is widely recognized as a driver of genome rearrangement4–6. This is primarily due to the presence of homeologous chromosomes, which are prone to mispairing at meiosis7,8. Crossovers between homeologs can create translocations of medium-to-large chromosomal segments, while gene conversion events can transfer smaller sequences from one homeolog to the other4. Numerous examples of intergenomic rearrangement in polyploids have been described7–11. Such rearrangements are thought to be less common in diploids, because meiotic mispairing is less likely to occur. Indeed, paleopolyploidy, and subsequent gradual reversion to diploidy, has recently become implicated in fostering genome reshuffling among lineages that are now considered diploid3,6,12,13. Nevertheless, translocations between non-homeologous chromosomes in the absence of polyploidy are also known, though even these are sometimes associated with homoploid hybridization – i.e., hybridization without polyploidization14,15.
The oats (Avena spp.) form a genus of diploid, tetraploid, and hexaploid grasses with a base chromosome number of seven16–18. Most are self-pollinating annuals. Like wheat, the Avena polyploids exhibit disomic inheritance and are often designated as allotetraploid or allohexaploid18,19. Four cytogenetically distinct genomes, designated A through D, are recognized16,18. However, the B and D genomes are known only from polyploid species, since no extant diploids with these genomes have been described.
The most widely cultivated oat species, A. sativa, is hexaploid and contains the genomes A, C, and D, as do the wild hexaploid oats. A well supported consensus genetic linkage map is available for this economically important species20 which maps linkage groups to physical chromosomes. However, separate mapping populations of A. sativa contain unique rearrangements between homeologous chromosomes, indicating ongoing genomic exchange in this hexaploid. Seven linkage groups share markers primarily with A-genome diploids, while seven others share markers primarily with the C genome diploids21. The remaining seven share markers with the AC(DC) tetraploids A. maroccana (=A. magna), A. murphyi, and A. insularis, which are thus the most likely ancestor(s) of the C and D genomes in hexaploid oat. Furthermore, through map-based haplotype complementation21, as well as through previous cytogenetic analysis (summarized by Sanz et al.22), it appears that translocations between subgenomes within hexaploid A. sativa are more prevalent between C and D chromosomes than between A and D chromosomes. The above results support a hypothesis whereby the C and D genomes co-existed in a tetraploid oat (likely A. insularis) prior to hybridization(s) with an A-genome diploid to form the ACD genome hexaploids21. Although patterns of homeology in A. sativa have been heavily disrupted by translocations, four triplets of A/C/D linkage groups appear to have retained substantial linear homeology20.
The A and C genome diploids are distinct evolutionary lineages, appearing as separate clades in phylogenies of the diploid Avena species23–26. Several sub-types have been designated within each group based on karyotype analysis and chromosome pairing16,18. The most common subtype among the A-genome diploids is the As genome subtype, which also shows the strongest cytogenetic similarity to the A genome of the hexaploid A. sativa21. Thus the most intensive study of A genome diploids has been focused on species with this genome subtype, and several mapping populations have been studied27–29. It remains unclear which C-genome diploid is the progenitor of A. sativa, and we know of no genetic mapping population in C genome diploids.
The B-genome of Avena is known only from four AB tetraploid species, the most abundant of which is A. barbata. There has been a long running debate over whether the B genome should be recognized as distinct from the A genome16,19, and by extension whether A. barbata is auto- or allo tetraploid30. Early chromosome studies identified partial homologous pairing of A and B genome chromosomes during meiosis in hybrids, although this often occurred in chains, which suggested rearrangement of the A vs B genomes30–33. While Southern blot experiments failed to discriminate the genomes34, FISH probes specific to the A genome were able to distinguish A and B genomes within A. barbata35,36. Most recently, Chew et al.37 argued against the recognition of a distinct B genome because multivariate clustering of GBS markers placed AB genome tetraploids near As genome diploids. However, a more distinct separation of AB tetraploids from the A-genome diploids is obtained by selecting GBS markers in a way that would avoid an ascertainment bias caused by a prevalence of A-genomes in the test population21. In any case, the tetraploid A. barbata exhibits disomic inheritance30,38, allowing construction of a partial linkage map using AFLP markers39. However, this map is clearly incomplete, since the total length is shorter than most maps of diploid oats29, and because it contains nineteen linkage groups for A. barbata’s 14 chromosomes.
In the present study we had three goals. First, we used Genotyping by Sequencing (GBS) markers to update the linkage maps from A. strigosa C.I. 3815 × A. wiestii C.I. 199429 and A. barbata39. Second, we examined clusters of similar loci that segregate within A. barbata to identify homeologous regions40 within this tetraploid, and determine whether the A and B subgenomes are distinct. Third, we identified markers segregating in both mapping populations as anchors to align homologous regions of the diploid and tetraploid genomes, and then extended this to GBS markers on the domestic A. sativa linkage map. This provides a comparative map across three ploidy levels each containing a copy of the A genome.
Methods
Crosses
We used the A. strigosa C.I. 3815 × A. wiestii C.I. 1994 F6:8 (henceforth SW) recombinant inbred line (RIL) population28. Seeds from 96 of the RILs, along with the parents, were obtained from Iowa State University and used for this study.
We also used an existing A. barbata mapping population39, derived from a cross between Californian ecotypes associated with moist and dry habitats41. Of the initial 196 RILs, 180 F7 lines were available for GBS. In addition, we genotyped several accessions of the mesic (moist), and xeric (dry) ecotypes, with one accession of each ecotype subjected to a ‘deep read’, so as to capture as many segregating alleles as possible and correctly assign polarity in the cross.
Genotyping by sequencing
Fresh tissue was grown from seed in the greenhouse (A. barbata) or growth cabinets (SW). Leaves were dried on silica, and DNA was extracted using QIAGEN DNEasy Mini kits. All RILs of both mapping populations were genotyped using a double restriction digest (PstI and MspI) GBS protocol42. Barcoded samples were pooled into 96-plex libraries and subjected to 100 bp single read runs (one per population) on a HiSeq2500 sequencing system. In addition, the A. barbata mapping population was also subjected to GBS by a single-digest protocol43 using PstI alone.
GBS analysis was performed using the Haplotag pipeline44 which is specifically designed to address problems of paralogous loci in polyploid species. Haplotag treats tag-level haplotypes, which may differ at multiple SNPs, as discrete alleles, ignoring the potential for very rare recombinations within their 64-base length. It first clusters similar tags based on nearest-neighbor joining of all tags that differ by three or fewer SNPs. It then screens all possible subsets of tags within each cluster to identify putative loci, subject to a set of population-based filtering criteria, then partitions tag-level haplotypes into the best available model of mutually-exclusive loci. We set the filtering criteria such that the maximum heterozygosity was limited to 15%, and that no more than 2 alleles could segregate at a locus within the same RIL population. However, since segregation distortion has been documented in the A. barbata map39, we filtered leniently for allele frequency departures from the expected 1:1 ratio.
Map Construction and error checking
We constructed framework maps from the most reliable subset of the loci identified by Haplotag. We included only those loci that were scored in >90% of the RILs, with <5% heterozygosity, and with no more than three base pairs difference between alleles. In A. barbata, loci were included in map construction only if they showed fixed differences between the parental accessions with no missing parental data. In the SW population, marker phase was sometimes ambiguous due to missing parental genotypes and/or parental heterogeneity. Thus, an in-house script was used to phase the markers based on their patterns of recombination in this population. Loci that were identified by Haplotag, but which did not meet the above criteria, were set aside as ‘secondary loci’, and placed on the map after framework construction.
We used MSTMap45 to infer the map, using the Kosambi distance function and estimating the recombination distance from a simple count of the number of crossovers. We discarded any linkage group shorter than 15 cM or with fewer than four markers. The probability threshold (e) for joining markers into a linkage group was decreased progressively in successive runs until the correct number of linkage groups was obtained (Supplementary Methods). Mis-identified genotypes can inflate map distances by appearing as double recombinants. We therefore screened the linkage map for such errors by removing any apparent double crossover involving fewer than three markers and less than 2 cM. Such genotype calls were converted to missing data (since they were likely in error) and the map was re-estimated (Supplementary Methods).
Presence-absence variants
To increase the number of mapped loci which might permit us to align the genomes, we screened the data for presence-absence variants (PAV’s). These came from two sources: (a) after loci had been identified by Haplotag, any tags that were ‘left over’ in the tag clusters were treated as potential PAVs (Supplementary Methods), and (b) the large number of ‘singleton’ tags which showed no sequence similarity to any other tags in the data set. PAVs from either source can increase the number of comparisons between mapping populations, but only the former could be used to identify paralogs/homeologs within genomes.
We filtered these tags stringently to identify high-quality PAVs. First, only those which showed fixed differences between the parents were retained. PAV’s needed to be present at >10 read depth in one of the parental accessions (and in the case of A. barbata, to be present in at least 3/5 accessions of one parent, and absent from all accessions of the other parent). Assuming a high degree of homozygosity in the inbred lines, Mendelian PAVs should be present in 50% of the lines. We therefore eliminated any tag present in fewer than 40% or more than 60% of the RILs, although this prevented identifying any PAVs in regions of segregation distortion.
We placed secondary loci and PAVs on the map at the point of lowest recombination fraction (Supplementary Methods). Because less stringently filtered loci and PAVs are likely to contain more error than the stringently filtered loci used to construct the map, we did not integrate these loci into the map, but any that showed <10% recombination to the closest marker were assigned to that position. These approximate map positions are probably accurate enough to inform our inferences of synteny within and across the linkage maps, while the additional markers increase our power to detect patterns. RFLP markers29 and AFLP markers39 were also positioned in this way, such that the new maps could be aligned with the original maps.
Homologs, paralogs and homeologs
To identify homologous tags, we used BLAST46 to find the closest sequence match of each tag between the set of mapped tags in A. barbata and SW. We analyzed homology at the tag (rather than locus) level, since most biallelic loci matched at only one allele, and because PAVs were included (e.g., similar PAVs segregating in both maps, or a PAV in one map matching an allele at a biallelic locus in the other). We extended this comparison to the GBS map of A. sativa20,47. Each tag was included in only that pair with the closest match, and we excluded tag pairs with more than three base pair mismatches (since this was the threshold for clustering tags in the Haplotag pipeline).
Paralogs and homeologs within A. barbata were identified in the Haplotag pipeline by cases where more than one segregating locus occurs within a cluster of tags (Supplementary Methods). If more than two segregating loci occurred in a tag cluster, two of those loci were chosen at random. These loci were then mapped independently of each other. Where multiple pairs of such loci showed synteny between separate linkage groups, we inferred these regions of the genome to be broadly homeologous.
Finally, we conducted a BLAST46 search of all tag sequences against the full genome sequences of barley, Brachypodium and rice, to determine whether syntenic chromosome regions could be identified. Matches were restricted to e < 10−15, and any tag with more than ten matches was eliminated.
Results
Map construction
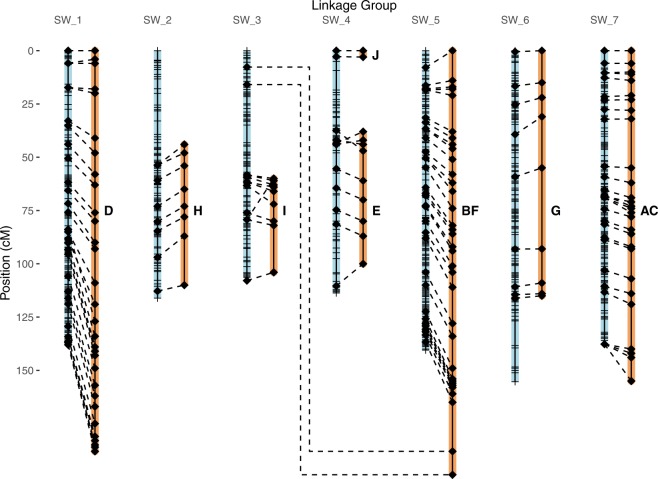
We identified 6651 segregating loci in the SW mapping population, of which 3170 passed stringent filtering for use in map construction (Supplementary Table S1). MSTMap returned seven linkage groups at e = 10−10, corresponding to the expected seven chromosomes. Initial map length was estimated at 1850 cM, but following removal of 2285 putative errors (0.75% error rate), this was reduced to 905 cM (Fig. 1). The corrected markers clustered into 754 unique bins with an average inter-marker distance of 1.2 cM, a maximum gap of 11.5 cM, and only 15 gaps of >5 cM. All but five of the 204 RFLP markers from the original map29 were placed on the new GBS map, and these placements aligned with the original linkage groups (Fig. 1).
Figure 1.
GBS linkage map of diploid A. strigosa (C.I. 3815) x A. wiestii (C.I. 1994) RIL population (blue, left), aligned to the RFLP map (orange, right) of Portyanko et al.29 from the same population. Linkage distances shown here were calculated after removal of likely GBS genotyping errors. Diamonds represent positions of common RFLP markers, while hatches indicated bins of other GBS markers.
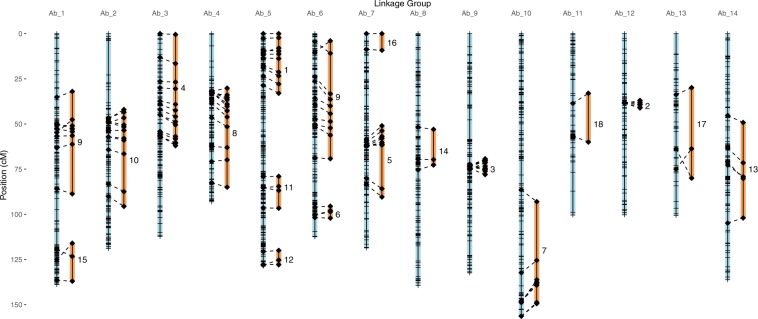
In A. barbata, the Haplotag pipeline identified 9148 segregating loci from the double-digest GBS library, of which 4015 passed stringent filtering for use in map construction. MSTMap returned 14 linkage groups for A. barbata at a threshold of e = 10−12. The initial map length of roughly 2500 cM was reduced to 1688 cM after the removal of 12,789 genotyping errors (error rate 1.8%). The 4015 markers clustered into 1529 bins, with an average inter-marker distance of 1.1 cM, but with somewhat more gaps than the SW map above. There is a gap of 20 cM on SW_2, and some 49 gaps of >5 cM (Fig. 2). All of the 129 AFLP markers of Gardner and Latta39 could be placed on the newer GBS map, allowing alignment of that partial map with the present map (Fig. 2).
Figure 2.
GBS linkage map (blue, left) of tetraploid A. barbata mapping population aligned to the AFLP map (orange, right) of Gardner and Latta39. Note that several linkage groups of the fragmentary AFLP map align to the same GBS chromosome. This map is derived from the double restriction digest (PstI-MspI) GbS library, with linkage distances calculated after removal of likely GBS genotyping errors. Diamonds represent position of AFLP markers, while hatches indicated bins of other GBS markers.
The single restriction digest GBS library obtained fewer markers - only 3134 segregating loci, of which 1730 passed stringent filtering. These markers had greater read depth (median 29/locus as opposed to 11/locus in the two-enzyme library). However, there was very little overlap in the loci revealed by the two library preps – only 369 biallelic marker loci were identified in both libraries (Supplementary Table S2). Stringently filtered loci from the single restriction digest library gave an independently constructed A. barbata linkage map, which also contained 14 linkage groups and which closely aligned with the double digest GBS map (Supplementary Fig. S1). Since the two independently derived maps are thus concordant, markers from the single-enzyme library were placed on the double enzyme map for all further comparisons
We also identified 8700 segregating PAVs in SW, and over 30,000 segregating PAVs in A. barbata. The vast majority of the secondary (less stringently filtered) biallelic loci and most of the PAVs could be placed on the maps at <10% recombination with the closest markers (Supplementary Methods). Compared with biallelic loci, however, the PAVs showed greater recombination with the mapped markers, consistent with the greater error in determining PAV genotypes. Nevertheless, with all loci included, more than 40,000 markers were mapped in A. barbata, and nearly 15,000 in SW.
Homologs, paralogs and homeologs
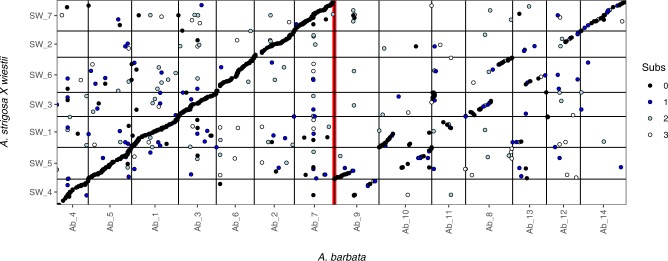
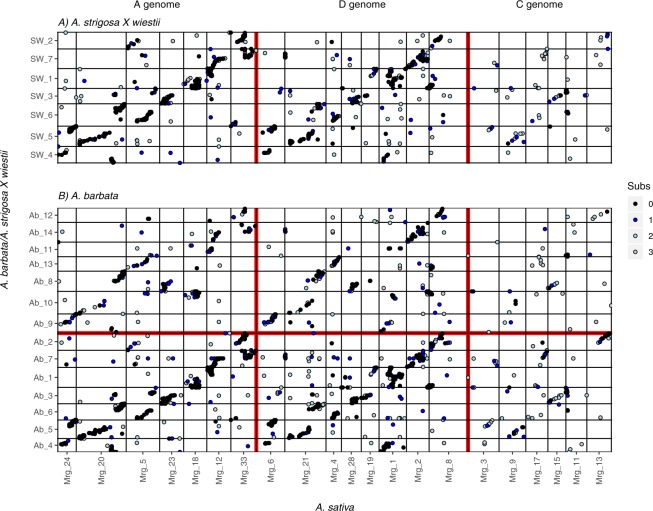
We identified a total of 3732 homologous tag pairs from 3100 loci across the SW and A. barbata mapping populations (Table 1). These homologous markers mapped widely across all seven of the SW linkage groups, but in A. barbata, most of these homologs mapped to only seven of the linkage groups (Fig. 3). Moreover, these homologies were syntenic across linkage groups, indicating a direct 1:1 correspondence between the seven chromosomes of the A-genome diploids, and half of the 14 chromosomes in the AB tetraploid A. barbata. We have therefore designated these seven linkage groups as the A-genome of A. barbata, and assigned them corresponding numbers, 1 through 7. (Fig. 3.) The remaining seven linkage groups (Ab_8-14) in A. barbata thus presumably represent the B genome. These share many fewer homologous tags, giving a lower Dice similarity, with the SW linkage groups, and a larger fraction of those that are shared have base pair mismatches (Table 1). However, those that are shared are consistent with the pattern of homeology within the A. barbata genome (below).
Table 1.
Number of putatively homologous tag pairs between subgenomes within linkage maps of diploid, tetraploid and hexaploid oats.
| A. strigosa x A. wiestii (2n = 2 × = 14) | A. barbata (2n = 4 × = 28) | Totalsa | ||
|---|---|---|---|---|
| A genome | A genome | B genome | ||
| A. strigosa × A. wiestii | ||||
| A genome | NA (NA) | 2420/815 (0.1114) | 155/342 (0.0251) | 22018 |
| A. sativa (2n = 6× = 42) | ||||
| A genome | 417/337 (0.0396) | 619/494 (0.0427) | 79/173 (0.0149) | 16042 |
| D genome | 188/270 (0.0215) | 222/432 (0.0231) | 90/197 (0.0150) | 20572 |
| C genome | 10/75 (0.0042) | 22/117 (0.0051) | 9/45 (0.0030) | 18680 |
| Totalsa | 22018 | 36075 | 17742 | |
The number of putative homologies is indicated separately for exact sequence matches, and for pairs showing 1–3 bp mismatches. Dice similarities, which account for the different number of tags in each genome are given in brackets.
aTotal number of markers from each subgenome. Thus, for example, out of a total of 22018 mapped tags in the A. strigosa x A. wiestii mapping population, and 36075 tags mapped to the A genome of A. barbata, 2420 identical tags and 815 pairs showing 1–3 base mismatches were observed. This gives a total of 3235 putatively homologous tags for a Dice similarity of 0.1114.
Figure 3.
Dot-plot showing alignment of linkage groups between diploid A. strigosa (C.I. 3815) × A. wiestii (C.I. 1994) RIL population and tetraploid A. barbata linkage maps. Dots are coloured to indicate the number of base substitutions (black = perfect match; blue, grey, and white represent 1, 2, or 3 mismatches, respectively) between matching Tag sequences. Linkage groups have been re-ordered to emphasize the pattern.
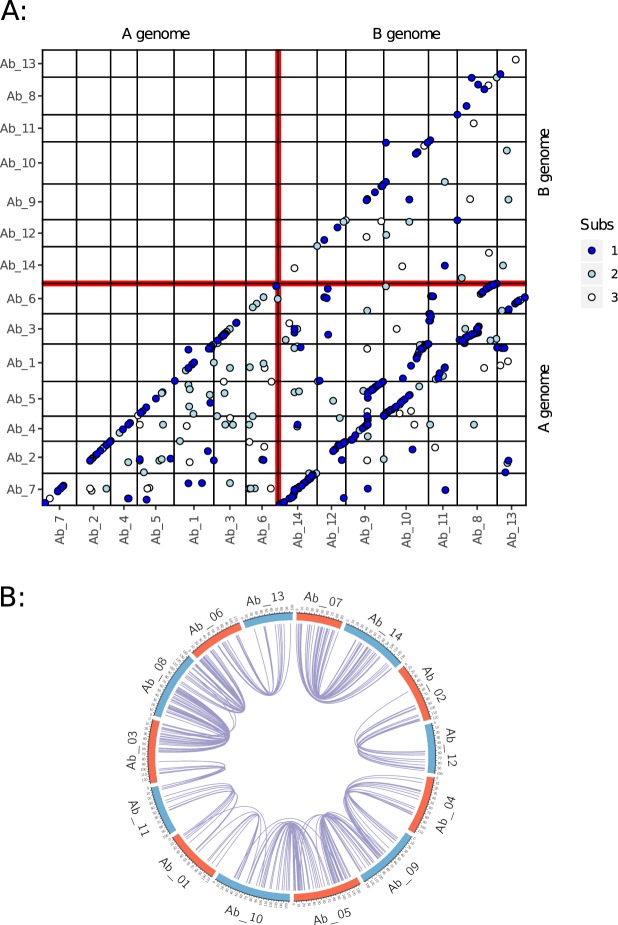
Within the A. barbata genome, there are 644 multi-locus tag clusters (in which two or more separate segregating loci or PAVs can be identified) which represent putative paralogs or homeologs. A randomization test (Supplementary Table S3) indicated that these pairs are not randomly distributed across the genome. First, locus pairs that map to the same linkage group are significantly over-represented. These typically map very close to each other (frequently in the same map bin, and rarely more than 2 cM apart - Fig. 4a) and are likely paralogs that resulted from small tandem repeats. More interestingly, several pairings of one linkage group from the A genome (as identified above), and one from the B-genome, have more paralogs than expected by chance alone. In these linkage groups, the paralogs show syntenic arrangements on the two linkage groups indicating large regions of homeology between the A and B genomes (Fig. 4a). In one instance, Ab_7 and Ab_14 show syntenic arrangement along the length of both linkage groups. However, most B genome chromosomes show substantial evidence of translocation when compared to A genome chromosomes. For example, Ab_4 is syntenic with one arm of Ab_9, the other arm of which is syntenic with Ab_5. This pattern continues through most of the A. barbata linkage groups (Fig. 4b) with alternating homeologies between the A and B genomes: 4-9-5-10-1-etc. The weak homology between the B-genome of A. barbata and the diploid A genome (Fig. 3) is largely consistent with this pattern of homeology within A. barbata. For example, Ab_14 shows fewer homologies to SW_7 than does its A genome homeolog Ab_7 (and fewer of these are identical sequence matches – Table 1), but both of these homeologous linkage groups are syntenic with the same linkage group in SW.
Figure 4.
Paralogs and homeologs within the A. barbata genome. (A) Dot-plot of all paralogous loci detected with GBS. Linkage groups have been re-ordered to emphasize the pattern, and colour represents the number of base pair substitutions between the tag sequences of paralogous loci (blue, grey, and white represent 1, 2, or 3 mismatches, respectively). (B) Circos58 plot highlighting the pattern among linkage groups having more paralogous markers than expected by chance. A and B genome assignments from Fig. 3 are coloured red and blue respectively. Paralogous loci are joined by blue curved lines.
Comparison to A. sativa
Given the strong synteny between SW and the A genome of A. barbata, it is not surprising that they both showed similar patterns of homology with A. sativa (Fig. 5). The largest number of homologies occurs between the A genomes of SW/barbata and the A genome of A. sativa, with less similarity to D and least to the C-genome (Table 1). A similar pattern occurs between the B genome and the subgenomes of A. sativa, but with less homology than for the SW/barbata A genome.
Figure 5.
Dot-plots showing alignment of the consensus linkage map from A. sativa20,47 with the diploid A. strigosa × A. wiesteii map and with the tetraploid A. barbata map. Linkage groups have been re-ordered to emphasize the pattern, and colour represents the number of base pair substitutions between the tag sequences of homologous loci (black = perfect match; blue, grey, and white represent 1, 2, or 3 mismatches, respectively). Genome assignments of linkage groups in A. sativa following Yan et al.21 are shown across the top, with linkage group assignments at the bottom. Groups Mrg18 and Mrg19 are shown as A and D genomes, respectively, although they contain large translocations from the hexaploid C genome21.
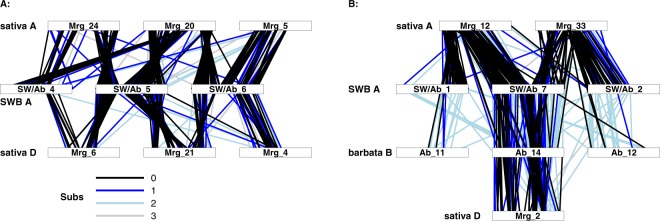
However, only a few linkage groups in A. sativa show synteny along their entire length with SW/barbata A-genome linkage groups (Fig. 5). For example, Mrg2 shows synteny to SW_7 and Ab_7, but most of the hexaploid linkage groups show apparent translocations, some of which are remarkably consistent between the A and D genomes of A. sativa. For example, alternate ends of linkage groups 4, 5 and 6 in SW/barbata appear to be rearranged as parts of Mrg24, 20 and 5 in the A genome of A. sativa, and as parts of Mrg 6, 21, and 4 in the D genome of A. sativa (Fig. 6A). Notably, these same pairs of hexaploid groups (Mrg24/6, 20/21, and 5/4) were identified as A/D genome homeologs by Chaffin et al.20. Overall, the D and A genomes of A. sativa show a parallel set of rearrangements compared to the SW/barbata genomes (Fig. 5).
Figure 6.
Example alignments of linkage groups from A. strigosa, A. wiestii and A. barbata against the A and D genomes of A. sativa. (A) alignment of LGs 4, 5, and 6 from the A. sativa × A. wiestei and A. barbata maps (labelled “SWB”) against the corresponding A and D genome LGs of A. sativa. Note that Mrg24 and Mrg6 are homeologs20 as are Mrg20 and Mrg21 as well as Mrg4 and Mrg5. (B) alignment of LG7 (A genome of SW and A. barbata), LG 14 (B genome of A. barbata), and Mrg2 (D genome of A. sativa) with Mrg12 and 33 both of the A genome of A. sativa. Linkage groups have been standardized to unit length, and line colour indicates the number of base pair differences between homologous tag sequences.
Similarly, the B genome of A. barbata shows homology to the A. sativa genome that parallels that of the SW/barbata A genome (Fig. 5). For example, A. barbata Ab_14 shows the same homology with A. sativa Mrg2 (D genome) as does its A genome homeolog, SW_7/Ab_7 (Fig. 6B). This synteny appears to have been rearranged in the A genome of A. sativa between Mrg12 and Mrg33 (Fig. 6B).
Only a faint signal of homology to the C genome is observed (Fig. 5), in that few pairs of linkage groups show more homologous tags than expected by chance (for example, SW_3/Ab_3 with A. sativa Mrg15). We note that as the number of putative homologs declines, indicating more dissimilar genomes, the quality of the sequence match declines in parallel, as indicated by a greater proportion of those homologs containing base pair mismatches (Table 1).
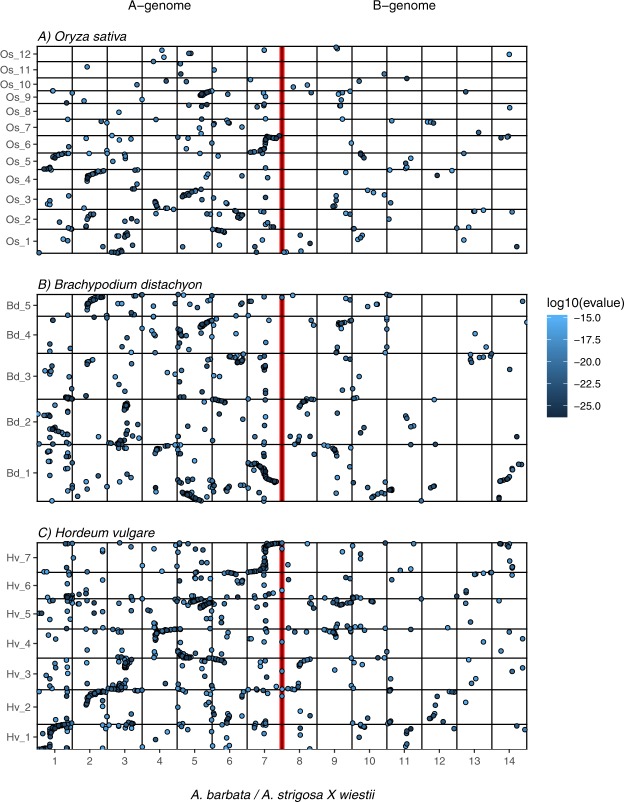
There is substantial synteny between barley chromosomes and several of the A genome linkage groups (Fig. 7). Although this has clearly been interrupted in some chromosomes, we have numbered the A genome linkage groups in SW and A. barbata to reflect this similarity with barley. Fewer areas of synteny are seen with the B-genome of A. barbata, in part because far fewer markers were identified in the B-genome. Some regions of synteny are seen for Brachypodium and rice, but since these are more distantly related to Avena than is barley, fewer regions of synteny are evident.
Figure 7.
Dot-plot comparison of the Avena barbata and A. strigosa × A. wiestii (SW) linkage maps with the rice (top), Brachypodium (middle) and barley (bottom) genomes. LG’s 1–7 include markers from both A. barbata and SW with blast hits in the target genomes. X axis in recombination units (cM) with linkage groups standardized to unit length. Y axis in base pairs. Blast hits were filtered to max e-value of 10−15, and tags matching more than 10 locations in the target genomes were removed.
Discussion
Our GBS data have permitted a considerable improvement in the maps of diploid and tetraploid Avena species. The greatly-increased density of these maps, together with the high numbers of secondary GBS loci and PAVs, will provide future opportunities for comparative mapping, and will allow integration of these maps with future genome sequencing efforts. Our GBS map (905 cM) allows some slight extension of SW_2, 3, 4 and 6, compared to the original RFLP map of A. strigosa X wiestii29 which covered some 862 cM. However, the improvement comes primarily in the much higher density of markers on the GBS map, since the RFLP map had numerous gaps of up to 40 cM.. A much greater improvement is seen in extending the tetraploid A. barbata map by some 2.5 fold compared to an earlier AFLP map, which Gardner and Latta39 acknowledged was incomplete.
We would expect that the tetraploid genome would be roughly twice the size of the diploid, and the A. barbata map does span slightly less than twice the length of the SW map. Polyploid Avena species have slightly less DNA than predicted from the composite diploid genomes48, an observation consistent with ‘genome downsizing’ in polyploids3,49. The relative sizes of the linkage maps (905 vs. 1688 cM) present a strkingly close match to the relative DNA contents of A. strigosa and A. wiestii (9.07 pg/2 C) versus A. barbata (16.45 pg/2 C). This would suggest that any genomic changes since polyploidization have not altered the overall frequency of crossovers per megabase of DNA.
The striking homology and synteny between the SW linkage map and Ab_1–7 of A. barbata strongly suggests that the A genome of A. barbata is closely related to the A genome diploids A. strigosa and A. wiestii. Seven Avena species share the As designation, and chloroplast phylogenies23,24,26 place A. barbata within this clade, indicating that an As genome diploid was the maternal ancestor of A. barbata. Multi-dimensional scaling of GBS markers21 also suggested a closer similarity of A. barbata to A. hirtula and A. atlantica than to A. strigosa or A. wiestii, and A. hirtula has been suggested as the A genome ancestor of A. barbata50. The close similarity between Ab_1–7 and SW_1–7 imply a recent divergence between A. barbata’s A genome and its diploid relatives (Fig. 8).
Figure 8.
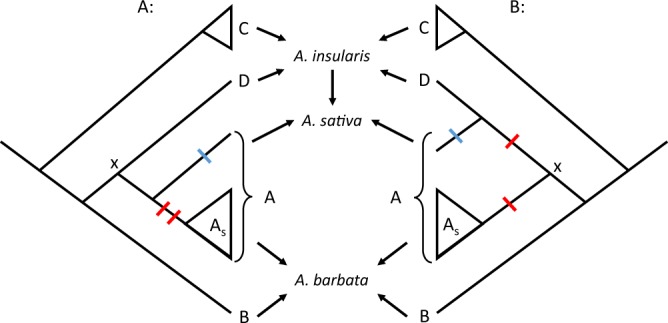
Two hypothetical relationships among the diploid ancestors of polyploid oats prior to the hybridization and polyploidization events (arrows) creating A. barbata, A. insularis and A. sativa. Left: The A genome diploid ancestor of A. sativa is more closely related to As diploids including A. strigosa and A. wiestii. Under this view, the ancestor at the node labelled “x” likely exhibited the genome arrangement seen in Mrg4/5, Mrg20/21, Mrg6/24, and a series of translocations (red hatches) in the common ancestor of As diploids led to the arrangement in SW_4–6 (Fig. 6a). Right: The A and D genome diploid ancestors of A. sativa are each other’s closest relatives. If this hypothesis is correct, then the contrasting arrangements in Mrg4/5, Mrg20/21, Mrg6/24 and SW_4–6 represent the accumulation of translocations (red hatches) as each lineage diverged from ancestor x. In either case, the unique arrangement of Mrg12 and Mrg33 in the A genome of A. sativa occurred (blue hatch) after that diploid ancestor diverged from the D genome ancestor.
The greatly reduced number of shared loci between the A. barbata B genome and the genome of SW (Fig. 3, Table 1) suggests a much more distant relationship. Moreover, extensive chromosomal rearrangements distinguish the A and B genomes of A. barbata, as evidenced by the fact that few pairs of linkage groups are syntenic along their entire length (Fig. 4). These findings imply a much earlier divergence of the B genome from the main A genome lineage (Fig. 8) Thus, our results make clear that A. barbata is an allopolyploid that arose from the hybridization of two species with distinct chromosome arrangements, rather than an autopolyploid, resulting from genome doubling within a single species.
Yet if the divergence between the A genome of A. barbata and its diploid ancestor was relatively recent, then the hybridization and polyploidy event that formed A. barbata must also be recent. While no B-genome diploid oat species has been identified, it must have been extant up until the time of this hybridization, though it may have been quite rare. We also note that the lower density of markers on the B genome chromosomes suggests the possibility of less polymorphism in loci on the B genome. This in turn suggests that the B genome diploid ancestor may have been a genetically homogeneous species that hybridized multiple times51 with a more common and genetically heterogeneous A genome species.
We find no evidence that exchanges between A (Ab_1–7) and B (Ab_8–14) genomes have occurred since the formation of polyploid A. barbata. Had they occurred, such exchanges would have created circumstances where different parts of a single linkage group in SW would show strongest homology to A vs. B genome linkage groups in A. barbata. No such case is seen in Fig. 3, although we recognize that small gene conversion events may fall below the resolution of our maps. By contrast, such inter-genomic exchanges are typical of hexaploid oat20,21 and other allopolyploids4,7,52. The lack of intergenomic translocations implies that the structural differences between the A and B genomes must have been present in their diploid ancestors prior to hybridization and polyploidization. Structural rearrangement of diploids has been documented in other grass species. Notably, rye (Secale cereale) exhibits several translocation events derived from the putative Triticeae ancestral karyotype14, and the diploid wheat relative Aegilops markgrafii exhibits rearrangements compared to the D genome of wheat15. Intriguingly, in both of these examples, phylogenetic analysis suggests homoploid hybridization in the ancestry of the rearranged diploids.
There is a striking pattern to the homeologies between the A and B genomes that might be called an ‘enchainment of translocations’ (Fig. 4b). This pattern has a close parallel in the A and C genomes of tetraploid Brassica napus12. In that system, the contrasting genome arrangements are present in the diploid ancestors, B. rapa and B. oleracea. Similarly, we contend that the different arrangements in the genomes of polyploid oats were present in the diploid ancestors, rather than being a product of polyploidy. Rearrangements in the genomes of Brassica diploids have been traced to an earlier round of polyploidization and re-diploidization some 5–20 mya12,53,54. However, there is currently no evidence suggesting paleopolyploidy in the lineage leading to modern oats (at least none more recent than the ancestral grass karyotype)55, though admittedly the genomic resources to detect such a paleopolyploidy event in Avena are not fully developed. Alternatively, we can speculate that a series of reciprocal translocations similar to the 4 AL-5AL-7BS cyclical translocation in wheat56 may have structured the A. barbata genome. In wheat an early 4 A/5A reciprocal translocation was followed by a subsequent 4A/7B exchange. These are non-homeologous exchanges, the first of which occurred in the diploid ancestor, Triticum urartu. The pattern in A. barbata appears to be longer, and not fully cyclical – a pattern which could arise if one of the translocations occurred between highly asymmetrical chromosomal blocks (sometimes termed “non-reciprocal”, though see Schubert and Lysak57). As in wheat, this pattern presumably formed from a succession of pairwise translocations, not all of which necessarily took place in the B genome. Rather, as the A and B genomes diverged from a common ancestral arrangement, translocations in both lineages likely led to the present pattern of homeology. Supplementary Fig. S2 presents one possible sequence for these translocations.
While the A genomes of SW and A. barbata are very similar, the A-genome of A. sativa is much less similar to either (Fig. 5, Table 1), consistent with suggestions that it is derived from a different sub-lineage within the A genome diploids, such as the Al genome of A. longiglumis21. Moreover, this A genome diploid ancestor of A. sativa likely diverged from the As genome diploids much earlier than the A genome diploid ancestor of A. barbata (See Fig. 8) Indeed, the D genome of A. sativa is only slightly less similar to SW and A. barbata than is the A. sativa A genome. The B-genome of A. barbata shows still fewer homologies to the A genome of SW (Table 1), suggesting that it is more distantly related (Fig. 8). It is plausible that the B and D genomes are derived from other sub-types of A-genome diploids. Of course, the C genome is least similar to any of the other genomes, which is consistent with phylogenetic analyses23,24,26 that consistently place the C-genome diploids as a separate clade (along with CC autotetraploid A. macrostachya).
We have inferred a recent hybrid origin of A. barbata because of the recent divergence between its A genome diploid ancestor and the other As genome diploids. The lack of intergenomic translocations in A. barbata (Fig. 3) is consistent with such a recent origin. By the same reasoning, an earlier divergence of A sativa’s A genome diploid ancestor implies that the hybridization event that formed A. sativa may be older than the one that formed A. barbata. An earlier formation of A. sativa than A. barbata may explain why intergenomic translocations are seen frequently in A. sativa20,21 but not A. barbata, simply because there has been less time for such exchanges to occur in A. barbata. In A. sativa, more translocations are seen between the C and D genomes than between either subgenome and A20,21. This is consistent with the fact that the CD tetraploid ancestor of A. sativa (likely A. insularis)21 must have evolved prior to the time A. sativa arose, giving the C and D genomes more time to undergo inter-genomic translocations.
Comparisons with A. sativa (Fig. 5) reveal similar patterns of enchained translocation to those seen in A. barbata, although these do not seem to extend across the A. sativa genome to the same extent as A. barbata. This may be in part because ongoing translocations between the homeologous genomes in the hexaploid20 have obscured the similarities of pattern within genomes. Remarkably, the A and D genomes of A. sativa often show the same rearrangements relative to the A genome of A. strigosa, A. wiestii and A. barbata (Figs 5, 6A). These appear to be parallel rearrangements within the A and D genomes, and thus are difficult to ascribe to intergenomic translocations following polyploidization. Instead, we find it more parsimonious to hypothesize that the distinct chromosome arrangement in the A and D genomes of A. sativa was present in the common ancestor of the diploid lineages that gave rise to those genomes (labelled “x” on Fig. 8). Since the A genome of A. sativa shows more homologies with the A genome of SW/barbata than does the D genome (Table 1), this suggests that the D genome diverged prior to the divergence of the A genome of A. sativa and SW/barbata (Fig. 8A). Under this view, the arrangement seen in the A genome of SW and A. barbata is derived from that of the A. sativa A genome ancestor, through successive translocations in SW_4,5,6. This hypothesis implies that a series of translocations took place along a relatively short branch of the phylogeny. Alternatively, if the common arrangement of the A and D genomes in A. sativa is more indicative of common ancestry than their Dice similarities to SW, then the diploid A and D genome ancestors of A. sativa may be more closely related to each other than either is to the SW/barbata A genome ancestor (Fig. 8B). In this case, the different arrangements in SW/barbata vs the A and D genomes of A. sativa would be the cumulative result of translocations along both of the lineages as they diverged from some unknown ancestral arrangement at node x (Fig. 8B).
It would of course, be valuable here to identify an ancestral genome arrangement in the common ancestor of the A, B and D genomes. This is beyond the scope of this study, but one suggestion is tempting. Linkage groups SW_7 and Ab_7 (A genome), Ab_14 (B genome) and Mrg2 (D genome) all show strong homology and are syntenic along their entire length. Indeed, we note that SW_7 and Ab_7 show strong collinearity with barley chromosome 7 (and extends even to Brachypodium Bd_1 and rice Os_6 – Fig. 7). This strongly suggests that this chromosome has an ancestral arrangement. By contrast, Mrg12 and Mrg33 from the A genome of A. sativa exhibit a unique rearrangement of this chromosome (Fig. 6B). Parsimony thus suggests that this rearrangement is autapomorphic to the A genome ancestor of A. sativa prior to hybridizing with A. insularis (Fig. 8). Testing this suggestion, and fully inferring the arrangement of the ancestral genome would likely require extensive sequence data across multiple ploidy levels.
What our maps do reveal is substantial karyotypic variation among oats. This diversity has frustrated efforts to ascribe a consistent chromosome nomenclature across the genus. We have been able to apply a consistent nomenclature to SW and half of the A. barbata linkage groups, but there is no 1:1 correspondence to the B genome, or to the subgenomes of A. sativa. Not only have we found evidence that the B genome is indeed a distinct entity with a distinct chromosome arrangement, we also find evidence that the A genome in A. sativa has a further different karyotype from the As diploid genome of SW and A. barbata. This diversity is most parsimoniously explained if we hypothesize that rearrangements took place within the diploid ancestors of oats prior to the formation of allopolyploids. In this view, these rearrangements do not result as a consequence of polyploidy. Since new sequencing technologies may soon permit routine de novo analysis of complete genomes, we suggest that the largely-unknown structure of most of the 30+ Avena species will provide a rich arena to test hypotheses regarding karyotype evolution across a polyploid series.
Supplementary information
Acknowledgements
We are grateful to Jesse Poland for carrying out the double digest GBS library preparation and to Andrew Sharpe for coordinating Illumina sequencing of the diploid oat samples. Single Enzyme libraries in A. barbata were prepared and sequenced at the Biotechnology Resource Center of the Institute of Biotechnology at Cornell University. Seeds of the A. strigosa X wiestii mapping population were provided by Dr. Michael Lee at Iowa State University, Ames, IA, USA. Funding for this work was provided in part by an NSERC Discovery Grant to RGL.
Author Contributions
R.G.L. collected the data and produced the linkage map of A. barbata, while W.A.B., C.P.W. and N.A.T. collected the data and produced the linkage map of A. strigosa X wiestii. R.G.L. and W.A.B. analyzed the data and aligned the maps. R.G.L. drafted the main manuscript, which was edited and reviewed by all authors.
Data Availability
GBS raw sequence read data are deposited at the NCBI short read archive under project numbers PRJNA517481 for the diploid the A. strigosa X wiestii mapping population, and PRJNA517323 for A. barbata. Genotype calls for each recombinant inbred line are included in Supplementary Data.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48639-7.
References
- 1.Wendel, J. F. Genome evolution in polyploids. Plant Mol. Biol.42, 225–249 (2000). 10.1023/A:1006392424384 [DOI] [PubMed] [Google Scholar]
- 2.Adams, K. & Wendel, J. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol.8, 135–141 (2005). 10.1016/j.pbi.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 3.Soltis, D. E., Visger, C. J., Marchant, D. B. & Soltis, P. S. Polyploidy: Pitfalls and paths to a paradigm. Am. J. Bot.103, 1146–1166 (2016). 10.3732/ajb.1500501 [DOI] [PubMed] [Google Scholar]
- 4.Gaeta, R. T. & Pires, J. C. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol.186, 18–28 (2010). 10.1111/j.1469-8137.2009.03089.x [DOI] [PubMed] [Google Scholar]
- 5.Wendel, J. F., Jackson, S. A., Meyers, B. C. & Wing, R. A. Evolution of plant genome architecture. Genome Biol.17, 37, 10.1186/s13059-016-0908-1 (2016). 10.1186/s13059-016-0908-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandakova, T. & Lysak, M. A. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant Biol.42, 55–65 (2018). 10.1016/j.pbi.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 7.Udall, J., Quijada, P. & Osborn, T. Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics169, 967–979 (2005). 10.1534/genetics.104.033209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicolas, S. D. et al. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics175, 487–503 (2007). 10.1534/genetics.106.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pontes, O. et al. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA101, 18240–18245 (2004). 10.1073/pnas.0407258102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chester, M. et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA109, 1176–1181 (2012). 10.1073/pnas.1112041109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmon, A., Flagel, L., Ying, B., Udall, J. A. & Wendel, J. F. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol.186, 123–134 (2010). 10.1111/j.1469-8137.2009.03093.x [DOI] [PubMed] [Google Scholar]
- 12.Cheng, F., Wu, J. & Wang, X. Genome triplication drove the diversification of Brassica plants. Horticulture Research1, 14024, 10.1038/hortres.2014.24 (2014). 10.1038/hortres.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pont, C. & Salse, J. Wheat paleohistory created asymmetrical genomic evolution. Curr. Opin. Plant Biol.36, 29–37 (2017). 10.1016/j.pbi.2017.01.001 [DOI] [PubMed] [Google Scholar]
- 14.Martis, M. M. et al. Reticulate evolution of the rye genome. Plant Cell25, 3685–3698 (2013). 10.1105/tpc.113.114553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danilova, T. V., Akhunova, A. R., Akhunov, E. D., Friebe, B. & Gill, B. S. Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant Journal92, 317–330 (2017). 10.1111/tpj.13657 [DOI] [PubMed] [Google Scholar]
- 16.Rajhathy, T. & Thomas, H. Cytogenetics of oats (Avena L.). (Genetic Society of Canada, Ottawa, 1974).
- 17.Baum, B. R. Oats: wild and cultivated. A monograph of the genus Avena L. (Poaceae). (Minister of Supply and Services, Ottawa, 1977).
- 18.Thomas, H. Cytogenetics of Avena. In: Oat science and technology. (eds Marshall, H. G. & Sorrells, M. E.) 473–507 (American Society of Agronomy, Crop Science Society of America, Madison, 1992).
- 19.Leggett, J. M. & Thomas, H. Oat evolution and cytogenetics. In: The Oat Crop. (ed. Welch, R. W.) 120–149 (Springer, 1995).
- 20.Chaffin, A. S. et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome9, 2, 10.3835/plantgenome2015.10.0102 (2016). 10.3835/plantgenome2015.10.0102 [DOI] [PubMed] [Google Scholar]
- 21.Yan, H. et al. High-density marker profiling confirms ancestral genomes of Avena species and identifies D-genome chromosomes of hexaploid oat. Theor. Appl. Genet.129, 2133–2149 (2016). 10.1007/s00122-016-2762-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanz, M. J. et al. A new chromosome nomenclature system for oat (Avena sativa L. and A. byzantina C. Koch) based on FISH analysis of monosomic lines. Theor. Appl. Genet.121, 1541–1552 (2010). 10.1007/s00122-010-1409-3 [DOI] [PubMed] [Google Scholar]
- 23.Fu, Y. Oat evolution revealed in the maternal lineages of 25 Avena species. Scientific Reports8, 4252, 10.1038/s41598-018-22478-4 (2018). 10.1038/s41598-018-22478-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, Y. et al. Phylogenetic investigation of Avena diploid species and the maternal genome donor of Avena polyploids. Taxon59, 1472–1482 (2010). 10.1002/tax.595012 [DOI] [Google Scholar]
- 25.Peng, Y. et al. Phylogenetic relationships in the genus Avena based on the nuclear Pgk1 gene. Plos One13, e0200047, 10.1371/journal.pone.0200047 (2018). 10.1371/journal.pone.0200047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, H. et al. Phylogenetic analysis of the genus Avena based on chloroplast intergenic spacer psbA-trnH and single-copy nuclear gene Acc1. Genome57, 267–277 (2014). 10.1139/gen-2014-0075 [DOI] [PubMed] [Google Scholar]
- 27.O’Donoughue, L. et al. An RFLP-based linkage map of oats based on a cross between two diploid taxa (Avena atlantica X A. hirtula). Genome35, 765–771 (1992). 10.1139/g92-117 [DOI] [Google Scholar]
- 28.Kremer, C. A., Lee, M. & Holland, J. B. A restriction fragment length polymorphism based linkage map of a diploid Avena recombinant inbred line population. Genome44, 192–204 (2001). 10.1139/g00-115 [DOI] [PubMed] [Google Scholar]
- 29.Portyanko, V., Hoffman, D., Lee, M. & Holland, J. A linkage map of hexaploid oat based on grass anchor DNA clones and its relationship to other oat maps. Genome44, 249–265 (2001). 10.1139/g01-003 [DOI] [PubMed] [Google Scholar]
- 30.Ladizinsky, G. & Zohary, D. Genetic relationships between diploids and tetraploids in series Eubarbatae of Avena. Canadian Journal of Genetics and Cytology10, 68–81 (1968). 10.1139/g68-010 [DOI] [Google Scholar]
- 31.Rajhathy, T. & Morrison, J. W. Chromosome morphology in the genus Avena. Canadian Jour Bot37, 331–337 (1959). 10.1139/b59-024 [DOI] [Google Scholar]
- 32.Holden, J. Species relationships in Avenae. Chromosoma20, 75–124 (1966). 10.1007/BF00331899 [DOI] [Google Scholar]
- 33.Sadasivaiah, R. S. & Rajhathy, T. Genome relationships in tetraploid Avena. Canadian Journal of Genetics and Cytology10, 655–669 (1968). 10.1139/g68-083 [DOI] [Google Scholar]
- 34.Katsiotis, A., Hagidimitriou, M. & HeslopHarrison, J. S. The close relationship between the A and B genomes in Avena L (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Annals of Botany79, 103–109 (1997). 10.1006/anbo.1996.0312 [DOI] [Google Scholar]
- 35.Irigoyen, M. et al. Discrimination of the closely related A and B genomes in AABB tetraploid species of Avena. Theor. Appl. Genet.103, 1160–1166 (2001). 10.1007/s001220100723 [DOI] [Google Scholar]
- 36.Badaeva, E. D., Shelukhina, O. Y., Goryunova, S. V., Loskutov, I. G. & Pukhalskiy, V. A. Phylogenetic relationships of tetraploid AB-genome Avena species evaluated by means of cytogenetic (C-Banding and FISH) and RAPD analyses. Journal of Botany2010, 742307, 10.1155/2010/742307 (2010). 10.1155/2010/742307 [DOI] [Google Scholar]
- 37.Chew, P. et al. A study on the genetic relationships of Avena taxa and the origins of hexaploid oat. Theor. Appl. Genet.129, 1405–1415 (2016). 10.1007/s00122-016-2712-4 [DOI] [PubMed] [Google Scholar]
- 38.Hutchinson, E. S., Hakim-Elahi, A., Miller, R. D. & Allard, R. W. The genetics of the diploidized tetraploid Avena barbata - acid-phosphatase, esterase, leucine aminopeptidase, peroxidase, and 6-phosphogluconate dehydrogenase loci. J. Hered.74, 325–330 (1983). 10.1093/oxfordjournals.jhered.a109803 [DOI] [Google Scholar]
- 39.Gardner, K. & Latta, R. Identifying loci under selection across contrasting environments in Avena barbata using quantitative trait locus mapping. Mol. Ecol.15, 1321–1333 (2006). 10.1111/j.1365-294X.2005.02835.x [DOI] [PubMed] [Google Scholar]
- 40.Glover, N. M., Redestig, H. & Dessimoz, C. Homoeologs: what are they and how do we infer them? Trends Plant Sci.21, 609–621 (2016). 10.1016/j.tplants.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clegg, M. T. & Allard, R. W. Patterns of genetic differentiation in slender wild oat species Avena barbata. Proc. Natl. Acad. Sci. USA69, 1820–1824 (1972). 10.1073/pnas.69.7.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poland, J. A., Brown, P. J., Sorrells, M. E. & Jannink, J. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. Plos One7, e32253, 10.1371/journal.pone.0032253 (2012). 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elshire, R. J. et al. A Robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. Plos One6, e19379, 10.1371/journal.pone.0019379 (2011). 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tinker, N. A., Bekele, W. A. & Hattori, J. Haplotag: Software for haplotype-based genotyping-by-sequencing analysis. G3-Genes Genomes Genetics6, 857–863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, Y., Bhat, P. R., Close, T. J. & Lonardi, S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet4, e1000212, 10.1371/journal.pgen.1000212 (2008). 10.1371/journal.pgen.1000212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul, S., Gish, W., Miller, W., Myers, E. & Lipman, D. Basic Local Alignment Search Tool. J. Mol. Biol.215, 403–410 (1990). 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 47.Bekele, W. A., Wight, C. P., Chao, S., Howarth, C. J. & Tinker, N. A. Haplotype-based genotyping-by-sequencing in oat genome research. Plant Biotechnology Journal16, 1452–1463 (2018). 10.1111/pbi.12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan, H. et al. Genome size variation in the genus Avena. Genome59, 209–220 (2016). 10.1139/gen-2015-0132 [DOI] [PubMed] [Google Scholar]
- 49.Leitch, I. & Bennett, M. Genome downsizing in polyploid plants. Biol. J. Linn. Soc.82, 651–663 (2004). 10.1111/j.1095-8312.2004.00349.x [DOI] [Google Scholar]
- 50.Allard, R. W., Garcia, P., Saenzdemiera, L. E. & Delavega, M. P. Evolution of multilocus genetic structure in Avena hirtula and Avena barbata. Genetics135, 1125–1139 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soltis, D. E. et al. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc.82, 485–501 (2004). 10.1111/j.1095-8312.2004.00335.x [DOI] [Google Scholar]
- 52.Szadkowski, E. et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol.186, 102–112 (2010). 10.1111/j.1469-8137.2010.03182.x [DOI] [PubMed] [Google Scholar]
- 53.Lysak, M., Koch, M., Pecinka, A. & Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res.15, 516–525 (2005). 10.1101/gr.3531105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet.43, 1035–1039 (2011). 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- 55.Murat, F. et al. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res.20, 1545–1557 (2010). 10.1101/gr.109744.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devos, K., Dubcovsky, J., Dvorak, J., Chinoy, C. & Gale, M. Structural evolution of wheat chromosomes 4a, 5a, and 7b and its impact on recombination. Theor. Appl. Genet.91, 282–288 (1995). 10.1007/BF00220890 [DOI] [PubMed] [Google Scholar]
- 57.Schubert, I. & Lysak, M. A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends in Genetics27, 207–216 (2011). 10.1016/j.tig.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 58.Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res19, 1639–1645 (2009). 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GBS raw sequence read data are deposited at the NCBI short read archive under project numbers PRJNA517481 for the diploid the A. strigosa X wiestii mapping population, and PRJNA517323 for A. barbata. Genotype calls for each recombinant inbred line are included in Supplementary Data.