Abstract
The retina is the light sensing tissue of the eye which contains multiple layers of cells required for the detection and transmission of a visual signal. Loss of the light-sensing photoreceptors leads to defects in visual function and blindness. Previously, we found that mosaic deletion of Kcnj13, and subsequent loss of the potassium channel Kir7.1, in mice leads to photoreceptor degeneration and recapitulates the human retinal disease phenotype (Zhong et al., 2015).
Kcnj13 expression in the retinal pigment epithelium (RPE) is essential for normal retinal electrophysiology, function, and survival. Mice with homozygous loss of Kcnj13 die at postnatal day 1 (P1), requiring a tissue-specific approach to study retinal degeneration phenotypes in adult mice. We used the CRISPR-Cas9 system to generate a floxed, conditional loss-of-function (cKO) Kcnj13flox allele to study the pathogenesis of Kcnj13 deficiency in the retina. To investigate if the Kcnj13 is required in the RPE for photoreceptor function and survival, we used Best1-cre, which is specifically expressed in the RPE. We observe complete loss of Kcnj13 expression in Cre-positive RPE cells. Furthermore, our findings show that widespread loss of Kcnj13 in the RPE leads to severe and progressive thinning of the outer nuclear layer and a reduced response to light. Finally, to detect Best1-cre expression in the RPE of live animals without sacrificing the animal for histology, we generated a Cre-reporter-containing Kcnj13 cKO mouse line (cKOR: Kcnj13flox/flox; Best1-cre; Ai9) which can be rapidly screened using retinal fluorescence microscopy. These findings provide new tools for studying the roles of Kcnj13 in retinal homeostasis.
Keywords: Leber Congenital Amaurosis, KCNJ13, Kir7.1, Photoreceptors, Retinal Degeneration, Conditional Knockout, RPE, Retinal Function
1. Introduction
Leber Congenital Amaurosis (LCA) is the most severe inherited retinal dystrophy, affecting 2–3 in 100,000 births worldwide (Shah, 2016). Patients with LCA typically exhibit various visual abnormalities including: severe vision impairment or blindness within the first year of life, diminished response to light, nystagmus, and a habitual oculo-digital reflex (Shah, 2016; Weleber et al., 2013). LCA is a genetically heterogeneous autosomal recessive disease and is associated with mutations in 22 genes, to date. Mutations affect genes involved in numerous cellular pathways essential for visual function, including phototransduction, protein trafficking, the visual cycle, photoreceptor outer segment phagocytosis, and photoreceptor development.
Mutations in Kcnj13 were identified and associated with the human retinal dystrophies LCA and Snowflake Vitreoretinal Degeneration (Hejtmancik et al., 2008; Kumar and Pattnaik, 2014; Pattnaik et al., 2015; Sergouniotis et al., 2011; Zhang et al., 2013). Kcnj13 encodes an inwardly rectifying potassium (Kir) channel subunit, KCNJ13 or Kir7.1, which is abundantly expressed in the apical membrane of the retinal pigment epithelium (RPE) (Krapivinsky et al., 1998; Kumar and Pattnaik, 2014; Kusaka et al., 2001; Shimura et al., 2001; Yang et al., 2003). The RPE is a pigmented epithelial cell monolayer in the eye which is essential for nourishing and maintaining the neural retina (Strauss, 2005). The RPE is involved in numerous retinal functions, including: 1) the phagocytosis and clearance of photoreceptor outer segments; 2) the recycling of all-trans-retinol into cis-11-retinal (the visual cycle); 3) formation of the blood-retinal barrier; 4) absorption of scattered light; and 5) the transport of nutrients and ions between the blood supply and the retina. Kir7.1 regulates potassium (K+) transport between the choriocapillaris/RPE and the subretinal space by facilitating K+ efflux into the extracellular microenvironment surrounding photoreceptors (Kumar and Pattnaik, 2014; Shahi et al., 2017). However, the molecular mechanisms underlying Kcnj13-associated retinal functions and how KCNJ13 dysfunction and/or loss promotes retinal degeneration are unknown.
Previously, we showed that somatic, mosaic loss of Kcnj13 in the mouse retina leads to photoreceptor degeneration phenotypes, including thinning of the outer nuclear layer (ONL), rhodopsin mislocalization, and reduced visual function (Zhong et al., 2015). We found that RPE cells without KCNJ13 protein survive and wild-type RPE cells can rescue nearby photoreceptors cells underneath mutant RPE cells, suggesting a non-cell-autonomous and indirect role in maintaining photoreceptor function and survival. We also observed that homozygous Kcnj13 null mice die postnatally, thus requiring a conditional approach to study Kcnj13 functions in the retina of adult mice.
Here we report the generation of an RPE-specific Kcnj13 loss-of-function (Kcnj13 cKO) mouse model which mimics LCA pathology. The Kcnj13flox allele was generated using the CRISPR/Cas9 system to insert loxP sites flanking exons 2 and 3 of the Kcnj13 gene, which include the entire coding region of the gene. We achieved RPE-specific deletion of Kcnj13 using the Cre recombinase mouse line Best1-cre (Iacovelli et al., 2011). Best1-cre drives Cre expression using the Bestrophin-1 promoter, which is variably expressed in 40–70% of RPE cells (Iacovelli et al., 2011). This approach enables mice to survive beyond the P1 lethal phase observed with null homozygotes, permitting the study of Kcnj13 in a tissue-specific manner. In this study, we used Kcnj13 cKO mutant mice (Kcnj13flox/flox; Best1-cre) to investigate retinal phenotypes associated with loss of Kcnj13 in the RPE. We observe thinning of the outer nuclear layer by 15 days after birth (P15) in Kcnj13 cKO mutant mice with broad Best1-cre expression, with severe thinning by 3 months of age. Photoreceptor degeneration observed in Kcnj13 cKO mutant mice coincides with strong defects in visual function, consistent with previously reported findings (Zhong et al., 2015). Our results demonstrate the utility of new genetic tools for studying Kcnj13 gene function in a tissue-specific manner.
2. Materials and methods
2.1. Generation of a Kcnj13 conditional allele using the CRISPR-Cas9 system
To achieve Cre-mediated deletion of Kcnj13, we generated a floxed Kcnj13 allele (Kcnj13flox) by introducing loxP inserts flanking exons 2 and 3 using the CRISPR-Cas9 system. As shown in Fig.1A and Suppl.Fig.1A, we selected the two gRNA sites (target sites #1 and #2) in the mouse Kcnj13 locus using an online CRISPR design tool (http://crispr.mit.edu/). We synthesized two 185-bp single-stranded donors (Suppl.Fig.1A, ssDonor#1 and ssDonor#2) to introduce loxP elements by homology-directed repair. The gRNAs were synthesized and cloned into pDR274 (Addgene, Cambridge, MA), a T7 promoter gRNA expression vector (Hwang et al., 2013). Following DraI digestion, the linearized expression vectors were purified using a QIAquick Gel Purification Kit (QIAGEN) and used as DNA templates to produce gRNAs with a MAXIscript T7 kit (Life Technologies). For Cas9 mRNA production, we used the T7 promoter- containing pX330 vector (Cong et al., 2013; Zhong et al., 2015), which was digested with NotI and then purified using a QIAquick Gel Purification Kit (QIAGEN). Subsequently, the linearized and purified vector was used as a DNA template to synthesize Cas9 mRNA using the mMESSAGE mMACHINE T7 Ultra Kit (Life Technologies). Cas9 mRNA and gRNAs were purified using RNA Clean & Concentrator-25 (ZYMO Research) and dissolved in RNase-free water. RNA concentrations were measured using a NanoDrop ND1000. Finally, the gRNAs and Cas9 mRNA were mixed with an equal volume of formamide, respectively, and the denatured mixtures were run on an agarose gel to evaluate RNA quality (not shown).
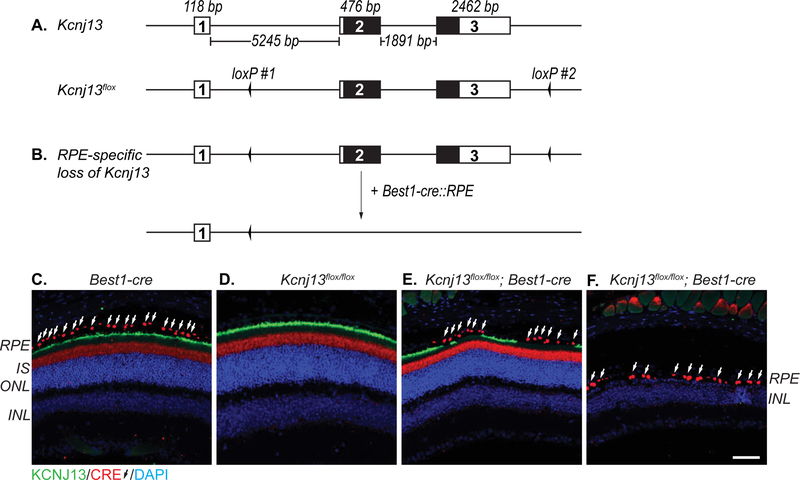
Figure 1. Structure of the mouse Kcnj13 conditional loss-of-function (Kcnj13flox) allele.
Schematic of Kcnj13 wild-type and conditional knockout (Kcnj13flox) alleles (A). The Kcnj13 locus spans approximately 10 kb, comprising of 3 exons. The protein coding sequences are found within exons 2 and exon 3. The Kcnj13flox allele was generated using the CRISPR/Cas9 system to insert two loxP sites targeting intronic sites flanking exons 2 and 3. One mouse containing both loxP sites in cis was recovered. Genotyping is performed using two sets of primers which flank each of the loxP site. Untranslated sequence (white region). Protein coding sequence (black region). Strategy to induce a RPE-specific loss of Kcnj13 (B). Mice containing the Kcnj13flox allele are crossed to the RPE-specific Best1-cre mouse line. Subsequent generations of mice are genotyped using both the Kcnj13flox and Best1-cre primer sets. Best1-cre is expressed in the RPE and KCNJ13 is localized to the apical membrane (C). We observed cross reactivity of the anti-Cre antibody with unknown antigen in the inner segment (IS) of the photoreceptors in wild-type (C,D) and Kcnj13 mutant retinae (E). Best1-cre induces loss of Kcnj13 in mice that are homozygous for the Kcnj13flox allele (E). Best1-cre expression ranges from 10% to 90% of RPE cells among animals. Homozygous Kcnj13flox mice without the Best1-cre allele (Kcnj13flox/flox) or have low Best1-cre penetrance are histologically and functionally wild-type (D,E). Kcnj13flox/flox; Best1-cre mice with high penetrance of Best1-cre show a retinal degeneration phenotype (F). Images were taken at 10x magnification. KCNJ13 (green; goat anti-Kir7.1), Cre (red, white arrows; rabbit anti-Cre), DAPI (blue). Retinal pigment epithelium (RPE); inner segment (IS); outer nuclear layer (ONL); inner nuclear layer (INL). Scale Bar (50 μm). Diagrams not drawn to scale.
According to the literature (Singh et al., 2015; Wang et al., 2013; Yang et al., 2013) and our previous injection experience (Zhong et al., 2015), a mixture of gRNAs, Cas9 mRNA, and single-stranded donor DNAs (12.5 ng/μl for each gRNA, 50 ng/μl Cas9 mRNA, and 25 ng/μl for each donor) were prepared using RNase-free water. The mixture was microinjected into the cytoplasm of C57BL/6 inbred zygotes. After injection, surviving zygotes were immediately transferred into oviducts of ICR albino pseudo-pregnant females. To confirm if the two loxP inserts were introduced into the intended sites, we performed genotyping using PCR on CRISPR founder mice, and selected founders carrying both loxP inserts. Next, we checked for germline co-transmission of the two loxP inserts by crossing selected founders to Best1-cre animals, to identify animals with Kcnj13 alleles containing cis-inserted loxP sites. To check if the loxP inserts affect Kcnj13 expression and if the Best1-cre induced excision could cause Kcnj13 loss-of-function, we performed Cre and Kcnj13 immunofluorescent staining on Kcnj13wt/wt; Best1-cre, Kcnj13flox/flox; Best1-cre and Kcnj13flox/flox mouse retinal paraffin sections. Subsequent generations of mice were genotyped for the Kcnj13flox allele using the following sets of primers: LoxP1 (Fwd: 5’-AAAATTTTACTTCTCTCAACTTCT-3’; Rev: 5’-AAACATTTTTGGTTTTGTTTT- 3’) and LoxP2 (Fwd: 5’-CAACTTAGATTTATGCTTGAAA-3’; Rev: 5’-AAATAGACATTGATGATGTTGTT-3’). Kcnj13 cKO mice are maintained with a mixed C57BL/6 and 129S6/SvEvTac background. All mice were maintained under 12-h light and 12-h dark cycles. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
Animals used in this study were screened post-mortem using KCNJ13/Cre immunostaining to assess extent of KCNJ13 and Cre expression. Animals with widespread RPE-specific Cre expression and loss of KCNJ13 immunoreactivity were selected for analyses. Animals with patchy Cre and KCNJ13 immunoreactivity were excluded from analyses. All histological and electroretinographic analyses were performed using the Kcnj13 cKO mice without fluorescent Cre-reporter allele. Number of animal used per group and analysis are included in the figure legends.
2.2. Generation and in vivo screening of Kcnj13 conditional knockout reporter (cKOR) mice
We crossed Kcnj13 cKO mice to Ai9 (tdtomato fluorescent protein) Cre-reporter mice to generate the Kcnj13 cKOR mouse mice with Cre-dependent retinal fluorescence (Suppl.Fig.3) (Madisen et al., 2010). Genotyping on subsequent generations was done using PCR with Kcnj13flox, Best1-cre, and Ai9 primers. Ai9 and Best1-cre alleles were genotyped as previously described (Iacovelli et al., 2011; Madisen et al., 2010). Kcnj13 cKOR mice are maintained with a mixed C57BL/6 and 129S6/SvEvTac background. TdTomato fluorescence was used as a proxy for Best1-cre expression. TdTomato fluorescence in the retina was evaluated in live animals using the Micron IV retinal fluorescence microscope (Phoenix Research Labs). A custom TRITC-optimized BrightLine BasicTM single-band exciter filter (25 mm housed, Cat# FF01–542/20–25, Semrock) and barrier (12.5 mm unhoused, Cat# FF01–620/52–12.5-D, Semrock) were installed on the Micron IV camera to allow visualization of TdTomato fluorescence. Mice were anesthetized by intraperitoneal injection of 22 mg/kg ketamine, 4.4 mg/kg xylazine and 0.37 mg/kg acepromazine. Both pupils were dilated, corneas anesthetized, and moistened for 1 min with a drop of tropicamide (1.0%), proparacaine (1.0%), and hypromellose (2.5%), respectively. The objective of the Micron IV camera was gently placed onto the cornea of the animal. The fundus was imaged using bright-field (10 gain; 6 FPS) and TdTomato (10 gain; 2 FPS) exciter and barrier sets.
2.3. Histological analysis and immunofluorescence
We performed hematoxylin and eosin (H&E) staining to examine retinal morphology and immunofluorescence (IF) staining for Cre and KCNJ13 to examine expression and localization in the RPE. Mice were sacrificed and eyes were fixed in Davidson’s fixative and processed for paraffin embedding. Serial sections (7–10 μm thick) were cut for approximately one-half of each eye and were processed for H&E and IF staining. For H&E staining, retinal sections were deparaffinized in xylenes for 20 mins, rehydrated in decreasing serial ethanol washes (100%, 95%, 70%, 50% ethanol in water) for 5 mins each prior to H&E staining. Rehydrated sections were stained with hematoxylin for 1–2 mins, washed in running tap water until clear, and then stained with eosin for 1–2 mins. Stained retinal sections were dehydrated in increasing serial ethanol washes (50% 1 min, 70% 1 min, 95% 5 min, 100% 5 min ethanol in water) and finally xylenes for 10 mins. Coverslips were mounted using Cytoseal mounting media (Cat# 8312–4, Thermo Fisher). For IF staining, antigen retrieval was performed by boiling sections in Tris- EDTA (TE) buffer (10 mM Tris, 1 mM EDTA, pH 9.1) for 30 minutes. After cooling at room temperature, sections were washed three times in 1x PBST buffer (0.3% Triton X-100/PBS) for 5 mins. Sections were then incubated in 10% donkey serum (in 1x PBST buffer) at room temperature for 1 hour and incubated with primary antibodies overnight at room temperature. After three washes in PBST for 5 mins each, sections were then incubated with secondary antibodies at room temperature for 2 hrs, followed by three PBST washes. Sections were counterstained for nuclei using DAPI-containing Prolong Gold Antifade mountant (Cat# P36935, Thermo Fisher Scientific). Fluorescence microscopy (Zeiss Axio Imager MRc5 with Apotome attachment and Zeiss Axio Observer) was used to capture 10x, 20x, and 40x objective magnification images.
2.4. Antibodies
Primary antibodies: 1:200 goat anti-Kir7.1 (C19, Santa Cruz Biotechnology) and 1:1000 rabbit anti-Cre (Novagen #69050–3, MilliporeSigma). Secondary antibodies (1:200): Alexa Fluor® 568 or 647 donkey anti-goat IgG and Alexa Fluor® 488 donkey anti-rabbit IgG.
2.5. Retinomorphometry
H&E stained retinae were imaged using a Zeiss Axio Observer inverted microscope to capture 20x objective magnification images. Individual images were processed into a single stitched image using Zen Digital Imaging Software (Blue edition, Zeiss). Retinomorphometric data was collected from H&E stained wild-type and Kcnj13 mutant retinae. The thickness of the outer nuclear layer (ONL) was measured at 4 equally spaced (300 μm) intervals, starting at the optic nerve (ON) and moving toward the distal ciliary body. At each interval, 3 measurements were taken, and mean value was calculated. A minimum of 3 mice were used per genotype. For comparing age and ONL thickness, mean ONL thickness was calculated, excluding ON values. Percent of ONL thickness relative to wild-type was calculated by dividing the mean Kcnj13 mutant ONL thickness by wild-type ONL thickness. ONL thickness was plotted using GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA).
2.6. Visual Function Analysis using Electroretinography (ERG)
Mice were dark-adapted overnight and then anesthetized by a single intraperitoneal injection of 22 mg/kg ketamine, 4.4 mg/kg xylazine and 0.37 mg/kg acepromazine. Both pupils were dilated with a drop of tropicamide (1.0%) and phenylephrine (2.5%) and then corneas were anesthetized with a drop of proparacaine (1.0%). After 1 min, excess fluid was removed and a drop of hypromellose (2.5%) was placed on each cornea to keep it moistened and provide a good contact between the cornea and the ERG electrode (Cat# N1530NNC, LKC Technologies). All tests were performed under a dim red light and a feedback-controlled heating pad was used to keep treated mice at a constant body temperature of 37.6 °C. ERG recordings using six flash intensities (−34, −32, −14, −4, 0 and 10 dB) were performed with the UTAS Visual Diagnostic System and EMWIN software (LKC Technologies, Gaithersburg, MD, USA). In this study, scotopic ERG recordings were performed at 15 days (P15), 21 days (P21), 1 (1M) and 2 (2M) months of age for wild-type and Kcnj13 mutant mice. Mice were light adapted for 2 mins after scotopic ERG recordings and photopic responses were recorded with 0, 10, and 25 dB flash intensities (Eblimit et al., 2017). Scotopic A-wave, scotopic B-wave, and photopic B-wave amplitudes were plotted using GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA). For ERG analysis, a minimum of 3 animals were used for each genotype. Amplitudes for 10 dB (25 cd*s/m2) flash intensity for each time point were plotted for scotopic A-wave and B- wave in order to compare ERG amplitudes across age. For comparing photopic amplitudes across age, 25 dB (790 cd*s/m2) was plotted instead.
2.7. Statistical Analysis
Data are shown as the mean ± SEM. A 2-way ANOVA with multiple comparisons and Sidak’s correction was performed between wild-type and Kcnj13 mutant groups to determine statistical significance. P>0.05 (ns = not significant); 0.05≥P>0.01 (*); 0.01≥P≥0.001 (**); P<0.001 (***).
3. Results
3.1. Generation of Kcnj13 conditional loss-of-function (cKO) mice
We previously generated Kcnj13 somatic mosaic mutant mice using the CRISPR/Cas9 system to delete the start codon of Kcnj13 (Zhong et al., 2015). However, homozygous Kcnj13 null animals die at P1, thus requiring a conditional knockout approach to study Kcnj13 function in older animals. To generate a Kcnj13 floxed allele (Kcnj13flox), we used the CRISPR/Cas9 system to insert loxP sites flanking exons 2 and 3 of the Kcnj13 gene (Fig.1A,Suppl.Fig.1A), which encodes the entire open reading frame. From a total of 19 founder mice born, 3 carried one loxP insertion at either target site #1 or #2, and one carried loxP insertions at both target sites, which was used for subsequent breeding with RPE-specific Best1-cre mice (Fig.1B, Suppl.Fig.1B) (Iacovelli et al., 2011). Germline co-transmission of both loxP insertions was observed, indicating the insertions occurred in cis. Importantly, Cre-induced excision was detected in the first generation of Kcnj13wt/flox; Best1-cre+ mice, indicating the loxP sites are functional even though one (loxP #1) had sustained a single base mutation (Fig.1B,Suppl.Fig1B,1C). We investigated if Best1-cre-mediated recombination of Kcnj13flox reduces KCNJ13 expression in the RPE by performing immunostaining for both Cre and KCNJ13 (Fig.1E,F). We detected patchy KCNJ13 expression in the RPE of Kcnj13flox/flox; Best1- cre retinae (Fig.1E), consistent with known Best1-cre expression patterns (Iacovelli et al., 2011). Photoreceptor degeneration was observed in Kcnj13flox/flox; Best1-cre retinae with widespread Cre expression and corresponding KCNJ13 loss (Fig.1F). In contrast to Kcnj13flox/flox; Best1-cre retinae, KCNJ13 is widely expressed throughout the apical membrane of the RPE of Kcnj13wt/wt; Best1-cre, Kcnj13flox/flox, and Kcnj13wt/flox; Best1-cre mice (Fig.1C,1D). This suggests that Kcnj13 loss and the photoreceptor degeneration phenotype closely correlate with Cre expression.
Based on these findings, Kcnj13wt/wt; Best1-cre, Kcnj13flox/flox and Kcnj13flox/flox; Best1-cre with low Cre expression are phenotypically wild-type; however, Kcnj13flox/flox; Best1-cre with low Cre expression are excluded from any analyses. Kcnj13flox/flox; Best1-cre with high Cre expression are phenotypically mutant. The extent of Cre and KCNJ13 expression was confirmed with immunostaining.
3.2. RPE-specific loss of Kcnj13 causes severe photoreceptor degeneration and defects in visual function
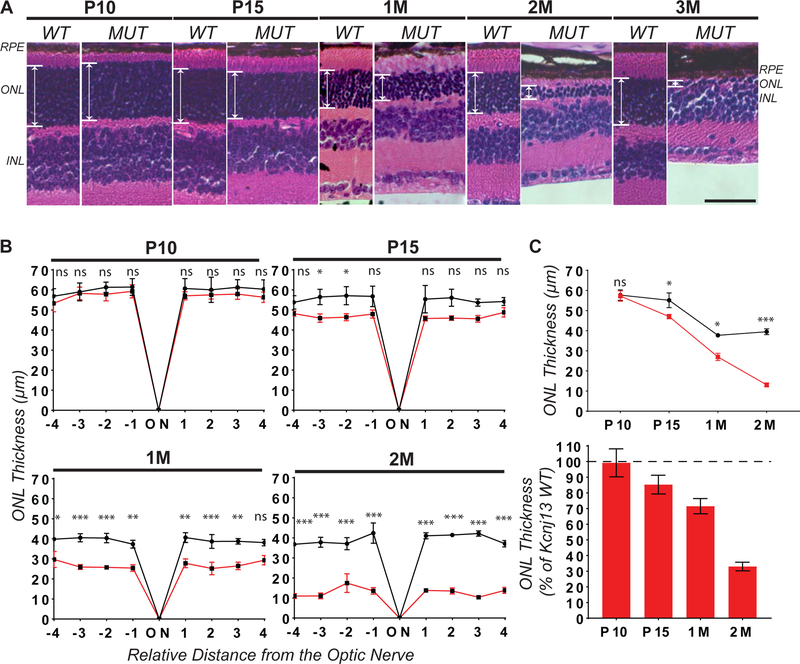
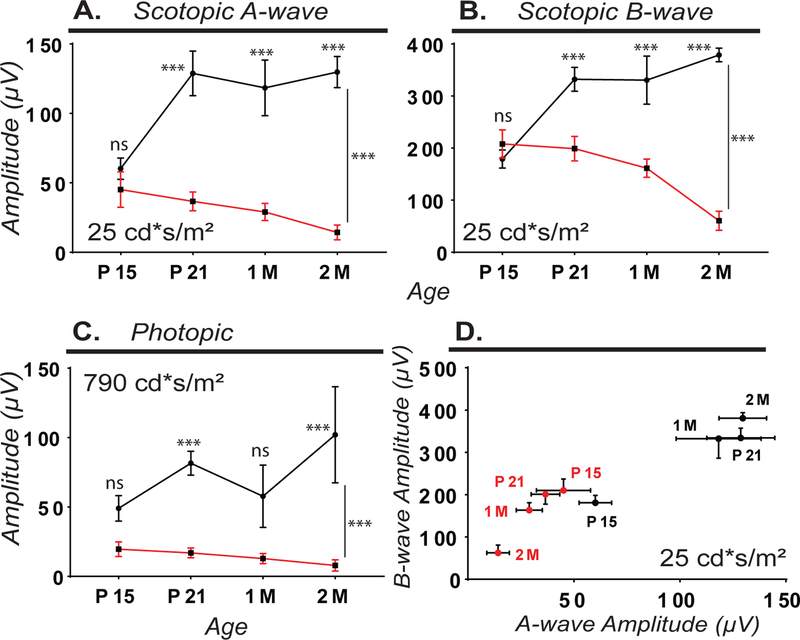
To determine if RPE-specific loss of Kcnj13 causes a retinal degeneration phenotype, we performed histological analysis of Kcnj13 mutant retinae using H&E staining at P10, P15, P21 and 1, 2, and 3 months of age. In animals with broad Cre expression, we observed a strong and progressive loss of photoreceptors between P15 and 2 months of age with a near complete loss by 3 months (Fig.2A). To quantitatively compare wild-type and Kcnj13 mutant retinae, we performed retinal morphometric analysis (Fig.2B). We observed significant thinning of the photoreceptor-containing outer nuclear layer (ONL) with ~15% loss by P15, ~10% loss by P21, ~30% loss by 1 month, and ~70% loss by 2 months compared to wild-type retinas (Fig.2B,2C). By 3 months, we observed nearly complete photoreceptor degeneration with only a single row of nuclei in the ONL (Fig.2A). To assess visual function, we performed electroretinography on dark-adapted wild-type and Kcnj13 mutant mice at P15, P21, 1 and 2 months of age (Fig.3). We observed significantly reduced scotopic A-wave (Fig.3A,D,Suppl.Fig.2A) and B-wave (Fig.3B,D,Suppl.Fig.2B) amplitudes in Kcnj13 mutant retinae in comparison with wild-type retinae between P21 and 2 months of age, suggesting that RPE-specific loss of Kcnj13 leads to severe visual defects, consistent with the observed photoreceptor degeneration. We also observed significantly reduced photopic cone responses between P15 and 2M, suggesting severe cone photoreceptor dysfunction (Fig.3C,Suppl.Fig.2C). These findings suggest loss of Kcnj13 in the RPE leads to early, progressive, and severe photoreceptor degeneration, with detectable thinning by 15 days of age and observable visual defects by 21 days of age.
Figure 2. Conditional loss of Kcnj13 in the RPE causes severe and progressive photoreceptor degeneration.
Hematoxylin and eosin (H&E) staining paraffin-embedded retinae obtained from wild-type (WT) and Kcnj13 conditional knockout (MUT) mice between 10 days (P10) and 3 months (3M) of age (A). Left panel: wild-type (WT) control mouse retina. Right panel: Kcnj13 mutant (MUT) mouse retina. Retinal Pigment Epithelium layer (RPE), Outer Nuclear Layer (ONL), Inner Nuclear Layer (INL). Scale Bar (20 μm). Images were taken at 20x magnification. Retinomorphometry (B) was used to quantify the thickness of the ONL at 4 equally spaced positions (300 μm) from the optic nerve (ON). Each point represents the mean of three wild-type (black lines) and Kcnj13 mutant (red lines) retinae. Mean ONL thickness and % of wild-type ONL thickness was determined at each age (C). Kcnj13 mutant retinae have significant and progressive thinning of the ONL, consistent with photoreceptor degeneration. Number of mice used: P10: WT=3, MUT=6; P15: WT=3, MUT=6; 1M: WT=3, MUT=4; 2M: WT=3, MUT=4. Statistical analyses were performed using 2-way ANOVA with multiple comparisons test and Sidak’s correction. Error Bars (SEM). P>0.05 (ns = not significant); 0.05≥P>0.01 (*); 0.01≥P>0.001 (**); P<0.001 (***).
Figure 3. Kcnj13 cKO mice exhibit scotopic and photopic visual function defects.
While wild-type mice had normal scotopic ERG responses, Kcnj13 mutant mice have significantly reduced A-wave (A) and B-wave (B) amplitudes at the strongest flash intensity tested (25 cd*s/m2) between P15 and 2 months of age. Kcnj13 mutant mice have significantly reduced photopic cone responses (at 790 cd*s/m2) between P21 and 2M (C). Comparison of B-wave and A-wave amplitudes in WT and Kcnj13 mutant mice (D). Horizontal error bar (A-wave SEM), vertical error bar (B-wave SEM). Wild-type is drawn in black and Kcnj13 mutant in red. Number of mice used: P15: WT=8, MUT=7; P21: WT=13, MUT=9; 1M: WT=6, MUT=7; 2M: WT=4, MUT=5. Statistical analysis was performed using 2-way ANOVA with multiple comparisons and Sidak’s correction. Error Bars (SEM). P>0.05 (ns = not significant); 0.05≥P>0.01 (*); 0.01≥P>0.001 (**); P<0.001 (***).
3.3. Kcnj13 cKO reporter mice express transgenic TdTomato fluorescent protein upon Cre- mediated excision
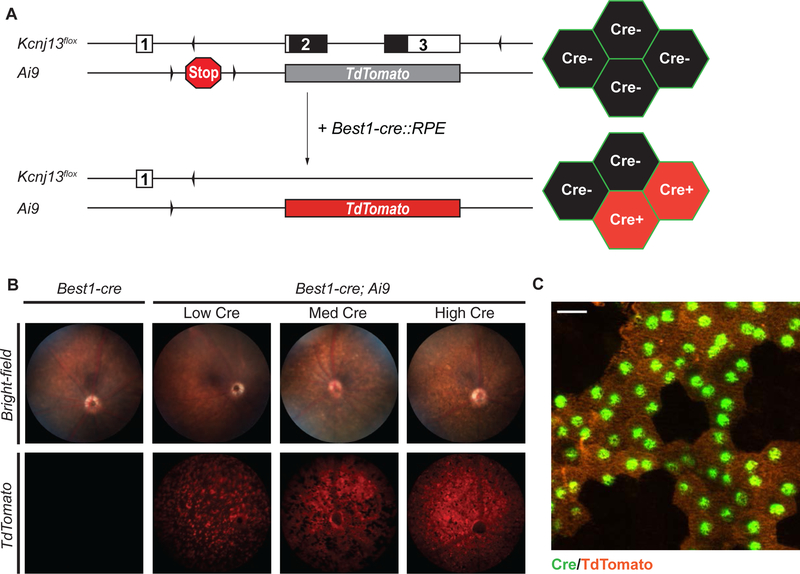
The variability of Best1-cre expression can create a significant obstacle for the study of Kcnj13 function due to the highly non-cell-autonomous manner of Kcnj13 action (Zhong et al., 2015). In particular, only mice with broad Cre expression are useful for phenotypic analyses. However, both non-invasive (i.e., ERGs) and histological approaches to determining whether any given animal of the correct genotype (Kcnj13flox/flox; Best1-cre) has broad Cre expression are time-consuming and laborious. To address this problem, we generated Kcnj13 conditional knockout reporter (Kcnj13 cKOR: Kcnj13flox/flox; Best1-cre; Ai9) mice (Fig.4A,Suppl.Fig.3) by crossing our Kcnj13 cKO mice with the Ai9 fluorescent Cre-reporter mouse line to assess Best1- cre expression in live animals using retinal fluorescence imaging (Madisen et al., 2010). The Ai9 Cre-reporter allele expresses TdTomato fluorescent protein upon Cre-mediated excision of the preceding loxP-stop-loxP sequence (Madisen et al., 2010). We observed TdTomato fluorescence in retinae of Best1-cre; Ai9 mice consistent with Best1-cre expression patterns, ranging from 10% to >70% of RPE cells expressing TdTomato, using live retinal fluorescence imaging (Fig.4B,C).
Figure 4. In vivo screening of mice for Best1-cre expression using Cre-reporter fluorescence.
Fluorescent Cre-reporter strategy used to identify Cre expression in live mice (A). Animals containing Kcnj13flox and Best1-cre alleles were crossed to the Ai9 mouse line. The Ai9 Cre-reporter allele comprises the TdTomato gene, with a floxed transcription termination cassette upstream, inserted into the ROSA26 locus. Cre expression mediates the removal of the stop sequence, resulting in expression of TdTomato in Cre-positive cells. Mice with both Best1-cre and Ai9 alleles have varying degree of detectable TdTomato expression in the RPE, consistent with variable Best1-cre expression (B). Control retinae from mice without Best1-cre or Ai9 show no detectable TdTomato fluorescence. Animals with widespread TdTomato fluorescence are selected for phenotypic studies. The RPE from the cKOR mice appear as a mosaic of TdTomato(+); Cre(+) and TdTomato(−); Cre(−) cells (C). Bright-field fundus images were taken at 6 frames per second (FPS), 10 gain. Fluorescence fundus images were taken at 2 (FPS), 10 gain. Immunofluorescence image was taken at 40x magnification. Scale bar: 20 μm. Cre (Green), TdTomato (Orange).
This suggests that not only can the Ai9 Cre-reporter allele reveal the extent of Best1-cre expression in the highly pigmented RPE by using TdTomato as a proxy, we can also use retinal fluorescence microscopy to quickly pre-screen the fundus of live animals for time-intensive, age- dependent assays, such as for gene therapy studies, without having to sacrifice them for histology to identify Cre penetrance.
4. Discussion
Mechanistic studies of the functional roles of Kcnj13 have been previously limited due to the homozygous P1 lethality of Kcnj13 null animals and the availability of a suitable conditional animal model for postnatal analyses. In the present study, we describe the first reported conditional Kcnj13 loss-of-function mouse model which uses Cre-mediated deletion to trigger loss of Kcnj13 in a tissue-specific manner. In conjunction with Best1-cre expression, Kcnj13 is specifically deleted in the RPE of the retina. We positioned the loxP sites upstream of exon 2 and downstream of exon 3, allowing for the deletion of the entire protein coding region of Kcnj13. In Kcnj13flox/flox; Best1-cre RPE cells, strong or complete loss of Kcnj13 is observed.
However, large patches of Kcnj13 mutant RPE cells are necessary to observe retinal phenotypes. In animals with widespread Cre expression and loss of Kcnj13, we observed severe and progressive thinning of the outer nuclear layer, accompanied by strong defects in visual function. At P15, we observed ~15% ONL thinning, however, substantial visual defects are not observed until P21. Between P21 and 3 months of age, we observed consistent thinning of the outer nuclear layer and strong visual defects. These findings demonstrate the importance of Kcnj13 in maintaining photoreceptor survival and visual function (Zhong et al., 2015).
Although Best1-cre provides RPE-specific deletion of floxed genes, one obstacle is the patchy Cre expression, which ranges from 10% to 90% of RPE cells and strongly affects the penetrance of the retinal phenotypes. We observed that about 45% of Kcnj13flox/flox; Best1-cre animals had a retinal phenotype. To address these obstacles, we combined a Cre-reporter allele with the Kcnj13 cKO allele to create the Kcnj13 cKO reporter (or cKOR) genotype, which can be used to identify the extent of Cre-expression using fundal TdTomato fluorescence in living mice. In conjunction with retinal fluorescence microscopy, we detect patchy TdTomato fluorescence consistent with known Best1-cre expression patterns. The Kcnj13 cKOR mouse model allows for two-dimensional pre-screening of live animals for strong TdTomato fluorescence prior to functional and histological studies. The ability to rapidly identify retinal Cre expression of mice with mosaic expression of Cre is essential for long-term or time-dependent studies, reducing animal usage, and reducing the need for time-consuming post-mortem KCNJ13/Cre screening. In addition, the mosaicism of juxtaposed wild-type and Kcnj13 mutant patches of RPE cells in low and medium Best1-cre expressing animals provide an internal positive control for studying RPE phenotypes. The Kcnj13 cKO and Kcnj13 cKOR mouse models also provide unique tools for investigating non-cell-autonomous and cell-autonomous roles of Kcnj13 in the retinal function and survival.
The emerging role of Kcnj13 as a key regulator of retinal electrophysiology has provided insight into the glial functions involved in RPE/PR homeostasis (Kumar and Pattnaik, 2014; Shahi et al., 2017). Kcnj13 expresses an inwardly rectifying potassium (Kir) channel, a class of K+ channels which have greater flow into the cell under experimental conditions due to inherent rectification properties (Wimmers et al., 2007). However, under physiological conditions, Kir channels conduct mainly outward K+ currents (Wimmers et al., 2007). Kir channels and K+ transport are essential for various cellular processes, including the maintenance of resting membrane potential by bringing the cellular membrane potential toward the K+ equilibrium potential (Wimmers et al., 2007). The subcellular localization of KCNJ13 with Na+/K+ ATPase at the apical membrane of the RPE, as well as in the choroid plexus, suggest an involvement of KCNJ13 in spatial “K+ recycling” processes between the RPE and the subretinal space, allowing for passive K+ diffusion down an established K+ concentration gradient (Hibino et al., 2010; Kusaka et al., 2001; Shahi et al., 2017; Strauss, 2005; Suzuki et al., 2003; Wimmers et al., 2007). Sub-retinal K+ homeostasis is essential for maintaining the excitability of photoreceptors (Reichhart and Strauss, 2014; Strauss, 2005; Wimmers et al., 2007). Light stimulation of photoreceptors decreases subretinal K+ from 5 to 2 mM (Strauss, 2005). The RPE is essential for restoring subretinal K+ to dark, unstimulated levels (Oakley and Green, 1976).
Changes to subretinal ion composition and RPE membrane potential are reflected by a delayed positive C-wave, which occurs at approximately 2 sec following light stimulation (Kumar and Pattnaik, 2014; Strauss, 2005). Recent findings have shown a decreased C-wave when Kir7.1 is reduced using RNAi and pharmacological inhibition, further supporting the potential role of Kcnj13 as an essential component of RPE/PR homeostasis and retinal electrophysiology.
In summary, we have generated new conditional Kcnj13 loss-of-function mouse models which can be used to study the molecular roles of Kcnj13 in the retina. The photoreceptor degeneration phenotype of the RPE-specific Kcnj13 cKO mouse models is consistent with previously observed pathology (Zhong et al., 2015). The Kcnj13 cKO and cKOR mouse models provide the opportunity to study the function of Kcnj13 in specific populations of cells and to gain insight into the molecular and functional roles of Kcnj13 in maintaining retinal homeostasis.
Supplementary Material
Acknowledgements
Dr. Andrew Groves, Dr. Joana Jankowski, Dr. Daryl Scott, Rogers Brown (Groves Lab), Mohammad Uddin (Jankowski Lab) and Valerie Jordan (Scott Lab) for generously donating Ai9 and 129S6/SvEvTac mice. Dr. Aiden Eblimit and Smriti Agrawal for generously providing technical training and support, equipment access, and donating CRX-cre mice for fundus imaging controls. This project was supported by the Mouse Embryonic Stem Cell Core (Director: Jason Heaney, Ph.D.) at Baylor College of Medicine.
Funding
This work was supported by grants from the Retina Research Foundation to Graeme Mardon and Rui Chen. Dany Roman was supported by grants from the National Eye Institute (T32EY007102; PI: Graeme Mardon).
Footnotes
Conflicts of Interest
The authors declare no conflict of interest for this study.
References
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblimit A, Agrawal SA, Thomas K, Anastassov IA, Abulikemu T, Mardon G, Chen R, 2017. Conditional loss of Spata7 in photoreceptors causes progressive retinal degeneration in mice. Exp Eye Res 166, 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejtmancik JF, Jiao X, Li A, Sergeev YV, Ding X, Sharma AK, Chan CC, Medina I, Edwards AO, 2008. Mutations in KCNJ13 cause autosomal-dominant snowflake vitreoretinal degeneration. Am J Hum Genet 82, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y, 2010. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev 90, 291–366. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR, 2013. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 8, e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovelli J, Zhao C, Wolkow N, Veldman P, Gollomp K, Ojha P, Lukinova N, King A, Feiner L, Esumi N, Zack DJ, Pierce EA, Vollrath D, Dunaief JL, 2011. Generation of Cre Transgenic Mice with Postnatal RPE-Specific Ocular Expression. Investigative Ophthalmology & Visual Science 52, 1378–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Eng L, Krapivinsky L, Yang Y, Clapham DE, 1998. A novel inward rectifier K+ channel with unique pore properties. Neuron 20, 995–1005. [DOI] [PubMed] [Google Scholar]
- Kumar M, Pattnaik BR, 2014. Focus on Kir7.1: physiology and channelopathy. Channels (Austin) 8, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaka S, Inanobe A, Fujita A, Makino Y, Tanemoto M, Matsushita K, Tano Y, Kurachi Y, 2001. Functional Kir7.1 channels localized at the root of apical processes in rat retinal pigment epithelium. J Physiol 531, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H, 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B 2nd, Green DG, 1976. Correlation of light-induced changes in retinal extracellular potassium concentration with c-wave of the electroretinogram. J Neurophysiol 39, 1117–1133. [DOI] [PubMed] [Google Scholar]
- Pattnaik BR, Shahi PK, Marino MJ, Liu X, York N, Brar S, Chiang J, Pillers DA, Traboulsi EI, 2015. A Novel KCNJ13 Nonsense Mutation and Loss of Kir7.1 Channel Function Causes Leber Congenital Amaurosis (LCA16). Hum Mutat 36, 720–727. [DOI] [PubMed] [Google Scholar]
- Reichhart N, Strauss O, 2014. Ion channels and transporters of the retinal pigment epithelium. Exp Eye Res 126, 27–37. [DOI] [PubMed] [Google Scholar]
- Sergouniotis PI, Davidson AE, Mackay DS, Li Z, Yang X, Plagnol V, Moore AT, Webster AR, 2011. Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause leber congenital amaurosis. The American Journal of Human Genetics 89, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah VS, 2016. Leber Congenital Amaurosis, in: Medina AC, Townsend HJ, Singh DA (Eds.), Manual of Retinal Diseases: A Guide to Diagnosis and Management. Springer International Publishing, Cham, pp. 125–128. [Google Scholar]
- Shahi PK, Liu X, Aul B, Moyer A, Pattnaik A, Denton J, Pillers DM, Pattnaik BR, 2017. Abnormal Electroretinogram after Kir7.1 Channel Suppression Suggests Role in Retinal Electrophysiology. Sci Rep 7, 10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura M, Yuan Y, Chang JT, Zhang S, Campochiaro PA, Zack DJ, Hughes BA, 2001. Expression and permeation properties of the K(+) channel Kir7.1 in the retinal pigment epithelium. J Physiol 531, 329–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E, 2015. A mouse geneticist’s practical guide to CRISPR applications. Genetics 199, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O, 2005. The retinal pigment epithelium in visual function. Physiol Rev 85, 845–881. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yasuoka Y, Shimohama T, Nishikitani M, Nakamura N, Hirose S, Kawahara K, 2003. Expression of the K+ channel Kir7.1 in the developing rat kidney: role in K+ excretion. Kidney Int 63, 969–975. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R, 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weleber RG, Francis PJ, Trzupek KM, Beattie C, 2013. Leber Congenital Amaurosis, in: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD,Fong CT, Mefford HC, Smith RJH, Stephens K (Eds.), GeneReviews(R), Seattle (WA). [Google Scholar]
- Wimmers S, Karl MO, Strauss O, 2007. Ion channels in the RPE. Prog Retin Eye Res 26, 263–301. [DOI] [PubMed] [Google Scholar]
- Yang D, Pan A, Swaminathan A, Kumar G, Hughes BA, 2003. Expression and localization of the inwardly rectifying potassium channel Kir7.1 in native bovine retinal pigment epithelium. Invest Ophthalmol Vis Sci 44, 3178–3185. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R, 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang X, Wang H, Sharma AK, Edwards AO, Hughes BA, 2013. Characterization of the R162W Kir7.1 mutation associated with snowflake vitreoretinopathy. Am J Physiol Cell Physiol 304, C440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Chen Y, Li Y, Chen R, Mardon G, 2015. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep 5, 8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.