Abstract
Chimeric antigen receptors (CARs) are synthetic antigen receptors that reprogram T cell specificity, function and persistence1. Patient-derived CAR T cells have demonstrated remarkable efficacy against a range of B-cell malignancies1–3, and the results of early clinical trials suggest activity in multiple myeloma4. Despite high complete response rates, relapses occur in a large fraction of patients; some of these are antigen-negative and others are antigen-low1,2,4–9. Unlike the mechanisms that result in complete and permanent antigen loss6,8,9, those that lead to escape of antigen-low tumours remain unclear. Here, using mouse models of leukaemia, we show that CARs provoke reversible antigen loss through trogocytosis, an active process in which the target antigen is transferred to T cells, thereby decreasing target density on tumour cells and abating T cell activity by promoting fratricide T cell killing and T cell exhaustion. These mechanisms affect both CD28- and 4-1BB-based CARs, albeit differentially, depending on antigen density. These dynamic features can be offset by cooperative killing and combinatorial targeting to augment tumour responses to immunotherapy.
Main
We modelled CAR therapy relapse by infusing limiting doses of CD19 CAR T cells in the well-established NALM6 acute lymphoblastic leukaemia (ALL) model in immunocompromised mice10–16 (Extended Data Fig. 1a). CARs encompassing CD28 or 4-1BB co-stimulatory domains (referred to as 19-28ζ or 19-BBζ, respectively) effectively controlled NALM6 cells at the dose of 0.4 × 106–1.0 × 106 CAR T cells, but allowed for frequent leukaemia relapse at the dose of 0.2 × 106 cells (Fig. 1a, Extended Data Fig. 1b–e). Although both types of CAR T cell showed limited evidence of exhaustion two weeks after infusion (Extended Data Fig. 1f, g), 19-BBζ cells were markedly exhausted by the time of relapse, whereas 19-28ζ cells were no longer detected (Fig. 1b, Extended Data Fig. 1h), consistent with clinical experience13–16 and CAR stress test models12. CD19 expression was reduced in progressing 19-BBζ-treated NALM6 cells, averaging 4,500 molecules per cell, down from the starting 11,000, which remain unchanged in 19-28ζ relapses and in untreated mice (Fig. 1c). The loss of CD19 occurred early on, as it was already present by day 14 and thus occurred in the presence of abounding CAR T cells (Fig. 1b, c and Extended Data Fig. 1f). The same patterns were found with CD19 CARs comprising single chain Fv (scFv) antibody fragments SJ25C116 or FMC6313–15 (Extended Data Fig. 2). Concurrent with decreased CD19 expression in tumour cells, a large fraction of CAR T cells stained positive for CD19 (Extended Data Fig. 3a). Notably, CD19 expression in the retrieved NALM6 cells was reversible after short-term culture (Fig. 1d). Because there was little variation in the expression of Cd19 mRNA (Extended Data Fig. 3b), these findings indicated that a reversible, post-transcriptional loss of CD19 occurred in the presence of CAR T cells. CD19 expression did not vary when fresh NALM6 cells were segregated from CAR T cells in transwells, but promptly decreased when T cells were co-cultured (Extended Data Fig. 3c, d). CD19 was not lost in co-cultures with untransduced T cells or T cells expressing a non-signalling CD19 CAR (Extended Data Fig. 3d). The transfer of CD19 protein from NALM6 cells to T cells thus displayed the hallmarks of CAR-mediated trogocytosis, as further compounded by inhibition with blockers of actin polymerization17 (Extended Data Fig. 3e). Co-culture with CD19-knockout NALM6 cells expressing a CD19–mCherry fusion molecule resulted in the detection of both mCherry and CD19 in T cells, demonstrating whole-protein CD19 membrane extraction (Fig. 1e). Loading of CD19–mCherry-expressing NALM6 cells with heavy amino acids and 19-28ζ cells with light amino acids and then the sorting of mCherry-positive trogocytosis-positive (trog+) and -negative (trog−) singlet T cells after brief co-culture unequivocally demonstrated CD19 peptides in the trog+ but not the trog− fraction (Fig. 1f, Extended Data Fig. 3f). CD81, which forms a complex with CD19, was also detected in trog+ but not trog− T cells (Fig. 1f), while concomitantly lost in the co-cultivated NALM6 cells (Extended Data Fig. 3g). By contrast, CD22 remained unchanged in NALM6 cells and was not detected by mass spectrometry in the T cells (Fig. 1f, Extended Data Fig. 3h, i).
Fig. 1: Trogocytic antigen extraction promotes tumour escape.
a, Tumour burden was monitored using bioluminescence image (average radiance, photons s−1 cm−2 sr−1) in mice bearing NALM6 cells after treatment with CD19 CAR T cells (n = 6–7 mice per group; two independent experiments are pooled for the 0.2 × 106 CAR T cell dose). NT, non-treated mice. b, Left, CAR T cell counts (n = 3–7 mice per group). Middle, EOMES/T-bet ratio (n = 3–5 mice per group). Right, expression of PD-1, LAG-3 and TIM-3 in CAR T cells (n = 3–5 mice per group). D32–D70, days 32–70. c, CD19 expression in NALM6 cells from mice treated with 19-28ζ or 19-BBζ CAR T cells (day 14; n = 3–7 mice per group; days 32–70; n = 3–9 mice per group). d, CD19 expression in ex vivo cultured NALM6 cells retrieved from 19-BBζ-treated mice at day 0 (D0; time of retrieval) or day 6 (D6) (n = 9 independent samples), or NALM6 cell in vitro (n = 4 independent samples). In b–d, cells were collected from bone marrow or extramedullary tumour sites of mice treated with 0.2 × 106 CAR T cells. e, Mean fluorescence intensity (MFI) of CD19 in NALM6 CD19–mCherry cells (middle) and CAR T cells (right; representative of three donors, n = 3 independent samples). UT, untransduced. 19-del denotes CAR T cells that lacks co-stimulatory and ζ-chain signalling domains. f, Heat map of protein expression (CD19, CD81 and CD22) in trog+ and trog− 19-28ζ cells and CD19–mCherry-expressing NALM6 cells. aa, amino acids. g, MFI of CD19 on blasts (left) and CAR+ T cells (right) derived from patients with ALL (ALL-1 and ALL-2) or with CLL (CLL-1 and CLL-2) (n = 4 patient samples). h, Percentage of CAR T cells stained with cell tracer dye and positive for IFNγ or granzyme B (GzmB) (n = 4 donors). i, Cytotoxic chromium (51Cr) release assay (representative of 6 donors, n = 3 independent samples). j, Percentage of CAR T cells co-expressing PD-1, LAG-3 and TIM-3 (n = 4 donors). NA, not available (no detectable cells). P values were determined by two-sided Mann–Whitney U-test (c, g, h, j) or two-sided Wilcoxon matched-pairs signed-rank test (d). Data are mean ± s.e.m.
CD19 trogocytosis occurred similarly after co-culture with NALM6, SUP-B15, Raji and CD19+ SK-OV-3 cells (Extended Data Fig. 4a) and, importantly, after the co-incubation of autologous 19-28ζ cells with primary samples from patients with ALL or chronic lymphocytic leukaemia (CLL) (Fig. 1g, Extended Data Fig. 4b). CAR-induced trogocytosis was observed with all other tested CAR targets, including CD22, B-cell maturation antigen (BCMA) and mesothelin (Extended Data Fig. 4c–e). The same antigen escape profile was achieved after targeting CD22 in vivo (Extended Data Fig. 5). NALM6 cells, which express approximately 2,000 CD22 molecules per cell at baseline levels, relapsed with expression of 750 CD22 molecules per cell after treatment with 0.2 × 106 22-BBζ cells (Extended Data Fig. 5c). Trogocytic target acquisition is thus a general feature of CAR T cells, probably applying to many if not all antigens.
Co-culture of sorted trog+ but not trog− CAR T cells with fresh 19-28ζ cells elicited the production of IFNγ and GzmB by 19-28ζ cells (Fig. 1h). When stably expressing CD19 at approximately equivalent levels to those detected after trogocytosis (Extended Data Fig. 6a), both 19-28ζ and 19-BBζ cells engaged in CD19+ T cell killing, more so in the former, consistent with their greater effector function12,18 (Fig. 1i). The culture of sorted trog+ and trog− T cells for 6 days revealed increasing expression of PD-1, LAG-3 and TIM-3 in trog+ cells, more so in 19-28ζ than 19-BBζ cells (Fig. 1j, Extended Data Fig. 6b). In T cells stably co-expressing CAR and CD19, those that did not succumb to T cell fratricide killing tended to acquire exhaustion markers (Extended Data Fig. 6c–e). CD19 trogocytosis was associated with diminished cell-surface CAR expression, both in vitro and in vivo, and co-localized intracellular CAR and CD19 (Extended Data Fig. 7).
The infusion of ‘fresh’ 19-BBζ T cells 10 days after the initial 19-BBζ treatment failed to rescue the relapse-prone mice (Fig. 2a), suggesting that CD19 density had already decreased to a level eluding 19-BBζ threshold efficacy. 19-28ζ CAR T cells, however, did rescue these relapses (Fig. 2a). Differential antigen sensitivity between the two CARs was confirmed in models using stable, graded CD19 levels (Fig. 2b, Extended Data Fig. 8). Mice bearing NALM6med or NALM6low tumours (obtained after monoalleleic or biallelic disruption of the Cd19 gene, respectively) showed diminished responses to low-dose CAR therapy, with 19-28ζ cells consistently inducing longer survival than 19-BBζ counterparts (Fig. 2b). These studies thus confirmed that a reduction of target antigen density alone can foster CAR T cell resistance. Together with the promotion of T cell exhaustion (Fig. 1j, Extended Data Fig. 6b–e) and fratricide T cell killing (Fig. 1h, i and Extended Data Fig. 6d), CAR target extraction is thus poised to promote antigen-low tumour relapse.
Fig. 2: Antigen density reduction differentially impacts CAR T cell activity.
a, b, Kaplan–Meier survival analysis. a, Mice bearing NALM6 cells were treated with 0.2 × 106 19-BBζ cells and infused 10 days later with 0.5 × 106 19-28ζ or 19-BBζ cells (data representative of three independent experiments). b, Mice bearing NALM6 cells, NALM6med cells or NALM6lowcells were treated with 0.2 × 106 CAR T cells (data pooled from two independent experiments). P values were determined by log-rank Mantel–Cox test.
Notably, 19-BBζ cells were able to control wild-type NALM6 when administered at a higher dose (Fig. 1a). We hypothesized that higher effector:target ratios may overcome clonal sensitivity thresholds. In microwells containing one CAR T cell and one NALM6 cell (1:1 E:T; Fig. 3a), 19-28ζ cells were more likely to lyse NALM6 cells than 19-BBζ cells (57% versus 39% over 24 h; Fig. 3b). The time to tumour cell death after the formation of a T cell–NALM6 cell conjugate showed that 19-28ζ cells killed NALM6 cells after 261 ± 18 min (mean ± s.e.m.), whereas 19-BBζ cells did so in 569 ± 35 min (mean ± s.e.m.) (Fig. 3b). The lesser killing ability of 19-BBζ cells did not result from poorer antigen recognition, as the frequency of non-lytic stable conjugate formation was not less but higher than measured with 19-28ζ cells, and the time spent in a non-lytic conjugate was greater (Fig. 3c).
Fig. 3: CAR T cell cooperativity augments insufficient clonal tumour-lytic potential.
a, Single-cell time-lapse imaging cytotoxicity assay. Time from the formation of CAR T cell–target cell conjugate (start point, T0) to target cell death (determined by positivity of the propidium iodide (PI) dye; TPI) recorded over 24 h in wells containing 1 CAR T cell and 1 tumour cell (1:1 effector:target (E:T) ratio). CTV, CellTrace Violet. b, Left, percentage of wells with target killing. Right, time from conjugate formation to propidium iodide influx in target cells (lytic conjugates). c, Left, percentage of non-lytic conjugate formation. Right, duration of non-lytic conjugates. d, Cytotoxic time-lapse imaging with two CAR T cells and one tumour cell (2:1 E:T). Time from first conjugate formation (start point, T0–1) to target cell death (TPI) is indicated as ΔT1. Time from second conjugate formation (start point, T0–2) to target cell death (TPI) is indicated as ΔT2. e, Percentage of target killing in wells at a 2:1 E:T ratio. f, Ratio of observed killing (Pobs.2/1) to estimated killing (Pest.2/1) in wells at a 2:1 E:T ratio. g, Percentage of target killing due to engagement of two CAR T cells with one tumour cell. h, Time from the formation of first conjugate (ΔT1) and second conjugates (ΔT2) to propidium iodide influx in target cells. In b, c, e–h, n = 5 donors; in b, c and h, time is indicated in minutes. i, Percentage of target killing in wells at 1:1 (left) and 2:1 (right) E:T ratios. WT denotes wild-type NALM6 cells. Med and Low denote NALM6med and NALM6low cells, respectively. n = 3 donors. In b, c, n ≥ 197, e–h n ≥ 242, i, n ≥ 70 (ratio 1:1) and n ≥ 42 (ratio 2:1) individual wells were examined. P values were determined by two-sided two-sample test of proportions (b (left), c (left), e, g and i), two-sided unpaired t-test (b (right), c (right) and h) or two-sided Mann–Whitney U-test (f). Data are mean ± s.e.m.
In microwells containing two CAR T cells and one tumour cell (2:1 E:T; Fig. 3d), the frequency of tumour lysis increased to about 75% for both 19-28ζ and 19-BBζ cells (Fig. 3e). Although this increase could be accounted for by additive killing with 19-28ζ cells, the gain seen with 19-BBζ cells exceeded simple additive effects (Fig. 3f, Extended Data Fig. 9a, b). This greater-than-expected tumour killing accorded with the observed frequency of conjugates comprising two T cells bound to the same tumour cell and the shorter time to target cell death measured for the second T cell conjugate (Fig. 3f–h). Microwell studies using the NALM6med and NALM6low target cells confirmed and extended these results, showing that 19-BBζ cells were more sensitive to antigen loss than 19-28ζ cells, and revealing cooperativity between 19-28ζ cells as the CD19 density decreased (Fig. 3i, Extended Data Fig. 9c, d). The possibility of cooperative killing previously noted in chronic infection models19 underscores the benefit of achieving higher effector:target ratios and the potential for population-based killing to transcend clonal limitations.
These considerations raised the possibility that combinatorial targeting, which has been proposed to pre-empt the immune escape of antigen-negative tumours5,7, could mitigate that of antigen-low tumours. We thus investigated combinatorial possibilities using our CD19-calibrated NALM6 lines (Extended Data Fig. 8d). Treating mice with established NALM6 cells with 0.2 × 106 19-ΒΒζ cells followed by a second infusion of 0.5 × 106 22-28ζ or 22-BBζ cells was ineffective (Fig. 4a), consistent with reduced expression of CD22 and poor CAR T cell expansion (Extended Data Fig. 5b, c). A higher CAR T cell dose (1 × 106) afforded a better survival benefit, more so with 22-28ζ cells (Fig. 4a). Dual targeting proved more effective in preventing antigen escape (0.2 × 106 CD19/CD22 CAR T cells, Fig. 4b) than sequential infusions (Fig. 4a). If CD19 were targeted with a BBζ CAR, targeting CD22 with a 28ζ CAR proved to be much more efficacious (Fig. 4b).
Fig. 4: Rational combinatorial targeting overcomes antigen-low tumour escape.
a–d, Kaplan–Meier survival analysis. a, Mice bearing NALM6 cells treated with 0.2 × 106 19-BBζ cells followed by infusion of 0.5 × 106 (left) or 1 × 106 (right) CD22 CAR T cells 10 days later. b–d, Mice bearing NALM6 cells (b), NALM6med cells (c) and NALM6low cells (d) treated with 0.2 × 106 single or dual-targeted CAR T cells. e, Summary of the outcomes of the combinatorial targeting strategies, based on relative antigen densities and CAR co-stimulatory design (based on median survival of treated mice in b–d). P values were determined by log-rank Mantel–Cox test.
Under lower CD19-antigen density conditions, the 19-28ζ and 22-ΒΒζ combination afforded the most durable responses (Fig. 4c, d), outperforming ΒΒζ CAR pairings. At the lowest CD19 level, 19-ΒΒζ CAR therapy could no longer be rescued by co-targeting the low-density CD22 target, regardless of which CD22 CAR was used. Thus, combinatorial targeting can reduce escape of tumours with low baseline or post-trogocytic target expression, provided that CAR co-stimulatory features are adapted to target antigen density (Fig. 4e).
Different antigen expression profiles—original, negative or diminished—are associated with escape from immunotherapy4,6,7,9,14. This study shows that tumour cells that are engaged but not killed by T cells are susceptible to trogocytic reduction of antigen density, with several possible consequences depending on antigen density, the effector:target ratio and CAR design (Extended Data Fig. 10). Understanding these dynamic features provides a framework for rational CAR T cell dosing and combinatorial targeting strategies, including target-adapted CAR co-stimulatory functions.
Methods
Data reporting
The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment, except where noted.
Cell culture
NIH/3T3, NALM6, SUP-B15, Raji, SK-OV-3 and A549 cell lines were obtained from ATCC. The KMS-12-BM cell line was obtained from DSMZ. NALM6, SUP-B15 and Raji cell lines were cultured in RPMI-1640 (Invitrogen) supplemented with 10% fetal bovine serum (FBS; HyClone), 10 mM HEPES (Invitrogen), L-glutamine 2 mM (Invitrogen), NEAA 1× (Invitrogen), 0.55 mM β-mercaptoethanol, 1 mM sodium pyruvate (Invitrogen). KMS-BM-12 cells were cultured in RPMI-1640 supplemented with 20% FBS. SK-OV-3 and A549 cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS. NIH-3T3 cells were cultured with DMEM supplemented with 10% fetal calf serum (FCS; HyClone). For proteomics quantification experiments, NALM6 cells transduced to express the CD19–mCherry fusion protein were grown in RPMI 1640 containing stable isotope 13C6-L-lysin (SILAC Protein Quantitation Kit-RPMI 1640, Thermo Scientific) for 7 days before processing to co-culture with CAR T cells. NALM6 cells were transduced with firefly luciferase-GFP to allow in vivo tumour burden imaging as described12. SK-OV-3 cells were transduced to express CD19. A549 cells were transduced to express MSLN20. NIH/3T3 cells expressing human CD19 served as alternative antigen-presenting cells in proliferation assays. All cells were routinely tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza).
Vector constructs
19-28ζ-LNGFR or 19-BBζ-LNGFR CARs contain the SJ25C1 or FMC63 CD19-specific scFv fragment13–15. 19-del-LNGFR, a CAR construct that lacks co-stimulatory and ζ-chain signalling domains, contains the SJ25C1 CD19-specific scFv. CD22-28ζ-DsRed and CD22-BBζ-DsRed CARs contain the m971 CD22-specific scFv7. BCMA-BBζ-DsRed CARs contain the 11D5-3 BCMA-specific scFv4. MSLN-BBζ-DsRed CARs contain the m912 MSLN-specific scFv20. CAR cDNAs were cloned in the SFG γ-retroviral vector. Cd19 cDNA was cloned in SFG γ-retroviral or pLM-lentiviral vector backbone under the control of the PGK-100 promoter. The CD19–mCherry fusion was generated by fusing the mCherry sequence at the C terminus of CD19 via a GS linker and cloned into the pLM-lentiviral vector backbone. The CAR–GFP fusion has been previously described21. All constructs were prepared using standard molecular biology techniques. Viral supernatants were prepared as previously described21.
T cell activation and transduction
Buffy coats from healthy donors were obtained from the New York Blood Center. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation and activated as previously described13. Purified PBMCs were cultured in RPMI-1640 medium supplemented with 10% FBS. Forty-eight hours after activation, T cells were transduced with retroviral supernatants by centrifugation on retronectin (Takara)-coated plates. Apheresis product and blood and bone marrow samples before and after CAR T cell infusion were obtained from patients that were consented and enrolled in phase I 19-28ζ CAR T cell clinical trials approved by the MSKCC Institutional Review Board (IRB). Clinical trials are registered under identifiers and at the https://clinicaltrials.govwebsite. CAR T cells from patients have been manufactured as previously described16,22.
T cell expansion
A total of 3 × 105 irradiated NIH/3T3 cells expressing human CD19 were plated on 24-well plates and used to stimulate 1 × 106 CAR T cells per ml as previously described12. Cells were maintained in RPMI-1640 medium supplemented with 10% FBS without addition of cytokines. For in vitro assays, CAR T cells were bead-sorted four days after T cell transduction and before co-culture with NIH/3T3 cells. CAR T cells were stained with LNGFR–PE (clone C40-1457, BD antibody followed by co-incubation with anti-PE beads (Miltenyi). All in vitro assays were performed 7 days after stimulation on CD19+ NIH/3T3 cells. T cells were enumerated using an automated cell counter (Nexcelom Bioscience).
Mouse systemic tumour model
Male or female 8–12-week-old NOD.Cg-PrkdcscidIl2rgtmWjl/SzJ (NSG) mice (Jackson Laboratory) were used, under a protocol approved by the MSKCC Institutional Animal Care and Use Committee, according to all relevant animal use guidelines and ethical regulations. A total of 0.5 × 106 FFLuc-GFP NALM6 cells were administered intravenously by tail vein injection (day −4). Four days later, 1 × 106, 0.4 × 106 or 0.2 × 106 CAR T cells were administered intravenously by tail vein injection (day 0). In some experiments, a second CAR T cell infusion was administered intravenously as indicated in the text. All in vivo assays were performed with bulk transduced CAR T cells except dual antigen targeting experiments. CAR T cells co-transduced with anti-CD19 CAR and anti-CD22 CAR were bead-sorted based on the expression of anti-CD19 CAR cells. Tumour burden was measured by Bioluminescence imaging used the Xenogen IVIS Imaging System (Xenogen). Living Image software (Xenogen) was used to analyse acquired bioluminescence data.
Flow cytometry
All antibodies were titrated. CAR expression was measured with Alexa-Fluor-647-conjugated goat anti-mouse Fab (Jackson ImmunoResearch) or biotinylated protein L (Thermo Scientific) followed by BV510–streptavidin (BD). The phenotype of primary cells and cell lines was determined using the following anti-human antibodies: CD19–PE, CD19–BUV395 (clone SJ25C1, BD), CD19–BV510 (clone SJ25C1, Biolegend), CD22–PE (clone S-HCL-1, BD), CD22–BV421 (clone HIB22, BD), CD81–BV605 (clone JS-81, BD), BCMA–BV421 (clone 19F2, BD), MSLN–Alexa-Fluor-700 (clone 420411, R&D Systems), CD3–BUV737 (clone UCHT, BD), LNGFR–PE, LNGFR–PE–Cy7, LNGFR–Alexa-Fluor-647 (clone C40–1457, BD), PD-1–BV711 (clone EH121, BD), LAG-3–BV650 (clone 11C3C65, Biolegend), TIM-3–BV785 (clone F38–2E2, Biolegend), T-bet–Alexa-Fluor-647 (clone 4B10, Biolegend), EOMES–PE (clone WD1928, eBioscience), CD45–BV711 (clone HI30, BD), CD5–BB515 (clone UCHT2, BD) and CD10–PE (clone HI10a, BD). Countbright beads (Invitrogen) were used to determine the absolute number of cells according to the manufacturer’s protocol. 7-AAD or DAPI was used to exclude dead cells. For fixed cells, eFluor506 fixable viability dye (eBioscience) was used. Fc Receptor Binding Inhibitor Antibody Human (eBioscience) and Fc block Mouse (Miltenyi) were used to block Fc receptors. For intracellular staining, cells were fixed and permeabilized using Intracellular Fixation and Permeabilisation Buffer set (eBioscience) according to the manufacturer’s protocol. Phycoerythrin Fluorescence Quantitation Kit (BD) was used according to the manufacturer’s protocol to determine the number of CD19 and CD22 molecules per cell. Data were collected using BD LSR-II and BD LSR-Fortessa cytometer. Data were analysed with FlowJo Software (Treestar). Cell sorting was performed by a BD FACSAria cell sorter.
Flow cytometric trogocytosis assay
A total of 1 × 105 CAR T cells were co-cultured with target cells in 96-well plates at a 1:1 ratio. After 1, 2 or 4 h of co-culture at 37 °C, cells were washed with FACS buffer containing PBS, 0.5 mM EDTA and 0.5% BSA. After the last wash, cells were suspended in 50 μl of FACS buffer and stained with antibodies. Cells were incubated with staining antibodies for 30 min at 4 °C. Following staining, cells were washed and then analysed by flow cytometry. 7-AAD or DAPI was used to exclude dead cells. Trogocytosis was measured by the surface loss of antigen on target cells and its acquisition on CAR T cells. For trogocytosis inhibition assays, CAR T cells were pre-treated with 1 μM latrunculin A (Sigma-Aldrich) at 37 °C for 15 min before co-incubation with target cells.
Cytotoxicity assays
Bulk cytotoxicity of CAR T cells was determined by standard chromium release (51Cr) assay or luciferase-based assay. For the 51Cr release assay, T cells expressing CD19 were loaded with 51Cr (PerkinElmer) for 1 h at 37 °C. CD19+ T cells were cultured with CAR T cells per well in 96-well plates at different effector:target cell (E:T) ratios for 4 h. Specific 51Cr release was calculated using the formula (51Cr release – spontaneous release)/(maximum release – spontaneous release) × 100. For luciferase-based cytotoxic assay, NALM6 cells expressing FFluc-GFP were co-cultured in 96-well plates with CAR T cells at different E:T ratios for 18 h. Target cells alone were used to determine maximal luciferase expression (relative light units; RLUmax). After 18 h of co-culture, 50 μl luciferase substrate (Bright-Glo, Promega) was added to each well. Emitted light was detected in a luminescence plate reader or Xenogen IVIS Imaging System (Xenogen), and quantified using Living Image software (Xenogen). Lysis was determined as (1 − (RLUsample)/(RLU)) × 100.
Single-cell cytotoxicity assay
For single cell CTL assays in micro-wells, chamber slides containing a micro-well grid (50 × 50 × 50 μm per well; polydimethylsiloxane) were prepared and submerged in 10% FBS in RPMI-1640 medium without phenol red23. Bulk 1 × 104 CAR+ T cells already loaded with tracer dye (CellTrace Violet; Invitrogen) were seeded with unlabelled NALM6 cells at a ratio of 1:1 in the presence of 1.5 μM of propidium iodide (Life Technologies) to enable visualization of cell death. Images were acquired with an AxioObserver.Z1 microscope (Carl Zeiss) using 20×/0.5 or 40×/0.6 objectives. Appropriate excitation and emission filters were chosen to image CellTrace Violet and propidium iodide staining and bright field. Fifteen positions (1 position = 36 micro-wells) for each chamber were imaged every 10 min for 24 h. Acquired images were processed using a custom macro written on ImageJ software. Each individual well was scanned for the presence of T cells. These wells were then analysed to identify wells containing a single target cell and one or two effector T cells (as indicated in the legend of Fig. 3) and interactions were recorded. In wells in which conjugate formation led to target death (lytic conjugates), the duration of interaction between E:T was recorded from the formation (start point, T0) to the interruption of the conjugate (end point, TPI). Abortive T cell contacts (non-lytic conjugates) lasting at least 20 min but not leading up to target cell lysis as reflected by propidium iodide positivity within the 24 h of observation. Interruption of the non-lytic conjugate can be due to either conjugate dissociation or T cell death. Contact events lasting for only 1 frame (10 min) were not included. All analyses were performed in a blinded fashion.
Expected killing frequencies in wells with two T cells (Pest.2/1) were calculated based on the percentage of killing observed in wells with a single target cell and one T cell (Pobs.1/1). The frequency of killing under the hypothesis of independence (that is, simply additive killing) was calculated using the following formula: Pest.2/1 = Pobs.1/1 + (Pobs.1/1 − (Pobs.1/1 ×Pobs.1/1)). Cooperative T cell killing is inferred if Pobs.2/1 > Pest.2/1.
CD19 gene targeting
NALM6 cells were used to generate NALM6med, NALM6low and CD19-knockout NALM6 cells. The gRNA sequence ‘CTAGTGGTGAAGGTGGAAGG’ was cloned into a PBS-gRNA-MCSV2 vector. NALM6 cells were transfected by electrotransfer of Cas9 mRNA and the PBS-gRNA-CD19-MCSV2 vector using an AgilePulse MAX system (Harvard Apparatus). For electroporation, 3 × 106 cells were mixed with 5 μg of Cas9 mRNA and 5 μg of PBS-gRNA-MCSV2 coding for CD19-targeted gRNA into a 0.2-cm cuvette. After electroporation, cells were seeded into culture medium and incubated at 37 °C, 5% CO2. Knockout efficiency for CD19 was assessed by flow cytometry and deep sequencing of the knockout site. The purification of edited cells was performed by FACS sorting. The NALM6med cell line was repeatedly sorted. NALM6low cells were subcloned using single-cell sorting.
Deep sequencing
DNA was extracted using the DNeasy Blood&Tissue kit (Qiagen) according to the manufacturer’s protocol. To confirm and track editing events in the CD19 locus, the region surrounding the site of interest was amplified using the primers (forward: GAGGCTCAGAGAGGGTAAG and reverse: GTGCCCCGGAGAGTCTG). Following end repair and A-tailing, standard Illumina-compatible forked adapters (IDT) were ligated to the amplicon using the Kapa Hyper Library Preparation kit (KK8504). After library preparation, the amplicons were sequenced on a Hiseq 4000, PE125 to a depth of approximately 100,000–1,000,000 reads. Sequencing data were analysed using the Crispresso pipeline and amplicons were automatically counted after trimming using a custom R program script.
Confocal microscopy
CAR T cells expressing a CAR–GFP fusion and NALM6 cells were seeded at 1:1 ratio onto poly-L-lysine-coated glass surface chamber slides. Cells were then co-cultured at 37 °C for 1 h. Cells were fixed by adding of 4% paraformaldehyde into the culture medium (final concentration 1%) and incubating for 15 min. After fixation cells were washed twice with PBS. Cells were stained using automated system Leica Bond RX Protocol F. Monoclonal mouse anti-CD19 (clone BT51E, Leica Microsystems), chicken polyclonal anti-GFP (Abcam) and rabbit polyclonal anti-CD3 (DAKO, Tyramide Alexa-Fluor-488 (Life Technologies), with Tyramide Alexa-Fluor-594 (Life Technologies), and AffiniPure Fab fragment rabbit anti-goat IgG-Alexa-Fluor-647 (Jackson Immunoreserach) were used. For nucleus staining, cells were incubated in 5 μg ml−1 DAPI/PBS solution for 5 min. Slides were mounted using Mowiol fluorescence mounting medium (Mowiol 4-88 Reagent-Calbiochem) prepared in glycerol and Tris-HCl buffer according to the manufacturer’s protocol. Cells were kept in the dark at −20 °C. For imaging, confocal z-stacks were taken at optimal imaging parameters with a LSM 880 confocal microscope with Airyscan with a 63× 1.4 NA oil immersion objective (Carl Zeiss Microimaging). ImageJ software was used to generate the figures.
Transcriptome analysis
Cells were sorted into TRIzol LS (Invitrogen) and then submitted to the Integrated Genomics Operation at MSKCC for RNA extraction. After ribogreen quantification and quality control on a bioAnalyser, 500 ng of total RNA underwent library preparation using the Truseq Stranded Total RNA library preparation chemistry (Illumina), with six cycles of PCR. Samples were barcoded and run on a Hiseq 2500 1T in a 50 bp/50 bp paired-end run, using the TruSeq SBS Kit v3 (Illumina). An average of 51 million paired reads were generated per sample and the percentage of mRNA bases was 58% on average. The output FASTQ data files are mapped to the target genome using the rnaStar aligner which maps reads genomically and resolves reads across splice junctions. We use the two-pass mapping method outlined in which the reads are mapped twice. The first mapping pass uses a list of known annotated junctions from Ensembl. Novel junctions found in the first pass are then added to the known junctions and a second mapping pass is performed (on the second pass the RemoveNoncanoncial flag is used). After mapping, we post-process the output SAM files using the PICARD tools to: add read groups, AddOrReplaceReadGroups, which in additional sorts the file and coverts it to the compressed BAM format. We then compute the expression count matrix from the mapped reads using HTSeq (https://htseq.readthedocs.io/en/release_0.11.1/) and one of several possible gene model databases. The raw count matrix generated by HTSeq are then processed using the R/Bioconductor package DESeq (www-huber.embl.de/users/anders/DESeq), which is used to both normalize the full dataset and analyse differential expression between sample groups.
Quantitative mass spectrometry
Cells were lysed (8 M urea in 200 mM EPPS, pH 8.4, lysis buffer containing Roche protease inhibitor cocktail), sonicated (1 min), centrifuged (8,000g for 10 min at 4 °C) then the supernatant transferred to new tubes and protein concentration determined by BCA (Pierce). Lysates were reduced with dithiothreitol (DTT; 37 °C, 1 h), alkylated with iodoacetamide (room temperature, 30 min, dark), and quenched with an additional 5 mM DTT (room temperature, 15 min, dark). In-solution Lys-C digestion (enzyme:substrate 1:100) was performed at 37 °C for 6 h, then the urea concentration diluted to 1.3 M with buffer and trypsin was added (1:100) and further incubated another 16 h at 37 °C. Enzyme activity was quenched with trifluoroacetic acid and samples were frozen at −80 °C. Based on the BCA assay, 10 μg aliquots were removed from each sample for fractionation.
To fractionate the mixture of peptides, a five-cutter method based on the StageTip technique of Rappsibler using C18 disk (3M Empore Solid Phase Extraction Disk, 2315) was used. Before loading of samples, StageTips were conditioned with 50 μl washes of acetonitrile, 50% acetonitrile in 2.5 mM ammonium bicarbonate, then 2.5 mM ammonium bicarbonate. After washing, fractions 1–5 were eluted with 30 μl of 8, 15, 22, 30 and 50% acetonitrile in 2.5 mM ammonium bicarbonate. Fractions were frozen, lyophilized to dryness, and reconstituted in 5 μl of 0.1% formic acid for liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis.
Fractions were analysed by Microcapillary LC coupled (Waters NanoAcquity, 100-μm i.d. × 10 cm C18 column, 1.7 μm BEH130, configured with a trap column) to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were eluted at 300 nl min−1 using a 4-h acetonitrile gradient (2–50% acetonitrile in water with 0.1% formic acid). The QE Plus was operated in automatic, data-dependent MS/MS mode with one MS full scan (380–1,800 m/z) at 70,000 mass resolution and up to ten MS/MS scans at 17,500 resolution, an isolation window of 1.5 AMU and normalized collision energy of 27. Automatic gain control was set to 3 × 106 for MS1 and 5 × 104 and 60 ms max IT for MS2.
MS data were processed using MaxQuant version 1.5.5.30 and the Uniprot human protein sequence database (downloaded 1 August 2017). Carbamidomethylation of C was set as a fixed modification and the following variable modifications allowed: oxidation (M), N-terminal protein acetylation, deamidation (N and Q), phosphorylation (S, T, Y). Search parameters specified an MS tolerance of 8 p.p.m., an MS/MS tolerance at 40 p.p.m. and full trypsin digestion, allowing for up to two missed cleavages. Peptides were required to be at least seven amino acids in length with 1% false discovery rates calculated at peptides level.
Statistics
All experimental data are presented as mean ± s.e.m. No statistical methods were used to predetermine sample size. Appropriate statistical tests were used to analyse data, as described in the figure legends. Statistical analysis was performed on GraphPad Prism 7 software. The function ‘prop.test’ in R (version 3.5.1) was applied to determine P values when test of proportions with continuity correction was used. Two-sample test of proportions (one-sided) were used to evaluate: H0: Pobs2/1 ≤ Pest2/1 versus H1: Pobs2/1 > Pest2/1. We considered the experiments as a whole and did not account for any batch or donor effect given the limited sample size. Evaluating the percentage of killing within each experimental donor could lead to inconsistent conclusions across donors and more importantly an inflated type 1 error rate. We note that our approach is more conservative by not accounting for the correlation within a donor and would probably result in increased power if we accounted for it. We opted to use a less stringent type 1 error rate of 10% on the aggregate data (significance P < 0.1). We define no cooperative effect (additive effect) to be the null hypothesis, that is, H0: Pobs2/1 ≤ Pest2/1, which assumes T cell killing events are independent in wells containing one target cells and two effector T cells.
Extended Data
Extended Data Fig. 1: In vitro and in vivo characterization of 19-28ζ and 19-BBζ CAR T cells.
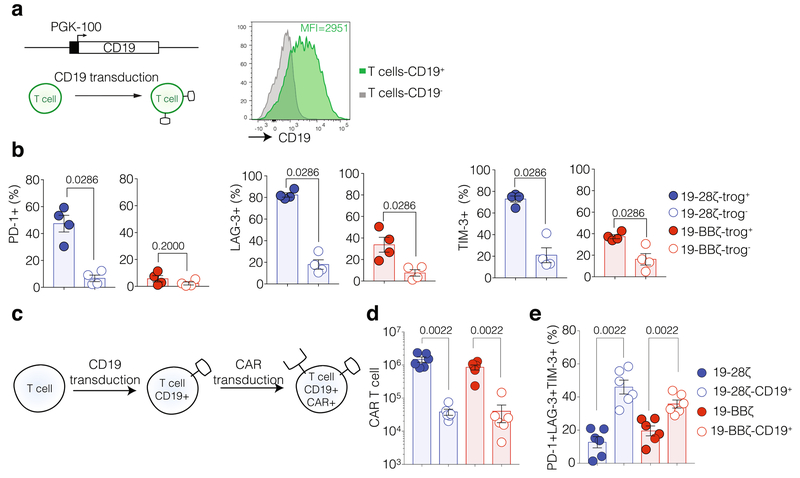
a, Schematic representation of the CD19 CARs. EC, extracellular domain; scFv, single chain variable region specific for anti-CD19; TM, transmembrane domain. b, c, Percentage of CAR+ T cells (left) and MFI of T cell surface CAR expression (right). b, CAR expression analysis performed 4 days after T cell transduction (n = 9 donors). c, CAR expression analysis performed 7 days after stimulation on NIH/3T3 cells expressing CD19 (n = 10 donors). d, Luciferase-based 18-h cytotoxic assay using NALM6 target cells at several E:T ratios (n = 3 donors). e, Kaplan–Meier survival analysis of NALM6 cell-bearing mice treated with 1 × 106, 0.4 × 106 or 0.2 × 106 transduced CAR T cells. Data are pooled from two independent experiments for the 0.2 × 106 CAR T cell dose. Corresponding bioluminescence imaging data are presented in Fig. 1a. f, Representative FACS profile of NALM6 and CAR T cells. NALM6 cells are GFP+; CAR T cells are LNGFR+. Absolute counts of NALM6 cells (left) and CAR T cells (right) at day 14 after infusion (n = 4 mice per group). g, Left, EOMES/T-bet ratio of CAR T cells. Right, fraction of CAR T cells expressing PD-1, LAG-3 and TIM-3 at day 14 after T cell infusion (n = 4 mice per group). In f and g, cells were isolated from the bone marrow of mice treated with 0.2 × 106 CAR T cells. h, Representative FACS profile of cells obtained by the time of relapse, 39 days after T cell infusion (19-28ζ, representative of 3 mice; 19-BBζ, representative of 9 mice). NA, not available (no detectable cells). P values were determined by two-sided Mann–Whitney U-test (b, c, f and g), or log-rank Mantel–Cox test (e). Data are mean ± s.e.m.
Extended Data Fig. 2: Leukaemic response and relapse patterns associated with SJ25C1 and FMC63 scFv-based CARs.
a, Schematic representation of the CD19 CARs using the scFv SJ25C1 or FMC63. b, c, Kaplan–Meier survival analysis of mice bearing NALM6 cells treated with 0.2 × 106transduced CAR T cells. n = 5–7 mice per group pooled from two independent experiments. (One of the groups shown in b is one of two experiments merged in Fig. 1a.) d, Representative FACS profile of cells isolated by the time of relapse (data are representative of n = 3 mice per group). e, f, Representative FACS profile of surface CD19 expression on NALM6 cells isolated by the time of relapse (data are representative of n = 3 mice per group). e, Mice treated with SJ25C1-28ζ or FMC63-28ζ. f, Mice treated with SJ25C1-BBζ or FMC63-BBζ. g, Representative FACS profile of PD-1, LAG-3 and TIM-3 surface expression on BBζ CAR T cells isolated from relapsed mice (data are representative of n = 3 mice per group). P values were determined by log-rank Mantel–Cox test (b, c).
Extended Data Fig. 3: CD19 trogocytosis in vitro and in vivo.
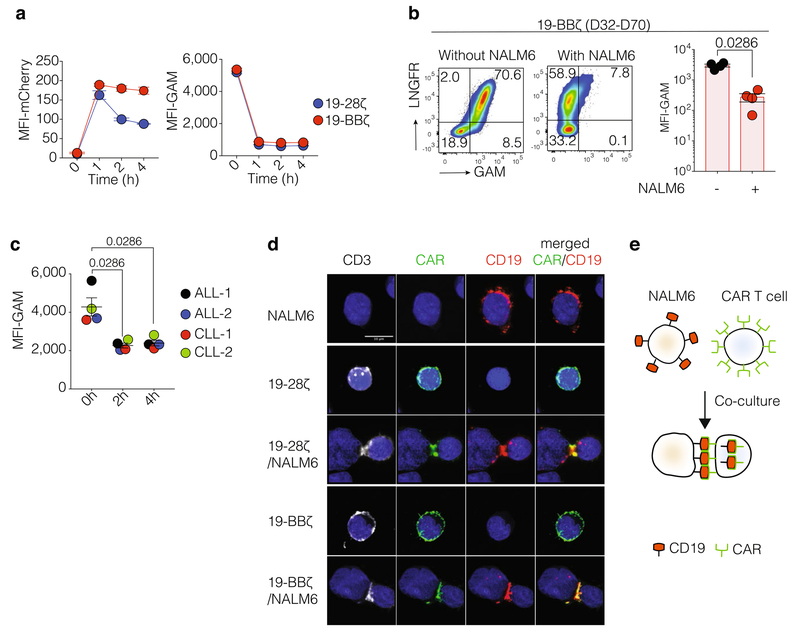
a, Left, representative FACS profile of CD19 staining on CAR T cells isolated by the time of relapse from bone marrow or extramedullary tumour sites of mice treated with 19-BBζ CAR T cells. Right, MFI of CD19 on CAR T cells (19-BBζ, n = 9 mice; control, n = 4 mice). b, Cd19 mRNA expression in NALM6 cells of relapsed mice treated with 19-BBζ CAR T cells (fold change D0 versus NALM6 in vitro = 0.97; adjusted P = 0.919; fold change D6 vs D0 = 0.625, adjusted P = 0.23; n = 7 independent samples). c, MFI of CD19 on the segregated NALM6 cells in a transwell assay. NALM6 cells alone were plated at the top of the plate and NALM6 cells with CAR T cells at the bottom. Data were collected on days 1, 4, 7 and 14 (n = 3 independent samples). d, Left, diagram illustrating the co-culture of NALM6 cells and CAR T cells. Percentage and MFI of CD19 on NALM6 cell surface (middle) and on CAR T cell surface (right). Data were collected at 0, 1, 2 and 4 h after co-culture gated on live singlet cells (n = 4 donors). e, MFI of CD19 on NALM6 cell surface after 2 h of co-culture with 19-28ζ (left) or 19-BBζ (right) CAR T cells treated with the F-actin inhibitor latrunculin A (n = 5 donors). f, Intensity of heavy amino acid CD19-derived peptides detected in the lysates of trog+ and trog− FACS-sorted cells after 1 h of co-culture with NALM6 cells expressing CD19–mCherry. CD19–mCherry-expressing NALM6 cells are used as a control (n = 2 donors). g, MFI of CD81 on NALM6 cell surface co-cultured with anti-CD19 CAR T cells (n = 3 donors). h, MFI of CD22 on NALM6 cell surface co-cultured with anti-CD19 CAR T cells (data are representative of three donors, n = 3 independent samples). i, MFI of CD22 on cell surface of residual NALM6 retrieved from mouse bone marrow 14 days after 19-BBζ CAR T cell treatment (0.2 × 106 CAR T cell dose, n = 3 mice). ns, non-significant. Pvalues were determined by two-sided Mann–Whitney U-test (a, e and i), binomial false discovery rate-adjusted (b), or two-way ANOVA (d). Data are mean ± s.e.m.
Extended Data Fig. 4: Trogocytosis occurs with several targets and cell types.
a, Top, MFI of CD19 on the cell surface of NALM6, SUP-B15, Raji and SK-OV-3-CD19+cells co-cultured with 19-BBζ CAR T cells. Bottom, MFI of CD19 on CAR T cell surface (n = 3 donors). b, MFI of CD19 on NALM6 cells co-cultured with 19-28ζ CAR T cells from patients with ALL and CLL (n = 4 patient samples). c, MFI of CD22 on NALM6 cell surface (top) and CD22 CAR T cells (bottom). BCMA CAR T cells are used as control. d, MFI of BCMA on KMS-12-BM cell surface (top) and BCMA CAR T cells (bottom). CD22 CAR T cells are used as a control. e, MFI of MSLN on A549 MSLN+ cell surface (top) and MSLN CAR T cells (bottom). CD19 CAR T cells are used as a control. In c–e, n = 3 independent samples, and data are representative of three donors. Data are mean ± s.e.m.
Extended Data Fig. 5: Therapeutic response and relapse patterns after CD22 CAR T cell treatment.
a, Tumour burden was monitored using bioluminescence imaging (average radiance (photons s−1 cm−2 sr−1)) in mice bearing NALM6–GFP luciferase treated with 0.2 × 106CAR T cells 4 days later (n = 7 mice per group). b, Absolute count of CAR T cells. c, Quantification of CD22 molecules on NALM6 cell surface. d, Expression of PD-1, LAG-3 and TIM-3 on CAR T cell surface. b–d, Cells were obtained from mice bone marrow at day 21 (relapse time, n = 5 mice per group). e, Quantification of CD22 molecules on NALM6 surface 6 days after ex vivo culture (CD22-28ζ and CD22-BBζ, n = 5 independent samples per group, NALM6, n = 3 independent samples) . NA, not available (no detectable cells). P values were determined by two-sided Mann–Whitney U-test. Data are mean ± s.e.m.
Extended Data Fig. 6: Functional effects of CD19 acquisition by CAR T cells.
a, Diagram illustrating transduction of T cells with retroviral vector encoding CD19 under a PGK-100 promoter. Representative flow cytometry profile of CD19 expression on transduced T cells (n = 6 donors). b, Percentage of PD-1, LAG-3 and TIM-3 expression in trog+ and trog− fractions (n = 4 donors). c, Diagram illustrating co-expression of CD19 and CAR on T cells. T cells were transduced with SFG vector expressing CD19 followed by CAR transduction 24 h later. After transduction, cells were left in culture for 6 days. d, Absolute count of CAR T cells (n = 6 donors). e, Percentage of T cells expressing of PD-1, LAG-3 and TIM-3 (n = 6 donors). P values were determined by two-sided Mann–Whitney U-test. Data are mean ± s.e.m.
Extended Data Fig. 7: Internalized CD19–CAR complex after trogocytosis.
a, Left, MFI of mCherry gated on CAR T cells after co-culture with NALM6 cells expressing CD19–mCherry. Right, MFI of goat anti-mouse IgG (GAM) gated on CAR T cells (n = 3 independent samples, data are representative of three donors). b, Left, representative FACS profile of CAR T cells stained with GAM and LNGFR in the presence or absence of NALM6 cells (data are representative of n = 4 mice per group). Right, MFI of GAM on CAR T cells isolated from mice bone marrow or extramedullary tumour sites by the time of relapse (n = 4 mice per group). c, MFI of GAM on CAR T cells co-cultured with autologous blasts derived from refractory/relapsed patients with ALL and CLL (n = 4 patient samples). d, Representative confocal microscopy images of NALM6 cell–CAR T cell conjugates. Data are representative of three donors (ten cells per experiments). e, Diagram illustrating CD19 trogocytosis by CAR T cells. Pvalues were determined by two-sided Mann–Whitney U-test. Data are mean ± s.e.m.
Extended Data Fig. 8: Generation of NALM6 cell lines with graded CD19 expression.
a, Genotype of edited CD19 locus in NALM6med (top) and NALM6low (bottom) cells. NALM6med and NALM6low cells were obtained by monoallelic and biallelic disruption of the Cd19 gene, respectively, using the CRISPR–Cas9 system. One of the alleles encodes a stop codon (asterisk); the second allele codes for a variant CD19 containing an amino acid substitution (Mut-1, 2 or 3aa). b, Prediction of the cleavage of CD19 protein sequence of the functional allele (Mut-3aa) in NALM6low cells (top) and the wild-type (WT) protein sequence (bottom) using signal 4.1 server. Cd19 gRNA was designed to target Cd19 exon 1, next to the site of cleavage of signal peptide. Data were obtained by deep sequencing. c, MFI of CD19 on the surface of NALM6 cells (NALM6wt), NALM6medcells, NALM6low cells and CD19-knockout NALM6 cells (NALM6KO) (n = 3 independent experiments). d, MFI of CD22 on the surface of NALM6 cells, NALM6med cells and NALM6low cell (n = 3 independent experiments). Data are mean ± s.e.m.
Extended Data Fig. 9: T cell cooperativity and antigen density determine CAR T cell killing capacity.
Tumour lysis frequency in wells containing a ratio of one CAR T cell:one tumour cell (R1:1 E:T) and one CAR T cell:two tumour cells (R2:1 E:T). a, c, d, NALM6 cells (a), NALM6med cells (c) and NALM6low cells (d). b, Estimated and observed killing percentage in wells containing one CAR T cell and two NALM6 cells (R2:1 E:T). Estimated percentage of killing is calculated as described in the Methods. P values were determined by two-sided two-sample test of proportions (a, c and d) and one-sided two-sample test of proportions (b), n = 5 donors (a), and n = 3 donors (c, d). In b, P < 0.1 is used as significance level (n = 5 donors). Data are mean ± s.e.m. Primary data in this figure are presented in Fig. 3.
Extended Data Fig. 10: Modelling of anticipated outcomes after single or combinatorial CAR T cell treatment schemas.
a–h, Antigen-positive relapse (a–d) versus tumour control scenarios (e–h) after single or combinatorial CAR T cell targeting. a, Antigen high relapse of residual tumour cells in the absence of CAR T cells. b, Antigen-low relapse in the presence of insensitive or exhausted CAR T cells. c, Tumour relapse after sequential combinatorial targeting failing against tumour cells with low antigen densities. Tumour rejection could overcome the relapse scenarios (a–c) by achieving a higher effector:target ratio (e–g), after higher T cell dose infusion or greater post-infusion expansion, operating through additive or cooperative tumour elimination. In d and h, combinatorial targeting is mediated by dual-targeted CAR T cells, which may still fail in the face of low antigen densities (d) depending on the co-stimulatory combinations (h). In all cases, low antigen density may be either constitutively low or actively lowered after CAR T cell-mediated trogocytosis. Antigen-negative escapes are not represented. Other, yet undiscovered, escape mechanisms may exist.
Acknowledgements
We thank R. Soni, B. Sénéchal, B. J. Safford, P. Vedantam, F. Tamzalit and G. Gunset for logistical and technical assistance. We also thank the SKI Cell Therapy and Cell Engineering, Molecular Cytology, Flow Cytometry, Integrated Genomics Operation, Microchemistry and Proteomics, Antitumor Assessment and Animal Core Facilities for their expert assistance. This work was supported by the Lake Road Foundation, the Lymphoma and Leukaemia Society, the Pasteur-Weizmann/Servier award and the NCI Cancer Center Support Grant P30 CA008748. SKI cores were in part supported by the Tow Foundation, Cycle for Survival, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology and NCI grant P30 CA08748. A.D. and T.G. were supported by fellowships from The Canadian Institutes of Health Research and the Alexander S. Onassis Public Benefit Foundation, respectively.
Reviewer information
Nature thanks Wolfgang Schamel and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Footnotes
Data availability
All in vivo data can be found in the Source Data. The RNA-sequencing data are available from Gene Expression Omnibus (GEO) under accession number GSE126753. The proteomics data are available from PRIDE database under accession number PXD012833. Other data generated for this manuscript are available from the corresponding author upon reasonable request.
Competing interests
Memorial Sloan Kettering has submitted patent applications based in part on results presented in this manuscript (PCT/US18/68134, which has been licensed to several companies; M. Hamieh, J.M.-S. and M.S. are listed among the inventors; US provisional application no. 62/807,181; M. Hamieh and M.S. are listed among the inventors). R.C.H. reports stock ownership in Merck. I.R. and M.S. report research funding from Juno Therapeutics, Fate Therapeutics, Takeda Pharmaceuticals and Atara Biotherapeutics. I.R. serves on the scientific advisory board of Flow Design and M.S. on those of Berkeley Lights and St Jude Children Research Hospital.
References
- 1.Sadelain M, Rivière I & Riddell S Therapeutic T cell engineering. Nature 545, 423–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June CH & Sadelain M Chimeric antigen receptor therapy. N. Engl. J. Med 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brudno JN & Kochenderfer JN Chimeric antigen receptor T-cell therapies for lymphoma. Nat. Rev. Clin. Oncol 15, 31–46 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Brudno JN et al. T cells genetically modified to express an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J. Clin. Oncol 36, 2267–2280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majzner RG & Mackall CL Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 8, 1219–1226 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Sotillo E et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 5, 1282–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fry TJ et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med 24, 20–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner R et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlando EJ et al. Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat. Med 24, 1504–1506 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Brentjens RJ et al. Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat. Med 9, 279–286 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Barrett DM et al. Noninvasive bioluminescent imaging of primary patient acute lymphoblastic leukemia: a strategy for preclinical modeling. Blood 118, e112–e117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Z et al. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell 28, 415–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DW et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maude SL et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turtle CJ et al. CD19 CAR-T cells of defined CD4+:CD8+composition in adult B cell ALL patients. J. Clin. Invest 126, 2123–2138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JH et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med 378, 449–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Martín N et al. T cell receptor internalization from the immunological synapse is mediated by TC21 and RhoG GTPase-dependent phagocytosis. Immunity 35, 208–222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priceman SJ et al. Co-stimulatory signaling determines tumor antigen sensitivity and persistence of CAR T cells targeting PSCA+ metastatic prostate cancer. OncoImmunology 7, e1380764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halle S et al. In vivo killing capacity of cytotoxic T cells is limited and involves dynamic interactions and t cell cooperativity. Immunity 44, 233–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adusumilli PS et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci. Transl. Med 6, 261ra151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyquem J et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brentjens RJ et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118, 4817–4828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Floc’h A et al. Annular PIP3 accumulation controls actin architecture and modulates cytotoxicity at the immunological synapse. J. Exp. Med 210, 2721–2737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]