Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignant tumors worldwide and the 5-year overall survival rate remains poor. Protein kinase, membrane associated tyrosine/threonine (PKMYT1) is overexpressed in several cancers and participate in tumor progression. However, the mechanism of PKMYT1 in ESCC is unclear.
Purpose
The objective of our study was to demonstrate the the expression and role of PKMYT1 in ESCC.
Patients and methods
We detected the expression of PKMYT1 in ESCC patients and analysed the correlation with overall survival time and disease-free survival time. Then we detected PKMYT1 expression in ESCC cell lines and immortalized human esophageal epithelial cell line. Down-regulated PKMYT1 was carried out in KYSE70 and KYSE450 cells to invetigate the mechanism of PKMYT1 in ESCC cells.
Results
PKMYT1 was up-regulated in tumor tissues and ESCC cell lines, and higher expression of PKMYT1 correlated with poorer overall survival in ESCC patients. Besides, in ESCC cell lines KYSE70 and KYSE450, knocking down PKMYT1 allowed more cells to skip G2/M checkpoint to complete mitosis, which promoted cell apoptosis, inhibited cell proliferation, and prevented the EMT phenotype in vitro. Meantime, we also observed that down-regulated PKMYT1 in ESCC cells suppressed AKT/mTOR signaling pathway. These results demonstrated PKMYT1 may act as an oncogene in ESCC.
Conclusion
PKMYT1 plays an crutial role in ESCC progression, downregulated PKMYT1 might inhibit the development of ESCC by AKT/mTOR signaling pathway, and might be a novel target in the treatment of ESCC.
Keywords: ESCC, PKMYT1, AKT, EMT, cell apoptosis
Introduction
Esophageal carcinoma (EC) is the ninth common carcinoma and the sixth leading cause of cancer-related death all over the world.1–3 The histological type of EC includes esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma, with ESCC accounting for the majority of EC patients in China.4–6 With the evolution of the multidisciplinary treatment, including novel surgical approaches, chemotherapy, radiotherapy, and molecular-targeted therapy, the 5-year overall survival rate remains poor.7 Thus, it is necessary and urgent to explore the pathogenesis of ESCC which might provide new insights into the treatment of ESCC.
Protein kinase, membrane associated tyrosine/threonine (PKMYT1) is a member of the Wee family of protein kinases. PKMYT1 acts as a negative regulator of the cell cycle by inactivating the CDK1-cyclinB complex through phosphorylation of Tyr14/Tyr15 and preventing cell cycle entering into mitosis at the G2/M transition.8,9 In cell cycle, there will be a large number of DNA mismatches and DNA defect when DNA is duplicated. Normal cells repair DNA at G1/S checkpoint, whereas tumor cells repair DNA and shun immature cells entry into mitosis to prevent mitotic catastrophe at G2/M checkpoint because they have abrogated G1/S checkpoint due to the mutation of P53.10,11 Therefore, PKMYT1 is more essential for tumor cells than normal cells.12 Nowadays, a new tendency for cancer therapy is to keep cell cycle with unrepaired DNA damage in premature mitosis, which induces apoptotic or non-apoptotic cell death.13,14 Previous studies have demonstrated that up-regulated PKMYT1 plays an important role in the progression of these malignancies such as hepatic carcinoma, colon cancer, glioblastoma.15–18 A high frequency of TP53 mutations has been found in ESCC patients,19,20 but how PKMYT1 participates in the pathogenesis and development of ESCC have not been fully investigated. In this study, we found that PKMYT1 was highly expressed in ESCC cell lines and ESCC patients, and high PKMYT1 expression induced poorer prognosis in patients with ESCC. Then, we investigated its biological functions in ESCC cells. Our results indicated that PKMYT1 might be a potential target in the treatment of ESCC.
Materials and methods
Patients
Totally, 60 ESCC patients tissues including paired tumor tissues and normal adjacent tissues (>5 cm from tumor) were collected in the First Affiliated Hospital of Zhengzhou University from February 2018 to October 2018. Fresh tissues were immediately frozen at −80°C for RNA extraction. Besides, we collected histopathological sections of 104 patients who underwent radical resection of esophageal cancer from 2014 to 2017 to perform immunohistochemistry. All patients in this subject had none preoperative treatments including chemotherapy and radiotherapy. This study was approved by the Ethics Committee Board of the First Affiliated Hospital of Zhengzhou University and all patients signed written informed consent. And all these assays were conducted in accordance with the Declaration of Helsinki.
Expression profiling and interactive analyses (GEPIA) database and Kaplan–Meier plotter database
GEPIA is a new Bioinformatics tool for analyzing the expression of mRNA by The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects (http://gepia.cancer-pku.cn/).21 We analyzed the mRNA expression of PKMYT1 in ESCC tumor tissues and normal tissues, then we also explored the correlation between PKMYT1 expression and TWIST, Ki67, and AKT expression in GEPIA. Kaplan–Meier Plotter was performed to analyze the correlation between the expression of PKMYT1 and survival time in breast cancer, lung cancer, and gastric cancer, and this database had 54,675 genes and 10,461 cancer samples, which included 5143 breast, 1186 ovarian, 2437 lung, and 1065 gastric cancer samples (http://kmplot.com/analysis/).22
Cell lines and cell culture
Both ESCC cell lines (KYSE450, KYSE70) and immortalized human esophageal epithelial cell line HET-1A were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All ESCC cells and HET-1A cells were cultured in RMPI1640 medium (Hyclone, SH30809.01B, Logan, UT, USA) supplemented with 10% fetal bovine serum (Hyclone, FBS, Logan, UT, USA), 100 units/mL penicillin, and 100 µg/mL streptomycin. All cells were maintained in humidified atmosphere with 5% carbon dioxide at 37°C.
siRNA and cell transfection
siRNAs targeting PKMYT1 were synthesized by GenePharma (Shanghai, China), the sequence of siRNAs was listed in Table 1. Lipofectamine 3000 (Invitrogen, Lipofectamine 3000, Carlsbad, CA, USA) was used for cell transfection according to the manufacturer’s protocol. We transfected ESCC cells in a six-hole plate, and 48 hrs after transfection, we collected ESCC cells for further assays.
Table 1.
Sequences of siRNAs
| Name | Sequence of siRNA (5ʹ–3ʹ) |
|---|---|
| PKMYT1-868 | CCUACGGAGAGGUCUUCAATTUUGAAGACCUCUCCGUAGGTT |
| PKMYT1-1396 | UCCUGGAAGUGGCAUGCAATTUUGCAUGCCACUUCCAGGATT |
| PKMYT1-NC | UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT |
Abbreviation: PKMYT1, protein kinase, membrane associated tyrosine/threonine.
RNA extraction, reverse transcription, and quantitative real-time PCR (qRT-PCR)
All RNA was isolated from cells or tissues using Trizol (Invitrogen, TRIzol RNA Isolation Reagents, Carlsbad, CA, USA) according to the manufacturer’s protocol. Whole cDNA was synthesized from 1 µg RNA using the PrimescriptTMRT reagent Kit (Takara, PrimescriptTMRT reagent Kit, Beijing, China). The level of PKMYT1 mRNA was assessed by qRT-PCR using FastStart Essential DNA Green Master (Roche, Penzberg, Upper Bavaria, Germany) according to the manufacturer’s instructions. Glyceraldehyde 3-phosphate dehydrogenase was used for normalization of data and these data were analyzed by 2−ΔΔCT. The primers sequences (Sangon Biotech, Shanghai, China) for the RT-PCR analyses are listed in Table 2.
Table 2.
Primers used for RT-qPCR
| Genes | Primers sequences |
|---|---|
| PKMYT1 | |
| Forward | 5ʹ-CATGGCTCCTACGGAGAGGT-3ʹ |
| Reverse | 5ʹ-ACATGGAACGCTTTACCGCAT-3ʹ |
| GAPDH | |
| Forward | 5ʹ-GGGT GTGAAC CATGAGAAGT-3′ |
| Reverse | 5ʹ-GGCATG GACTGTGGTCATGA-3′ |
Abbreviations: PKMYT1, protein kinase, membrane associated tyrosine/threonine; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Cell cycle
After the transfection, 1×106 were collected for flow cytometry, cell cycle detection kit (KeyGEN BioTECH, KGA512, Nanjing, Jiangsu Province, China) was used in this assay according to the manufacturer’s protocol. All data were analyzed by Modfit software.
CCK8
CCK8 assay was measured by Cell Counting Kit-8 (Dojindo, CCK-8, Shanghai, China). Transfected ESCC cells were seeded in 96-well plate for 1000 cells per well. Cells were cultured for 24, 48, 72, 96 hrs, and 10 μL of CCK-8 was added into each well. The absorbance was read at 450 nm (Thermo Scientific, Multiskan MK3, Waltham, MA, USA) after incubation for 4 hrs at 37°C.
Colony formation
In the colony formation assay, 2000 cells were plated in six-well plate and cultured for 2 weeks at 37°C in humidified atmosphere with 5% carbon dioxide. Four percent paraformaldehyde was used to fix cell colonies, and cell colonies were stained in 0.5% crystal violet for 20 mins. Then the cell colonies were counted and analyzed.
Cell apoptosis
One hundred thousand ESCC cells were plated into 24-well plates and harvested 24 hrs later. Cell apoptosis was detected using propidium iodide/Annexin-V-FITC (Biolegend, San Diego, CA, USA), then flow cytometry was performed, and Flowjo software (Tree Star Software, San Carlos, CA, USA), USA) was used to analyze the results. All the assays were performed in triplicate.
Migration and invasion
The migration assay was performed by using 8 μm transwell chamber (Corning Costar corporation, Corning, NewYork, NY, USA). 1×105 ESCC cells were plated into the upper chamber with serum-free medium, and medium containing 10% fetal bovine serum was added into the lower chamber. After incubating for 48 hrs, upper chamber cells were scraped off and the migration cells were fixed in 4% paraformaldehyde, and then stained in 0.5% crystal violet. The migration cells were counted under a microscope (Olympus Corporation, IX73, Tokyo, Japan). Matrigel (Corning Life Science, C-Matrigel, NewYork, NY, USA) was used to measure the invasion ability of ESCC cells, 50 μL matrigel was plated in the upper chamber, the other procedures were the same as migration assay.
Wound healing
When there was 80–90% confluence in six-well plates, 200 μL pipette was used to scratch the cells, and then PBS was used to remove floating cells and detached cells. Serum-free medium was added into plates, and photos were photographed using a microscope in 0, 12, and 24 hrs to analyze cell migration.
Western blot
All the protein was collected by using RIPA lysis buffer (Beyotime Biotechnology, P0013, Shanghai, China). Protein was quantified using Coomassie blue staining in microplate spectrophotometer (Thermo Scientific, Multiskan MK3, Waltham, MA, USA) at 594 nm. Identical amount of protein was separated into 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then the gels were transferred onto nitrocellulose membrane (GE Healthcare Life Sciences, Amersham™ Protran™ 0.45 NC, Chicago, IL, USA) after electrophoresis. Membranes were blocked by 5% skim milk for 2 hrs. Then, the membranes were incubated with primary antibodies (E-cadherin (ECAD), N-cadherin (NCAD), Vimentin (VIM), and β-actin were purchased from Proteintech group, Wuhan, China; PKMYT1, AKT, mTOR, S6, Phospho-AKT, Phospho-mTOR, Phospho-S6, and Twist1 were purchased from Cell Signaling Technology, Danvers, MA, USA) in 5% bovine serum albumin overnight at 4°C. After rewarming to room temperature for 1 hr, the membranes were incubated with secondary antibody for 1 hrat 37°C. We used enhanced chemiluminescence (Beyotime Biotechnology, Shanghai, China) to detect the blots with ChemiDoc MP (Bio-Rad Laboratories, ChemiDoc MP, Hercules, CA, USA) and the images were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Immunofluorescence
Two thousand cells were seeded into 24-well plates, and then fixed in 4% paraformaldehyde and permeabilized by 0.5% Triton X-100. Five percent goat serum was used to block cells for 1 hr, and cells were incubated with primary antibodies (ECAD, NCAD, VIM) overnight at 4°C. Then, the cells were incubated with secondary antibodies (Biolegend, San Diego, CA, USA) for 1 hr, and then stained with 4ʹ,6-diamidino-2-phenylindole (DAPI) (Biolegend, San Diego, CA, USA). Images were taken under a fluorescence microscope and analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry (IHC)
Patients tissues were fixed with formalin and embedded in paraffin. After dewaxing and hydration, tissue sections were incubated with antigen retrieval solution. Then, the tissue sections were treated with 3% H2O2 and 5% goat serum. Sections were incubated with primary antibody (PKMYT1, 1:50) at 4°C overnight. After rewarming to room temperature for 1 hr, the sections were marked with secondary biotinylated antibody, and diaminobenzidine was used as a chromogen. Then, we used hematoxylin for nuclear counterstain. Subsequently, the sections were observed by microscope and all images were at 200× magnification and assessed by two pathologists independently using a semi-quantitative immunoreactivity score (IRS). According to the proportion of positive tumor cells examined, the tissues were scored as 0 (no positive tumor cells), 1 (0–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). Staining intensity was scored as 0 (no staining), 1 (week staining), 2 (intermediate staining), or 3 (strong staining). IRS was evaluated as staining intensity × proportion of positive tumor cells that ranging between 0 and 12. Patients with a total score<4 were considered low PKMYT1 expression and total score ≥4 was considered as high PKMYT1 expression. We divided these patients into two groups, high PKMYT1 group (n=50), and low PKMYT1 group (N=54).
Statistical analysis
Statistical analysis was performed by using SPSS 22.0 (IBM Corporation, Armonk, NY, USA) program. The results of RT-PCR, apoptosis, migration, and invasion were analyzed with Student’s t-test. Chi-square tests were conducted for analyzing the associations of PKMYT1 and clinicopathological data. The overall survival rate and disease-free survival rate were analyzed by Kaplan–Meier curve with the log-rank test method. Cox regression models were used to analyze the clinicopathological data. P<0.05 was considered to indicate statistically significant differences.
Results
Expression of PKMYT1 in ESCC and its association with clinical-pathological features
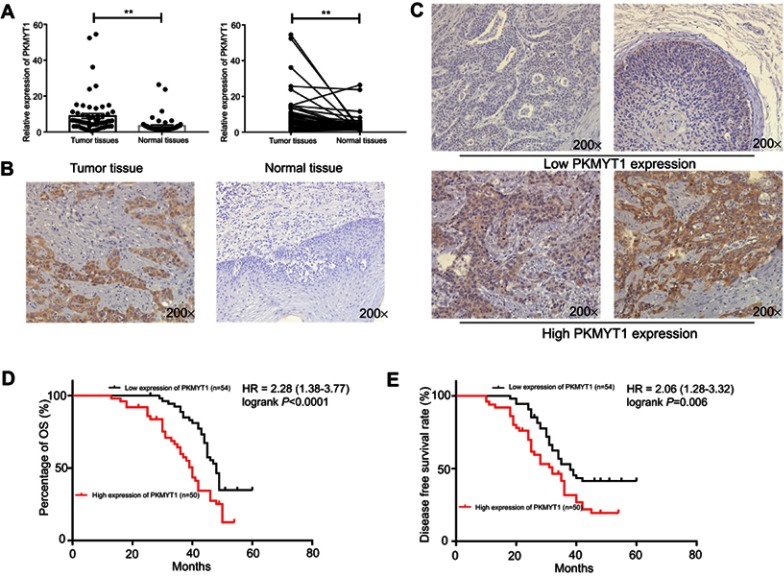
To investigate the expression of PKMYT1 in ESCC patients, RT-PCR and IHC assays were carried out with ESCC patients’ paired tissues. The results indicated that the expression levels of PKMYT1 in tumor tissues were higher than the one in the paired normal adjacent tissues (Figure 1A and B). We then analyzed the expression of PKMYT1 in ESCC cell lines and HET-1A cells by RT-PCR, showing that PKMYT1 expression in ESCC cells was significantly higher than HET-1 cells (Figure 3A). These results demonstrated that PKMYT1 was up-regulated in ESCC. Then, we analyzed the correlation between PKMYT1 and clinicopathological parameters of patients. The results revealed that the protein expression of PKMYT1 in ESCC tissues was associated with lymph nodes metastasis (P=0.018), tumor differentiation (P=0.018), and pathological stage (P=0.014). However, there were no correlations between PKMYT1 with tumor invasion depth, age, cigarette or alcohol intake, and gender (Table 3).
Figure 1.
PKMYT1 is up-regulated and associated with prognosis in patients with ESCC. (A) The RT-qPCR results indicated that PKMYT1 is up-regulated in ESCC tumor tissues compared with normal tissues. (B) The IHC results indicated PKMYT1 was up-regulated in ESCC tumor tissues than normal adjacent tissues. (C) According to the scores and staining of IHC, IHC score (Left upper:0; Right upper:2; Left lower:8; Right lower:12), we divided these patients to two groups. (D, E) High expression of PKMYT1 related to lower overall survival time and disease-free survival time. **P<0.01.
Abbreviations: PKMYT1, protein kinase, membrane associated tyrosine/threonine; ESCC, esophageal squamous cell carcinoma; IHC, immunohistochemistry; OS, overall survival.
Figure 3.
PKMYT1 is overexpressed in ESCC cell lines, and silenced PKMYT1 contributes to decreased G2/M ratio in cell cycle, inhibits proliferation in ESCC cells and promotes cell apoptosis. (A) The RT-qPCR results indicated that PKMYT1 was up-regulated in ESCC cell lines than normal cell line. (B) PKMYT1 was inhibited by siRNAs. (C) Down-regulated the expression of PKMYT1 in ESCC cells decreased G2/M ratio in cell cycle. (D, E) CCK8 assays (D) and colon assays (E) indicated that down-regulated the expression of PKMYT1 inhibited cell proliferation in ESCC cells.(F) Flow cytometry showed inhibited the expression of PKMYT1 contributed to high apoptosis rate in ESCC cells. (G) Western blotting indicated that the expression of apoptosis-related proteins was changed, the expression of Bcl-2 was decreased but the expression of Bax and cleaved caspase 3 was increased. *P<0.05, **P<0.01, *** P<0.001,****P<0.0001.
Abbreviations: PKMYT1, protein kinase, membrane associated tyrosine/threonine; ESCC, esophageal squamous cell carcinoma; NC, negative control.
Table 3.
The correlation between PKMYT1 expression and clinicopathological factors in ESCC patients
| Clinic factors | All cases | PKMYT1 expression | P-value | |
|---|---|---|---|---|
| High (n=50) | Low (n=54) | |||
| Age (years) | 0.327 | |||
| <65 | 52 | 28 | 24 | |
| ≥65 | 52 | 22 | 30 | |
| Gender | 0.416 | |||
| Male | 67 | 30 | 37 | |
| Female | 37 | 20 | 17 | |
| Cigarette intake | 0.325 | |||
| Yes | 48 | 26 | 22 | |
| No | 56 | 24 | 32 | |
| Alcohol intake | 0.436 | |||
| Yes | 53 | 28 | 25 | |
| No | 51 | 22 | 29 | |
| Tumor invasion depth | 0.326 | |||
| T1+T2 | 54 | 23 | 31 | |
| T3+T4 | 50 | 27 | 23 | |
| Lymph nodes metastasis | 0.018 | |||
| Positive | 51 | 31 | 20 | |
| Negative | 53 | 19 | 34 | |
| Differentiation | 0.018 | |||
| Poor | 60 | 35 | 25 | |
| Moderate/Well | 44 | 15 | 29 | |
| TNM stage | 0.014 | |||
| I-II | 52 | 18 | 34 | |
| III-IV | 52 | 32 | 20 | |
Note: The bold values mean that the P-value<0.05.
Abbreviations: ESCC, esophageal squamous cell carcinoma; PKMYT1, protein kinase, membrane associated tyrosine/threonine.
PKMYT1 with clinical outcomes in patients with ESCC
We used Kaplan–Meier method and log-rank test to assess whether the expression of PKMYT1 was related to the prognosis of ESCC patients. The results of IHC demonstrated that high PKMYT1 expression was significantly related to poor overall survival in patients with ESCC (Figure 1C and D), and high PKMYT1 expression also related to shorter disease-free time (Figure 1E). Then, we used Cox proportional hazards model with univariate and multivariate analysis to verify whether PKMYT1 can be used as an independent risk factor, these results indicated that high PKMYT1 expression was an independent risk factor (P=0.014) in patients with ESCC (Table 4). All these results indicated that PKMYT1 associated with the prognosis of patients with ESCC.
Table 4.
Univariate and multivariate Cox regression analysis of the relative risk of death according to the expression of PKMYT1
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR(95% CI) | P-value | HR(95% CI) | P-value | |
| Age | 1.769(1.083–2.889) | 0.023 | 1.595(0.929–2.74) | 0.091 |
| Gender | 0.706(0.420–1.187) | 0.189 | 1.506(0.894–2.537) | 0.124 |
| Tumor invasion depth | 1.544(0.950–2.510) | 0.079 | 2.007(1.167–3.452) | 0.012 |
| Differentiation grade | 3.309(1.870–5.857) | 0.000 | 2.973(1.587–5.570) | 0.001 |
| Lymph nodes metastasis | 8.966(5.037–15.959) | 0.000 | 10.725(4.664–24.662) | 0.000 |
| TNM stage | 7.828(4.445–13.785) | 0.000 | 1.938(0.473–7.935) | 0.358 |
| PKMYT1 expression | 2.376(1.453–3.883) | 0.001 | 1.698 (1.02–2.827) | 0.042 |
Note: The bold values mean that the P-value<0.05.
Abbreviation: PKMYT1, protein kinase, membrane associated tyrosine/threonine.
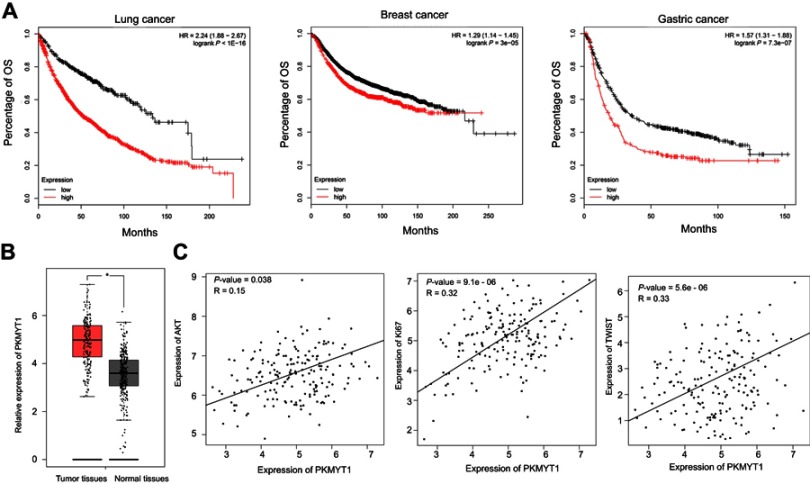
Also, we performed in-silico analysis of microarray gene expression data using an online survival analysis tool (data from http://kmplot.com/analysis/) to assess the prognostic effect of PKMYT1 in lung cancer, breast cancer, and gastric cancer, showing that high expression of PKMYT1 was related to poor prognosis (Figure 2A). Moreover, using a web server for cancer and normal gene GEPIA, we found that PKMYT1 expression was significantly higher in ESCC tumor tissues than in normal tissues (Figure 2B). The expression of PKMYT1 associated with some important genes of ESCC, such as Ki67, TWIST,23,24 AKT (Figure 2C).25,26
Figure 2.
PKMYT1 associates with prognosis in several cancers and correlates with several crucial functional genes in ESCC. (A) Data from Kaplan–Meier Plotter showed that overexpressed of PKMYT1 related to poorer prognosis in several cancers (Copyright © 2018. Balazs Gyorffy. Data from Nagy A, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):922752). (B) PKMYT1 was up-regulated in tumor tissues than normal tissues (Data from Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W10221). (C) PKMYT1 associated with several biomarkers which promoted tumorigenesis and metastasis in ESCC (Data from Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W10221). *P<0.05.
Abbreviations: PKMYT1, protein kinase, membrane associated tyrosine/threonine; ESCC, esophageal squamous cell carcinoma; OS, overall survival.
Effect of PKMYT1 knockdown on cell cycle, cell proliferation, and cell apoptosis
To further explore the function of PKMYT1, KYSE70, KYSE450 cell lines were chosen for the subsequent experiments. Two different siRNAs were designed and transfected into those ESCC cells to knockdown PKMYT1 in ESCC cells (Figure 3B). Forty-eight hours after the transfection, cell cycle assays indicated that knocked down PKMYT1 expression reduced the G2/M phase in cell cycle (Figure 3C), and CCK-8 assays showed that knockdown of PKMYT1 significantly suppressed cell proliferation in KYSE450 cells compared with negative control group (Figure 3D). This observation was further supported by reduced numbers of colony formation upon PKMYT1 knockdown (Figure 3E). Additionally, we observed that the negative control group had lower apoptosis rate than siRNA groups in KYSE450 cells (Figure 3F), and Western blot analysis indicated that the expression of Bcl-2 was decreased, but the expression of Bax and cleaved caspase3 wasincreased (Figure 3G). Similar results were obtained when performing these analyses in KYSE70 cells following PKMYT1 knockdown. All these assays showed that inhibited the expression of PKMYT1 promoted ESCC cell apoptosis and reduced the proliferation of ESCC cells.
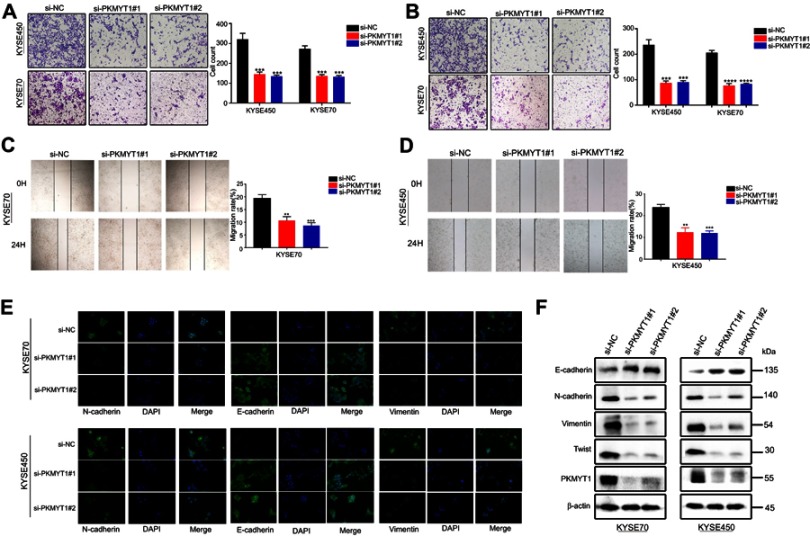
Effect of PKMYT1 knockdown on invasion and migration and PKMYT1 implication in establishing the mesenchymal phenotype ESCC cells
Transwell assays were used to investigate the effects of knockdown PKMYT1 on the invasion and migration capability in ESCC cells. The transwell assays showed that there were fewer cells migrated in siRNA group than negative group in KYSE450 and KYSE70 cells (Figure 4A and B), indicating that PKMYT1 knockdown could inhibit the migration and invasion capacities of ESCC cells. Similarly, the wound healing assays revealed that the siRNA groups had less migrated cell proportions compared to the control groups in both ESCC cell lines (Figure 4C and D). All these results indicated that PKMYT1 might involve in metastasis of esophageal cancer. To further investigate how PKMYT1 affect metastasis capability of ESCC cells, we detected epithelial-mesenchymal transition (EMT) phenotype, an important pathway in tumor progression and metastasis. The necessary markers of EMT phenotype NCAD, ECAD, and VIM were detected by immunofluorescence. In KYSE450 cells, the protein level of ECAD was up-regulated while NCAD, and VIM levels were reduced in the siRNA groups. Parallel results were observed in KYSE70 cells (Figure 4E). Western blot was also used to analyze the protein levels of these EMT markers, and we found that siRNA groups had higher ECAD expression, while expressions of NCAD, VIM, and TWIST were lower in siRNA groups (Figure 4F). All these results revealed that PKMYT1 could accelerate ESCC cells migration and invasion by EMT.
Figure 4.
Down-regulated PKMYT1 in ESCC cells inhibits the migration and invasion abilities of ESCC cells. (A) Transwell migration assays indicated that the migration abilities of KYSE70 and KYSE450 cells were weakened in siRNA groups. (B) Invasion assays showed silencing PKMYT1 inhibited the invasion ability.(C, D) Wound healing assay showed that inhibited the expression of PKMYT1 induced weaken migration ability (E) Western blot showed the change of EMT-related biomarkers. The expression of E-CAD was increased in siRNA groups while the decreased expression of NCAD, VIM, and TWIST. (F) Immunofluorescence showed that after transfection, increased expression of ECAD and decreased expression of NCAD and VIM in siRNA groups than NC group, which meant down-regulated the expression of PKMYT1 inhibited EMT progression in ESCC cells. **P<0.01, *** P<0.001,****P<0.0001.
Abbreviations: PKMYT1, protein kinase, membrane associated tyrosine/threonine; ESCC, esophageal squamous cell carcinoma; ECAD, E-Cadherin; NCAD, N-Cadherin; VIM, vimentin.
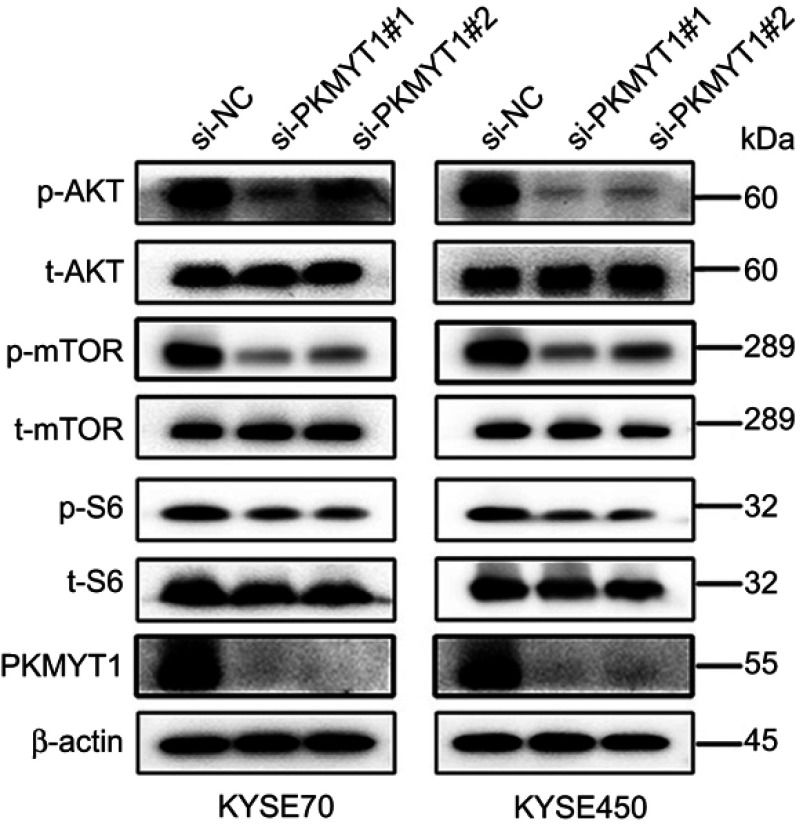
Down-regulated PKMYT1 suppressed Akt/mTOR signaling pathway in ESCC cells
To investigate the further mechanisms of how down-regulated PKMYT1 inhibited ESCC cells proliferation and migration, we detected the expressions of Akt/mTOR signaling pathway in ESCC cells by Western blot. Western blot confirmed that down-regulated PKMYT1 significantly decreased the expressions of p-Akt, p-mTOR, and p-S6 (Figure 5), suggesting down-regulated PKMYT1 might inhibit Akt/mTOR signaling pathway in ESCC cells and thus weakened the basic biological functions in ESCC cells in vitro.
Figure 5.
Knockdown of PKMYT1 expression suppressed the AKT/mTOR pathway. The expression of p-AKT, p-mTOR, and p-S6 was decreased in siRNA groups than NC group.
Abbreviations: PKMYT1, protein kinase, membrane associated tyrosine/threonine; NC, negative control.
Discussion
ESCC is one of the most malignant tumors with high recurrence and high mortality.5,6 Due to the unique anatomical structure of esophagus, metastasis in early-stage ESCC patients and recurrence after surgery are common, which result in the lower 5-year overall survival than the ones in other gastrointestinal cancers.27 Hence, it is necessary to explore molecular mechanism and discover novel therapeutic targets for the ESCC treatment.
PKMYT1 plays a negative role in the G2/M phase of cell cycle and has a great impact on the normal mitosis of tumor cells.9,12 However, to our knowledge, there is no report on its expression and specific role in ESCC. In this study, we found that PKMYT1 was highly expressed in ESCC patients and ESCC cell lines. To investigate the potential role of PKMYT1 in ESCC, we down-regulated PKMYT1 expression in vitro by siRNA and we found that it could inhibit the proliferation of ESCC cells. This is in line with previous findings in other tumors, such as hepatocellular carcinoma and colorectal cancer.15–18 We hypothesized that aberrant PKMYT1 expression was involved in abnormal proliferation and tumorigenesis in ESCC. Recent research has shown that abnormal apoptosis is associated tumor initiation and progress of ESCC.28 The key regulator in apoptosis-related pathway is Bcl-2 family, which has two representative molecules Bcl-2 and Bax, and the Bax/Bcl-2 ratio determines whether cells enter an apoptotic state or not. Our assays demonstrated that down-regulated PKMYT1 induced higher rate of cell apoptosis.29,30 Furthermore, we also found that the expression ratio of the apoptosis-related proteins Bcl-2/Bax decreased by Western blot. In cancer cells, G2 checkpoint is important for DNA repair because of the defective G1 checkpoint mechanism which is caused by a p53 mutation.10,11 We speculated that down-regulated PKMYT1 in ESCC cells might result in DNA-deficient in ESCC cells, thereby contributing to a large amount of apoptosis. Therefore, we consider that PKMYT1 inhibition might play an anti-tumor role in ESCC cells by inducing apoptosis, which might be a potential therapy in the treatment of ESCC.31
The characteristics of invasion and metastasis of malignant tumors are the main factors related to tumor-specific death.32,33 The EMT is one of the most important events associated with tumor metastasis, and EMT is closely related to disease progression and tumor metastasis of ESCC.34,35 The biomarkers of EMT changed during tumor progression and metastasis, such as the downregulation of ECAD and upregulation of NCAD, VIM, and TWIST.36,37 Substantial evidence has indicated the importance of EMT in cancer proliferation, metastasis, and chemoresistance which makes it a potential therapeutic target for tumor treatment.38 In our assays, we detected decreased migration and invasion abilities in ESCC cell lines after PKMYT1 knockdown. Moreover, PKMYT1 knockdown also triggered abnormal expression of several other molecular, such as increased expression of ECAD and decreased expression of NCAD, VIM, and Twist, which was also in line with previous studies.16,18 This indicated that PKMYT1 might promote the EMT progression in ESCC cells. Multiple reports have demonstrated that Twist, VIM, NCAD, and ECAD were independent risk factors in ESCC and associated with ESCC patients’ prognosis.23,24,39 Thus, PKMYT1 may participate in EMT of ESCC cells via these related molecular network.
AKT/mTOR signaling pathway has a crucial impact on tumor proliferation, apoptosis, and metastasis,40,41 and several studies indicated AKT/mTOR signaling pathway could facilitate the progression of ESCC.26,42 Recent reports have shown that AKT can promote the tumor progress by activating TWIST.43,44 We observed that reduced invasion and metastasis capacity and the changed expression of EMT-related biomarkers and decreased expression of p-AKT, p-mTOR, and p-S6 in ESCC cells due to PKMYT1 knockdown. Therefore, we speculated that PKMYT1 suppression could inhibit the expression of TWIST by inhibiting the activation of the AKT pathway, thus hindering the migration and metastasis of cells and promoting the apoptosis in ESCC cells. However, we only constructed a temporary knockdown model in this study, further research was required to investigate the role of PKMYT1 in ESCC by using stable knockdown and overexpression models in ESCC cells.
Recent investigations have identified cell cycle checkpoints as potential therapeutic targets. The WEE1 inhibitor MK1775 is currently being in clinical trials to treat various solid tumors combined with chemotherapy.45,46 Several PKMYT1 inhibitors have been already developed, including the well-known tyrosine kinase inhibitors dasatinib and bosutinib, the pyridopyrimidine derivatives PD-0166285, PD-173952, PD-173955, and PD-180970 also have been identified by applying different approaches,47–49 and more novel molecular inhibitors are underway.50,51
In conclusion, we firstly found that PKMYT1 was significantly up-regulated in ESCC tumor tissues and cell lines, and its higher expression was associated with poor overall survival in ESCC patients. The down-regulated of PKMYT1 expression in ESCC cells could inhibit cell proliferation, promote cell apoptosis, and restrict the EMT phenotype in vitro. Besides, the PKMYT1 downregulation also suppressed the Akt/mTOR/S6 signaling pathway in ESCC cells. These results demonstrated that PKMYT1 acted as an oncogene that might promote tumor progression by the AKT/mTOR pathway, and might be a potential target in ESCC treatment.
Acknowledgments
The paper was supported by the National Natural Science Foundation of China (NSFC) (No. 81802857) and the Key Research Project of Henan Provincial Colleges and Universities (No. 19A320062).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390(10110):2383–2396. doi: 10.1016/S0140-6736(17)31462-9 [DOI] [PubMed] [Google Scholar]
- 2.Fitzmaurice C, Dicker D, Pain A, et al.; Global Burden of Disease Cancer C. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 5.Zeng H, Zheng R, Zhang S, et al. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thorac Cancer. 2016;7(2):232–237. doi: 10.1111/1759-7714.12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371(26):2499–2509. doi: 10.1056/NEJMra1314530 [DOI] [PubMed] [Google Scholar]
- 7.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149(7):1700–1715. doi: 10.1053/j.gastro.2015.08.054 [DOI] [PubMed] [Google Scholar]
- 8.Zhu JY, Cuellar RA, Berndt N, et al. Structural basis of Wee Kinases functionality and inactivation by diverse small molecule inhibitors. J Med Chem. 2017;60(18):7863–7875. doi: 10.1021/acs.jmedchem.7b00996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booher RN, Holman PS, Fattaey A. Human Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997;272(35):22300–22306. doi: 10.1074/jbc.272.35.22300 [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Simpson ER, Brown KA. p53: protection against tumor growth beyond effects on cell cycle and apoptosis. Cancer Res. 2015;75(23):5001–5007. doi: 10.1158/0008-5472.CAN-15-0563 [DOI] [PubMed] [Google Scholar]
- 11.Kastan MB, Canman CE, Leonard CJ. P53, cell cycle control and apoptosis: implications for cancer. Cancer Metastasis Rev. 1995;14(1):3–15. [DOI] [PubMed] [Google Scholar]
- 12.Chow JP, Poon RY. The CDK1 inhibitory kinase MYT1 in DNA damage checkpoint recovery. Oncogene. 2013;32(40):4778–4788. doi: 10.1038/onc.2012.504 [DOI] [PubMed] [Google Scholar]
- 13.Blagosklonny MV. Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle. 2007;6(1):70–74. doi: 10.4161/cc.6.1.3682 [DOI] [PubMed] [Google Scholar]
- 14.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJ, Wurdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res. 2011;17(13):4200–4207. doi: 10.1158/1078-0432.CCR-10-2537 [DOI] [PubMed] [Google Scholar]
- 15.Toledo CM, Ding Y, Hoellerbauer P, et al. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep. 2015;13(11):2425–2439. doi: 10.1016/j.celrep.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Wu J, Wang S, et al. PKMYT1 promoted the growth and motility of hepatocellular carcinoma cells by activating beta-catenin/TCF signaling. Exp Cell Res. 2017;358(2):209–216. doi: 10.1016/j.yexcr.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 17.Jeong D, Kim H, Kim D, et al. Protein kinase, membrane associated tyrosine/threonine 1 is associated with the progression of colorectal cancer. Oncol Rep. 2018;39(6):2829–2836. doi: 10.3892/or.2018.6371 [DOI] [PubMed] [Google Scholar]
- 18.Wang XM, Li QY, Ren LL, et al. Effects of MCRS1 on proliferation, migration, invasion, and epithelial mesenchymal transition of gastric cancer cells by interacting with Pkmyt1 protein kinase. Cell Signal. 2019;59:171–181. doi: 10.1016/j.cellsig.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 19.Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–95. doi: 10.1038/nature13176 [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, et al. Integrated genomic characterization of oesophageal carcinoma. Nature.2017;541(7636):169–175. doi: 10.1038/nature20805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. doi: 10.1007/s10549-016-4013-7 [DOI] [PubMed] [Google Scholar]
- 23.Forghanifard MM, Rad A, Farshchian M, et al. TWIST1 upregulates the MAGEA4 oncogene. Mol Carcinog. 2017;56(3):877–885. doi: 10.1002/mc.22541 [DOI] [PubMed] [Google Scholar]
- 24.Zang C, Liu X, Li B, et al. IL-6/STAT3/TWIST inhibition reverses ionizing radiation-induced EMT and radioresistance in esophageal squamous carcinoma. Oncotarget. 2017;8(7):11228–11238. doi: 10.18632/oncotarget.14495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Kang L, Zhang H, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene. 2019;38:5250–5264. doi: 10.1038/s41388-019-0790-x [DOI] [PubMed] [Google Scholar]
- 26.Shigaki H, Baba Y, Watanabe M, et al. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19(9):2451–2459. doi: 10.1158/1078-0432.CCR-12-3559 [DOI] [PubMed] [Google Scholar]
- 27.Hiripi E, Jansen L, Gondos A, et al. Survival of stomach and esophagus cancer patients in Germany in the early 21st century. Acta Oncol. 2012;51(7):906–914. doi: 10.3109/0284186X.2012.673732 [DOI] [PubMed] [Google Scholar]
- 28.Nicholson DW, Thornberry NA. Apoptosis. Life and death decisions. Science. 2003;299(5604):214–215. doi: 10.1126/science.1081274 [DOI] [PubMed] [Google Scholar]
- 29.Renault TT, Dejean LM, Manon S. A brewing understanding of the regulation of Bax function by Bcl-xL and Bcl-2. Mech Ageing Dev. 2017;161(Pt B):201–210. doi: 10.1016/j.mad.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 30.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13(8):1351–1359. doi: 10.1038/sj.cdd.4401987 [DOI] [PubMed] [Google Scholar]
- 31.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15(1):49–63. doi: 10.1038/nrm3722 [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 33.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168(4):670–691. doi: 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Han S, Wu S, Bai Y, Zhang N, Wei L. Dual role of twist1 in cancer-associated fibroblasts and tumor cells promoted epithelial-mesenchymal transition of esophageal cancer. Exp Cell Res. 2019;375(2):41–50. doi: 10.1016/j.yexcr.2019.01.002 [DOI] [PubMed] [Google Scholar]
- 35.Zhang CY, Li RK, Qi Y, et al. Upregulation of long noncoding RNA SPRY4-IT1 promotes metastasis of esophageal squamous cell carcinoma via induction of epithelial-mesenchymal transition. Cell Biol Toxicol. 2016;32(5):391–401. doi: 10.1007/s10565-016-9341-1 [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Tillo E, Liu Y, de Barrios O, et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69(20):3429–3456. doi: 10.1007/s00018-012-1122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andl CD, Fargnoli BB, Okawa T, et al. Coordinated functions of E-cadherin and transforming growth factor beta receptor II in vitro and in vivo. Cancer Res. 2006;66(20):9878–9885. doi: 10.1158/0008-5472.CAN-05-4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaufhold S, Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J Exp Clin Cancer Res. 2014;33:62. doi: 10.1186/s13046-014-0062-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen HF, Chan YP, Wong ML, et al. Upregulation of twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol. 2007;60(5):510–514. doi: 10.1136/jcp.2006.039099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song M, Bode AM, Dong Z, Lee MH. AKT as a therapeutic target for cancer. Cancer Res. 2019;79(6):1019–1031. doi: 10.1158/0008-5472.CAN-18-2738 [DOI] [PubMed] [Google Scholar]
- 41.O’Hayre M, Degese MS, Gutkind JS. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr Opin Cell Biol. 2014;27:126–135. doi: 10.1016/j.ceb.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hildebrandt MA, Yang H, Hung MC, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27(6):857–871. doi: 10.1200/JCO.2008.17.6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vichalkovski A, Gresko E, Hess D, Restuccia DF, Hemmings BA. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29(24):3554–3565. doi: 10.1038/onc.2010.115 [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 2015;9(4):317–324. doi: 10.1080/19336918.2015.1016686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Do K, Wilsker D, Ji J, et al. Phase I study of single-agent AZD1775 (MK-1775), a Wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol. 2015;33(30):3409–3415. doi: 10.1200/JCO.2014.60.4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leijen S, van Geel RM, Pavlick AC, et al. Phase I study evaluating WEE1 inhibitor AZD1775 as monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J Clin Oncol. 2016;34(36):4371–4380. doi: 10.1200/JCO.2016.67.5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis MI, Hunt JP, Herrgard S, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1046–1051. doi: 10.1038/nbt.1990 [DOI] [PubMed] [Google Scholar]
- 48.Ren LH, Peng JJ, Xu XL, Ye HM, Jia KY. [High maintenance dose of clopidogrel improves long-term clinical outcomes in patients with elective percutaneous coronary intervention]. Zhonghua Yi Xue Za Zhi. 2012;92(6):408–410. [PubMed] [Google Scholar]
- 49.Rohe A, Gollner C, Wichapong K, et al. Evaluation of potential Myt1 kinase inhibitors by TR-FRET based binding assay. Eur J Med Chem. 2013;61:41–48. doi: 10.1016/j.ejmech.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 50.Najjar A, Platzer C, Luft A, et al. Computer-aided design, synthesis and biological characterization of novel inhibitors for PKMYT1. Eur J Med Chem. 2019;161:479–492. doi: 10.1016/j.ejmech.2018.10.050 [DOI] [PubMed] [Google Scholar]
- 51.Platzer C, Najjar A, Rohe A, Erdmann F, Sippl W, Schmidt M. Identification of PKMYT1 inhibitors by screening the GSK published protein kinase inhibitor set I and II. Bioorg Med Chem. 2018;26(14):4014–4024. doi: 10.1016/j.bmc.2018.06.027 [DOI] [PubMed] [Google Scholar]
- 52.Nagy A, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]