Abstract
Colorectal cancer is a complex disease driven by well-established mutations such as APC and other yet to be identified pathways. The GTPase Rab11 regulates endosomal protein trafficking, and previously we showed that loss of Rab11 caused intestinal inflammation and hyperplasia in mice and flies. To test the idea that loss of Rab11 may promote cancer progression, we have analyzed archival human patient tissues and observed that 51 out of 70 colon cancer tissues had lower Rab11 protein staining. By using the Drosophila midgut model, we have found that loss of Rab11 can lead to three changes that may relate to cancer progression. First is the disruption of enterocyte polarity based on staining of the FERM domain protein Coracle. Second is an increased proliferation due to an increased expression of the JAK-STAT pathway ligand Upd3. Third is an increased expression of ImpL2, which is an IGFBP7 homolog and can suppress metabolism. Furthermore, loss of Rab11 can act synergistically with the oncoprotein RasV12 to regulate these cancer-related phenotypes.
Keywords: colon cancer, Drosophila, ImpL2, intestine, metabolism, Rab11, Ras
THE intestine has a high rate of cell turnover and tissue homeostasis is supported by intestinal stem cells (ISCs) (Tan and Barker 2014; Beumer and Clevers 2016; Perochon et al. 2018). With the identification of different stem cell populations such as the Bmi1+ and Lgr5+ ISCs, the mammalian intestine is a robust system to study stem cell-mediated homeostasis and related diseases. Colorectal cancer progression involves multiple events including mis-regulation of highly conserved signaling pathways and loss of genome stability (Armaghany et al. 2012; Cancer Genome Atlas Network 2012; Dienstmann et al. 2017). Histologically identical tumors may have different responses to treatment, suggesting that colorectal cancer is a heterogeneous disease (Kanthan et al. 2012; Sussman et al. 2012; Vaiopoulos et al. 2012; Welch and Robertson 2016). A better understanding of the molecular basis should provide better individual treatments.
Adult Drosophila midgut is equivalent to the mammalian stomach and small intestine. The Drosophila midgut contains resident ISCs, which are the major dividing cells that support homeostasis and regeneration (Micchelli and Perrimon 2006; Ohlstein and Spradling 2006; Amcheslavsky et al. 2009; Jiang et al. 2009). Asymmetric division of midgut ISCs generates precursors called enteroblasts (EBs) that differentiate into enterocytes (ECs) for food absorption, or precursors called preenteroendocrine cells (preEEs) that differentiate into enteroendocrine cells (EEs) for hormone secretion. The conserved Delta-Notch signaling is essential for regulation of ISC-EB asymmetry, as well as EC vs. EE fate (Bardin et al. 2010; Kapuria et al. 2012; Guo and Ohlstein 2015; Chen et al. 2018). Meanwhile, Wnt, BMP, Insulin, EGF, Hedgehog, and JAK-STAT pathway ligands produced from surrounding cells and tissues, including EBs, ECs, EEs, visceral muscle, tracheal cells and neurons, participate in the regulation of ISC division and subsequent differentiation under normal homeostasis or stress-stimulated regeneration (Lin et al. 2008; Amcheslavsky et al. 2009; Jiang et al. 2009, 2016; O’Brien et al. 2011; Cordero et al. 2012; Osman et al. 2012; Guo et al. 2013, 2016; Li et al. 2013; Zhou et al. 2013; Tian and Jiang 2014; Han et al. 2015; Tian et al. 2015).
In a previous report, we have shown that loss of Rab11a in the mouse intestine leads to hyperplasia, concomitant with increased expression of inflammatory cytokines including IL6 (Yu et al. 2014). The Drosophila midgut after loss of Rab11 also shows hyperplasia and increased expression of the JAK-STAT pathway ligand Unpaired 3 (Upd3), which has structural similarity to IL6 (Oldefest et al. 2013; Yu et al. 2014). Here we report the analysis of Rab11 protein in human colon cancer samples, revealing that the protein staining was consistently lower, particularly in samples from older patients. Experiments using the Drosophila midgut suggest that loss of Rab11 interacts with the Ras and Wnt pathways to promote cancer-related phenotypes.
Materials and Methods
Colon tissue immunofluorescence staining
Frozen sections of normal colon and adenocarcinoma colon tissues of deidentified patients (number followed by N for normal and T for tumor) were obtained from the Tissue and Tumor Bank of the University of Massachusetts Medical School Cancer Center; the sample number and clinical information are listed in an excel file (https://figshare.com/s/7ec190b0f62a930ea7dc). For immunofluorescence staining, frozen section slides were removed from storage at −80° and fixed with 4% formaldehyde immediately. The tissues on the slides were then permeabilized by incubating twice with 0.1% Triton X-100 in phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) for 30 min each time, followed by blocking for 1.5 hr in a buffer containing 5% normal horse serum, 0.5% BSA, 0.1% Triton X-100 in PBS. Rab11 antibody from rabbit (1:100, , #3539; Cell Signaling), β-catenin monoclonal antibody from mouse (1:5000, #610153; BD transduction), IGFBP7 antibody from rabbit (1:200, HPA002196; Sigma, St. Louis, MO), and phospho-Stat3 (Y705) antibody from rabbit (1:100, #9145; Cell Signaling) were diluted with blocking buffer and incubated with the tissue sections overnight at 4°. The sections were then washed four times with 0.5% BSA and 0.1%Triton X-100 in PBS for 20 min each time, followed by incubation with secondary antibodies for 2 hr at room temperature. Alexa Fluor488 goat anti-rabbit (A11070; Life Technologies) was used to detect the signals for Rab11, and Alexa Fluor568 goat anti-mouse (A11019; Life Technologies) was used to detect the signals for β-catenin. The slide sections were washed four times again with 0.1% Triton X-100 in PBS after secondary antibody incubation and covered in Vectorshield containing DAPI (H-1200; Vector Laboratories, Burlingame, CA) for microscopy.
Colon tissue image acquisition and analysis
Images of immunofluorescence-stained frozen colon tissue sections were visualized and acquired by using a Nikon Spinning Disk Confocal microscope. Slides were first observed under a 10× objective lens to locate malignant areas. Representative pictures were then taken under a 20× objective lens. To quantify the epithelial Rab11 staining signals from normal samples, multiple region of interest (ROI) were selected from each confocal image so that each ROI contained either a hemicrypt or cross-sections of crypts, but the luminal or stromal areas were not included. A few regions that contained no cells were also selected to represent the background value for each sample slide. We decided to collect the signal of multiple ROI from 12 different normal patient samples, and the average value of all these images is set as the normal expression level. Tumor samples usually had more disorganized morphology (Supplemental Material, Figure S1), and ROI were selected to cover equivalent tissue area and empty areas were not included. A few regions that contained no tissue on each slide were also selected to represent the background value for each tumor sample slide. Region gray levels were gathered for each ROI, and the average would become the value for that tumor sample, after subtracting the average background value. β-catenin double staining was performed for each slide. The staining for β-catenin had been consistently detected in both normal and cancer epithelia cells, and frequently we used the β-catenin staining to identify epithelia cell clusters in advanced cancer, which has highly disorganized morphology. To assess the ratio of tumor relative to normal for Rab11, the quantified signals from each tumor sample were averaged and compared to the average normal expression level as described above (https://figshare.com/s/7ec190b0f62a930ea7dc).
Statistics
Student’s t-test was performed to test the significance when comparing two groups of results. The P value * is <0.05 and ** is <0.01 when indicated. The error bar in Figures 3, 4, and 5 graphs represents the SEM. The P values in Table 1 were based on comparison of the expression levels of all ROI in the tissue samples of the whole cancer patient group as specified and the expression levels of all ROI in the 12 individual normal tissues (https://figshare.com/s/7ec190b0f62a930ea7dc).
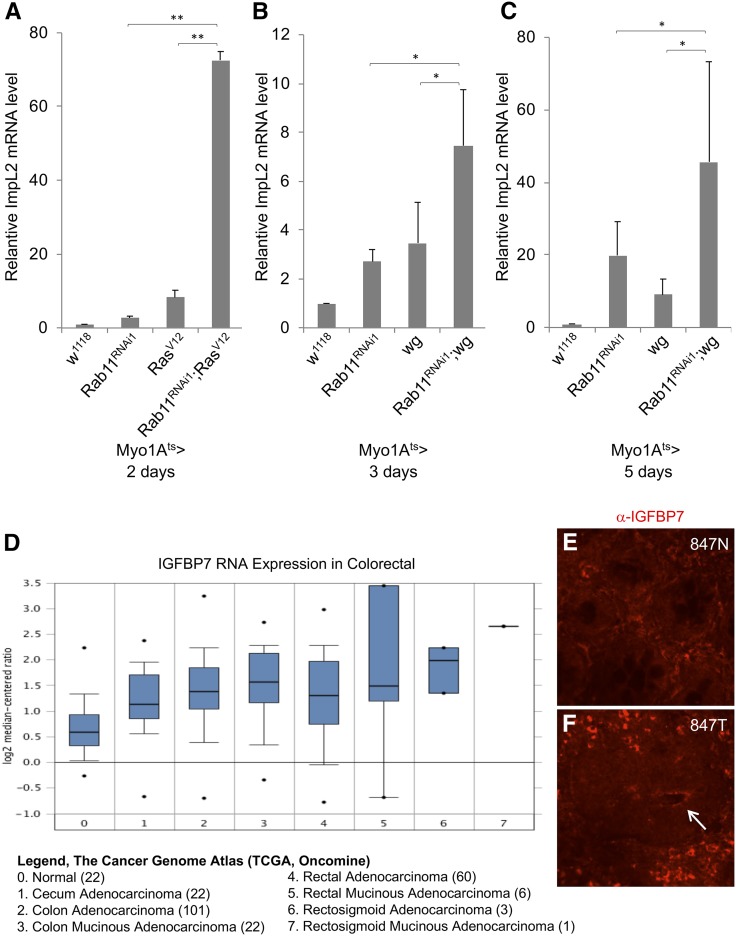
Figure 3.
Synergistic interaction of Rab11 RNAi and RasV12 leads to increased expression of ImpL2. (A–C) RNA expression analysis of ImpL2 by qPCR of total RNA isolated from midguts of flies harboring the indicated UAS-Rab11dsRNA, UAS-RasV12, or UAS-wingless (wg) expression constructs. The driver used was Myo1Ats > GFP and the days of incubation at 29° was as indicated, to allow transgene expression. The expression level of ImpL2 RNA in control fly guts was set as 1 in each panel, and the other samples were calculated as fold increase relative to the control. The P value * is <0.05 and ** is <0.01 when indicated. (D) IGFBP7 RNA expression levels in human colorectal adenocarcinoma. Data were collected from www.oncomine.org using data set of The Cancer Genome Atlas (TCGA) – Colon and Rectum Adenocarcinoma Gene Expression Data. (E and F) Confocal images of IGFBP7 antibody immunofluorescence staining of human normal colon and cancer tissue sections. The number represents the sample number of the UMMS tissue bank deidentified patients, with N for normal and T for tumor. The arrow in the tumor tissue indicates possibly higher staining of IGFBP7 surrounding an apical/luminal space.
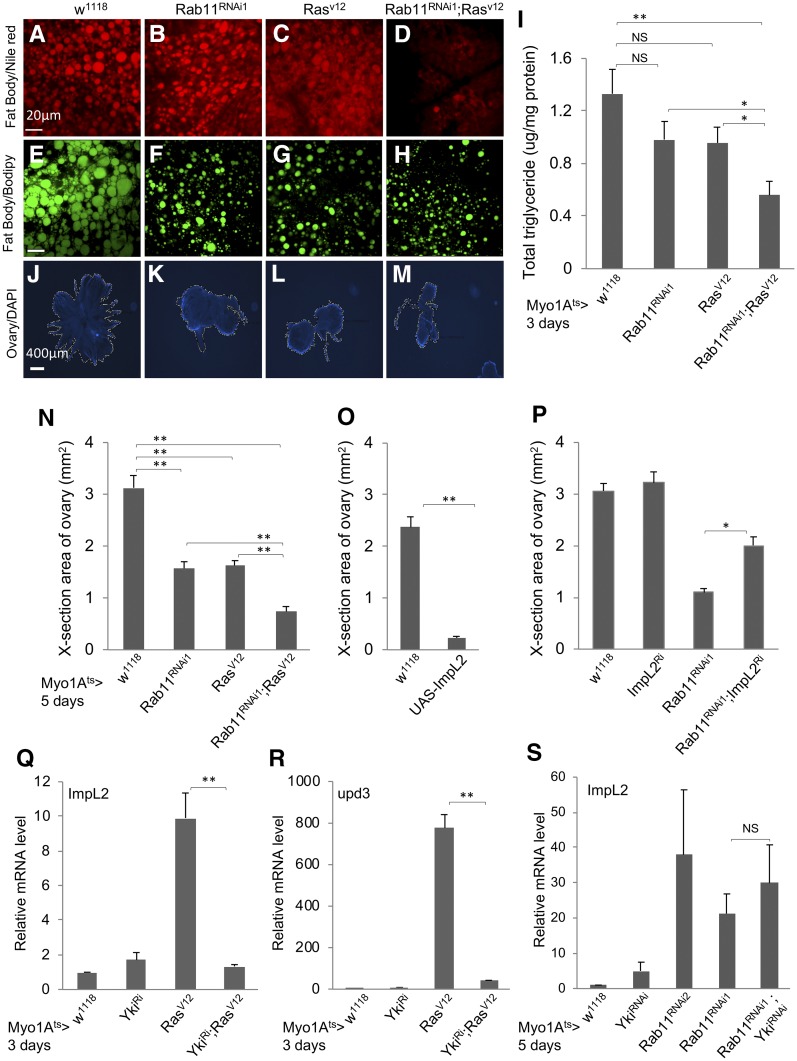
Figure 4.
Loss of Rab11 and expression of ImpL2 in midgut regulate the metabolism of neighboring tissues. (A–D) The Myo1Ats > GFP flies were crossed with flies of genotypes indicated at the top of the images. The resulting adult flies were shifted to 29° for 5 days and the abdomens were used for Nile Red staining for lipid droplets. These are higher magnifications of the images shown in Figure S3, A–D. (E–H) Flies from crosses the same as A–D were used for a parallel Bodipy staining for lipid droplets. Bar, 20 μm. (I) Whole flies from crosses the same as A–D were used for homogenization, and the extracts were used for total triglyceride measurement and also total protein measurement, which was used as the internal control for normalization. The normalized value for total triglyceride is plotted as shown. (J–M) DAPI-stained fluorescent images of ovaries dissected from adult flies with the same genotypes, as indicated at the top. (N) Quantification of the cross-section areas of the ovaries as shown in J–M. More than 10 images of each genotype were used to measure the area, and the average area of each genotype is plotted as shown. (O) Measurement of ovary cross-section areas from control flies and flies containing an overexpression construct of ImpL2 complementary DNA (cDNA). The expression was driven by the Myo1Ats > GFP in ECs, and the temperature shift was for 5 days before ovary dissection and image analysis. (P) A similar assay for ovary cross-section areas, using flies containing the combination of Rab11 RNAi and ImpL2 RNAi constructs. The expression of the transgenic dsRNA constructs was driven by Myo1Ats > GFP, and the temperature shift was for 5 days before ovary dissection and image analysis. (Q–S) Expression of ImpL2 or upd3 RNA by qPCR of total RNA from dissected guts of flies containing the yki RNAi, Rab11 RNAi, RasV12 overexpression, or combinations of the constructs. The driver was Myo1Ats > GFP and the temperature shift was for 3 or 5 days as indicated before gut dissection and RNA preparation. The RNA expression in each set of experiments was normalized to that of rp49 and set as 1 for the Myo1Ats > crossed with w1118 control samples. The expression level in other samples was plotted as fold increase.
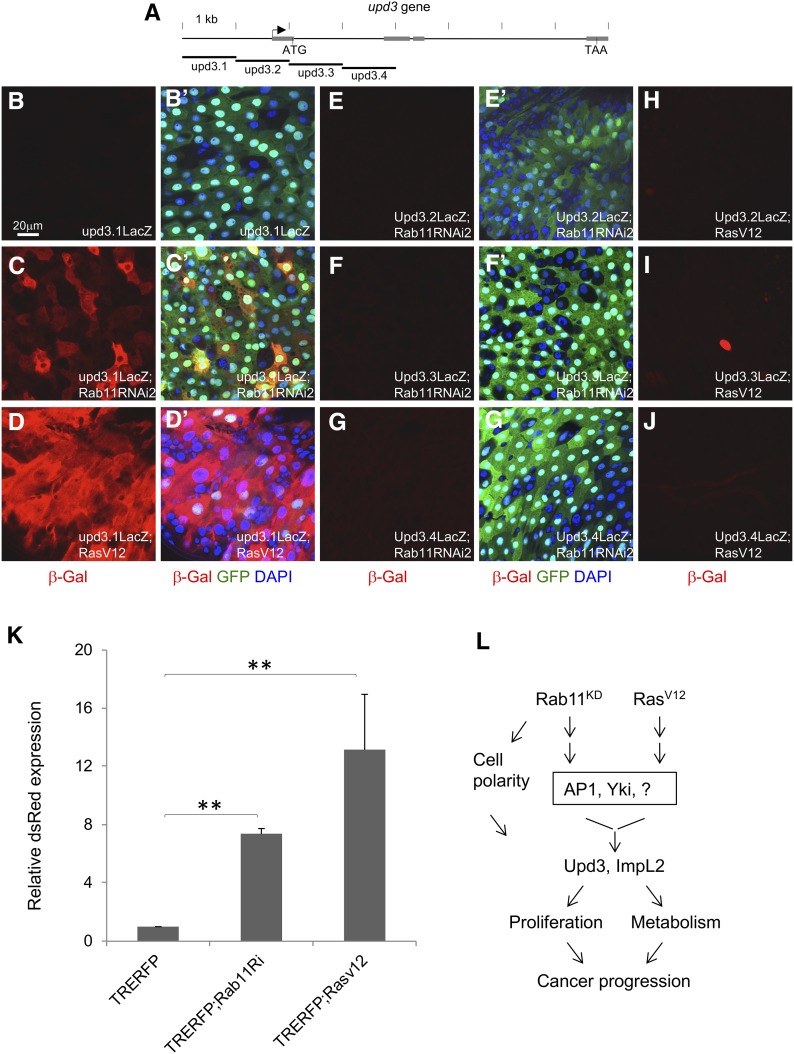
Figure 5.
Multiple regulatory components act downstream of Rab11 and RasV12. (A) A diagram showing the genomic locus of upd3. The arrow indicates predicted transcription start site. The gray boxes are exons, and the start and stop codons as ATG and TAA, respectively, are indicated. The four genomic elements labeled upd3.1-upd3.4, each containing ∼1 kb of DNA, were used to generate transgenic lacZ reporter flies (Jiang et al. 2009). (B–J) Each of the four reporters was crossed with either Rab11 RNAi or RasV12 overexpression flies, and then with the Myo1Ats > GFP driver. The resulting progenies were used for temperature shift for 3 days and then gut dissection. Antibodies for β-galactosidase were used for tissue staining and shown as read in the images. (B’–G’) are same corresponding images showing the 3-color combination. (H–J) No corresponding 3-color combinations are shown here because there was no significant β-galactosidase staining and they were similar to those shown in E’–G’. The color blue is DAPI, green is GFP and red is β-galactosidase. (K) Flies containing the AP1 reporter TRE-RFP were crossed to Rab11 RNAi or RasV12 flies and the resulting progenies were crossed with the Myo1Ats > GFP driver. After temperature shift for 3 days, the flies were used for gut dissection, RNA extraction, and qPCR quantification of dsRed mRNA expression. The expression in the control flies was set as 1, and the other samples were plotted as fold increase. (L) A diagram summarizes the results presented in this report. The working model is that loss of Rab11 function or activation of Ras can lead to activation of AP1, Yki, and other possible transcriptional regulators. The pathways converge onto the promoter elements of target genes including upd3 and ImpL2 to control their expression in the midgut. Upd3 expression increases intestine stem cell proliferation and ImpL2 expression reduced the metabolism of surrounding tissue, and together with the loss of cell polarity may promote cancer progression.
Table 1. Rab11 expression in colon cancer tissues of patients grouped by various clinical parameters.
| Number of tissues (70 total) | Number of tissues with lower Rab11 | Median Rab11 T/N | P value against normal | |
|---|---|---|---|---|
| Stage | ||||
| I | 15 | 9 (60%) | 0.637 | 0.0459 |
| II | 25 | 20 (80%) | 0.455 | 0.0225 |
| III | 26 | 19 (73%) | 0.518 | 0.0239 |
| IV | 4 | 3 (75%) | 0.375 | 0.0346 |
| N0M0, N0Mx | 41 | 30 (73%) | 0.489 | 0.0237 |
| N1M0, N2M0 | 12 | 8 (67%) | 0.591 | 0.148 |
| N1M1, N2M1 | 4 | 3 (75%) | 0.375 | 0.0346 |
| N1Mx, N2Mxa | 13 | 10 (77%) | 0.456 | 0.00563a |
| T1 | 6 | 3 (50%) | 0.615 | 0.247 |
| T2 | 11 | 8 (73%) | 0.626 | 0.016 |
| T3a | 37 | 31 (84%) | 0.455 | 0.01a |
| T4 | 16 | 9 (56%) | 0.616 | 0.108 |
| N0 | 41 | 30 (73%) | 0.489 | 0.024 |
| N1 | 15 | 11 (73%) | 0.456 | 0.025 |
| N2 | 14 | 10 (71%) | 0.554 | 0.023 |
| M0 | 34 | 25 (74%) | 0.562 | 0.055 |
| M1 | 4 | 3 (75%) | 0.375 | 0.035 |
| Mxa | 32 | 23 (72%) | 0.438 | 0.009a |
| Age | ||||
| <50 | 12 | 6 (50%) | 0.647 | 0.129 |
| 50–70 | 29 | 21 (72%) | 0.492 | 0.0276 |
| >70a | 29 | 24 (83%) | 0.455 | 0.00914a |
| Sex | ||||
| F | 45 | 33 (73%) | 0.462 | 0.0164 |
| M | 25 | 18 (72%) | 0.498 | 0.0294 |
The clinical information of the patients is listed in an excel file https://figshare.com/s/7ec190b0f62a930ea7dc. The relative expression level in tumor tissues was calculated as fractions over that of average normal samples (T/N) as described in Materials and Methods and listed in the excel file https://figshare.com/s/7ec190b0f62a930ea7dc. The 70 samples were then grouped by various clinical information and their median expression was calculated as shown. The top part of the table is grouped by tumor stage I–IV, or NM designation represents Node and Metastasis. Mx is not known, or grouped by tumor stage by TNM pathology designation, and presented as T1–T4, N0–N2, and M0–Mx. The other table entries are grouped by the age of patient at time of diagnosis and by sex.
The subgroup with the lowest P value and significant reduction of Rab11 protein level.
Drosophila stocks and genetics
Fly stocks were maintained at room temperature (∼22°) in yeast extract/cornmeal/molasses/agar food medium. w1118, Myo1A-Gal4ts > UAS-GFP or Myo1A-Gal4ts > UAS-GFP X w1118 flies were used as the wild-type control in different experiments. Transgenic and RNAi fly stocks were obtained from the Vienna Drosophila Research Center (VDRC, labeled as V) or Bloomington Stock Center (labeled as B), or otherwise cited. The stocks included Rab11S25N dominant negative (B9792), Rab11 RNAi (RNAi1 is V108382, RNAi2 is V22198, RNAi3 is B27730), UAS-Rasv12 (B64195), UAS-ImpL2 and ImpL2 RNAi as described in Kwon et al. (2015), UAS-Wg-HA (B108487), UAS-Yki-act (gift from N. Perrimon), YkiN+C RNAi (gift from J. Jiang), Yki RNAi (B31965), UAS-TRERFP (gift from A. Bergmann), and upd3.1LacZ-upd3.4LacZ (gift from H. Jiang).
Drosophila transgenic expression for various assays
Newly eclosed flies were kept in vials with regular food medium at room temperature until 5–8 days old, and were shifted to 29° for 1–5 days as indicated in the individual figures for optimal results to inactivate the tubulin-Gal80ts repressor and to enable Gal4-dependent UAS-dsRNA or transgene expression. Female flies were used for tissue dissection for various assays including immunofluorescence staining and RNA isolation for quantitative PCR. Feeding experiments were performed as previously described (Amcheslavsky et al. 2009). Approximately 25 flies of each genotype were kept at 29° for 5 days on regular food and then starved for 2 hr in empty plastic vials before feeding with 5% sucrose with food dye, plus any ingredients indicated, dropped onto a Whatman paper in the bottom of plastic vials. Where appropriate, flies were dissected to check percentages of flies with food dye in the gut at different times.
Drosophila immunofluorescence confocal microscopy
Female flies were dissected for analysis routinely because of their bigger size. Fly gastrointestinal tract was pulled from the posterior end directly into 4% formaldehyde in PBS (Mallinckrodt Chemicals) for fixation for 1 hr, followed by permeabilization with 0.1% Triton X-100/PBS for 0.5 hr and blocking in 5% normal horse serum, 0.5% BSA, 0.1% Triton X-100, 1XPBS for 1 hr. Primary antibody diluted in blocking buffer: anti-Coracle (1:00, mouse monoclonal, Hybridoma Bank C566.9), anti β-galactosidase (1:50,000, rabbit polyclonal, Cappel; MP Biomedicals), and anti-Phospho-Histone3 (1:1000, rabbit polyclonal; Millipore, Bedford, MA) were incubated overnight at 4°. Subsequent washes and secondary antibody incubation were in 0.5% BSA, 0.1% Triton X-100, 1XPBS. Secondary antibodies used were goat anti-mouse IgG conjugated to Alexa 568 (A-11032; Life Technologies) or goat anti-rabbit IgG conjugated to Alexa 555 (A-11008; Life Technologies). After three washes in 0.1% Triton X-100, 1XPBS, guts were mounted in Vectorshield DAPI (Vector Laboratories)/PBS solution (1:1) for microscopy analysis. Images were taken using a Nikon Spinning Disk Confocal microscope (UMass Medical School Imaging Core Facility) or light microscope Nikon E600 equipped with Normaski and fluorescent optics. The acquisition and processing software for the confocal microscope is MetaMorph (Molecular Devices), and single optical sections are shown as images in the figures, and Spot and NIS Elements digital imaging systems for the light microscope.
Drosophila metabolism measurement
Total body triglycerides measurement was adapted from a previous publication (Kwon et al. 2015). Briefly, batches of eight female flies for each testing genotype (Myo1Ats > GFP, Myo1Ats > Rab11Ri, Myo1Ats > RasV12 or Myo1Ats > Rab11Ri, RasV12) were homogenized using pestles in 500-μl PBS with 0.2% Triton X-100 and 1% protease inhibitor (G653A; Promega, Madison, WI). The homogenates were heat-inactivated at 70° for 5 min, followed by centrifugation at 14,000 rpm for 10 min. Supernatants were transferred into new sets of tubes, and 10 μl was taken to proceed to total triglycerides measurement (TR0100; Sigma-Aldrich) or Bicinchoninic Acid (BCA) protein assay measurement (500–0202; BioRad, Hercules, CA) following the manufacturer’s protocol . A Softmax Pro plate reader was used to measure the samples in a 96-well plate format. Total triglyceride level was normalized to the protein level for comparison.
For Nile Red staining, fly cuticles were dissected in 4% formaldehyde/PBS and stained in Nile Red solution (N1142; Life Technologies). The Nile Red stock solution was prepared in acetone at 2.5 mg/ml and kept frozen. The cuticles were stained in 0.25 mg/ml Nile Red for 0.5 hr that was diluted 10× from the stock solution just before use. The stained cuticles were mounted and observed with confocal microscopy for epidermal lipid visualization.
For lipid droplet Bodipy staining, fly cuticles were dissected in cold PBS and fixed in 4% formaldehyde/PBS for 30 min at room temperature. Tissues were then washed three times with 1× PBS and incubated for 30 min in 1:1000 dilution with 1 mg/ml BODIPY 493/503 in PBS just before use (D3922; Invitrogen, Carlsbad, CA), and then rinsed twice with distilled water . The stained cuticles were mounted in 80% glycerol. Images were taken using a confocal microscope for epidermal lipid droplet visualization.
For fly ovary size measurement, 8 to 10 female flies of each genotype of interest were dissected at 5 days post temperature shift. Ovaries were fixed in 4% formaldehyde and permeabilized with 0.1% Triton X-100 in PBS for 0.5 hr and mounted in Vectorshield DAPI/PBS solution (1:1). Cross-section images were taken and analyzed with Nikon NIS Elements software.
Differential expression gene analysis
Total RNA was extracted from 20 guts of flies from Myo1Ats > GFP control and >Rab11 RNAi (V108382), respectively. The flies were shifted to 29° for 5 days before dissection. Three independent biological samples were used for total RNA isolation. The RNA was sequenced using either the Applied Biosystems’ Ion Torrent platform (PrimBio) or the Illumina HiSeq 2500 (Ribobio, Guangzhou, China). Fastqc (version 0.10.1) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used on the fastq sequences for all six samples to generate sequence quality reports. The raw sequences (from Ion Torrent and Illumina platforms) were mapped to the reference genome Drosophila melanogaster Ensembl BDGP5 (Adams et al. 2000) using the aligner STAR (version 2.4.2a) (Dobin et al. 2013). HTseq-count (version 0.6.1) (Anders et al. 2015) was used to count uniquely mapped reads at the gene level. DESEQ2 (Love et al. 2014) was used to perform statistical analysis to determine differentially expressed genes between the control samples and the knockdown samples. Expression of Rab11 and Upd3 were validated using the IGV tool (version 2.3.60) (Thorvaldsdottir et al. 2013). Differentially expressed genes are those with Padj value <0.05 (N = 2018). Some of the most highly differentially expressed genes are presented in Table 2 (top part). The differentially expressed genes were used for the pathway analysis using Enrichr (Chen et al. 2013; Kuleshov et al. 2016). The BioCarta 2015 database was used for pathway analysis in Table 2 (lower part). The RNA sequencing data can be accessed from GEO (accession number GSE92705).
Table 2. RNA sequencing analysis of adult midguts after Rab11 RNAi in enterocytes.
| gene ID | baseMean | log2 fold upregulated | gene ID | baseMean | log2 fold downregulated |
|---|---|---|---|---|---|
| CG4250 | 478.99 | 6.18 | CG4830 | 817.12 | −6.04 |
| CG7017 | 161.88 | 5.66 | CG12057 | 36140.03 | −5.41 |
| upd3* | 273.83 | 5.1 | lectin-24A | 200.99 | −5.21 |
| CG16959 | 597.67 | 4.9 | CG34215 | 206.9 | −5.12 |
| CG7910 | 316.87 | 4.6 | Diedel3 | 6166.5 | −4.71 |
| mus209 | 219.47 | 4.41 | Jon25Biii | 120864.62 | −4.4 |
| rho | 208.49 | 4.2 | Jon25Bii | 13713.75 | −4.39 |
| Pen | 184.02 | 4.19 | CG15263 | 601.32 | −4.35 |
| CG13117 | 2513.47 | 4.14 | kek1 | 144.87 | −4.27 |
| Act42A | 17906.75 | 4.13 | CG1946 | 973.53 | −4.26 |
| ImpL2* | 3375.39 | 3.9 | LpR1 | 1306.48 | −4.23 |
| CG31344 | 323.87 | 3.89 | CG7916 | 31481.39 | −4.22 |
| CG6933 | 299.19 | 3.75 | Jon25Bi | 23529.52 | −4.03 |
| spz | 126.44 | 3.68 | Gal | 3227.75 | −3.9 |
| rempA | 147.94 | 3.66 | mag | 24607.21 | −3.87 |
| Mip | 184.76 | 3.66 | CG12374 | 191299.71 | −3.84 |
| CG32284 | 213.98 | 3.57 | CG17571 | 19784.03 | −3.77 |
| Cht4 | 6618.66 | 3.55 | CG12942 | 615.92 | −3.75 |
| CG13488 | 240.88 | 3.54 | CG5107 | 23701.36 | −3.71 |
| Jupiter | 1591.73 | 3.46 | Mal-A7 | 32489.84 | −3.61 |
| LysX | 4204.24 | 3.39 | Ugt37b1 | 704.47 | −3.52 |
| cv-2 | 864.39 | 3.32 | CG11911 | 25670.12 | −3.48 |
| Mmp2 | 346.94 | 3.3 | CG11912 | 693.16 | −3.32 |
| Sox21a | 439.24 | 3.23 | CG8997 | 38550.87 | −3.3 |
| RnrL | 247.67 | 3.14 | CG15534 | 8786.17 | −3.28 |
| GstD2 | 1017.37 | 3.1 | CG33965 | 764.32 | −3.2 |
| Pvf1 | 1977.45 | 3.04 | CG11162 | 287.18 | −3.19 |
| Cyp305a1 | 153.03 | 3.03 | ninaD | 1779.54 | −3.18 |
| Ets21C | 281.76 | 3.01 | CG5150 | 8290.16 | −3.18 |
| AnnX | 716.87 | 2.99 | CG9394 | 5022.04 | −3.03 |
| Dpt | 174.98 | 2.9 | CG30272 | 451.84 | −3.02 |
| Biological pathway | P value | ||||
|---|---|---|---|---|---|
| CDK regulation of DNA replication | 5.67E−10 | ||||
| Proteasome complex | 0.001897 | ||||
| The igf-1 receptor and longevity | 0.005583 | ||||
| Ras signaling pathway | 0.009665 | ||||
| Mechanisms of transcriptional repression by DNA methylation | 0.004766 | ||||
| Pten-dependent cell cycle arrest and apoptosis | 0.04322 | ||||
| Gamma-aminobutyric acid receptor life cycle pathway | 0.04322 | ||||
Adult flies containing the dsRNA construct driven by Myo1A-Gal4 and tubulin-Gal80ts were used for temperature shift for 5 days and then gut dissection. Total RNA was isolated and used to generate libraries for RNA sequencing. Top part of table shows a selection of top genes that are up- or downregulated after Rab11 RNAi. The expression of upd3 and ImpL2 is highlighted in the left column. Lower part of table shows the most related pathways that involve the changed gene expression after Rab11 RNAi. Analysis was done using differential expression gene list of Rab11 RNAi at Enrichr (http://amp.pharm.mssm.edu/Enrichr/; database BioCarta_2015). Full list of genes that show significant changes are at https://figshare.com/s/9c15d272ffca6ba900c4.
Data availability statement
Strains and plasmids are available upon request.
Supplemental materials include Figures S1–S3 and Table S1.
RNAseq fastq files deposited in GEO (accession number GSE92705).
Cancer tissue clinical information and image analysis excel file in https://doi.org/10.6084/m9.figshare.6985223.
Full list of genes in Tale 2 top part excel file in https://doi.org/10.6084/m9.figshare.7505612. Supplemental material available at FigShare: https://doi.org/10.6084/m9.figshare.8340950.
Results
Rab11 protein expression is reduced in many human colon cancer samples
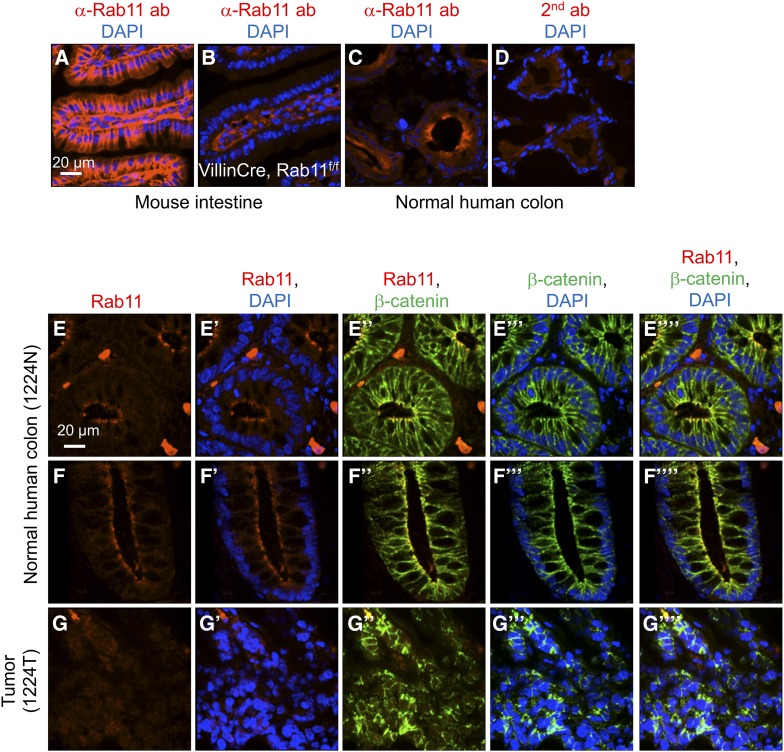
We used immunofluorescence staining to assess the expression of Rab11 protein in deidentified human colon cancer patient samples from the University of Massachusetts Medical School Cancer Center Tissue Bank. By using villin-cre-directed Rab11a knockout mouse intestinal tissue, we show that this Rab11 antibody exhibited the expected specificity because the strong signal was lost in the epithelium but not the stroma (Figure 1, A and B). In normal human colon tissue, Rab11 staining was observed with higher signal at the apical border proximal to the lumen (Figure 1, C and D), consistent with a previous report (Talmon et al. 2012). As a marker for intestinal epithelial cells, β-catenin antibody was included in the double staining. In comparison, β-catenin had a more homogeneous signal along the apical–basal axis in the same normal colonic epithelial cells (Figure 1, E and F).
Figure 1.
Rab11 expression in mammalian intestine and colon. (A–D) Immunofluorescence staining of tissue sections using a Rab11 antibody, or secondary antibody alone in D. (A) Wild-type mouse small intestine, (B) Rab11flox/f;lox; villinCre mouse intestine, (C) normal human colon, and (D) normal human colon staining with secondary antibody alone. Red color is Rab11 staining, and blue is DAPI for DNA staining. (E–G) Immunofluorescence staining of colon tumor and matching normal tissue from a deidentified patient. The tissue sections were double stained using antibodies for Rab11 (shown in red) and β-catenin (shown in green). Blue is DAPI staining. E and F are images of different parts of the same slide, showing horizontal sections and longitudinal sections, respectively, of the colonic crypts. G is an image of a horizontal section of the tumor tissue. The different color combinations, as indicated, of the same region are shown in the parallel panels (E–E””, F–F””, and G–G””). The number 1224N and 1224T represents the sample number of the UMMS tissue bank deidentified patients, with N for normal and T for tumor.
Frozen tissue sections from deidentified patients were examined for Rab11 and β-catenin double immunofluorescence staining. Our final analysis included 70 colon cancer samples that had morphology matching the clinical diagnosis and showed reproducible staining (Figure 1, F and G and Figure S1). After initial analysis, we decided to use an average staining quantification of 12 normal tissues as our reference normal expression level for Rab11. For tumor samples, the cancer area was identified by tissue morphology as well as β-catenin staining that consistently illuminated both normal and cancer epithelia (Figure 1, E–G). The Rab11 signal was captured by confocal imaging, and more than seven small cancer areas (ROI) within each confocal image were quantified, except one sample had only three small areas quantified because it had a limited cancer area (clinical information and quantification data available at https://figshare.com/s/7ec190b0f62a930ea7dc).
Among the 70 cancer samples that were successfully analyzed, 51 showed lower levels of Rab11 protein staining (Table 1). The expression levels fell in the range of 17.6–221.1% of the average normal level, with the median expression level being 49.1% (Table 1). Based on the available clinical information, cancer patients with stage 3 or early metastasis classification, which is N1M1 or N1MX, appear to have a more consistently (lowest P value) lower level of Rab11 protein expression (Table 1). While the sex of the patients did not seem to show any difference, patients that were 70 years or older correlated most consistently (lowest P value) with reduced Rab11 expression (Table 1). For those samples that showed higher expression, we speculate that the timing of tumor development, the different genetic defects such as microsatellite instability (hyper-mutators), and the specific combination of gene mutations (P53, KRAS, APC mutations) may affect the final Rab11 protein level.
We performed similar quantitative analyses using two other antibodies, the β-catenin antibody and the phosho-Stat3 antibody (Table S1). The quantification for β-catenin staining revealed only a modest reduction (median 87.3% of normal level) when compared to that of Rab11 (median 49.1% of normal). The phospho-Stat3 antibody revealed an overall increase (median 113.9% of normal), consistent with a positive correlation of Stat activity in cancer (Groner and von Manstein 2017; Taher et al. 2018; Trivedi and Starz-Gaiano 2018). Therefore, the observed reduction of Rab11 expression is likely specific. A larger scale analysis should help to improve this idea.
The apical localization of Coracle is defective after loss of Rab11 in Drosophila midgut
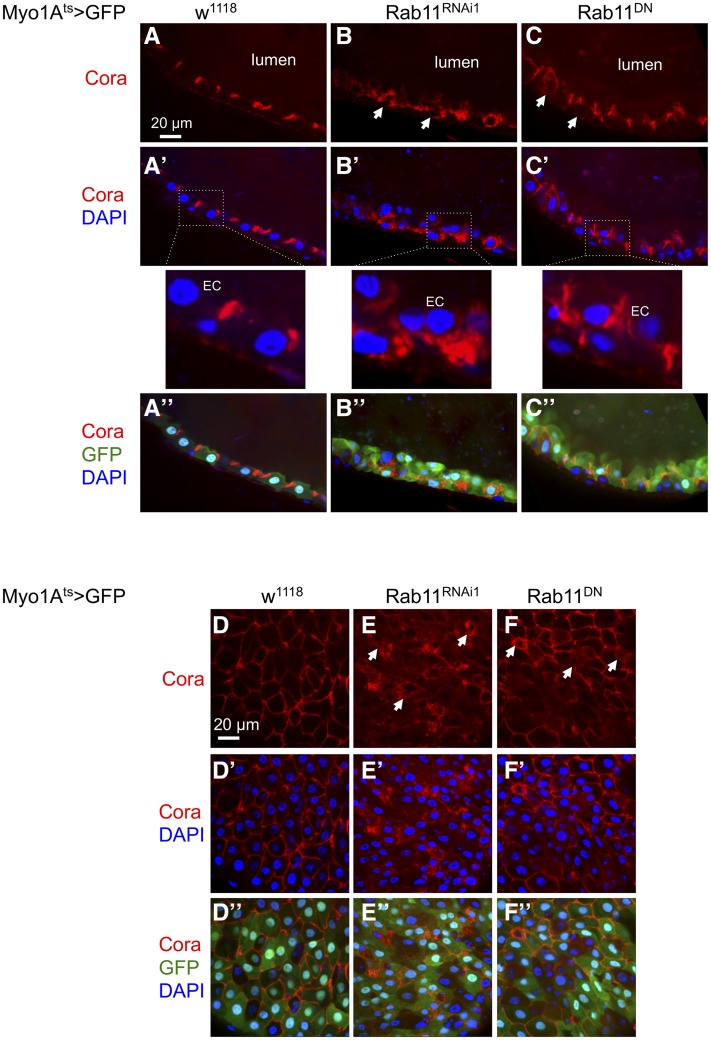
We used Drosophila midgut as a model to investigate the consequences of reducing Rab11 expression. Rab11 loss-of-function mutants are homozygous lethal (Sasikumar and Roy 2009; Mateus et al. 2011). Therefore, we used the adult midgut EC driver Myo1Ats> (Myo1A promoter-Gal4; tubulin-Gal80ts; UAS-CD8GFP) for spatial and temporal control of expression of the Rab11 RNAi and Rab11 S25N dominant negative constructs (Zhang et al. 2007; Jiang et al. 2009; Yu et al. 2014). The Rab11 RNAi1 line (V108382, Materials and Methods) was used in most of these experiments, with the transgene located on the second chromosome, while two other RNAi lines were used to confirm the phenotype whenever possible. Normal ECs in Drosophila midgut have a well-defined basal-apical polarity, and many polarity proteins and junction proteins are localized to the apical side toward the lumen (Jiang et al. 2009; Liang et al. 2017). One of these polarity proteins Coracle can be detected as well-defined, tight staining near the apical/luminal side of ECs (Figure 2A) (Laprise et al. 2009; Sollier et al. 2015). In the Myo1A > Rab11 RNAi guts, however, the Coracle staining exhibited a disorganized pattern, with concomitant ISC proliferation increase and epithelial cell number increase as previously shown (Yu et al. 2014). Much of the staining in mutants appeared to be throughout the cytoplasm of many ECs; this increased cytoplasmic staining led to clear definition of unstained nuclei that was not observed in the control cells (Figure 2, B and E, arrows). Similar expression of a dominant negative Rab11 mutant in ECs caused a similar, albeit milder phenotype of diffused apical and increased cytoplasmic staining (Figure 2, C and F, arrows). Therefore, loss of Rab11 causes disruption of epithelial cell polarity, and this phenotype is reminiscent of that shown in the mammalian CACO2 intestinal culture cells with Rab11a knockdown (Knowles et al. 2015).
Figure 2.
The apical protein Coracle is mislocalized after loss of Rab11. Drosophila midgut images after staining with Coracle (Cora) antibody. The control flies were Myo1Ats > GFP (Myo1A-Gal4, UAS-GFP, tubulin-Gal80ts) crossed with w1118 (w−). The other samples were the Myo1A driver crossed with UAS-Rab11 dsRNA (Rab11RNAi1) or a UAS-Rab11DN construct (dominant negative, Rab11 S25N mutation). The resulting flies were used for midgut dissection and staining. (A–C) Sagittal views of the midguts; (D–F) surface views of the midguts. Different color combinations (A’, B’, C’, A“, B’, C“, D’, E’, F’, D“, E’, F’, color combination indicated to the left) are shown for each genotype, with Cora antibody staining shown in red, GFP shown in green, and DAPI staining for DNA shown in blue. The white arrows in B, C, E, and F indicate examples of ECs showing mislocalized, cytoplasmic Cora staining. The lumen is as indicated, and for the midgut epithelium the luminal side is the apical side of the ECs.
Expression profiling of the Drosophila midgut suggests multiple defects after loss of Rab11
We further investigate the defects in the midgut after loss of Rab11 by RNA sequencing and differential gene expression analysis. Total RNA from three sets of midgut samples from Myo1Ats > w1118 as wild type or Rab11 RNAi as mutant were subjected to sequencing. The results indicate that >2000 genes had statistically significant change of RNA expression in the midgut (GEO project number GSE92705). Importantly, upd3 RNA was among the highly changed gene products (Table 2, top part), demonstrating that the profiling was consistent with previous experimental results (Yu et al. 2014). Pathway and ontology analysis suggest that some potentially interesting components including the Ras signaling and IGF-1 pathways are mis-regulated after loss of Rab11 (Table 2, lower part). Among the 2450 genes on the list (https://figshare.com/s/9c15d272ffca6ba900c4), most house-keeping genes such as tubulin and actin are not included.
Among the genes that exhibited a higher fold change, ImpL2 attracted our attention because of its function in regulating metabolism in recent Drosophila cancer models (Figueroa-Clarevega and Bilder 2015; Kwon et al. 2015). ImpL2 is a secreted factor that binds to insulin-like peptides and inhibits insulin signaling. ImpL2 secreted from cancer tissues can inhibit the metabolism of the whole animal in different Drosophila cancer models involving Scribble or Yorkie, leading to a tissue-wasting phenotype resembling that of cancer patients at late stages. Therefore, our gene profiling results suggest a new possibility that loss of Rab11 not only promotes intestinal tissue proliferation but also causes metabolic changes.
Loss of Rab11 interacts with oncogenic Ras to increase ImpL2 expression
By quantitative PCR, we detected highly increased ImpL2 RNA expression after 5 days of Rab11 RNAi in adult ECs, while the expression was mildly increased after 2–3 days (compare first two columns in Figure 3, A–C). Multiple pathways including Ras, Wnt, and EGF can promote ISC division in the Drosophila midgut (Guo et al. 2016; Jiang et al. 2016). We tested some of these pathways and found that the expression of the constitutively active GTP-bound mutant RasV12 (Lee et al. 1996) showed the highest synergy with Rab11 RNAi to increase ImpL2 RNA expression when the level was only mildly increased in the early time points of Rab11 RNAi (Figure 3A). This RasV12 mutant has been used in both intestine and other tissues of Drosophila for various cancer models (Jiang et al. 2011; Figueroa-Clarevega and Bilder 2015). Meanwhile, the coexpression of the Wingless/Wnt pathway also led to a modest increase of ImpL2 expression after a longer time (Figure 3, B and C).
An ImpL2 homolog in human is the IGFBP7. We plotted the expression of IGFBP7 in various colon cancer samples as listed in the public domain of The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Network 2012). There is a consistently, albeit marginally, higher level of the transcripts in various colon cancer samples (Figure 3D). We have also used a commercially available IGFBP7 antibody to perform staining on 12 pairs of colon cancer and matching normal tissues. The results, as shown in Figure 3, E and F and Figure S2, appear to show slightly higher staining around the apical/luminal side in some cancer samples (arrows in the images). However, more investigation is required to confirm whether there is significant change of this protein in cancer tissues.
Loss of Rab11 and expression of ImpL2 in midgut regulate the metabolism of neighboring tissues
To investigate the metabolic phenotypes regulated by ImpL2, we quantified the fat body associated lipid, the total triglyceride of whole flies, and the ovary size. These assays were used successfully to assess the metabolic defects induced by expression of oncoproteins in both adult gut and larval imaginal discs (Figueroa-Clarevega and Bilder 2015; Kwon et al. 2015). Staining of lipid by Nile Red showed that lower levels of lipid were present in the fat tissues underneath the female adult epidermis, especially after coexpression of Rab11 dsRNA and RasV12 (Figure 4, A–D and Figure S3, A–D). Similar staining using Bodipy revealed smaller lipid droplets in the single and double expression combinations (Figure 4, E–H). Quantitative assay of total triglyceride revealed that while expression of Rab11 dsRNA or RasV12 individually had modest effects, the coexpression of the two constructs led to a significant decrease of total triglyceride in whole animals (Figure 4I). Meanwhile, the ovary size of female flies is highly dependent on the metabolic activity of the animal. Measurement of ovary size showed a significant decrease after expression of Rab11 dsRNA or RasV12 individually. Moreover, the coexpression of the two constructs led to a more substantial decrease (Figure 4, J–M, quantified in Figure 4N).
We found that in the Rab11 dsRNA- or RasV12-expressing flies there was no significant change of food dye ingestion (Figure S3E), no change of expression of two major insulin-like peptides DILP2 and DILP3 in fly heads (Figure S3F), and no decrease of overall body weight (Figure S3G). Therefore, we conclude that the expression of Rab11 dsRNA or RasV12 did not affect metabolism through reduction of food intake.
We also investigated the direct effect of ImpL2 by expression of a UAS-ImpL2 construct in midgut ECs using the Myo1A-Gal4 driver. This manipulation caused a highly reduced ovary size (Figure 4O). Lastly, we performed a genetic suppression experiment by using an ImpL2 RNAi construct expressing in ECs. Even though ImpL2 RNAi by itself had no significant effect on ovary size, the inclusion of this ImpL2 RNAi in the Rab11 RNAi background caused a significant although not complete rescue of the ovary size (Figure 4P). Unfortunately, we were not able to test the ImpL2 RNAi in the combined Rab11 RNAi and RasV12 background due to the many transgenic chromosomes involved. Nonetheless, the results together suggest that increased expression of ImpL2 in the midgut after loss of Rab11 contributes to reduction of animal metabolism.
Multiple regulatory components act downstream of Rab11 and RasV12
The Rab11 GTPase has been shown to regulate endosomal trafficking and probably can control more than one cargo or pathway during trafficking (Guichard et al. 2014; Zhang and Gao 2016). One of the better-studied transcriptional regulators in midgut homeostasis is Yorkie (Yki), which is a growth-promoting factor that can activate upd3 expression but is normally restricted by the upstream Ste20 kinases Hippo and Misshapen (Ren et al. 2010; Li et al. 2014). Previous reports have shown that Yki expression in midgut EBs or ECs can lead to highly increased target gene expression and ISC proliferation (Li et al. 2014; Kwon et al. 2015). Therefore, we combined yki RNAi with Rab11 RNAi or RasV12 in midgut ECs and examined ImpL2 expression. The yki RNAi caused strong suppression of RasV12-induced expression of ImpL2, as well as that of upd3 (Figure 4, M and N). However, we were not able to obtain significant suppression of Rab11 RNAi-induced ImpL2 expression (Figure 4O), suggesting that multiple pathways are involved and Yki is only one of the downstream components.
We used the previously generated upd3-promoter-lacZ reporters (Jiang et al. 2009) to assess the response to Rab11 RNAi and RasV12. Four consecutive promoter fragments of ∼1 kb each surrounding the upd3 transcription start site were fused with the LacZ reporter (Figure 5A), and transgenic flies containing each of these reporter constructs were crossed with either Rab11 RNAi or RasV12 flies. By β-galactosidase protein staining, we observed an increased expression of the reporter upd3.1-lacZ, in both the Rab11 RNAi and RasV12 background (Figure 5, B–D). The other three promoter reporters did not show such increased staining (Figure 5, E–J). These results suggest that the Rab11 RNAi and RasV12 pathways can regulate a common promoter element, or separate elements that are located within this 1-kb region. A recent report also suggests that Hippo, TGF-β, and MAPK pathways regulate multiple target elements on the complex upd3 promoter (Houtz et al. 2017).
We also tested whether the two pathways can converge on another promoter element. The JNK pathway has been shown to function within ECs to activate growth of the midgut (Jiang et al. 2009). The TRE-driven dsRed reporter contains four head-to-tail concatenated copies of an artificial tetradecanoylphorbol acetate response element (TRE) that constitute an optimal Jun/Fos (AP-1) heterodimer binding site, fused upstream of a basal promoter followed by the DsRed (Chatterjee and Bohmann 2012). When this reporter was crossed to the Rab11 RNAi and RasV12 background, detectable expression was observed (Figure 5K), demonstrating that AP1 activation of a target promoter can happen downstream of the two pathways. These results again suggest that the pathways acting downstream of Rab11 RNAi and RasV12 can converge onto similar regions of target promoters (Figure 5L).
Discussion
Our results illustrate that the loss of Rab11-induced inflammation in the mammalian intestine as previously reported may have the biological effect of enhancing cancer progression, perhaps through collaboration with other pathways (Figure 5L). There are lower levels of Rab11 protein staining in most colon cancer samples examined, and the reduction is more consistently found in patients of older age and with early metastasis. Because of the limited availability of good quality cancer and matching normal tissues, we elected to use the method of comparing each tumor sample to an average of 12 normal samples. With this consideration in mind, we focused on the trend of correlation based on clinical groups, rather than individual patients. While there may be individual variations, the sum of all analyses should provide good support for the interpretation. Moreover, the analysis of two other proteins, the membrane associated protein β-catenin and the transcription factor phosphorylated-Stat, shows that neither exhibited as significant a decrease in protein staining as that of Rab11, therefore further supporting our conclusion.
Another concern is that inactivating Rab11 causes defects in apical–basal polarity, therefore making it difficult to measure the abundance of proteins that normally accumulate at these sites, such as Rab11 itself. As shown for β-catenin in cancer tissues and Coracle in mutant fly guts, these two proteins remained rather easily detectable. This suggests that not all apical proteins have as substantial a reduction of protein levels as that occurring in Rab11. The genomic sequences shown in the TCGA database suggest that the human Rab11 gene is not consistently mutated, and the RNA level is only slightly reduced in patient samples. Therefore, the lower Rab11 protein expression we observed in patient tissue sections may represent a combination of transcriptional and post-transcriptional defects.
While metabolic changes in the Rab11 mutant may be predictable based on substantial previous findings regarding Rab11 (Takahashi et al. 2012; Puri et al. 2018), to demonstrate the regulation of ImpL2 as a downstream event is a new finding and provides an important link to the observed metabolic defects. Rab11 has a well-established function in regulating intracellular endosomal trafficking. For instance, the movement of E-cadherin through the cell membrane depends on Rab11 function in an infection model (Guichard et al. 2014). Knockout of Rab11a in the mouse intestinal epithelium and knockdown of Rab11a in Caco2 cells caused mislocalization of a number of apical proteins including E-cadherin, Syntaxin3, Ezrin, and ERM (Knowles et al. 2015). Therefore, the loss of apical-basal polarity is a conserved mechanism, and such a defect may exacerbate the oncogenic transformation of the intestinal epithelium. Whether the downstream components Coracle and Yki are direct cargos that ultimately control metabolism requires further experiments.
Recent reports in both mammalian cells and Drosophila imaginal discs have demonstrated that AP1 and Yki can synergistically activate target genes (Zanconato et al. 2015; Liu et al. 2016). By various assays, we demonstrate the functional importance of Yki downstream of RasV12. Moreover, we show that a upd3-promoter-lacZ and an AP1-reporter are activated by both pathways. However, we have not been able to demonstrate a functional requirement of Yki downstream of Rab11, nor have we been able to show suppression of upd3 or ImpL2 expression when various Fos, Jun, or JNK RNAi or dominant negative constructs were crossed to Rab11 RNAi and RasV12. It is possible that a complex downstream event after loss of Rab11 that can interact with multiple oncogenic pathways leads to the midgut phenotypes (Figure 5L). Further assays to identify various components that mediate the development of the phenotypes after loss of Rab11 may provide novel insights into cancer progression.
Acknowledgments
P.W.S. was a visiting scientist from Sanofi-Genzyme Corp., 153 2nd Ave., Waltham, MA 02451. We thank Juerg Straubhaar and Sharvari Gujja for help with bioinformatics, the Bloomington Drosophila Stock Center for TRiP lines and stocks. Transgenic RNAi fly stocks were also obtained from the Vienna Drosophila Resource Center (VDRC). We thank Young Kwon, Jin Jiang, Huaqi Jiang, Norbert Perrimon, and Andreas Bergmann for Drosophila stocks; and Karl Simin of the University of Massachusetts Medical School (UMMS) Cancer Center Tissue Bank for advice. Y.T.I. is a member of the UMass DERC (DK32520), and a member of the UMass Center for Clinical and Translational Science (UL1TR000161). W.W., B.Z., and Y.T.I. are members of the Guangdong Innovative Research Team Program (No. 201001Y0104789252) and members of the Science and Technology Program of Guangzhou (No. 201704030044). N.G. is supported by a National Institutes of Health (NIH) grant (DK102934). Y.T.I. is supported by NIH grants (GM107457, DK083450). The authors declare no competing or financial interests.
Author contributions: Y.N., N.G., and Y.T.I. conceived the project and designed the experiments; Y.N., S.Y., Q.L., A.A., and P.W.S. carried out the experiments; Z.J. designed the colon cancer experiment and analyzed the cancer tissue staging; W.W., B.Z., and Y.J.K.E. performed RNA sequencing and bioinformatics analysis; Y.N. and Y.T.I. wrote the manuscript, and all authors amended the manuscript.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.6084/m9.figshare.8340950, https://doi.org/10.6084/m9.figshare.6985223, and https://doi.org/10.6084/m9.figshare.7505612.
Communicating editor: D. Andrew
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A., Jiang J., Ip Y. T., 2009. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4: 49–61. 10.1016/j.stem.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaghany T., Wilson J. D., Chu Q., Mills G., 2012. Genetic alterations in colorectal cancer. Gastrointest. Cancer Res. 5: 19–27. [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Perdigoto C. N., Southall T. D., Brand A. H., Schweisguth F., 2010. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137: 705–714. 10.1242/dev.039404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J., Clevers H., 2016. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143: 3639–3649. 10.1242/dev.133132 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network , 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N., Bohmann D., 2012. A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One 7: e34063 10.1371/journal.pone.0034063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., et al. , 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu N., Wang C., Huang P., Huang H., et al. , 2018. Transient Scute activation via a self-stimulatory loop directs enteroendocrine cell pair specification from self-renewing intestinal stem cells. Nat. Cell Biol. 20: 152–161 (erratum: Nat. Cell Biol. 20: 991). 10.1038/s41556-017-0020-0 [DOI] [PubMed] [Google Scholar]
- Cordero J. B., Stefanatos R. K., Scopelliti A., Vidal M., Sansom O. J., 2012. Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 31: 3901–3917. 10.1038/emboj.2012.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstmann R., Vermeulen L., Guinney J., Kopetz S., Tejpar S., et al. , 2017. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 17: 79–92 (erratum: Nat. Rev. Cancer 17: 268). 10.1038/nrc.2016.126 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Clarevega A., Bilder D., 2015. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev. Cell 33: 47–55. 10.1016/j.devcel.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner B., von Manstein V., 2017. Jak Stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol. Cell. Endocrinol. 451: 1–14. 10.1016/j.mce.2017.05.033 [DOI] [PubMed] [Google Scholar]
- Guichard A., Nizet V., Bier E., 2014. RAB11-mediated trafficking in host-pathogen interactions. Nat. Rev. Microbiol. 12: 624–634. 10.1038/nrmicro3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Ohlstein B., 2015. Stem cell regulation. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science 350: aab0988 10.1126/science.aab0988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Driver I., Ohlstein B., 2013. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J. Cell Biol. 201: 945–961. 10.1083/jcb.201302049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Lucchetta E., Rafel N., Ohlstein B., 2016. Maintenance of the adult Drosophila intestine: all roads lead to homeostasis. Curr. Opin. Genet. Dev. 40: 81–86. 10.1016/j.gde.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Pan C., Liu C., Lv X., Yang X., et al. , 2015. Gut-neuron interaction via Hh signaling regulates intestinal progenitor cell differentiation in Drosophila. Cell Discov. 1: 15006 10.1038/celldisc.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtz P., Bonfini A., Liu X., Revah J., Guillou A., et al. , 2017. Hippo, TGF-β, and Src-MAPK pathways regulate transcription of the upd3 cytokine in Drosophila enterocytes upon bacterial infection. PLoS Genet. 13: e1007091 10.1371/journal.pgen.1007091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G., et al. , 2009. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137: 1343–1355. 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Grenley M. O., Bravo M. J., Blumhagen R. Z., Edgar B. A., 2011. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8: 84–95. 10.1016/j.stem.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Tian A., Jiang J., 2016. Intestinal stem cell response to injury: lessons from Drosophila. Cell. Mol. Life Sci. 73: 3337–3349. 10.1007/s00018-016-2235-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthan R., Senger J. L., Kanthan S. C., 2012. Molecular events in primary and metastatic colorectal carcinoma: a review. Pathol. Res. Int. 2012: 597497 10.1155/2012/597497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuria S., Karpac J., Biteau B., Hwangbo D., Jasper H., 2012. Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLoS Genet. 8: e1003045 10.1371/journal.pgen.1003045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles B. C., Weis V. G., Yu S., Roland J. T., Williams J. A., et al. , 2015. Rab11a regulates syntaxin 3 localization and microvillus assembly in enterocytes. J. Cell Sci. 128: 1617–1626. 10.1242/jcs.163303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., et al. , 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Song W., Droujinine I. A., Hu Y., Asara J. M., et al. , 2015. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 33: 36–46. 10.1016/j.devcel.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P., Lau K. M., Harris K. P., Silva-Gagliardi N. F., Paul S. M., et al. , 2009. Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase form a novel group of epithelial polarity proteins. Nature 459: 1141–1145. 10.1038/nature08067 [DOI] [PubMed] [Google Scholar]
- Lee T., Feig L., Montell D. J., 1996. Two distinct roles for Ras in a developmentally regulated cell migration. Development 122: 409–418. [DOI] [PubMed] [Google Scholar]
- Li Q., Li S., Mana-Capelli S., Roth Flach R. J., Danai L. V., et al. , 2014. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev. Cell 31: 291–304. 10.1016/j.devcel.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Han L., Shi L., Lin X., 2013. Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev. Cell 24: 133–143. 10.1016/j.devcel.2012.12.010 [DOI] [PubMed] [Google Scholar]
- Liang J., Balachandra S., Ngo S., O’Brien L. E., 2017. Feedback regulation of steady-state epithelial turnover and organ size. Nature 548: 588–591. 10.1038/nature23678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Xu N., Xi R., 2008. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455: 1119–1123. 10.1038/nature07329 [DOI] [PubMed] [Google Scholar]
- Liu X., Li H., Rajurkar M., Li Q., Cotton J. L., et al. , 2016. Tead and AP1 coordinate transcription and motility. Cell Rep. 14: 1169–1180. 10.1016/j.celrep.2015.12.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus A. M., Gorfinkiel N., Schamberg S., Martinez Arias A., 2011. Endocytic and recycling endosomes modulate cell shape changes and tissue behaviour during morphogenesis in Drosophila. PLoS One 6: e18729 10.1371/journal.pone.0018729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C. A., Perrimon N., 2006. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439: 475–479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- O’Brien L. E., Soliman S. S., Li X., Bilder D., 2011. Altered modes of stem cell division drive adaptive intestinal growth. Cell 147: 603–614. 10.1016/j.cell.2011.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A., 2006. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439: 470–474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Oldefest M., Nowinski J., Hung C. W., Neelsen D., Trad A., et al. , 2013. Upd3 – an ancestor of the four-helix bundle cytokines. Biochem. Biophys. Res. Commun. 436: 66–72. 10.1016/j.bbrc.2013.04.107 [DOI] [PubMed] [Google Scholar]
- Osman D., Buchon N., Chakrabarti S., Huang Y. T., Su W. C., et al. , 2012. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell Sci. 125: 5944–5949. 10.1242/jcs.113100 [DOI] [PubMed] [Google Scholar]
- Perochon J., Carroll L. R., Cordero J. B., 2018. Wnt signalling in intestinal stem cells: lessons from mice and flies. Genes (Basel) 9: E138 10.3390/genes9030138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C., Vicinanza M., Ashkenazi A., Gratian M. J., Zhang Q., et al. , 2018. The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P–RAB11A. Dev Cell 45: 114–131.e8. 10.1016/j.devcel.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Wang B., Yue T., Yun E. Y., Ip Y. T., et al. , 2010. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. USA 107: 21064–21069. 10.1073/pnas.1012759107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikumar S., Roy J. K., 2009. Developmental expression of Rab11, a small GTP-binding protein in Drosophila epithelia. Genesis 47: 32–39. 10.1002/dvg.20441 [DOI] [PubMed] [Google Scholar]
- Sollier K., Gaude H. M., Chartier F. J., Laprise P., 2015. Rac1 controls epithelial tube length through the apical secretion and polarity pathways. Biol. Open 5: 49–54. 10.1242/bio.015727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D. A., Santaolalla R., Strobel S., Dheer R., Abreu M. T., 2012. Cancer in inflammatory bowel disease: lessons from animal models. Curr. Opin. Gastroenterol. 28: 327–333. 10.1097/MOG.0b013e328354cc36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher M. Y., Davies D. M., Maher J., 2018. The role of the interleukin (IL)-6/IL-6 receptor axis in cancer. Biochem. Soc. Trans. 46: 1449–1462. 10.1042/BST20180136 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Kubo K., Waguri S., Yabashi A., Shin H. W., et al. , 2012. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125: 4049–4057. 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- Talmon G., Holzapfel M., DiMaio D. J., Muirhead D., 2012. Rab11 is a useful tool for the diagnosis of microvillous inclusion disease. Int. J. Surg. Pathol. 20: 252–256. 10.1177/1066896911430959 [DOI] [PubMed] [Google Scholar]
- Tan D. W., Barker N., 2014. Intestinal stem cells and their defining niche. Curr. Top. Dev. Biol. 107: 77–107. 10.1016/B978-0-12-416022-4.00003-2 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A., Jiang J., 2014. Intestinal epithelium-drived BMP controls stem cell self-renewal in Drosophila adult midgut. eLife 3: e01857 10.7554/eLife.01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A., Shi Q., Jiang A., Li S., Wang B., et al. , 2015. Injury-stimulated Hedgehog signaling promotes regenerative proliferation of Drosophila intestinal stem cells. J. Cell Biol. 208: 807–819. 10.1083/jcb.201409025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi S., Starz-Gaiano M., 2018. Drosophila Jak/STAT signaling: regulation and relevance in human cancer and metastasis. Int. J. Mol. Sci. 19: E4056 10.3390/ijms19124056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiopoulos A. G., Kostakis I. D., Koutsilieris M., Papavassiliou A. G., 2012. Colorectal cancer stem cells. Stem Cells 30: 363–371. 10.1002/stem.1031 [DOI] [PubMed] [Google Scholar]
- Welch H. G., Robertson D. J., 2016. Colorectal cancer on the decline – why screening can’t explain it all. N. Engl. J. Med. 374: 1605–1607. 10.1056/NEJMp1600448 [DOI] [PubMed] [Google Scholar]
- Yu S., Nie Y., Knowles B., Sakamori R., Stypulkowski E., et al. , 2014. TLR sorting by Rab11 endosomes maintains intestinal epithelial-microbial homeostasis. EMBO J. 33: 1882–1895. 10.15252/embj.201487888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., et al. , 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17: 1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Schulze K. L., Hiesinger P. R., Suyama K., Wang S., et al. , 2007. Thirty-one flavors of Drosophila rab proteins. Genetics 176: 1307–1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gao N., 2016. RAB and RHO GTPases regulate intestinal crypt cell homeostasis and enterocyte function. Small GTPases 7: 59–64. 10.1080/21541248.2016.1159274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Rasmussen A., Lee S., Agaisse H., 2013. The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Dev. Biol. 373: 383–393. 10.1016/j.ydbio.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request.
Supplemental materials include Figures S1–S3 and Table S1.
RNAseq fastq files deposited in GEO (accession number GSE92705).
Cancer tissue clinical information and image analysis excel file in https://doi.org/10.6084/m9.figshare.6985223.
Full list of genes in Tale 2 top part excel file in https://doi.org/10.6084/m9.figshare.7505612. Supplemental material available at FigShare: https://doi.org/10.6084/m9.figshare.8340950.