Abstract
Divergent selection works when an allele establishes in the subpopulations in which it is adaptive, but not in the ones in which it is deleterious. While such a locally adaptive allele is maintained, the target locus of selection works as a genetic barrier to gene flow or a barrier locus. The genetic divergence (or FST) around the barrier locus can be maintained, while in other regions of the genome, genetic variation can be mixed by gene flow or migration. In this work, we consider theoretically the evolutionary process of a barrier locus, from its birth to stable preservation. Under a simple two-population model, we use a diffusion approach to obtain analytical expressions for the probability of initial establishment of a locally adaptive allele, the reduction of genetic variation due to the spread of the adaptive allele, and the process to the development of a sharp peak of divergence (genomic island of divergence). Our results will be useful to understanding how genomes evolve through local adaptation and divergent selection.
Keywords: speciation, population genetics, diffusion theory, migration, gene flow, divergent selection
A genomic island of divergence could arise when a locally adapted allele establishes in a certain subpopulation (e.g., Wu 2001; Turner et al. 2005; Nosil 2012). This local establishment could be stably maintained by divergent selection if the allele confers sufficient benefit in the subpopulations in which it is adaptive, but not in the ones in which it is deleterious. Such a locus works as a genetic barrier to gene flow, or a barrier locus, because migrants are maladaptive. Due to recombination, the genomic region that is affected by divergent selection is limited, thereby creating a peak of divergence along the chromosome (i.e., a genomic island of divergence). Further development of multiple barrier loci in the genome might initiate ecological speciation (Turner et al. 2005; Nosil 2012). Here, we are interested in the evolutionary dynamics of a barrier locus, from its establishment via a partial local sweep, through the emergence of a peak of divergence, to its stable preservation.
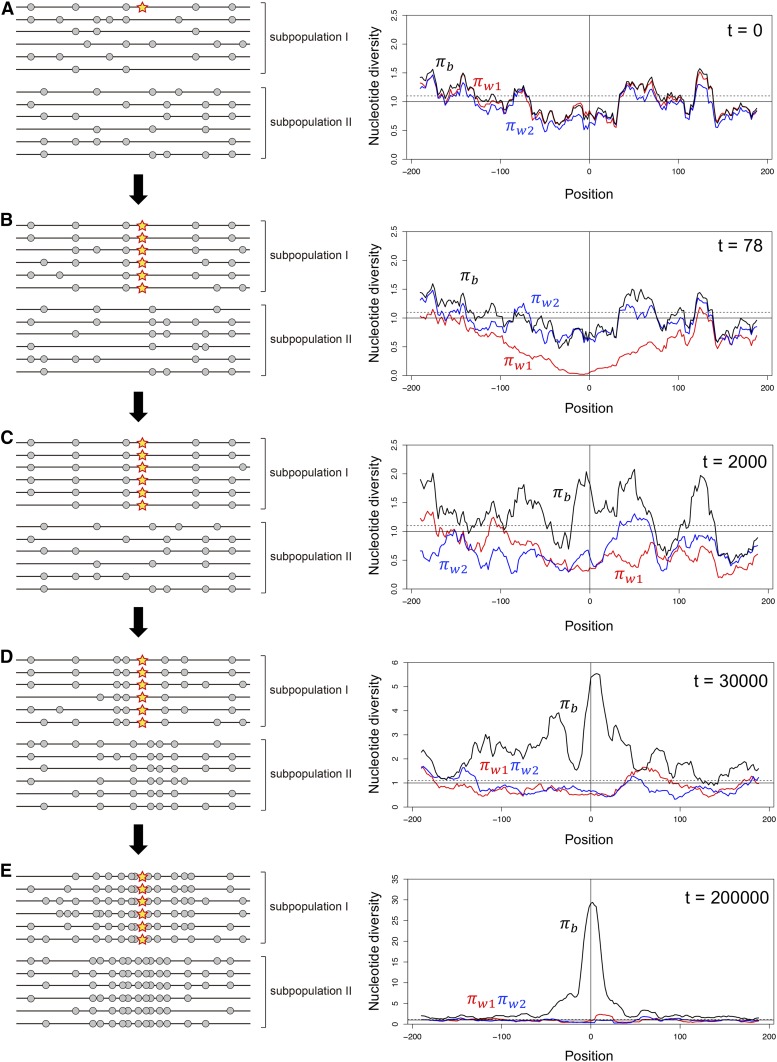
We consider the process theoretically by dividing into three phases—establishment, consolidation and equilibrium—as illustrated in Figure 1. We consider a simple situation with two subpopulations: I and II. Assuming a relatively high migration rate between them, the levels of polymorphism within the two subpopulations are similar to each other (measured by the heterozygosities, and , for subpopulations I and II). In the meantime, the population divergence (measured by , the heterozygosity between the two subpopulations) is very low (Figure 1A). Then, a de novo mutation (star in Figure 1A) arises in subpopulation I, in which the mutation is advantageous, whereas it is maladaptive (or deleterious) in subpopulation II. In the establishment phase, the mutation spreads in subpopulation I and nearly fixes (Figure 1B), but its frequency in subpopulation II is low because it should be selected against if migrated into subpopulation II. In a strict sense, this is not a fixation that can be treated mathematically as an absorbing state, because migration keeps providing maladaptive alleles. Therefore, after Kimura (1954), we hereafter use the terminology of “quasi-fixation” for this nearly fixed state. The quasi-fixation should occur quickly, and a partial local selective sweep occurs in subpopulation I (Figure 1B), thereby establishing a barrier locus. Around the barrier locus, it is typical to observed a “block” of region with low genetic variation in subpopulation I, with a slightly elevated genetic divergence (FST). The consolidation phase starts after the initial establishment of the barrier locus, during which the block of low genetic variation gradually shrinks in length over time by recombination and migration, while new mutations accumulate and the divergence between two subpopulations increases particularly near the barrier locus (Figure 1C). Then, at the end, a stable sharp peak of divergence arises in the equilibrium phase (Figure 1D). The equilibrium shape of the peak of divergence is determined mainly by the balance between selection intensity and the rates of recombination and migration.
Figure 1.
Illustrating the evolution of a barrier locus in a simple two-population model with fairly high migration between them. (A) A locally adaptive de novo mutation arises in subpopulation I at position 0. A typical pattern of polymorphism is shown in left. The star is the locally adaptive mutation, and gray circles are neutral polymorphism in the surrounding region. The right panel shows the spacial distributions of nucleotide diversity obtained by a simulation. The simulation considers two subpopulations with population sizes are , between which symmetric migration is allowed at rate We assume selection intensity and . The entire simulated region is 400 kb if a population recombination rate of = 0.001 per site is assumed. See Appendix A for details about the simulation. The polymorphism levels within the two populations ( and ) are in red and blue, and divergence between the two populations is in black. , and can be considered as the averages of , and in a 20-kb window. The y-axis is adjusted such that under neutrality (the solid line) and the broken line exhibits , (i.e., = the genomic average). (B) The mutation quasi-fixes in subpopulation I, causing a drastic reduction in . (C) Migration shuffles polymorphisms in the two subpopulations, while selection works to maintain the quasi-fixation of the mutation. (D) The divergence gradually increases around the mutation, and (E) a clear peak of divergence arises. It should be noted that the star is fixed in the sample (left panes in B–E), but it does not necessarily mean that it is fixed in subpopulation I because there should be maladaptive immigrant alleles at a low frequency.
The scope of this work is to provide a unified and comprehensive theoretical understanding of the evolution of a new peak of divergence, from its birth to stable preservation in equilibrium. We use a simple two-population model, where migration is allowed between subpopulations I and II. Suppose a de novo mutation arises that confers a selective advantage specific to subpopulation I, which is the initial state of our system. Under this model, we derive the following:for the establishment phase,
The establishment probability of the de novo mutation, that is, the probability that the mutation quasi-fixes in subpopulation I.
The expected reduction of genetic variation within subpopulations I and II after the quasi-fixation (i.e., partial local sweep).
for the consolidation phase,
3. the evolutionary dynamics at both the barrier locus and the linked neutral sites since the quasi-fixation.
and for the equilibrium phase,
4. The expected shape of the peak of divergence at equilibrium.
Several theoretical works have focused on a specific part of these aspects. For (1) the establishment probability, perhaps the most flexible, useful theoretical framework was introduced by Barton (1987) in a general multiple-island-model. By using a diffusion approximation, Barton (1987) derived a partial differential equation for the establishment probability. Essentially the same result was obtained by Pollak (1966), who used a branching process, and the establishment probability was derived from a probability generating function. Barton’s differential equation was solved, and closed forms of the establishment probability have been available only in several specific situations in continuous habitat models. In a one-dimensional continuous habitat model, Barton (1987) solved his partial differential equation analytically, assuming two forms of fitness gradient (linear and pocket). Kirkpatrick and Peischl (2013) used a branching process, from which they obtained a partial differential equation that is similar to that of Barton (1987). Then, the authors successfully incorporated changes in fitness gradient along time, and derived an approximate establishment probability.
In discrete population models, Barton’s general formula (and also Pollak’s one) is difficult to handle, and has not been fully explored even in a simple two-population model with symmetric migration. Therefore, the currently available theoretical results are not based on Barton’s differential equation, and have some limitations. In a continent-island model with migration, Aeschbacher and Bürger (2014) solved the establishment probability of a locally beneficial mutation linked to another locally beneficial mutation that was already established, where mathematical treatment is facilitated by unidirectional migration (see also Yeaman et al. 2016). Yeaman and Otto (2011) obtained an approximate establishment probability by using a heuristic approach that is a combination of the leading eigenvalue of the transition matrix of deterministic process and Kimura’s formula of fixation probability (Kimura 1962). As shown in their paper, this formula well describes the establishment probability when a de novo mutation arises in the adapted subpopulation (i.e., subpopulation I in our model), but it does not work when it arises in the maladapted subpopulation (i.e., subpopulation II in our model). Recently, Tomasini and Peischl (2018) provided an approximate establishment probability by assuming a slightly supercritical branching process. Their formula works well under the assumption of slightly supercritical approximation, namely, the leading eigenvalue of the transition matrix of deterministic model is not large, but it may not work well when the selection intensity in the adapted subpopulation is very large.
In this work, we derive a closed form formula of the establishment probability in a two-population model with bidirectonal migration along the formulation of Barton (1987). We extend Barton’s derivation with simultaneous quadratic equations and solve them allowing unequal subpopulation sizes. Our formula is more general than previous ones (Yeaman and Otto 2011; Tomasini and Peischl 2018); it works with strong selection and it allows that a de novo mutation can arise either subpopulation I or II.
To the best of our knowledge, there is no theoretical work on the hitch-hiking process of a partial local sweep in a two-population model. With regard to a single population model, many studies investigated the reduction of polymorphism theoretically due to a selective sweep. These studies considered a selected site and a linked neutral site, and assumed that a sufficiently advantageous mutation arises and goes to fixation in the population. Along this fixation, they derived how much polymorphism can be reduced at the linked site. Maynard Smith and Haigh (1974) first obtained the reduction of polymorphism, where the stochastic effect of genetic drift at the linked site was ignored. The model was extended to include the stochastic effect by using a coalescent approach (Kaplan et al. 1989) and by using a diffusion method (Stephan et al. 1992; see also Barton 1998; Etheridge et al. 2006). Durrett and Schweinsberg (2004) used a different approach for a faster approximate simulation of a selective sweep and derived some analytical expressions (see also Schweinsberg and Durrett 2005).
There are several theoretical studies on a sweep in multi-population models available, but these considered a fixation across multiple subpopulations, not a local fixation. In a model with multiple subpopulations, Slatkin and Wiehe (1998) and Santiago and Caballero (2005) considered the process where a beneficial mutation fixes in the entire population through weak migration. Kim and Maruki (2011) allowed stronger migration, and derived an analytical expression in a two-population model. Our interest is different from these studies in that we consider a locally beneficial mutation that can quasi-fix only in the subpopulation in which it is beneficial (not in the entire population). We here extend the theory of Stephan’s diffusion model (Stephan et al. 1992) to a two-population model, and consider how much polymorphism can be reduced locally at a linked site after a partial local sweep.
We then turn to the evolutionary dynamics at both the barrier locus and the linked neutral sites after the completion of the partial local sweep. We here consider this process after a local sweep as described in Figure 1. A local sweep creates a “block” of a fairly long region with almost no genetic variation within the subpopulation in which the new mutation is adaptive (i.e., subpopulation I in our model). In this work, given an arbitrary configuration of genetic variation after a local sweep, we obtain, analytically, the moments of allele frequency at a linked site, with which we describe how a genomic island decays. Yeaman et al. (2016) investigated a similar problem in a different situation, where an genomic island evolves due to the clustering of two barrier loci. In their model, considering a secondary contact, erosion starts when there already are a large number of fixed sites that spread over the genome, and islands appear because selection works to maintain divergence at selected site(s), while losing divergence in other regions through homogenization by migration. By using the structured coalescent, they obtained the expected spatial distribution of FST (in terms of relative coalescent time) around selected sites as a function of the time since the secondary contact. They also considered the scenario where a de novo mutation broadens a genomic island that has been created by a barrier locus, and revealed the final shape of a two-barrier island is the same as the genomic island under the secondary-contact scenario. However, their derivation did not consider the effect of selective sweep of the de novo mutation. It should be noted that, because our derivation accepts any arbitrary initial allele frequency at a linked site, it can be applied to any situation, not only that after a secondary contact but also that after a local sweep.
In the equilibrium phase, the balance between selection, migration, recombination, and mutation holds. Theoretical treatment at equilibrium is relatively straightforward, and there are several theoretical studies on the spatial distribution of FST (Charlesworth et al. 1997; Akerman and Bürger 2014; Yeaman et al. 2016). Under our framework for the consolidation phase, essentially the same result can be provided as a special case, with time going to infinity.
Model and Results
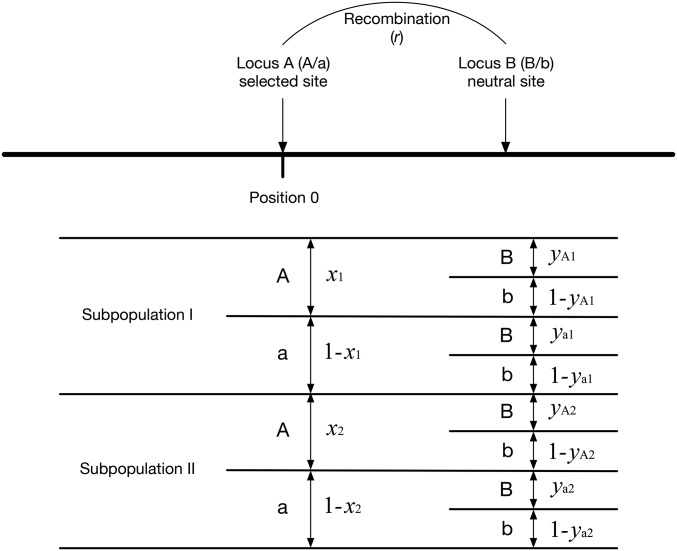
We consider a two-population model with discrete generations and monoecious diploid individuals that mate at random. The diploid population sizes of subpopulations I and II are assumed to be constant at and , respectively. As illustrated in Figure 1, we are specifically interested in selection for local adaptation in subpopulation I. We consider a genomic region encompassing a selected site at position 0, which is referred to as locus A (Figure 2). At locus A, two alleles (A/a) are allowed with no recurrent mutation between them. Allele A confers a selection coefficient in subpopulation I and in subpopulation II (we assume and ). Additive selection is assumed so that the fitness of individuals with AA, Aa, and aa are given by , , and 1 in subpopulation I, and , and 1 in subpopulation II. Selection works only at this selected site, and all remaining sites are assumed to be neutral. For the following derivation under a two-locus model, we consider a secondary neutral site (locus B), at which two alleles (B/b) are allowed with recurrent mutation between them (Figure 2). The mutation rate from allele B to allele b is u, and that from allele b to allele B is v. The recombination rate between the two loci, A and B, is r.
Figure 2.
Two-locus model used in this work. Locus A targeted by divergent selection is placed at position 0, and a linked neutral locus B can be placed at an arbitrary position. The frequencies of allele A and allele a at locus A, and those of allele B and allele b at locus B in the two subpopulations are illustrated.
The system starts when a de novo mutation (allele A) arises in a single individual either in population I or II, where allele a is fixed in both subpopulations. Therefore, the initial state is or , where and are frequencies of the new allele A in subpopulations I and II, respectively. Throughout this article, we assume strong selection and weak migration so that maladapted individuals are rare in each subpopulation once the initial establishment is achieved.
Establishment probability
We derive the establishment probability of a new de novo allele using the general framework of Barton (1987), who derived a simultaneous quadratic equation from the diffusion theory. This section focuses only on the selected locus A (see Figure 2), at which we are interested in the probability that allele A quasi-fixes in subpopulation I. Following Haldane (1927), we approximate the establishment probability by the probability that the new mutation increases in frequency and escapes from immediate extinction. This is because, under the assumption of strong selection, the behavior of such a mutation is almost deterministic once it escapes from extinction by genetic drift.
Let be the establishment probability when the frequencies of allele A are and in the two subpopulations. By using an analogous procedure to Barton (1987), we derive and , the establishment probability when the new allele arises in subpopulations I and II, respectively. According to the diffusion theory, F satisfies the Kolmogorov backward equation:
| (1) |
where is the proportion of immigrant individuals just after migration in subpopulation I (II). To keep the subpopulation sizes constant, we assume , and we ignore higher order terms of (i.e., , ). This is reasonable because of the assumption that the establishment probability is determined mainly at low frequencies. Because the extinction probabilities of individual mutations are independent, we can write F as
| (2) |
where is the extinction probability of a new mutant in subpopulation i; therefore, is determined as . After substitution of Equation 2 into Equation 1, one can show that solutions to Equation 1 can be obtained by solving the following system of equations:
| (3) |
which corresponds to equation 4b in Barton (1987). Equation 3 can be rearranged to
| (4) |
| (5) |
where . Equation 4 can be solved by using the solution of a cubic equation. Equations 4 and 5 have, at most, one solution that fulfills and . The condition where Equations 4 and 5 have such a solution is or , which corresponds to the situation where the deterministic growth rate of the mutant allele is positive (see Appendix B for details).
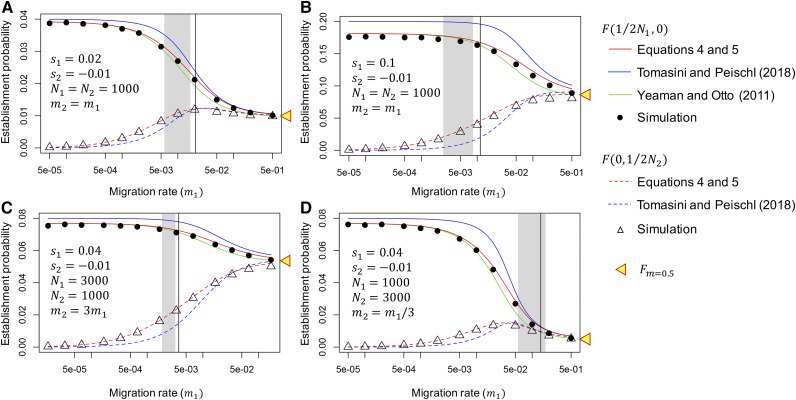
Figure 3 shows the establishment probability from Equations 4 and 5 as a function of migration rate. We first consider a symmetric model , and two selection intensities ( and ) are assumed, while is fixed (Figure 3, A and B). The establishment probability can be computed when a locally adaptive mutation arises either in subpopulation I or II, represented as and , respectively. We performed a forward simulation to check the performance of our analytical result (Appendix A). For each parameter set, we ran 1,000,000 independent replications of the simulation, and counted the number of replications where the new allele A was preserved in 10,000 generations. The establishment probability was then obtained as the proportion of such replications. Therefore, it includes replications where two alleles (A and a) coexisted (case C) and those where allele A is completely fixed in both subpopulations (case F). The proportion of case C in the established replications decreases with increasing migration rate (see below).
Figure 3.
Establishment probability as a function of migration rate. (A) Weak selection ( and ) and (B) strong selection ( and ) are assumed in a symmetric model . (C and D) Asymmetric population settings are considered ( in C and in D). Our result in red is compared with those of Yeaman and Otto (2011) and Tomasini and Peischl (2018), together with the result of our forward simulation. The establishment probability for a mutation that arises in subpopulation I is shown by solid lines and closed circles, and that for a mutation that arises in subpopulation II is shown by broken lines and open triangles. The establishment probability at the high migration limit is shown by a yellow triangle. In each panel, a gray region is placed such that the proportion of the replications where two alleles (A and a) coexisted in the left, while in the right. The vertical line presents the critical migration rate, above which allele A fixes in the entire population, obtained by Equation 6 (see text for detail).
Our result (red in Figure 3) is in excellent agreement with the simulation result: is approximately when the migration rate is very low, consistent with the prediction in a single population model (Kimura 1957). As the migration rate increases, decreases and increases, and they become similar to each other. With a very high migration rate , the two subpopulations can be considered as a single random-mating population, and the fixation probability of a single mutation is mainly determined by the average selection coefficient, , namely, where (Nagylaki 1980). Indeed, in our simulations, allele A was fixed in both populations in almost all established cases (). In each panel in Figure 3, a gray region is placed such that in the left, while in the right. It has been demonstrated that, under a deterministic model, the condition where two alleles (A and a) coexist is and
| (6) |
(Nagylaki and Lou 2008). The critical migration rate predicted by this equation is shown by the vertical lines in Figure 3, which roughly agrees with the line of (see Yeaman and Otto 2011). This indicates that the pattern dramatically changes in a short range of , and the left side is the scope of this article. Similar results were also obtained in asymmetric models ( in Figure 3C and in Figure 3D).
Figure 3 quantitatively compares our analytical results with those of previous studies (Yeaman and Otto 2011; Tomasini and Peischl 2018). It is found that from Yeaman and Otto (2011) is almost as good as ours, but unfortunately was not provided by Yeaman and Otto (2011). It seems that Tomasini and Peischl (2018) overestimates and underestimates .
Reduction of genetic variation due to a selective sweep
When a new locally adaptive mutation (aA) arises and quasi-fixes in subpopulation I, genetic variation in the surrounding region in subpopulation I should be reduced dramatically due to the hitch-hiking effect. In this section, we consider a two-locus model as defined in Figure 2. We derive the degree of reduction in heterozygosity at a linked neutral site (locus B) in subpopulation I, , by extending the diffusion approach of Stephan et al. (1992), who investigated the effect of hitch-hiking in a single population model with no population structure.
Overview of Stephan’s diffusion approach:
We first briefly introduce the approach of Stephan et al. (1992), which provides the basis of our derivation below. The expected reduction of heterozygosity at locus B for a single population model with diploid size N is denoted by . With the assumption of strong selection, Stephan et al. (1992) assumed that the behavior of the frequency (x) of the beneficial allele A with selection coefficient, s, follow a deterministic function:
| (7) |
where selection is additive. This deterministic treatment works once the frequency of allele A exceeds a certain threshold such that it escapes from immediate extinction by genetic drift, as mentioned in the previous section. This treatment makes the following derivation much easier because the dynamics can be described by a two-dimensional diffusion equation. It should be noted that x with no subscript denotes the frequency of allele A in the single population model, whereas, in our two-population model, the frequencies of allele A in subpopulations I and II are denoted by and , respectively (see Figure 2). We consider another biallelic neutral locus (B/b), and the recombination rate between this neutral locus and the selected locus is assumed to be r. is the frequency of allele B among A-chromosomes, and is the frequency of allele B among a-chromosomes. Then, the expected changes of an arbitrary function is described as the following ordinary differential equation:
| (8) |
where L is a differential operator of the Kolmogorov backward equation:
| (9) |
By using this formula, Stephan et al. (1992) solved the first and second moments of and after a sweep, from which the expected reduction of heterozygosity at the linked site can be computed numerically. With some approximation, Stephan et al. (1992) further obtained a nice closed form of the solution:
| (10) |
In this work, we found that this equation somehow undervalues the effect of random genetic drift at the linked neutral locus, perhaps due to the approximation of Stephan et al. (1992). We noted that, in Equation 10, goes to in the limit of , whereas heterozygosity should decrease by genetic drift by a factor of per generation, even in the absence of the hitch-hiking effect. Here, we consider the expected reduction of heterozygosity along the quasi-fixation as , because the fixation time is approximately given by
This equation means that the expected reduction of heterozygosity due to genetic drift, , is not negligible compared to . To correct for this factor, we add this into Equation 10:
| (11) |
We found that this heuristic approach is in very good agreement with the numerical solution obtained by directly computing Equation 9.
Local sweep in the two-population model:
In this work, we extend Stephan derivation (Stephan et al. 1992) to the two-population model defined above. We first consider the dynamics of the new mutant allele frequency at the selected locus (position 0) in subpopulation I. The major difference from the corresponding formula in Stephan et al. (1992) (i.e., Equation 7) is that the effect of migration should be considered in the two-population model. Because allele A is very rare in subpopulation II under the assumption of strong selection and low migration, we can ignore migrants with A allele from subpopulations II to I. Then, the dynamics of can be approximated by a deterministic function:
| (12) |
We set the time such that when the mutation arises, and when the mutation quasi-fixes. We next consider the neutral locus B (B/b). As illustrated in Figure 2, is the frequency of haplotype A-B among A-chromosomes in subpopulation I (II), and is the frequency of haplotype a-B among a-chromosomes in subpopulation I (II). We assume that is very small throughout the sweep process. Then, the expected changes of an arbitrary function is described as the following ordinary differential equation:
| (13) |
where L is a differential operator of the Kolmogorov backward equation. Following Ohta and Kimura (1969), we obtain L for our model as
| (22) |
This equation is derived such that several terms are added to Equation 9 for incorporating random genetic drift within subpopulation II (first term on the third line) and the effect of migration. The second term of (second line) is for migration from subpopulation II to subpopulation I, and the term of (third line) is for migration from subpopulation I to subpopulation II. Due to the assumption of strong selection, migrant A-chromosomes from subpopulation I to subpopulation II should be selected out immediately. Therefore, the migration rate of locus B can be effectively considered as the product of migration rate and the probability that at least one recombination event occurs before selection purges allele A, :
| (15) |
(Bengtsson 1985). Then, Equation 13 directly allows us to compute the first and second moments of and after the quasi-fixation of allele A (i.e., and . We obtain heterozygosity within each subpopulation ( and ) and between them at as
| (16) |
from which the expected reduction of heterozygosity is obtained as
| (17) |
Generally, involves the initial frequencies, and . However, it should be noted that their quantitative effect on is not large unless and are not very similar.
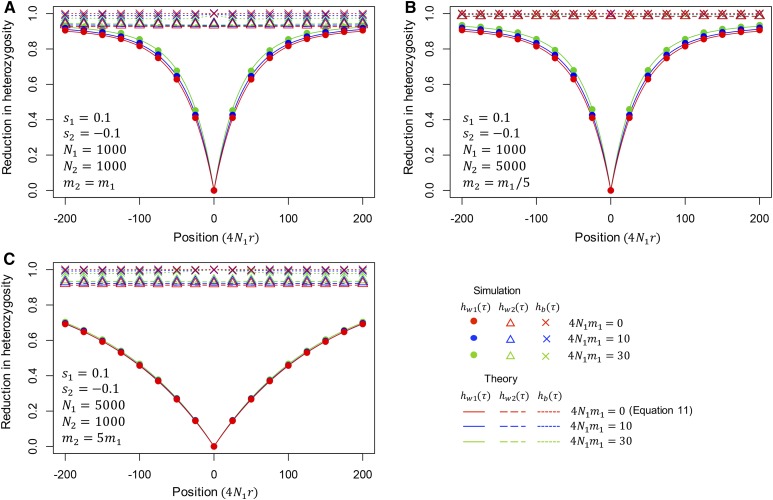
Figure 4 shows the effect of migration on the reduction in heterozygosity. The plot in red is the case of no migration, where our result is essentially identical to that of Stephan et al. (1992), and the plots in blue and green are for migration cases. We consider three pairs of population sizes, in A, in B, and in C. For each parameter set, filled circles represent the average over 100,000 replications of forward simulation (see Appendix A). In Figure 4, , and are plotted such that before the sweep, so that directly corresponds to . In all cases, our theoretical result from Equation 14 is in excellent agreement with the simulation results. It is found that the effect of a local partial sweep seems to be only on subpopulation I, and there is almost no effect on the variation in subpopulation II. Moving away from the selected site at position 0, is larger for a higher migration rate. This is because that migration brings standing variation maintained in subpopulation II into subpopulation I, thereby increasing the polymorphism level in subpopulation I. We observed is slightly elevated around the selected site at position 0. If we assume roughly approximates FST, where is heterozygosity when the two subpopulations are merged together, it can be said that a local sweep creates a relatively wide block of region with elevated FST, which can be considered as an initial peak of divergence.
Figure 4.
The expected reduction of heterozygosity after a local partial sweep in the two-population model. Position is shown in from the selected site. Theoretical results for and computed from (14)–(17) by assuming for convenience, but very similar results were obtained for other values of and . In the case of no migration (red), our results is identical to Stephan et al. (1992) (i.e., Equation 11). Results for three parameter sets are shown: (A) s_1=0.1, s_2=-0.1,N_1=1000, N_2=1000, m_2=m_1, (B) s_1=0.1, s_2=-0.1,N_1=1000, N_2=5000, m_2=m_1/5, (C) s_1=0.1, s_2=-0.1,N_1=5000, N_2=1000, m_2=5m_1.
Consolidation of a barrier locus with a peak of divergence
When a new locally adaptive mutation (aA) quasi-fixes in subpopulation I, a block of region with elevated FST arises, where genetic variation in subpopulation I is dramatically reduced (Figure 1B). In this section, by using the two-locus model defined in Figure 2, we consider the process after this state, but our derivation is flexible enough to plug in any initial state.
We use a similar diffusion approach to the previous section but we focus on the behavior of and . The expected changes of an arbitrary function is described as the following ordinary differential equation:
| (18) |
where L is a differential operator of the Kolmogorov backward equation, which is given by
| (19) |
The two terms in the first line of Equation 19 are for random genetic drift within subpopulations I and II, and the terms in the second line describes the deterministic change of the frequency of allele B due to mutation and migration. As well as the previous section, we use the effective migration rate (Bengtsson 1985):
| (20) |
where is the relative selection coefficient of maladapted individuals in subpopulation I. is defined by Equation 15. We consider the dynamics of the first and second order moments, and put . By using Equation 18, we derive a differential equation for as follows:
| (21) |
where is the matrix given by
| (14) |
and . See Appendix C for details. By solving Equation 21, is given by
| (23) |
where is the identity matrix of size 5 (Appendix C). at equilibrium is given by . Our solution at equilibrium is well consistent with previous studies (Charlesworth et al. 1997; Yeaman et al. 2016) that used the coalescent approach (see Appendix D for a proof).
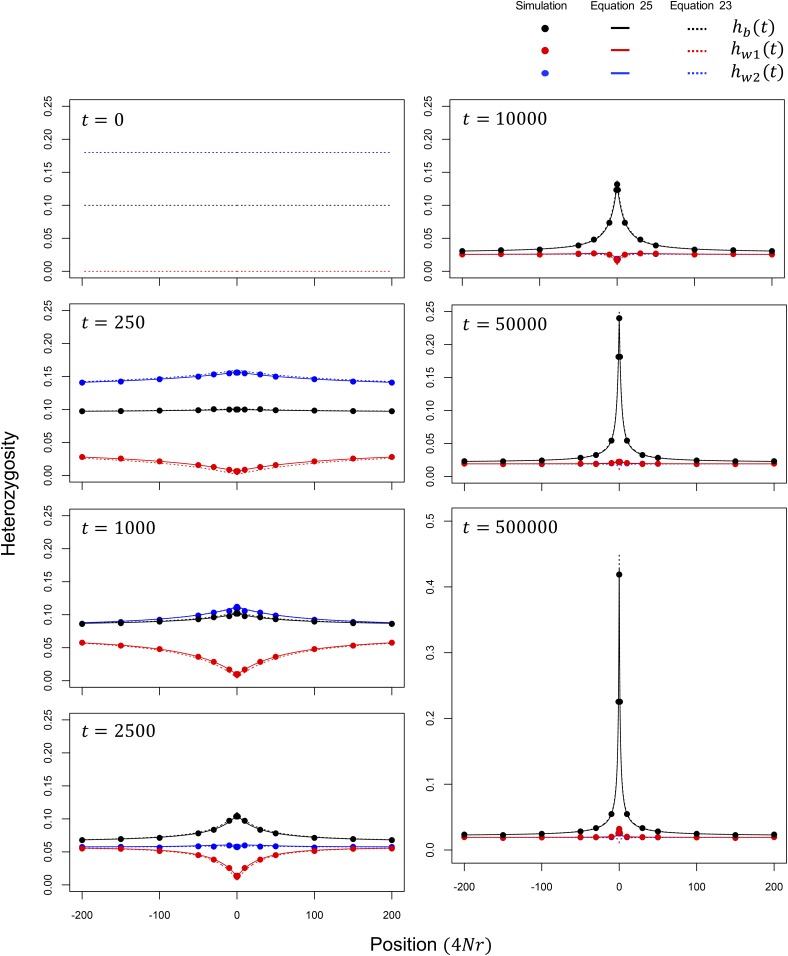
Figure 5 compares our theoretical results from Equation 23 (broken lines) with simulation results (closed circles). are assumed to represent a strong selection case. As the initial condition , we set , and , representing a situation after a local sweep in subpopulation I. Equation 23 describes how a sharp peak of divergence grows along time. As time goes, and become closer to each other, and eventually reaches their equilibrium values (t10,000). also decreases except for a short region surrounding the selected site. The rate of erosion (decrease of ) increases moving away from the selected site. At the selected site, gradually increases and eventually develops a sharp peak, and simultaneously and also exhibit a small peak that can be created by migration between two subpopulations. It reaches an equilibrium after a significant amount of time, where the selection-migration balance holds so that the shape of the peak does not change much.
Figure 5.
Temporal change of heterozygosity after a local sweep in subpopulation I. The spacial distributions of , and are shown for seven time points (t = 0, 250, 1000, 2500, 10,000, 50,000, and 500,000 generations after a sweep). Position in the simulated regions is shown in from the selected site. , and are assumed. Theoretical results from Equations 23 and 25 are shown by broken and solid lines, respectively. Simulation results (closed circles) are the averages over 50,000 replications of forward simulations.
Figure 5 shows that Equation 23 (broken lines) is consistent with the simulation results, but the agreement could be further improved if we account for the presence of locally maladapted allele, i.e., allele A (a) in subpopulation I (II). At migration-selection equilibrium, alleles A and a are present in subpopulation I and II at an expected frequency of and , respectively. Even though these frequencies are small under our assumption of weak migration relative to selection, we show in the following that the approximation in Equation 23 can be improved by accounting for them. Let us focus on the fate of a single neutral allele at the neutral locus linked to an immigrant locally maladaptive allele. We ask how long such a neutral immigrant allele survives on the locally maladaptive background. The linked neutral allele will either be eliminated by selection against the locally maladapted allele in its background, or it recombines off its deleterious background onto a locally beneficial background. The expected time until elimination by selection or recombination in subpopulations I and II are, respectively, given by
| (24) |
Therefore, the expected numbers of neutral alleles from the other subpopulation with the maladapted allele is and in subpopulations I and II, respectively.
Let the frequencies of B in subpopulations I and II including those on the locally maladapted background and . Accounting for the presence of locally maladaptive alleles, the first- and second-order moments of are:
| (25) |
See Appendix C for details. Figure 5 shows that Equation 25 fits the simulation results better than Equation 23. A notable improvement is seen in for a narrow region around the selected site. Because Equation 23 ignores the presence of maladaptive alleles (assuming their immediate death), Equation 23 predicts a small dip, but our simulation demonstrated rather that a small peak arises. This small peak of is well described by the improved Equation 25.
Discussion
In the early stages of ecological speciation with gene flow, divergent selection is required to maintain phenotypes that are adaptive to each niche (Wu 2001; Turner et al. 2005; Nosil 2012). Each target locus of divergent selection works as a barrier locus to migration, because maladaptive migrants should be selected out in a short time. Such a barrier locus can be formed if a locally adaptive mutation arises and becomes established in subpopulations where it is adaptive. This quasi-fixation of a locally adaptive mutation causes a local partial sweep, thereby creating a block of region with elevated F. Then, while divergent selection maintains the mutation, recombination shuffles genetic variation in the linked regions and mutations accumulate around the barrier locus. Through this process, a sharp peak of divergence develops in a narrow region around the barrier locus.
This article considers theoretically the evolutionary behavior of a barrier locus, from its initial establishment to stable preservation. The process was divided into three phases: establishment, consolidation and equilibrium (Figure 1). We obtained (1) the establishment probability of a locally adaptive mutation, (2) the expected reduction of genetic variation within subpopulations I and II after a partial local sweep, (3) the evolutionary dynamics at both the barrier locus and the linked neutral sites since the sweep, and (4) the expected shape of the peak of divergence around the barrier locus at equilibrium.
For (1), we derived a closed-form formula of the establishment probability along the formulation of Barton (1987). Our simulations showed that our theoretical results for and outperform the previous approximations, although Yeaman and Otto (2011)’s heuristic approach is almost as good as ours. Because we focused on divergent selection so that allele A is quasi-fixed in subpopulation I, whereas allele a is quasi-fixed in subpopulation II, we assumed and . However, as shown in Figure 3, it is possible that either allele A or a could fix in the entire population, even if and hold, although it might take an extremely long time. In contrast, Gavrilets and Gibson (2002) and Whitlock and Gomulkiewicz (2005) obtained the probability of such eventual fixation in the entire population. These studies and ours can be understood in a single framework as follows. Assuming and , the establishment of allele A first occurs and is maintained quite stably for a long time, but with time going toward infinity, allele A could fix in the entire population most likely when the average selection coefficient is positive, while allele a could likely fix when is negative. This is why our formula of the establishment probability (Equation 2) is the same as the numerator of the fixation probability when is positive (equations 7 and 8 in Gavrilets and Gibson 2002 and equation 6 in Whitlock and Gomulkiewicz 2005). On the other hand, the establishment probability significantly differs from the fixation probability of Gavrilets and Gibson (2002) and Whitlock and Gomulkiewicz (2005) when is negative because such a mutation hardly goes to eventual fixation, although it can be maintained as a quasi-fixed state for a sufficiently long time.
For (2), we extended the diffusion method of Stephan et al. (1992) to our two-population model. Because the beneficial allele A quasi-fixes only in one subpopulation, the process is very similar to that of a single population model (Stephan et al. 1992), except that migration between two subpopulations has some effect. Our theoretical result (see Figure 4) demonstrated a relatively minor effect of migration; with an increasing migration rate, the level of polymorphism in subpopulation I increases because migration brings genetic variation from subpopulation II.
For (3) and (4), we considered the evolutionary dynamics at both the barrier locus and the linked neutral sites since the quasi-fixation, followed by the development of a stable peak of divergence around the barrier locus. This process to equilibrium can be described by a single Equation 25. Furthermore, Equation 25 is flexible enough to plug in any initial state, such as a secondary contact of already diverged subpopulation. To demonstrate this, in Supplemental Material, Figure S1, we compare the pattern after a local sweep (left panels) and that after a secondary contact (right panels) (see also Appendix E for details). After a secondary contact, is already high across the genome, and gradually decreases, but selection works to keep divergence around the selected site, thereby creating a peak of divergence. After a very long time (i.e., in equilibrium), the shape of the peak becomes identical to that after a sweep, as pointed out by Yeaman et al. (2016). We further performed simulations to investigate how robust our derivation is when the selection intensity is reduced (although we assumed strong selection). The results with 10 times lower selection intensity are shown in Figure S2. This selection intensity is fairly weak, and close to the lower limit to maintain the quasi-fixation state of the two alleles. Yet, Equation 25 is in fairly good agreement with the simulation results, although the performance of Equation 23 is not very good. This is because the frequency of maladaptive alleles is not negligible with a reduced selection intensity.
We thus developed analytical expressions for the evolutionary behavior of a barrier locus, from its emergence to development of a peak of divergence. In the early stages of ecological speciation, it is possible that multiple barrier loci develop and genomic islands of divergence arise, but this does not necessarily mean that the emergence of genomic islands of divergence always results in speciation. It is possible that genomic islands of divergence could disappear due to environmental changes, or by chance, and no speciation occurs. To achieve speciation, many other forces would be necessary, including emergence of additional islands (Feder et al. 2012a,b; Via 2012; Aeschbacher and Bürger 2014; Yeaman et al. 2016), further divergence on a genomic-scale possible due to a reduction in migration rate, and environmental changes. More theoretical studies are needed to fully understand the process to ecological speciation.
Acknowledgments
We thank S. Takuno and three anonymous reviewers for helpful comments. This work was supporter by JSPS (Japan Society for the Promotion of Science) and SOKENDAI (The Graduate University for Advanced Studies).
Appendix
Appendix A: Forward Simulation
Here, we describe the setting and assumptions of our forward simulations. A model with two subpopulations (I and II) is used. Subpopulations I and II consist of and haploids. We are interested in how DNA sequence evolves at the population level around a selected locus. We considered a genomic region encompassing a selected locus at the center, and assumed the infinite-site model for simulating patterns of nucleotide polymorphisms (e.g., Figure 1). For other simulations, we consider a two-locus model with the selected locus and a linked neutral locus. The recombination rate between the two loci is r. The fitness of an individual is determined by the allelic state at the selected locus: The fitness of an individual with allele A and a are, respectively, and 1 in subpopulation I, and and 1 in subpopulation I. Every generation, migration is allowed such that individuals are swapped between the two subpopulations, Then, to construct a new population in the next generation, and individuals are chosen randomly from the current subpopulations I and II, respectively, where their fitness is taken into account. No recurrent mutation is allowed at this site in order to trace the fate of the mutation (unless otherwise mentioned). In contrast, at the linked neutral locus, recurrent mutation is allowed at rate μ per generation. Heterozygosities within and between ( and ) subpopulations can be scored at any arbitrary time point.
Appendix B: The Solution of Equations 4 and 5
First, we present a proof that there is, at most, one solution that fulfills and , and the condition on which such a solution exists is or . Then, we give a closed expression of the solution.
For and to satisfy and , and are needed. Note that because the migration rate and population size are always positive. Although, in this work, we consider only the case of , Equations 4 and 5 may also work in the case of . Therefore, here, we present the proof that allows . We set and note that the first derivative of is . We discuss the complementary following three cases.
-
From Equation 5, , then is needed. Because the x-coordinate of the vertex of , , is not greater than a, increases monotonically when . Noting that , there is only one solution to .
-
and
From Equation 5, , then is needed. Because and the x-coordinate of the vertex of , , is smaller than 0, > 0 when . Therefore, whether has a solution or not in (0, ) depends on the sign of . If , i.e., , there is no solution. Otherwise, there is only one solution.
-
and
From Equation 5, , then is needed. Because the x-coordinate of the vertex of , , is smaller than 0, increases monotonically when . Noting that , there is only one solution.
Noting that and is negative when , the condition on which one solution exists is rearranged to or . This is the same as the condition where a deterministic model,
| (B1) |
has a positive growth rate. In other words, the matrix in Equation B1 has at least one positive eigenvalue.
Next, we present a closed form of . From the above proof, if there is a nonzero real root of which fulfills and , the root is the largest real root of . Therefore, by using the solution of cubic equation, can be expressed as
| (B2) |
where . In the above expression, we assume the range of principal value of as .
Appendix C: Derivation of Equations 21, 23, and 25
Here, we describe the derivation of Equations 21, 23, and 25 in more detail. By applying Equation 18 to and , we can derive the time derivative of moments of and as follows:
| (C3) |
By setting , and defining as Equation 22, Equation C3 can be rearranged in a matrix form (Equation 21). Then, by using the solution of a linear differential equation with constant coefficients, the solution of Equation 21 is given by
| (C4) |
The solution 23 is further improved by accounting for neutral immigrant alleles linked to maladaptive alleles. To do so, we derive the expected time of a neutral immigrant allele until its elimination by selection or recombination as Equation 24. The expected frequencies of such an allele are and in subpopulations I and II, respectively. and , denote the frequencies of B in subpopulations I and II including those on the locally maladapted background. Then, and can be approximated by
| (C5) |
By using Equation C5 and taking expectations, the first and second-order moments of and are given by Equation 25.
Appendix D: Comparison Between Diffusion and Coalescent at Equilibrium Phase
In the main text, we show that replacing the migration rate in the neutral diffusion equation by the effective migration rate well approximates the effect of linkage with the locus under divergent selection. In a neutral model, heterozygosity at equilibrium in a structured population is already well studied by the coalescent theory under the infinite-site model (reviewed in Wakeley 2009). In this work, we alternatively used the forward diffusion approach because the diffusion approach can be applied to more general conditions. In this Appendix, we show our diffusion result at equilibrium is consistent with that of the coalescent theory.
We attempt to derive the expected heterozygosity under the infinite-site setting along our diffusion-based derivation. In practice, we first consider a K-allele model, and then the results will be transformed to the infinite-site model. Let B allele be one of the alleles at the locus. We put and as frequency of allele B in subpopulation I and II, respectively. In the following derivation, we assume and . The differential operator of the Kolmogorov backward equation is as follows,
| (D1) |
At the equilibrium, we derive the moments up to the second order as
| (D2) |
where , and . In the limit to the infinite-allele model, that is, and , the expected heterozygosity within and between subpopulation goes to
| (D3) |
This result under the infinite-allele setting can be transformed to the infinite-site mode: If we put and n goes to , and are described as
| (D4) |
which is identical with the result from the coalescent theory (Charlesworth et al. 1997; Yeaman et al. 2016).
Appendix E: Comparing the Scenarios of Local Partial Sweep and Secondary Contact
We compute Equation 23 for a scenario of secondary contact, and compared with the results of a local partial sweep shown in Figure 5. For a secondary-contact scenario, we assume that already diverged two subpopulation have merged so that there are a number of fixed sites between the two subpopulations. To make a realization of this situation, we set and , and the other parameters are identical to those used for Figure 5 (i.e., and ). Figure S1 compares the patterns after a local sweep (left panels) and after a secondary contact (right panels). After a secondary contact, is already high across the genome, and gradually decreases but selection works to keep divergence around the selected site, thereby creating a peak of divergence. In equilibrium, the shape of the peak becomes identical to that after a sweep, in agreement with Yeaman et al. (2016).
Figure S2 shows the results with identical parameter sets to those for Figure S1 except for the selection intensity. The purpose is to check how robust our derivation is when the selection intensity is reduced. We here set and . The results with 10 times lower selection intensity are shown in Figure S1. This selection intensity is fairly weak, and it is close to the lower limit to maintain the quasi-fixation state of the two alleles.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8188019.
Communicating editor: G. Coop
Literature Cited
- Aeschbacher S., Bürger R., 2014. The effect of linkage on establishment and survival of locally beneficial mutations. Genetics 197: 317–336. 10.1534/genetics.114.163477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman A., Bürger R., 2014. The consequences of gene flow for local adaptation and differentiation: a two-locus two-deme model. J. Math. Biol. 68: 1135–1198. 10.1007/s00285-013-0660-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1987. The probability of establishment of an advantageous mutant in a subdivided population. Genet. Res. 50: 35–40. 10.1017/S0016672300023314 [DOI] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. 10.1017/S0016672398003462 [DOI] [Google Scholar]
- Bengtsson B. O., 1985. The flow of genes through a genetic barrier, pp. 31–42 in Evolution: Essays in Honor of John Maynard Smith, edited by Greenwood J. J., Harvey P. H., Slatkin M. Cambridge University Press, New York. [Google Scholar]
- Charlesworth B., Nordborg M., Charlesworth D., 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174. 10.1017/S0016672397002954 [DOI] [PubMed] [Google Scholar]
- Durrett R., Schweinsberg J., 2004. Approximating selective sweeps. Theor. Popul. Biol. 66: 129–138. 10.1016/j.tpb.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Etheridge A., Pfaffelhuber P., Wakolbinger A., 2006. An approximate sampling formula under genetic hitchhiking. Ann. Appl. Probab. 16: 685–729. 10.1214/105051606000000114 [DOI] [Google Scholar]
- Feder J. L., Egan S. P., Nosil P., 2012a The genomics of speciation-with-gene-flow. Trends Genet. 28: 342–350. 10.1016/j.tig.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Feder J. L., Gejji R., Yeaman S., Nosil P., 2012b Establishment of new mutations under divergence and genome hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 461–474. 10.1098/rstb.2011.0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S., Gibson N., 2002. Fixation probabilities in a spatially heterogeneous environment. Popul. Ecol. 44: 51–58. 10.1007/s101440200007 [DOI] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection, part v: selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. 10.1017/S0305004100015644 [DOI] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Maruki T., 2011. Hitchhiking effect of a beneficial mutation spreading in a subdivided population. Genetics 189: 213–226. 10.1534/genetics.111.130203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1954. Process leading to quasi-fixation of genes in natural populations due to random fluctuation of selection intensities. Genetics 39: 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1957. Some problems of stochastic processes in genetics. Ann. Math. Stat. 28: 882–901. 10.1214/aoms/1177706791 [DOI] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Peischl S., 2013. Evolutionary rescue by beneficial mutations in environments that change in space and time. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120082 10.1098/rstb.2012.0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J. M., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. 10.1017/S0016672300014634 [DOI] [PubMed] [Google Scholar]
- Nagylaki T., 1980. The strong-migration limit in geographically structured populations. J. Math. Biol. 9: 101–114. 10.1007/BF00275916 [DOI] [PubMed] [Google Scholar]
- Nagylaki T., Lou Y., 2008. The dynamics of migration-selection models, pp. 117–170 in Tutorials in Mathematical Biosciences IV: Evolution and Ecology (Lecture Notes in Mathematics). Springer, Berlin: 10.1007/978-3-540-74331-6_4 [DOI] [Google Scholar]
- Nosil P., 2012. Ecological Speciation. Oxford University Press, Oxford: 10.1093/acprof:osobl/9780199587100.001.0001 [DOI] [Google Scholar]
- Ohta T., Kimura M., 1969. Linkage disequilibrium at steady state determined by random genetic drift and recurrent mutation. Genetics 63: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E., 1966. On the survival of a gene in a subdivided population. J. Appl. Probab. 3: 142–155. 10.2307/3212043 [DOI] [Google Scholar]
- Santiago E., Caballero A., 2005. Variation after a selective sweep in a subdivided population. Genetics 169: 475–483. 10.1534/genetics.104.032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg J., Durrett R., 2005. Random partitions approximating the coalescence of lineages during a selective sweep. Ann. Appl. Probab. 15: 1591–1651. 10.1214/105051605000000430 [DOI] [Google Scholar]
- Slatkin M., Wiehe T., 1998. Genetic hitch-hiking in a subdivided population. Genet. Res. 71: 155–160. 10.1017/S001667239800319X [DOI] [PubMed] [Google Scholar]
- Stephan W., Wiehe T. H., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. 10.1016/0040-5809(92)90045-U [DOI] [Google Scholar]
- Tomasini M., Peischl S., 2018. Establishment of locally adapted mutations under divergent selection. Genetics 209: 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W., Nuzhdin S. V., 2005. Genomic islands of speciation in anopheles gambiae. PLoS Biol. 3: e285 10.1371/journal.pbio.0030285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S., 2012. Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 451–460. 10.1098/rstb.2011.0260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction. Roberts & Company, Greenwood Village, CO. [Google Scholar]
- Whitlock M. C., Gomulkiewicz R., 2005. Probability of fixation in a heterogeneous environment. Genetics 171: 1407–1417. 10.1534/genetics.104.040089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-I., 2001. The genic view of the process of speciation. J. Evol. Biol. 14: 851–865. 10.1046/j.1420-9101.2001.00335.x [DOI] [Google Scholar]
- Yeaman S., Otto S. P., 2011. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65: 2123–2129. 10.1111/j.1558-5646.2011.01277.x [DOI] [PubMed] [Google Scholar]
- Yeaman S., Aeschbacher S., Bürger R., 2016. The evolution of genomic islands by increased establishment probability of linked alleles. Mol. Ecol. 25: 2542–2558. 10.1111/mec.13611 [DOI] [PubMed] [Google Scholar]