Abstract
Activation of the Saccharomyces cerevisiae HO promoter is highly regulated, requiring the ordered recruitment of activators and coactivators and allowing production of only a few transcripts in mother cells within a short cell cycle window. We conducted genetic screens to identify the negative regulators of HO expression necessary to limit HO transcription. Known repressors of HO (Ash1 and Rpd3) were identified, as well as several additional chromatin-associated factors including the Hda1 histone deacetylase, the Isw2 chromatin remodeler, and the corepressor Tup1. We also identified clusters of HO promoter mutations that suggested roles for the Dot6/Tod6 (PAC site) and Ume6 repression pathways. We used ChIP assays with synchronized cells to validate the involvement of these factors and map the association of Ash1, Dot6, and Ume6 with the HO promoter to a brief window in the cell cycle between binding of the initial activating transcription factor and initiation of transcription. We found that Ash1 and Ume6 each recruit the Rpd3 histone deacetylase to HO, and their effects are additive. In contrast, Rpd3 was not recruited significantly to the PAC site, suggesting this site has a distinct mechanism for repression. Increases in HO expression and SWI/SNF recruitment were all additive upon loss of Ash1, Ume6, and PAC site factors, indicating the convergence of independent pathways for repression. Our results demonstrate that multiple protein complexes are important for limiting the spread of SWI/SNF-mediated nucleosome eviction across the HO promoter, suggesting that regulation requires a delicate balance of activities that promote and repress transcription.
Keywords: gene regulation, coactivator, corepressor
EUKARYOTIC gene expression is a highly regulated process requiring a variety of protein factors that ensure proper temporal and cell type control. Activated transcription is initiated by DNA-binding factors that recognize specific sequences in promoters and recruit multi-subunit coactivator complexes (Weake and Workman 2010). These coactivators contain enzymes that can post-translationally modify histones, alter the position of nucleosomes, or evict histones from the DNA, and thus positively influence transcription. Multi-subunit corepressors oppose the action of coactivators, also largely through histone modifications and altered nucleosome positioning, in this case to make the chromatin environment less favorable to expression. A delicate balance between activating and repressing processes is therefore a necessary component of complex gene regulation.
The yeast Saccharomyces cerevisiae provides a useful model system for studying this balance due to its ease of genetic manipulation. In both yeast and higher eukaryotes, the chromatin state is a key determinant of expression level, and chromatin remodelers and modifiers are evolutionarily conserved (Li et al. 2007; Weake and Workman 2010). However, most yeast genes have promoters that are relatively simple compared with those of more complex organisms. A notable exception is the HO gene (Stillman 2013), which has been extensively studied because it displays more sophisticated regulatory mechanisms typically seen in higher eukaryotes.
The HO gene encodes an endonuclease that initiates mating type interconversion in yeast by cleaving the mating type (MAT) locus, initiating MAT allele replacement via gene conversion (Strathern et al. 1982). Transcription of HO is highly regulated both temporally and by cell type, as inappropriate expression could lead to additional double-stranded DNA breaks that would be deleterious to the cell (Stillman 2013). Yeast divides asymmetrically, giving rise to a larger mother cell and a smaller daughter cell from each mitotic division. Only haploid mother cells express the HO gene and are capable of switching their mating type, an evolutionary adaptation that allows mother/daughter pairs to mate and produce diploid progeny (Jensen et al. 1983; Nasmyth 1983). HO expression is also restricted to a narrow window in the cell cycle in late G1, with only a few transcripts produced per cell cycle (Nasmyth 1983; Miura et al. 2008).
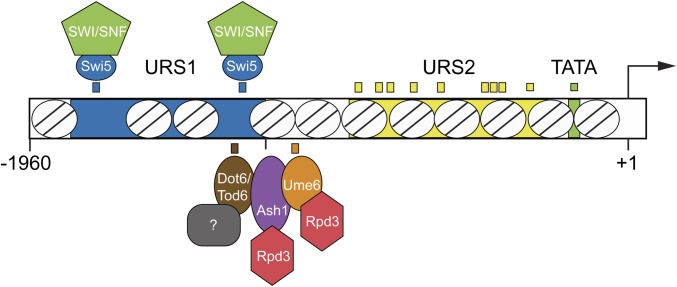
Activation of HO expression is highly complex, involving sequential, ordered recruitment of several transcription factors and coactivators (Cosma et al. 1999; Bhoite et al. 2001; Takahata et al. 2009b). The HO promoter is unusually large relative to others in yeast, with a distance of 3 kb between the HO translation start site and the 3′ end of the upstream ORF. Binding sites for transcription factors have been identified in sequences up to ∼ −1800 relative to the HO translation start site (Breeden and Nasmyth 1987; Stillman et al. 1988; Tebb et al. 1993), and a regulatory long noncoding RNA that originates at −2700 affects HO promoter memory under specific conditions (Yu et al. 2016). Two upstream regulatory sequences (URS) are required for expression of HO, URS1 and URS2 (Nasmyth 1985). The Swi5 activator binds to two sites, A and B, within nucleosome-depleted regions (NDRs) of URS1, at positions −1800 and −1300 (Figure 1A) (Stillman et al. 1988; Tebb et al. 1993; Jiang and Pugh 2009; Brogaard et al. 2012). The SCB binding factor (SBF), consisting of Swi4 and Swi6, associates with nine sites in URS2 (−900 to −200) (Taba et al. 1991); these sites are occluded by nucleosomes which must be evicted prior to binding of SBF (Takahata et al. 2009b; Yarrington et al. 2016).
Figure 1.
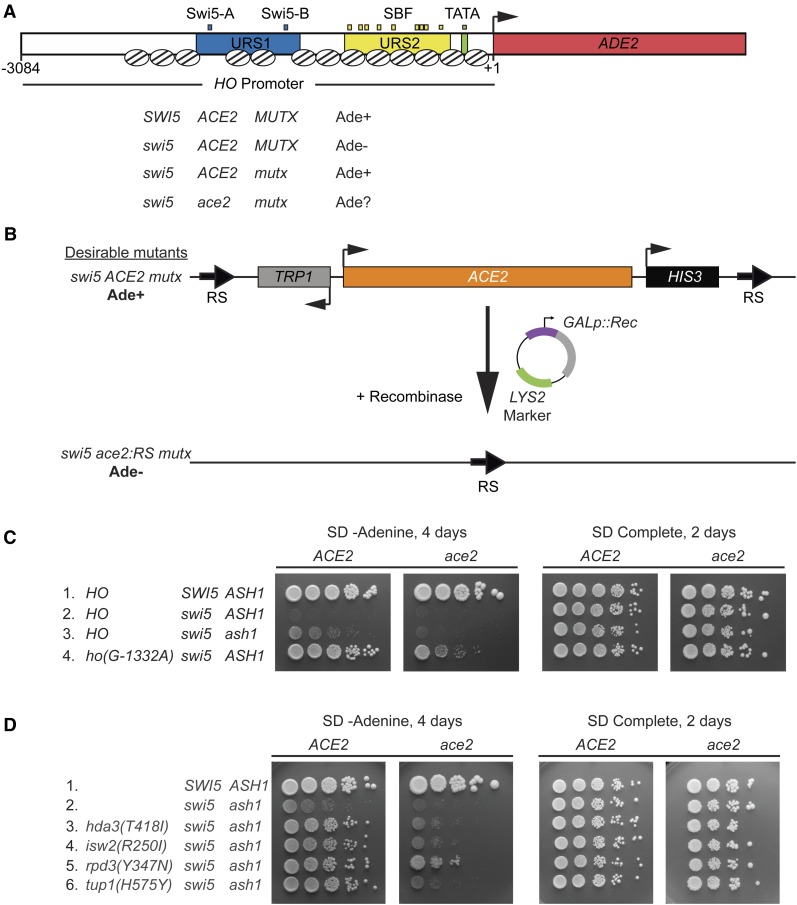
Genetic screen for mutants that allow activation of HO-ADE2 in the absence of Swi5. (A) A schematic of the HO-ADE2 reporter is shown. The ADE2 ORF (red) replaced the HO ORF, preserving the 3084 possible nucleotides of the HO promoter, including Upstream Regulatory Sequences 1 (URS1; blue) and 2 (URS2; yellow). Small blue boxes = Swi5 binding sites; small yellow boxes = SBF binding sites; small green box = TATA. The arrow indicates the position of the ATG (+1). Well-defined nucleosome positions are shown throughout the region from ∼ −2700 to +1 as ovals with lines. A strain that is SWI5 ACE2 and wild type for other factors involved in HO expression (MUTX) grows on media lacking adenine (Ade+), due to expression of the HO-ADE2 reporter. The strain used in the initial screen was swi5 mutant and phenotypically Ade−, since HO-ADE2 is very weakly expressed (swi5 ACE2 MUTX). Mutants identified in the screen (mutx) allowed HO-ADE2 to be expressed in the swi5 strain, making them Ade+ (swi5 ACE2 mutx). The mutx mutants identified in the screen were also tested for their ability to grow in the absence of adenine when lacking ACE2 (swi5 ace2 mutx). (B) A schematic of the ACE2 popout strategy is shown. The endogenous ACE2 locus was modified to introduce RS recombinase target sites as well as TRP1 and HIS3 markers flanking ACE2. Mutant strains from the screen (swi5 ACE2 mutx) were transformed with a plasmid encoding the recombinase under the control of a galactose-inducible promoter (GALp::Rec). Subsequent growth on media containing galactose and selection for Trp- cells with 5-FAA allowed for isolation of strains lacking the ACE2 gene (swi5 ace2:RS mutx). Assessment of the growth on -Ade was used to determine dependence of the Ade+ phenotype on the ACE2 gene. Strains that were Ade+ with ACE2 but Ade− with ace2:RS were kept for further analysis. (C and D) Shown are 10-fold serial dilutions, demonstrating the level of growth of wild-type and mutant strains on media lacking adenine (SD -Adenine, left) and complete media (SD Complete, right). Strain genotypes are listed at the far left, and both the original strains (ACE2) and ace2:RS popout strains (ace2) are shown.
The initiating event for HO activation is the binding of Swi5 to URS1, which occurs when Swi5 enters the nucleus during anaphase (Nasmyth et al. 1990; Cosma et al. 1999). Swi5 recruits three coactivator complexes, the SWI/SNF chromatin remodeler, the SAGA complex containing the Gcn5 histone acetyltransferase, and Mediator; these coactivators are interdependent upon one another for full recruitment to HO (Cosma et al. 1999; Bhoite et al. 2001; Mitra et al. 2006). Following recruitment of coactivators, Swi5 is rapidly degraded (Tebb et al. 1993; Cosma et al. 1999). Remodeling of nucleosomes by SWI/SNF proceeds as a cascade of nucleosome evictions, first at URS1, then at the left end of URS2, and finally at the right half of URS2, allowing SBF to bind to its sites within URS2 (Takahata et al. 2009b; Yarrington et al. 2015). SBF also recruits the SWI/SNF, SAGA, and Mediator coactivator complexes to extend the region of nucleosome eviction to the TATA region, ultimately allowing association of RNA polymerase. However, the final steps of activation are delayed until very late G1 by the association of inhibitors Whi5 and Stb1 with SBF, and their recruitment of the Rpd3 histone deacetylase complex to URS2 (Costanzo et al. 2004; de Bruin et al. 2004; Takahata et al. 2009a). At the end of G1, the Cdc28 cyclin-dependent kinase (CDK) hyper-phosphorylates Whi5; Whi5 then leaves the nucleus and no longer represses HO transcription.
The unusually complex and regulated nature of HO promoter activation suggests it has a high barrier to transcription, maintained by potentially multiple repressive mechanisms. Early genetic screens identified the Sin3 and Rpd3 subunits of a histone deacetylase complex as negative regulators of HO (Nasmyth et al. 1987; Sternberg et al. 1987; Dorland et al. 2000), and subsequently Ash1 was identified as a critical determinant of daughter-specific repression of HO (Bobola et al. 1996; Sil and Herskowitz 1996). Ash1, a GATA-family DNA-binding protein that associates with the HO promoter, recruits the Rpd3 deacetylase to URS1, opposing the action of the Gcn5 coactivator and making the nucleosomes more restrictive to transcription (Maxon and Herskowitz 2001; Mitra et al. 2006; Takahata et al. 2011). We reasoned that the HO promoter likely has other mechanisms of repression to ensure that activation is terminated quickly, allowing only a few transcripts to be produced prior to rebinding of Swi5 in the next cell cycle. To identify other proteins that negatively affect HO expression, we employed a series of genetic screens designed to uncover mutations that would increase HO expression. We wanted to design a strain in which HO expression was driven by a very weak activator, such that it would be possible to easily identify increases in HO expression using a reporter system. We could then look for suppressor mutations that would allow the weak activator to function better, reasoning that these mutations may be in the types of negative factors we wished to identify. To design such a screen, we made use of the fact that the Swi5 pioneer transcription factor at HO has a paralogue, Ace2, that under normal circumstances fails to activate HO expression at a level that allows mating type switching.
The Swi5 and Ace2 proteins have nearly identical DNA-binding domains and bind to the same sequences in vitro, but in vivo they activate different genes (Dohrmann et al. 1992, 1996; Voth et al. 2007). The HO gene is one example that is activated by Swi5 but not by Ace2 (Dohrmann et al. 1992). The similarity between Swi5 and Ace2 is low outside the zinc finger DNA-binding domain, and thus it is likely that the two factors interact differently with proteins, including coactivators. Swi5 may have the ability to recruit multiple factors that do not associate with Ace2 but are critical for overcoming the repression complexes at HO. We reasoned that by screening for mutants that would allow Ace2 to activate HO, we might identify inhibitory proteins that are important for restricting HO expression.
We therefore conducted a series of screens to identify mutants that would allow Ace2 to activate the HO gene in the absence of Swi5. The screens revealed a variety of HO promoter mutations as well as several protein factors previously shown to have roles in chromatin modification and transcriptional regulation. Combinations of mutants yielded higher levels of HO expression than single mutants. Thus, HO requires a host of both positive and negative factors that overlap functionally to collectively produce optimal coordination of transcription. Recruitment of Rpd3 appears to be critical, as at least two factors independently bring this protein to the HO promoter. The reduced repression observed in the mutants is likely to be the result of unopposed coactivator function. Consistent with this, we demonstrate that the mutants lead to additive increases in the level of association of SWI/SNF with HO.
Materials and Methods
Strain construction
All yeast strains used are listed in Supplemental Material, Table S1 and are isogenic in the W303 background (leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15) (Thomas and Rothstein 1989). Standard genetic methods were used for strain construction (Rothstein 1991; Sherman 1991; Knop et al. 1999; Storici et al. 2001). Plasmids are listed in Table S2. Oligos and further details regarding plasmid and strain construction are available upon request.
Strain DY10171 for the Ace2 popout genome-wide screen (HO-ADE2HO-CAN1swi5::LEU2 RS::TRP1::ACE2::HIS3::RS ade2::HphMX) was constructed from DY10085 (HO-ADE2HO-CAN1swi5::LEU2ade2::HphMX) in a two-step process using plasmids containing RS::TRP1::ACE2 (M5011) and ACE2::HIS3::RS (M5048). The RS::TRP1::ACE2 and ACE2::HIS3::RS plasmids were constructed in yeast using standard PCR and homologous recombination. The RS::TRP1::ACE2::HIS3::RS cassette was introduced into additional strains (for the swi5ash1 genome-wide screen, as well as the HO promoter targeted screens) using standard genetic crosses. Further details on construction of strains for the targeted HO promoter screens are provided below in the description of these screens.
The ho(G-1332A) strain DY10941 was constructed by first recovering the mutant ho(G-1332A) promoter from the screen strain using homologous recombination into linearized plasmid M4915 (HO in YCplac22) to generate M5181 (ho(G-1332A)-ADE2 in YCplac22) and then transforming strain DY7874 ho[URA3(-1496 to -1222 deleted)]::KanMX(3′) to replace the ho[URA3] with the ho(G-1332A) promoter (Rothstein 1991). All other promoter mutants were constructed using a similar replacement strategy. The relevant portion of the HO promoter was amplified from each mutant strain and used to replace a URA3 or URA3-KanMX cassette within the HO promoter by homologous recombination (see Table S1 for strains used; DY14815, DY14198, and DY14840). Some promoter mutant strains had a KanMX marker present 3′ to the HO gene. Those that lacked this marker were subsequently transformed with a fragment from plasmid M4531 (HO::KanMX(3′) for tagging HO at the 3′ end with KanMX), so that mutant HO alleles could be followed in crosses. HO promoter mutations in all strains constructed via crosses were confirmed by quantitative PCR (qPCR) melting curve analysis (Wittwer et al. 2003) or by sequencing to verify the promoter mutation had not been separated from the 3′ marker by recombination during meiosis. The ho(G-1332A, G-1142A) double mutant was constructed using a two-step PCR in which ho(G-1332A) and ho(G-1142A) were PCR amplified from DY10941 and DY15937, respectively, forming overlapping products that were used for a final full-length PCR with the outermost primers. The PCR product containing both mutations was then transformed into strain DY14839 (ho[URA3(K. lactis)::KanMX(inserted at -1200)] ash1::LEU2swi5::TRP1) to replace the URA3(K. lactis)::KanMX marker. The strains used in Figure 10 have mutations at all nine SBF sites within URS2 [9xSBFmut is the combination of the LX4 and RX5 mutations described in Yarrington et al. (2016)], to eliminate any recruitment of Rpd3 to the URS2 region by SBF.
Figure 10.
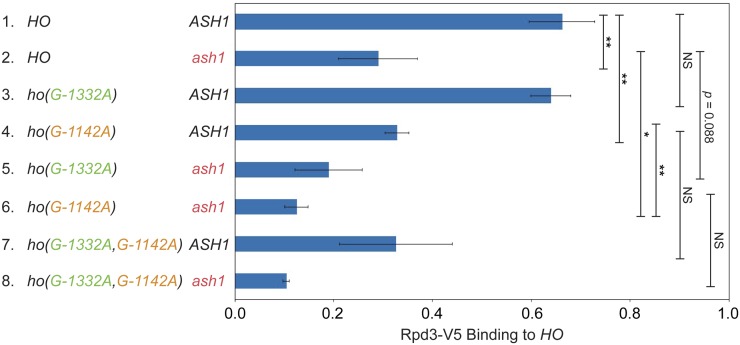
Rpd3 binding is diminished by ash1 and ho(G-1142A) mutants. Binding of Rpd3-V5 to the HO promoter in single and combination mutant strains was determined by ChIP, followed by qPCR with primers that amplify HO sequence from −1295 to −1121. The strains in this experiment have mutations at all nine SBF sites within URS2 (9xSBFmut), to eliminate any recruitment of Rpd3 to the URS2 region by Rpd3. Rpd3-V5 enrichment for each sample was normalized to that of the promoter of INO1, and to the corresponding input sample and graphed relative to wild-type enrichment. Strain genotypes are indicated on the left. Error bars show the SD of three biological replicates. * P < 0.05, ** P < 0.01.
Standard crosses were used to isolate the tup1(H575Y) allele from the strain in which the screen was conducted, using qPCR melt curve analysis (Wittwer et al. 2003) to differentiate the tup1(H575Y) allele from the wild-type TUP1 allele. Whole gene replacements of DOT6, TOD6, and UME6 were constructed in diploids using standard PCR integration methods to amplify markers and replace the coding regions (Longtine et al. 1998). Diploids were sporulated, and resulting haploids were used in subsequent standard genetic crosses to obtain all necessary haploid genotypes.
The DOT6-V5 and ASH1-V5 alleles were constructed by PCR amplifying and C-terminally integrating a V5 epitope tag with a HIS3MX marker from pZC03 (pFA6a-TEV-6xGly-V5-HIS3MX), provided by Zaily Connell and Tim Formosa (plasmid #44073; Addgene). The UME6-FLAG allele was constructed by PCR amplifying and C-terminally integrating a 3x FLAG::URA3::3x FLAG cassette from ZM467 (Moqtaderi and Struhl 2008).
The TOR1-1 fpr1::NATMX RPL13A-2 x FKBP12::TRP1 strain (Haruki et al. 2008) used for constructing Anchor Away strains was obtained from EUROSCARF (Y40343). The strain has two copies of FKBP12 fused to the C-terminus of RPL13A, with a downstream TRP1 marker, integrated at the PMA1 locus. Rpl13A is a ribosomal protein that shuttles in and out of the nucleus and is therefore a suitable anchor for nuclear proteins. TOR1-1 is a dominant allele of TOR1 that allows growth in the presence of rapamycin. For Anchor Away experiments, the strain was first modified to integrate an FRB Rapamycin binding protein at the C-terminus of CKS1, TUP1, or UME6 by PCR amplifying the FRB domain and KanMX marker from plasmid pFA6a-FRB-KanMX (EUROSCARF P30578). The resulting strain was then used to construct additional genotypes using standard genetic crosses.
Genetic screens: genome-wide
For each of the swi5 and swi5ash1 Ace2-popout screens (using strains DY10171 and DY13589, respectively), independent cultures were grown to saturation overnight to allow formation of spontaneous mutants. Cells were plated at a low dilution onto synthetic complete medium lacking adenine with 2% dextrose (SD-Ade) to select for those that allowed activation of HO-ADE2. After 2–3 days at 30°, Ade+ colonies were streaked to new SD-Ade plates alongside the wild-type starting strain. Mutants displaying stronger growth than the wild type were tested for Ace2 dependence in the following manner. Strains were transformed with plasmid M5053, a YCp-LYS2 plasmid containing the Zygosaccharomyces rouxii recombinase under the control of a galactose-inducible promoter, and selected on SD-Lys media (Matsuzaki et al. 1990). Three transformants from each were patched to SD-Lys and then replica plated to synthetic complete medium lacking lysine with 2% galactose (SG-Lys) to activate transcription of the Z. rouxii recombinase. From SG-Lys, clones of each transformant were obtained on synthetic complete medium containing 2% dextrose and 5-fluoroanthranilic acid (SD+5-FAA) (Toyn et al. 2000), and colonies were then chosen from SD+5-FAA and tested for growth on SD-Leu (as a control for growth; strains are Leu+ due to either the swi5::LEU2 or ash1::LEU2 markers), SD-His (to confirm excision of the RS::TRP1::ACE2::HIS3 cassette) and SD-Ade. The popout strains lacking Ace2 were then transformed with M2291, a YCp plasmid with ACE2, and confirmed to be Ade+ again with the reintroduction of Ace2. Strains that were Ade+ before popout (RS::TRP1::ACE2::HIS3), Ade− after popout (ace2::RS), and Ade+ after transformation with the ACE2 plasmid (ace2::RS, ACE2 YCp) were kept for further testing.
Each mutant strain was backcrossed to an appropriate strain marked with a drug resistance gene at the 3′ end of HO-ADE2 (strains DY10061 for swi5 and DY13857 for swi5ash1) to determine whether a single mutation was responsible for the phenotype, to assess possible linkage to HO and ACE2 (using the TRP1 and HIS3 markers surrounding ACE2 and the HO 3′ marker), and to generate MATa and MATα strains for complementation and dominance testing. A cross to an ace2 strain was used to confirm the Ace2 dependence of each mutation (strains DY10174 for swi5 and DY13855 for swi5ash1). Strains that were suspected to contain multiple mutations based on the initial backcross underwent additional backcrosses to separate mutations, and any of these that failed to show Ace2 dependence by a cross to an ace2 strain were discarded. Each mutation was also determined to be either dominant or recessive by crossing to a MAT deletion strain (MATΔ; DY9341 for swi5 strains and DY13891 for swi5ash1 strains) to produce MATa/MATΔ pseudodiploids that could be tested for growth on SD-Ade. HO is repressed in a MATa/MATα diploid by the a1/α2 heterodimer, but HO is expressed in a MATa/MATΔ pseudodiploid (Jensen et al. 1983). To generate strains for complementation testing, a drug resistance gene (KanMX or NatMX) was PCR amplified from pFA6-KanMX4 or pAG25-NATMX4 (Wach et al. 1994; Goldstein and McCusker 1999) and used to replace the MATα gene in specific mutants by homologous recombination. Complementation testing was performed by scoring the Ade+ phenotype of MATa/MATΔ pseudodiploids generated by mating pairs of mutants.
The genes responsible for the Ade+ phenotype in mutants from the Ace2-popout screens were determined using multiple methods. HO and ACE2 mutants were identified by linkage analysis, using the TRP1 and HIS3 markers surrounding ACE2 and the drug resistance markers at the 3′ end of HO. The HO promoter or ACE2 gene was then recovered from the mutant strain by allele rescue (Rothstein 1991) into a linearized plasmid (M4915, HO in YCplac22 and M2291, ACE2 in YCplac33), followed by Sanger sequencing to determine the exact mutation(s). To identify the gene mutated in the 17 alleles comprising a single complementation group from the swi5 screen, we first used chromosome loss-based gene mapping (Reid et al. 2008) to determine that the relevant gene was located on chromosome XI. Since ASH1 is located on chromosome XI and was known to be a repressor of HO transcription, we then specifically tested ASH1 by plasmid complementation using a YIp-URA3 plasmid with ASH1 (M5404).
Affected genes for eight of the mutants from the swi5ash1 screen were determined by Illumina whole genome sequencing. Yeast DNA was isolated using a combination protocol involving lyticase treatment and precipitation/extraction of proteins with potassium acetate and phenol, followed by further purification using QIAGEN DNeasy columns (69504; Supplementary Protocol for Yeast, starting with addition of AL Buffer). A pellet from a 50-ml culture of cells at approximately OD 1.0 was resuspended in an equal volume of 20 mM Tris, pH 8.0, 100 mM EDTA, and 0.5 M 2-mercaptoethanol, and cells were incubated with 500 units of lyticase for 30 min at 37°, vortexing intermittently. Lysis buffer (100 mM Tris, pH 8.0, 50 mM EDTA, 3% SDS, 1 mg/ml proteinase K) was added, and the mixture was incubated for 15 min at 65° and cooled. Proteins were precipitated using 5 M potassium acetate, followed by phenol-chloroform extraction and ethanol precipitation. DNA was treated with RNaseA and then purified with a QIAGEN DNeasy Blood and Tissue Kit (69504; see above). Purified DNA was sonicated and used for Illumina library preparation and sequencing with an Illumina GAIIx 75 cycle single-end run (University of Utah High Throughput Genomics Facility).
High-throughput sequences from each mutant strain were aligned to the S. cerevisiae S288C sequence (SGD release R63-1-1) using Novocraft novoalign (http://www.novocraft.com, version 2.7). Variants within each strain were identified using samtools (https://github.com/samtools/, release 0.1.19) mpileup function and bcftools (part of samtools 0.1.19). Variants were filtered with vcfutils.pl (part of samtools 0.1.19). Common and unique variants between the mutant strains were identified using the script intersect_SNPs.pl (biotoolbox-legacy, https://github.com/tjparnell/biotoolbox-legacy). Unique variants were annotated with locate_SNPs.pl (biotoolbox-legacy), and variants not eliciting a codon change were discarded. Variants present in at least 50% of the reads from a given strain were evaluated for quality by viewing the alignments in a genome browser. Following analysis, each mutant produced a list of one to five candidate genes, for which all high-quality reads showed the mutation. Candidates were then tested by plasmid complementation and/or linkage analysis. Mutants identified by whole genome sequencing include tup1(H575Y), hda3(T418I), isw2(R250*), three independent frameshift mutations within ITC1, and two mutations within SFP1 (one in the 5′ UTR and one stop mutation; see also Table 1).
Table 1. Mutants identified in screens for factors that increase HO-ADE2 expression in the absence of Swi5.
| Gene | Screen | Number of mutants | How identified | Nature of mutation(s) |
|---|---|---|---|---|
| ACE2 | swi5 | 1 | Link; Seq | H675* |
| ASH1 | swi5 | 17a | Comp | ND |
| CKS1 | swi5 ash1 | 1 | Library; Link | Q81*, Q121R |
| HO | swi5 | 2 | Link; Seq | G-1332A (2) |
| swi5 ash1 | 3 | Link; Seq | G-1358C | |
| G-1332A | ||||
| G-1332T | ||||
| HDA1 | swi5 ash1 | 2 | Comp; Link | ND |
| HDA3 | swi5 ash1 | 2 | WG Seq (1) Comp; Link (2) | T418I |
| ND | ||||
| ITC1 | swi5 ash1 | 3 | WG Seq; | FS at 66, |
| Comp; Link | 517, 771 | |||
| ISW2 | swi5 ash1 | 7 | WG Seq (1); | R250* |
| Comp; Link (7) | ND (6) | |||
| RPD3 | swi5 ash1 | 1 | Comp; Link | Y347N |
| SFP1 | swi5 ash1 | 2 | WG Seq; Comp; Link | G-455T Q163stop |
| TUP1 | swi5 ash1 | 1 | WG Seq; | H575Y |
| Comp; Link |
= Stop. Link, linkage analysis; Seq, sanger sequencing; Comp, complementation analysis (plasmid); ND, not determined; WG Seq, whole genome sequencing; FS, frameshift.
Two of the 17 mutants each had two mutations, one in the ash1 gene and a second that was Ace2-independent and therefore discarded.
After mutations in some strains were identified by genomic sequencing, strains with unidentified mutations were tested for alleles of the same genes (hda1, hda3, isw2, rpd3) by plasmid complementation testing and linkage analysis (Table 1). For most of these mutants, the gene was not sequenced to determine the precise nature of the mutation. CKS1 was identified by screening the ATCC library #37415 (15- to 20-kb inserts in YCp50, URA3) for a complementing clone, which was then sequenced to determine the relevant genes. Plasmid complementation then identified CKS1 as the gene responsible for complementation of the mutant phenotype, and this was confirmed by linkage analysis.
Genetic screens: HO promoter
Strains for the targeted HO promoter screen were designed to allow for integration of a mutagenized HO promoter PCR product at the HO-ADE2 locus. A portion of the HO promoter in HO-ADE2 was replaced or deleted in these strains, rendering the promoter nonfunctional. Mutagenic PCR of a plasmid containing the HO promoter generated fragments that spanned from −1827 to −268, which were used to replace the promoter in the nonfunctional HO-ADE2 strains, allowing for a functional HO-ADE2 to be produced by homologous recombination.
Three strains were constructed and used for screens, one with an internal deletion within the HO promoter (swi5 only; DY13862) and two with a replacement of a portion of the HO promoter with the sup4-o transfer RNA (tRNA) gene (swi5; DY13863 and swi5ash1; DY13925). The precursor strain for all three was constructed by replacing a portion of the HO promoter from −1496 to −1222 with a URA3 marker (strain DY13832; using M2026, ho[URA3(−1496 to −1222 deleted)] disruptor). Subsequently, the ho[URA3] was replaced with either an HO promoter with an internal deletion (strain DY13862; using M346, deletion of HO promoter from −1496 to −1130) or an HO promoter containing a sup4-o tRNA gene (strain DY13863; using M3208, where sup4-o replaces HO sequences from −1496 to −1222). The sup4-o is an ochre suppressor tRNA that allows read-through of the can1-100 nonsense allele, making the strains effectively CAN1+ (Nasmyth 1985). Can1 is a transporter that confers sensitivity to the drug canavanine, a toxic analog of arginine. Replacement of part of the HO promoter with sup4-o thereby rendered the strains canavanine sensitive, and subsequent homologous recombination with mutant ho promoter PCR sequence to eliminate sup4-o allowed the strains to become resistant to canavanine.
For each screen, multiple HO promoter PCR reactions were each transformed separately to allow for later determination of mutants that had arisen independently during PCR. Growth on SD-Ade was used to select colonies that displayed stronger activation of HO-ADE2 than the wild-type HO promoter. Ade+ colonies from screens with strains containing a sup4-o in the HO-ADE2 promoter were also tested for growth on canavanine to confirm that the promoter had been replaced. Candidates were then tested for Ace2-dependence of the Ade+ phenotype using the strategy for Ace2 popout and add-back described above. For those that showed Ace2-dependence, genomic DNA was isolated, and a region spanning the replaced portion of the HO promoter was PCR amplified and subjected to Sanger sequencing to identify mutations. A number of strains had >1 mutation. For most, we were able to deduce the mutation likely causing the Ade+ phenotype by comparison with other strains that contained only a single mutation in either an identical nucleotide or one in close proximity (Table 3). Others were determined by testing single mutations for effects on HO expression in the endogenous context (Table 4).
Table 3. HO promoter mutants identified in screens for suppressors of swi5 or ash1 swi5.
| Mutation position | Screen | Number independent mutants | Nature of mutation | Additional mutations |
|---|---|---|---|---|
| −1762 | 5 | 1 | G to C | None |
| 1 | G to A | None | ||
| −1734a | 5 | 1 | A to G | None |
| 1 | A to T | −1399 | ||
| −1442 | 5 | 1 | T to C | None |
| −1416 | 5 | 3 | T to C | None |
| 1 | T to C | −737, −544 | ||
| −1412 | 5 | 1 | T to C | −1363, −1297 |
| −1402 | 5 | 1 | A to G | −479 |
| −1401 | 5 | 2 | T to C | None |
| −1399a | 5 | 1 | T to C | −1734 |
| −1358 | 2 | 1 | G to C | None |
| −1338 | 5 | 2 | T to C | None |
| −1335 | 5 | 1 | G to A | None |
| −1334 | 5 | 1 | A to G | None |
| 1 | A to G | −1514 | ||
| −1333 | 3 | 1 | T to G | None |
| 5 | 11 | T to C | None | |
| 5 | 1 | T to C | −1073 | |
| −1332 | 1 | 2 | G to A | None |
| 2 | 1 | G to A | None | |
| 2 | 1 | G to T | None | |
| 3 | 5 | G to A | None | |
| 5 | 1 | G to A | None | |
| 1 | G to A | −1104 | ||
| −1331 | 5 | 1 | A to T | None |
| 2 | A to G | None | ||
| −1328 | 5 | 1 | T to A | None |
| −1144 | 3 | 1 | G to A | −1384 |
| 3 | 1 | G to T | −1317 | |
| 5 | 1 | G to A | −825 | |
| −1143 | 5 | 1 | C to T | None |
| −1142 | 5 | 1 | G to A | −759 |
| 1 | G to A | −697, −640 | ||
| −1141 | 5 | 1 | T to A | −1554 |
| 1 | G to A | −646 | ||
| −1139 | 3 | 1 | T to C | −531 |
| −1138 | 5 | 1 | A to G | None |
Screen 1 = genome-wide screen with HO-ADE2 swi5 ASH1; Strain DY10171. Screen 2 = genome-wide screen with HO-ADE2 swi5 ash1; Strain DY13589. Screen 3 = promoter mutagenesis screen with ho(−1496 to −1130 deleted)-ADE2 swi5 ASH1; Strain DY13862. Screen 4 = promoter mutagenesis screen with ho[sup4-o(−1496 to −1222 deleted)]-ADE2 swi5 ASH1; Strain DY13863. Screen 5 = promoter mutagenesis screen with ho[sup4-o(−1496 to −1222 deleted)]-ADE2 swi5 ash1; Strain DY13925.
Mutant is listed twice, one for each of the two mutations.
Table 4. Effect of HO mutants on expression in swi5 ash1.
| Mutation position | Cluster | Nucleotide | Fold change over swi5 ash1a | No.b | Ace2 dependent?c |
|---|---|---|---|---|---|
| −1762 | NA | G | 1.3 ± 0.0 | 2 | Mostly |
| −1734 | NA | A | 3.6 ± 0.9 | 2 | Yes |
| −1442 | NA | T | 3.0 ± 0.2 | 2 | Mostly |
| −1416 | 1 | T | 3.4 ± 0.9 | 6 | Mostly |
| −1401 | 1 | T | 3.4 ± 1.0 | 8 | Yes |
| −1399 | 1 | T | 1.7 ± 0.1 | 2 | Mostly |
| −1358 | NA | G | 2.2 ± 0.3 | 2 | Mostly |
| −1338 | 2 | T | 3.1 ± 1.2 | 2 | Yes |
| −1335 | 2 | G | 3.4 ± 0.5 | 2 | Mostly |
| −1334 | 2 | A | 5.3 ± 1.4 | 4 | Mostly |
| −1333 | 2 | T | 4.1 ± 0.7 | 4 | Mostly |
| −1332 | 2 | G | 6.4 ± 2.3 | 25 | Mostly |
| −1331 | 2 | A | 4.2 ± 2.4 | 4 | Mostly |
| −1328 | 2 | T | 4.7 ± 1.0 | 2 | Mostly |
| −1144 | 3 | G | 2.4 ± 0.4 | 4 | Yes |
| −1143 | 3 | C | 2.4 ± 0.3 | 4 | Mostly |
| −1142 | 3 | G | 4.3 ± 1.2 | 18 | Mostly |
| −1141 | 3 | G | 2.6 ± 0.5 | 4 | Mostly |
| −1138 | 3 | A | 2.8 ± 0.6 | 2 | Mostly |
NA = Not Applicable.
Fold change = HO RNA level in swi5 ash1 ho(mut) divided by HO RNA level in swi5 ash1 HO. Average fold change is shown, ± SD.
No. = mumber of biological replicates.
Ace2 dependence determined by measuring and comparing HO RNA levels in swi5 ash1 ace2 vs. swi5 ash1 isogenic strains.
Growth assays
For plate spot dilution assays, liquid cultures of the indicated strains were grown to saturation, serially diluted in 10-fold increments, spotted onto SD-Ade or SD Complete media for the number of days indicated, and photographed.
RNA expression and chromatin immunoprecipitation (ChIP) analysis
For logarithmic cell collection (OD660 of 0.6–0.8), cells were grown at 30° in YPA medium (1% yeast extract, 2% bactopeptone, 0.002% adenine) supplemented with 2% dextrose (Sherman 1991). Cell cycle synchronization was performed by galactose withdrawal and readdition with a GALp::CDC20 strain grown at 25° in YPA medium containing 2% galactose and 2% raffinose (Bhoite et al. 2001). Synchrony was confirmed by microscopic analysis of budding indices and analysis of cell-cycle-regulated messenger RNAs (mRNAs) (data not shown). For Anchor Away experiments, cells were grown to an OD660 of 0.3–0.4 in YPA media containing 2% glucose at 30°. The cultures were then split into two, and rapamycin (1μg/ml final concentration, dissolved in ethanol; LC Laboratories; +Rapamycin) or ethanol alone (−Rapamycin) was added to the media for the final 2 hr of growth prior to collection of cells at an OD660 of 0.8–1.0.
RNA was isolated from either logarithmically growing cells or synchronized cells, and HO mRNA levels were measured by reverse transcription quantitative PCR (RT-qPCR), as described previously (Voth et al. 2007). HO RNA expression was normalized to that of RPR1. RPR1 encodes the RNA component of RNase P and is transcribed by RNA polymerase III. Most genetic manipulations that affect RNA Pol II transcription do not affect transcription of RPR1. For logarithmic cells, normalized HO RNA expression values were graphed relative to wild-type expression.
ChIPs were performed as described (Bhoite et al. 2001; Voth et al. 2007), using mouse monoclonal antibodies to the V5 epitope (SV5-Pk1; Abcam), the FLAG epitope (M2; Sigma, St. Louis, MO), or the Myc epitope (4A6; Upstate) and antibody-coated magnetic beads (Pan Mouse IgG beads; Life Technologies). Cells from either logarithmically growing cells or synchronized cells were cross-linked in 1% formaldehyde for 20 min at room temperature and quenched with 125 mM glycine. ChIP signals for relevant target genes within each sample were first normalized to either an expected negative reference control or a known positive reference control, and then to their respective input controls. Normalization to the negative reference controls (IGR-I intergenic region of chromosome I and CDC2 coding region) yielded a high degree of variability between ChIP experiments due to normalization to a very small number. However, this normalization was useful for observing the presence of a ChIP signal over the background from a strain lacking the V5 or FLAG epitope tag (Figure 5, C–E, Figure 6B, Figure 7, Figure 8C, and Figures S1, S2, S4, and S5). Normalization to a positive reference resulted in less variability among samples. Positive references used include: GCD10 for Dot6 (the promoter region of GCD10, shared with that of NOP2, has two PAC sites), INO1 for Ume6 and Rpd3 (INO1 is a well-established target of Ume6; Slekar and Henry 1995; Jackson and Lopes 1996), CLN3 for Ash1 (CLN3 is bound by Ash1 by ChIP analysis; Di Talia et al. 2009; Zapata et al. 2014), CTS1 for Swi2, and EGT2 for Swi5 (Figure 5B, Figure 6A, Figure 8, A and B, Figure 10, Figure 11, and Figure S7). For figures using a negative reference control, all values were graphed relative to the No Tag control. For figures using a positive reference control, all values were graphed relative to the wild-type control.
Figure 5.
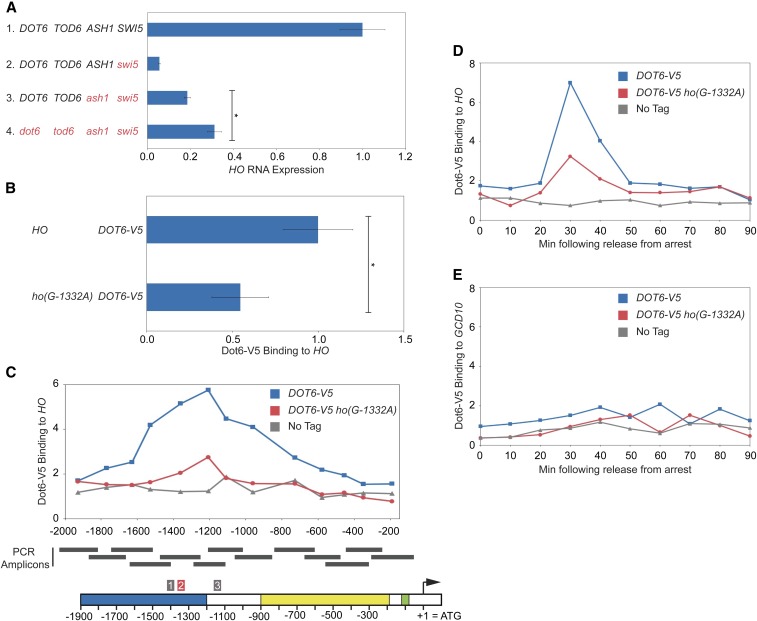
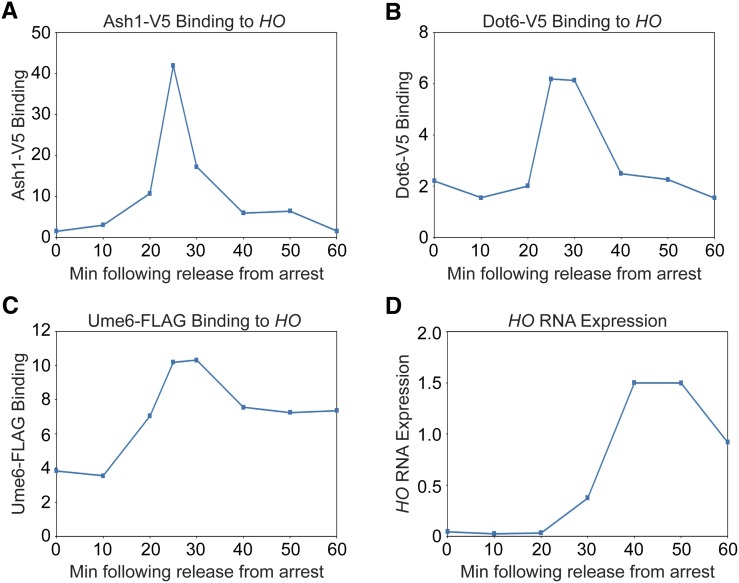
Dot6 associates with the PAC motif in the HO promoter and may influence HO expression. (A) HO mRNA levels were measured, normalized to RPR1, and expressed relative to wild type. Genotypes are indicated at the left. Error bars represent the SD of three to four biological replicates for each strain. * P < 0.05. (B) Binding of Dot6-V5 to HO wild-type and ho(G-1332A) promoters from heat-shocked cells was determined by ChIP, followed by qPCR with primers that amplify HO sequence from −1471 to −1250. Dot6-V5 enrichment at HO was normalized to enrichment at the GCD10 promoter and to the corresponding input sample and then graphed relative to wild type. Error bars show the SD of three biological replicates. * P < 0.05. (C) Single samples of HO wild type (blue) and ho(G-1332A) (red) were chosen from B and used for qPCR with primers that span the length of URS1 and URS2 in ∼150-bp increments. Positions of the PCR amplicons are indicated by the gray bars. Points on the graph correspond to the midpoints of these amplicons, with the x-axis showing position across the HO promoter. A schematic of the HO promoter is shown at the bottom, with positions of Clusters 1, 2, and 3 indicated. Cluster 2 contains the PAC motif and is highlighted red. For each primer set, Dot6 enrichment was normalized to an intergenic region on chromosome I (IGR-I) and to input. A “No Tag” control is shown to indicate the background level of binding (gray). (D) Binding of Dot6-V5 to the HO wild-type (blue) and ho(G-1332A) (red) promoters was measured in cells containing the GALp::CDC20 allele that had been synchronized by galactose withdrawal and readdition. The 0 min time point represents the G2/M arrest, before release with galactose addition. Cells were harvested at the indicated time points following release (x-axis), and samples were processed for ChIP analysis. Dot6 enrichment at the HO promoter was measured using primers that span from −1295 to −1121 and was normalized to IGR-I and to input. Note that HO expression begins to rise at ∼30 min after release, and peaks at 50 min (see Figure 7). (E) Same as D, using primers to the GCD10 promoter, normalized to IGR-I and to input.
Figure 6.
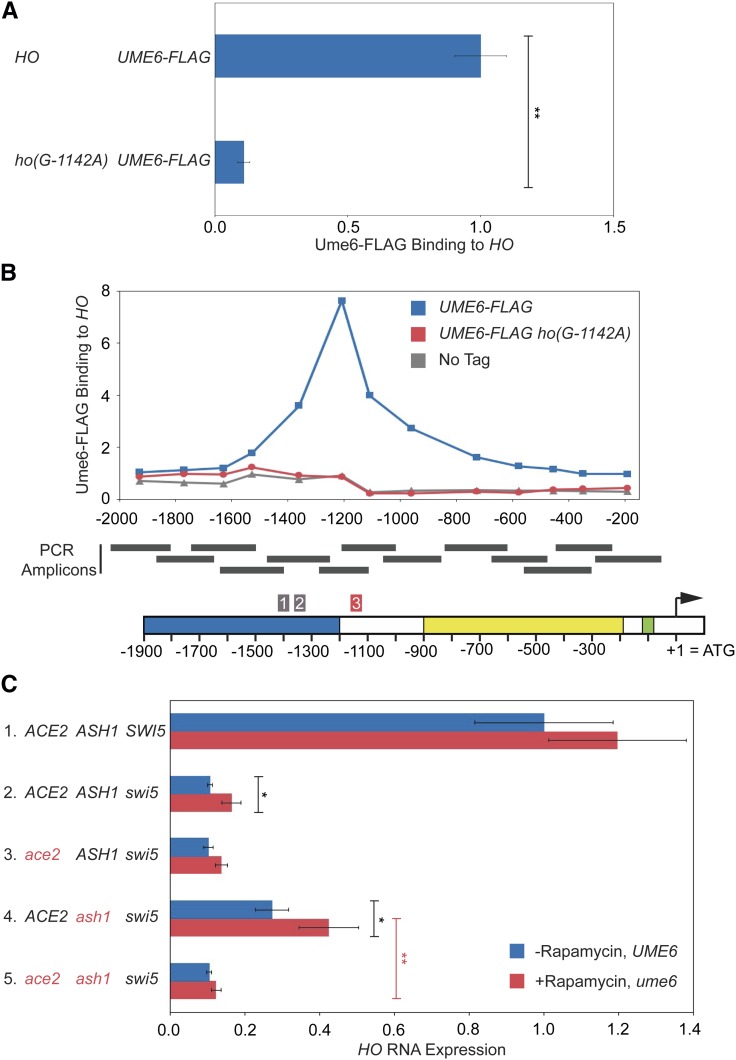
Ume6 associates with its consensus motif in the HO promoter and influences HO expression. (A) Binding of Ume6-FLAG to HO wild-type and ho(G-1142A) promoters was determined by ChIP, followed by qPCR with primers that amplify HO sequence from −1295 to −1121. Ume6-FLAG enrichment for each sample was normalized to enrichment at the promoter of INO1 and to the corresponding input sample and then graphed relative to wild type. Error bars show the SD of three biological replicates. ** P < 0.01. (B) Single samples of HO wild type (blue) and ho(G-1142A) (red) were chosen from A and used for qPCR with primers that span the length of URS1 and URS2 in ∼150-bp increments. Positions of the PCR amplicons are indicated by the gray bars. Points on the graph correspond to the midpoints of these amplicons, with the x-axis showing position across the HO promoter. A schematic of the HO promoter is shown at the bottom, with positions of Clusters 1, 2, and 3 indicated. Cluster 3 contains the Ume6 consensus motif and is highlighted red. For each primer set, Ume6 enrichment was normalized to an intergenic region on chromosome I (IGR-I) and to input. A “No Tag” control is shown to indicate the background level of binding (gray). (C) HO mRNA levels were measured, normalized to RPR1, and expressed relative to wild type. Error bars represent the SD of at least three biological replicates for each strain. See Materials and Methods section for a full description of genotypes and growth conditions. Cultures were split into two, and one half was untreated (−Rapamycin, UME6; blue bars), while the other half was treated with 1 μg/ml rapamycin for 2 hr (+Rapamycin, ume6; red bars). * P < 0.05, ** P < 0.01.
Figure 7.
The Ash1, Dot6, and Ume6 repressors associate with the HO promoter before the peak of HO expression. (A–C) Binding of Ash1-V5 (A), Dot6-V5 (B), and Ume6-FLAG (C) to the HO promoter during a cell cycle arrest and release experiment. Cells containing the GALp::CDC20 allele were synchronized by galactose withdrawal and readdition. The 0 min time point represents the G2/M arrest, before release by addition of galactose. Cells were harvested at the indicated time points following release (x-axis), and samples were processed for ChIP analysis. Enrichment at the HO promoter was measured using primers that span from −1295 to −1121 and was normalized to IGR-I and to input. Graphs show one representative experiment from each strain for comparison. Figure S4 shows three replicates for each strain. (D) A representative example of HO mRNA expression measured over the course of the synchrony experiment and normalized to RPR1 (using a Dot6-V5 strain).
Figure 8.
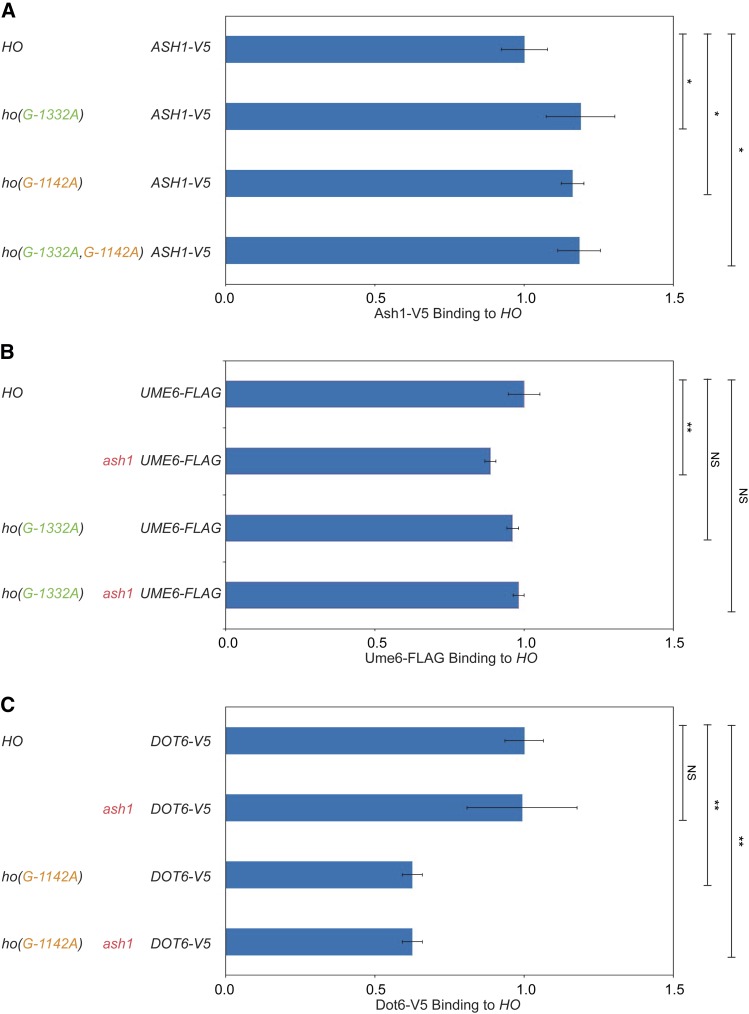
Ash1, Dot6, and Ume6 are largely independent of one another for association with the HO promoter. Binding of Ash1-V5, Dot6-V5, and Ume6-FLAG to the HO promoter in wild type and in the absence of one or both of the other factors or sites was determined by ChIP. Each sample was normalized to a reference control and to its corresponding input and graphed relative to wild-type enrichment. Strain genotypes are indicated at the left. Error bars show the SD of four biological replicates. * P < 0.05, ** P < 0.01. (A) Ash1-V5 enrichment at HO −1295 to −1121 is shown normalized to binding at the promoter of the CLN3 gene. (B) Ume6-FLAG enrichment at HO −1295 to −1121 is shown normalized to binding at the promoter of the INO1 gene. (C) Dot6-V5 enrichment at HO −1295 to −1121 in heat-shocked cells is shown normalized to binding at the intergenic region IGR-I.
Figure 11.
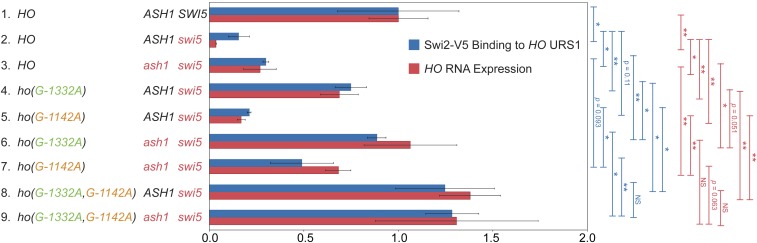
Swi2 binding is restored in ash1, ho(G-1332A), and ho(G-1142A) mutants. A comparison of Swi2-V5 binding at the HO promoter as determined by ChIP (blue) with HO RNA expression (red) in mutant strains is shown. Cells for ChIP and RNA originated from the same cultures that were split upon harvesting. Strain genotypes are indicated on the left. Error bars show the SD of three biological replicates. The qPCR following ChIP was performed using primers that amplify HO sequence from −1429 to −1158. Swi2-V5 enrichment for each sample was normalized to that of the promoter of CTS1, and to the corresponding input sample and graphed relative to wild-type enrichment. HO expression was normalized to RPR1 and expressed relative to wild type. * P < 0.05, ** P < 0.01.
qPCR experiments for both RNA and ChIP analysis were run on a Roche Lightcycler 480, and concentrations were determined using wild-type complementary DNA (cDNA) or ChIP input for in-run standard curves via the E-method (Tellmann 2006). Error bars represent the SD of at least three biological samples. The Student’s t-test was used to determine significance of changes in HO expression and factor binding between different genotypes. For all comparisons mentioned in the Results and Discussion, P-values are indicated in the figures. Primers for all experiments are available upon request. For ChIP tiling PCR across the HO promoter, ChIP time courses with Ash1, Dot6 and Ume6, and time course HO expression, a single sample was shown in the main figures for simplicity (Figure 5C, Figure 6B, and Figure 7). Triplicate biological samples for the time course ChIP and RNA experiments are shown in Figure S4. The time course Dot6 ChIP in wild type and the ho(G-1332A) mutant shown in Figure 5, D and E was repeated in multiple independent experiments; one is shown for simplicity (note that for wild type, additional experiments are shown in Figure S4).
Data availability
Strains and plasmids are available upon request. Table S1 lists the strains and Table S2 lists the plasmids used in this study. Seven Supplemental Figures and two Supplemental Tables were uploaded to the GSA Figshare portal. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8171276.
Results and Discussion
Genetic screen to identify mutants that allow Ace2 to activate the HO gene
We used an HO-ADE2 reporter to perform a genetic screen to identify mutations that would allow Ace2 to activate HO expression in the absence of the Swi5 activator. ADE2 encodes an enzyme necessary for biosynthesis of adenine and has been used previously as a reporter for expression of genes in yeast (Jansen et al. 1996). When all ADE genes are wild type, the cells biosynthesize adenine and are capable of growth on media that lacks adenine. Cells that have an ade2 mutation do not synthesize adenine and become dependent on its presence in the media; hence, they fail to grow on media lacking adenine (-Ade). Previous studies have demonstrated that ADE2 is a useful model for HO expression, as modest levels of HO-ADE2 transcription allow robust growth on -Ade media, while low transcript levels do not confer growth on -Ade, and intermediate levels allow for slow growth (Jansen et al. 1996). Therefore, it is possible to observe alterations in expression of HO-ADE2 by the ability of the strains to grow on media lacking adenine.
The ADE2 open reading frame was integrated at the genomic HO locus, replacing the HO ORF, to construct the HO-ADE2 reporter (Figure 1A). All 5′ and 3′ regulatory sequences necessary for HO expression are present, notably the entire 3084 nucleotides of the possible promoter region extending to the ORF of the upstream gene. Our screen was performed in an HO-ADE2swi5ACE2 strain. This strain has very low HO expression, due to the absence of the Swi5 activator; hence, the cells are phenotypically Ade− (swi5ACE2 MUTX; Figure 1A). We screened for spontaneous mutants that would confer some level of growth on -Ade media (Ade+; swi5ACE2 mutx; Figure 1A).
We anticipated that mutations in a variety of general chromatin and transcription factors could increase HO expression by affecting the nucleosome structure at URS2, which could in turn allow premature binding and/or activation by SBF, regardless of whether Ace2 promoted transcription. To avoid obtaining hits in such generic processes, which should be independent of Ace2, we devised a “popout” strategy to quickly remove ACE2 from the mutants to compare phenotypes in the presence and absence of Ace2 (Figure 1B). The ACE2 locus was modified by introducing Z. rouxii RS recombinase target sites and TRP1 and HIS3 marker genes flanking ACE2. The 31 nucleotide RS sites are targets for the Z. rouxii recombinase, thereby “popping out” the ACE2 gene as well as the two markers in a small percentage of cells (Matsuzaki et al. 1990; Roca et al. 1992). Cells lacking the TRP1-ACE2-HIS3 cassette were selected on 5-fluoroanthranilic acid (5-FAA), which selectively kills cells that express TRP1 (Toyn et al. 2000). The subsequent strains lacking the TRP1-ACE2-HIS3 cassette were then retested for their growth on -Ade to determine the level of HO-ADE2 expression (swi5ace2 mutx; Figure 1A). This allowed us to identify and discard those whose increased HO-ADE2 expression was Ace2-independent.
We obtained 124 mutants from the initial genetic screen, 20 of which remained after further testing (Table 1, swi5 screen). Most were eliminated because their Ade+ phenotype was not dependent on Ace2, as determined either by the popout recombinase test or by a genetic cross to introduce an ace2 deletion allele. A small subset of the mutants (∼20) were removed because the phenotype of these strains did not remain consistently Ade+ upon repeated further testing. We speculate that these strains became aneuploid due to a mutation in the gene encoding Ume6, a factor that we later found to be involved in negative regulation of HO (see below).
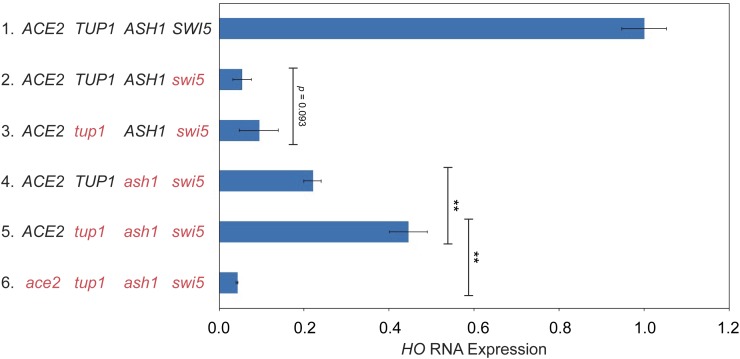
Complementation testing revealed that 17 of the 20 desirable mutants likely had alterations in the same gene. We identified the mutated gene in the 17 strains as ASH1, the previously known repressor and determinant of mother-cell-specific HO expression (Table 1, swi5 screen) (Bobola et al. 1996; Sil and Herskowitz 1996). The identification of ASH1 mutants in our screen was not surprising, given that both Ash1 and Ace2 are largely daughter-specific proteins (Bobola et al. 1996; Colman-Lerner et al. 2001). The Ash1 protein is translated predominately in daughter cells, as the ASH1 mRNA is transported through the bud neck and anchored to the distal tip of the daughter cell via the She proteins (Bertrand et al. 1998; Munchow et al. 1999). Ace2 accumulates selectively in the daughter cell nucleus due to masking of its nuclear export sequence (NES) by the daughter-specific protein Cbk1 (Mazanka et al. 2008). The Ash1 protein therefore presents a barrier to Ace2 primarily in daughter cells, so removal of Ash1 could allow Ace2 to inappropriately activate HO in daughter cells, increasing the number of cells expressing HO without altering the function of the promoter. However, the level of HO-ADE2 activation in a swi5ash1 mutant was still very weak relative to wild type (Figure 1C, line 3 ACE2).
The three remaining Ace2-dependent mutants were identified through linkage analysis; one had a mutation in the ACE2 gene itself, and two had mutations in the HO promoter (Table 1, swi5 screen). The ace2(H675*) allele is a truncation within the C2HC zinc finger of Ace2 and likely reduces its interaction with partner proteins needed for activation of target genes, including ASH1 (data not shown). The two HO promoter mutants each have substitutions of “G” to “A” at position −1332 relative to the HO start site of translation. This ho(G-1332A) mutation is within an NDR, 24 nucleotides upstream of the Swi5 Site B. The ho(G-1332A) promoter mutants had a much stronger Ade+ phenotype than the ash1 mutants, with a growth level approaching that of wild type (Figure 1C, line 4 ACE2). The ho(G-1332A) position could be located within a binding site for Ash1, or for another protein complex that negatively affects HO expression.
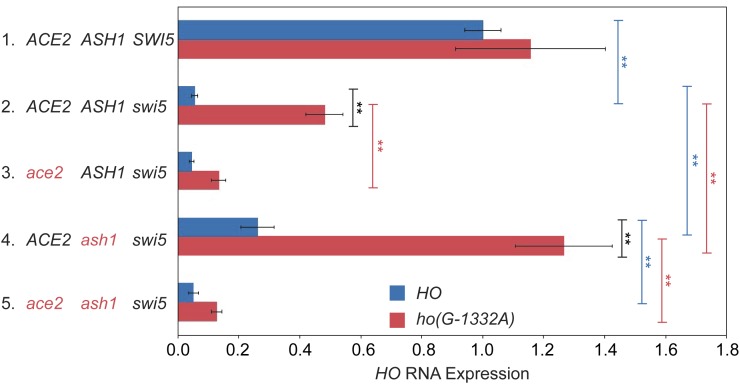
We next examined whether deletion of ash1 or the ho(G-1332A) mutation could affect endogenous HO gene expression in a native context, i.e., without the ADE2 reporter. We also constructed a swi5ash1ho(G-1332A) strain to determine whether the ash1 and ho(G-1332A) mutations were additive. If the ho(G-1332) position is part of a binding site for Ash1, we expect the effect of the two mutations to be similar, and the two mutations together would not have a stronger effect than either single mutation. HO expression measured using RT-qPCR was severely reduced in the swi5 mutant, to 5% of wild type (Figure 2, line 2 blue). The ash1 null increased expression of HO above swi5 by fourfold, demonstrating suppression, just as had been observed in the HO-ADE2 strain (compare lines 2 and 4 blue). The ho(G-1332A) mutant showed much more robust suppression, up to approximately half of the normal wild-type level of HO expression, nearly 10-fold over that of swi5 alone (compare lines 2 red and blue). The combination of ash1 and ho(G-1332A) was additive, raising HO expression to over 100% (line 4 red). Thus, it is likely that the protein(s) bound at ho(G-1332) repress HO through a mechanism independent of Ash1. Similar to the spot dilution assays, the suppression by ash1 and ho(G-1332A) is mostly, but not completely, dependent upon Ace2, because ace2 mutants showed significantly reduced HO expression (compare lines 2 and 3 red and lines 4 and 5 blue).
Figure 2.
An ho(G-1332A) mutant and an ash1 null additively suppress swi5 for endogenous HO expression. HO mRNA levels were measured, normalized to RPR1, and expressed relative to wild type, set at 1.0. Data for strains containing wild-type HO (blue) and ho(G-1332A) (red) promoters are shown for each genotype on the left. Error bars represent the SD of six biological replicates for each strain genotype. ** P < 0.01.
Multiple negative regulators contribute to repression of the HO gene
The ho(G-1332A) mutants from our first genetic screen suggested that other, Ash1-independent repressors regulate the HO promoter. In the context of the initial screen, it may have been difficult to identify these genes, due to the necessity of the weak Ace2 activator to overcome the abundance of Ash1 in daughter cells. We therefore repeated the screen in a swi5ash1 mutant. HO-ADE2 expression in the swi5ash1 strain is still much weaker than in wild type (Figure 1C, lines 1–3 ACE2), which allowed us to observe an additive increase in HO expression when another negative regulator was removed. From this screen, 152 mutants were obtained, 22 of which remained after further testing. Mutations in a number of repressive chromatin/transcription factors were identified, including Isw2, Itc1, Hda1, Hda3, Rpd3, and Tup1 (Table 1, swi5ash1 screen). Ten mutants were in the Isw2 ATP-dependent chromatin remodeling complex, either in the catalytic Isw2 itself or in the Itc1 subunit. Isw2 associates with a large number of sites genome-wide, both via direct recruitment through transcription factors and indirect looping to ectopic sites, and Isw2 has been implicated in repression of many genes (Fazzio et al. 2001; Whitehouse et al. 2007; Yadon and Tsukiyama 2013). The Isw2 complex appears to localize near NDRs and promotes the movement of nucleosomes into the NDR to repress transcriptional initiation; it also plays a role in repression of noncoding RNAs (Whitehouse et al. 2007; Yadon et al. 2010). Five mutants were components of histone deacetylase complexes, four from the Hda1 complex (either Hda1 or Hda3) and one in Rpd3. These HDACs are recruited specifically to individual promoters via DNA-binding proteins (e.g., Rpd3 to HO via Ash1), and they also act globally in a more nonspecific repressive role (Kurdistani and Grunstein 2003). A point mutation in the Tup1 corepressor protein was also identified in the screen. Tup1 is recruited to a wide variety of promoters in yeast via specific DNA-binding proteins and is thought to inhibit expression by masking the activation domain of transcription factors that recruit SWI/SNF and other coactivators (Smith and Johnson 2000; Malave and Dent 2006; Wong and Struhl 2011).
The mutants identified in this screen displayed substantially stronger growth on -Ade media than swi5ash1 alone, similar to the effects of swi5ho(G-1332A) (Figure 1C, line 4 ACE2 and 1D, lines 3–6 ACE2). Suppression was Ace2-dependent for most mutants, but rpd3(Y347N) showed weak growth even in the absence of Ace2 (Figure 1D, lines 3–6 ace2). This is not surprising, given that Rpd3 is an important component of repression of URS2 at the HO promoter, and relief of this repression is not expected to be Ace2-dependent. There were likely a number of additional rpd3 mutants initially identified in the screen that were eliminated due to their Ace2-independent effect on repression at URS2. The particular rpd3 point mutant that showed some Ace2-dependence may therefore have an effect on Rpd3 repression at both URS1 and URS2. The observation that rpd3(Y347N) is additive with ash1 for suppression of swi5 suggests there could be an additional factor that recruits Rpd3 to URS1.
Two surprising mutants were also identified in the screen, cks1 and sfp1 (Table 1, swi5ash1 screen). The connection between Cks1 and Sfp1 and repression of HO transcription is not clear, and it may be indirect by virtue of their effect on other genes that regulate HO. Cks1 is a small, regulatory phospho-adaptor protein involved in cell cycle regulation that associates with the CDK (the S. cerevisiae equivalent of Cdk1) and modulates its activity (Hadwiger et al. 1989; Tang and Reed 1993; Reynard et al. 2000; McGrath et al. 2013). In addition, Cks1 associates with the Paf1 complex, serving as an adaptor to allow recruitment of the proteasome during induction of specific genes, including GAL1 (Pan et al. 2013). The 19S proteasome plays a role in evicting nucleosomes from chromatin during the course of GAL1 induction (Chaves et al. 2010). It is possible that Cks1 could play a similar role during the activation of HO expression. Alternatively, this cks1 mutation could alter the cell cycle and thus influence HO expression, or it could affect the expression of factors that regulate HO.
Sfp1 is an activator of ribosomal protein (RP) and ribosome biogenesis (Ribi) genes and is one of several proteins activated through the TOR pathway that determine cell size and regulate the cellular response to nutrients and stress (Jorgensen et al. 2002, 2004; Fingerman et al. 2003; Marion et al. 2004). A recent study demonstrated that Sfp1 also acts as a glucose-regulated repressor at G1/S genes and associates with these promoters via an interaction with Swi4 (Albert et al. 2019). We and others were unable to detect binding of Sfp1 at the HO promoter in log phase cells (B. Albert and D. Shore, personal communication). It is possible that Sfp1 binding is transient during the cell cycle, making detection of binding in log phase cells difficult. Alternatively, the effect of sfp1 mutants on HO expression could be indirect, due to changes in the cell cycle, regulation of factors that influence HO expression, or through its ability to influence Sch9, another protein that responds to TOR and phosphorylates Dot6 (Loewith and Hall 2011), a DNA-binding repressor that we later found to be involved in HO regulation (see below).
We constructed strains with disruptions for many of the genes that were identified from the swi5ash1 screen, and then examined the effect of the null mutations on expression of the endogenous HO gene in a swi5ash1 mutant. Deletions of hda1, hda3, and isw2 had a very modest ability to suppress swi5ash1, allowing a small but reproducible increase (1.5- to 2-fold) in HO expression (Table 2). Here, the observed effect on HO-ADE2 expression seems to be greater than that on endogenous HO expression, as the growth on -Ade appeared to increase more substantially. One possibility is that there is a nonlinear correlation between growth on -Ade and the actual level of HO-ADE2 expression. Alternatively, these mutants are global regulators of chromatin in yeast and as such, affect the expression of many genes. Thus, it is difficult to interpret the effects on HO expression if these mutants also affect other regulators of HO expression. These pleiotropic effects could dampen the observed effect on HO expression, and the effects could be different in observing a growth phenotype vs. direct measurement of mRNA levels. Finally, it is possible that the alleles isolated in the screen are not nulls, and thus could show more specific and distinct effects than a null (this was true for tup1; see below). Though mutations affecting Hda1 and Isw2 did not show strong effects on HO RNA, the modest suppression is consistent with these factors contributing in some way to repression of HO. Both have been linked to the Tup1 corepressor, and it is possible that their effect at HO is mediated through Tup1 (Wu et al. 2001; Green and Johnson 2004; Zhang and Reese 2004; Fleming et al. 2014). Suppression by an rpd3 null was stronger than that observed for the other chromatin modulators (threefold above the swi5ash1 strain), but, as with the rpd3 allele isolated in the screen, was only partially dependent on Ace2, likely due to its effects on both Swi5/Ace2-mediated activation at the URS1 region of the promoter and on SBF at the URS2 region.
Table 2. Effect of mutants on expression of HO in swi5 ash1.
| Gene | Fold change over swi5 ash1a | No.b | Ace2 Dependent?c |
|---|---|---|---|
| CKS1d | 1.7 ± 0.1 | 4 | No |
| HDA1 | 2.0 ± 0.5 | 17 | Mostly |
| HDA3 | 1.4 ± 0.4 | 13 | Yes |
| ISW2 | 1.5 ± 0.4 | 19 | Yes |
| RPD3 | 3.0 ± 1.8 | 19 | Partial |
| SFP1e | ND | NA | NA |
| TUP1f | 2.2 ± 0.7 | 16 | Yes |
| UME6d | 1.7 ± 0.4 | 6 | Mostly |
| DOT6 TOD6 | 1.5 ± 0.2 | 10 | Yes |
ND = Not Determined.
NA = Not Applicable.
Fold change = HO RNA level in swi5 ash1 mutx divided by HO RNA level in swi5 ash1 MUTX. Average fold change is shown, ± SD.
No. = number of biological replicates.
Ace2 dependence determined by measuring and comparing HO RNA levels in swi5 ash1 ace2 vs. swi5 ash1 isogenic strains.
Anchor Away.
Could not obtain accurate results; see text.
H575Y point mutant.
We attempted to assess the effects of Cks1 and Sfp1 on HO expression using null mutations. An sfp1 null caused changes in expression of a wide variety of genes, making it too difficult to determine an appropriate normalization control for RT-qPCR. The cks1 null strain was extremely slow growing, and RNA results were inconsistent between experiments. We therefore used Anchor Away as an alternative to measure HO transcription while transiently depleting the nucleus of Cks1 (Haruki et al. 2008). Using this strategy, we found that loss of Cks1 did increase HO expression in a swi5 mutant, but this effect was not dependent on Ace2 (Table 2). This could suggest a generic role for Cks1 and the proteasome in eviction of nucleosomes at URS2.
We found that HO expression was very low in tup1 null strains, even with wild-type SWI5. Flow cytometry revealed that both the tup1 null and an Anchor Away depletion caused a delay in G1 prior to the advent of HO expression (data not shown), making measurement of HO transcription uninterpretable. We therefore used the point mutant isolated in the screen, an H575Y substitution, for endogenous HO RNA analysis. The tup1(H575Y) mutant displays normal growth, suggesting some critical targets of Tup1 are not affected by this mutation. However, tup1(H575Y) increased suppression of swi5 additively with ash1, at a level exceeding that of the null mutants for hda1, hda3, and isw2 (Table 2; Figure 3, compare lines 4 and 5). Expression of HO in the tup1(H575Y) ash1swi5 mutant was Ace2 dependent (Figure 3, compare lines 5 and 6).
Figure 3.
The tup1(H575Y) point mutant suppresses an ash1 swi5 mutant for endogenous HO expression. HO mRNA levels were measured, normalized to RPR1, and expressed relative to wild type. Genotypes are indicated at the left. Error bars represent the SD of three biological replicates for each strain. ** P < 0.01.
Additional HO promoter mutants suggest the presence of binding sites for negative regulators
Three HO promoter mutants were also identified from the swi5ash1 screen, two changes at −1332 (the position identified in the swi5 screen) and one at −1358 (Table 3). Since these genome-wide screens may not have been saturated for HO promoter mutations, we next performed directed screens in which we mutagenized the region of the HO promoter from −1826 to −268, encompassing both URS1 and URS2, and integrated the mutant versions into a complete HO-ADE2 reporter. Many additional HO promoter mutants were obtained (Table 3). All mutations were in the URS1 region of the promoter, which was not surprising, given that we were selecting for mutants that were dependent upon the Ace2 activator and therefore required relief of repression in URS1. A total of seven additional independent ho(G-1332A) mutants were identified, highlighting the importance of −1332 in HO regulation. Other mutations were identified in nucleotides in close proximity to ho(G-1332A), further suggesting the presence of a binding site for a factor at this position.
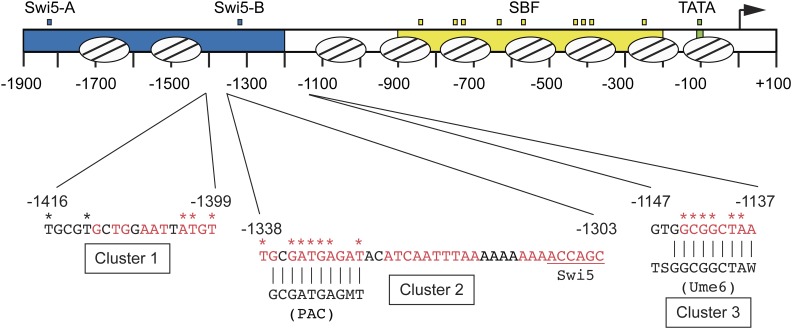
Listing the mutants by position within the promoter revealed not only the cluster of mutations encompassing ho(G-1332A) (Cluster 2; containing mutations at positions −1338 to −1328) but also two additional clusters, Cluster 1 (with mutations from −1416 to −1399) and Cluster 3 (with mutations from −1144 to −1138) (Figure 4 and Table 3). Only a few mutations fell outside these clusters (positions −1762, −1734, and −1442), and several sites were identified multiple times, suggesting the screen was near saturation. Isolates with multiple mutations were either compared to other hits from the screen or tested further as single mutations to determine the likely cause of suppression (Table 3 and Table 4). Cluster 1 contains mutations at five positions in the HO promoter, from −1416 to −1399, where red indicates evolutionarily conserved nucleotides (Figure 4). We searched for known recognition motifs for DNA-binding proteins within this region and tested candidate proteins by ChIP analysis but were unable to identify a protein disrupted by these mutations. Clusters 2 and 3 encompass known regulatory motifs, as detailed below.
Figure 4.
Clusters of mutations within the HO promoter that suppress swi5 ash1. A schematic of 1900 nucleotides of the HO promoter is shown, with nucleosome positions indicated by ovals with lines. URS1 (blue), URS2 (yellow), and TATA (green) are indicated. Small blue boxes = Swi5 binding sites; small yellow boxes = SBF binding sites; small green box = TATA. The arrow indicates the position of the ATG (+1). Nucleotide sequences are shown for three clusters of mutations identified in screens for HO mutants that allow activation of HO-ADE2 in a swi5 or swi5 ash1 mutant. Red nucleotides are conserved across species of Saccharomyces, and asterisks indicate the positions of the mutations. Cluster 2 contains a PAC motif and Cluster 3 contains a Ume6 binding site, with the sequence of the consensus site shown for each. Vertical lines indicate positions of HO sequence that match the PAC or Ume6 consensus sites. The sequence downstream of Cluster 2 was extended to show the relative position of the −1300 Swi5 binding site, underlined in red.
Strains were constructed with individual HO promoter mutations in the native HO context, and RNA expression was measured. These cis mutations had a stronger effect on HO expression than the trans mutants identified in the previous screens, with the promoter mutations generally causing a two- to sixfold increase in expression compared to the swi5ash1 control (Table 4). Additionally, these HO promoter mutations were mostly Ace2-dependent. We speculate that off-target pleiotropic effects caused by eliminating important chromatin/transcription regulatory factors diminished the specific effect of these mutations at the HO locus, while promoter mutations had only local effects.
The HO promoter contains a PAC motif that is bound by the Dot6 protein
Mutations within Cluster 2 were the most frequently identified in our screens and displayed the strongest effects on HO expression in a swi5ash1 mutant (Figure 4 and Table 4). The altered nucleotides in Cluster 2 mutants are all conserved among species of Saccharomyces. Cluster 2 includes a perfect match to a polymerase A/C (PAC) motif (GCGATGAGMT), and seven of our mutations are in or adjacent to this motif. PAC sites were first identified within promoters of ribosome biogenesis (Ribi) genes, a group of ∼300 genes that encode various factors required for ribosome assembly and translation (Hughes et al. 2000; Jorgensen et al. 2004; Wade et al. 2006). Two paralogous repressive factors, Dot6 and Tod6, bind to the PAC sites in Ribi genes and reduce transcription via recruitment of the Rpd3 complex (Huber et al. 2011). Association of Dot6/Tod6 with Ribi promoters is modulated via phosphorylation by Sch9 in response to environmental conditions through the TOR and PKA pathways (Lippman and Broach 2009; Huber et al. 2011). A number of non-Ribi genes also have PAC sites within their promoters, raising the possibility that Dot6 and/or Tod6 may be general factors that are employed to repress other genes in different contexts (Lippman and Broach 2009). Since the HO promoter has a perfect match to the PAC motif, and the strongest mutations we identified are within the core PAC site, we investigated whether Dot6 and/or Tod6 might play a role in regulation of HO expression.
We constructed a dot6tod6swi5ash1 strain to determine whether loss of Dot6 and Tod6 could increase HO expression above the level observed in a swi5ash1 mutant. Expression of HO in the dot6tod6swi5ash1 mutant was ∼30% of wild type, an increase of 1.5- to 1.7-fold above that of swi5ash1 (Table 2; Figure 5A, compare lines 3 and 4). This is a very modest effect, although similar to what was observed with hda1, hda3, and isw2, and could suggest that like these other global transcription and chromatin factors, null mutations in DOT6 and TOD6 do not recapitulate the effect of an HO promoter mutation, possibly due to additional global effects. While the RNA results are not striking, they are consistent with the possibility that Dot6/Tod6 may bind to HO and influence its expression.
To determine whether Dot6 and/or Tod6 associate with the HO promoter, we performed ChIP analysis using Dot6-V5 and Tod6-V5 strains both in normal growth conditions and in conditions of heat shock stress. The heat shock conditions allowed us to use a Ribi gene as a positive control to demonstrate that the ChIP was successful. We were unable to detect a reliable ChIP signal for either Tod6-V5 or Tod6-HA under normal growth conditions, either at Ribi targets or at HO. However, we detected Dot6-V5 bound to multiple Ribi targets as well as to HO under heat shock conditions (Figure 5B and Figure S1, A and B). Using primers that span the URS1 and URS2 regions of the HO promoter in ∼150-bp increments, we observed a broad peak of Dot6 binding centered over the region of the promoter that contains the PAC site (Figure 5C). This binding was significantly diminished, but not completely eliminated, by the ho(G-1332A) mutation (Figure 5 and Figure S1), suggesting that Dot6 is capable of binding to HO at the PAC site and that the ho(G-1332A) mutation affects its binding. However, we were unable to reproducibly detect binding of Dot6 to HO under the conditions in which we typically monitor HO expression, i.e., non-heat-shock stress. This suggests that Dot6 may bind to HO only transiently at a specific point in the cell cycle or that Dot6 does not tightly associate with the promoter and can therefore only reliably be detected when Dot6 has an increased capacity to bind chromatin during stressful conditions.
We performed Dot6-V5 ChIP in cells synchronized using a GALp::CDC20 arrest and release protocol to investigate the possibility that Dot6 binds to HO at a specific time in the cell cycle. This type of cell synchrony has been used extensively to monitor sequential factor binding and expression at the HO promoter during the cell cycle (Bhoite et al. 2001; Takahata et al. 2009b). The Swi5 transcription factor binds at 20 min following release from G2/M arrest, and HO expression occurs at 50 min. We observed a strong peak of Dot6 binding at 30 min following release from the arrest, a time that is coincident with binding of the Ash1 repressor (Figure 5D; see also Figure 7). Similar to the ChIP results in heat shock conditions, the ho(G-1332A) mutation diminished but did not eliminate binding of Dot6. Importantly, a peak was not observed during the cell cycle time course at the Ribi target genes we tested (Figure 5E; the GCD10 promoter is shown as an example). These results support the idea that Dot6 can bind to the HO promoter in the absence of an environmental stress and that it could be the factor responsible for the change in HO expression observed in the ho(G-1332A) mutant. It remains an open possibility that Tod6 could also associate with HO and influence its expression. The fact that we did not obtain mutations in DOT6 as part of either screen may suggest that both DOT6 and TOD6 need to be mutated to observe an effect on HO-ADE2 expression.
Ume6 binds to the HO promoter and negatively regulates its expression
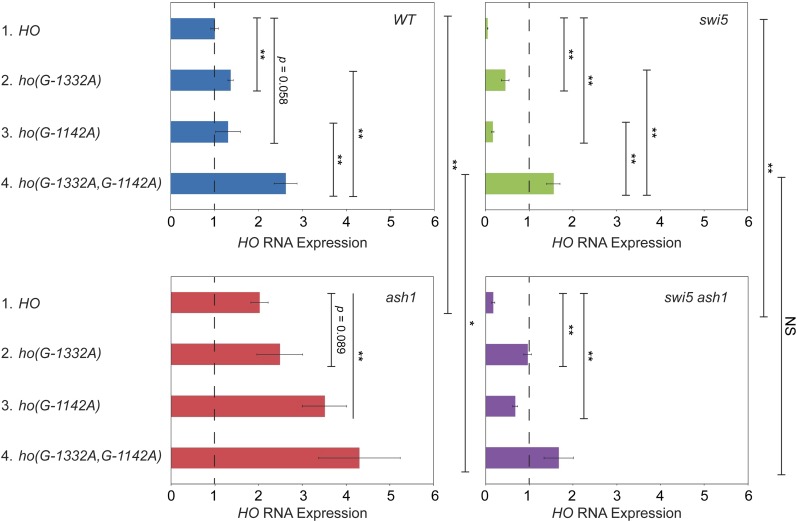
Cluster 3 mutations (−1144 to −1138) also have strong effects on HO expression (Table 4). This region of the promoter contains a match to the consensus Ume6 binding site (Park et al. 1992; Williams et al. 2002; Doniger et al. 2005). Ume6 is a DNA-binding factor that was originally identified as a repressor of meiotic genes (Strich et al. 1994). Significantly, this screen also identified SIN3 and RPD3, which repress HO expression (Dorland et al. 2000). Ume6 localizes to many sites genome-wide and recruits both the Rpd3 histone deacetylase complex and the Isw2 chromatin remodeling complex (Kadosh and Struhl 1997; Rundlett et al. 1998; Goldmark et al. 2000). We examined whether Ume6 binds to the HO promoter using a Ume6-FLAG tagged strain. Strong binding of Ume6 to the HO promoter was observed, and this binding was eliminated by the ho(G-1142A) mutation, one of the mutations identified in the screen (Figure 6A and Figure S2A). Examination of binding across the HO promoter revealed a very distinct peak of Ume6 binding centered at −1140, as expected based on the position of the Ume6 consensus site (Figure 6B).
No mutations in the UME6 gene were identified in either of our suppressor screens. However, in both screens, we eliminated a number of Ade+ mutants whose Ade+ phenotype dissipated with continued passage, and we now suggest these strains could have had a mutation in UME6. Yeast cells with a ume6 mutation have a strong growth defect, and in many cases this defect can be suppressed by chromosome duplication (Fazzio et al. 2001). Hence, ume6 mutants could have been initially Ade+ due to the loss of a negative regulator that inhibits HO-ADE2 transcription, but then became Ade− due to the aneuploidy. Many of the mutants we eliminated also displayed multiple colony sizes and variable Ade+ phenotypes, consistent with some type of genomic instability.
To test this, we disrupted UME6 in diploid cells and then sporulated to isolate fresh haploid ume6 mutants. However, it was problematic to directly examine the effect of a ume6 null on HO expression, as we had inconsistent results for biological replicates within an experiment and between experiments. There was also variation in colony size in the haploid ume6 strains, suggesting rapid aneuploidy. We therefore used Anchor Away as an alternate strategy to deplete the nucleus of Ume6. INO1, a known Ume6 target gene (Slekar and Henry 1995; Jackson and Lopes 1996), displayed greatly increased expression after Ume6 was depleted by addition of rapamycin to the medium (Figure S3). Under these conditions, we also observed suppression of both swi5 and swi5ash1 for HO expression (Figure 6C; compare lines 2 red vs. blue and 4 red vs. blue). Consistent with the effects of other global negative regulators on HO expression, the 1.6-fold increase in transcription was very modest but reproducible. Importantly, the effect of Ume6 depletion in the ash1swi5 strain was Ace2-dependent (compare lines 4 and 5 red). We conclude that Ume6 is an important repressor of HO expression, but binding site mutations more accurately demonstrate its significance than loss of the protein, which causes global effects on transcription.
Ash1, Dot6, and Ume6 associate with the HO promoter largely independently, but with similar timing
Our screens identified three DNA-binding repressor proteins that influence HO expression: Ash1, Dot6, and Ume6. All of these factors are substoichiometric members of the Rpd3 complex and have been demonstrated to recruit Rpd3 to target genes. At the HO promoter, Ash1, Dot6, and Ume6 could be redundant with one another, such that loss of one would not have a substantial effect on expression. Alternatively, they could all be necessary for complete repression, either acting independently or in an interdependent fashion.
To begin to determine whether Ash1, Dot6, and Ume6 play similar or distinct roles in repression at HO, we examined the timing of binding of each protein to the HO promoter in cells synchronized by GALp::CDC20 arrest and release. All three proteins bound to HO at ∼ 25–30 min following release from the arrest, a time point following Swi5 activator binding (20 min) but preceding HO expression (50 min) (Figure 7). Ash1 and Dot6 binding peaked at 25 min and then diminished back to their original levels before HO expression started. While Ume6 associated with the promoter at approximately the same time as Ash1 and Dot6, binding did not diminish as much as for Ash1 and Dot6, and significant amounts of Ume6 remained bound beyond the peak of HO expression.
The similar timing of association of Ash1, Dot6, and Ume6 with the HO promoter in synchronized cells suggested the possibility that one or more of these factors might be interdependent upon one another for binding. We therefore examined binding of each protein in the absence of the other two factors by ChIP analysis. We used an ash1 null to examine the effect of loss of Ash1 and promoter mutants ho(G-1332A) and ho(G-1142A) to cause the specific reduction or loss of Dot6 and Ume6, respectively, at the HO promoter.
Ash1-V5 association with the HO promoter did not decrease in either an ho(G-1332A) or in an ho(G-1142A) mutant, suggesting that Ash1 binding does not require Dot6 or Ume6 (Figure 8A). Instead, we observed a small increase in binding of Ash1 in these mutant strains. This slight additional binding could suggest that Dot6 and Ume6 binding are very weakly inhibitory to association of Ash1. Ume6-FLAG association with the HO promoter did not change significantly in the ho(G-1332A) mutant, and was reduced modestly by an ash1 mutation (Figure 8B). Thus, Ume6 binding is completely independent of Dot6 and largely independent of Ash1.
The enrichment of Dot6 was decreased in both the ho(G-1142A) and ash1ho(G-1142A) mutants, suggesting that the presence of Ume6 may have some influence on the level of association of Dot6 with the HO promoter (Figure 8C). Some Dot6 could be brought to HO via Ume6 if Dot6 comes along as part of the Ume6-containing Rpd3 complex (Carrozza et al. 2005), and this may explain the very broad peak of Dot6 binding across the HO promoter (Figure 5C) in contrast to the narrower peak of Ume6 binding (Figure 6B). This possibility is supported by the observation that the ho(G-1332A) mutation, which reduced but did not eliminate Dot6 binding, shifted residual Dot6 binding toward the transcription start site, near the Ume6 site (Figure 5C). Similar results were observed when we mutated the entire PAC site (data not shown). Alternatively, perhaps the presence of Ume6 affects the positioning of nucleosomes in such a way that Dot6 association is altered in the absence of Ume6. This possibility could explain how Ume6 could affect Dot6 binding at its PAC site despite the nearly 200 nucleotides between the binding sites for these two factors. Overall, these results suggest that while all three proteins are capable of some level of association with the HO promoter independent of the others, the presence of Ume6 does appear to affect binding of Dot6 through a mechanism that is currently not understood.
The ash1, ho(G-1332A), and ho(G-1142A) mutations are additive for effects on HO expression but not for loss of Rpd3 recruitment
The lack of strong dependency of Ash1, Dot6, and Ume6 upon one another for binding to the HO promoter suggests they may each be required for full repression of HO expression. We therefore examined whether the absence of each of these factors was additive for their relief of HO repression at URS1. For these experiments, we continued to use the ash1 null mutant, along with ho(G-1332A) for reduction in Dot6 (and possibly Tod6) and ho(G-1142A) for elimination of Ume6.
We measured HO expression in all combinations of ash1, ho(G-1332A), and ho(G-1142A), both in SWI5+ (Figure 9, left graphs) and in swi5 strains (Figure 9, right graphs). All three single mutants caused an increase in HO expression in otherwise wild-type cells (Figure 9, lines 2 and 3 blue and line 1 red). The ash1 mutant had the strongest effect, with an approximately twofold increase, likely due to a gain of expression of HO in daughter cells (line 1 red). The ho(G-1332A) (Dot6) and ho(G-1142A) (Ume6) mutants were additive with one another, increasing HO expression to 2.6-fold higher than wild type (compare line 4 blue to lines 2 and 3 blue), which was increased even higher in the triple ash1ho(G-1332A, G-1142A) mutant (4.3-fold higher than wild type; compare line 4 red to line 4 blue). The ho(G-1142A) mutant showed significant additivity with ash1, while the additivity of ho(G-1332A) and ash1 was less pronounced (compare lines 2 and 3 red to line 1 red).
Figure 9.
Combining the ash1, ho(G-1332A), and ho(G-1142A) mutations shows additive suppression of ash1 swi5. HO mRNA levels were measured, normalized to RPR1, and expressed relative to wild type. Each graph shows results for the genotype indicated (wild type in blue, swi5 in green, ash1 in red, ash1 swi5 in purple). The type of HO promoter for each bar is shown at the left (HO wt, ho(G-1332A), ho(G-1142A) and ho(G-1332A, G-1142A)). The dotted line indicates the level of HO expression in a SWI5 ASH1 strain with the wild-type HO promoter. Error bars represent the SD of four biological replicates for each strain. * P < 0.05, ** P < 0.01.
All three single mutants suppressed swi5 for expression of HO (Figure 9, right graphs). The strongest was ho(G-1332A), which increased expression ninefold over that of a swi5 mutant (45% of wild type; compare lines 1 and 2 green); ho(G-1142A) and ash1 had more modest effects (16% and 18% of wild type; compare line 3 green and line 1 purple to line 1 green). Each of the ho promoter mutants was additive with ash1 for suppression of swi5 (compare lines 2 and 3 purple to line 1 purple). Interestingly, the ho(G-1332A) and ho(G-1142A) mutants were also strongly additive with one another for suppression of swi5, with a value 150% of wild type (compare line 4 green with lines 2 and 3 green); this value did not increase substantially with the addition of the ash1 mutant (line 4 purple). These results demonstrate that mutations that reduce or eliminate Ash1, Dot6, and Ume6 localization are generally additive for relief of HO repression, suggesting the three proteins and their associated complexes act independently. However, in the absence of the Swi5 activator, loss of Ash1 is additive with either single promoter mutant but not with the double promoter mutant. This suggests that if the proteins at the PAC site and at the Ume6 site have been reduced or removed, elimination of Ash1 has little additional effect on increasing HO expression in a swi5 mutant. The same is not true for loss of Ash1 and the PAC-associated proteins or Ash1 and the Ume6 site-associated proteins; in both of these cases, removal of the third complex further relieves repression.
The additivity of the Ash1, PAC, and Ume6 site mutants thus appears to be different in the presence and absence of Swi5, for reasons that are not clear. Mother and daughter cells could differ in their responses to loss of the repressors. Ash1 protein levels are vastly different in the two cell types, and the effect of no Ash1 in mother cells may be different from that in daughter cells. Swi5 has the potential to activate HO in both mother and daughter cells, whereas in the absence of Swi5, Ace2 can activate HO only in daughter cells. Perhaps the additivity of all three mutants (Ash1, PAC, Ume6) is only seen in a SWI5+ strain because Swi5 normally activates ASH1 expression (Bobola et al. 1996).
Interpretation of these results is also somewhat complicated by the observation that Ume6 appears to contribute positively to Dot6 binding at the HO promoter (Figure 8C). The additivity of the ho(G-1332A) and ho(G-1142A) mutants could partly be explained if the amount of Dot6 association with HO in the ho(G-1332A, G-1142A) double mutant is lower than in the ho(G-1332A) mutant alone. However, Ume6 binding becomes undetectable in the ho(G-1142A) mutant, while Dot6 binding is simply reduced, suggesting that the effect of ho(G-1142A) is likely mostly due to the absence of Ume6.
The additivity of ash1, ho(G-1332A), and ho(G-1142A) mutants for relief of HO repression suggests that Ash1, Dot6, and Ume6 may independently recruit the Rpd3 complex to contribute to repression. To examine this possibility, we performed Rpd3-V5 ChIP in strains with single mutants and mutant combinations to determine whether recruitment of Rpd3 by these factors is redundant or additive (Figure 10 and Figure S5A). At the URS2 region of HO, SBF recruits Whi5 and Stb1, which in turn recruit Rpd3 (Takahata et al. 2009a), raising the possibility that Rpd3 bound at URS2 could affect the results of our Rpd3 ChIP assay at URS1. To circumvent this problem, we used strains with mutations that disrupt binding at all nine SBF sites within URS2. Both ash1 and ho(G-1142A) single mutants diminished recruitment of Rpd3 at URS1, but neither alone eliminated binding (compare lines 2 and 4 to line 1). When combined, ash1 and ho(G-1142A) reduced Rpd3 binding further (compare line 6 to lines 2 and 4), suggesting Ash1 and Ume6 recruit Rpd3 independently, with each contributing separately to its full recruitment and repression. In contrast, the ho(G-1332A) mutant did not appear to substantially affect Rpd3 binding either on its own or in combination with ash1, ho(G-1142A) or both (compare lines 3 vs. 1, 5 vs. 2, 7 vs. 4, 8 vs. 6). Thus, Ash1 and Ume6 are important for recruitment of Rpd3 to URS1 of the HO promoter, but surprisingly, Dot6 does not appear to contribute in a significant way. The observation that ho(G-1142A) and ho(G-1332A) do not have the same effect on Rpd3 binding at HO supports the suggestion that the effect of ho(G-1142A) on HO expression (Figure 9) is not mediated by the partial loss of Dot6 but rather by the substantial loss of Ume6 binding. If the predominant effect of both mutations was partial loss of Dot6, both should display the same consequence for Rpd3 binding.
The discrepancy between the strong effect on HO expression observed in an ho(G-1332A) mutant (Figure 9 and Table 4) and the apparent negligible effect on Rpd3 recruitment could suggest that Dot6 plays a separate, yet undetermined, repressive role at HO. Dot6 and/or Tod6 could repress non-Ribi genes such as HO using an Rpd3-independent mechanism. Alternatively, the Rpd3 that associates with HO via Dot6 may be difficult to measure at HO if the Dot6 interaction itself is more transient than Ash1 and Ume6, as suggested by the lack of consistent detection of Dot6 by ChIP in asynchronous, non-heat-shocked cells.
Negative regulators additively contribute to repression of HO by decreasing SWI/SNF recruitment
The crucial importance of URS1 for HO expression is recruitment of the SWI/SNF remodeling complex, which causes waves of nucleosome eviction along the promoter, uncovering the SBF sites within URS2 and ultimately allowing recruitment of RNA polymerase and transcription of the gene. We therefore examined whether mutation of each of these repressors would additively increase SWI/SNF recruitment in the context of a swi5 mutant. ChIP assays were performed to measure Swi2-V5 recruitment to URS1 in a set of mutant strains, and cells were collected from the same cultures for parallel analysis of HO RNA expression (Figure 11). Swi2-V5 recruitment and HO expression were both very low in a swi5 strain, relative to wild type (compare line 2 to line 1), suggesting that the Ace2 activator is unable to promote high HO expression because it does not recruit sufficient SWI/SNF for nucleosome eviction to uncover the SBF sites in URS2. Each single mutant representing reduction or loss of Ash1, Dot6, and Ume6 increased SWI/SNF recruitment to a level that closely matched the corresponding level of HO expression increase (compare lines 3–5 to line 2). Binding of Swi2-V5 to URS2 was weaker than at URS1 in all strains (Figure S6A), as expected (Takahata et al. 2009b). Similar trends of changes in Swi2-V5 binding between each mutant and wild type were observed at URS1 and URS2 (Figure S6B), as expected if SWI/SNF is recruited to URS1 and spreads downstream to URS2. Ace2 is thus more capable of recruiting SWI/SNF to URS1 in the absence of the repressors, restoring the cascade of nucleosome eviction that extends into URS2 to allow SBF binding and HO transcription.
The ho(G-1332A) and ho(G-1142A) mutations were each additive with ash1 for recruitment of Swi2-V5 and HO expression (Figure 11, compare line 6 to lines 3 and 4 and line 7 to lines 3 and 5). The ho(G-1332A, G-1142A) double mutant was also strongly additive for an increase in Swi2-V5 binding, but Swi2-V5 binding did not increase further in the ho(G-1332A, G-1142A) ash1 triple mutant (compare line 8 to lines 4 and 5 and line 9 to line 8). These SWI/SNF binding and HO expression results are consistent with the HO expression data in Figure 9, and further illustrate that if both complexes at the PAC site and Ume6 site are reduced or eliminated in the absence of Swi5, HO expression is increased to a point in which an ash1 mutation has no further effect. Additionally, the observation that ho(G-1332A) had strong effects on Swi2-V5 recruitment but not on Rpd3-V5 recruitment is consistent with the idea that proteins bound at the PAC site employ an Rpd3-independent mechanism for repression.
Concluding remarks: a large number of different repressors is necessary for complex regulation
Our Swi2 ChIP results suggest that the purpose of the Ash1, Ume6, and PAC site complexes is to limit the spread of SWI/SNF-mediated nucleosome eviction from URS1 to URS2. This could ensure that the URS2 and TATA regions of the promoter are not opened prematurely or kept open longer than necessary to achieve the appropriate number of HO transcripts. The observation that Ash1, Dot6, and Ume6 associate with the promoter after the initiating event of Swi5 binding, but before the rise in HO RNA levels, suggests that their effect is likely on recruitment of SWI/SNF itself rather than on recruitment of the Swi5 activator, which would in turn affect the level of SWI/SNF. Consistent with this idea, we have found that Swi5 levels at the HO promoter do not change in response to Ash1 levels or HO promoter mutants (Figure S7).
Extending this view to the larger picture of repression at the HO promoter, we suggest that the primary role of the cohort of negative regulators is to restrict the recruitment and/or action of coactivator complexes. The number and diversity of regulators suggest there are multiple mechanisms to accomplish this goal, targeting different coactivators at a range of locations along the promoter. The Rpd3 complex brought by Ash1 and Ume6 would limit the action of the Gcn5 histone acetyltransferase within the SAGA coactivator. The Hda1 complex, for which we also identified mutants in our screen, likely has a similar function. Because SAGA levels affect recruitment of SWI/SNF, these histone deacetylase complexes could, by extension, also affect the levels of SWI/SNF, as we observed in our Swi2 ChIP experiments. The Isw2 chromatin remodeler (also likely to play a role at HO since mutants were identified in our screen) may more directly oppose SWI/SNF action. Ume6 is known to associate with Isw2 throughout the genome, and thus Ume6 could serve as a recruitment site for both Rpd3 and Isw2. Tup1 could also fit within this model, as studies have demonstrated connections between Tup1 and both Hda1 and Rpd3, and it has been shown that Tup1 inhibits SWI/SNF recruitment (Wong and Struhl 2011). Additionally, Tup1 could affect recruitment or function of the Mediator complex, a coactivator that is important at HO, but for which a clear connection to any of the other repressors identified in our screens has not been established.
The majority of Ash1 binding within the HO promoter is within the nucleosome just upstream of the linker containing the Ume6 site (data not shown; manuscript in preparation). The position of Ash1 and Ume6 at the right end of URS1 is consistent with a role in limiting the spread of nucleosome eviction into URS2. Why would HO require two methods of recruiting Rpd3 to the promoter at similar locations? One possible reason could be the differential levels of repression in mother vs. daughter cells. In daughter cells, strong repression is necessary to keep the gene off. However, in mother cells, a more nuanced mechanism of repression is required to ensure an adequate but not excessive level of nucleosome eviction spreading into URS2. Since Ash1 and Ume6 associate with HO after Swi5 has bound, it seems likely that the Swi5 activator binds to the HO promoter in both mothers and daughters. Recruitment of coactivators to URS1 and subsequent nucleosome eviction at URS1 would thereby occur in both cell types, but the much higher concentration of Ash1-Rpd3 complex in daughters would prevent nucleosome eviction from ultimately spreading into URS2 and promoting gene activation. In contrast, the small amount of Ash1 in mother cells, in combination with Ume6, would serve as an effective, but not absolute, restraint on the ability of SWI/SNF and SAGA to evict nucleosomes.
The PAC site is located near a Swi5 binding site within the NDR upstream of the Ash1/Ume6 sites. The observation that this site does not affect Rpd3 levels suggests that it limits coactivator recruitment by a currently unknown mechanism, different from that of Ash1 and Ume6. The PAC site is particularly intriguing, since it is highly conserved and substitutions here have the strongest effects of all of our mutations on HO expression. There is much more to explore related to the PAC site and its influence on HO expression, including whether Tod6 and/or other proteins also bind to this site and what types of repressor complexes may be recruited there. The observation that Dot6 binds at HO at a time during the cell cycle when it is not bound to Ribi genes suggests there may be a mechanism that operates specifically at HO (or HO and a class of other genes) to allow Dot6 binding at this time. Dot6 association with chromatin is suggested to be influenced by its phosphorylation (Huber et al. 2011). One possible idea is that a phosphatase recruited to HO may transiently dephosphorylate Dot6, allowing Dot6 binding at a time when it would not associate with Ribi genes.
Our screens thus identified many contributors to repression at HO: the DNA-binding proteins Ash1, Ume6, and Dot6, as well as other regulatory complexes that oppose transcription, including the Rpd3, Hda1, and Isw2 complexes, the Tup1 corepressor, and a possible additional complex located at the position of the Cluster 1 mutations. These proteins appear to associate with a variety of locations within the HO promoter, including the nucleosome and linker downstream of Swi5 Site B at the right end of URS1 (Ash1 and Ume6), within the NDR that includes Swi5 Site B (Dot6), and within the NDR that includes Swi5 Site A (putative complex at Cluster 1) (Figure 12). The question then becomes why the HO promoter requires such a large quantity and diversity of repressors. Many complex genes, such as HO, require a series of coactivators with different activities to achieve the appropriate level of transcription. It is not surprising that a parallel set of corepressors could be needed to provide balance, restricting the action of coactivators both temporally and by cell type. While corepressor complexes were initially thought to be exclusively negative in terms of their effect on transcription, increasing evidence suggests they can be required for resetting the promoter for subsequent expression and are therefore an integral part of the activation process (Wang et al. 2009; Dovey et al. 2010; Wong and Struhl 2011). A delicate balance as well as coordination between activating and repressing factors and processes is a necessary part of complex gene regulation. Further investigation of the repressor complexes at the HO promoter may serve to clarify and understand their specific roles and their relationship to one another and to the coactivator complexes.
Figure 12.
Model: HO expression is regulated by multiple transcriptional promoting and repressing factors. A model is shown indicating the approximate positions of some of the factors associated with URS1 that influence HO expression. DNA elements URS1 (blue), URS2 (yellow), and the TATA element (green) are shown. The arrow indicates the position of the ATG (+1). Well-defined nucleosome positions are shown throughout the region from ∼ −1960 to +1 as ovals with lines. Factors that activate HO expression are shown on top and include Swi5 (blue) and the SWI/SNF complex (green). Factors that repress HO expression are shown on the bottom and include Dot6/Tod6 (brown), Ash1 (purple), Ume6 (orange), and the Rpd3 complex (red). A gray shape with a “?” represents unknown factors recruited by Dot6/Tod6 that contribute to repression. Known binding sites for factors are shown as small boxes: blue = Swi5, yellow = SBF, green = TATA, brown = Dot6/Tod6 (PAC element), orange = Ume6.
Acknowledgments
We thank Tim Formosa and Laura McCullough for comments on the manuscript, Tim Parnell for help with the bioinformatics, and Tim Formosa and members of the Stillman lab for helpful advice throughout the course of this project. We thank Zaily Connell, Tim Formosa, Mark Gartenberg, and Kevin Struhl for plasmids, and Benjamin Albert and David Shore for personal communication. We thank undergraduate students Tom Engar and Jon Navar who contributed to development of the genetic screen. This work was supported by National Institutes of Health grant GM121079, awarded to D.J.S.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8171276.
Communicating editor: A. Hinnebusch
Literature Cited
- Albert B., Tomassetti S., Gloor Y., Dilg D., Mattarocci S., et al. , 2019. Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes Dev. 33: 288–293. 10.1101/gad.322040.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., et al. , 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2: 437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Bhoite L. T., Yu Y., Stillman D. J., 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15: 2457–2469. 10.1101/gad.921601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N., Jansen R. P., Shin T. H., Nasmyth K., 1996. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84: 699–709. 10.1016/S0092-8674(00)81048-X [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K., 1987. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48: 389–397. 10.1016/0092-8674(87)90190-5 [DOI] [PubMed] [Google Scholar]
- Brogaard K., Xi L., Wang J. P., Widom J., 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501. 10.1038/nature11142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., et al. , 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731: 77–87. 10.1016/j.bbaexp.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Chaves S., Baskerville C., Yu V., Reed S. I., 2010. Cks1, Cdk1, and the 19S proteasome collaborate to regulate gene induction-dependent nucleosome eviction in yeast. Mol. Cell. Biol. 30: 5284–5294. 10.1128/MCB.00952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin T. E., Brent R., 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107: 739–750. 10.1016/S0092-8674(01)00596-7 [DOI] [PubMed] [Google Scholar]
- Cosma M. P., Tanaka T., Nasmyth K., 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311. 10.1016/S0092-8674(00)80740-0 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa J. L., Tang X., Millman J. S., Schub O., et al. , 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913. 10.1016/j.cell.2004.05.024 [DOI] [PubMed] [Google Scholar]
- de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., III, Wittenberg C., 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898. 10.1016/j.cell.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Di Talia S., Wang H., Skotheim J. M., Rosebrock A. P., Futcher B., et al. , 2009. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 7: e1000221 10.1371/journal.pbio.1000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann P. R., Butler G., Tamai K., Dorland S., Greene J. R., et al. , 1992. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6: 93–104. 10.1101/gad.6.1.93 [DOI] [PubMed] [Google Scholar]
- Dohrmann P. R., Voth W. P., Stillman D. J., 1996. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol. Cell. Biol. 16: 1746–1758. 10.1128/MCB.16.4.1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger S. W., Huh J., Fay J. C., 2005. Identification of functional transcription factor binding sites using closely related Saccharomyces species. Genome Res. 15: 701–709. 10.1101/gr.3578205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorland S., Deegenaars M. L., Stillman D. J., 2000. Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154: 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey O. M., Foster C. T., Cowley S. M., 2010. Emphasizing the positive: a role for histone deacetylases in transcriptional activation. Cell Cycle 9: 2700–2701. 10.4161/cc.9.14.12626 [DOI] [PubMed] [Google Scholar]
- Fazzio T. G., Kooperberg C., Goldmark J. P., Neal C., Basom R., et al. , 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21: 6450–6460. 10.1128/MCB.21.19.6450-6460.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman I., Nagaraj V., Norris D., Vershon A. K., 2003. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2: 1061–1068. 10.1128/EC.2.5.1061-1068.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. B., Beggs S., Church M., Tsukihashi Y., Pennings S., 2014. The yeast Cyc8-Tup1 complex cooperates with Hda1p and Rpd3p histone deacetylases to robustly repress transcription of the subtelomeric FLO1 gene. Biochim. Biophys. Acta 1839: 1242–1255. 10.1016/j.bbagrm.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T., 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433. 10.1016/S0092-8674(00)00134-3 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Green S. R., Johnson A. D., 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15: 4191–4202. 10.1091/mbc.e04-05-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Mendenhall M. D., Reed S. I., 1989. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell. Biol. 9: 2034–2041. 10.1128/MCB.9.5.2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., Laemmli U. K., 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Huber A., French S. L., Tekotte H., Yerlikaya S., Stahl M., et al. , 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 30: 3052–3064. 10.1038/emboj.2011.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. D., Estep P. W., Tavazoie S., Church G. M., 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296: 1205–1214. 10.1006/jmbi.2000.3519 [DOI] [PubMed] [Google Scholar]
- Jackson J. C., Lopes J. M., 1996. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 24: 1322–1329. 10.1093/nar/24.7.1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. P., Dowzer C., Michaelis C., Galova M., Nasmyth K., 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell 84: 687–697. 10.1016/S0092-8674(00)81047-8 [DOI] [PubMed] [Google Scholar]
- Jensen R. E., Sprague G. F., Jr, Herskowitz I., 1983. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc. Natl. Acad. Sci. USA 80: 3035–3039. 10.1073/pnas.80.10.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Pugh B. F., 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 10: R109 10.1186/gb-2009-10-10-r109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M., 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400. 10.1126/science.1070850 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., et al. , 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18: 2491–2505. 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371. 10.1016/S0092-8674(00)80217-2 [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., et al. , 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- Kurdistani S. K., Grunstein M., 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4: 276–284. 10.1038/nrm1075 [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L., 2007. The role of chromatin during transcription. Cell 128: 707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Lippman S. I., Broach J. R., 2009. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. USA 106: 19928–19933. 10.1073/pnas.0907027106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Hall M. N., 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Malave T. M., Dent S. Y., 2006. Transcriptional repression by Tup1-Ssn6. Biochem. Cell Biol. 84: 437–443. [DOI] [PubMed] [Google Scholar]
- Marion R. M., Regev A., Segal E., Barash Y., Koller D., et al. , 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101: 14315–14322. 10.1073/pnas.0405353101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H., Nakajima R., Nishiyama J., Araki H., Oshima Y., 1990. Chromosome engineering in Saccharomyces cerevisiae by using a site-specific recombination system of a yeast plasmid. J. Bacteriol. 172: 610–618. 10.1128/jb.172.2.610-618.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxon M. E., Herskowitz I., 2001. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl. Acad. Sci. USA 98: 1495–1500. 10.1073/pnas.98.4.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazanka E., Alexander J., Yeh B. J., Charoenpong P., Lowery D. M., et al. , 2008. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 6: e203 10.1371/journal.pbio.0060203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath D. A., Balog E. R., Koivomagi M., Lucena R., Mai M. V., et al. , 2013. Cks confers specificity to phosphorylation-dependent CDK signaling pathways. Nat. Struct. Mol. Biol. 20: 1407–1414. 10.1038/nsmb.2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D., Parnell E. J., Landon J. W., Yu Y., Stillman D. J., 2006. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell. Biol. 26: 4095–4110. 10.1128/MCB.01849-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Yoshida M., Uematsu C., Kito K., et al. , 2008. Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics 9: 574 10.1186/1471-2164-9-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Struhl K., 2008. Expanding the repertoire of plasmids for PCR-mediated epitope tagging in yeast. Yeast 25: 287–292. 10.1002/yea.1581 [DOI] [PubMed] [Google Scholar]
- Munchow S., Sauter C., Jansen R. P., 1999. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112: 1511–1518. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., 1983. Molecular analysis of a cell lineage. Nature 302: 670–676. 10.1038/302670a0 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., 1985. At least 1400 base pairs of 5′-flanking DNA is required for the correct expression of the HO gene in yeast. Cell 42: 213–223. 10.1016/S0092-8674(85)80117-3 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Stillman D., Kipling D., 1987. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell 48: 579–587. 10.1016/0092-8674(87)90236-4 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Adolf G., Lydall D., Seddon A., 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell 62: 631–647. 10.1016/0092-8674(90)90110-Z [DOI] [PubMed] [Google Scholar]
- Pan Y. R., Sun M., Wohlschlegel J., Reed S. I., 2013. Cks1 enhances transcription efficiency at the GAL1 locus by linking the Paf1 complex to the 19S proteasome. Eukaryot. Cell 12: 1192–1201. 10.1128/EC.00151-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. D., Luche R. M., Cooper T. G., 1992. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 20: 1909–1915. 10.1093/nar/20.8.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R. J., Sunjevaric I., Voth W. P., Ciccone S., Du W., et al. , 2008. Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes. Genetics 180: 1799–1808. 10.1534/genetics.108.087999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard G. J., Reynolds W., Verma R., Deshaies R. J., 2000. Cks1 is required for G(1) cyclin-cyclin-dependent kinase activity in budding yeast. Mol. Cell. Biol. 20: 5858–5864. 10.1128/MCB.20.16.5858-5864.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J., Gartenberg M. R., Oshima Y., Wang J. C., 1992. A hit-and-run system for targeted genetic manipulations in yeast. Nucleic Acids Res. 20: 4671–4672. 10.1093/nar/20.17.4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194: 281–301. 10.1016/0076-6879(91)94022-5 [DOI] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Suka N., Turner B. M., Grunstein M., 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392: 831–835. 10.1038/33952 [DOI] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. 10.1016/0076-6879(91)94004-V [DOI] [PubMed] [Google Scholar]
- Sil A., Herskowitz I., 1996. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84: 711–722. 10.1016/S0092-8674(00)81049-1 [DOI] [PubMed] [Google Scholar]
- Slekar K. H., Henry S. A., 1995. SIN3 works through two different promoter elements to regulate INO1 gene expression in yeast. Nucleic Acids Res. 23: 1964–1969. 10.1093/nar/23.11.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Johnson A. D., 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25: 325–330. 10.1016/S0968-0004(00)01592-9 [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Stern M. J., Clark I., Herskowitz I., 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48: 567–577. 10.1016/0092-8674(87)90235-2 [DOI] [PubMed] [Google Scholar]
- Stillman D. J., 2013. Dancing the cell cycle two-step: regulation of yeast G1-cell-cycle genes by chromatin structure. Trends Biochem. Sci. 38: 467–475. 10.1016/j.tibs.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A., 1988. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 7: 485–494. 10.1002/j.1460-2075.1988.tb02836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Lewis L. K., Resnick M. A., 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773–776. 10.1038/90837 [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., et al. , 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31: 183–192. 10.1016/0092-8674(82)90418-4 [DOI] [PubMed] [Google Scholar]
- Strich R., Surosky R. T., Steber C., Dubois E., Messenguy F., et al. , 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8: 796–810. 10.1101/gad.8.7.796 [DOI] [PubMed] [Google Scholar]
- Taba M. R. M., Muroff I., Lydall D., Tebb G., Nasmyth K., 1991. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 5: 2000–2013. 10.1101/gad.5.11.2000 [DOI] [PubMed] [Google Scholar]
- Takahata S., Yu Y., Stillman D. J., 2009a The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 28: 3378–3389. 10.1038/emboj.2009.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Yu Y., Stillman D. J., 2009b FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell 34: 405–415. 10.1016/j.molcel.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Yu Y., Stillman D. J., 2011. Repressive chromatin affects factor binding at yeast HO (homothallic switching) promoter. J. Biol. Chem. 286: 34809–34819. 10.1074/jbc.M111.281626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Reed S. I., 1993. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 7: 822–832. 10.1101/gad.7.5.822 [DOI] [PubMed] [Google Scholar]
- Tebb G., Moll T., Dowser C., Nasmyth K., 1993. SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev. 7: 517–528. 10.1101/gad.7.3.517 [DOI] [PubMed] [Google Scholar]
- Tellmann G., 2006. The E-Method: a highly accurate technique for gene-expression analysis. Nat. Methods 3: i–ii. 10.1038/nmeth894 [DOI] [Google Scholar]
- Thomas B. J., Rothstein R., 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630. 10.1016/0092-8674(89)90584-9 [DOI] [PubMed] [Google Scholar]
- Toyn J. H., Gunyuzlu P. L., White W. H., Thompson L. A., Hollis G. F., 2000. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast 16: 553–560. [DOI] [PubMed] [Google Scholar]
- Voth W. P., Yu Y., Takahata S., Kretschmann K. L., Lieb J. D., et al. , 2007. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 26: 4324–4334. 10.1038/sj.emboj.7601859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- Wade C. H., Umbarger M. A., McAlear M. A., 2006. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast 23: 293–306. 10.1002/yea.1353 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D. E., Barski A., et al. , 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031. 10.1016/j.cell.2009.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake V. M., Workman J. L., 2010. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11: 426–437. 10.1038/nrg2781 [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Rando O. J., Delrow J., Tsukiyama T., 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035. 10.1038/nature06391 [DOI] [PubMed] [Google Scholar]
- Williams R. M., Primig M., Washburn B. K., Winzeler E. A., Bellis M., et al. , 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99: 13431–13436. 10.1073/pnas.202495299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer C. T., Reed G. H., Gundry C. N., Vandersteen J. G., Pryor R. J., 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49: 853–860. 10.1373/49.6.853 [DOI] [PubMed] [Google Scholar]
- Wong K. H., Struhl K., 2011. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 25: 2525–2539. 10.1101/gad.179275.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Suka N., Carlson M., Grunstein M., 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7: 117–126. 10.1016/S1097-2765(01)00160-5 [DOI] [PubMed] [Google Scholar]
- Yadon A. N., Tsukiyama T., 2013. DNA looping-dependent targeting of a chromatin remodeling factor. Cell Cycle 12: 1809–1810. 10.4161/cc.25114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon A. N., Van de Mark D., Basom R., Delrow J., Whitehouse I., et al. , 2010. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol. Cell. Biol. 30: 5110–5122. 10.1128/MCB.00602-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrington R. M., Rudd J. S., Stillman D. J., 2015. Spatiotemporal cascade of transcription factor binding required for promoter activation. Mol. Cell. Biol. 35: 688–698. 10.1128/MCB.01285-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrington R. M., Goodrum J. M., Stillman D. J., 2016. Nucleosomes are essential for proper regulation of a multigated promoter in Saccharomyces cerevisiae. Genetics 202: 551–563. 10.1534/genetics.115.183715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Yarrington R. M., Chuong E. B., Elde N. C., Stillman D. J., 2016. Disruption of promoter memory by synthesis of a long noncoding RNA. Proc. Natl. Acad. Sci. USA 113: 9575–9580. 10.1073/pnas.1601793113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata J., Dephoure N., Macdonough T., Yu Y., Parnell E. J., et al. , 2014. PP2ARts1 is a master regulator of pathways that control cell size. J. Cell Biol. 204: 359–376. 10.1083/jcb.201309119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Reese J. C., 2004. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J. 23: 2246–2257. 10.1038/sj.emboj.7600227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Table S1 lists the strains and Table S2 lists the plasmids used in this study. Seven Supplemental Figures and two Supplemental Tables were uploaded to the GSA Figshare portal. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8171276.