Abstract
Construction of genetic linkage maps has become a routine step for mapping quantitative trait loci (QTL), particularly in animal and plant breeding populations. Many multiparental populations have recently been produced to increase genetic diversity and QTL mapping resolution. However, few software packages are available for map construction in these populations. In this paper, we build a general framework for the construction of genetic linkage maps from genotypic data in diploid populations, including bi- and multiparental populations, cross-pollinated (CP) populations, and breeding pedigrees. The framework is implemented as an automatic pipeline called magicMap, where the maximum multilocus likelihood approach utilizes genotypic information efficiently. We evaluate magicMap by extensive simulations and eight real datasets: one biparental, one CP, four multiparent advanced generation intercross (MAGIC), and two nested association mapping (NAM) populations, the number of markers ranging from a few hundred to tens of thousands. Not only is magicMap the only software capable of accommodating all of these designs, it is more accurate and robust to missing genotypes and genotyping errors than commonly used packages.
Keywords: genetic map construction, hidden Markov model (HMM), multiparent advanced generation intercross (MAGIC), nested association mapping (NAM), cross-pollinated (CP), multiparental populations, MPP
THE construction of a genetic linkage map consists of grouping, ordering, and spacing genetic markers in experimental crosses. Genetic maps provide insights and clues for understanding genetic processes such as recombination, chromosome arrangement, and genome evolution. Although available physical maps can provide marker grouping and ordering, genetic maps can validate physical maps, improve de novo genome assemblies (Fierst 2015; Song et al. 2016), supply intermarker genetic distances, and include the markers that cannot be localized on physical maps. Most importantly, genetic maps allow more powerful strategies for mapping QTL, particularly in animal and plant breeding populations (e.g., Paterson et al. 1988).
Traditional biparental mapping populations are produced from two inbred lines. Many software packages exist for map construction in these populations. Examples include MapMAKER (Lander and Green 1987), JoinMAP (Stam 1993; Van Ooijen 2006), CarthaGène (Schiex and Gaspin 1997), Neighbor Mapping (Ellis 1997), R/qtl (Broman et al. 2003), RECORD (Van Os et al. 2005), MadMapper (Kozik and Michelmore 2006), AntMap (Iwata and Ninomiya 2006), MSTmap (Wu et al. 2008), THREaD Mapper (Cheema et al. 2010), and MapDisto (Lorieux 2012). Hidden Markov models (HMM) with the Lander and Green algorithm (Lander et al. 1987) have been used by MapMAKER, CarthaGène, and R/qtl, where hidden inheritance vectors describe the gene flow from founders to offspring in a breeding pedigree.
Many multiparental populations have recently been produced, where multiple inbred founders are crossed for some generations to increase genetic diversity and QTL mapping resolution. Examples include Kover et al. (2009), Dell’Acqua et al. (2015), Pascual et al. (2015), and Liller et al. (2017), and see the review by Huang et al. (2015) for more examples in crops. Currently, mpMap (Huang and George 2011) is the package most commonly used for multiparental populations, but see also the recently released R/qtl2 (Broman et al. 2019). However, the mpMap package is limited to funnel scheme multi-way recombinant inbred lines (RILs; e.g., Dell’Acqua et al. 2015; Pascual et al. 2015), and does not allow markers with missing founder genotypes. In the funnel scheme, the founders of each line are randomly permuted, and each line is produced by an intercross scheme that combines all founder genomes through several generations of pairwise crosses prior to repeated generations of selfing.
CP populations are often used for QTL mapping when only outbred founders are practical. Examples of software packages for CP populations include OneMap (Margarido et al. 2007), FsLinkageMap (Tong et al. 2010), JoinMAP (Van Ooijen 2011), HighMap (Liu et al. 2014), HetMapps (Hyma et al. 2015), and Lep-MAP3 (Rastas 2017). Various HMMs have been used by all these packages except HetMapps. See also CRI-MAP for map construction in a large multi-generation pedigree but with some available information arbitrarily excluded (Green et al. 1990).
The primary aim of this paper is to build a general framework for genetic map construction in diploid populations, which can be applied to all of the above mapping populations including bi- and multiparental populations and CP populations. The population design information can be specified by either a breeding pedigree or a series of mating schemes from one generation to the next. The algorithm magicMap builds on our previously developed HMM with hidden states being the ancestral origins of genotypes (Zheng et al. 2014, 2015, 2018a,b; Zheng 2015). The algorithm is computationally efficient, because the state space in our HMM is proportional to the number of inbred founders in the case of homozygous populations, whereas the state space in the packages such as MapMAKER and CarthaGène increases exponentially with pedigree size.
The second aim is to analyze the increasing available genotyping-by-sequencing (GBS) data. Low coverage sequencing is often used to cut costs, resulting in many missing genotypes and errors. To increase the robustness of map construction to missing genotypes and genotyping errors, we integrate map construction with founder linkage phasing, genotype imputation, and correction, and develop an efficient multilocus likelihood maximization. The maximum likelihood gives the marginal probability of marker data by integrating all hidden states, which has been shown to be more robust to missing genotypes than simplified two-point statistics such as the sum of adjacent recombination fractions (Hackett and Broadfoot 2003; Tong et al. 2010). The criterion using two-point statistics has been commonly used in the packages such as RECORD, MSTmap, mpMap, and HighMap because of high computational efficiency.
Most software packages, such as MSTmap and Lep-MAP3, start with two-locus linkage analysis, and then group markers using hard thresholds such as the logarithm of the odds (LOD) score or recombination fraction. However, the grouping is very sensitive to the input hard thresholds, and the desired number of groups cannot always be obtained by varying the thresholds because of missing data and genotyping errors. Another aim of magicMap is to develop an automatic pipeline for map construction to allow for extensive evaluations, in particular, we increase the robustness of grouping by combining several clustering methods, and removing the dependence on input thresholds.
We evaluate magicMap by simulated data in three types of populations: the F2, the CP, and the funnel scheme eight-way RIL, which differ in the number of founders and the homozygosity of founders or offspring to show the wide applicability of our method. We study the sensitivity of magicMap to missing genotypes and genotyping errors, and compare magicMap with one of the commonly used methods in each type of population: MSTmap in biparental populations, Lep-MAP3 in the CP, and mpMap in the eight-way RIL. Furthermore, we evaluate magicMap by eight real datasets: one biparental population, one CP, four MAGIC, and two NAM, the number of markers ranging from a few hundred to tens of thousands (Table 1), and compare magicMap with MSTmap, Lep-MAP3, and the available genetic and physical maps.
Table 1. The running time (seconds) for map construction in the real datasets.
| Population | #founders | #offspring | #markers | #groups | missing f | magicMap |
|---|---|---|---|---|---|---|
| Arabidopsis RIL | 2 | 148 | 846 | 5 | 0.006 | 572a |
| Apple CP | 2 | 87 | 1,903 | 17 | 0.050 | 1,508b |
| Arabidopsis MAGIC | 19 | 703 | 1,228 | 5 | 0.020 | 8,339 |
| Barley MAGIC | 5 | 916 | 357 | 7 | 0.015 | 2,355 |
| Tomato MAGIC | 8 | 238 | 1,482 | 12 | 0.052 | 2,551 |
| Maize MAGIC | 8 | 303 | 41,473 | 10 | 0.081 | 243,532 |
| Maize US-NAM | 26 | 4699 | 1,144 | 10 | 0.078 | 47,723 |
| Maize EU-NAM | 11 | 841 | 34,223 | 10 | 0.021 | 308,352 |
25 sec for MSTmap.
253 sec for Lep-MAP3.
Methods
Overview of magicMap
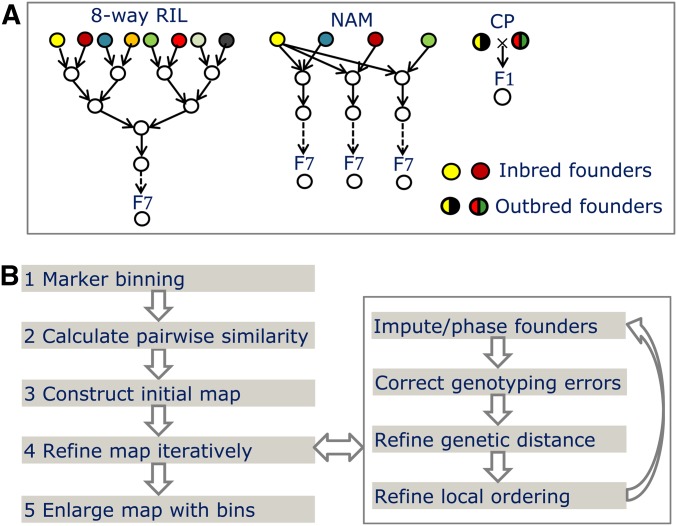
Figure 1 shows the workflow of magicMap for map construction in multiparental populations. The inputs include the genotypic data of founders and sampled offspring at a set of single nucleotide polymorphism (SNP) markers, and the breeding design in terms of pedigree or mating schemes since the founder population. The map construction consists of five stages: (1) group cosegregating markers into bins, (2) calculate similarity matrix by independence test and two-locus linkage analysis, (3) construct an initial map by spectral clustering and ordering, (4) refine the map by iteratively improving intermarker distances and local ordering using simulated annealing, and (5) enlarge the refined map by reintroducing binned markers. The map refinement also includes the founder genotype imputation or the founder linkage phasing in the case of outbred founders, and offspring error correction. We describe the five stages in the following, see Supplemental Material, File S1 for the details of the algorithm magicMap and Table S1 for a list of symbols and their brief explanations.
Figure 1.
Overview of magicMap for map construction in multiparental populations. (A) Examples of multiparental crosses. The cross schemes of the tomato MAGIC and the maize MAGIC are similar to the eight-way cross. (B) Workflow of the algorithm magicMap.
Marker binning
A large proportion of markers may become cosegregating, because of a limited number of recombination events accumulated during the generations between founders and offspring. Two markers become cosegregating if there are no recombination events between them, and they have the same offspring genotypes if they have the same founder genotypes.
An adjacency matrix is first produced based on the observed genotypes. The matrix element is set to 1 if offspring at markers i and j have the same observed genotypes except missing data and allelic coding, and otherwise it is set to 0. An undirected graph can be generated from the adjacency matrix. The marker binning is based on the partitioning of the resulting graph, according to clique rather than connectivity, since a marker with many missing data would be connected to many other markers. A marker bin corresponds to a clique, a maximal set of markers where the corresponding subgraph is a complete graph, that is, all pairs of markers within the bin are connected. In a bin, the marker with least missing genotypes represents the bin, and we delete those markers with less than one-half observed genotypes being in common with the representative marker.
Pairwise similarity
We calculated the similarity matrix among all pairs of representative markers. We first performed independence tests, and then pairwise linkage analysis, since the former does not require parental genotypes and is thus less sensitive to segregation distortion. The independence test is a likelihood ratio test called the G-test, and the test statistic is given by
where is the observed count of offspring with genotype at marker 1 and genotype at marker 2, and is the expected count under the null hypothesis of two markers being independent. The G-test statistic follows a chi-square distribution. To compare with the linkage analysis, the resulting P-value is transformed into a LOD score.
In a two-locus linkage analysis, offspring are assumed to be independent, conditional on parental haplotypes. The likelihood for offspring genotypic data at two markers is a function of recombination fraction r, and parental haplotypes at marker 1 and at marker 2, where the hidden ancestral origins are integrated out (File S1). The parental haplotypes account for missing genotypes in founders and possible unknown parental phases. The likelihood implicitly depends on allelic error probabilities and in founders and offspring, respectively. Conditional on a combination of parental haplotypes and , we estimate r using the maximum likelihood approach, and take the estimate with the largest likelihood among all combinations. We calculate the linkage LOD score under the null model of ; the recombination fraction is scaled so that its maximum is 1. Thus, a pair of markers has zero linkage LOD score if it has a flat or noninformative likelihood function.
To save computational time, we performed linkage analysis only for those pairs of markers with the independence LOD scores larger than a threshold . To save storage space, the results of linkage analyses are saved only if the linkage LOD scores are no less than . The similarity matrix S is given by one minus the scaled recombination fraction matrix, where the similarities for unsaved pairs of markers are set to zero.
Initial map construction
The construction of an initial genetic map can be divided into three steps: (1) cosegregation binning based on the results of two-locus linkage analyses; (2) marker grouping by the spectral clustering, where markers are treated as nodes of a graph, and the nodes are then mapped to a low-dimensional space so that they can be easily clustered; and (3) marker ordering by the spectral ordering, where the markers are treated as nodes of a connected graph and the nodes are then mapped to a one-dimensional space [i.e., Fielder vector (Fiedler 1973, 1989)] so that they can be easily ordered. We choose the spectral clustering and ordering to construct an initial map, mainly because they are fast and their results are independent of input marker ordering. In the following, we focus on preparing the input weight matrix, see File S1 for the details of the spectral clustering and ordering.
Cosegregation binning:
Cosegregation binning is the same as marker binning in the first stage but with a different adjacency matrix A. The matrix element is set to 1 if the estimated recombination fraction between markers i and j is zero and the corresponding linkage LOD score is larger than , and otherwise it is set to zero.
Spectral clustering:
We group the representative markers by the spectral clustering approach (Shi and Malik 2000; von Luxburg 2007). The inputs of the spectral clustering include the number of groups to construct and a weight matrix. Let and be the LOD score thresholds for linkage analysis and independence test, respectively. We obtain the input weight matrix from the similarity matrix by setting the matrix element to zero if its linkage LOD score and its independence LOD score .
The matrix is transformed into a similarity graph; two markers are connected only if the weight between them is positive. Assuming that , we estimate the threshold LOD score from the connectivity of the similarity graph. For a given LOD score threshold, we delete the connected components with the number of markers being <5. We estimate the threshold by maximizing it while minimizing the number of deleted markers and keeping the remaining graph connected.
Spectral ordering:
Within a linkage group, we order markers by the spectral ordering approach (Ding and He 2004). The input of the spectral ordering is a weight matrix. For a linkage group, we recalculate the weight matrix, rather than use the weight matrix from the spectral clustering. We obtain the weight matrix by reducing the noise in the similarity matrix in two ways. First, a similarity matrix element is set to zero if its linkage LOD score or its independence LOD score . By default, the LOD thresholds are the same as those for marker grouping. Second, we make the resulting matrix further sparse by taking only the nearest neighbors for each marker, where the closeness between two markers is measured by the similarity. Here, is set to be the square root of the number of markers in the group, and it is increased to a value until the corresponding similarity graph is connected.
Map refinement
The initial linkage map was refined via simulated annealing (Kirkpatrick et al. 1983). A proposal genetic map differs from the current map in either one intermarker distance or local ordering within a small window. The two maps are compared in term of log marginal likelihood, which is obtained from the standard forward-backward algorithm of the HMM by integrating all latent ancestral origins (Rabiner 1989). The proposal map is accepted with probability , where is the change of log likelihood by the proposal map (positive for a “good” map), and T is the current temperature. See File S1 for an efficient calculation of in the HMM.
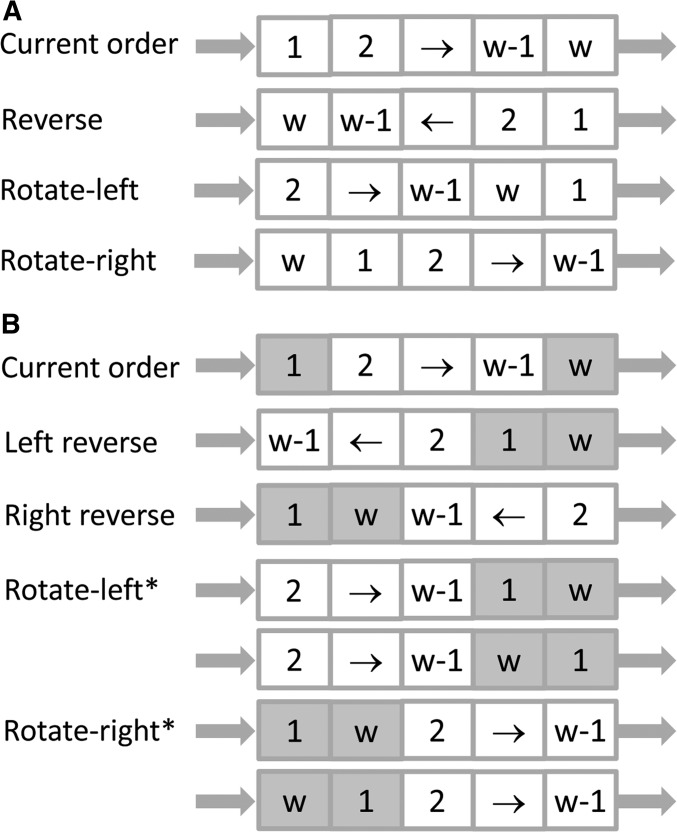
The proposal of marker ordering is critical for map refinement. Besides the commonly used ordering proposal in a window of fixed size (Figure 2A), we develop a new ordering proposal based on the neighborhood obtained in two-locus linkage analyses. According to the pairwise similarity matrix, 10 nearest neighbors for each marker are saved in the initial map. In a neighbor-based update window, the rightmost marker is randomly chosen among the 10 neighbors of the leftmost marker, and the two markers become neighbors in the proposal (Figure 2B).
Figure 2.
Illustration of the proposals for local ordering in simulated annealing. The arrows denote ordered markers. (A) Update window of fixed size. The rightmost marker is chosen based on a prefixed window size. (B) Neighbor-based update window. The rightmost marker is randomly chosen among the leftmost marker’s neighbors, which are determined from two-locus linkage analyses.
Differing from the standard simulated annealing, we introduce an additional temperature parameter, , so that heating iterations focus on ordering improvement and freezing iterations focus on distance improvement. Above , temperature T decreases linearly to (α < 1) after each iteration, the marker ordering is updated by sliding a window from left to right along chromosomes using both types of proposals (Figure 2), and the intermarker distance is updated by a log-normal proposal distribution that is parametrized so that the accept ratio being ∼0.44 (Gelman et al. 2013). Below , we speed up the algorithm convergence by decreasing temperature T to after each iteration, update marker ordering using only the proposals with fixed window sizes (Figure 2A), and update intermarker distances by maximizing the likelihood using the Brent’s numerical method (Brent 1973).
The updates of intermarker distances and marker ordering are conditional on founder haplotypes. We performed founder imputation for inbred founders, and both founder imputation and linkage phasing for outbred founders. In addition, we performed genotyping error correction for offspring in heterozygous populations. To save computational time, we perform these updates in every third iteration, and the algorithms have been described (Zheng et al. 2018a).
Chromosome direction is reversed in every iteration, primarily to overcome the asymmetry of the neighbor-based update windows. After five iterations of refining, the skeleton map consists of representatives of cosegregation bins, and all markers within each bin are introduced and set to the same position as the representative, and the algorithm continues to refine the enlarged map. The algorithm stops if the increase of marginal likelihood is <1 or the number of freezing iterations reaches . The map refinements for linkage groups are carried out in parallel.
Map enlargement
The refined map is enlarged in the last stage by setting all binned markers in the first stage to the same positions as their representatives. The markers within each bin are permuted so that the input marker ordering has no effect.
Data simulation
We simulate genotypic data as three types of population: F2, CP, and eight-way RIL with four generations of selfing. The eight-way RIL population has a funnel scheme so that each offspring is produced, with the eight inbred founders being randomly permuted. For each population type, we consider three population sizes: small, medium, and large. They are set to 50, 100, and 200 for the F2 and the CP, and 200, 400, and 800 for the eight-way RIL.
The simulation accounts for missing genotypes and allelic errors in founders and offspring. We set the following parameter setup as a baseline: founder allelic error probability , offspring allelic error probability , missing fraction of founder genotypes , and missing fraction of offspring genotypes . To study the effect of allelic errors, we vary 0, 0.01, 0.02, 0.05, 0.075, and 0.1, keeping the other parameters constant. To study the effects of missing genotypes, we vary 0, 0.1, 0.2, 0.35, 0.5, and 0.75, keeping the other parameters constant. There are, in total, 108 combinations of population type, population size, allelic error probability , and missing fraction f. Each dataset is simulated by dropping founder haplotypes on a simulated breeding pedigree. An offspring gamete is produced by chromosomal crossovers between two parental chromosomes, where the number of crossovers in a gamete follows a Poisson distribution, with the mean being the chromosome length in Morgan, and the positions of crossovers are distributed uniformly across the chromosome (see Zheng et al. 2015, 2018a for detailed descriptions).
The founder haplotypes for each population type are specified as follows. The four parental haplotypes for the two outbred parents of CP are provided by the first four founders of the Arabidopsis MAGIC (Kover et al. 2009). There remain 715 markers after removing monomorphic markers, and their genetic map is set by multiplying the physical map with a constant recombination frequency of 4.17 cM/Mbp. The same set of markers and their genetic map are used for the F2 and the eight-way RIL. The two founder haplotypes of F2 are set to be polymorphic, and the eight founder haplotypes of the eight-way RIL are set to the first eight founders of the Arabidopsis MAGIC (Kover et al. 2009).
Algorithm evaluation and map accuracy
We evaluate magicMap by simulated data and eight real datasets in various mapping populations (Table 1), see File S1 for the preparation of the real datasets. The algorithm magicMap is compared with MSTmap in biparental populations, with Lep-MAP3 in the CP, and with mpMap in the eight-way RIL. We do not compare with the popular JoinMap because it is nonscriptable for extensive evaluations through computationally automatic construction. See Wu et al. (2008) for the comparisons of MSTmap with RECORD, CarthaGène, and JoinMap, Rastas et al. (2016) for the comparisons of Lep-MAP2 with HighMap and JoinMap, and Rastas (2017) for the comparisons of Lep-MAP3 with Lep-MAP2 and MSTmap.
The genetic map estimated from magicMap is compared with the true map in the simulation or the other map, in terms of grouping, ordering, and spacing. For marker grouping, isolated markers such as singletons are pooled as a single group, and we calculate grouping accuracy by the pair counting F-measure ranging from 0 to 1, a symmetric measure of the similarity between two groupings (Pfitzner et al. 2009).
The accuracy of ordering within a linkage group is measured by Kendall τ, a nonparametric correlation coefficient. Since the chromosome direction is nonidentifiable, we can always obtain non-negative τ by reversing estimated ordering. The overall ordering accuracy can be obtained by averaging over linkage groups of the estimated map.
The performance of marker spacing within a linkage group is summarized by the estimation of total chromosome length.
Algorithm implementation and setting
The algorithm magicMap is implemented as an automatic pipeline, so that it exports a final genetic map from the required input marker data and population design information. On the other hand, the five stages of magicMap are implemented independently as functions, and they are linked by input and output text files, so that each function can be rerun or called by other software packages.
We set up magicMap with the following parameters: , , , , , , , , α, and , where the LOD score thresholds and are estimated internally by magicMap. The many other options are used mainly for testing magicMap, and it is generally unnecessary to change them. We set allelic error probabilities , and smaller values may be used for stringent filtered marker data. A pair of markers are dissimilar or noninformative if either the linkage LOD or independence LOD is no greater than the LOD score threshold , which is set to 1 and should be smaller than and . We set by so that cosegregation binning in the initial map construction is not performed by default, and it should be set to be a positive value to increase computational efficiency in the case of high density marker data.
For map refinement, we set a low initial temperature, and a small cooling constant, to increase computational efficiency. A high value between 5 and 10 may be set in the case of a bad initial map, but too high value may destroy the ordering of a good initial map, and a large cooling constant α may then be needed. We set freezing temperature and the maximum number of freezing iterations ; the map length usually becomes stable after several freezing iterations. A reasonably good map can often be obtained from a fast map refinement by setting two heating and three freezing generations; the default setting takes ∼20 iterations. By default, the parental imputation and phasing are performed if the missing fraction in founders is ≤0.05 or founders are outbred, offspring imputation is not performed, and the error correction in offspring is performed only for heterozygous populations (see also Discussion).
We use the default parameter values for magicMap, except that for the maize MAGIC and the maize EU-NAM datasets is estimated internally to be 10 or larger so that the resulting number of bins is no less than one-third of the number of the markers in two-locus analyses; a large is used to ensure cosegregating. For marker grouping using MSTmap and Lep-MAP3, we estimate cut off P-value or LOD score threshold roughly so that the grouping accuracy is high for the simulated data or the grouping is close to that of magicMap for the real data. Haldane’s mapping function is used for MSTmap, Lep-MAP3, and mpMap. See File S1 for the running setups for magicMap, MSTmap, Lep-MAP3, and mpMap.
Data availability
The algorithm magicMap is currently implemented in Mathematica 11.0 (Wolfram Research Inc. 2016), and has been included as a function in the RABBIT software. RABBIT is available from the web site: https://github.com/chaozhi/RABBIT.git, and is offered under the GNU Affero general public license, version 3 (AGPL-3.0). A Mathematica license is required to run the RABBIT software. Example scripts for simulating genotypic data and constructing linkage map are included.
The marker data for the Arabidopsis RIL are available from http://www.atgc.org/XLinkage/MadMapper/ath_sfp_map_example/, and the physical positions of the markers were obtained from Arabidopsis annotation version 4, TIGR release May 2003 (http://elp.ucdavis.edu/data/analysis/sfp_map/sfp_map.html). The marker data for the Arabidopsis MAGIC are available at http://mtweb.cs.ucl.ac.uk/mus/www/magic/. The marker data for the maize US-NAM are available at https://www.panzea.org/. The marker data for the apple CP, the barley MAGIC, the tomato MAGIC, the maize MAGIC, and EU-NAM are available in the supplementary materials of Bauer et al. (2013), Gardner et al. (2014), Dell’Acqua et al. (2015), Pascual et al. (2015), and Liller et al. (2017), respectively. The available physical maps from the corresponding marker data were also downloaded and used for evaluating magicMap. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8243180.
Results
Simulation evaluation
Marker grouping:
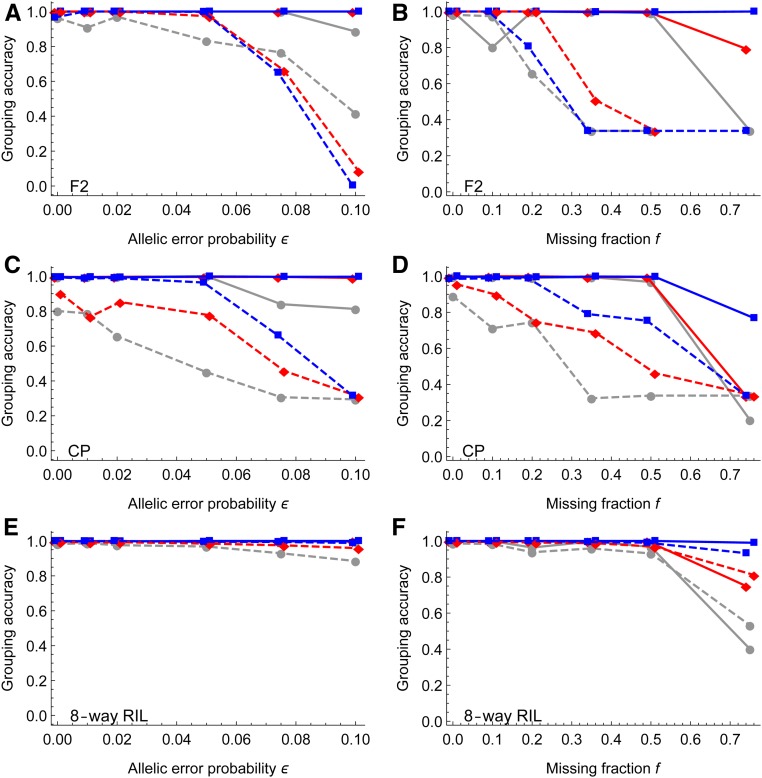
Figure 3 shows that the grouping of magicMap is more robust to genotyping errors and missing genotypes than MSTmap, Lep-MAP3, and mpMap in the F2, the CP, and the eight-way RIL, respectively. In the medium and large populations, the grouping accuracies of magicMap are close to 1, except for the large missing fraction, . In contrast, the grouping accuracies of MSTmap and Lep-MAP3 drop dramatically when or , and the grouping accuracies of mpMap in the eight-way RIL gradually decrease with ϵ and f.
Figure 3.
Simulation evaluation of marker grouping. The left panels show the effects of allelic error probability, and the right panels for the effects of missing fraction. The solid lines denote the results of magicMap, and the dashed lines denote the results of the alternative methods, which are MSTmap, Lep-MAP3, and mpMap for the F2 (A and B), the CP (C and D), and the eight-way RIL (E and F), respectively. The gray circles (○), red diamonds , and blue rectangles (□) refer to small, medium, and large population sizes, respectively; the x-coordinates are jittered to avoid overlapping.
The worse performance of MSTmap and Lep-MAP3 is partly because both methods are based on a single hard threshold in two-locus linkage analysis, and partly because it becomes difficult to correctly estimate the number of groups from a decreasing amount of accurate genotypic data; magicMap and mpMap require an input for the number of groups. The outperformance of magicMap over mpMap is because magicMap deletes isolated markers, whereas mpMap groups all markers by hierarchical clustering, and because magicMap selects informative eigenvectors before performing hierarchical clustering; the outperformance of mpMap over magicMap at for small and medium population sizes is probably because magicMap deleted too many markers.
Marker ordering:
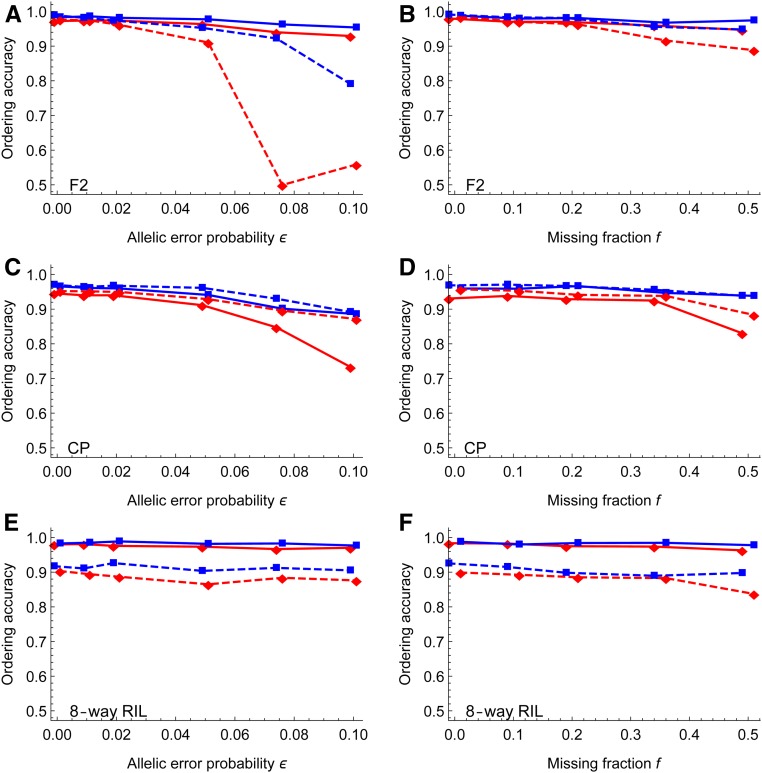
Figure 4, A and B show that the ordering of magicMap is more robust to genotyping errors and missing genotypes than MSTmap in the F2. Figure 4, C and D show that Lep-MAP3 is slightly more accurate than magicMap, probably because magicMap does not delete markers during ordering while the fraction of markers that are deleted by Lep-MAP3 increases up to with ϵ and f (see Figure S1). Figure 4, E and F show that the ordering of both magicMap and mpMap is robust to genotyping errors and missing genotypes in the eight-way RIL, but magicMap is always more accurate than mpMap. See Figure S2 for the illustrative comparisons between estimated ordering and the true ordering.
Figure 4.
Simulation evaluation of marker ordering. The left panels show the effects of allelic error probability, and the right panels show the effects of the missing fraction. The solid lines denote the results of magicMap, and the dashed lines denote the results of the alternative methods, which are MSTmap, Lep-MAP3, and mpMap for the F2 (A and B), the CP (C and D), and the eight-way RIL (E and F), respectively. The red diamonds and blue rectangles (□) refer to medium and large population sizes, respectively.
Figure S3 shows that the ordering improvement of map refinement over spectral ordering increases with the number of founders in the mapping populations. The amount of relative improvement increases from in the biparental F2, to in the CP (pseudo 4-way cross), and to in the eight-way RIL. This is probably because the similarity matrix of spectral ordering calculated from the pairwise linkage analysis is less accurate when there are more founders.
Figure S4 shows that the inclusion of error correction during map refinement increases the ordering accuracy when there are at least some genotyping errors . The error correction slightly decreases the ordering accuracy for the datasets with various missing fractions and . The error correction slightly decreases the ordering accuracy in the eight-way RIL, because the erroneous heterozygous genotypes have been deleted and the error probability from one homozygous genotype to the other is very small .
Marker spacing:
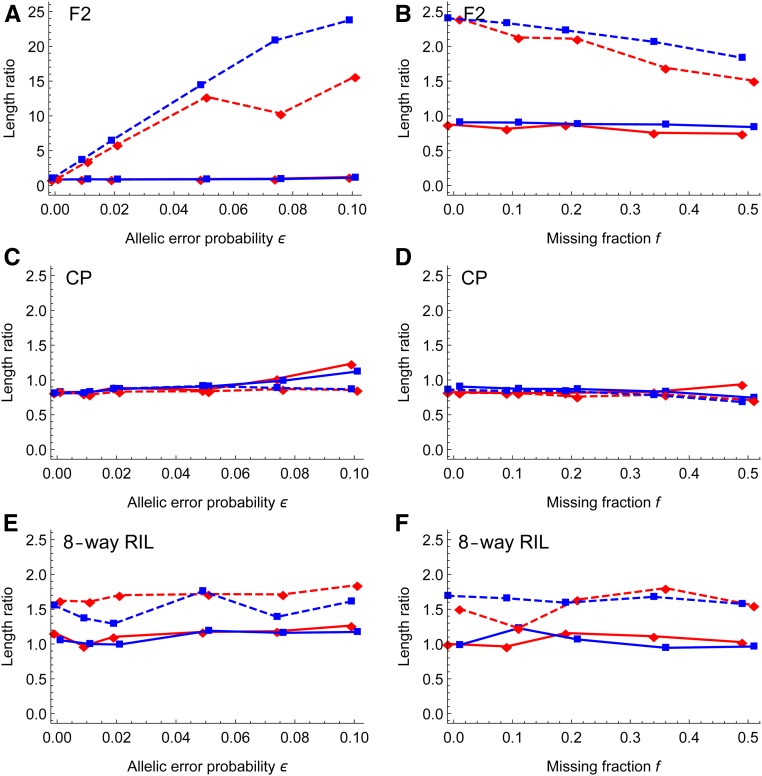
Figure 5 shows that the spacing of magicMap is more robust to genotyping errors and missing genotypes than MSTmap in the F2 and mpMap in the eight-way RIL; the ratio of estimated map length to the true value is always ∼1.0 for magicMap. The ratio from MSTmap in the F2 increases up to ∼25 with ϵ increasing up to 0.1, while it decreases gradually with f. The ratio from mpMap in the eight-way RIL is ∼1.5, almost independent of ϵ and f. In the CP, both magicMap and Lep-MAP3 are robust to genotyping errors and missing genotypes, although the ratio from magicMap increases slightly with ϵ.
Figure 5.
Simulation evaluation of marker spacing. The y-axis denotes the ratio of estimated chromosome length to the true value. The left panels show the effects of allelic error probability, and the right panels show the effects of the missing fraction. The solid lines denote the results of magicMap, and the dashed lines denote the results of the alternative methods, which are MSTmap, Lep-MAP3, and mpMap for the F2 (A and B), the CP (C and D), and the eight-way RIL (E and F), respectively. The red diamonds and blue rectangles (□) refer to medium and large population sizes, respectively.
The large inflation of map length by MSTmap is because its estimation of intermarker distance is based on two-locus linkage analysis, while magicMap, Lep-MAP3, and mpMap use various multilocus approaches. For magicMap in the CP, the slight increase of map length with may be because the ordering accuracy decreases with (Figure 4C). Similarly for mpMap in the eight-way RIL, the large inflation of map length may be due to the low ordering accuracy.
Figure S5 shows that the inclusion of the error correction prevents the inflation of map length in the F2 and CP, but it slightly deflates the map length in the eight-way RIL. This is because most erroneous heterozygous genotypes in the homozygous populations have been deleted beforehand and the error correction may change small recombinant haplotypes into nonrecombinant.
Evaluation by real data
Biparental RIL:
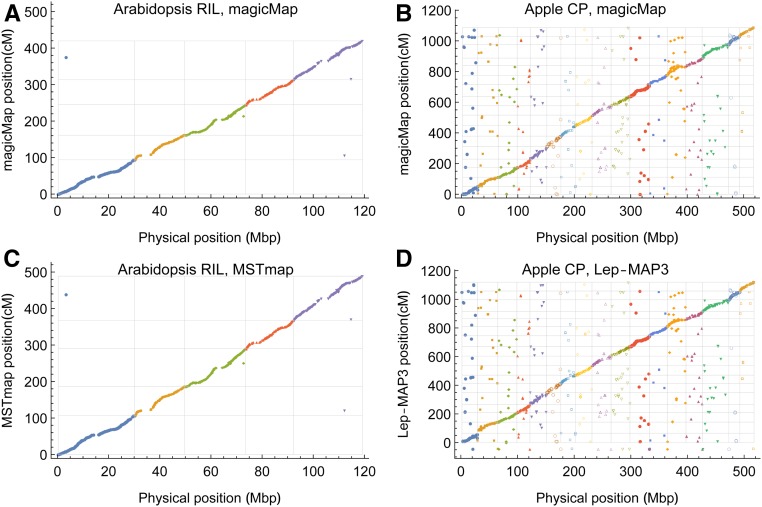
The high quality data of 864 single feature polymorphic markers in the biparental population were obtained by stringent filtering criteria (West et al. 2006). Figure 6, A and C show the comparisons among magicMap, MSTmap, and the physical map in the Arabidopsis RIL (West et al. 2006). For both magicMap and MSTmap, two markers are inconsistently grouped, and two markers have a large inconsistency of ordering. The Kendall’s τ coefficients are 0.978 between magicMap and the physical map, 0.980 between MSTmap and the physical map, and 0.986 between magicMap and MSTmap. The total genetic length of 419 cM obtained by magicMap is shorter than the 489 cM by MSTmap.
Figure 6.
Evaluation of magicMap performances by the real Arabidopsis 2-way RIL (A and C) and the apple CP (B and D). The gray grid lines denote the chromosome boundaries, and the dots with negative y-values denote the ungrouped markers. (A and B) Genetic map constructed by magicMap vs. the physical map. (C) Genetic map constructed by MSTmap vs. the physical map. (D) Genetic map constructed by Lep-MAP3 vs. the physical map.
CP population:
The CP marker data were prepared by Gardner et al. (2014) for using JoinMap; the number of markers is decreased by two orders of magnitude after several stringent filtering steps. Figure 6, B and D show the comparisons among magicMap, Lep-MAP3, and the physical map in the apple CP (Gardner et al. 2014). The physical map is derived from the “Golden Delicious” v1.0 reference genome (Velasco et al. 2010); the two outbred parents in the apple CP are “Golden Delicious” and “Scarlet Spur.” In comparison with the physical map, 362 markers are inconsistently grouped by magicMap, and 360 markers by Lep-MAP3, similar to 364 by JoinMap4 (Gardner et al. 2014). Excluding the inconsistently grouped markers, the Kendall’s τ coefficients are 0.893 between magicMap and the physical map, 0.902 between Lep-MAP3 and the physical map, and 0.897 between magicMap and Lep-MAP3. The total genetic length is estimated to be 1086 cM by magicMap, shorter than the 1120 cM by Lep-MAP3 and the 1272 cM by JoinMap (Gardner et al. 2014).
MAGIC populations:
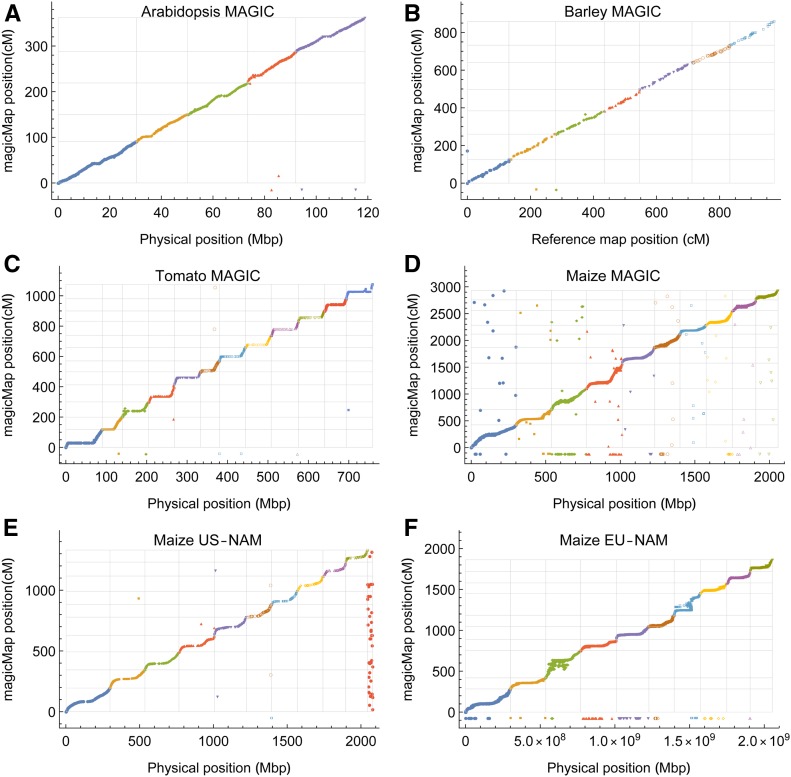
Figure 7, A–D show the comparisons between magicMap and the physical maps in the Arabidopsis MAGIC (Kover et al. 2009), the barley MAGIC (Liller et al. 2017), the tomato MAGIC (Pascual et al. 2015), and the maize MAGIC (Dell’Acqua et al. 2015); the reference map provided by Liller et al. (2017) is based on the POPSEQ map (Ariyadasa et al. 2014). There are only several ungrouped or inconsistently grouped markers out of markers in the Arabidopsis, the barley, and the tomato MAGIC. And there are 161 ungrouped and 87 inconsistently grouped markers out of 41,473 markers in the maize MAGIC. The Kendall’s τ between magicMap and the physical or reference maps are 0.985, 0.961, 0.961, and 0.958 for the Arabidopsis, the barley, the tomato, and the maize MAGIC, respectively.
Figure 7.
Evaluation of magicMap performances by the real multiparental populations. Panels (A–F) refer to the Arabidopsis MAGIC, the barley MAGIC, the tomato MAGIC, the maize MAGIC, the maize US-NAM, and the maize EU-NAM, respectively. The gray grid lines denote the chromosome boundaries, and the dots with negative y-values denote the ungrouped markers. The dots in the last column of the grid in (E) the denote unmapped markers in the physical map.
Pascual et al. (2015) constructed the genetic map of the tomato MAGIC using mpMap and the physical map. The total genetic length obtained from mpMap is 2156 cM, around twice as long as the biparental maps (Sim et al. 2012). In comparison, the genetic length from magicMap is 1075 cM.
Dell’Acqua et al. (2015) derived the genetic map of the maize MAGIC using the physical map and the biparental map (Ganal et al. 2011), and the resulting genetic map length is 1711 cM. For magicMap, the genetic map length increases from 1637 cM for randomly selected 2000 markers to 2929 cM for the full dataset of 41,473 markers, while the ordering accuracy slightly decreases from 0.970 to 0.958 (Figure S6).
NAM populations:
Figure 7, E and F show the comparisons between magicMap and the physical maps in the maize US-NAM (McMullen et al. 2009) and the maize EU-NAM (Bauer et al. 2013). There are two ungrouped and seven inconsistently grouped markers out of 1144 markers in the US-NAM, and there are 364 ungrouped markers out of 34,223 markers in the EU-NAM. The Kendall’s τ between magicMap and the physical maps are 0.993 and 0.923 for the US-NAM and the EU-NAM, respectively.
McMullen et al. (2009) constructed the genetic map for the US-NAM using MAPMAKER 3.0 (Lander and Green 1987). The Kendall’s τ between magicMap and MAPMAKER map is 0.995. The genetic map length obtained from MAPMAKER is 1399 cM, similar to the 1330 cM from magicMap.
Giraud et al. (2014) constructed the genetic map for the EU-NAM using the physical map. The resulting genetic map length is 1344 cM. For magicMap, the genetic length increases from 1506 cM for randomly selected 2000 markers to 1863 cM for the full dataset of 34,223 markers, while the ordering accuracy stays ∼0.92 (Figure S6).
Running time:
Table 1 shows the running time of map construction for the eight real datasets on a standard desktop computer with 32 GB memory; a large memory is required for the large marker data with tens of thousands of markers. The MSTmap package is applicable only to the Arabidopsis RIL data and Lep-MAP3 is applicable only to the apple CP data, and they are faster than magicMap.
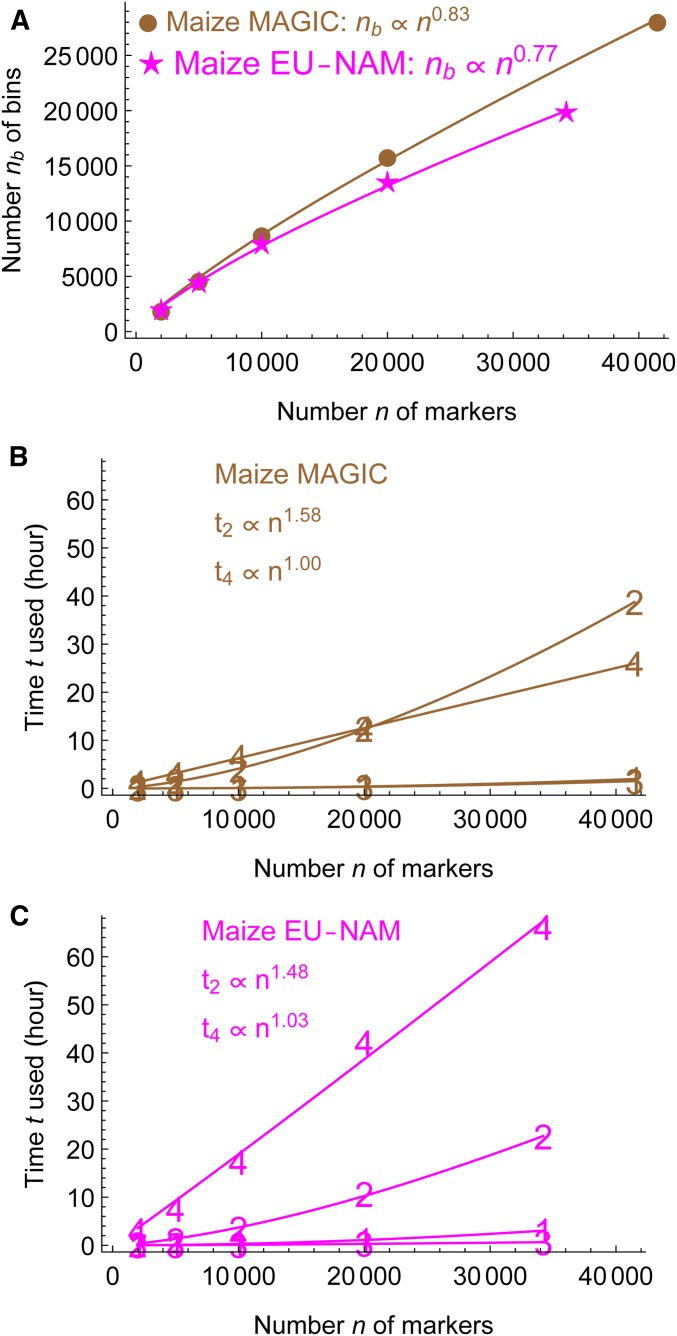
Figure 8 shows that magicMap spent > of computational time in the second stage of two-locus analysis and the fourth stage of map refinement in the maize MAGIC and the maize EU-NAM. Because the number of bins in the first stage increases sublinearly with the input number n of markers, the time used in the second stage increases subquadratically with n, and the time used in the fourth stage increases linearly with n.
Figure 8.
Scaling of magicMap running time with the number of markers in the maize MAGIC and the maize EU-NAM. The denotes the computational time used in the ith stage of magicMap, and n for the input number of markers randomly chosen from the full dataset. (A) Number of bins resulting from the first stage of marker binning. (B) Computational times for each of the first four stages of magicMap in the maize MAGIC. (C) Computational times for the maize MAGIC.
Discussion
Comparisons with previous algorithms
Marker grouping:
The evaluations by simulation and real data show that the magicMap grouping is robust, because it combines several different ways to reduce the impact of data noises. First, noisy connections and isolated markers are deleted by using a low threshold that is internally estimated. Second, the spectral clustering algorithm discards noninformative eigenvectors. Lastly, hierarchical clustering groups markers using the selected eigenvectors. In comparison, most software packages such as JoinMap, MSTmap, and Lep-MAP3 use a high threshold for grouping, the current mpMap changes into hierarchical clustering, and HighMap searches hierarchical clustering under many different hard thresholds.
Spectral ordering:
The magicMap ordering algorithm combines spectral ordering and local ordering improvement via simulated annealing. Before spectral ordering using a similarity matrix, we keep only some nearest neighbors of each marker because of the linear structure of chromosomes and the potential noise in distant neighbors. Spectral ordering results in a good long-range ordering, probably because the Fiedler vector (Fiedler 1973, 1989), the eigenvector associated with the nonzero smallest eigenvalue, reflects the global property of the graph Laplacian obtained from similarity matrix.
Cheon et al. (2016) described a Laplacian ordering approach for the loci-ordering via the Fiedler vector, which requires an input of similarity matrix and does not perform marker spacing. The authors grouped markers from a sparse similarity matrix that is obtained by choosing appropriately k nearest neighbors for each marker. Such grouping is still sensitive to the choice of k according to our preliminary simulation studies. For each linkage group, Cheon et al. (2016) then performed a similar spectral ordering after rechoosing appropriately k nearest neighbors, whereas magicMap chooses k in a very loose way.
Iterative refining:
From the initial map that is constructed based on two-locus analysis, map refinement is often necessary, which is also computationally intensive. Many packages such as mpMap and MapDisto rely on rippling for improving local ordering, which is inefficient because of simultaneously comparing all possible permutations within a small sliding window (of size ). The algorithm magicMap performs map refinement via the simulated annealing with a low initial temperature , whereas a much high initial temperature is often required in the simulated annealing for map construction (Jansen et al. 2001; Hackett et al. 2003).
In the iterative improvement of magicMap, a proposal of local ordering is based on operations such as reversion (2-opt) and node insertion that are commonly used in traveling salesman problems (TSP) (Reinelt 1994), as do the most software packages for map construction such as CarthaGène, MSTmap, and Lep-MAP3. The update windows in these TSP-like proposals are often randomly chosen (see Figure 2A). However, magicMap includes neighbor-based update windows (Figure 2B), which turn out to be very effective so that it is more likely to accept the proposal within a larger window.
Both marker spacing and local ordering are updated in the iterative improvement of magicMap, so that the multilocus likelihood of the genetic map increases with decreasing annealing temperature. For a given marker ordering, we update the intermarker distances one by one by the maximum likelihood approach, which has advantages over the expectation-maximization (EM) algorithms (Lander et al. 1987; Tong et al. 2010; Rastas et al. 2013). First, we estimate genetic distances directly instead of recombination fractions, because, generally, no explicit mapping function exists, even under the assumption of no genetic interference. Second, our spacing algorithm converges within a few iterations, much more rapidly than the EM algorithms.
Guidelines for using magicMap
Assumptions:
The algorithm magicMap takes several key assumptions from the previous HMM framework (Zheng et al. 2014, 2015, 2018a,b; Zheng 2015). First, we assume that there is no genetic interference. However, it is not expected to be important under a high marker density. Second, we assume that there is no segregation distortion. Our calculation of similarity matrix accounts for independent tests that are robust to segregation distortion, although we did not study the effects of segregation distortion on map construction. It is suggested to delete markers with severe distortion by using the chi-squared test with a low significant level (e.g., 0.01 divided by the number of markers), or by deleting markers with minor allele frequency below a certain threshold, since severe distortion may be caused by genotyping errors in founders or offspring.
Last, but not least, sampled offspring are assumed to be independent, conditional on phased founder haplotypes, which substantially reduces the computational load. Population designs with a very small population size in an intermediate generation would generate strong dependencies among offspring, resulting in biased estimations of recombination fractions and difficulties in grouping markers. It is thus recommended to use bottleneck generation as the founder population.
Missing data:
Unlike other methods such as mpMap, magicMap allows missing genotypes in founders. However, it is suggested to delete markers with too many missing founder genotypes (e.g., ), since the number of founder haplotypes increases exponentially with the number of missing genotypes. By default, magicMap does not impute missing offspring genotypes, because the genetic map length would otherwise be increasingly underestimated with missing fraction. This indicates that offspring imputation by a maximum likelihood approach suppresses recombination events.
Genotypic errors:
Hackett and Broadfoot (2003) have reported that the maximum likelihood criterion results in a substantial inflation of map length in the presence of genotyping errors, which has been confirmed in our results (Figure S5). We have solved this problem by the error correction algorithm (Zheng et al. 2018a), where suspicious genotyping errors are corrected during iterative map refinement. In the simulation studies, the error correction can increase the accuracies of the map length in heterozygous populations, whereas it slightly underestimates the map length in homozygous populations where most heterozygous errors have been removed (see Figures S4 and S5); the map deflation may because genotyping errors and short recombinant segments are confounded. Figure S6 shows that the genetic map length gradually increases with the number of markers increasing up to tens of thousands, in contrast to only ∼1000 markers in the simulation evaluation.
The increase in map length with the number of markers may be because of the increasing number of genotyping errors even with a small genotypic error probability, because of the difficulties in ordering the increasing number of markers within cosegregation bins, or because of the increasing number of detected recombination events. Since the recombinant segments from higher marker density data contain more markers, and they become less likely confounded with genotyping errors, it is suggested to perform the error correction in heterozygous populations and in homozygous populations with a large number of markers (e.g., >5000).
Limitations:
One main limitation of magicMap is that it is computationally intensive, especially in the stage of map refinement. One solution is to translate magicMap into the Julia language, a high-level high performance dynamic language for technical computing (Bezanson et al. 2017). Currently, the translation of the functions such as magicImpute into Julia is under progress; these functions are called by magicMap. In addition, we can improve magicMap for map integration in multiple mapping populations (e.g., the NAM), where the HMM adopts a joint state space for all founders; a more efficient way is to account for the population structure since the state space of the HMM in a subpopulation is usually much smaller than the joint state space.
Conclusion
We have demonstrated the generality of magicMap for genetic map construction, in the sense that it is not restricted to specific breeding designs, and is applicable to both inbred and outbred founders. Furthermore, we have shown that magicMap is more accurate and robust to missing genotypes and genotyping errors than commonly used packages.
Acknowledgments
C.Z. thanks Vincent Garin for help on preparing the marker data of the maize EU-NAM, Pasi Rastas for help on using Lep-MAP3, B. Emma Huang for help on using mpMap, and four anonymous reviewers for valuable comments. This research was supported by the Stichting Technische Wetenschappen (STW), Technology Foundation, which is part of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek, Netherlands Organisation for Scientific Research, and which is partly funded by the Ministry of Economic Affairs. The specific grant number was STW-Rijk Zwaan project 12425.
Author contributions: C.Z. designed the study, created the model, developed the software and algorithm, and wrote the first draft of the manuscript. M.P.B. and F.A.v.E. provided critical feedback and helped shape the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8243180.
Communicating editor: M. Sillanpää
Literature Cited
- Ariyadasa R., Mascher M., Nussbaumer T., Schulte D., Frenkel Z., et al. , 2014. A sequence-ready physical map of barley anchored genetically by two million single-nucleotide polymorphisms. Plant Physiol. 164: 412–423. 10.1104/pp.113.228213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103 10.1186/gb-2013-14-9-r103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanson J., Edelman A., Karpinski S., Shah V. B., 2017. Julia: a fresh approach to numerical computing. SIAM Rev. 59: 65–98. 10.1137/141000671 [DOI] [Google Scholar]
- Brent R. P., 1973. Algorithms for Minimization Without Derivatives. Courier Corporation, Chelmsford, MA. [Google Scholar]
- Broman K., Wu H., Sen S., Churchill G., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Gatti D. M., Simecek P., Furlotte N. A., Prins P., et al. , 2019. R/qtl2: software for mapping quantitative trait loci with high-dimensional data and multiparent populations. Genetics 211: 495–502. 10.1534/genetics.118.301595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema J., Ellis T. H. N., Dicks J., 2010. THREaD Mapper Studio: a novel, visual web server for the estimation of genetic linkage maps. Nucleic Acids Res. 38: W188–W193. 10.1093/nar/gkq430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon M., Kim C., Chang I., 2016. Uncovering multiloci-ordering by algebraic property of Laplacian matrix and its Fiedler vector. Bioinformatics 32: 801–807. 10.1093/bioinformatics/btv669 [DOI] [PubMed] [Google Scholar]
- Dell’Acqua M., Gatti D. M., Pea G., Cattonaro F., Coppens F., et al. , 2015. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biol. 16: 167–189. 10.1186/s13059-015-0716-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., He X., 2004. Linearized cluster assignment via spectral ordering, pp. 30–37 in Proceedings of the Twenty-first International Conference on Machine Learning, ICML ’04 ACM, New York. 10.1145/1015330.1015407 [DOI] [Google Scholar]
- Ellis T. H. N., 1997. Neighbour mapping as a method for ordering genetic markers. Genet. Res. 69: 35–43. 10.1017/S0016672397002632 [DOI] [Google Scholar]
- Fiedler M., 1973. Algebraic connectivity of graphs. Czech. Math. J. 23: 298–305. [Google Scholar]
- Fiedler M., 1989. Laplacian of graphs and algebraic connectivity. Banach Center Publications 25: 57–70. 10.4064/-25-1-57-70 [DOI] [Google Scholar]
- Fierst J. L., 2015. Using linkage maps to correct and scaffold de novo genome assemblies: methods, challenges, and computational tools. Front. Genet. 6: 220–227. 10.3389/fgene.2015.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal M. W., Durstewitz G., Polley A., Berard A., Buckler E. S., et al. , 2011. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One 6: e28334 10.1371/journal.pone.0028334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. M., Brown P., Cooke T. F., Cann S., Costa F., et al. , 2014. Fast and cost-effective genetic mapping in apple using next-generation sequencing. G3 (Bethesda) 4: 1681–1687. 10.1534/g3.114.011023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Stern H. S., Carlin J. B., Dunson D. B., Vehtari A., et al. , 2013. Bayesian Data Analysis. Chapman and Hall/CRC, Boca Raton, FL. [Google Scholar]
- Giraud H., Lehermeier C., Bauer E., Falque M., Segura V., et al. , 2014. Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the Flint and Dent heterotic groups of maize. Genetics 198: 1717–1734. 10.1534/genetics.114.169367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P., Falls K., Crooks S., 1990. Documentation for CRI-MAP, Version 2.4. Washington University School of Medicine, St. Louis. [Google Scholar]
- Hackett C. A., Broadfoot L. B., 2003. Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90: 33–38. 10.1038/sj.hdy.6800173 [DOI] [PubMed] [Google Scholar]
- Hackett C. A., Pande B., Bryan G. J., 2003. Constructing linkage maps in autotetraploid species using simulated annealing. Theor. Appl. Genet. 106: 1107–1115. 10.1007/s00122-002-1164-1 [DOI] [PubMed] [Google Scholar]
- Huang B. E., George A. W., 2011. R/mpMap: a computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 27: 727–729. 10.1093/bioinformatics/btq719 [DOI] [PubMed] [Google Scholar]
- Huang B. E., Verbyla K. L., Verbyla A. P., Raghavan C., Singh V. K., et al. , 2015. MAGIC populations in crops: current status and future prospects. Theor. Appl. Genet. 128: 999–1017. 10.1007/s00122-015-2506-0 [DOI] [PubMed] [Google Scholar]
- Hyma K. E., Barba P., Wang M. H., Londo J. P., Acharya C. B., et al. , 2015. Heterozygous mapping strategy (hetmapps) for high resolution genotyping-by-sequencing markers: a case study in grapevine. PLoS One 10: e0134880 10.1371/journal.pone.0134880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H., Ninomiya S., 2006. Antmap: constructing genetic linkage maps using an ant colony optimization algorithm. Breed. Sci. 56: 371–377. 10.1270/jsbbs.56.371 [DOI] [Google Scholar]
- Jansen J., de Jong A. G., van Ooijen J. W., 2001. Constructing dense genetic linkage maps. Theor. Appl. Genet. 102: 1113–1122. 10.1007/s001220000489 [DOI] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Vecchi M. P., 1983. Optimization by simulated annealing. Science 220: 671–680. 10.1126/science.220.4598.671 [DOI] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozik, A., and R. Michelmore, 2006 MadMapper and CheckMatrix–python scripts to infer orders of genetic markers and for visualization and validation of genetic maps and haplotypes in Proceedings of the Plant and Animal Genome XIV Conference, San Diego, Livingston, NJ: Scherago International. [Google Scholar]
- Lander E. S., Green P., 1987. Construction of multilocus genetic-linkage maps in humans. Proc. Natl. Acad. Sci. USA 84: 2363–2367. 10.1073/pnas.84.8.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., et al. , 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. 10.1016/0888-7543(87)90010-3 [DOI] [PubMed] [Google Scholar]
- Liller C. B., Walla A., Boer M. P., Hedley P., Macaulay M., et al. , 2017. Fine mapping of a major QTL for awn length in barley using a multiparent mapping population. Theor. Appl. Genet. 130: 269–281. 10.1007/s00122-016-2807-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. Y., Ma C. X., Hong W. G., Huang L., Liu M., et al. , 2014. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS One 9: e98855 10.1371/journal.pone.0098855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M., 2012. MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30: 1231–1235. 10.1007/s11032-012-9706-y [DOI] [Google Scholar]
- Margarido G. R. A., Souza A. P., Garcia A. A. F., 2007. OneMap: software for genetic mapping in outcrossing species. Hereditas 144: 78–79. 10.1111/j.2007.0018-0661.02000.x [DOI] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P., Li H. H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. 10.1126/science.1174320 [DOI] [PubMed] [Google Scholar]
- Pascual L., Desplat N., Huang B. E., Desgroux A., Bruguier L., et al. , 2015. Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13: 565–577. 10.1111/pbi.12282 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Lander E. S., Hewitt J. D., Peterson S., Lincoln S. E., et al. , 1988. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721–726. 10.1038/335721a0 [DOI] [PubMed] [Google Scholar]
- Pfitzner D., Leibbrandt R., Powers D., 2009. Characterization and evaluation of similarity measures for pairs of clusterings. Knowl. Inf. Syst. 19: 361–394. 10.1007/s10115-008-0150-6 [DOI] [Google Scholar]
- Rabiner L., 1989. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77: 257–286. 10.1109/5.18626 [DOI] [Google Scholar]
- Rastas P., 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 33: 3726–3732. 10.1093/bioinformatics/btx494 [DOI] [PubMed] [Google Scholar]
- Rastas P., Paulin L., Hanski I., Lehtonen R., Auvinen P., 2013. Lep-MAP: fast and accurate linkage map construction for large SNP datasets. Bioinformatics 29: 3128–3134. 10.1093/bioinformatics/btt563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., Calboli F. C. F., Guo B. C., Shikano T., Merila J., 2016. Construction of ultradense linkage maps with Lep-MAP2: stickleback F-2 recombinant crosses as an example. Genome Biol. Evol. 8: 78–93. 10.1093/gbe/evv250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinelt G., 1994. The Traveling Salesman: Computational Solutions for TSP Applications. Springer-Verlag, New York. [Google Scholar]
- Schiex T., Gaspin C., 1997. Cartagene: constructing and joining maximum likelihood genetic maps in Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology, Porto Carras, Halkidiki, Greece. [PubMed] [Google Scholar]
- Shi J., Malik J., 2000. Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22: 888–905. 10.1109/34.868688 [DOI] [Google Scholar]
- Sim S. C., Durstewitz G., Plieske J., Wieseke R., Ganal M. W., et al. , 2012. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS One 7: e40563 10.1371/journal.pone.0040563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q. J., Jenkins J., Jia G. F., Hyten D. L., Pantalone V., et al. , 2016. Construction of high resolution genetic linkage maps to improve the soybean genome sequence assembly glyma1.01. BMC Genomics 17: 33–43. 10.1186/s12864-015-2344-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P., 1993. Construction of integrated genetic-linkage maps by means of a new computer package - Joinmap. Plant J. 3: 739–744. 10.1111/j.1365-313X.1993.00739.x [DOI] [Google Scholar]
- Tong C. F., Zhang B., Shi J. S., 2010. A hidden Markov model approach to multilocus linkage analysis in a full-sib family. Tree Genet. Genomes 6: 651–662. 10.1007/s11295-010-0281-2 [DOI] [Google Scholar]
- Van Ooijen J., 2006. Joinmap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma BV, Wageningen, The Netherlands. [Google Scholar]
- Van Ooijen J. W., 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. 93: 343–349. 10.1017/S0016672311000279 [DOI] [PubMed] [Google Scholar]
- Van Os H., Stam P., Visser R. G. F., Van Eck H. J., 2005. RECORD: a novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 112: 30–40. 10.1007/s00122-005-0097-x [DOI] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., et al. , 2010. The genome of the domesticated apple (Malus domestica Borkh.). Nat. Genet. 42: 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- von Luxburg U., 2007. A tutorial on spectral clustering. Stat. Comput. 17: 395–416. 10.1007/s11222-007-9033-z [DOI] [Google Scholar]
- West M. A. L., van Leeuwen H., Kozik A., Kliebenstein D. J., Doerge R. W., et al. , 2006. High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis. Genome Res. 16: 787–795. 10.1101/gr.5011206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research, Inc. , 2016. Mathematica, Version 11.0. Wolfram Research, Inc., Champaign, IL. [Google Scholar]
- Wu Y. H., Bhat P. R., Close T. J., Lonardi S., 2008. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4: e1000212 10.1371/journal.pgen.1000212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., 2015. Modeling X-linked-linked ancestral origins in multiparental populations. G3 (Bethesda) 5: 777–801. 10.1534/g3.114.016154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Boer M. P., van Eeuwijk F. A., 2014. A general modeling framework for genome ancestral origins in multiparental populations. Genetics 198: 87–101. 10.1534/genetics.114.163006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Boer M. P., van Eeuwijk F. A., 2015. Reconstruction of genome ancestry blocks in multiparental populations. Genetics 200: 1073–1087. 10.1534/genetics.115.177873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Boer M. P., van Eeuwijk F. A., 2018a. Accurate genotype imputation in multiparental populations from low-coverage sequence. Genetics 210: 71–82. 10.1534/genetics.118.300885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Boer M. P., van Eeuwijk F. A., 2018b. Recursive algorithms for modeling genome blocks in a fixed pedigree. G3 (Bethesda) 8: 3231–3245. 10.1534/g3.118.200340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The algorithm magicMap is currently implemented in Mathematica 11.0 (Wolfram Research Inc. 2016), and has been included as a function in the RABBIT software. RABBIT is available from the web site: https://github.com/chaozhi/RABBIT.git, and is offered under the GNU Affero general public license, version 3 (AGPL-3.0). A Mathematica license is required to run the RABBIT software. Example scripts for simulating genotypic data and constructing linkage map are included.
The marker data for the Arabidopsis RIL are available from http://www.atgc.org/XLinkage/MadMapper/ath_sfp_map_example/, and the physical positions of the markers were obtained from Arabidopsis annotation version 4, TIGR release May 2003 (http://elp.ucdavis.edu/data/analysis/sfp_map/sfp_map.html). The marker data for the Arabidopsis MAGIC are available at http://mtweb.cs.ucl.ac.uk/mus/www/magic/. The marker data for the maize US-NAM are available at https://www.panzea.org/. The marker data for the apple CP, the barley MAGIC, the tomato MAGIC, the maize MAGIC, and EU-NAM are available in the supplementary materials of Bauer et al. (2013), Gardner et al. (2014), Dell’Acqua et al. (2015), Pascual et al. (2015), and Liller et al. (2017), respectively. The available physical maps from the corresponding marker data were also downloaded and used for evaluating magicMap. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.8243180.