Abstract
Saccharomyces cerevisiae lives in boom and bust nutritional environments. Sophisticated regulatory systems have evolved to rapidly cope with these changes while preserving intracellular homeostasis. Target of Rapamycin Complex 1 (TorC1), is a serine/threonine kinase complex and a principle nitrogen-responsive regulator. TorC1 is activated by excess nitrogen and downregulated by limiting nitrogen. Two of TorC1’s many downstream targets are Gln3 and Gat1—GATA-family transcription activators—whose localization and function are Nitrogen Catabolite Repression- (NCR-) sensitive. In nitrogen replete environments, TorC1 is activated, thereby inhibiting the PTap42-Sit4 and PTap42-PP2A (Pph21/Pph22-Tpd3, Pph21,22-Rts1/Cdc55) phosphatase complexes. Gln3 is phosphorylated, sequestered in the cytoplasm and NCR-sensitive transcription repressed. In nitrogen-limiting conditions, TorC1 is downregulated and PTap42-Sit4 and PTap42-PP2A are active. They dephosphorylate Gln3, which dissociates from Ure2, relocates to the nucleus, and activates transcription. A paradoxical observation, however, led us to suspect that Gln3 control was more complex than appreciated, i.e., Sit4 dephosphorylates Gln3 more in excess than in limiting nitrogen conditions. This paradox motivated us to reinvestigate the roles of these phosphatases in Gln3 regulation. We discovered that: (i) Sit4 and PP2A actively function both in conditions where TorC1 is activated as well as down-regulated; (ii) nuclear Gln3 is more highly phosphorylated than when it is sequestered in the cytoplasm; (iii) in nitrogen-replete conditions, Gln3 relocates from the nucleus to the cytoplasm, where it is dephosphorylated by Sit4 and PP2A; and (iv) in nitrogen excess and limiting conditions, Sit4, PP2A, and Ure2 are all required to maintain cytoplasmic Gln3 in its dephosphorylated form.
Keywords: Gln3, nitrogen catabolite repression, PP2A, rapamycin, Sit4, TorC1, Ure2
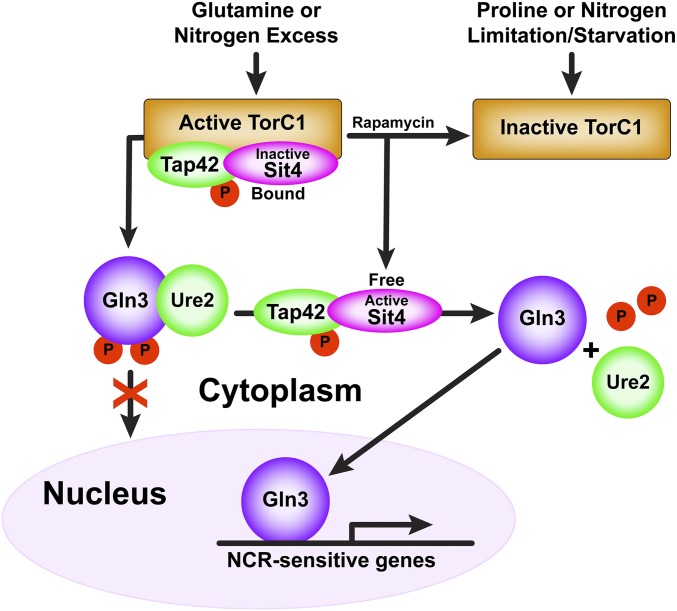
SACCHAROMYCES CEREVISIAE lives in an ever-changing and, at times, hostile, boom and bust nutritional environment. Sophisticated regulatory systems have evolved to rapidly cope with these changes while preserving intracellular homeostasis. Target of Rapamycin Complex 1 (TorC1), is a serine/threonine kinase complex that is activated/upregulated by excess nitrogen, and inhibited/downregulated by limiting nitrogen growth conditions (Figure 1) (Beck and Hall 1999; Hughes Hallett et al. 2014; González and Hall 2017). Two of the many downstream targets regulated by TorC1 are Gln3 and Gat1—GATA-family transcription activators responsible for expression of many Nitrogen Catabolite Repression- (NCR-) sensitive genes (Cooper 1982; Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Bertram et al. 2000). Among these genes are most of those that function to transport, degrade, and interconvert poor nitrogen sources scavenged from adverse nitrogen environments (Cooper 1982, 2002).
Figure 1.
Simplified early description of TorC1 responses to replete and limiting nitrogen conditions, and, in turn, its downstream regulation of Gln3 by regulating Sit4 activity.
In a nitrogen-replete environment, Gln3 is cytoplasmic (bound in a Gln3-Ure2 complex) and NCR-sensitive transcription is repressed (Figure 1). In nitrogen-limiting conditions, Gln3 relocates to the nucleus and activates/derepresses transcription (Hofman-Bang 1999; Magasanik and Kaiser 2002; Cooper 2004; Broach 2012; Ljungdahl and Daignan-Fornier 2012; Conrad et al. 2014; Swinnen et al. 2014; González and Hall 2017; Zhang et al. 2018). In the presence of excess nitrogen, TorC1 is active, binds to, and phosphorylates, Tor Associated Protein 42 (Tap42). Phosphorylated PTap42 in turn forms complexes with the Sit4 and PP2A phosphatases (Figure 1) (DiComo and Arndt 1996; Jiang and Broach 1999). PTap42-Sit4 and PTap42-PP2A complexes bound to TorC1 are enzymatically inactive (Yan et al. 2006). TorC1 also phosphorylates Gln3 in vitro and an intact TorC1 kinase domain is required to sequester Gln3 within the cytoplasm (Figure 1) (Bertram et al. 2000). PP2A is a heterotrimer, Pph21/22-Tpd3-Ccd55/Rts1, in which Pph21/22 are redundant catalytic subunits (Zabrocki et al. 2002; Zaman et al. 2009).
When TorC1 is inhibited with rapamycin, or nitrogen-limitation/starvation, the PTap42-Sit4 and PTap42-PP2A complexes dissociate from TorC1 (Figure 1) (Wang et al. 2003; Yan et al. 2006). Thus freed, they dephosphorylate Gln3, which dissociates from the cytoplasmic Gln3-Ure2 complex, enters the nucleus, and activates NCR-sensitive transcription (Figure 1) (Blinder et al. 1996; Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Bertram et al. 2000; Kulkarni et al. 2001). Treating cells with the glutamine synthetase inhibitor methionine sulfoximine (Msx) also elicits nuclear Gln3 localization (Crespo et al. 2002; Kulkarni et al. 2006; Georis et al., 2011a; Tate and Cooper 2013).
The requirements of Sit4 and PP2A for nuclear Gln3 localization, however, are dependent on how nitrogen limiting conditions are established (Tate and Cooper 2013). For rapamycin-treated, glutamine-grown cells, both Sit4 and PP2A are required (Beck and Hall 1999; Bertram et al. 2000; Tate et al. 2006a, 2009, 2010; Tate and Cooper 2013). Parenthetically, even though Sit4 can dephosphorylate Gln3 in rapamycin-treated, glutamine-grown cells lacking PP2A, Gln3 cannot enter the nucleus (Tate et al. 2009). In limiting nitrogen or short-term nitrogen starvation (1–4 hr depending on the strain assayed), only Sit4 is required (Tate and Cooper 2013). Finally, neither Sit4 nor PP2A is required for nuclear Gln3 localization after long-term nitrogen starvation that correlates with G-1 arrest of the cell cycle (∼8–10 hr of starvation), or in cells treated with Msx (Tate and Cooper 2013).
The course of Gln3 once it is in the nucleus correlates with the glutamine concentration (Rai et al. 2015). When glutamine levels are highest, i.e., when glutamine is the sole nitrogen source, or a glutamine analog (GAGM, l-glutamic acid-γ-monohydroxamate) is provided, Gln3 can exit from the nucleus in the absence of binding to its GATA-target sequences situated in the promoters of NCR-sensitive genes (Rai et al. 2015). In contrast, when glutamine levels are lowered, by adding other amino acids along with glutamine, employing amino acids other than glutamine as sole nitrogen sources or inhibiting glutamine synthetase with Msx, Gln3 cannot exit the nucleus unless it is also capable of binding to its GATA targets (Rai et al. 2015). In this context, it is important to note that PP2A is situated in both the cytoplasm and nucleus (Georis et al. 2011). Further, PP2A is required for Gln3 to bind to some of its target GATA sequences and mediate transcription, even in a ure2Δ mutant where Gln3 is constitutively nuclear and PP2A is not required for nuclear Gln3 localization (Tate and Cooper 2008; Georis et al. 2011a,b).
Although there is much evidence in support of Sit4 and PP2A participation in NCR-sensitive Gln3 regulation, when nitrogen is limiting and TorC1 is downregulated, there are at least two critical paradoxical observations that remain unexplained by the scenario described above and in Figure 1: (i) Sit4 more extensively dephosphorylates Gln3-Myc13 in repressive glutamine medium than in derepressive proline medium (Tate et al. 2006); and (ii) Msx elicits nuclear Gln3-Myc13 localization, but with increased rather than decreased phosphorylation (Tate et al. 2005; Kulkarni et al. 2006). Rapamycin treatment also elicits nuclear Gln3 localization, but, in that case, Gln3 is dephosphorylated. Neither of these paradoxical observations can be accommodated by the scenario of Gln3 regulation by TorC1 and the phosphatases it regulates (Figure 1).
These paradoxical observations motivated us to reinvestigate the roles of Sit4 and PP2A phosphatases in Gln3 regulation. In the most desirable situation, we would have followed the phosphorylation/dephosphorylation of specific Gln3 residues. This, however, was not technically possible, due to the extreme lability of the highly disordered Gln3 molecule, its small quantities within the cell, and high content (∼20%) of serine/threonine residues. In several previous studies, the phosphorylation levels of particular Gln3 residues have been reported to change (Urban et al. 2007; Huber et al. 2009; Breitkreutz et al. 2010; Soulard et al. 2010). However, those results could not be substantiated by genetic modification of the residues identified in that manner (Rai et al. 2013, 2014). Therefore, we chose to assay gross Gln3 phosphorylation to monitor Sit4 and PP2A activities under various conditions and intracellular locations of Gln3.
Our experiments have generated surprising and provocative results that add new dimensions to our understanding of nitrogen-responsive transcription factor regulation and explain some of the existing paradoxical observations: (i) Sit4 and PP2A dephosphorylate Gln3 not only when TorC1 is inactive, but also, and more importantly, when TorC1 is activated. (ii) Gross Gln3 phosphorylation levels are greater when it is in the nucleus than when it is in the cytoplasm. (iii) In nitrogen-replete conditions, Gln3 relocates from the nucleus to the cytoplasm. Arriving in the cytoplasm, Gln3 is dephosphorylated in a Sit4-and PP2A-dependent manner. (iv) Sit4, PP2A, and Ure2 are all required to maintain cytoplasmic Gln3 in its dephosphorylated form.
Materials and Methods
Strains and culture conditions
The S. cerevisiae strains and Gln3 plasmids that we used appear in Table 1 and Table 2, respectively. Transformants were prepared by the lithium acetate method (Ito et al. 1983), and used as quickly as possible after transformation (5 days or fewer). Cultures (50 ml) were grown to mid-log phase (A600nm = 0.4–0.5) in Yeast Nitrogen Base (YNB; Difco, without amino acids or ammonia) minimal medium. The indicated nitrogen sources were provided at a final concentration of 0.1%. Leucine (120 μg/ml), histidine (20 μg/ml), tryptophan (20 μg/ml), and uracil (20 μg/ml) were added as needed to cover auxotrophic requirements. Cells were treated with 200 ng/ml of rapamycin for 15 min or 2 mM methionine sulfoximine (Msx) for 30 min (Georis et al. 2011).
Table 1. Strains used in this work.
| Strain | Pertinent genotype | Complete genotype | Reference |
|---|---|---|---|
| TB123 | Wild Type | MATa, leu2-3, 112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX] | Beck and Hall (1999) |
| JK9-3da | Wild Type | MATa, leu2-3,112, ura3-52, trp1, his4, rme1, HMLa | Beck and Hall (1999) |
| TB138-1a | ure2Δ | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, ure2::URA3, GLN3-MYC13[KanMX] | Beck and Hall (1999) |
| RR215 | ure2Δ | MATa, leu2-3,112, ura3-52, trp1, his4, rme1, HMLa, ure2::[KanMX] | Rai et al. (2013) |
| TB136-2a | sit4Δ | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, GLN3-MYC13[KanMX], sit4::kanMX | Beck and Hall (1999) |
| FV029 | sit4Δ | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, sit4::natMX | Georis et al. (2008) |
| FV239 | pph21Δ,pph22Δ | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa, pph21::natMX, pph22::[kanMX] | Georis et al. (2011) |
| 03705d | pph21Δ,pph22Δ | MATα, leu2-3, 112, ura3-52, rme1, trp1, his4, HMLα, GLN3-MYC13[KanMX], pph21::[kanMX], pph22::kanMX | Tate et al. (2009) |
| FV071 | ure2Δ, sit4Δ | MATa, leu2-3,112, ura3-52, rme1, trp1, his4, GAL+, HMLa, ure2::natMX, GLN3-MYC13[KanMX], sit4::kanMX | Georis et al. (2008) |
| FV165 | ure2Δ, pph21Δ,pph22Δ | MATα leu2-3,112 ura3-52 trp1 his3 rme1 HMLα GLN3-MYC13[KanMX] pph21::kanMX pph22::kanMX ure2::natMX | Georis et al. (2011) |
Table 2. Plasmids used in this work.
| Plasmida | Characteristic | Reference |
|---|---|---|
| pRR536 | Gln31–730 (full-length wild type) | Rai et al. (2013) |
| pRR752 | Gln3L64D,L67R,L71D,F73D-Myc13 (Gln3 NES abolished) | Rai et al. (2015) |
Plasmids were constructed in CEN-based vectors. Gln3 transcription was driven by the native, wild type GLN3 promoter.
In evaluating the data derived from our experiments, it is important to recognize that ammonia is not as repressive a nitrogen source as glutamine in the strains we used, but considerably more so than proline (Cooper 1982).
Transfer of cells to nitrogen-free medium
To measure the effects of short- and long-term nitrogen starvation, the cells to be tested were grown to an A600nm = 0.45–0.50 in YNB-glutamine medium. At that cell density, a 5 ml untreated control sample was collected and processed for indirect immunofluorescence microscopy as described below. The remainder of the culture (45 ml) was gently, but rapidly, harvested by filtration (using type HA, 0.45-μm Millipore filter), washed twice with 12.5 ml of prewarmed, preaerated nitrogen-free YNB medium and resuspended in 50 ml prewarmed, preaerated, nitrogen-free YNB medium containing only supplements needed to cover auxotrophic requirements of the strain(s). Transfer of cells took between 20 and 30 sec. Sampling of the culture was then continued at the indicated time points or collected for western blot analysis as described.
Cell preparation and imaging for indirect immunofluorescence microscopy of Gln3-Myc13
Cell collection and fixation for indirect immunofluorescence microscopy were performed using modifications of the method of Schwartz et al. (1997). In the current protocol, cells were fixed at 30° for 80 min after the addition of 0.55 ml of 1 M potassium phosphate buffer (pH 6.5) and 0.5 ml of 37% formaldehyde to a 5-ml aliquot of the desired culture. After fixation, the samples were washed and resuspended in 0.1 M potassium phosphate buffer (pH 6.5) containing 1.2 M sorbitol. Zymolyase-20T (Sunrise Scientific Products) was used for digestion of cell walls with (i) the addition of β-mercaptoethanol (10 mM final concentration) to the Zymolyase-20T digestion mixture, and (ii) adjustment and optimization of the digestion times depending on the strain and growth conditions used (Tate et al. 2006). Cells were then plated onto 0.1% polylysine coated slides (poly-l-lysine hydrobromide; Sigma) and incubated overnight at 4° in phosphate buffer (pH 7.5) containing 0.5% bovine serum albumin (Sigma) and 0.5% Tween 20. Antibody incubations and subsequent washes were performed using this phosphate buffer. Primary antibody labeling of Gln3-Myc13 was performed using 9E10 (c-myc) monoclonal antibody (sc-40; Santa Cruz Biotechnology) at a dilution of 1:1000, followed by secondary labeling with Alexa Fluor 594 goat anti-mouse IgG antibody (A11032; Invitrogen Molecular Probes) at a dilution of 1:200. (Feller et al. 2013)
Primary images were collected at room temperature using a Zeiss Axio Imager.M2 microscope with a 63×/1.40 Plan-Aprochromat oil objective, Zeiss Axio camera, and Zeiss Axiovision 4.8.1 software. Only primary .zvi images were used for scoring of intracellular localization of Gln3-Myc13.
Image processing
Microscopic images for illustrative presentation were prepared by converting original .zvi files to .tif files and processed using Adobe Photoshop and Illustrator programs. Level settings (shadow and highlight only) were altered where necessary to avoid changes or loss in cellular detail relative to that observed in the microscope. All changes were applied uniformly to the image presented and were similar from one image to another. Midtone, gamma settings were never altered. These processed images were used for illustrative purposes only, not for scoring Gln3-Myc13 intracellular distributions (Feller et al. 2013).
Determination of intracellular Gln3-Myc13 distribution
Intracellular Gln3-Myc13 distributions were scored manually in 200 or more cells for each data point. Unaltered, primary .zvi image files viewed with Zeiss AxioVision 4.8.1 software were exclusively used for scoring purposes. Cells containing the tagged proteins were classified into one of three categories: cytoplasmic (cytoplasmic fluorescent material only; red histogram bars), nuclear-cytoplasmic (fluorescent material appearing in both the cytoplasm and colocalizing with DAPI-positive material, DNA; yellow bars), or nuclear (fluorescent material colocalizing only with DAPI-positive material; green bars). Representative “standard” images and detailed descriptions for scoring cells in each of the three categories appear in Figure 2 of Tate et al. (2009) with descriptions of how the criteria were applied.
Microscopic images accompanying the histograms were subjectively chosen on the basis that they exhibited intracellular Gln3-Myc13 distributions as close as possible to those observed by quantitative scoring. However, identifying a field that precisely reflected the more quantitative scoring data were sometimes difficult unless Gln3-Myc13 was situated in a single cellular compartment.
Statistical analyses
The precision of our scoring has been repeatedly documented with overall SD <10% for N = 7–10 experiments performed over 9 months in some cases, and up to 3 years in others (Tate et al. 2006, 2010, 2018; Rai et al. 2013, 2014). Precision decreases ∼5% when Gln3 is more or less equally distributed in all three scoring categories. In the present work, all histograms are presented as averages of multiple biological replicates. Standard deviations (presented as error bars) were calculated for each data point with the number of biological replicates indicated in each figure legend.
The statistical significance of individual data points in Figure 5 was analyzed with a three-way ANOVA with cell type, condition-time, and location as the main effects with all possible two- and three-way interactions included in the model. A total of 18 planned contrasts were made using a significance level of 0.05. SAS version 9.4 was used for the statistical analysis.
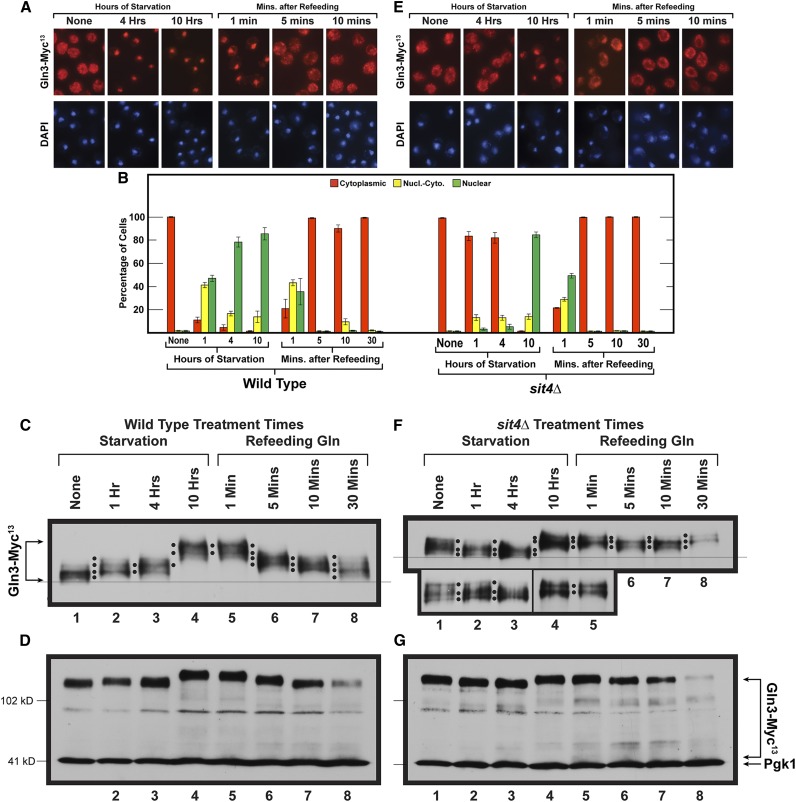
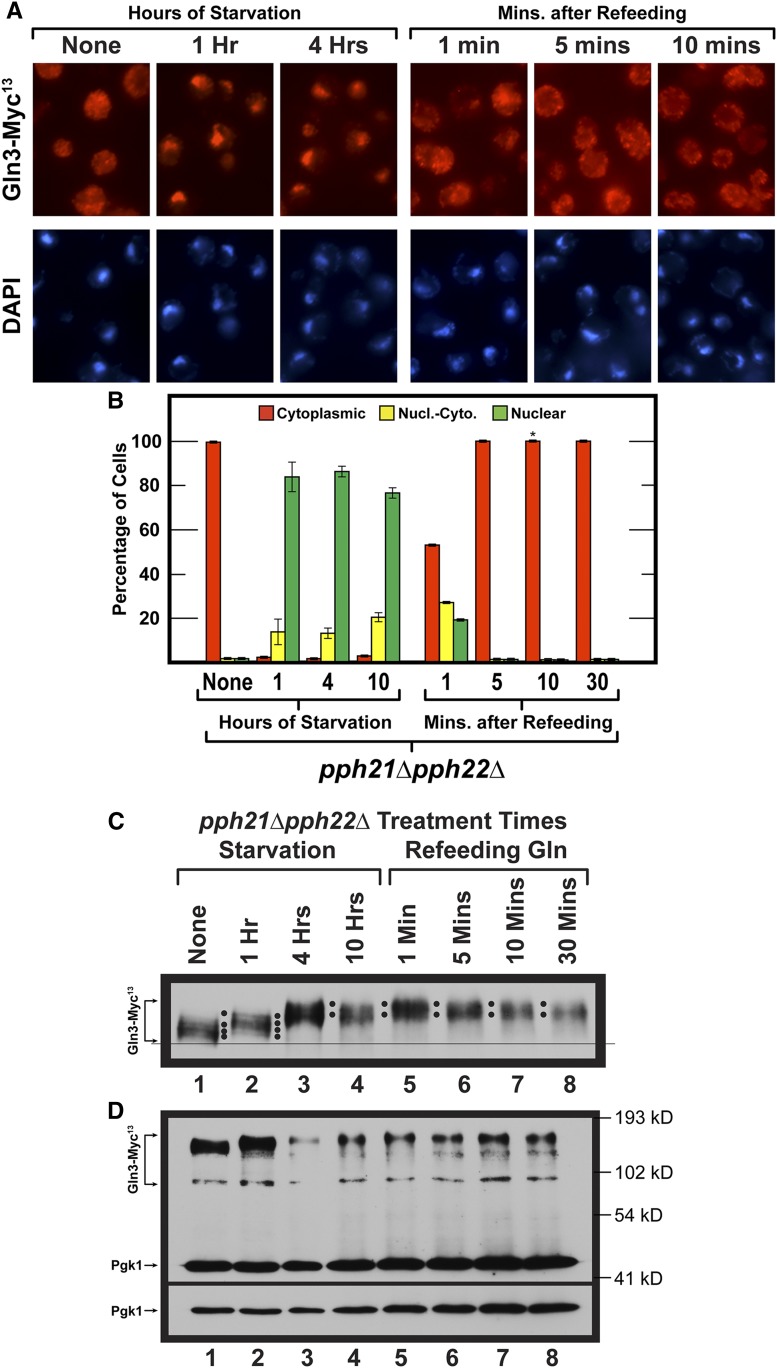
Figure 5.
Time course of Gln3-Myc13 intracellular localization and Gln3-Myc13 phosphorylation in short- and long-term nitrogen starvation of wild-type (A and B left side, C and D) and sit4Δ mutant (B right side, E–G) cells followed by the refeeding of excess nitrogen. Wild-type (TB123) and sit4Δ mutant (TB136-2a) cells were pregrown in YNB-glutamine medium, then transferred to nitrogen-free medium and sampled as described for 1, 4, and 10 hr. After 10 hr of starvation, glutamine was added to a final concentration of 0.1%, and sampling continued at the indicated times. Wild-type and sit4Δ cultures were processed for indirect immunofluorescence imaging as described in Materials and Methods. Data in (B), left side are averages and SD derived from seven biological replicates of starvation, and three biological replicates for refeeding. For sit4Δ cells, four and two biological replicates, respectively, were used for the starvation and refeeding portions of the experiments. For each time point, data derived from wild type (B left side) vs. sit4Δ (B right side) were compared to ascertain whether there were significant, mutant-dependent differences in the outcomes. Since the number of cells scored in each sample varied (average = 248; SD = 29), it was necessary to generate P values by comparing the number of cells in each cellular compartment of wild-type cells (cytoplasmic, nuclear-cytoplasmic, nuclear) at each time point to those in each parallel cellular compartment of sit4Δ cells at each of the parallel time points using a three-way ANOVA with cell type, condition-time, and location as the main effects with all possible two- and three-way interactions included in the model. The calculated P values were: None (0.9999, 0.9999, 0.9999); Starvation 1 hr (<0.0001, <0.0001, <0.0001); Starvation 4 hr (<0.0001, 0.3000, <0.0001); Starvation 10 hr 0.9145, 0.9999, 0.8439; Refeeding 1 min (0.9284, 0.0108, 0.0139); Refeeding 5 min (0.8574, 0.999, 0.9999); Refeeding 10 min (0.0932, 0.1137, 0.9687); Refeeding 30 min (0.9046, 0.9522, 0.9999), respectively. Localizations were considered to be different if the observed P values were <0.05. Upper panels (C and F) data derived from a six percent acrylamide gel, whereas the lower panels (D and G) data derived from a parallel sample of the same culture, but electrophoresed for a shorter time in a 7% gel to maintain the loading control, Pgk1, within the gel. Molecular weight standards are indicated.
Cell collection for western blot or qRT-PCR analysis
Cultures were grown to mid-log phase (A600nm = 0.4–0.5) as described above. Once the desired OD600nm was reached, or following treatment, the cells were harvested by filtration (using type HA, 0.45-μm Millipore filter), quickly scraped from the filter, placed in a sterile 1.5-ml microcentrifuge tube, and flash-frozen by submerging the microcentrifuge tube and cells in liquid nitrogen for 20–30 sec. The total time for cell harvest to the point of submersion in liquid nitrogen was 25–35 sec (this time is designated as 1 min in the figures). The tube, still containing liquid nitrogen, was then quickly transferred to −80° until further processing of the cells was performed. All western blots are representative of two to five biological replicates.
Protein extraction and western blot analyses
Extracts for western blots were prepared following the method of Liu et al. (2008). Total protein was extracted by lysing cells in a solution of 0.3 N NaOH, 1.2% β-mercaptoethanol (final concentrations), on ice for 10 min. Protein was then precipitated with trichloroacetic acid (TCA) at a final concentration of 8%, for an additional 10 min on ice. Precipitated protein pellets were then resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer and the extract neutralized with 1 M unbuffered Tris. Crude extracts were then boiled, protein resolved by SDS-PAGE (6 or 7% polyacrylamide) and transferred to nitrocellulose membrane (Bio-Rad) in non-SDS containing buffer. Membranes were blocked for 1 hr at room temperature with 5% milk in 1× TTBS (20 mM Tris-HCL pH 7.5, 0.05% Tween20, 0.5 M NaCl). Membranes were then incubated overnight at room temperature with 9E10 (c-myc) monoclonal antibody (sc-40; Santa Cruz Biotechnology) at a dilution of 1:1000 and/or PGK1 monoclonal antibody (22C5D8; Invitrogen) at a dilution of 1:4000 in 1× TBS (20 mM Tris-HCL pH 7.5, 0.5 M NaCl) plus 0.25% gelatin. Membranes were washed with 1× TBS and incubated with goat anti-mouse IgG (H+L)-horseradish peroxidase conjugate antibody (Bio-Rad) at a dilution of 1:10,000 for 1 hr in 1× TBS containing 0.005% Tween20 and 0.25% gelatin. Membranes were wash with 1× TBS containing 0.025 Tween20 buffer. Immunoreactive species were detected using the SuperSignal West Pico Chemiluminescent Substrate kit (ThermoScientific) following the manufacturer instructions and results recorded on Classic blue autoradiography Film BX (Midwest Scientific).
The conditions of electrophoresis and western blot loadings were designed to resolve and analyze phosphorylated Gln3-Myc13 species rather than to quantitate the amounts of Gln3-Myc13 present under the various conditions we used. To this end, we attempted to load the electrophoresis lanes such that they contained relatively equivalent Gln3-Myc13 signals, and extended the time of electrophoresis thereby increasing species resolution by permitting them to migrate further into the 6% gel. To achieve signals of similar intensity, it was necessary to increase the amount of protein by approximately twofold for samples taken beyond 1 hr of starvation irrespective of whether wild-type or mutant cells were employed. This necessity derives from the fact that Gln3 is predicted to be a very highly disordered protein that is exquisitely susceptible to proteolysis (Tate et al. 2018). This sensitivity becomes increasingly severe the longer the cells are subjected to adverse growth conditions, and irrespective of whether or not optimal growth conditions are then restored for a short time. More highly cross-linked (7%) gels run for shorter times were used to demonstrate Gln3-Myc13 degradation products. While Gln3-Myc13 degradation products and loading standards are resolved and visualized under these conditions, Gln3-Myc13 phosphorylation profiles are not. Western blots were biologically replicated from two to four times.
A fine line has been placed across the bottom of the blot images. This is to facilitate assessment of differences in the mobilities of the most rapidly migrating Gln3-Myc13 species in each condition. Black dots have also been placed between some of the lanes to facilitate identification, comparison and/or emphasis of individual Gln3-Myc13 species in the lanes adjacent to the dots. Black dots between one pair of lanes cannot always be moved a priori to a new pair of lanes and remain accurate markers of the species’ mobilities in the second pair of lanes.
Quantitative RT-PCR analysis
Cultures (50 ml) were grown and harvested by flash freezing as described above (Tate et al. 2005). Total RNA was extracted using the RNeasy Mini Kit (Qiagen), following the manufacturer’s instructions for purification of total RNA from yeast—mechanical disruption of cells (Rai et al. 2015). Two modifications were made to this protocol from our previous report: (i) cells were broken with glass beads (0.45 μm) using a BeadBug homogenizer (Benchmark Scientific): 4000 rpm, 4° for 30-sec intervals followed by 30 sec in an ice water bath, and (ii) on-column RNase-free DNase I treatment was performed for 1 hr instead of 40 min. Quality of the total RNA was analyzed on an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nanochip by the University of Tennessee Health Science Center (UTHSC) Molecular Resource Center. Complementary DNAs (cDNAs) were generated using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s recommended protocol using both Oligo(dT)18 and random hexamer primers (provided with the kit) for synthesis. Samples were prepared for quantification with LightCycler 480 SYBR Green I Master Mix (KAPABiosystems) using the manufacturer’s protocol. Quantification and subsequent analysis of cDNAs were performed on a Roche LightCycler 480 Real Time PCR System using LightCycler 480 software version 1.5. GDH2 and TBP1 primer sequences were as described previously (Georis et al. 2008, 2011a,b) .
Data availability
Strains and plasmids will be provided upon request, but only for noncommercial purposes. Commercial and commercial-development uses are prohibited. Materials provided may not be transferred to a third party without written consent. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
The roles of the Sit4 and PP2A (Pph21/22 catalytic subunits) phosphatases in NCR-sensitive Gln3 localization and function have been intensely investigated from the vantage point of dephosphorylating Gln3 and/or Ure2 associated with their dissociation and the entry of Gln3 into the nucleus in response to rapamycin treatment (Figure 1) (Beck and Hall 1999; Bertram et al. 2000; Yan et al. 2006). However, the phosphorylation levels of Gln3 as it transits into, is situated within, and relocates out of, the nucleus have received little attention. Gln3 is generally accepted to be dephosphorylated by these phosphatases under nitrogen-limiting derepressive conditions where it is nuclear, and phosphorylated by TorC1 kinase in nitrogen-replete repressive conditions where Gln3 is cytoplasmic (Figure 1). While significant evidence supports this elegant model, several paradoxical observations led us to hypothesize that functional participation of PP2A and Sit4 in Gln3 regulation might actually be more complex than previously appreciated. Experiments presented below test and investigate this hypothesis.
Prior to investigating the functions of Sit4 and PP2A, it was necessary to establish steady-state levels of Gln3 phosphorylation when it was restricted to specific cellular compartments. Achieving this objective was greatly aided by the fact that four conditions exist where Gln3 is accepted to be highly or completely nuclear: following treatment with methionine sulfoximine (Msx), a glutamine synthetase inhibitor; in a gln3 nuclear export deficient mutant (pRR752, Gln3L64D,L67R,L71D,F73D-Myc13, hereafter referred to as the gln3 NES mutant or NES Gln3-Myc13 for the protein); during long-term nitrogen starvation; and in a ure2Δ mutant. Conversely, three conditions exist where Gln3 is accepted to be effectively sequestered in the cytoplasm: in nitrogen-replete conditions; in a sit4Δ mutant; or following rapamycin treatment of a pph21Δ,pph22Δ mutant.
Sit4 and PP2A dephosphorylate Gln3-Myc13 when TorC1 is active in ammonia medium
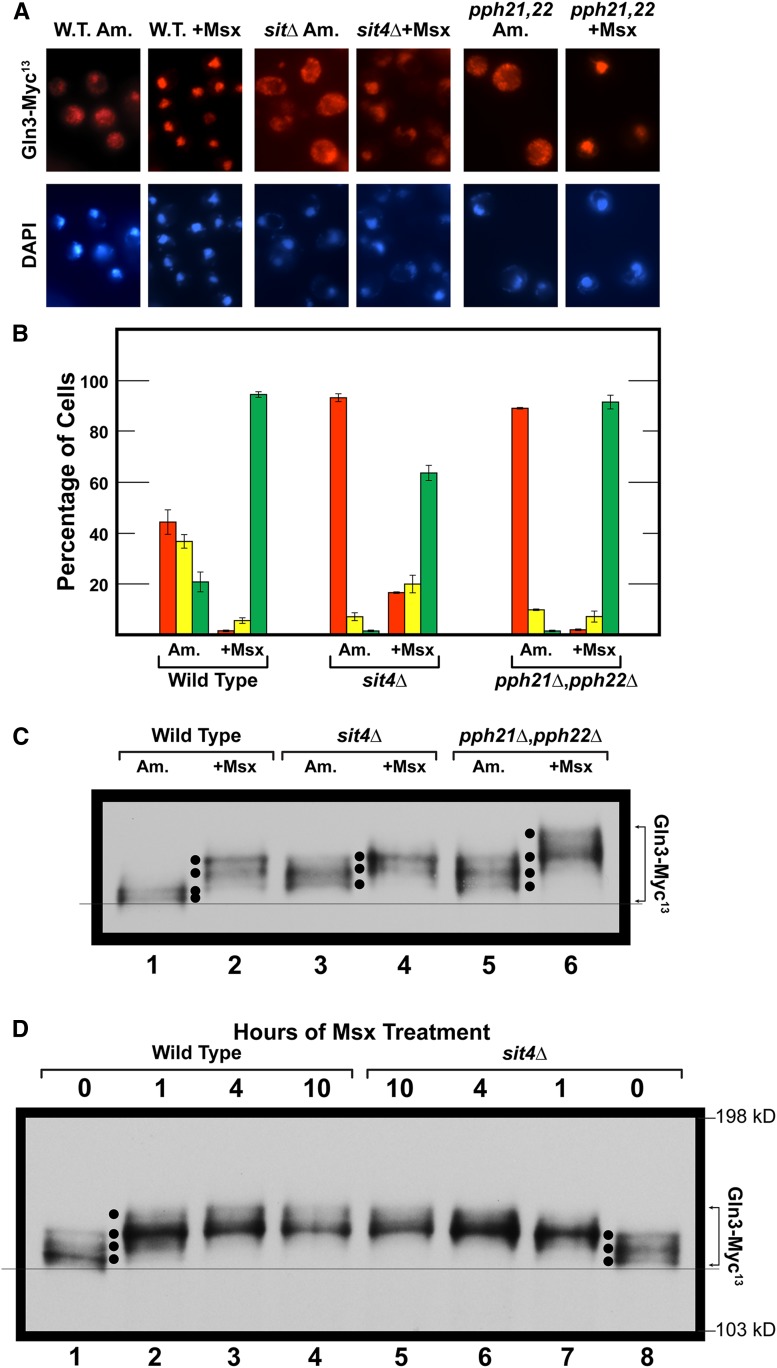
Our first objective was to correlate intracellular Gln3-Myc13 localization with its degree of phosphorylation and determine the effects of abolishing Sit4 and PP2A on these Gln3 phosphorylation levels. To this end, we determined Gln3-Myc13 intracellular distribution in ammonia-grown wild-type cells. Under this condition, Gln3-Myc13 exhibited a tripartite intracellular distribution, being more or less equally distributed in all three of the scoring categories, cytoplasmic, nuclear cytoplasmic, and nuclear (Figure 2, A and B, wild type). The loss of either Sit4 or PP2A phosphatase was sufficient to almost completely prevent this nuclear-cytoplasmic and nuclear Gln3-Myc13 localization (Figure 2, A and B, sit4Δ and pph21Δ,pph22Δ).
Figure 2.
Effects of abolishing Sit4 or PP2A (Pph21/Pph22) activities on intracellular Gln3 localization and phosphorylation. (A and B) Wild type (JK9-3da), sit4Δ (FV029), and pph21Δ,pph22Δ (FV239) mutant cells, each transformed with wild type Gln3-Myc13 plasmid pRR536, were cultured in YNB-ammonia medium (Am.) to an A600nm ∼0.5. Methionine sulfoximine (+Msx; 2 mM) was then added to each culture, and, 30 min later, samples were collected and processed for indirect immunofluorescence microscopy. Gln3-Myc13 intracellular localization was then scored in images of each sample. The data are averages of four biological replicates for wild type, and three for each of the mutants, with SD of the measurements indicated as error bars. Illustrative images of the semiquantitative data shown by the histograms (B) are presented in (A). The details of cell processing for indirect immunofluorescence, image collection, and Gln3-Myc13 intracellular localization are described in Materials and Methods. Red bars indicate Gln3-Myc13 localized to only the cytoplasm; yellow bars indicate Gln3-Myc13 was located in both the cytoplasm and nucleus; green bars indicate that Gln3-Myc13 was localized exclusively to the nucleus (colocalizing with DAPI positive staining). (C). Wild-type and mutant cells were cultured as described in (A and B). Samples were then prepared and western blot analyses performed as described in Materials and Methods. In this and subsequent figures black dots have been used exclusively between lanes to facilitate identification, comparison and/or emphasis of individual Gln3-Myc13 species in the lanes adjacent to the dots. Black dots between one pair of lanes cannot always be moved a priori to a new pair of lanes, and remain accurate markers of the species’ mobilities in the second pair of lanes. The fine line across the bottom of the blot was placed to facilitate assessment of differences in the mobilities of the most rapidly migrating Gln3-Myc13 species in each condition. Species with slower mobility are indicative of increased phosphorylation, whereas those with faster mobility are indicative of decreased phosphorylation. (D) Wild type (TB123) and sit4Δ mutant (TB136-2a) cells were grown as described in (A–C). Untreated cells are designated as “0 hr of Msx treatment” (lanes 1 and 8). At the designated times thereafter, samples of each culture were collected and processed for western blot analysis.
The electrophoretic mobility of cytoplasmic Gln3-Myc13 slowed markedly in untreated sit4Δ and pph21Δ,pph22Δ (PP2A) mutant cells relative to wild type (Figure 2C, lanes 1, 3, 5; note the position of the fine reference line and black dots, see figure legend for an explanation). This behavior has been repeatedly shown to derive from increased phosphorylation (Tate et al. 2009; Rai et al. 2013, 2014, 2016). Increased Gln3-Myc13 phosphorylation observed when a particular phosphatase is abolished by deleting its cognate gene is evidence that it participated in maintaining Gln3-Myc13 at the wild type dephosphorylated level when that phosphatase was present.
The data in Figure 2C indicated that Sit4 and PP2A phosphatases were both active in untreated, ammonia-grown cells and were required to maintain Gln3-Myc13 at the hypo-phosphorylated level observed in the untreated wild type. The critical importance of this observation is that both Sit4 and PP2A actively dephosphorylated Gln3-Myc13 in ammonia, a relatively repressive nitrogen source, where TorC1 is activated. According to the generally accepted paradigm, when TorC1 is activated, cytoplasmic Gln3-Myc13 should be hyper-phosphorylated and Sit4 and PP2A inactive (Figure 1). Hence, a priori, we expected Gln3-Myc13 phosphorylation to be unaffected by abolishing Sit4 and PP2A activities under these growth conditions (DiComo and Arndt 1996; Beck and Hall 1999; Jiang and Broach 1999; Binda et al. 2009, 2010). Our experimental observations were just the opposite. Both phosphatases were dephosphorylating Gln3-Myc13 when TorC1 was activated.
Gln3-Myc13 is nuclear, and hyper-phosphorylated in response to Msx treatment
When the above three strains were treated with Msx for 30 min, Gln3-Myc13 relocated almost completely to the nucleus, where it was highly phosphorylated (Figure 2, A–C, lanes 1, 2, 4, 6). This contrasts remarkably with the hypo-phosphorylation of Gln3-Myc13 in rapamycin-treated cells, where Gln3-Myc13 also relocates to the nucleus (Figure 1 and wild type in Figure 3, C, D, and F). Therefore, although Gln3-Myc13 became nuclear when cells were treated with Msx or rapamycin, their phosphorylation profiles were diametrically opposite.
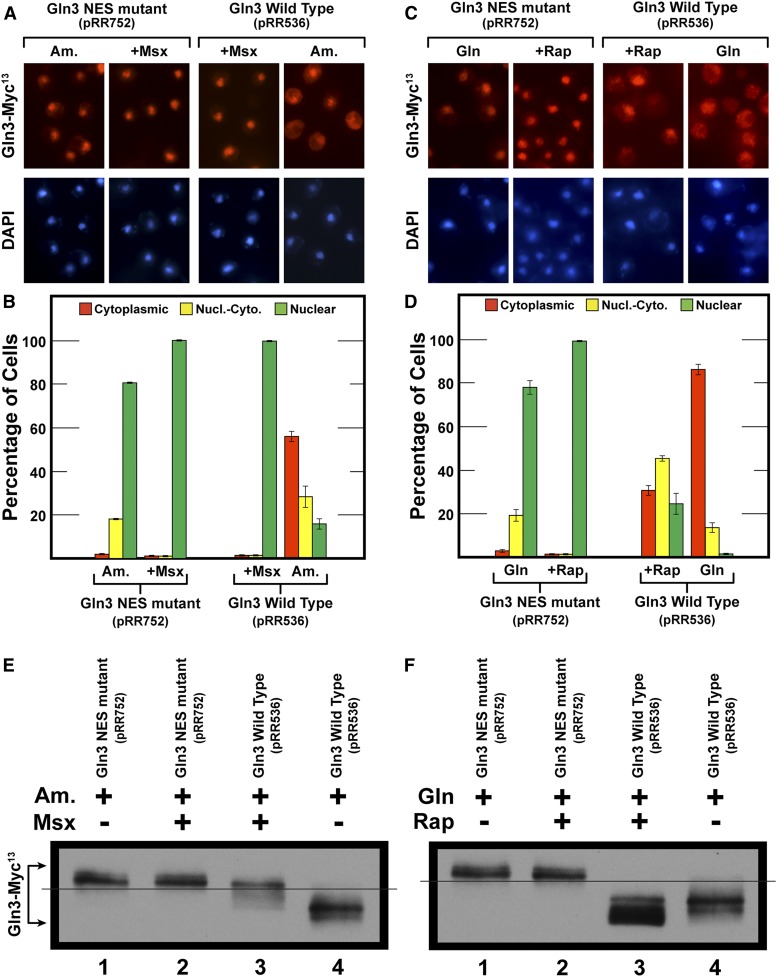
Figure 3.
The loss of nuclear export results in constitutively nuclear Gln3 that is hyperphosphorylated. (A–D) Wild-type strain JK9-3da was transformed with Cen II-based plasmids containing either wild-type Gln3-Myc13 (pRR536) or the gln3 NES mutant (pRR752). Cells for (A and B) were cultured and treated as described in Figure 2, A and B. Cells for (C and D) were cultured in YNB-glutamine medium (Gln) to A600 nm ∼0.5. Cultures were then treated for 15 min with 200 nM rapamycin (+Rap). Sample preparation and data collection were as described in Materials and Methods and Figure 2, A and B. Data in (A–D) are averages of two and five biological replicates, respectively. SD of the measurements are indicated as error bars. (E and F) Wild-type (strain JK9-3da) transformants containing plasmid pRR536 or pRR752 were cultured as in (A–D). Samples were then collected and processed for western blot analyses as described in Materials and Methods.
Treating wild type and sit4Δ mutant cells with Msx did not yield measurable differences in hyper-phosphorylated Gln3-Myc13 (Figure 2C, lanes 2 and 4). However, when pph21Δ,pph22Δ cells were treated with Msx, the degree of Gln3-Myc13 phosphorylation increased significantly beyond that observed with Msx-treated wild type or sit4Δ cells (Figure 2C, lanes 2, 4, 6). Since Gln3-Myc13 was already highly nuclear in the Msx-treated wild type, sit4Δ and pph21Δ,pph22Δ cells, one possible interpretation of this observation is that increased Gln3-Myc13 phosphorylation observed in the pph21Δ,pph22Δ mutant occurred in the nuclei of the Msx-treated cells. This latter observation correlates positively with PP2A being detected both in the cytoplasm and nucleus (Georis et al. 2011). The influence of PP2A activity on Gln3 regulation will be further described later in this work.
To ensure that the above observations with the sit4Δ mutant did not derive from the kinetics of Msx action, we followed the time course of Gln3-Myc13 phosphorylation in ammonia-grown wild-type and sit4Δ cells treated with Msx. Except for untreated cells (zero time points), the profiles of increasing phosphorylation in Msx-treated, wild-type, and sit4Δ strains were indistinguishable (Figure 2D, lanes 2–7). In untreated wild-type cells (zero time point), the most rapidly migrating (hypo-phosphorylated) Gln3-Myc13 species predominated (Figure 2D, lane 1, bottom black dot). In contrast, in untreated sit4Δ cells, the two most slowly migrating (hyper-phosphorylated) species predominated (Figure 2D, lanes 1 vs. 8, upper two black dots). This, in sharp contrast with the accepted scenario depicted in Figure 1, again indicated that Sit4 was functioning in the ammonia-grown cells where TorC1 was activated.
Gln3-Myc13 is nuclear, and hyper-phosphorylated in a gln3 nuclear export mutant
Given the observation that Msx-elicited nuclear Gln3-Myc13 was hyper-phosphorylated, we determined whether similar results were obtained when Gln3-Myc13 was restricted within the nucleus by another, independent mechanism. To this end, we assayed Gln3-Myc13 phosphorylation in a gln3 nuclear export sequence defective mutant where Gln3-Myc13 is constitutively nuclear due to the loss of its export function (Figure 3, A and B) (Rai et al. 2015).
NES-Gln3-Myc13 was equivalently hyper-phosphorylated irrespective of whether or not the cells were treated with Msx (Figure 3E, lanes 1, 2). In contrast, wild type Gln3-Myc13 phosphorylation and nuclear localization increased upon Msx-treatment, but only to the same level as observed in the gln3 NES mutant (Figure 3, A, B, and E, lanes 1, 2 vs. 3, 4). These data continued to support the contention that Gln3 was more phosphorylated in the nucleus than in the cytoplasm, just the opposite of that previously presumed in the literature (Figure 1) (Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Bertram et al. 2000; Broach 2012; Ljungdahl and Daignan-Fornier 2012; Zhang et al. 2018).
With Gln3-Myc13 being constitutively nuclear in the gln3 NES mutant, we did not expect its localization or phosphorylation to be affected when the cells were treated with rapamycin, and, indeed, that is what occurred (Figure 3, C, D, and F, lanes, 1 and 2). In the wild-type control experiment, where Gln3-Myc13 was cytoplasmic (Figure 3, C and D, W.T. Gln), Gln3-Myc13 phosphorylation decreased in rapamycin-treated cells as expected (Figure 1 and Figure 3F, lanes 3, 4).
Expectations of Gln3 behavior in response to short- and long-term nitrogen starvation
A major contribution to the TorC1-regulatory paradigm was the demonstration that nitrogen starvation and rapamycin treatment elicit similar outcomes from downstream reporter proteins (Figure 1). Supporting that view, both treatments increase nuclear Gln3-Myc13 localization (Tate and Cooper 2013). By analogy, these responses have previously led to the expectation that Gln3-Myc13 phosphorylation would decrease in both cases as well (Figure 1). Therefore, our next objective was to test that expectation.
As the data in Figure 4 are evaluated, it is important to be aware that the duration of nitrogen starvation (usually 1–1.5 hr in the literature) is not considered as an important determinant of downstream outcomes. When cells are nitrogen-starved short-term, i.e., up to 4–5 hr (depending on the strain assayed), Gln3-Myc13 relocation to the nucleus is highly Sit4-dependent (Tate and Cooper 2013). The same phosphatase requirement is observed for nuclear Gln3-Myc13 localization when growth is limited by provision of a poor nitrogen source such as proline (Tate and Cooper 2013) (Figure 1). In contrast, when nitrogen starvation is extended long-term, i.e., to the point that cells arrest in G-1 phase (8–10 hr), Gln3-Myc13 relocates almost completely to the nucleus, but in a Sit4-independent manner (Tate and Cooper 2013).
Figure 4.
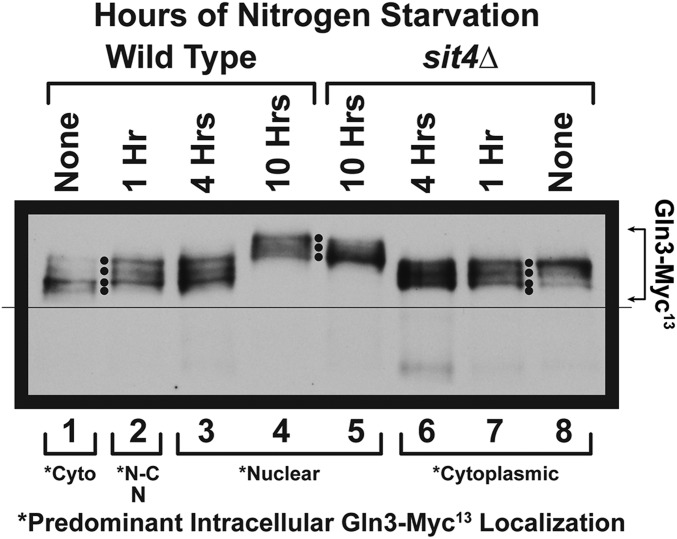
Time course of Gln3-Myc13 phosphorylation in glutamine-grown wild type and sit4Δ cells. Wild type (TB123) and sit4Δ mutant (TB136-2a) cells were cultured in YNB-glutamine medium to A600nm =∼0.5. At that time the cells were gently collected on a Millipore filter, washed twice, and transferred to nitrogen-free YNB medium as described in Materials and Methods. Samples were then collected at the times indicated, and processed for western blot analysis. Predominant intracellular localizations of Gln3-Myc13 as previously reported (Tate and Cooper 2013); Cyto, cytoplasmic; N-C, nuclear-cytoplasmic, N, nuclear.
Sit4 dephosphorylates Gln3-Myc13 when TorC1 is highly activated
To evaluate Gln3-Myc13 phosphorylation, during short- and long-term nitrogen starvation, we followed it in glutamine-pregrown wild type cells before and after 1, 4, or 10 hr of nitrogen starvation. At the initial time point of the experiment, i.e., glutamine-grown, steady-state, unstarved, wild type and sit4Δ cells, a clear difference in Gln3-Myc13 phosphorylation was noted (Figure 4, lanes 1 vs. 8, “None,” note the four black dots). Gln3-Myc13 phosphorylation in sit4Δ cells markedly increased relative to wild type with a majority of the phosphorylation occurring in a single, more slowly migrating, species (Figure 4, lane 1, bottom two black dots vs.. lane 8, uppermost black dot). This was the second instance where the loss of Sit4 led to increased Gln3-Myc13 phosphorylation in nitrogen-replete medium, this time with highly repressive glutamine as the nitrogen source. In this condition, TorC1 is known to be highly active and the TorC1-bound PTap42-Sit4 complex inactive (DiComo and Arndt 1996; Beck and Hall 1999; Jiang and Broach 1999; Binda et al. 2009, 2010; Broach 2012; Ljungdahl and Daignan-Fornier 2012; Zhang et al. 2018). Nonetheless, the loss of Sit4 still elicited an increase in Gln3-Myc13 phosphorylation, indicating that Sit4 was active despite TorC1 activation—a major departure from the scenario in Figure 1.
Gln3-Myc13 dephosphorylation during short- and long-term nitrogen starvation of wild-type and sit4Δ cells
In the early stages of starvation (1 and 4 hr), Gln3-Myc13 phosphorylation increased in wild-type, but decreased in sit4Δ cells relative to unstarved levels (Figure 4, lanes 1–3 vs. 6–8). Surprisingly, the phosphorylation profiles in wild-type and sit4Δ cells at these times were indistinguishable from one another (Figure 4, lanes 2 and 3 vs. 6 and 7). These results indicated that, as long as nitrogen starvation was not extended too long, the cell recognized a decrease in nitrogen availability, and responded by triggering a change in Gln3-Myc13 phosphorylation levels. In wild-type unstarved cells, where Gln3-Myc13 was hypo-phosphorylated, the onset of starvation substantially increased Gln3-Myc13 phosphorylation. However, in unstarved sit4Δ cells, where Gln3-Myc13 phosphorylation was already significantly higher than in the wild type (lanes 1 vs. 8), the onset of starvation decreased Gln3-Myc13 phosphorylation relative to unstarved cells (lanes 6–8). In other words, there was a marked Gln3 response to derepressive, nitrogen-limiting conditions (early starvation); however, Sit4 was not responsible for it as would normally be expected from the view expressed in Figure 1.
At 10 hr of starvation, Gln3-Myc13 was nuclear and much more highly phosphorylated (decreased mobility) in both wild type and a sit4Δ mutant, albeit less so in the sit4Δ strain (Figure 4, lanes 4 and 5). Note the lowest mobility species is less apparent in lane 5 relative to lane 4 (three black dots between lanes 4 and 5). Hence, for a third condition, and, in contrast with literature expectations (Figure 1), highly nuclear Gln3-Myc13 positively correlated with its hyper-phosphorylation, despite the fact that, under these conditions, TorC1 activity is low, and hence Sit4 and PP2A phosphatases would be highly active.
Localization of Gln3-Myc13 during nitrogen-starvation and refeeding
The above observations made a clear prediction that merited testing. If Gln3-Myc13 phosphorylation was dictated by/correlated with its intracellular location, and was much higher in the nuclei of long-term nitrogen-starved cells, it stood to reason that refeeding excess nitrogen should elicit exit of Gln3-Myc13 from the nucleus along with its dephosphorylation occurring before or after it arrived in the cytoplasm. To test this prediction, we again nitrogen starved glutamine-pregrown wild-type and sit4Δ cells for 1, 4, and 10 hr, then added glutamine to the cultures and continued sampling at 1, 5, 10, or 30 min thereafter (Figure 5).
Gln-Myc13 was completely cytoplasmic in both wild type and sit4Δ growing in glutamine medium (Figure 5, A, B, and E “None”). At 1 and 4 hr after the onset of nitrogen starvation, Gln3-Myc13 relocated to the nuclei of wild-type, but not sit4Δ, cells at these times (P < 0.0001), supporting a previous report that Sit4 is required for Gln3 to relocate to the nuclei of cells subjected to short-term nitrogen starvation (Tate and Cooper 2013). By 10 hr of starvation Gln3-Myc13 was largely nuclear in both wild type and sit4Δ cells.
Phosphorylation levels in nitrogen-starved wild-type cells were the same as observed in Figure 4, lanes 1–4. Gln3-Myc13 phosphorylation was greater at 1 and 4 hr of starvation relative to unstarved cells. (Figure 5C). However, Gln3-Myc13 phosphorylation was much higher and completely nuclear at 10 hr (Figure 5, A and B, left side and Figure 5C, lane 4). It is important to emphasize that, in wild-type cells, Sit4 and PP2A are both present.
Within 1 min after glutamine was added to the starved wild-type cells, Gln3-Myc13 began exiting from the nucleus. However, at that time, there were few, if any, cells where it was completely cytoplasmic (Figure 5, A and B). Correlating with this observation, there was also little, if any, detectable change in Gln3-Myc13 phosphorylation for the first minute following glutamine addition (Figure 5C, lanes 4 vs. 5, two black dots). By 5 min, however, Gln3-Myc13 was completely cytoplasmic, and remained so for the duration of the experiment (Figure 5, A and B left side). In comparison, after 1 min of refeeding, Gln3-Myc13 phosphorylation decreased continuously, nearly reaching the level that existed in the initial, unstarved cells 30 min after glutamine was added to the cultures (Figure 5C, lanes 1, 5–8).
Three important conclusions derive from this refeeding experiment: (i) Gln3-Myc13 continued to be dephosphorylated long after it was completely cytoplasmic, arguing that a substantial portion, if not all, of the Gln3-Myc13 dephosphorylation occurred in the cytoplasm. (ii) These data added further support to the contention that Gln3 was phosphorylated more in the nucleus than in the cytoplasm. (iii) Gln3-Myc13 was being dephosphorylated under conditions where TorC1 was being activated by excess nitrogen. All three conclusions are counterintuitive with the paradigmatic scenario in Figure 1.
We noted that Gln3-Myc13 signals diminished especially by the conclusion of the experiment, and that proteolytic degradation products were prominent (Figure 5D). It is somewhat puzzling that degradation occurred at times when a good nitrogen supply had been restored in the cultures (Figure 5D, lanes 5–8). In fact, by 30-min postrefeeding, the Gln3-Myc13 signals had decreased significantly. We do not presently know the source of this effect. However, it is important to note in this context that proteases produced in large quantities during starvation do not disappear nearly as rapidly as does the cessation of NCR-sensitive gene expression and the exit of Gln3-Myc13 from the nucleus. This is due to the length of mRNA and protein half-lives.
Dephosphorylation of Gln3-Myc13 on its return to the cytoplasm is partially Sit4-dependent
Our next objective was to ascertain whether or not Sit4 played any role in the dephosphorylation we observed upon refeeding starved wild-type cells. To that end, we repeated the starvation-refeeding experiment shown in Figure 4 with a sit4Δ, where only PP2A remained active (Figure 5). Gln3-Myc13 was completely cytoplasmic at 1 and 4 hr of nitrogen starvation, but was predominantly nuclear after 10 hr of starvation (Figure 5, A and B right side). Gln3-Myc13 phosphorylation did not detectably increase during short-term (1 and 4 hr) starvation as occurred in the wild type (Figure 5C, lanes 1–3); in fact, it decreased relative to unstarved sit4Δ cells during this time frame, as previously observed in Figure 4. At 10 hr of starvation, Gln3-Myc13 phosphorylation again increased dramatically, correlating with its high nuclear localization (Figure 5F, lane 4).
We then determined the effect of abolishing Sit4 on Gln3-Myc13 phosphorylation levels when these starved cells were refed a highly repressive nitrogen source (Gln). The course of Gln3-Myc13 relocation to the cytoplasm and initial phase of dephosphorylation were similar to that of wild-type cells. In the first minute following glutamine addition, there was only limited relocation to the cytoplasm, and no detectable Gln3-Myc13 dephosphorylation. Although the difference between the number of wild type and sit4Δ cells containing cytoplasmic Gln3-Myc13 was not statistically significant (P value, 0.9284) at 1 min post-refeeding, the differences in nuclear-cytoplasmic and nuclear distributions in wild-type and sit4Δ cells were moderately significant (P values of 0.0108 and 0.0139). By 5 min postrefeeding, Gln3-Myc13 was totally cytoplasmic, and dephosphorylation had commenced [Figure 5B (right side), Figure 5, E and F, lanes 4–7]. However, in contrast with the wild type, Gln3-Myc13 dephosphorylation proceeded no further, i.e., the dephosphorylation levels at 5 and 10 min were indistinguishable (Figure 5F, lanes 6 and 7). At 30 min postrefeeding, Gln3-Myc13 phosphorylation was even greater than at 10 min (Figure 5F, lanes 7 and 8). Moreover, the level of dephosphorylation did not proceed nearly as far as it had in the wild-type strain (Figure 5C, lanes 1 and 8 vs. Figure 5F, lanes 1 and 8).
We have included additional images of Gln3-Myc13 mobilities (from two other experiments) under starvation, and 1 min after refeeding to facilitate distinguishing the various phosphorylated species from one another (Figure 5F, bottom two panels). Also as occurred with wild-type cells, one can see the proteolysis of Gln3-Myc13 in the overexposed films (Figure 5G). Low molecular weight Gln3 products become detectable 4 hr after the onset of starvation, and reach a maximum at ∼5 min after glutamine has been added to the starved cultures (Figure 5G, lanes 3–6). Thereafter, these products decrease, and, by 30 min postrefeeding, both the products and much of the high molecular weight Gln3-Myc13 are gone (Figure 5G, lanes 7 and 8). It is pertinent that Gln3-Myc13 proteolysis occurred in both wild-type and sit4Δ cells.
From these data, we concluded that Sit4 was, at minimum, responsible for a portion of the Gln3-Myc13 dephosphorylation that occurred after it exited from the nucleus. This conclusion correlated with our inability to demonstrate Sit4-Myc13 in the nucleus (Georis et al. 2011). Further, and more importantly, Sit4 participated in Gln3-Myc13 dephosphorylation under conditions where TorC1 was highly active. The latter conclusion again differs distinctly from that previously accepted in the literature, where Sit4 is thought to be inactive in nitrogen replete medium (Figure 1).
Ure2 and Sit4 collaborate to maintain hypo-phosphorylated Gln3-Myc13
The fourth and last approach that we used to test the correlation between Gln3 localization and hyper-phosphorylation employed ure2Δ and sit4Δ mutants. The rationale here was that, in a ure2Δ mutant, Gln3-Myc13 is completely nuclear (See Figure 6, A and B of Georis et al. 2008; Figure 6A Tate et al. 2017; Figure 12, Tate et al. 2018), whereas in a sit4Δ, it remains staunchly cytoplasmic, irrespective of whether the cells are provided with a repressive or derepressive nitrogen source or treated with rapamycin (Blinder et al. 1996; Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999; Bertram et al. 2000; Kulkarni et al. 2001; Broach 2012; Ljungdahl and Daignan-Fornier 2012; Zhang et al. 2018). These two deletions cleanly localize Gln3-Myc13 to different cellular compartments in multiple different nutritional environments.
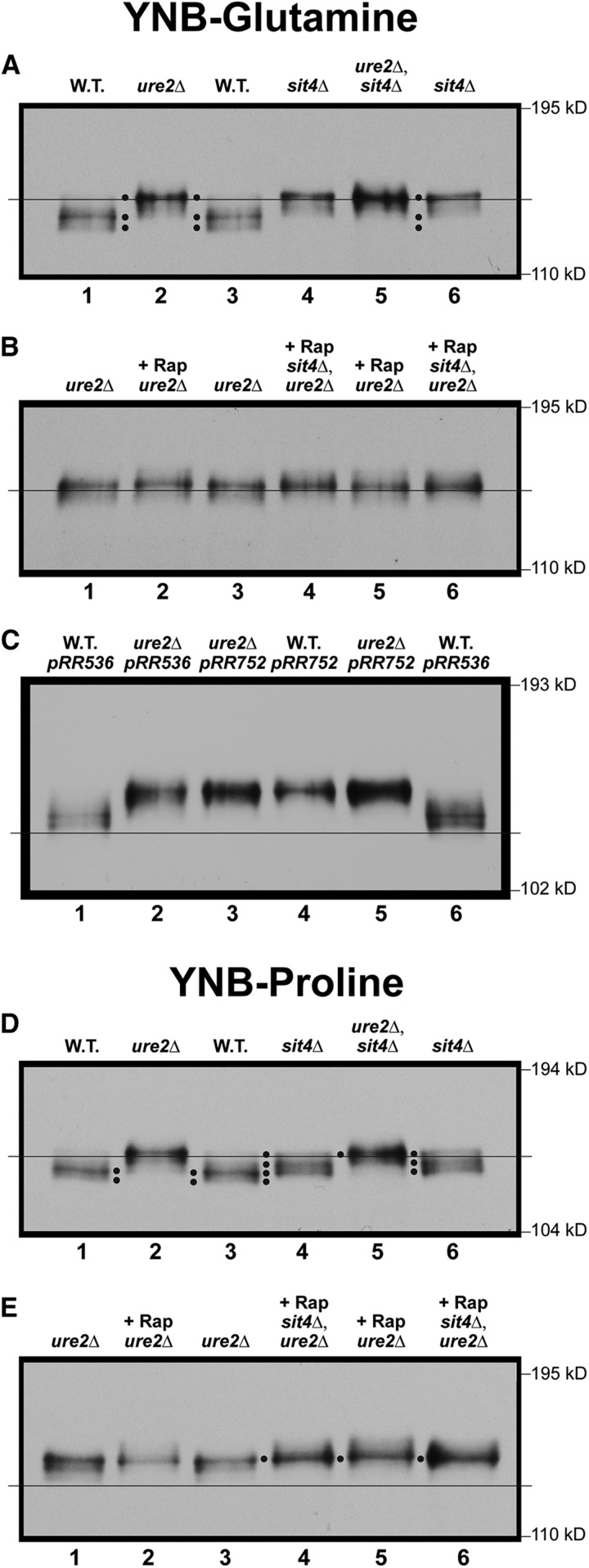
Therefore, to determine the Gln3-Myc13 localization/phosphorylation relationships, we compared them in glutamine-grown wild-type, ure2Δ, and sit4Δ strains. Gln3-Myc13 mobility decreased in the glutamine-grown ure2Δ cells (Figure 6A, lanes 1–3). In other words, Gln3-Myc13 restricted to the nuclei of ure2Δ cells was more highly phosphorylated than when it was sequestered in the cytoplasm.
Figure 6.
Gln3 phosphorylation levels in glutamine- or proline-grown wild type, sit4Δ single and sit4Δ,ure2Δ double-mutant cells in the presence (+Rap) or absence of rapamycin treatment. Wild type (TB123), sit4Δ (TB136-2a), ure2Δ (TB138-1a), and ure2Δ,sit4Δ (FV071) strains were grown to an A600nm ∼0.5 in YNB-glutamine (A–C) or YNB-proline (D and E). (B and E) Untreated cultures were sampled without addition of rapamycin or following 20 min incubation with 200 ng/ml rapamycin (+Rap). (C) Wild type (JK9-3da) or ure2Δ mutant (RR215) were transformed with wild-type (pRR536) or gln3 NES mutant (pRR752) plasmids and grown as described in (A). All samples were processed for western blot analysis as described in Materials and Methods.
When we repeated this experiment with a sit4Δ, where the URE2 gene is notably wild type and Gln3-Myc13 sequestered in the cytoplasm (Figure 1), Gln3-Myc13 mobility also decreased, detectably to the same position observed with the ure2Δ mutant (Figure 6A, lanes 2–4 and 6). Gln3-Myc13 mobility in the ure2Δ,sit4Δ double mutant exhibited the same mobility as the ure2Δ or sit4Δ single mutants (Figure 6A, lanes 2, 4–6).
Beyond the positive correlation of nuclear Gln3-Myc13 localization and its hyper-phosphorylation demonstrated with the ure2Δ strain, the striking and unexpected conclusion of the above experiments was that Ure2 and Sit4 functioned together to maintain Gln3-Myc13 at the hypo-phosphorylated level observed in repressed, glutamine-grown, wild-type cells. Neither protein alone was sufficient to achieve this result. It is important to recognize that in a sit4Δ single mutant, Gln3-Myc13 is efficiently sequestered in the cytoplasm. Therefore, the decreased Gln3-Myc13 mobility (increased phosphorylation) observed in the sit4Δ single mutant cannot be accounted for as deriving from Gln3-Myc13 being nuclear and Sit4 cytoplasmic.
Since increased Gln3-Myc13 phosphorylation in the sit4Δ mutant was clearly a cytoplasmic event, results with the ure2Δ,sit4Δ double mutant could possibly be interpreted in two different ways: Gln3 being nuclear in the double mutant and Sit4 localizing only to the cytoplasm (Tate et al. 2006), it could be argued that Sit4 did not influence nuclear Gln3-Myc13 phosphorylation levels, because Gln3-Myc13 being nuclear in the ure2Δ,sit4Δ double mutant was inaccessible to Sit4 (Tate et al. 2006). However, it could be alternatively argued that Ure2 is required to maintain Gln3-Myc13 in the cytoplasm, where it is accessible to Sit4. Hence, the requirement of both proteins to maintain Gln3-Myc13 in its hypo-phosphorylated form.
Rapamycin treatment had no demonstrable effect irrespective of whether assayed in the ure2Δ or ure2Δsit4Δ double mutants (Figure 6B).
In both ure2Δ and gln3 NES mutants, Gln3-Myc13 localizes to the nucleus. One as a result of loss of the negative regulation of Ure2, and the other due to the inability of Gln3-Myc13 to exit from the nucleus. We, therefore, wanted to ascertain whether Gln3-Myc13 phosphorylation was the same or different in the two mutants. We compared Gln3-Myc13 phosphorylation in a glutamine-grown strain containing the gln3 NES mutation (pRR752) alone or in combination with a ure2Δ mutation. Gln3-Myc13 exhibited the same electrophoretic mobility whether derived from the gln3 NES, ure2Δ or double mutant strain (Figure 6C). This again demonstrated that: (i) the nuclear localization of Gln3-Myc13 correlated with its hyper-phosphorylation even though two different methods were used to sequester it in that location. (ii) There were no additional effects of the two mutations together.
Since Sit4 had been demonstrated above to function during growth under highly repressive conditions, did derepressive conditions, (proline medium), where TorC1 is downregulated and Gln3 more nuclear, yield the same or different results? Gln3-Myc13 was again more phosphorylated in a proline-grown ure2Δ mutant than in wild type, just as occurred in the repressive glutamine medium (Figure 6D, lanes 1–3). It was also more phosphorylated in the sit4Δ mutant relative to wild type, but to a lesser degree than in ure2Δ cells (Figure 6D, lanes 1–4). Only one of four Gln3-Myc13 species observed in sit4Δ cells comigrated with the hyper-phosphorylated Gln3-Myc13 species observed in the ure2Δ mutant. The remaining three species, which accounted for the bulk of the Gln3-Myc13 in the sit4Δ mutant, migrated more rapidly (Figure 6D, lanes 3 and 4, four black dots). Nonetheless, the least phosphorylated Gln3-Myc13 species in sit4Δ cells was still more highly phosphorylated than seen in the wild type.
Therefore, both Sit4 and Ure2 were again required to maintain Gln3 at its wild-type hypo-phosphorylated level, this time in derepressive, proline medium. These data also suggested that Sit4 performed less Gln3-Myc13 dephoshorylation in proline than in glutamine medium. Interestingly, the profile of Gln3-Myc13 phosphorylation was similar to that observed after 4 hr of nitrogen starvation in wild-type and sit4Δ cells (Figure 3A). This observation correlates with the fact nuclear Gln3-Myc13 localization elicited by short-term nitrogen starvation and growth in a poor nitrogen source (proline) required active Sit4 (Tate and Cooper 2013). As occurred in glutamine medium, rapamycin treatment did not demonstrably affect Gln3-Myc13 phosphorylation in either strain (Figure 6E).
Three important conclusions collectively emanated from the above experiments: (i) Gln3-Myc13 was much more highly phosphorylated in the nucleus of a ure2Δ mutant than in the cytoplasm. (ii) Sit4 actively dephosphorylated Gln3-Myc13 in highly repressive, nitrogen-replete conditions where TorC1 was activated. (iii) Remarkably, both Sit4 and Ure2 were required to maintain Gln3-Myc13 in its hypo-phosphorylated form in the cytoplasm. None of these conclusions are accommodated by the scenario in Figure 1.
PP2A dephosphorylates both cytoplasmic and nuclear Gln3-Myc13 when TorC1 is up- and downregulated
Previous work demonstrated that, in addition to Sit4, PP2A phosphatase (Pph21/22-Tpd3-Cdc55/Rts1) is required for Gln3 to relocate to the nucleus when glutamine-grown cells are treated with rapamycin (Tate et al. 2009). Therefore, we queried whether loss of PP2A activity generated the same outcomes as that of Sit4 in glutamine-grown cells. To that end, we performed western blot assays analogous to those in Figure 6.
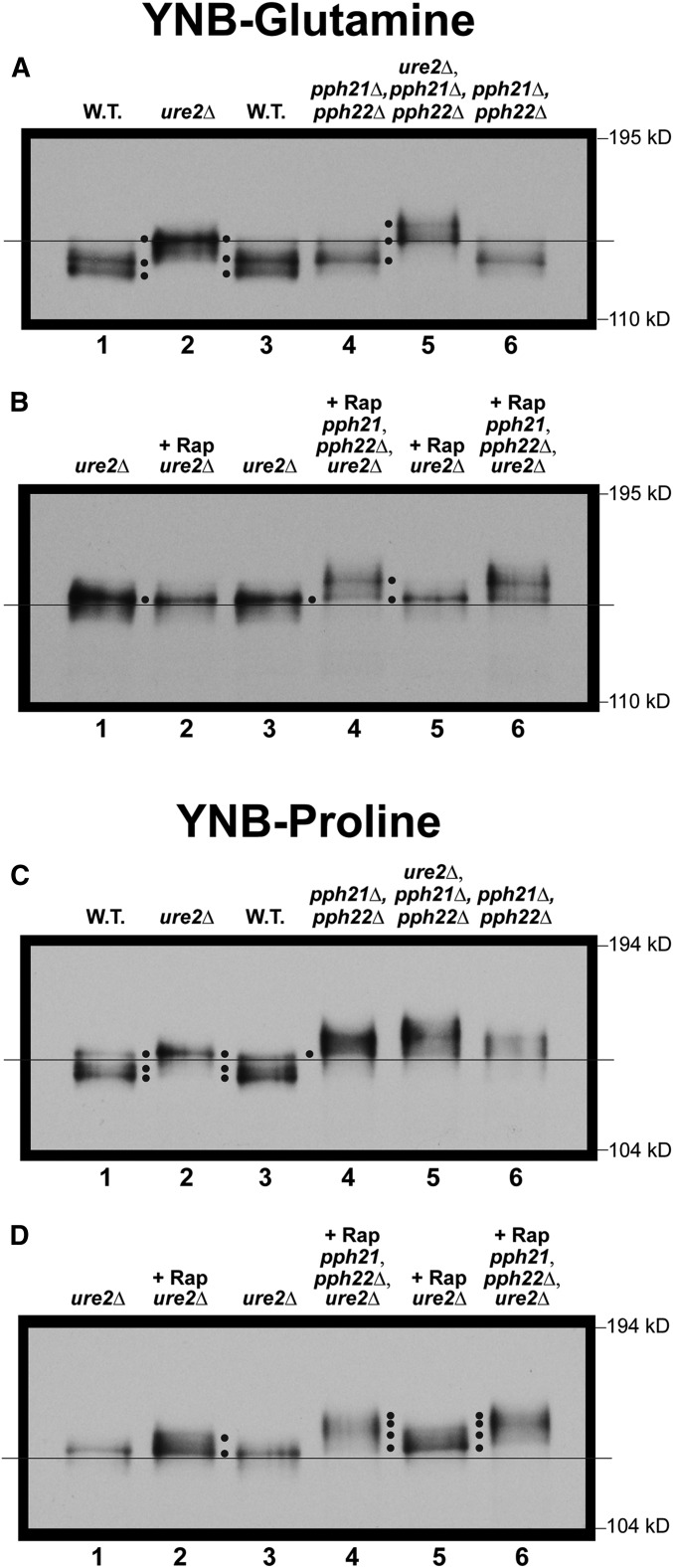
There was a clear decrease in Gln3-Myc13 mobility relative to wild type, i.e., phosphorylation increased, when PP2A catalytic activity was abolished in a glutamine-grown pph21Δ,pph22Δ (Figure 7A, lanes 1, 3, 4 and 6). These data indicated that, like Sit4, PP2A dephosphorylated cytoplasmic Gln3-Myc13, even though TorC1 was highly activated by a replete nitrogen supply—a completely unexpected result since PP2A is downregulated when TorC1 is most active (Figure 1). Unlike the sit4Δ in Figure 6, however, this increase did not mirror that of the ure2Δ; it was less (Figure 7A, cf. lanes 2 and 4). Yet it was clear that both Ure2 and PP2A were required to maintain Gln3-Myc13 at the hypo-phosphorylated level observed in the wild type glutamine-grown cells (Figure 7A, lanes 2–4).
Figure 7.
Gln3 phosphorylation levels in glutamine- or proline-grown wild type, ure2Δ, or pph21Δ,pph22Δ double-mutant or pph21Δ,pph22Δ,ure2Δ triple-mutant cells in the presence or absence of rapamycin treatment. Wild type (TB123), and pph21Δ,pph22Δ (03705d), ure2Δ (TB138-1a) and pph21Δ,pph22Δ,ure2Δ (FV165) mutant strains were grown to an A600nm = ∼0.5 in YNB-glutamine (A and B) or YNB-proline (C and D). The untreated cultures were sampled without addition of rapamycin (A and C) or following 20 min incubation with 200 ng/ml rapamycin (+Rap) (B and D). The cultures were then sampled, and the samples processed for western blot analysis.
Importantly, when the experiment was carried out in a ure2Δ,pph21Δ,pph22Δ triple mutant, where Gln3-Myc13 is nuclear (Georis et al. 2011), the level of Gln3-Myc13 mobility decreased significantly further than observed in the ure2Δ alone (Figure 7A, lanes 4–6). This was also a quite different result from that observed with the sit4Δ mutant, and indicated that PP2A dephosphorylated Gln3-Myc13 after its dissociation from Ure2 or within the nucleus. As occurred with the sit4Δ mutant, rapamycin had little effect on the outcomes with glutamine-grown ure2Δ and ure2Δ,pph21Δ,pph22Δ mutants (Figure 7B).
When the experiments in Figure 7, A and B were repeated in proline medium, the pph21Δ,pph22Δ mutant not only yielded a Gln3-Myc13 profile exhibiting greater phosphorylation than the wild type, but also even more than observed in the ure2Δ mutant (Figure 7C, lanes 1–4). In the ure2Δ,pph21Δ,pph22Δ triple mutant, the response was much like that in glutamine, i.e., Gln3-Myc13 phosphorylation was additively greater than observed with either the ure2Δ or pph21Δ,pph22Δ mutant alone (Figure 7C, lanes 4–6). This was consistent with previous findings that (i) PP2A more actively dephosphorylates Gln3 in derepressive proline than repressive glutamine medium (Tate et al. 2009), and (ii) PP2A locates to both the nucleus and cytoplasm (Georis et al. 2011). Therefore, as with Sit4, PP2A performed multiple roles, but additionally, likely in different cellular compartments as well.
There was another interesting difference between the Gln3-Myc13 phosphorylation profiles derived from glutamine- vs. proline-grown ure2Δ cells. Rapamycin had no demonstrable effect on Gln3-Myc13 mobility when the ure2Δ mutant was cultured in glutamine medium (Figure 7B, lanes 1–3). In contrast, when proline was employed as the nitrogen source, rapamycin treatment decreased Gln3-Myc13 mobility beyond that observed with untreated ure2Δ cells (Figure 7D, lanes 1–3). These data suggested that PP2A dephosphorylated Gln3-Myc13 more extensively when both PP2A and PTap42-PP2A were active after Gln3-Myc13 was dissociated from Ure2, or was situated in the nucleus.
Abolishing PP2A increases phosphorylation of Gln3-Myc13 sequestered in the nucleus
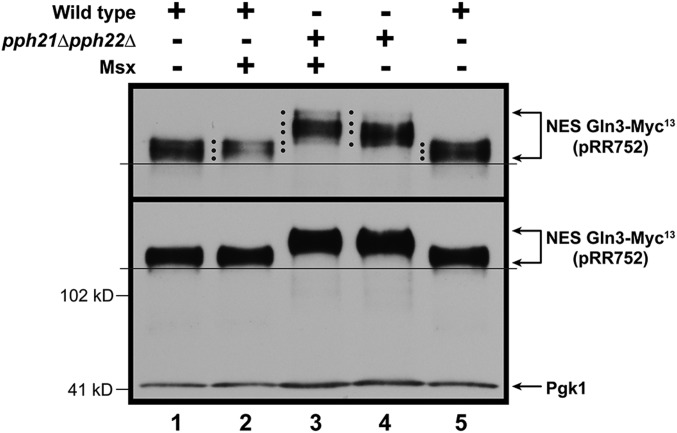
If PP2A was indeed dephosphorylating nuclear Gln3-Myc13, one might expect to see an effect of abolishing PP2A in cells where Gln3 was confined to the nucleus, i.e., in a transformant containing the gln3 NES mutant (pRR752). To test this possibility, we first assayed Gln3-Myc13 phosphorylation in untreated and Msx-treated, ammonia-grown wild-type transformants containing the gln3 NES mutant (pRR752). As demonstrated earlier, Msx treatment did not further increase Gln3-Myc13 phosphorylation relative to the untreated cells (Figure 8 lanes 1 and 2). This was not unexpected, because Gln3-Myc13 was nuclear in both conditions. However, when this experiment was repeated employing a pph21Δ,pph22Δ transformation recipient, Gln3-Myc13 phosphorylation increased significantly relative to the wild type recipient (Figure 8, lanes 3 and 4). Further, if Msx was added to the pph21Δ,pph22Δ transformant, there was one minor, additional, Gln3-Myc13 species with a slower mobility than observed in the untreated cells (Figure 8, lanes 3 and 4, top black dot). Also, the most rapidly migrating NES Gln3-Myc13 species observed in lane 4 is largely absent from lane 3 in which Msx was added to the pph21Δ,pph22Δ mutant (lanes 3 and 4, bottom black dot). These data support the contention that PP2A is quite likely dephosphorylating Gln3-Myc13 within the nucleus.
Figure 8.
Effects of abolishing PP2A when Gln3 is restricted to the nucleus. Wild type (JK9-3da) and pph21Δ,pph22Δ (FV239) were transformed with pRR752 (NES Gln3-Myc13), and grown in ammonia medium in the presence or absence of Msx for 30 min. Sample collection and western blot analysis were as described in Materials and Methods. Upper panel data derived from a six percent acrylamide gel, whereas the lower panel data derived from a parallel sample of the same culture, but electrophoresed for a shorter time in a 7% gel to retain the loading control, Pgk1, within the gel.
Note that the lower panel of Figure 8 depicts a film derived from the same samples as used in the upper panel, but resolved under electrophoretic conditions where there is insufficient resolution of the various Gln3-Myc13 species (higher percentage gel and shorter time of electrophoresis), but the entire blot, including the Pgk1 loading standard and potential Gln3 degradation products, are visible. This blot exhibits very little, if any, proteolytic NES Gln3-Myc13 degradation products compared with the blots in Figure 5, in which the cells were subjected to starvation.
PP2A-dependent phosphorylation as Gln3-Myc13 moves into, and out of, the nucleus
The observations that PP2A dephosphorylated Gln3-Myc13 when TorC1 was activated or inhibited, predicted that we should be able to follow Gln3-Myc13 dephosphorylation as Gln3 cycled into and out of the nucleus, as observed with Sit4. To test this prediction, we nitrogen-starved a pph21Δ,pph22Δ mutant culture for 1, 4, and 10 hr, as previously done with the wild type and sit4Δ mutant strains in Figure 5. Note that nuclear Gln3-Myc13 localization did not exhibit a requirement for PP2A upon short- or long-term nitrogen starvation (Figure 9, A and B). We then added glutamine to the starved culture. Gln3-Myc13 substantially relocated from the nucleus to the cytoplasm within 1 min after being refed, and was completely cytoplasmic within 5 min and thereafter.
Figure 9.
(A–C) Time course of Gln3-Myc13 intracellular localization and Gln3-Myc13 phosphorylation during short- and long-term nitrogen starvation of pph21Δ,pph22Δ cells followed by refeeding of excess nitrogen (0.1% glutamine final concentration). These experiments were performed, and the data evaluated and presented as described in Figure 5, except that a pph21Δ,pph22Δ mutant (03705d) was used in place of the sit4Δ mutant strain. Data are averages, and SD of four biological replicates for starvation and two for refeeding. (D) A film that was exposed for a shorter period of time to facilitate evaluation of the loading control, Pgk1.
The mobility of Gln3-Myc13 species decreased much more rapidly and continuously in the pph21Δ,pph22Δ mutant than it had in sit4Δ cells (Figure 9, lanes 1–4 vs. Figure 5F, 1–4). By 4 hr, Gln3-Myc13 mobility in the pph21Δ,pph22Δ mutant decreased to the same level as fully starved cells at 10 hr. In contrast, 10 hr were required for Gln3-Myc13 mobility to reach this minimum in the sit4Δ mutant, i.e., greatest degree of phosphorylation. Also the loss of PP2A greatly diminished the ability of the cell to dephosphorylate Gln3-Myc13 after being refed glutamine (Figure 9, lanes 5–8). Even at 30 min postrefeeding, Gln3-Myc13 was still much more phosphorylated than in untreated cells (Figure 9, lanes 1 and 8). These data indicated that (i) PP2A was much more responsive than Sit4 to the onset of nitrogen starvation, i.e., NCR-sensitivity, and (ii) it was much more responsible for overall Gln3 dephosphorylation than Sit4 when Gln3 departed from the nucleus under conditions where TorC1 was activated.
Effects of nitrogen starvation and refeeding on NCR-sensitive gene expression in wild-type and mutant cells
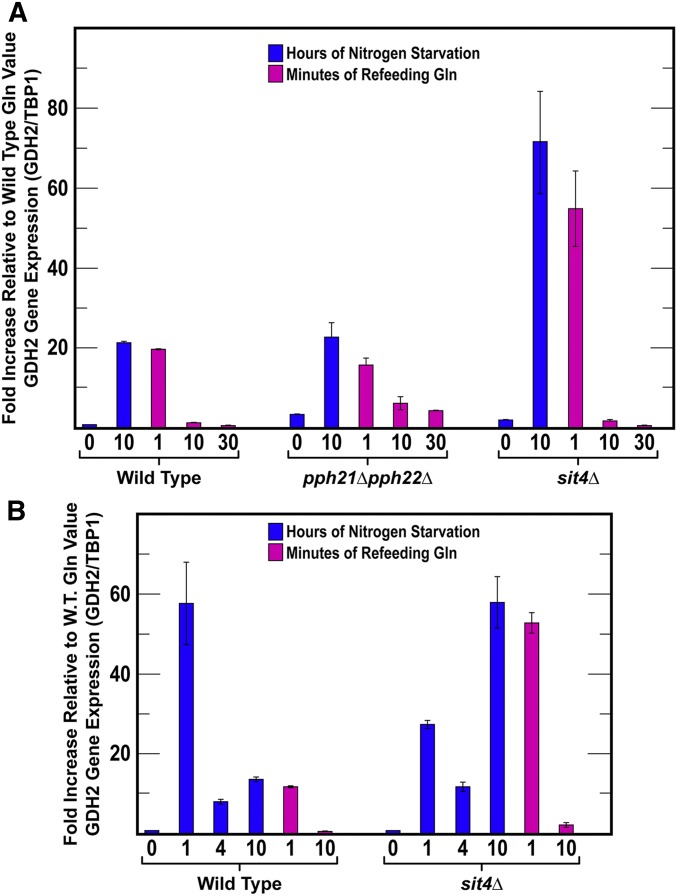
Finally, it is reasonable to ask how the phosphorylation data correlate with NCR-sensitive gene expression. To answer this, we used qRT-PCR to assess the effects of sit4Δ and pph21Δ,pph22Δ mutations on the expression of a highly NCR-sensitive gene: GDH2. It is important to recognize the following constraints when attempting to interpret data from this comparison. Sit4 and PP2A are required neither for nuclear Gln3 localization in response to long-term nitrogen starvation nor the imposition of NCR. However Sit4 is required for a response to short-term nitrogen starvation.
We initially compared GDH2 mRNA levels in wild-type and mutant cells cultured with a rich nitrogen source, glutamine, and then transferred them to nitrogen-free medium for 10 hr. We then added glutamine and sampled the cultures again at 1, 10, and 30 min as in previous experiments.
GDH2 mRNA in wild type and pph21Δ,pph22Δ increased to similar levels following 10 hr of nitrogen starvation (Figure 10A). They also similarly decreased a small amount at 1 min postrefeeding. Thereafter, mRNA levels in the pph21Δ,pph22Δ culture declined more slowly than in the wild type. Although this correlated to an extent with the decreased phosphorylation observed in the mutant, it was not sufficient to conclude existence of a cause-effect relationship between the two parameters. These were not surprising results because PP2A is required neither for nuclear Gln3 localization in response to long-term nitrogen starvation nor the imposition of NCR (note the steady state levels prior to starvation).
Figure 10.
(A and B) GDH2 expression in response to short- and long-term nitrogen starvation and refeeding excess nitrogen. The format of the experiment was the same as in Figure 5. GDH2 and TBP1 mRNA concentrations were determined as described in Materials and Methods. Data presented are the averages of three biological replicates.
GDH2 mRNA levels in the sit4Δ mutant differed markedly from those in wild-type and pph21Δ,pph22Δ cells (Figure 10A). After 10 hr of nitrogen starvation, sit4Δ cells contained three to four times as much GDH2 mRNA as the other two strains at the same duration of starvation. Loss of GDH2 mRNA in the sit4Δ mutant paralleled that in wild type. Suspicious that something had gone awry during starvation, we repeated the sit4Δ mutant experiment monitoring mRNA levels at 1, 4, and 10 hr of starvation (Figure 10B). After 1 hr of starvation, the sit4Δ mutant contained about half as much GDH2 mRNA as wild type. Recall that Sit4 is highly required for nuclear Gln3 localization during short-term nitrogen starvation. At 4 hr starvation, wild-type and sit4Δ mRNA levels were similar. However, at 10 hr of starvation, GDH2 mRNA in the sit4Δ mutant was again around four times that in the wild type. These data argued that, while Sit4 played a measurable and necessary role in GDH2 expression during short-term starvation, somewhat as expected, other factors were clearly contributed to regulation this expression as well.
Discussion
For nearly two decades, Sit4 and PP2A have been accepted to be regulated by TorC1 (Figure 1 and Figure 11). When TorC1 is activated, the phosphatases are inhibited (Figure 11, purple background). Conversely, when TorC1 is inactive, the phosphatases become active, and dephosphorylate downstream targets, e.g., Gln3 (yellow background). The present work demonstrates that Sit4 and PP2A are active irrespective of whether TorC1 is activated or inactive (blue and yellow background). An implied corollary of the TorC1 regulatory model has been that nuclear Gln3 is dephosphorylated (Figure 1). In contrast, we show that, except in rapamycin-treated cells, nuclear Gln3 is more highly phosphorylated than when it is in the cytoplasm (Figure 11, blue background). Further, upon relocating to the cytoplasm in response to excess nitrogen, Gln3 is dephosphorylated by Sit4 and PP2A. Finally, we show that Sit4 and PP2A are required, along with Ure2, to maintain Gln3 in a dephosphorylated form when situated in the cytoplasm of cells grown under highly repressive conditions (Figure 11, blue background).
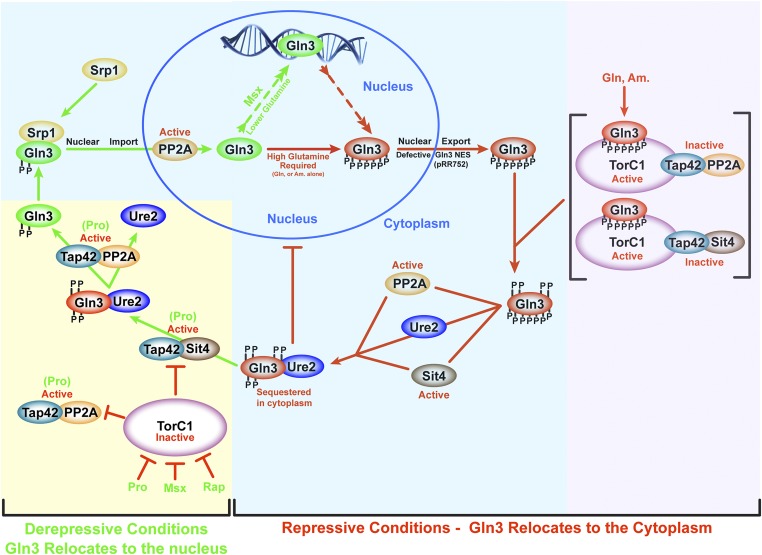
Figure 11.
Schematic working summary of data investigating PP2A and Sit4 phosphatases in the regulation of Gln3 under repressive [glutamine (Gln), ammonia (Am.) as nitrogen source; red arrows or bars] or derepressive [proline (Pro) as nitrogen source, or following rapamycin or Msx treatment; green arrows)] conditions. The phosphate groups indicated in the diagram are only illustrative of relative degrees of phosphorylation. It is important to emphasize that the specific order of the depicted reactions are not known beyond the limits of the data and conclusions presented in the Discussion. Yellow and purple background highlighting indicates TorC1-dependent phosphatase regulation, whereas blue background highlighting indicatesTorC1-independent phosphatase regulation.
These conclusions derive from a systematic investigation of the functional roles played by the Sit4 and PP2A phosphatases in NCR-sensitive Gln3 regulation. The data obtained give a more complete and detailed view of cellular compartment-specific Gln3 phosphorylation, and how Sit4 and PP2A participate in achieving this. Not only does the work elucidate new roles for these phosphatases under conditions not previously reported, it also opens a new dimension to the study of nitrogen-responsive Gln3 regulation.
However, limitations must also be acknowledged with respect to how far the data may be prudently interpreted. The experiments we described measured gross Gln3-Myc13 phosphorylation rather than the phosphorylation of specific Gln3 residues—an objective that is not yet technically attainable for us due to the lability of the disordered Gln3 molecule itself, and the fact that ∼20% of its residues (146 of 730) are serine or threonine. Therefore, one cannot, at this point, safely speculate about the residue-specific changes in the Gln3 molecule as it cycles into and out of the nucleus. That being said, this is not a limitation to interpreting the presence or absence of functioning phosphatases in comparison to Figure 1. Here, we used the Gln3 molecule as a reporter, thereby permitting determination of the conditions under which Sit4 and PP2A function, as well as their relation to Ure2 in maintaining hypo-phosphorylated Gln3 in excess nitrogen.
A second limitation must also be acknowledged. Figure 11 depicts orders of the regulatory reactions. In some cases, these orders have not been unambiguously demonstrated.
Sit4 and PP2A function when TorC1 is both inactive and highly active
Likely the most important and striking outcome of our experiments was the finding of previously undiscovered Sit4-and PP2A functions in the regulation of Gln3. As is now well known, both phosphatases mediate Gln3-Myc13 dephosphorylation when TorC1 is inactive. Remarkably, however, Sit4 and PP2A also dephosphorylate Gln3 in nitrogen-replete medium, where TorC1 is highly activated (Figure 11, yellow and blue backgrounds). Moreover, this TorC1-independent dephosphorylation more than likely occurs in the cytoplasm after Gln3 has exited from the nucleus (Figure 11, blue background). Supporting this idea, following the addition of excess nitrogen, Gln3 completely exits from the nuclei of nitrogen-starved cells in a much shorter time than it takes Gln3-Myc13 to be dephosphorylated.
Further support for the location of Gln3 dephosphorylation derives from the phenotypes of the sit4Δ and pph21Δ,pph22Δ mutants. The sit4Δ cells do not reach the Gln3-Myc13 dephosphorylation levels observed in unstarved cells even after 30 min of refeeding them excess nitrogen. Keep in mind that 5 min was sufficient for Gln3-Myc13 to fully relocate to the cytoplasm. When the PPH21,PPH22 genes were deleted, a similar result was obtained, except that much less dephosphorylation occurred after refeeding the starved pph21Δ,pph22Δ cells. Yet, when both phosphatases were present, as in the wild type, dephosphorylation decreased to the level observed in the initial, unstarved cells.
How might one reconcile the above conclusion, conceding that activated TorC1 inhibits Sit4 and PP2A, as repeatedly reported in the literature? The most likely explanation is that Sit4 and PP2A exist in multiple forms, both free and complexed with Tap42 (Figure 11). Di Como and Arndt reported that <5% of Sit4 and <2% of Pph21 is bound to Tap42 (DiComo and Arndt 1996). It is the PTap42-Sit4 and PTap42-PP2A complexes that are inhibited by activated TorC1 (DiComo and Arndt 1996; Beck and Hall 1999; Jiang and Broach 1999; Wang et al. 2003; Yan et al. 2006). The remaining free Sit4 and PP2A are still available to mediate other regulatory functions. We speculate that it is the free Sit4 and PP2A that dephosphorylate Gln3 after it exits from the nucleus. This reasoning explains the early paradoxical observation that Sit4-dependent Gln3 dephosphorylation is much greater in repressive, nitrogen-rich medium than in derepressive, nitrogen-poor medium (Tate et al. 2006). PP2A, on the other hand, functions in both repressive and derepressive conditions, albeit much more so in derepressive conditions (Tate and Cooper 2008).
Finally, both PTap42-Sit4 and PTap42-PP2A remain functional when TorC1 is inhibited via rapamycin treatment, or when cells are provided with a poor nitrogen source. We speculate that PTap42-PP2A functions downstream of PTap42-Sit4 because rapamycin is incapable of eliciting Sit4-dependent nuclear Gln3-Myc13 localization in a pph21Δ,pph22Δ mutant (Figure 11, yellow background) (Tate et al. 2009).
Sit4, PP2A and Ure2 are all required to maintain hypo-phosphorylated, cytoplasmic Gln3
Not only did we find that Sit4 and PP2A dephosphorylate Gln3-Myc13 in nitrogen-replete conditions, but, equally surprising, they and Ure2 are all required to maintain Gln3-Myc13 in its cytoplasmic hypo-phosphorylated form (Figure 11, blue background). One interpretation of these results is that Ure2 is required to maintain Gln3 within the cytoplasm, and the phosphorylation levels we observe in the glutamine-grown cells derive from Sit4 and PP2A functioning only when Gln3-Myc13 is cytoplasmic. That interpretation, however, does not answer the question of whether the phosphatases act prior to, or after, Gln3 forms a Gln3-Ure2 complex. Both are realistic possibilities; Zheng’s laboratory has shown that Ure2 binds to both phosphorylated and dephosphorylated Gln3 (Bertram et al. 2000). Two additional observations, however, impact on this question: (i) Ure2 exhibits greater affinity for dephosphorylated than phosphorylated Gln3 (Bertram et al. 2000), and (ii) overproduced Ure2 immuno-precipitates a small amount of a more slowly migrating species of Gln3 (Blinder et al. 1996). We speculate that, when its concentration is abnormally high, Ure2 may successfully compete with Sit4 and PP2A for Gln3 binding before the phosphatases have a chance to dephosphorylate it, thus accounting for the more slowly migrating species.
PP2A dephosphorylates Gln3 in both the cytoplasm and nucleus
In contrast with Sit4, which demonstrably dephosphorylates Gln3 only in the cytoplasm, PP2A likely dephosphorylates Gln3 in both the cytoplasm and nucleus. The clearest evidence for this conclusion is that, in Msx-treated pph21Δ,pph22Δ, the ure2Δ,pph21Δ,pph22Δ triple mutant, and the gln3 NES mutant, the loss of PP2A results in much higher levels of Gln3-Myc13 phosphorylation than observed in the untreated wild type or ure2Δ mutant strain. This interpretation positively correlates with our earlier results demonstrating that: (i) Pph21-Myc13 and Pph22-Myc13 reside in both the cytoplasm and nucleus; and (ii) PP2A is required for Gln3 to bind to some, but not all, NCR-sensitive promoter GATA sequence targets (Georis et al. 2011 a,b). These observations of intranuclear PP2A function, and the fact that Gln3-Myc13 is highly phosphorylated when restricted to the nucleus, are consistent with the proposed existence of two forms of nuclear Gln3-Myc13 (Rai et al. 2015). When glutamine levels are high, Gln3 can exit from the nucleus, even though it cannot bind to its target gene promoters, whereas, when glutamine is lowered, the exit of Gln3-Myc13 is prevented if DNA binding is abolished (Figure 11, blue background) (Rai et al. 2015).
Gene expression is not always a good reporter of transcription factor localization and function
The gene expression experiment in this work was performed to assess its correlation with Gln3 phosphorylation. At face value, the experiment was, at best, only a partial success. PP2A is not required for nuclear Gln3 localization during short- or long-term nitrogen starvation, and this correlated with GDH2 expression. On the other hand, Sit4 is required for nuclear Gln3 localization, and its loss could be detected in diminished GDH2 expression. Also, GDH2 expression correlated well with the rapid exit of Gln3 from the nucleus in the absence of PP2A and Sit4. GDH2 levels in the initial response to nitrogen starvation, and the final level after long-term nitrogen starvation, however, do not correlate well. This, too, is not unexpected because Gln3 phosphorylation and nuclear localization are only two of the parameters that contribute to overall gene expression. Additionally, six promoter elements are situated in the GDH2 promoter, and GDH2 expression is regulated by intracellular ammonia concentrations as well as NCR (Miller and Magasanik 1991; Tate and Cooper 2003). It is the complexity of transcriptional regulation that motivated us to depend increasingly on intracellular Gln3 localization as our preferred reporter to investigate nitrogen-responsive regulation. Although control of Gln3 localization is itself complicated, it is more straightforwardly associated to nitrogen-responsive regulation than NCR-sensitive transcription.
Questions still remain
Although we have learned a good deal more about Sit4 and PP2A function in the regulation of Gln3 from the present experiments, there are still questions to be answered and additional paradoxical observations that remain to be explained. For example, PP2A has been shown to dephosphorylate Gln3 in an NCR-sensitive manner (Tate et al. 2009). At what stage of Gln3 relocating to the nucleus does this occur? After Gln3 dissociates from Ure2? After Gln3 enters the nucleus? How does one explain that rapamycin elicits Sit4-dependent dephosphorylation and nuclear localization of Gln3, when all other conditions that elicit nuclear Gln3 localization increase its phosphorylation? Why is rapamycin-elicited Gln3 dephosphorylation insufficient to bring about nuclear Gln3 localization when PP2A activity is abolished (Tate et al. 2009)? Finally, how do Gln3 residue-specific and overall phosphorylation/dephosphorylation profiles correlate with one another? Clearly much remains to be done before we fully understand the complex control of Gln3 responding to multiple nitrogen-responsive regulatory mechanisms. However, using the tools at hand, each new finding moves us closer to that objective.
Acknowledgments
The authors express their gratitude to members of the University of Tennessee Health Science Center (UTHSC) Molecular Resource Center for providing the equipment and assistance for RNA and qRT-PCR analyses. This work was supported by National Institutes of Health (NIH) grant GM-35642-27 and the University of Tennessee Harriet S. Van Vleet Chair of Excellence.
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Beck T., Hall M. N., 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. 10.1038/45287 [DOI] [PubMed] [Google Scholar]
- Bertram P. G., Choi J. H., Carvalho J., Ai W., Zeng C., et al. , 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275: 35727–35733. 10.1074/jbc.M004235200 [DOI] [PubMed] [Google Scholar]
- Binda M., Péli-Gulli M. P., Bonfils G., Panchaud N., Urban J., et al. , 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35: 563–573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Binda M., Bonfils G., Panchaud N., Péli-Gulli M. P., De Virgilio C., 2010. An EGOcentric view of TORC1 signaling. Cell Cycle 9: 221–222. 10.4161/cc.9.2.10585 [DOI] [PubMed] [Google Scholar]
- Blinder D., Coschigano P. W., Magasanik B., 1996. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J. Bacteriol. 178: 4734–4736. 10.1128/jb.178.15.4734-4736.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., et al. , 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046. 10.1126/science.1176495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., 2012. Nutritional control of growth and development in yeast. Genetics 192: 73–105. 10.1534/genetics.111.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J., 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13: 3271–3279. 10.1101/gad.13.24.3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M., Schothorst J., Kankipati H. N., Van Zeebroeck G., Rubio-Texeira M., et al. , 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38: 254–299. 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., 1982. pp. 39–99 in Molecular Biology of the Yeast Saccharomyces. Metabolism and Gene Expression, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Cooper T. G., 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26: 223–238. 10.1111/j.1574-6976.2002.tb00612.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., 2004. pp. 225–257 in Nutrient-induced Responses in Eukaryotic Cells. Topics in Current Genetics, edited by Winderickx J., Taylor P. M. Springer Verlag, Berlin: 10.1007/978-3-540-39898-1_10 [DOI] [Google Scholar]
- Crespo J. L., Powers T., Fowler B., Hall M. N., 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99: 6784–6789. 10.1073/pnas.102687599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C. J., Arndt K. T., 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10: 1904–1916. 10.1101/gad.10.15.1904 [DOI] [PubMed] [Google Scholar]
- Feller A., Georis I., Tate J. J., Cooper T. G., Dubois E., 2013. Alterations in the Ure2 αCap domain elicit different GATA factor responses to rapamycin treatment and nitrogen limitation. J. Biol. Chem. 288: 1841–1855. 10.1074/jbc.M112.385054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I., Tate J. J., Cooper T. G., Dubois E., 2008. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J. Biol. Chem. 283: 8919–8929. 10.1074/jbc.M708811200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I., Tate J. J., Cooper T. G., Dubois E., 2011a Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J. Biol. Chem. 286: 44897–44912. 10.1074/jbc.M111.290577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I., Tate J. J., Feller A., Cooper T. G., Dubois E., 2011b Intranuclear function for protein phosphatase 2A: Pph21 and Pph22 are required for rapamycin-induced GATA factor binding to the DAL5 promoter in yeast. Mol. Cell. Biol. 31: 92–104. 10.1128/MCB.00482-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Hall M. N., 2017. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36: 397–408. 10.15252/embj.201696010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J. S., Kuruvilla F. G., Tong J. K., Shamji A. F., Schreiber S. L., 1999. Rapamycin-modulated transcription defines the subset of nutrient- sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96: 14866–14870. 10.1073/pnas.96.26.14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman-Bang J., 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 12: 35–73. 10.1385/MB:12:1:35 [DOI] [PubMed] [Google Scholar]
- Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., et al. , 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23: 1929–1943. 10.1101/gad.532109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett J. E., Luo X., Capaldi A. P., 2014. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198: 773–786. 10.1534/genetics.114.168369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A., 1983. Transfomation of intact yeast cells treated with alkali ions. J. Bacteriol. 53: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Broach J. R., 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18: 2782–2792. 10.1093/emboj/18.10.2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A. A., Abul-Hamd A. T., Rai R., El Berry H., Cooper T. G., 2001. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J. Biol. Chem. 276: 32136–32144. 10.1074/jbc.M104580200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A. A., Buford T. D., Rai R., Cooper T. G., 2006. Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae. FEMS Yeast Res. 6: 218–229. 10.1111/j.1567-1364.2006.00031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Thornton J., Spirek M., Butow R., 2008. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 28: 551–563. 10.1128/MCB.00929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl P. O., Daignan-Fornier B., 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190: 885–929. 10.1534/genetics.111.133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18. 10.1016/S0378-1119(02)00558-9 [DOI] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B., 1991. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 6229–6247. 10.1128/MCB.11.12.6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Tate J. J., Nelson D. R., Cooper T. G., 2013. gln3 mutations dissociate responses to nitrogen limitation (nitrogen catabolite repression) and rapamycin inhibition of TorC1. J. Biol. Chem. 288: 2789–2804. 10.1074/jbc.M112.421826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Tate J. J., Shanmuganatham K., Howe M. M., Cooper T. G., 2014. A domain in the transcription activator Gln3 specifically required for rapamycin responsiveness. J. Biol. Chem. 289: 18999–19018. 10.1074/jbc.M114.563668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Tate J. J., Shanmuganatham K., Howe M. M., Nelson D., et al. , 2015. Nuclear Gln3 import is regulated by nitrogen catabolite repression whereas export is specifically regulated by glutamine. Genetics 201: 989–1016. 10.1534/genetics.115.177725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Tate J. J., Cooper T. G., 2016. Multiple targets on the Gln3 transcription activator are cumulatively required for control of its cytoplasmic sequestration. G3 (Bethesda) 6: 1391–1408. 10.1534/g3.116.027615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K., Richards K., Botstein D., 1997. BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell 8: 2677–2691. 10.1091/mbc.8.12.2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., et al. , 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21: 3475–3486. 10.1091/mbc.e10-03-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen E., Ghillebert R., Wilms T., Winderickx J., 2014. Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 14: 17–32. 10.1111/1567-1364.12097 [DOI] [PubMed] [Google Scholar]
- Tate J. J., Cooper T. G., 2003. Tor1/2 regulation of retrograde gene expression in Saccharomyces cerevisiae derives indirectly as a consequence of alterations in ammonia metabolism. J. Biol. Chem. 278: 36924–36933. 10.1074/jbc.M301829200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Cooper T. G., 2008. Formalin can alter the intracellular localization of some transcription factors in Saccharomyces cerevisiae. FEMS Yeast Res. 8: 1223–1235. 10.1111/j.1567-1364.2008.00441.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Cooper T. G., 2013. Five conditions commonly used to down-regulate tor complex 1 generate different physiological situations exhibiting distinct requirements and outcomes. J. Biol. Chem. 288: 27243–27262. 10.1074/jbc.M113.484386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Rai R., Cooper T. G., 2005. Methionine sulfoximine treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J. Biol. Chem. 280: 27195–27204 (erratum: J. Biol. Chem. 282: 13139). 10.1074/jbc.M504052200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Feller A., Dubois E., Cooper T. G., 2006aSaccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J. Biol. Chem. 281: 37980–37992. 10.1074/jbc.M606973200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Rai R., Cooper T. G., 2006b Ammonia-specific regulation of Gln3 localization in Saccharomyces cerevisiae by protein kinase Npr1. J. Biol. Chem. 281: 28460–28469. 10.1074/jbc.M604171200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Georis I., Feller A., Dubois E., Cooper T. G., 2009. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J. Biol. Chem. 284: 2522–2534. 10.1074/jbc.M806162200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Georis I., Dubois E., Cooper T. G., 2010. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 285: 17880–17895. 10.1074/jbc.M109.085712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Buford D., Rai R., Cooper T. G., 2017. General amino acid control and 14–3-3 proteins Bmh1/2 are required for nitrogen catabolite repression-sensitive regulation of Gln3 and Gat1 localization. Genetics 205: 633–655. 10.1534/genetics.116.195800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Rai R., Cooper T. G., 2018. More than one way in: three Gln3 sequences required to relieve negative Ure2 regulation and support nuclear Gln3 import in Saccharomyces cerevisiae. Genetics 208: 207–227. 10.1534/genetics.117.300457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., et al. , 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26: 663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang X., Jiang Y., 2003. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 14: 4342–4351. 10.1091/mbc.e03-02-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Shen X., Jiang Y., 2006. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 25: 3546–3555. 10.1038/sj.emboj.7601239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabrocki P., Van Hoof C., Goris J., Thevelein J. M., Winderickx J., et al. , 2002. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol. Microbiol. 43: 835–842. 10.1046/j.1365-2958.2002.02786.x [DOI] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2009. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 2.1–2.55. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du G., Zhou J., Chen J., 2018. Regulation of sensing, transportation, and catabolism of nitrogen sources in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 82: pii: e00040-17. 10.1128/MMBR.00040-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids will be provided upon request, but only for noncommercial purposes. Commercial and commercial-development uses are prohibited. Materials provided may not be transferred to a third party without written consent. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.