Abstract
Maternally transmitted Wolbachia infect about half of insect species, yet the predominant mode(s) of Wolbachia acquisition remains uncertain. Species-specific associations could be old, with Wolbachia and hosts codiversifying (i.e., cladogenic acquisition), or relatively young and acquired by horizontal transfer or introgression. The three Drosophila yakuba-clade hosts [(D. santomea, D. yakuba) D. teissieri] diverged ∼3 MYA and currently hybridize on the West African islands Bioko and São Tomé. Each species is polymorphic for nearly identical Wolbachia that cause weak cytoplasmic incompatibility (CI)–reduced egg hatch when uninfected females mate with infected males. D. yakuba-clade Wolbachia are closely related to wMel, globally polymorphic in D. melanogaster. We use draft Wolbachia and mitochondrial genomes to demonstrate that D. yakuba-clade phylogenies for Wolbachia and mitochondria tend to follow host nuclear phylogenies. However, roughly half of D. santomea individuals, sampled both inside and outside of the São Tomé hybrid zone, have introgressed D. yakuba mitochondria. Both mitochondria and Wolbachia possess far more recent common ancestors than the bulk of the host nuclear genomes, precluding cladogenic Wolbachia acquisition. General concordance of Wolbachia and mitochondrial phylogenies suggests that horizontal transmission is rare, but varying relative rates of molecular divergence complicate chronogram-based statistical tests. Loci that cause CI in wMel are disrupted in D. yakuba-clade Wolbachia; but a second set of loci predicted to cause CI are located in the same WO prophage region. These alternative CI loci seem to have been acquired horizontally from distantly related Wolbachia, with transfer mediated by flanking Wolbachia-specific ISWpi1 transposons.
Keywords: cytoplasmic incompatibility, horizontal gene transfer, introgression, transposable elements, WO phage
ENDOSYMBIOTIC Wolbachia bacteria infect many arthropods (Bouchon et al. 1998; Hilgenboecker et al. 2008), including about half of all insect species (Werren and Windsor 2000; Weinert et al. 2015). Wolbachia often manipulate host reproduction, facilitating spread to high frequencies within host species (Laven 1951; Yen and Barr 1971; Turelli and Hoffmann 1991; Rousset et al. 1992; O’Neill et al. 1998; Weeks et al. 2007; Kriesner et al. 2016; Turelli et al. 2018). In Drosophila, reproductive manipulations include cytoplasmic incompatibility (CI) and male killing (Hoffmann et al. 1986; Hoffmann and Turelli 1997; Hurst and Jiggins 2000). CI reduces the egg hatch of uninfected females mated with Wolbachia-infected males, and recent work has demonstrated that WO prophage–associated loci cause CI (Beckmann and Fallon 2013; Beckmann et al. 2017, 2019; LePage et al. 2017). Although reproductive manipulations are common, some Wolbachia show little or no reproductive manipulation (e.g., wMel in Drosophila melanogaster, Hoffmann 1988; Hoffmann et al. 1994; Kriesner et al. 2016; wMau in D. mauritiana, Giordano et al. 1995; Meany et al. 2019; wAu in D. simulans, Hoffmann et al. 1996; wSuz in D. suzukii and wSpc in D. subpulchrella, Hamm et al. 2014; Cattel et al. 2018). These Wolbachia presumably spread by enhancing host fitness in various ways, with some support for viral protection, fecundity enhancement, and supplementation of host nutrition (Weeks et al. 2007; Hedges et al. 2008; Teixeira et al. 2008; Brownlie et al. 2009; Gill et al. 2014; Martinez et al. 2014; Moriyama et al. 2015; Kriesner and Hoffmann 2018). Better understanding of Wolbachia effects, transmission and evolution should facilitate using Wolbachia for biocontrol of human diseases by either transforming vector populations with virus-blocking Wolbachia (e.g., McMeniman et al. 2009; Hoffmann et al. 2011; Schmidt et al. 2017; Ritchie 2018) or using male-only releases of CI-causing Wolbachia to suppress vector populations (Laven 1967; O’Connor et al. 2012).
There is a burgeoning literature on Wolbachia frequencies and dynamics in natural populations (e.g., Kriesner et al. 2013; Kriesner et al. 2016; Cooper et al. 2017; Bakovic et al. 2018; Meany et al. 2019), but fewer studies elucidate the modes and timescales of Wolbachia acquisition by host species (O’Neill et al. 1992; Rousset and Solignac 1995; Huigens et al. 2004; Baldo et al. 2008; Raychoudhury et al. 2009; Ahmed et al. 2015; Schuler et al. 2016; Turelli et al. 2018). Sister hosts could acquire Wolbachia from their most recent ancestors. Such cladogenic acquisition seems to be the rule for the obligate Wolbachia found in filarial nematodes (Bandi et al. 1998); and there are also examples in at least two insect clades, Nasonia wasps (Raychoudhury et al. 2009) and Nomada bees (Gerth and Bleidorn 2016). In contrast, infections can be relatively young and acquired through introgressive or horizontal transfer (Raychoudhury et al. 2009; Schuler et al. 2016; Conner et al. 2017; Turelli et al. 2018). Comparisons of host nuclear and mitochondrial genomes with the associated Wolbachia genomes enable discrimination among cladogenic, introgressive, and horizontal acquisition (Raychoudhury et al. 2009; Turelli et al. 2018; see Supplemental Material, Figure S1). Concordant nuclear, mitochondrial, and Wolbachia cladograms—including consistent divergence-time estimates for all three genomes—support cladogenic acquisition and codivergence of host and Wolbachia lineages. Concordant Wolbachia and mitochondrial phylogenies and consistent Wolbachia and mitochondrial divergence-time estimates that are more recent than nuclear divergence support introgressive acquisition. In this case, mitochondrial and Wolbachia relationships may or may not recapitulate the host phylogeny. Finally, if Wolbachia diverged more recently than either nuclear or mitochondrial genomes, horizontal transfer (or paternal transmission; Hoffmann and Turelli 1988; Turelli and Hoffmann 1995) is indicated. This is often associated with discordance between host and Wolbachia phylogenies (O’Neill et al. 1992; Rousset and Solignac 1995; Turelli et al. 2018).
Introgressive Wolbachia acquisition may be common in Drosophila. About half of all closely related Drosophila species have overlapping geographical ranges and show pervasive evidence of reinforcement (Coyne and Orr 1989, 1997; Yukilevich 2012; Nosil 2013), indicating that hybridization must be common (Turelli et al. 2014). Several instances of sporadic contemporary hybridization and interspecific gene flow have been documented in the genus (Carson et al. 1989; Shoemaker et al. 1999; Jaenike et al. 2006; Kulathinal et al. 2009; Garrigan et al. 2012; Brand et al. 2013; Matute and Ayroles 2014; Lohse et al. 2015), but only two stable hybrid zones (HZs) have been well described, and both involve D. yakuba-clade species. In West Africa, D. yakuba hybridizes with endemic D. santomea on the island of São Tomé, and with D. teissieri on the island of Bioko (Lachaise et al. 2000; Comeault et al. 2016; Cooper et al. 2018). The ranges of D. yakuba and D. teissieri overlap throughout continental Africa, but contemporary hybridization has not been observed outside of Bioko (Cooper et al. 2018). Genomic analyses support both mitochondrial and nuclear introgression in the D. yakuba clade (Lachaise et al. 2000; Bachtrog et al. 2006; Llopart et al. 2014; Turissini and Matute 2017; Cooper et al. 2018), and the Wolbachia infecting all three species (wSan, wTei, and wYak) are at intermediate frequencies and identical with respect to commonly used typing loci (Lachaise et al. 2000; Charlat et al. 2004; Cooper et al. 2017).
Horizontal Wolbachia transmission has been repeatedly demonstrated since its initial discovery by O’Neill et al. (1992) (e.g., Baldo et al. 2008; Schuler et al. 2016), and it may be common in some systems (e.g., Huigens et al. 2000; Huigens et al. 2004; Ahmed et al. 2015; Li et al. 2017). Phylogenomic analyses indicate recent horizontal Wolbachia transmission, on the order of 5000–27,000 years, among relatively distantly related Drosophila species of the D. melanogaster species group (Turelli et al. 2018), but there is little evidence for nonmaternal transmission within Drosophila species (Richardson et al. 2012; Turelli et al. 2018). Horizontal Wolbachia transfer could occur via a vector (Vavre et al. 2009; Ahmed et al. 2015) and/or shared food sources during development (Huigens et al. 2000; Li et al. 2017). Rare paternal Wolbachia transmission has been documented in D. simulans (Hoffmann and Turelli 1988; Turelli and Hoffmann 1995). However, the most common mode of Wolbachia acquisition by Drosophila species (and most other hosts) remains unknown, and all three modes seem plausible in the D. yakuba clade (Lachaise et al. 2000; Bachtrog et al. 2006; Cooper et al. 2017).
Distinguishing acquisition via introgression vs. horizontal or paternal transmission requires estimating phylogenies and relative divergence times of mitochondrial DNA (mtDNA) and Wolbachia (Raychoudhury et al. 2009; Conner et al. 2017; Turelli et al. 2018). To convert sequence divergence to divergence-time estimates requires understanding relative rates of divergence for mtDNA, nuclear genomes, and Wolbachia. If each genome followed a constant-rate molecular clock, taxa with cladogenic Wolbachia transmission, such as Nasonia wasps (Raychoudhury et al. 2009) and Nomada bees (Gerth and Bleidorn 2016), would provide reference rates for calibration. However, like Langley and Fitch (1974), Turelli et al. (2018) found significantly varying relative rates of Wolbachia and mtDNA divergence. This variation, which we document across Drosophila, confounds attempts to understand Wolbachia acquisition, as discussed below.
The discovery of weak CI in the D. yakuba clade (Cooper et al. 2017) motivates detailed comparative analysis of loci associated with CI (CI factors or cifs) in WO prophage regions of Wolbachia genomes. Beckmann and Fallon (2013) first associated wPip_0282 and wPip_0283 proteins in wPip-infected Culex pipiens with Wolbachia-modified sperm. Later work confirmed that these proteins induce toxicity and produce rescue when expressed/co-expressed in Saccharomyces cerevisiae (Beckmann et al. 2017), lending support to a toxin-antidote model of CI (Beckmann et al. 2019; but see Shropshire et al. 2019). Homologs of these genes in wMel (WD0631 and WD0632) recapitulate CI when transgenically expressed in D. melanogaster (LePage et al. 2017), and transgenic expression of WD0631 in D. melanogaster rescues CI (Shropshire et al. 2018). A distantly related WO prophage–associated pair, present in wPip, wPip_0294 and wPip_0295, causes similar toxicity/rescue in S. cerevisiae (Beckmann et al. 2017), and causes CI when placed transgenically into a D. melanogaster background (M. Hochstrasser, unpublished data). We adopt Beckmann et al. (2019)’s nomenclature, which assigns names based on enzymatic activity of the predicted toxin [deubiquitylase (DUB) and nuclease (Nuc)], with superscripts denoting focal Wolbachia strains when needed. Specifically, we refer to wPip_0282-wPip_0283 and the wPip_0294-wPip_0295 pairs as cidA-cidBwPip and cinA-cinBwPip, respectively; and we refer to WD0631-WD632 as cidA-cidBwMel. This distinguishes cid (CI-inducing DUB) from cin (CI-inducing Nuc) pairs, with the predicted antidote and toxin denoted “A” and “B,” respectively (Beckmann et al. 2019). We acknowledge ongoing disagreement in the literature and direct readers to Beckmann et al. (2019) and Shropshire et al. (2019) for details. However, none of these debates on terminology or mechanism affect our findings.
Here, we use host and Wolbachia genomes from the D. yakuba clade to demonstrate introgressive and horizontal Wolbachia acquisition. General concordance of mitochondrial and Wolbachia phylogenies indicates that horizontal acquisition is rare within this clade. However, tests involving divergence-time estimates are complicated by varying relative rates and patterns of Wolbachia, mtDNA, and nuclear sequence divergence, as illustrated by data from more distantly related Drosophila (Drosophila 12 Genomes Consortium et al. 2007). Finally, we demonstrate that cid loci underlying CI in closely related wMel (LePage et al. 2017) are disrupted in all D. yakuba-clade Wolbachia. However, these Wolbachia also contain a set of cin loci, absent in wMel, but very similar to those found in wPip (Beckmann et al. 2017), a B-group Wolbachia strain that diverged 6–46 MYA from A-group wYak and wMel (Werren et al. 1995; Meany et al. 2019). This is the first discovery of two sets of loci implicated in CI co-occurring within the same prophage region. Several analyses implicate Wolbachia-specific insertion sequence (IS) transposable elements, specifically ISWpi1, in the horizontal transfer of these loci (and surrounding regions) between distantly related Wolbachia. Horizontal movement of incompatibility factors between prophage regions of Wolbachia variants adds another layer to what is already known about horizontal movement of prophages within and between Wolbachia variants that themselves move horizontally between host species.
Materials and Methods
Genomic data
The D. yakuba-clade isofemale lines included in our study were sampled over several years in West Africa (Comeault et al. 2016; Cooper et al. 2017; Turissini and Matute 2017). Each line within each species used in our analyses exhibits little nuclear introgression (<1%) (Turissini and Matute 2017), and no hybrids were included. Reads from D. yakuba (N = 56), D. santomea (N = 11), and D. teissieri (N = 13) isofemale lines were obtained from the data archives of Turissini and Matute (2017) and aligned to the D. yakuba nuclear and mitochondrial reference genomes (Drosophila 12 Genomes Consortium et al. 2007) with bwa 0.7.12 (Li and Durbin 2009), requiring alignment quality scores of at least 50. Because many of the read archives were single end, all alignments were completed using single-end mode for consistency.
mtDNA:
Consensus mtDNA sequences for each of the 80 lines were extracted with samtools v.1.3.1 and bcftools v.1.3.1 (Li 2011). Coding sequences for the 13 protein-coding genes were extracted, based on their positions in the D. yakuba reference. We also extracted the 13 protein-coding genes from additional unique D. yakuba (N = 28) and D. santomea (N = 15) mitochondrial genomes (Llopart et al. 2014), and from the D. melanogaster reference (Hoskins et al. 2015). Genes were aligned using MAFFT v.7 and concatenated (Katoh and Standley 2013). Lines identical across all 13 genes were represented by one sequence for the phylogenetic analyses.
Wolbachia:
Reads from each of the 80 Turissini and Matute (2017) lines were aligned using bwa 0.7.12 (Li and Durbin 2009) to the D. yakuba reference genome (Drosophila 12 Genomes Consortium et al. 2007) combined with the wMel reference genome (Wu et al. 2004). We calculated the average depth of coverage across the wMel genome. We considered lines with <1× coverage uninfected and lines with >10× coverage infected. No lines had between 1× and 10× coverage. To test our genomic analyses of infection status, we used a polymerase chain reaction (PCR) assay on a subset of lines. We extracted DNA using a “squish” buffer protocol (Gloor et al. 1993), and infection status was determined using primers for the Wolbachia-specific wsp gene (Braig et al. 1998; Baldo et al. 2006). We amplified a Drosophila-specific region of chromosome 2L as a positive control (Kern et al. 2015) (primers are listed in Table S1). For each run, we also included a known Wolbachia-positive line and a water blank as controls.
To produce a draft Wolbachia “pseudoreference” genome for each species, we first determined the isofemale line from each species that produced the greatest average coverage depth over wMel. We trimmed the reads with Sickle v.1.33 (Joshi and Fass 2011) and assembled with ABySS v.2.0.2 (Jackman et al. 2017). K values of 51, 61…91 were tried. Scaffolds with best nucleotide BLAST matches to known Wolbachia sequences with E-values <10−10 were extracted as the draft Wolbachia assembly. For each species, the assembly with the highest N50 and fewest scaffolds was kept as our Wolbachia pseudoreference genome for that host species, denoted wYak, wSan, and wTei (Table S2). To assess the quality of these three draft assemblies, we used BUSCO v.3.0.0 to search for homologs of the near-universal, single-copy genes in the BUSCO proteobacteria database (Simão et al. 2015). For comparison, we followed Conner et al. (2017) and performed the same search on the reference genomes for wMel (Wu et al. 2004), wRi (Klasson et al. 2009), wAu (Sutton et al. 2014), and wHa and wNo (Ellegaard et al. 2013) (Table S3).
Using these draft Wolbachia pseudoreference genomes, reads from all other genotypes were aligned to the D. yakuba reference (nuclear and mitochondrial) plus the species-specific Wolbachia draft assembly with bwa 0.7.12 (Li and Durbin 2009). Consensus sequences were extracted with samtools v.1.3.1 and bcftools v.1.3.1 (Li 2011).
To test whether the choice of Wolbachia pseudoreference influenced the Wolbachia draft sequence obtained for each infected line, we arbitrarily selected three infected lines of each host species and aligned reads from each of those lines independently to our wYak, wSan, and wTei pseudoreferences. This resulted in 27 alignments (nine per host species). Among the three Wolbachia consensus sequences generated for each line, we assessed single-nucleotide variants between different mappings of the same line within loci used for downstream analyses. Irrespective of the pseudoreference used, we obtained identical Wolbachia sequences. This indicates the robustness of our approach to generating population samples of draft Wolbachia genomes, without having a high-quality Wolbachia reference genome from any of the host species. Because Llopart et al. (2014) released only assembled mitochondrial genomes, we could not assess the Wolbachia infection status of their lines.
Loci for phylogenetic and comparative-rate analyses
Wolbachia genes:
For each phylogenetic analysis of Wolbachia data, the draft genomes were annotated with Prokka v.1.11 (Seemann 2014), which identifies homologs to known bacterial genes. To avoid pseudogenes and paralogs, we used only genes present in a single copy and with identical lengths in all of the sequences analyzed. Genes were identified as single copy if they uniquely matched a bacterial reference gene identified by Prokka v.1.11. By requiring all homologs to have identical length in all of our draft Wolbachia genomes, we removed all loci with indels across any of our sequences.
Given that many loci accumulate indels over time, the number of loci included in our phylogenetic analyses depended on the number of strains included. We first estimated phylograms for the A-group Wolbachia from the D. yakuba-clade, wMel from D. melanogaster (Wu et al. 2004), wInc from D. incompta (Wallau et al. 2016), wSuz from D. suzukii (Siozios et al. 2013), wAna from D. anannasae (Choi et al. 2015), Wolbachia that infect Nomada bees (wNFe, wNPa, wNLeu, and wNFa; Gerth and Bleidorn 2016), and Wolbachia that infect D. simulans (wRi, wAu, and wHa; Klasson et al. 2009; Ellegaard et al. 2013; Sutton et al. 2014). We also analyzed distantly related B-group Wolbachia: wNo from D. simulans (Ellegaard et al. 2013), wPip_Pel from C. pipiens (Klasson et al. 2008), and wAlbB from Aedes albopictus (Mavingui et al. 2012). To increase our phylogenetic resolution within the D. yakuba Wolbachia clade (by increasing the number of loci), we estimated a phylogram that included only D. yakuba-clade Wolbachia and wMel. Our phylogram with both A- and B-group Wolbachia included 146 genes (containing 115,686 bp), and our phylogram that included only D. yakuba-clade Wolbachia and wMel included 643 genes (containing 644,586 bp).
We also constructed an absolute chronogram to estimate divergence between D. yakuba-clade Wolbachia strains. To illustrate how divergence-time estimates vary with changing patterns of molecular divergence, we estimated two additional Wolbachia chronograms: one that considered only the wMel reference plus the two most-diverged wMel lines included in Richardson et al. (2012), and one that included these three wMel variants and our D. yakuba-clade Wolbachia. Our chronogram that included only D. yakuba-clade Wolbachia included 678 genes (containing 695,118 bp), our chronogram that included only wMel Wolbachia included 692 genes (containing 709,599 bp), and our chronograms with wMel plus D. yakuba-clade Wolbachia included 621 genes (containing 624,438 bp). Independent estimates were obtained using relaxed-clock analyses with (2,2) and (7,7) branch-rate priors. We also estimated divergence between D. yakuba-clade Wolbachia and wMel using a strict-clock analysis, corresponding to Γ(n,n) as n .
mtDNA and nuclear genes from diverse Drosophila:
To assess variation in the relative rates of divergence of mitochondrial and nuclear genes, we analyzed the canonical 12 Drosophila genomes (Drosophila 12 Genomes Consortium et al. 2007) plus D. suzukii (Chiu et al. 2013). We excluded D. sechellia and D. persimilis which show evidence of introgression with D. simulans and D. pseudoobscura, respectively (Kulathinal et al. 2009; Brand et al. 2013; Schrider et al. 2018). Coding sequences for 20 nuclear genes used in the analyses of Turelli et al. (2018) (aconitase, aldolase, bicoid, ebony, Enolase, esc, g6pdh, GlyP, GlyS, ninaE, pepck, Pgi, Pgm1, pic, ptc, Tpi, Transaldolase, white, wingless, and yellow) were obtained from FlyBase for each species. The genes were aligned with MAFFT v.7 (Katoh and Standley 2013). Coding sequences for the 13 protein-coding mitochondrial genes in the inbred reference strains were also obtained from FlyBase for each species and were aligned with MAFFT v.7 and concatenated.
Phylogenetic analyses:
All of our analyses used RevBayes v.1.0.9 (Höhna et al. 2016), following the procedures of Turelli et al. (2018). For completeness, we summarize those methods below. For additional details on the priors and their justifications, consult Turelli et al. (2018). Four independent runs were performed for each phylogenetic tree we estimated, and in all cases, the runs converged to the same topologies. Nodes with posterior probability <0.95 were collapsed into polytomies.
Wolbachia phylograms:
We estimated a phylogram for A- and B-group Wolbachia and for only D. yakuba-clade and wMel Wolbachia using the same methodology as Turelli et al. (2018). We used a GTR + Γ model with four rate categories, partitioning by codon position. Each partition had an independent rate multiplier with prior Γ(1,1) [i.e., Exp(1)], as well as stationary frequencies and exchangeability rates drawn from flat, symmetrical Dirichlet distributions [i.e., Dirichlet(1,1,1...)]. The model used a uniform prior over all possible topologies. Branch lengths were drawn from a flat, symmetrical Dirichlet distribution, thus they summed to 1. Since the expected number of substitutions along a branch equals the branch length times the rate multiplier, the expected number of substitutions across the entire tree for a partition is equal to the partition’s rate multiplier.
Wolbachia chronograms:
We first created a relaxed-clock relative chronogram with the root age fixed to 1 using the GTR + Γ model, partitioned by codon position, using the same birth-death prior as Turelli et al. (2018). Each partition had an independent rate multiplier with prior Γ(1,1), as well as stationary frequencies and exchangeability rates drawn from flat, symmetrical Dirichlet distributions. The branch-rate prior for each branch was (2,2), normalized to a mean of 1 across all branches (Table S4). We also tried a strict-clock tree and a relaxed-clock tree with branch-rate prior Γ(7,7), which produced no significant differences. We used the scaled distribution Γ(7,7) × 6.87 × 10−9 to model substitutions per third-position site per year. This transforms the relative chronogram into an absolute chronogram. This scaled distribution was chosen to replicate the upper and lower credible intervals of the posterior distribution estimated by Richardson et al. (2012), assuming 10 Drosophila generations per year, normalized by their median substitution-rate estimate. Branch lengths in absolute time were calculated as the relative-branch length times the third-position rate multiplier divided by the substitutions per third-position site per year estimate above.
We illustrate how divergence-time estimates depend on changing patterns of Wolbachia molecular evolution observed over different timescales, specifically the relative rates of third-site substitutions vs. first- and second-site substitutions. We compare divergence times estimated for variants within D. melanogaster-group host species, with a timescale of only hundreds or thousands of years, with the same divergence times estimated when more distantly related Wolbachia, with divergence times over tens of thousands of years, are included in the analyses. We present three separate analyses: one using only D. yakuba-clade Wolbachia variants, a second using only wMel variants analyzed by Richardson et al. (2012), and a third simultaneously analyzing both sets of data.
Drosophila phylogeny for 11 reference species:
We estimated a phylogram from our 20 nuclear loci using the GTR + Γ model, with partitioning by gene and codon position. The model was identical to the Wolbachia phylogram model above except for the partitioning (there are too few Wolbachia substitutions to justify partitioning by gene).
mtDNA phylogeny for the 11 reference species:
We estimated a phylogram from the 13 protein-coding mitochondrial loci using the GTR + Γ model, with partitioning only by codon position. The model was identical to the Wolbachia phylogram model above.
mtDNA chronogram for the D. yakuba clade:
We estimated a relative chronogram for the D. yakuba-clade mitochondria from the 13 protein-coding loci using the GTR + Γ model, partitioning only by codon position. To test the sensitivity of our results to priors, we ran a strict clock, a relaxed clock with a Γ(7,7) branch-rate prior, and a relaxed clock with a Γ(2,2) branch-rate prior, corresponding to increasing levels of substitution-rate variation across branches. The model was identical to the Wolbachia chronogram model above, except that we did not transform it into an absolute chronogram.
Ratios of divergence rates for mtDNA vs. nuclear loci:
To quantify ratios of mtDNA to nuclear substitution rates, we estimated relative substitution rates for host nuclear genes vs. mtDNA using the GTR + Γ model. The (unrooted) topology was fixed to the consensus topology for the nuclear and mitochondrial data. The data from 20 nuclear loci were partitioned by locus and codon position, the mtDNA data were partitioned only by codon position. All partitions shared the same topology, but the nuclear partitions were allowed to have branch lengths different from the mtDNA partitions. The sum of the branch lengths for each partition was scaled to 1. Assuming concurrent nuclear-mtDNA divergence (because we used only species showing no evidence of introgression), we imposed the same absolute ages for all nodes of the nuclear and mtDNA chronograms. For each nuclear locus, the third-position rate ratio for the mtDNA vs. nuclear genomes was calculated as follows: (mitochondrial branch length × mitochondrial third-position rate multiplier)/(nuclear branch length × nuclear third-position rate multiplier). We summarized the relative rates of mtDNA vs. nuclear substitutions along each branch using the arithmetic average of the 20 ratios obtained from the individual nuclear loci.
Introgressive vs. horizontal Wolbachia transfer–concordance of phylograms:
Horizontal transfer of Wolbachia was initially detected by discordance between Wolbachia and host phylogenies (O’Neill et al. 1992). When considering samples within species and closely related species, a comparison of mitochondrial and Wolbachia phylogenies provides a natural test for horizontal transmission. We first look for significant discrepancies between strongly supported nodes in the mitochondrial vs. Wolbachia phylogenies. To test for less obvious differences, we follow Richardson et al. (2012) and compute Bayes factors to assess the support for models that assume that mitochondrial vs. Wolbachia follow the same topology vs. distinct phylogenies. For these calculations, we omit D. santomea line Quija 37 due to its clear discordance.
We calculated the marginal likelihood in RevBayes for two models: one with a shared mitochondrial and Wolbachia phylogeny, and another with independent topologies. The mtDNA and Wolbachia were each partitioned by codon position, for a total of six partitions. In the shared phylogeny model, all six share the same topology; in the independent model, mitochondria and Wolbachia have separate topologies. All priors were the same as the Wolbachia phylogram model. Lines that were identical across the mtDNA and Wolbachia were collapsed into one sample. We ran two independent replicates of each model with 50 stepping stones per run. The Bayes factor is computed as the difference between the marginal likelihoods of each model.
Introgressive vs. horizontal Wolbachia transfer–relative rates:
Within species, the phylogenies of mitochondria and Wolbachia are fully resolved. Thus we consider an alternative approach for distinguishing between introgressive vs. horizontal transfer that depends on estimating divergence times for Wolbachia genomes and host mtDNA. This is complicated by variation in the relative rates of mtDNA vs. Wolbachia divergence and by systematic changes over time in the relative rates of substitutions at the three codon positions for Wolbachia. Following the procedures in Turelli et al. (2018), we estimated the mtDNA:Wolbachia third-position substitution-rate ratio for each branch in the D. yakuba clade. For each analysis, we computed the marginal likelihood of the model where all branches shared the same ratio and the same model, except allowing different ratios on each branch. We then calculated Bayes factors (i.e., differences in the log of the marginal likelihoods) to determine which model was favored. We repeated these analyses after including three wMel-infected D. melanogaster lines with the D. yakuba-clade data set. Because D. yakuba-clade species and D. melanogaster do not produce fertile hybrids (Sánchez and Santamaria 1997; Turissini et al. 2017), introgressive transfer of Wolbachia is not possible between these species. Hence, including wMel provides a control for our proposed test of introgressive transfer. For these controls, we calculated the mitochondrial vs. Wolbachia substitution-rate ratio for the interspecific branches separately, regardless of whether the shared rate ratio model was favored.
Wolbachia loci associated with CI
Wolbachia that infect all three D. yakuba-clade hosts cause weak intra-and interspecific CI (Cooper et al. 2017), but the genetic basis of CI in this clade remains unknown. We used tBLASTN to search for cif homologs in each of our D. yakuba-clade Wolbachia assemblies, querying cidA-cidB variants and the cinA-cinBwRi pair (wRi_006720 and wRi_006710) found in wRi and some wRi-like Wolbachia (Beckmann and Fallon 2013; Turelli et al. 2018), and the cinA-cinBwPip pair (Beckmann and Fallon 2013; Beckmann et al. 2017; LePage et al. 2017; Lindsey et al. 2018). We identified both cid and cin homologs, but we found no close matches to the cinA-cinBwRi pair (denoted Type II cif loci by LePage et al. 2017) in any of our genomes. For all of the samples, we extracted consensus sequences from our assemblies and alignments for the cidA-cidBwYak-clade and cinA-cinBwYak-clade gene pairs. The genes were aligned with MAFFT v.7 (Katoh and Standley 2013). We examined variation in these regions relative to cidA-cidBwMel and cinA-cinBwPip, respectively.
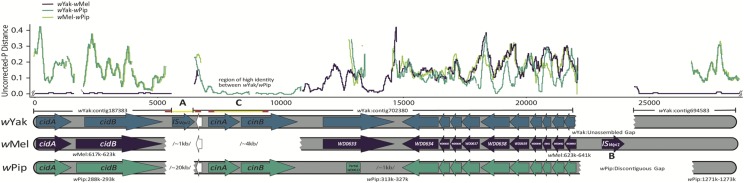
Because we unexpectedly identified homologs of cinA-cinB in all wYak-clade Wolbachia, we took additional measures to understand the origin and placement of these loci in the genomes. The cinA-cinBwYak-clade open reading frames (ORFs) are located on a ∼11,500 bp scaffold in the fragmented wYak assembly (wYak scaffold “702380” from ABySS output; we use quotes to indicate names assigned by ABySS). The first ∼4000 bp of this scaffold contain cinA and cinB genes that have ∼97% identity with those in wPip. We performed a BLAST search using all contigs in the wYak assembly as queries against the wMel genome (Camacho et al. 2009). The cinA-cinBwYak-clade scaffold was placed on the wMel genome (∼617,000–623,000) adjacent to and downstream of the wYak contig containing cidA-cidBwYak-clade loci (wYak contig “187383”). However, only ∼7000 bp of the 11,500 bp of the scaffold containing cinA-cinBwYak-clade align to this region of wMel. The ∼4000 bp sequence that contains the ORFs for cinA-cinBwYak-clade has no significant hit against the wMel genome.
To verify the placement of the scaffold containing cinA-cinBwYak-clade, we first performed targeted assembly using the cinA-cinBwYak-clade contig and the adjacent contigs as references (Hunter et al. 2015). Iterative targeted assembly often extends assembled scaffolds (Langmead and Salzberg 2012). Reads are mapped to the target of interest, and the mapped reads are then assembled using SPADES (Bankevich et al. 2012; Nurk et al. 2013). The newly assembled scaffolds serve as the target in a subsequent round of mapping. The procedure is repeated until no new reads are recruited to the assembled scaffold from the previous round. The extended scaffolds enabled us to merge flanking wYak contigs downstream of the cinA-cinBwYak-clade scaffold but failed to connect the fragment with the cidA-cidBwYak-clade contig. We used PCR to amplify the intervening region (Table S1), and Sanger sequencing to evaluate this product.
Subsequent mapping of paired-end reads to the merged scaffolds confirmed the correct order and orientation of the contigs containing the cidA-cidBwYak-clade and cinA-cinBwYak-clade genes. Observed, uncorrected, pairwise distances between focal regions in wYak, wMel, and wPip and between focal regions in wYak, wPip, wAlbB, and wNleu were calculated using a sliding window (window size = 200 bp, step size = 25 bp).
Data availability
The Wolbachia assemblies (BioProject PRJNA543889, accession numbers GCA_005862115.1, GCA_005862095.1, and GCA_005862135.1) and Sanger sequences (MK950151 and MK950152) are archived in GenBank. All three assemblies were manually corrected using Sanger sequence to accurately represent variation in the cin region. Relevant genetic data and scripts have been uploaded to DRYAD (10.5061/dryad.8n1n677). Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8301035.
Results
Unidirectional introgression of D. yakuba mitochondria into D. santomea
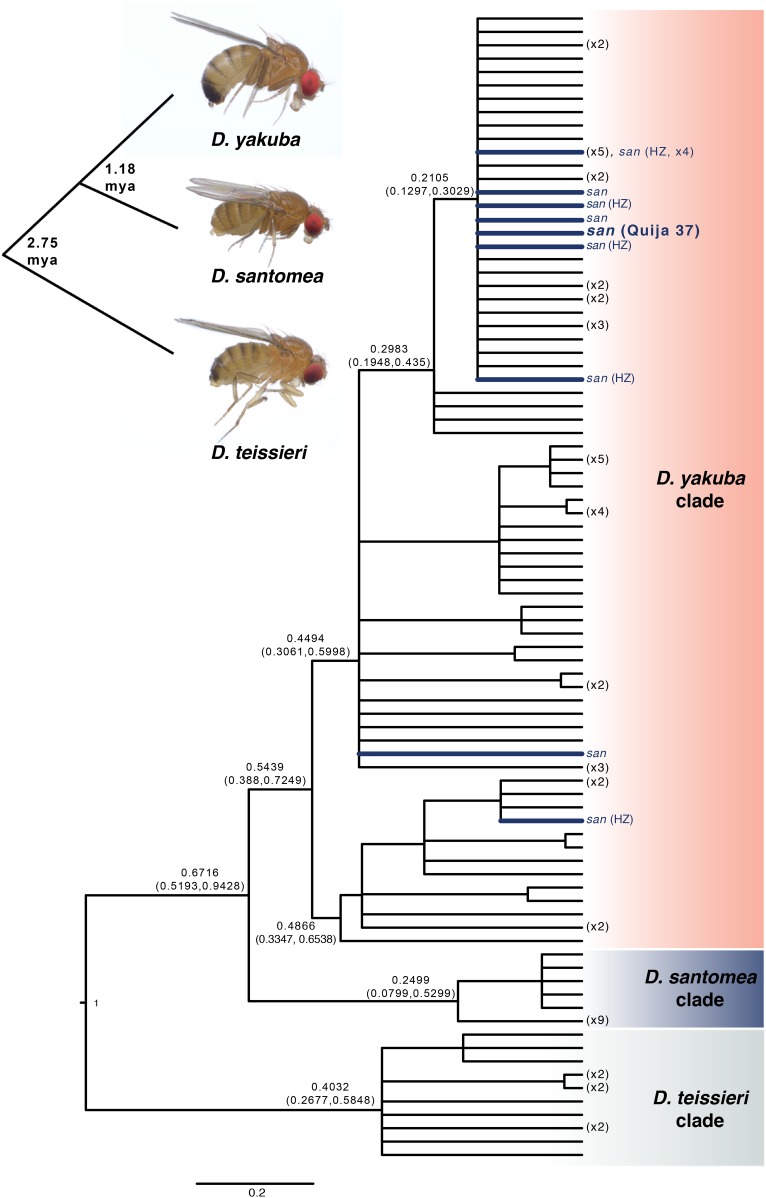
Previous analyses suggest that D. yakuba and D. santomea carry very similar mtDNA due to ongoing hybridization and introgression (Lachaise et al. 2000; Bachtrog et al. 2006; Llopart et al. 2014). However, our mtDNA relative chronogram (Figure 1), based on mitochondrial whole-proteome data sampled from throughout the ranges of all three species, supports three mtDNA clades that largely agree with the nuclear topology; D. teissieri mtDNA sequences are outgroup to sister D. yakuba and D. santomea mtDNA (Figure 1). Consistent with introgression, 12 out of 26 D. santomea isofemale lines have D. yakuba-like mtDNA (indicated by blue branches and letters in Figure 1). Yet, our samples showed no evidence of D. santomea mtDNA introgressed into D. yakuba and no mtDNA introgression involving D. teissieri. The 12 D. santomea isofemales with introgressed D. yakuba-like mtDNA were sampled from both within (N = 8) and outside (N = 4) the well-described Pico de São Tomé HZ (Llopart et al. 2014; Turissini and Matute 2017). The D. santomea HZ samples are indicated by “HZ” in Figure 1 in blue, and they are found throughout the clade that includes all D. yakuba mtDNA. This suggests that hybridization occurs in other areas of the island or that introgressed D. santomea genotypes migrate outside of the HZ.
Figure 1.
Bayesian relative chronogram for the D. yakuba clade mtDNA. The root age was fixed to 1. Nodes with posterior probability <0.95 were collapsed into polytomies, numbers at the tips show the number of genotypes collapsed. Node labels are the point estimates and 95% credible intervals for the relative ages. Tips and branches showing D. santomea individuals with D. yakuba mtDNA are colored blue, those sampled from the Pico de São Tomé hybrid zone are denoted by HZ. D. santomea line Quija 37 showing evidence of horizontal or paternal Wolbachia transmission (or paternal mtDNA transmission) is in bold blue. The chronogram was estimated from the 13 protein-coding mitochondrial genes. A representation of host nuclear relationships with photos of host females at the tips is inset, with the divergence times estimated by Turissini and Matute (2017) superimposed.
Previous analysis of nuclear genomes (Turissini and Matute 2017) also found more introgression from D. yakuba into D. santomea when populations of D. yakuba from near the Gulf of Guinea, Cameroon, and Kenya were included; however, excluding Cameroon and Kenya indicated similar amounts of introgression in each direction. Matings between D. santomea females and D. yakuba males are rare relative to the reciprocal cross (Coyne et al. 2002; Matute 2010). When these matings do occur, F1 females produce fewer progeny than do F1 females produced by D. yakuba females and D. santomea males (Matute and Coyne 2010). F1 hybrids produced by D. santomea females also have a shortened life span (Matute and Coyne 2010) due to copulatory wounds inflicted by D. yakuba males during mating (Kamimura and Mitsumoto 2012). These observations are consistent with our finding of preferential introgression of D. yakuba mitochondria into D. santomea backgrounds.
Wolbachia frequencies and draft genomes
As expected, we found Wolbachia in all three D. yakuba-clade species sampled by Turissini and Matute (2017). For D. yakuba, 21 out of 56 lines were infected, yielding an infection frequency of P = 0.36, with 95% binomial confidence interval (0.25–0.51). For D. santomea, 10 out of 11 were infected, yielding P = 0.91 (0.59–1.0); and for D. teissieri, 11 out of 13 were infected, yielding P = 0.85 (0.55–0.98). Additional frequency estimates are reported in Cooper et al. (2017), which found that Wolbachia frequencies vary through time and space in West Africa. All PCR tests for Wolbachia matched our coverage-based genomic analyses of infection status.
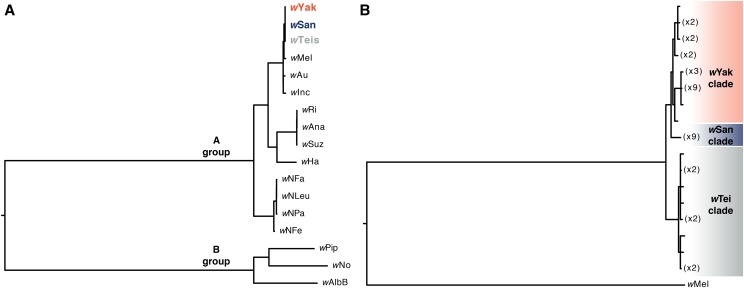
The average coverage across the genome, calculated as the total number of bases aligned to the wMel reference divided by its length, was 1940 for our wYak pseudoreference genome (yak-CY17C), 40 for wSan (san-Quija630.39), and 489 for wTei (teis-cascade_4_2). These pseudoreference genomes were included in the phylogram that includes A-group and B-group Wolbachia (Figure 2), and they cluster with the Wolbachia from their respective hosts (Figure 3), as expected if introgression is rare. In general, the wSan data are lower quality due to the relatively small size of the D. santomea libraries (Turissini and Matute 2017). The scaffold count, N50, and total assembly sizes are reported in Table S2.
Figure 2.
(A) Bayesian phylogram placing the D. yakuba-clade Wolbachia in the context of other A-group, and also B-group, Wolbachia. wPip, wNo, and wAlbB belong to Wolbachia group B, while D. yakuba clade-Wolbachia, wMel, wAu, wInc, wRi, wAna, wSuz, wHa, and Nomada Wolbachia belong to group A. The phylogram was estimated with 146 single-copy genes of identical length in each genome, spanning 115,686 bp. The limited number of genes across both A-group and B-group Wolbachia meeting our criteria precluded resolving relationships of D. yakuba-clade Wolbachia. (B) Bayesian phylogram of wYak, wSan, wTei, and outgroup wMel. The phylogram was estimated with 643 single-copy genes of identical length in each genome, spanning 644,586 bp. These more extensive data provide increased phylogenetic resolution of the D. yakuba-clade Wolbachia. For both phylograms, nodes with posterior probability <0.95 were collapsed into polytomies.
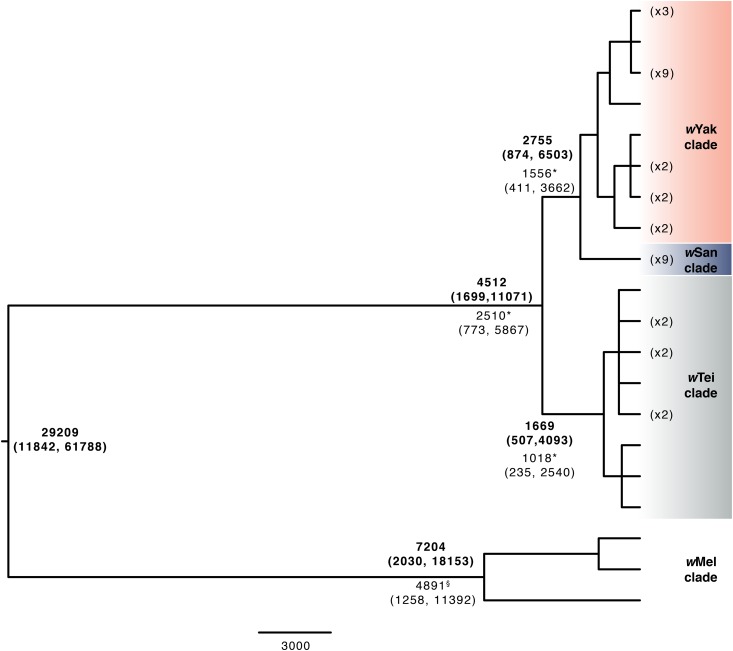
Figure 3.
Bayesian chronogram for the D. yakuba-clade Wolbachia plus three wMel genomes with absolute age estimates based on the calibration in Richardson et al. (2012) in bold. The chronogram was estimated with 621 single-copy genes of identical length in all of the genomes, spanning 624,438 bp. Nodes with posterior probability <0.95 were collapsed into polytomies. Node labels are the point estimates and 95% credible intervals for the ages. We also constructed two other chronograms: one that included only D. yakuba-clade Wolbachia and one that included only wMel Wolbachia. The divergence estimates for the chronogram including only D. yakuba-clade Wolbachia (*) and only wMel Wolbachia (§) are superimposed to illustrate that the inclusion of both groups in the chronogram alters the absolute divergence estimates within each group. Our chronogram with only D. yakuba-clade Wolbachia included 678 genes (containing 695,118 bp), and our chronogram including only wMel Wolbachia included 692 genes (709,599 bp).
Introgressive and horizontal Wolbachia transfer among hosts
Our phylogram that includes A-group and B-group Wolbachia places the three weak-CI-causing D. yakuba-clade Wolbachia together and sister to wMel in the A group (Figure 2A). In contrast, D. simulans carries diverse Wolbachia that do (wRi, wHa, wNo) and do not (wAu) cause CI, spanning Wolbachia groups A (wRi, wHa, wAu) and B (wNo). All wSan, wYak, and wTei Wolbachia variants included in our analysis are identical at the MLST loci often used to type Wolbachia variants (Baldo et al. 2006; Cooper et al. 2017). Because of the relatively small number of genes that meet our inclusion criteria (146 genes across 115,686 bp), the resulting phylogram does not resolve relationships among wSan, wYak, and wTei (Figure 2A). Our phylogram that includes only D. yakuba-clade and wMel Wolbachia (based on 643 genes across 644,586 bp) resolves these relationships, placing wTei variants outgroup to sister wSan and wYak in distinct clades (Figure 2B). We observe 0.0039% third-position pairwise differences between wTei and wYak and between wTei and wSan, and 0.0017% between wSan and wYak. The third-position pairwise differences between any of the D. yakuba-clade Wolbachia and wMel is 0.11%. For reference, the third-position pairwise difference between wYak and wRi is 2.9%, whereas wRi and wSuz differ by only 0.014% (Turelli et al. 2018).
Our absolute chronogram that includes wMel and D. yakuba-clade Wolbachia also indicates three monophyletic groups (Figure 3), whose topology––[wTei, (wYak, wSan)]––agrees with the nuclear topology of the hosts (Turissini and Matute 2017). However, one D. santomea line, D. santomea Quija 37, sampled from the south side of São Tomé (Matute 2010), carries Wolbachia identical to the other eight wSan samples across 695,118 bp, yet its mtDNA is nested in the D. yakuba mtDNA cluster (bold blue in Figure 1). This single example of Wolbachia-mtDNA phylogenetic discordance can be explained by either horizontal Wolbachia transfer or paternal transfer of Wolbachia or mtDNA. While rare, paternal Wolbachia transmission has been observed in other Drosophila (Hoffmann and Turelli 1988; Turelli and Hoffmann 1995), as has paternal transmission of mtDNA (Kondo et al. 1990). The only other Wolbachia-screened D. santomea isofemale line with introgressed mtDNA was Wolbachia uninfected.
In Figure 3, we present alternative estimates of the divergence times for Wolbachia within and between D. yakuba-clade species, within D. melanogaster, and between the D. yakuba clade and D. melanogaster. The estimates from the chronogram that includes all of the indicated Wolbachia variants are in bold. To demonstrate how these time estimates depend on the sequences included in the analyses (through estimates of relative divergence for each codon position), estimates that included only D. yakuba-clade Wolbachia or only wMel variants are superimposed and not in bold. These estimates were all obtained using a relaxed-clock analysis with a Γ(2,2) branch-rate prior [results using (7,7) and a strict clock are mentioned below]. When considering only the D. yakuba-clade Wolbachia (Figure 3), we estimate the root at 2510 years (95% credible interval 773–5867 years) and the wYak-wSan split at 1556 years (95% credible interval 411–3662 years). The wMel chronogram estimates the most-recent common ancestor (MRCA) of wMel at 4890 years ago (95% credible interval 1258–11,392 years). Our wMel result is consistent with the Richardson et al. (2012) estimate of 8008 years (95% credible interval 3263–13,998). The alternative branch-rate (7,7) prior has little effect on these divergence-times estimates, with credible intervals that overlap with those produced using a Γ(2,2) branch-rate prior (Figure 3 and Table S5). In contrast, Figure 3 shows that including more divergent variants influences our time estimates, although the credible intervals are overlapping in each case. Table S4 shows how estimates of the relative amounts of the divergence for the three codon positions vary across the data sets. The underlying model for divergence times assumes constant relative rates at the three positions. Hence, changes in relative rates affect divergence-time estimates.
As noted above, our estimates of the time of the MRCA for Wolbachia within the D. yakuba clade or within D. melanogaster are relatively insensitive to whether the branch-rate prior is Γ(2,2) or Γ(7,7). The choice of genes used to estimate divergence also has little effect—maximal distance between D. yakuba-clade strains was identical for analyses that included only the D. yakuba-clade (N = 678 genes) or wYak-clade plus wMel (N = 621 genes) gene sets (wYak-wTei distance = 0.0035%). The estimated divergence time between wMel and the D. yakuba-clade Wolbachia is less robust, although again the credible intervals generated using alternative branch-rate priors are overlapping (Figure 3 and Table S5). A strict-clock analysis produces an estimate of 72,612, with 95% credible interval (23,412–132,276), again overlapping with our relaxed-clock estimates. An alternative point estimate can be obtained from pairwise differences. Averaging over the D. yakuba-clade sequences, we observe an average third-position difference of 0.107% between the D. yakuba-clade Wolbachia and wMel, or 0.0535% from tip to root. The “short-term evolutionary rate” of divergence within wMel, estimated by Richardson et al. (2012), produces a point estimate of 78,000 years, which overlaps with the credible interval of our relaxed-tree estimate with a Γ(7,7) branch-rate prior (Table S5). This molecular-clock estimate closely agrees with our strict-clock chronogram analysis (Figure S2). Given that these estimates are much shorter than the divergence time between their reproductively isolated hosts, horizontal Wolbachia transmission must have occurred. As discussed below, the horizontal transmission probably involved intermediate hosts.
mtDNA and nuclear concordance across Drosophila:
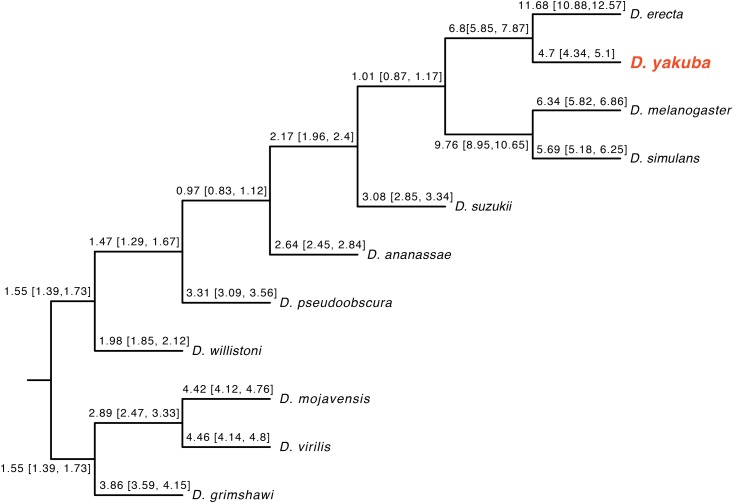
The topologies of the nuclear and mtDNA trees for the 11 reference Drosophila species are completely concordant (Figure 4) and agree with the neighbor-joining result presented in Drosophila 12 Genomes Consortium et al. (2007). We find the average ratio of mtDNA to nuclear substitution rates across branches vary from ∼1× to 10×, consistent with the results of Havird and Sloan (2016) across Diptera.
Figure 4.
A Bayesian phylogram, with branch lengths fixed to 1, displaying relative rate variation for mtDNA vs. nuclear loci. Branches are labeled with quartiles of the estimated third-position mtDNA to third-position nuclear substitution rate. The nuclear substitution rate was taken as the arithmetic mean third-position rate over the 20 loci used. The mtDNA and nuclear phylograms are topologically identical when run separately.
mtDNA and Wolbachia concordance in the D. yakuba clade:
D. santomea line Quija 37 unexpectedly carries mitochondria belonging to the clade associated with D. yakuba but has wSan Wolbachia. This discordance requires horizontal or paternal transmission. Apart from this, we find no phylogenetic discordance between strongly supported nodes (i.e., posterior probability at least 0.95) in the mitochondrial and Wolbachia trees (Figure S3). For a more refined test of concordance, we removed the Quija 37 line from our mitochondrial and Wolbachia data and computed Bayes factors. The shared topology model was favored by a Bayes factor of e55, indicating strong support for concordance of the mitochondrial and Wolbachia phylogenies.
Variation in the ratio of mtDNA:Wolbachia substitution rates:
Following the methods of Turelli et al. (2018) to look for different mitochondrial vs. Wolbachia branch lengths, we also found no evidence of variation in the ratio of mtDNA:Wolbachia substitution rates within the D. yakuba clade (median third-position ratio = 223, quartiles = 186 and 268); the model with all branches sharing the same ratio is favored over the model where each branch has its own ratio by a Bayes factor of e7 (Figure S4). For comparison, Turelli et al. (2018) found a median ratio of 566 within D. suzukii and 406 within wRi-infected D. ananassae-subgroup species. These differences in relative rates are broadly compatible with the 10-fold variance in relative rates noted above for nuclear vs. mtDNA divergence across Drosophila species. Hence, our data seem compatible with Wolbachia transmission via introgression within the D. yakuba clade.
Although this result seems to strongly support purely maternal transmission of Wolbachia within species and introgressive transfer between species (with more recent introgression between the sister species D. yakuba and D. santomea), this interpretation is severely weakened by our “control” analysis that includes wMel-infected D. melanogaster (for which introgression is impossible). Including the D. melanogaster data, the constant-ratio model was still favored over variable ratios by a Bayes factor of e54 (median third-position ratio = 297, quartiles = 276 and 323). We discuss the implications of this anomalous result below. The key observation is that significantly declining rates of substitution for Wolbachia [and mtDNA, see Ho et al. (2005)] over time, together with heterogeneity of relative rates (as illustrated in Figure 4), limit the power of relative substitution ratios to differentiate between introgression and horizontal transmission.
Transposon-mediated transfer of CI factors independent of WO phage
In contrast to wMel, which contains only the cidA-cidB gene pair (LePage et al. 2017), the D. yakuba-clade Wolbachia, which also belong to Wolbachia group A, have both cidA-cidB and a cinA-cinB pair homologous to CI loci originally identified in group-B wPip (Beckmann et al. 2017). The A and B Wolbachia groups diverged ∼ 6–46 MYA (Werren et al. 1995; Meany et al. 2019). Among our wYak-clade sequences, there are no single-nucleotide variants within any of the cif loci. The cidBwYak-clade locus has an inversion from amino acids 37–103 relative to the same region in cidBwMel in every wYak-clade Wolbachia variant we analyzed. This introduces several stop codons, which might render this gene nonfunctional. On the other hand, RNA polymerase should still transcribe a complete polycistronic transcript. Therefore translation of an N-terminally truncated CidBwYak-clade protein cannot be ruled out. With the exception of a 236 bp tandem duplication in cinBwYak-clade (Figure 5 and Figure 6), the sequence differences between cinAwYak-clade and cinBwYak-clade regions compared to cinAwPip and cinBwPip homologs are 1.35 and 0.59%, respectively. In contrast, the average difference between wYak and wPip genomes across all 194 genes (161,655 bp) present in single copy and with identical lengths in both genomes is 11.60%. Conversely, outside of the prophage regions, wMel and wYak differ by ∼1%. This is consistent with data that indicate WO phage regions have a different evolutionary history than the bulk of the Wolbachia genome (LePage et al. 2017; Lindsey et al. 2018). The 236 bp tandem duplication in the cinBwYak-clade introduces a frameshift in the transcript at position 588. It is unclear whether the cinBwYak-clade protein retains functionality.
Figure 5.
The gene structure and observed, uncorrected, pairwise distances among wYak, wMel, and wPip Wolbachia spanning the region of the WO prophage island where the hypothesized transfer occurred between the ISWpi1 element “A” present in wYak and the ISWpi1 sequence labeled “B” in wMel. Over the region shown, wYak, wSan, and wTei are identical. The difference between wYak and wMel is <1% between the contigs (wYak “187383” and wYak “694583”) flanking the region containing the cinA-cinB homologs. These homologs assembled on the first ∼4000 bp of the contig wYak “702380,” but they do not appear to exist in wMel. A BLAST search against the NCBI nucleotide collection using contig wYak “702380” as a query indicated the cinA-cinB homologs share high identity with group-B wPip Wolbachia (∼97%). The remainder of the wYak contig is approximately equally divergent from both wMel and wPip, consistent with the entire region being transferred from an unknown Wolbachia variant that had acquired the cinA-cinB homologs. Under this scenario, ISWpi1 elements mediate the transfer to wYak via excision of elements “A” and “B” along with the intervening DNA from the donor and subsequent homologous recombination between IS sequences “A” and “B” and WO phage island genes in wYak. The ISWpi1 element “A” present in wYak, but not in either wMel or wPip, was spanned using Sanger sequencing with primers anchored on unique sequence in the flanking contigs (Sanger sequences are denoted by yellow lines with primer locations denoted by red flanking regions, Table S1). Additionally, the presence of a 236 bp tandem duplication in cinB (“C”) was confirmed using Sanger sequencing. The ISWpi1 sequence labeled “B” is assembled in wMel and corresponds to wMel#9 (Cordaux 2008) but could not be assembled or spanned with Sanger sequencing in the wYak genome. White gene models (arrows) indicate a partial transposase gene present in the WO prophage island, truncated in wYak. Pairwise differences were calculated using a sliding window (window size = 200 bp, step size = 25 bp).
Figure 6.
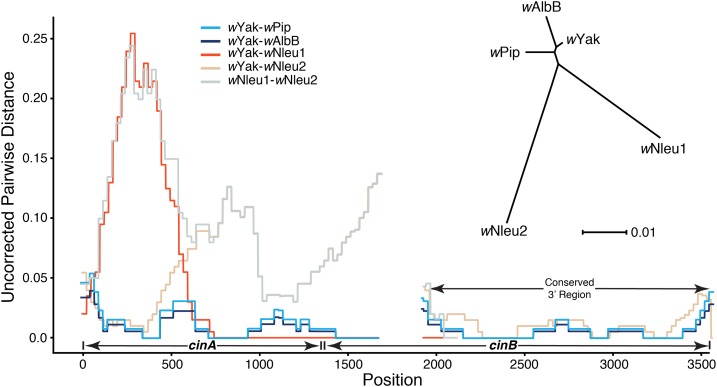
The observed pairwise differences between cinA and cinB copies in wYak-clade Wolbachia and those in wAlbB and wPip, not including flanking regions, are shown. We also plot the pairwise differences between the two copies of these loci found in Nomada Wolbachia (denoted wNleu1 and wNleu2); we chose wNleu to represent the Nomada-clade Wolbachia because this strain contains the longest assembled contigs for both cinA-cinB copies. Homologous cinA-cinB copies have relatively low divergence (inset unrooted tree), with the highest divergence in the 5′ end of region. The most highly diverged region among any of the copies is the first ∼750 bp of wNleu1, with pairwise differences between wNleu1 and wYak reaching ∼25% in some windows, with similar divergence between wNleu1 and wNleu2 in this same region. Pairwise differences abruptly change 750 bp from the start of the cinA gene with the remaining ∼1150 bp of the assembled region having a single difference from the wYak sequence (only ∼1900 bp assembled into a single contig in the wNleu1 copy). This pattern suggests a recent transfer/recombination event from the same unknown donor of the wYak cinA-cinB copy. The gap at position 1705–1940 represents a tandem duplication present in wYak. The unrooted tree was generated using RAxML 8.2.9 (Stamatakis 2014), and the representative cinA-cinB gene copies were obtained using BLAST to search all Wolbachia genomes in the NCBI assemblies database (https://www.ncbi.nlm.nih.gov/assembly/?term=Wolbachia).
The absence of cin loci in wMel, combined with the similarity of this region between wYak and distantly related wPip, led us to assess how these Wolbachia acquired cin loci. Targeted assembly extended the scaffold wYak “702380” containing the cin loci (Figure 5) but did not definitively place it relative to the wMel genome. BLAST searches indicated that contig wYak “187383” was likely flanked by wYak “702380.” PCR primers designed in both contigs amplified the intervening region (labeled “A” in Figure 5), confirming we have discovered the first WO prophage with two sets of cif loci. Subsequent Sanger sequencing revealed that this region contains an ISWpi1 element, found in many Wolbachia genomes, but not present at this location in the wMel reference sequence (Cordaux 2008).
IS elements encode a transposase gene that mediates their movement (Chandler and Mahillon 2002). ISWpi1 elements are related to the IS5 family, which seems to be restricted to Wolbachia (Duron et al. 2005; Cordaux 2008). ISWpi1 elements occur in more than half of the Wolbachia strains that have been evaluated, including wYak and wMel (Cordaux 2008); and these elements occur at variable copy number, potentially facilitating horizontal transmission of ISWpi1 elements and their intervening sequence between Wolbachia variants/strains (Cordaux 2008). Following placement of contig wYak “702380” relative to wMel, we aligned these regions and calculated pairwise differences along the chromosome using a sliding window.
In addition to containing the cinA-cinB loci, absent in wMel, contig wYak “702380” is on average ∼10% different from wMel. In contrast, most of wYak is <1% different from wMel (for instance, across the 650,559 bp used in our phylogenetic analyses, wMel and wYak differ by only 0.09%). Downstream of the cinA-cinBwYak-clade region, targeted assembly enabled us to join several more contigs. The junctions were corroborated by mapping paired-end reads (Langmead and Salzberg 2012) and visually inspecting the resulting bam files around joined contigs for reads spanning the new junctions and for concordant read-pair mappings. However, our attempts to fully bridge the gap downstream of the cinA-cinBwYak-clade genes via targeted assembly, scaffolding, and PCR were unsuccessful (see “wYak unassembled gap” in Figure 5). The unassembled region in wYak contains an ISWpi1 element in wMel, Mel #9 (labeled “B” in Figure 5; Cordaux 2008). Although not part of our wYak assembly, homologs of this ISWpi1 element appear in assemblies of this region of the WO prophage in several other A-group Wolbachia (Cordaux 2008; Bordenstein and Bordenstein 2016), specifically wInc from D. incompta (Wallau et al. 2016) and wRi from D. simulans (see Figure 2). In both, we find orthologs to Mel#9 ISWpi1 with corresponding flanking sequence assembled, indicating that this IS element probably occurs in this unassembled region of wYak, wSan, and wTei. The wYak sequence between these two ISWpi1 elements is highly diverged from wMel in comparison to the rest of the wYak genome (∼10% difference vs. 1%). It therefore seems plausible that this region experienced a horizontal transfer event in the ancestor of wYak, wTei, and wSan, mediated by the flanking ISWpi1 elements.
We conjectured that horizontal transfer occurred via the excision of the two ISWpi1 elements and the intervening DNA from the donor followed by homologous recombination within the IS elements. To assess the plausibility of this scenario, we used BLAST to compare cinA-cinBwPip genes against all published Wolbachia genomes in the NCBI assembly database (https://www.ncbi.nlm.nih.gov/assembly/?term=Wolbachia). We found homologs of the cinA-cinBwPip in the group-A Wolbachia associated with Nomada bees (Gerth and Bleidorn 2016). An unrooted tree of the ORFs for cinA and cinB (Figure 6) indicates that these genes in wYak, wSan, and wTei are more similar to cinA-cinB from group-B wPip and wAlbB than to cinA-cinB found in the fellow group-A Wolbachia associated with Nomada bees (wNFe, wNPa, wNLeu, and wNFa), which each harbor two different cinA-cinB copies. This indicates that cinA-cinBwYak-clade were acquired via a horizontal transfer event across Wolbachia groups B and A that is independent of the event(s) that placed cinA-cinB in the Wolbachia associated with Nomada bees, suggesting repeated transfers of cin loci.
The two cinA-cinB copies (denoted wNLeu1 and wNLeu2 in Figure 6) in Nomada Wolbachia are nearly as distinct from each other as they are from the homologs in wPip, wAlbB, and wYak (∼7% diverged from these strains, Figure 6). However, among the four Wolbachia-infected Nomada species, the orthologs are very similar, with cinAwNLeu1 having only 0–0.15% pairwise differences among the four strains and cinAwNLeu2 having 0–0.56% pairwise differences (reconstructions for cinB gene copies were more complicated as the cinBwNLeu1 copy fails to assemble into a single contig on the three-prime end). This pattern suggests that wNLeu1 and wNLeu2 cin copies were acquired by the common ancestor of the four Nomada Wolbachia strains analyzed, followed by cladogenic transfer across Nomada species (Gerth and Bleidorn 2016). The highly fragmented assemblies of the four Nomada Wolbachia strains, with duplicate copies confounding assembly, make it difficult to determine the relative positions of the cinA and cinB copies and if they are likewise flanked by ISWpi1 elements.
To determine whether these genes were potentially moved by ISWpi1 elements into the Wolbachia of the D. yakuba clade and Nomada, we searched each genome using both the cinA-cinBwYak-clade contig and the flanking ISWpi1 elements. Long repeated elements like ISWpi1 (916 bp) break most short-read assemblies. Despite this, there is often a small fragment of the element, the length of the short read, on either end of the broken contig, indicative of the repeat element being responsible for terminating contig extension. We looked for the footprint of these elements at the edges of the contigs on which the cin genes were found. We found ISWpi1 elements in the region flanking both cinAwNLeu1 and cinAwNLeu2 copies, consistent with our wYak assembly in which we verified the ISWpi1 element with Sanger sequencing. These data support a role for ISWpi1 in the acquisition of the cinA-cinB genes by the Wolbachia in the D. yakuba clade and the Nomada bees. We conjecture that future work will fully confirm ISWpi1 in the horizontal movement of incompatibility loci between Wolbachia.
Discussion
Our results indicate introgressive and horizontal Wolbachia acquisition in the D. yakuba clade. Evidence for horizontal Wolbachia transfer here and elsewhere (Turelli et al. 2018) suggests that double infections must be common, even if ephemeral. Such double infections provide an opportunity for ISWpi1 transposable elements to mediate horizontal transfer of incompatibility loci among divergent Wolbachia. Importantly, our results highlight that incompatibility factors may move independently of prophage, as evidenced by our discovery of the first prophage documented to have two sets of cif loci. We discuss these conclusions below.
mtDNA introgression
Our relative mitochondrial chronogram provides strong support for three mitochondrial clades, including a monophyletic D. teissieri clade that is outgroup to two sister clades: one clade consisting of mitochondria from 14 D. santomea individuals, and the other contains all D. yakuba mitochondria plus mitochondria from 12 D. santomea individuals. Our results suggest less mitochondrial introgression in the D. yakuba clade than a past report that used sequence data from only two mitochondrial loci (COII and ND5, 1777 bp) and reported a clade with mitochondria sampled from each species represented (Figure 3 in Bachtrog et al. 2006) (using sequence data from only COII and ND5, we can replicate this result, indicating data from additional loci are needed to add resolution). Our results also agree with recent work that demonstrated little nuclear introgression in the D. yakuba clade (Turissini and Matute 2017).
Our results broadly agree with the results produced by Llopart et al. (2014), who assessed mitochondrial introgression between D. santomea and D. yakuba using whole mitochondrial genomes. They generated a neighbor-joining tree that produced a clade consisting of all D. yakuba individuals and 10 D. santomea individuals, and this clade is sister to a clade with 6 D. santomea individuals; one D. santomea haplotype is outgroup to all other haplotypes included in their analysis (Figure 3 in Llopart et al. 2014). Nested within the mixed D. yakuba clade, Llopart et al. (2014) identified a “HZ clade” that includes D. yakuba individuals sampled from São Tomé and from continental Africa and D. santomea individuals sampled from both within and outside the Pico de São Tomé HZ. The sister D. santomea clade also contains both HZ and non-HZ individuals. Thus, their analysis and ours provide support for hybridization within and outside of the HZ, leading us to question both the existence of a HZ clade and the claim that these species “share the same mitochondrial genome” (Llopart et al. 2014). Instead, both our results and theirs suggest unidirectional introgression of D. yakuba mitochondria into D. santomea; we find 59% of D. santomea individuals having D. yakuba-like mitochondria and they found 46%.
Llopart et al. (2014) used a strict molecular clock to estimate the mitochondrial MRCA of D. santomea and D. yakuba at 10,792–17,888 years by calibrating their tree with the D. yakuba-D. erecta split, estimated at 10.4 MYA (Tamura et al. 2004). A high level of mitochondrial saturation over time, with an expected value of 1.44 substitutions per synonymous site for the D. yakuba-D. erecta split, could influence this estimate (Llopart et al. 2014). Moreover, Ho et al. (2005) demonstrated that the mtDNA substitution rate resembles an exponential curve, with high short-term substitution rates that approach the mutation rate, then slowing to a long-term rate after ∼1–2 MY of divergence, far younger than the inferred D. yakuba-D. erecta nuclear and mtDNA divergence. Hence, using the slow long-term D. yakuba-D. erecta calibration is likely to underestimate the more rapid rates of divergence experienced by D. yakuba-clade mtDNA, inflating divergence-time estimates (Ho et al. 2005). If we assume that Wolbachia and mitochondria were transferred by introgression, which our analyses support, our estimates in Figure 3 suggest that D. santomea and D. yakuba mtDNA diverged more recently, with our point estimates ranging from ∼1500 to 2800 years.
Wolbachia placement, divergence, and acquisition
Wolbachia placement:
Despite their similarity, wSan, wTei, and wYak form monophyletic clades with wTei outgroup to sisters wSan and wYak, recapitulating host relationships (Figure 1; Turissini and Matute 2017). wMel is sister to the D. yakuba-clade strains in the A group (Figure 2), which also includes D. simulans strains (wRi, wAu, and wHa), wAna, wSuz, and the Nomada Wolbachia (wNFa, wNLeu, wNPa, and wNFe). Our phylogram (Figure 2A) places wAlbB outgroup to wNo and wPip strains that diverged from A-group Wolbachia ∼6–46 MYA (Meany et al. 2019).
Wolbachia divergence:
Our chronogram analyses (Figure 3) estimate that D. yakuba-clade Wolbachia and three wMel variants diverged ∼29,000 years ago, and that wTei split from sisters wSan and wYak ∼2500–4500 years ago, with wSan and wYak diverging ∼1600–2800 years ago. We estimate that the two most divergent wMel variants from Richardson et al. (2012) and the reference wMel genome split ∼4900–7200 years ago, indicating more divergence among wMel variants than among D. yakuba-clade Wolbachia strains. All of these results depend on the calibration provided by Richardson et al. (2012) and the relative accuracy of the underlying models of molecular evolution, which assume constant relative rates of change across data partitions. For the deepest divergence in Figure 3, between D. yakuba-clade Wolbachia and wMel, we find that the estimated time depends on the variance in our prior distribution for substitution rates across branches, with a strict clock putting the divergence at ∼73,000 years rather than 29,000 years obtained with the most variable prior. Despite this uncertainty, the quantitative differences of our Wolbachia divergence-time estimates do not alter the qualitative conclusion that these Wolbachia did not codiverge with these hosts, which split several million years ago.
Our findings here and in Turelli et al. (2018) suggest that for several Drosophila species, their current Wolbachia infections have been in residence for only hundreds to tens of thousands of years. Bailly-Bechet et al. (2017) estimated Wolbachia residence times using data from >10,000 arthropod specimens spanning over 1000 species. However, they analyzed DNA sequences from only part of the fast-evolving fbpA Wolbachia locus and the host CO1 mtDNA locus. From an initial model-based meta-analysis, they concluded that “... most infections are very recent ...,” consistent with our results. However, they also fit a more complex model with “short” and “long” timescales for acquisition and loss, conjecturing that short-term rates were associated with imperfect maternal transmission. Focusing on long-time rates, they concluded that Wolbachia infections persisted in lineages for ∼7 MY on average, whereas lineages remained uninfected for ∼9 MY. For Drosophila, such long infection durations would imply that Wolbachia-host associations often persist through speciation [see Coyne and Orr (1997) and Turelli et al. (2014) for estimates of speciation times in Drosophila, generally 105–106 years]. Extrapolating from limited Wolbachia sequence data, Hamm et al. (2014) conjectured that cladogenic Wolbachia transmission might be common among Drosophila, but this extrapolation is refuted by our genomic analyses. The long Wolbachia durations proposed Bailly-Bechet et al. (2017) depend on their conjecture that their long-term rate estimates accurately reflect acquisition and loss of Wolbachia infections across species. This is worth testing with additional analyses of Wolbachia and mitochondrial and nuclear genomes from a broad range of arthropods.
Wolbachia acquisition––introgression vs. horizontal:
Our divergence-time estimates for the D. yakuba-clade Wolbachia vs. their hosts preclude cladogenic acquisition. Unlike introgression, horizontal (or paternal) Wolbachia acquisition should produce discordance between phylogenies inferred for Wolbachia and the associated mitochondria. With the notable exception of D. santomea line Quija 37, which has mitochondria belonging to the clade associated with D. yakuba, but has wSan Wolbachia, we find no evidence of discordance between the estimated mitochondrial and Wolbachia phylogenies. Hence our data indicate that acquisition by introgression is far more common than horizontal transmission between closely related species, consistent with data on acquisition of wRi-like Wolbachia in the D. melanogaster species group (Turelli et al. 2018). Similarly, consistent with extensive data from D. melanogaster (Richardson et al. 2012) and D. simulans (Turelli et al. 2018) and smaller samples from D. suzukii and D. ananassae (Turelli et al. 2018), we find only one possible example of paternal transmission or horizontal transmission within D. yakuba-clade species.
We also investigated an alternative approach to distinguish between introgressive and horizontal Wolbachia acquisition by estimating substitution ratios for mtDNA vs. Wolbachia (Turelli et al. 2018). Because we could estimate this ratio on each branch, we conjectured that this approach might have greater resolving power than our incompletely resolved mitochondrial and Wolbachia phylogenies. We expect higher ratios with horizontal transmission because mtDNA would have been diverging longer than recently transferred Wolbachia. However, this approach assumes that mtDNA and Wolbachia substitution rates remain relatively constant. This is contradicted by the finding of Ho et al. (2005) that mtDNA substitution rates decline substantially with increasing divergence time, reaching an asymptote after ∼1–2 MY. To calibrate their Wolbachia substitution rate estimates, Richardson et al. (2012) used an experimentally observed mitochondrial mutation rate in D. melanogaster (6.2 × 10−8 mutations per third-position site per generation) that extrapolates to 62% third-position divergence per million years. This extrapolation is nonsensical as a long-term substitution rate. As summarized by Ho et al. (2005), typical mtDNA substitution rates are 0.5–1.5% per coding site per million years. Nevertheless, using the ratio of short-term rates for mtDNA and Wolbachia, Richardson et al. (2012) produced an estimate of the long-term Wolbachia substitution rate that agrees with independent estimates from Nasonia wasps (Raychoudhury et al. 2009) and Nomada bees (Gerth and Bleidorn 2016) derived from much longer divergence times (assuming cladogenic Wolbachia acquisition) (Conner et al. 2017). This paradox is resolved if Wolbachia molecular evolution is not subject to the dramatic slowdown in rates seen for mtDNA.
The apparent difference between mtDNA molecular evolution (dramatic slowdown over longer timescales; Ho et al. 2005) and Wolbachia molecular evolution (relative constancy, as inferred from similar rates of differentiation over very different timescales) suggests why our relative-rate test does not reject introgressive transmission of Wolbachia between D. melanogaster and the D. yakuba clade, even though it is clearly impossible. A roughly 50-fold slowdown in mtDNA substitution rates over the timescale of the divergence of D. melanogaster from the D. yakuba clade, relative to the rate of differentiation within the D. yakuba clade, produces comparable mtDNA-Wolbachia substitution ratios for comparisons within the D. yakuba clade and between D. melanogaster and the D. yakuba clade. Because of this complication and our conjecture that relative rates of mtDNA vs. Wolbachia substitutions over longer periods are likely to mirror the ten-fold differences we see for mtDNA vs. nuclear genes, phylogenetic discordance between mitochondria and Wolbachia is clearly a much more robust indicator of horizontal (or paternal) Wolbachia transmission. Nevertheless, additional examples of cladogenic Wolbachia acquisition are needed to better understand relative rates and patterns of Wolbachia, mtDNA and nuclear differentiation over different timescales.
Our divergence-times estimate of the D. yakuba-clade Wolbachia vs. their hosts precludes cladogenic transmission; and our phylogenetic analyses suggest that these species share very similar Wolbachia because of introgression, as originally argued by Lachaise et al. (2000). However, under either introgressive or horizontal transfer of Wolbachia, we expect the donor species Wolbachia sequences would appear paraphyletic when analyzed jointly with the Wolbachia from the recipient. Paraphyly allowed Turelli et al. (2018) to infer that D. simulans likely obtained its Wolbachia from D. ananassae and D. subpulchrella likely obtained its Wolbachia from D. suzukii. Paraphyly is generally expected soon after gene flow stops between populations. As noted by Hudson and Coyne (2002; Figure 1), the timescale expected to produce reciprocal monophyly for mitochondria (and Wolbachia) under a neutral model of molecular evolution is on the order of the effective size of the species. Our results in Figure 3 indicate that, at least for our small samples, reciprocal monophyly for the Wolbachia in these three species has been achieved within a few thousand years. This suggests that reciprocal monophyly has been accelerated by species-specific selective sweeps within the Wolbachia or mitochondria of these species. This conjecture may be testable from estimates of host fitness using transinfected vs. native Wolbachia.
IS transposable elements mediate horizontal transfer of incompatibility loci between divergent Wolbachia
Wolbachia in all three D. yakuba-clade hosts cause both intra- and interspecific CI (Cooper et al. 2017), despite originally being characterized as non-CI causing (Charlat et al. 2004; Zabalou et al. 2004). CI is relatively weak, and its strength can vary among wTei variants and D. teissieri backgrounds (Table 3 in Cooper et al. 2017). Differences in CI among Wolbachia variants has also been demonstrated in interspecific backgrounds where wTei caused stronger CI in a D. simulans background (97.2 ± 1.3 SE, percent embryo mortality) than either wYak (26.5 ± 4.2 SE, percent embryo mortality) or wSan (24.0 ± 4.1 SE, percent embryo mortality) (Zabalou et al. 2008). Both wYak and wSan induced CI in D. simulans comparable in intensity to that found by Cooper et al. (2017; Figure 3) in their original hosts. Surprisingly, Zabalou et al. (2008) found that the strength of CI induced by wTei in D. simulans even eclipsed that of wRi (89.8 ± 4.5 SE, percent embryo mortality). These results must be reconciled with the fact that loci known to underlie CI do not vary within or among D. yakuba-clade Wolbachia variants we examined. The nearly complete CI induced by wTei in D. simulans may depend on CI-causing factors yet to be identified or differences in gene expression.
In each D. yakuba-clade Wolbachia variant included in our analyses, we find a disruption of cidBwYak-clade with an inversion from amino acids 37–103 relative to the same region in sister wMel. The inversion introduces multiple stop codons that could render this gene nonfunctional. Fixation of loss-of-function mutations in CI-causing loci is consistent with theoretical analyses showing that selection on Wolbachia within host lineages does not act to increase or maintain CI (Prout 1994; Turelli 1994; Haygood and Turelli 2009); indeed, we have also recently observed a single mutation that disrupts cidB in non-CI causing wMau Wolbachia that infect D. mauritiana on the island of Mauritius (Meany et al. 2019). In both wMau and the D. yakuba-clade Wolbachia, we find fixation of defects in the putative toxin gene. We expect that future genomic analyses will produce additional examples.
All D. yakuba-clade Wolbachia genomes included in our analysis harbor cinA-cinB loci originally discovered in the wPip strain that diverged from A-group Wolbachia, including the D. yakuba-clade variants, ∼6–46 MYA (Meany et al. 2019). cin loci are also present in B-group wAlbB that infects Ae. albopictus and in A-group wNFe, wNPa, wNLeu, and wNFa Wolbachia that infect Nomada bees. cin loci are absent from wMel, but the wYak contig containing these loci is ∼10% diverged from wMel, while observed divergence between wYak and wMel across the rest of the genome is <1%. The wYak-clade cin loci share ∼97% similarity with the divergent B-group wPip strain. wYak-clade cin loci are more similar to cinA-cinB from the B-group Wolbachia wPip and wAlbB than to those in A-group Nomada Wolbachia strains, which have two sets of cin loci that are as diverged from each other as they are from these regions in wYak and in B-group Wolbachia. These observations suggest independent horizontal transfer of cin loci into wYak and Nomada Wolbachia.
Our results indicate that independent of prophage movement, ISWpi1 element paralogs can move incompatibility loci via the excision of flanking ISWpi1 elements, followed by homologous recombination within the elements. Horizontal Wolbachia acquisition is common in Drosophila (Turelli et al. 2018) and other species (O’Neill et al. 1992), suggesting that double infections, which could provide the opportunity for ISWpi1-mediated transfer of incompatibility loci, may be common, even if transient (a second infection need not become stably transmitted for horizontal gene transfer via ISWpi1 elements to occur). In contrast, phage particles or virions could be introduced by a vector and provide the opportunity for ISWpi1-mediated transfer (Ahmed et al. 2015; Brown and Lloyd 2015), without the presence of a double Wolbachia infection. Determining whether the insertion of these loci was derived from a prophage region of the Wolbachia genome, or from a phage genome encapsulated in a phage particle, remains an open question. While the ISWpi1 element in wMel (Mel #9 labeled “B” in Figure 5; Cordaux 2008) is not part of our wYak assembly, homologs of this element are present in assemblies of several other A-group Wolbachia including wInc and wRi (Cordaux 2008; Bordenstein and Bordenstein 2016). We predict this element occurs in the unassembled region of our wYak assembly. Footprints of ISWpi1 elements in the region flanking the cinA genes for both copies of the gene in the Nomada Wolbachia provide further support for our hypothesis. Long-read–based Wolbachia assemblies from many infected host systems will elucidate the role of ISWpi1 elements in horizontal transfer of CI loci. Overall, the ecology of horizontal Wolbachia transmission is crucial to understanding Wolbachia acquisition, and the transfer and dynamics of CI loci are crucial to understanding Wolbachia evolution.
Acknowledgments
We thank Tim Wheeler for help in the laboratory and for taking pictures of host species. John Beckmann, Amelia Lindsey, Emily Delaney, Sylvain Charlat and three anonymous reviewers provided helpful comments. Research reported in this publication was supported by the National Institute Of General Medical Sciences of the National Institutes of Health (NIH) under award numbers R35GM124701 to B.S.C., R01GM121750 to D.R.M., and R01GM104325 to M.T. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8301035.
Communicating editor: M. Johnston
Literature Cited
- Ahmed M. Z., Li S.-J., Xue X., Yin X.-J., Ren S.-X., et al. , 2015. The intracellular bacterium Wolbachia uses parasitoid wasps as phoretic vectors for efficient horizontal transmission. PLoS Pathog. 11: e1004672 10.1371/journal.ppat.1004672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Thornton K., Clark A., Andolfatto P., 2006. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution 60: 292–302. 10.1111/j.0014-3820.2006.tb01107.x [DOI] [PubMed] [Google Scholar]
- Bailly-Bechet M., Martins-Simoes P., Szollosi G. J., Mialdea G., Sagot M.-F., et al. , 2017. How long does Wolbachia remain on board. Mol. Biol. Evol. 34: 1183–1193. 10.1093/molbev/msx073 [DOI] [PubMed] [Google Scholar]
- Bakovic V., Schebeck M., Telschow A., Stauffer C., Schuler H., 2018. Spatial spread of Wolbachia in Rhagoletis cerasi populations. Biol. Lett. 14: 20180161 (erratum: Biol. Lett. 14: 20180683). 10.1098/rsbl.2018.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L., Hotopp J. C. D., Jolley K. A., Bordenstein S. R., Biber S. A., et al. , 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72: 7098–7110. 10.1128/AEM.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L., Ayoub N. A., Hayashi C. Y., Russell J. A., Stahlhut J. K., et al. , 2008. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 17: 557–569. 10.1111/j.1365-294X.2007.03608.x [DOI] [PubMed] [Google Scholar]
- Bandi C., Anderson T. J., Genchi C., Blaxter M. L., 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. Biol. Sci. 265: 2407–2413. 10.1098/rspb.1998.0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. F., Fallon A. M., 2013. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 43: 867–878. 10.1016/j.ibmb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. F., Ronau J. A., Hochstrasser M., 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2: 17007 10.1038/nmicrobiol.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. F., Bonneau M., Chen H., Hochstrasser M., Poinsot D., et al. , 2019. The toxin-antidote model of cytoplasmic incompatibility: genetics and evolutionary implications. Trends Genet. 35: 175–185. 10.1016/j.tig.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S. R., Bordenstein S. R., 2016. Eukaryotic association module in phage WO genomes from Wolbachia. Nat. Commun. 7: 13155 10.1038/ncomms13155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon D., Rigaud T., Juchault P., 1998. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. Biol. Sci. 265: 1081–1090. 10.1098/rspb.1998.0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig H. R., Zhou W., Dobson S. L., O’Neill S. L., 1998. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand C. L., Kingan S. B., Wu L., Garrigan D., 2013. A selective sweep across species boundaries in Drosophila. Mol. Biol. Evol. 30: 2177–2186. 10.1093/molbev/mst123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. N., Lloyd V. K., 2015. Evidence for horizontal transfer of Wolbachia by a Drosophila mite. Exp. Appl. Acarol. 66: 301–311. 10.1007/s10493-015-9918-z [DOI] [PubMed] [Google Scholar]
- Brownlie J. C., Cass B. N., Riegler M., Witsenburg J. J., Iturbe-Ormaetxe I., et al. , 2009. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5: e1000368 10.1371/journal.ppat.1000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson H. L., Kaneshiro K. Y., Val F. C., 1989. Natural hybridization between the sympatric Hawaiian species Drosophila silvestris and Drosophila heteroneura. Evolution 43: 190–203. 10.1111/j.1558-5646.1989.tb04217.x [DOI] [PubMed] [Google Scholar]
- Cattel J., Nikolouli K., Andrieux T., Martinez J., Jiggins F., et al. , 2018. Back and forth Wolbachia transfers reveal efficient strains to control spotted wing Drosophila populations. J. Appl. Ecol. 55: 2408–2418. 10.1111/1365-2664.13101 [DOI] [Google Scholar]
- Chandler M., Mahillon J., 2002. Insertion sequences revisited, pp. 305–366 in Mobile DNA II, edited by Craig N., Craigie R., Gellert M., Lambowitz A. ASM Press, Washington, DC: . 10.1128/9781555817954.ch15 [DOI] [Google Scholar]
- Charlat S., Ballard J. W. O., Merçot H., 2004. What maintains noncytoplasmic incompatibility inducing Wolbachia in their hosts: a case study from a natural Drosophila yakuba population. J. Evol. Biol. 17: 322–330. 10.1046/j.1420-9101.2003.00676.x [DOI] [PubMed] [Google Scholar]
- Chiu J. C., Jiang X., Zhao L., Hamm C. A., Cridland J. M., et al. , 2013. Genome of Drosophila suzukii, the spotted wing Drosophila. G3 (Bethesda) 3: 2257–2271. 10.1534/g3.113.008185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Bubnell J. E., Aquadro C. F., 2015. Population genomics of infectious and integrated Wolbachia pipientis genomes in Drosophila ananassae. Genome Biol. Evol. 7: 2362–2382. 10.1093/gbe/evv158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeault A. A., Venkat A., Matute D. R., 2016. Correlated evolution of male and female reproductive traits drive a cascading effect of reinforcement in Drosophila yakuba. Proc. Biol. Sci. 283: 20160730 10.1098/rspb.2016.0730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner W. R., Blaxter M. L., Anfora G., Ometto L., Rota-Stabelli O., et al. , 2017. Genome comparisons indicate recent transfer of wRi-like Wolbachia between sister species Drosophila suzukii and D. subpulchrella. Ecol. Evol. 7: 9391–9404. 10.1002/ece3.3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B. S., Ginsberg P. S., Turelli M., Matute D. R., 2017. Wolbachia in the Drosophila yakuba complex: pervasive frequency variation and weak cytoplasmic incompatibility, but no apparent effect on reproductive isolation. Genetics 205: 333–351. 10.1534/genetics.116.196238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B. S., Sedghifar A., Nash W. T., Comeault A. A., Matute D. R., 2018. A maladaptive combination of traits contributes to the maintenance of a Drosophila hybrid zone. Curr. Biol. 28: 2940–2947.e6. 10.1016/j.cub.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., 2008. ISWpi1 from Wolbachia pipientis defines a novel group of insertion sequences within the IS5 family. Gene 409: 20–27. 10.1016/j.gene.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 1989. Patterns of speciation in Drosophila. Evolution 43: 362–381. 10.1111/j.1558-5646.1989.tb04233.x [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 1997. “Patterns of speciation in Drosophila” revisited. Evolution 51: 295–303. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Kim S. Y., Chang A. S., Lachaise D., Elwyn S., 2002. Sexual isolation between two sibling species with overlapping ranges: Drosophila santomea and Drosophila yakuba. Evolution 56: 2424–2434. 10.1111/j.0014-3820.2002.tb00168.x [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, . A. G. Clark, Eisen M. B., Smith D. R., Bergman C. M., Oliver B., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Duron O., Lagnel J., Raymond M., Bourtzis K., Fort P., et al. , 2005. Transposable element polymorphism of Wolbachia in the mosquito Culex pipiens: evidence of genetic diversity, superinfection and recombination. Mol. Ecol. 14: 1561–1573. 10.1111/j.1365-294X.2005.02495.x [DOI] [PubMed] [Google Scholar]
- Ellegaard K. M., Klasson L., Näslund K., Bourtzis K., Andersson S. G. E., 2013. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 9: e1003381 10.1371/journal.pgen.1003381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Kingan S. B., Geneva A. J., Andolfatto P., Clark A. G., et al. , 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22: 1499–1511. 10.1101/gr.130922.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth M., Bleidorn C., 2016. Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat. Microbiol. 2: 16241 10.1038/nmicrobiol.2016.241 [DOI] [PubMed] [Google Scholar]
- Gill A. C., Darby A. C., Makepeace B. L., 2014. Iron necessity: the secret of Wolbachia’s success? PLoS Negl. Trop. Dis. 8: e3224 10.1371/journal.pntd.0003224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano R., O’Neill S. L., Robertson H. M., 1995. Wolbachia infections and the expression of cytoplasmic incompatibility in Drosophila sechellia and D. mauritiana. Genetics 140: 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnsonschlitz D. M., Nassif N. A., Phillis R. W., et al. , 1993. Type-1 repressors of P-element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm C. A., Begun D. J., Vo A., Smith C. C. R., Saelao P., et al. , 2014. Wolbachia do not live by reproductive manipulation alone: infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol. Ecol. 23: 4871–4885. 10.1111/mec.12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird J. C., Sloan D. B., 2016. The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial vs. nuclear genomes. Mol. Biol. Evol. 33: 3042–3053. 10.1093/molbev/msw185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R., Turelli M., 2009. Evolution of incompatibility-inducing microbes in subdivided host populations. Evolution 63: 432–447. 10.1111/j.1558-5646.2008.00550.x [DOI] [PubMed] [Google Scholar]
- Hedges L. M., Brownlie J. C., O’Neill S. L., Johnson K. N., 2008. Wolbachia and virus protection in insects. Science 322: 702 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K., Hammerstein P., Schlattmann P., Telschow A., Werren J. H., 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281: 215–220. 10.1111/j.1574-6968.2008.01110.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. Y., Phillips M. J., Cooper A., Drummond A. J., 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22: 1561–1568. 10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., 1988. Partial cytoplasmic incompatibility between two Australian populations of Drosophila melanogaster. Entomol. Exp. Appl. 48: 61–67. 10.1111/j.1570-7458.1988.tb02299.x [DOI] [Google Scholar]
- Hoffmann A. A., Turelli M., 1988. Unidirectional incompatibility in Drosophila simulans: inheritance, geographic variation and fitness effects. Genetics 119: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Turelli M., 1997. Cytoplasmic incompatibility in insects, pp. 42–80 in Influential Passengers: Inherited Microorganisms and Arthropod Reproduction, edited by O’Neill S. L., Hoffmann A. A., Werren J. H. Oxford University Press, New York. [Google Scholar]
- Hoffmann A. A., Turelli M., Simmons G. M., 1986. Unidirectional incompatibility between populations of Drosophila simulans. Evolution 40: 692–701. 10.1111/j.1558-5646.1986.tb00531.x [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Clancy D. J., Merton E., 1994. Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics 136: 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Clancy D., Duncan J., 1996. Naturally-occurring Wolbachia infection in Drosophila simulans that does not cause cytoplasmic incompatibility. Heredity 76: 1–8. 10.1038/hdy.1996.1 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Montgomery B. L., Popovici J., Iturbe-Ormaetxe I., Johnson P. H., et al. , 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476: 454–457. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- Höhna S., Landis M. J., Heath T. A., Boussau B., Lartillot N., et al. , 2016. RevBayes: bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 65: 726–736. 10.1093/sysbio/syw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Coyne J. A., 2002. Mathematical consequences of the genealogical species concept. Evolution 56: 1557–1565. 10.1111/j.0014-3820.2002.tb01467.x [DOI] [PubMed] [Google Scholar]
- Huigens M. E., Luck R. F., Klaassen R. H., Maas M. F., Timmermans M. J., et al. , 2000. Infectious parthenogenesis. Nature 405: 178–179. 10.1038/35012066 [DOI] [PubMed] [Google Scholar]
- Huigens M. E., de Almeida R. P., Boons P. A., Luck R. F., Stouthamer R., 2004. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc. Biol. Sci. 271: 509–515. 10.1098/rspb.2003.2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. S., Lyon R. T., Sarver B. A. J., Hardwick K., Forney L. J., et al. , 2015. Assembly by reduced complexity (ARC): a hybrid approach for targeted assembly of homologous sequences. bioRxiv. 10.1101/014662 (Preprint posted January 31, 2015). [Google Scholar]
- Hurst G. D. D., Jiggins F. M., 2000. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg. Infect. Dis. 6: 329–336. 10.3201/eid0604.000402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman S. D., Vandervalk B. P., Mohamadi H., Chu J., Yeo S., et al. , 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27: 768–777. 10.1101/gr.214346.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J., Dyer K. A., Cornish C., Minhas M. S., 2006. Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol. 4: e325 [corrigenda: PLoS Biol. 5: e3 (2007)]. 10.1371/journal.pbio.0040325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N. A., Fass J. N., 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. Available at: https://scholar.google.com/scholar?cluster=6706699853004034677&hl=en&oi=scholarr
- Kamimura Y., Mitsumoto H., 2012. Lock-and-key structural isolation between sibling Drosophila species. Entomol. Sci. 15: 197–201. 10.1111/j.1479-8298.2011.00490.x [DOI] [Google Scholar]
- Katoh K., Standley D. M., 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A. D., Barbash D. A., Chang Mell J., Jensen A., Hupalo D., 2015. Highly constrained intergenic Drosophila ultraconserved elements are candidate ncRNAs. Genome Biol. Evol. 7: 689–698. 10.1093/gbe/evv011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Walker T., Sebaihia M., Sanders M. J., Quail M. A., et al. , 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25: 1877–1887. 10.1093/molbev/msn133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Westberg J., Sapountzis P., Näslund K., Lutnaes Y., et al. , 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. USA 106: 5725–5730. 10.1073/pnas.0810753106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo R., Satta Y., Matsuura E. T., Ishiwa H., Takahata N., et al. , 1990. Incomplete maternal transmission of mitochondrial DNA in Drosophila. Genetics 126: 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P., Hoffmann A. A., 2018. Rapid spread of a Wolbachia infection that does not affect host reproduction in Drosophila simulans cage populations. Evolution 72: 1475–1487. 10.1111/evo.13506 [DOI] [PubMed] [Google Scholar]
- Kriesner P., Hoffmann A. A., Lee S. F., Turelli M., Weeks A. R., 2013. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9: e1003607 10.1371/journal.ppat.1003607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriesner P., Conner W. R., Weeks A. R., Turelli M., Hoffmann A. A., 2016. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 70: 979–997. 10.1111/evo.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal R. J., Stevison L. S., Noor M. A. F., 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5: e1000550 10.1371/journal.pgen.1000550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D., Harry M., Solignac M., Lemeunier F., Benassi V., et al. , 2000. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from São Tomé. Proc. Sci. Biol. 267: 1487–1495. 10.1098/rspb.2000.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Fitch W. M., 1974. An examination of the constancy of the rate of molecular evolution. J. Mol. Evol. 3: 161–177. 10.1007/BF01797451 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven H., 1951. Crossing experiments with Culex strains. Evolution 5: 370–375. 10.1111/j.1558-5646.1951.tb02795.x [DOI] [Google Scholar]
- Laven H., 1967. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 216: 383–384. 10.1038/216383a0 [DOI] [PubMed] [Google Scholar]
- LePage D. P., Metcalf J. A., Bordenstein S. R., On J., Perlmutter J. I., et al. , 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543: 243–247. 10.1038/nature21391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-J., Ahmed M. Z., Lv N., Shi P.-Q., Wang X.-M., et al. , 2017. Plantmediated horizontal transmission of Wolbachia between whiteflies. ISME J. 11: 1019–1028. 10.1038/ismej.2016.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey A. R. I., Rice D. W., Bordenstein S. R., Brooks A. W., Bordenstein S. R., et al. , 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol. Evol. 10: 434–451 [corrigenda: Genome Biol. Evol. 11: 1320 (2019)]. 10.1093/gbe/evy012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A., Herrig D., Brud E., Stecklein Z., 2014. Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea. Mol. Ecol. 23: 1124–1136. 10.1111/mec.12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse K., Clarke M., Ritchie M. G., Etges W. J., 2015. Genome‐wide tests for introgression between cactophilic Drosophila implicate a role of inversions during speciation. Evolution 69: 1178–1190. 10.1111/evo.12650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Longdon B., Bauer S., Chan Y.-S., Miller W. J., et al. , 2014. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 10: e1004369 10.1371/journal.ppat.1004369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., 2010. Reinforcement of gametic isolation in Drosophila. PLoS Biol. 8: e1000341 10.1371/journal.pbio.1000341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., Ayroles J. F., 2014. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol. 27: 1057–1068. 10.1111/jeb.12391 [DOI] [PubMed] [Google Scholar]
- Matute D. R., Coyne J. A., 2010. Intrinsic reproductive isolation between two sister species of Drosophila. Evolution 64: 903–920. 10.1111/j.1558-5646.2009.00879.x [DOI] [PubMed] [Google Scholar]
- Mavingui P., Valiente Moro C., Tran-Van V., Wisniewski-Dye F., Raquin V., et al. , 2012. Whole-genome sequence of Wolbachia strain wAlbB, an endosymbiont of tiger mosquito vector Aedes albopictus. J. Bacteriol. 194: 1840 10.1128/JB.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C. J., Lane R. V., Cass B. N., Fong A. W. C., Sidhu M., et al. , 2009. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323: 141–144. 10.1126/science.1165326 [DOI] [PubMed] [Google Scholar]
- Meany M. K., Conner W. R., Richter S. V., Bailey J. A., Turelli M., et al. , 2019. Loss of cytoplasmic incompatibility and minimal fecundity effects explain relatively low Wolbachia frequencies in Drosophila mauritiana. Evolution 73: 1278–1295. 10.1111/evo.13745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M., Nikoh N., Hosokawa T., Fukatsu T., 2015. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. MBio 6: e01732–15 10.1128/mBio.01732-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P., 2013. Degree of sympatry affects reinforcement in Drosophila. Evolution 67: 868–872. 10.1111/j.1558-5646.2012.01817.x [DOI] [PubMed] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A., Korobeynikov A., et al. , 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, pp. 158–170 in Research in Computational Molecular Biology, edited by Deng M., Jiang R., Sun F., Zhang X. Springer, Berlin, Heidelberg. [Google Scholar]
- O’Connor L., Plichart C., Sang A. C., Brelsfoard C. L., Bossin H. C., et al. , 2012. Open release of male mosquitoes infected with a Wolbachia biopesticide: field performance and infection containment. PLoS Negl. Trop. Dis. 6: e1797 10.1371/journal.pntd.0001797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. L., Giordano R., Colbert A. M., Karr T. L., Robertson H. M., 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 89: 2699–2702. 10.1073/pnas.89.7.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. L., Hoffmann A. A., Werren J. H., 1998. Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford University Press, Oxford. [Google Scholar]
- Prout T., 1994. Some evolutionary possibilities for a microbe that causes incompatibility in its host. Evolution 48: 909–911. 10.1111/j.1558-5646.1994.tb01371.x [DOI] [PubMed] [Google Scholar]
- Raychoudhury R., Baldo L., Oliveira D. C. S. G., Werren J. H., 2009. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63: 165–183. 10.1111/j.1558-5646.2008.00533.x [DOI] [PubMed] [Google Scholar]
- Richardson M. F., Weinert L. A., Welch J. J., Linheiro R. S., Magwire M. M., et al. , 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8: e1003129 10.1371/journal.pgen.1003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S. A., 2018. Wolbachia and the near cessation of dengue outbreaks in Northern Australia despite continued dengue importations via travellers. J. Travel Med. 25: tay084. [DOI] [PubMed] [Google Scholar]
- Rousset F., Solignac M., 1995. Evolution of single and double Wolbachia symbioses during speciation in the Drosophila simulans complex. Proc. Natl. Acad. Sci. USA 92: 6389–6393. 10.1073/pnas.92.14.6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., Bouchon D., Pintureau B., Juchault P., Solignac M., 1992. Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. Biol. Sci. 250: 91–98. 10.1098/rspb.1992.0135 [DOI] [PubMed] [Google Scholar]
- Sánchez L., Santamaria P., 1997. Reproductive isolation and morphogenetic evolution in Drosophila analyzed by breakage of ethological barriers. Genetics 147: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. L., Barton N. H., Rašić G., Turley A. P., Montgomery B. L., et al. , 2017. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 15: e2001894 10.1371/journal.pbio.2001894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Ayroles J., Matute D. R., Kern A. D., 2018. Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14: e1007341 10.1371/journal.pgen.1007341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler H., Köppler K., Daxböck-Horvath S., Rasool B., Krumböck S., et al. , 2016. The hitchhiker’s guide to Europe: the infection dynamics of an ongoing Wolbachia invasion and mitochondrial selective sweep in Rhagoletis cerasi. Mol. Ecol. 25: 1595–1609. 10.1111/mec.13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T., 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Shoemaker D. D., Katju V., Jaenike J., 1999. Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution 53: 1157–1164. 10.1111/j.1558-5646.1999.tb04529.x [DOI] [PubMed] [Google Scholar]
- Shropshire J. D., On J., Layton E. M., Zhou H., Bordenstein S. R., 2018. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 115: 4987–4991. 10.1073/pnas.1800650115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire J. D., Leigh B., Bordenstein S. R., Duplouy A., Riegler M., et al. , 2019. Models and nomenclature for cytoplasmic incompatibility: caution over premature conclusions – a response to Beckmann et al. Trends Genet. 35: 397–399. 10.1016/j.tig.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Siozios S., Cestaro A., Kaur R., Pertot I., Rota-Stabelli O., et al. , 2013. Draft genome sequence of the Wolbachia endosymbiont of Drosophila suzukii. Genome Announc. 1: e00032-13. 10.1128/genomeA.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton E. R., Harris S. R., Parkhill J., Sinkins S. P., 2014. Comparative genome analysis of Wolbachia strain wAu. BMC Genomics 15: 928 10.1186/1471-2164-15-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S., 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol, Biol, Evol, 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Teixeira L., Ferreira A., Ashburner M., 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6: e1000002 10.1371/journal.pbio.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., 1994. Evolution of incompatibility-inducing microbes and their hosts. Evolution 48: 1500–1513. 10.1111/j.1558-5646.1994.tb02192.x [DOI] [PubMed] [Google Scholar]
- Turelli M., Hoffmann A. A., 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 353: 440–442. 10.1038/353440a0 [DOI] [PubMed] [Google Scholar]
- Turelli M., Hoffmann A. A., 1995. Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics 140: 1319–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Lipkowitz J. R., Brandvain Y., 2014. On the Coyne and Orr-igin of species: effects of intrinsic postzygotic isolation, ecological differentiation, X chromosome size, and sympatry on Drosophila speciation. Evolution 68: 1176–1187. 10.1111/evo.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Cooper B. S., Richardson K. M., Ginsberg P. S., Peckenpaugh B., et al. , 2018. Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr. Biol. 28: 963–971.e8. 10.1016/j.cub.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini D. A., Matute D. R., 2017. Fine scale mapping of genomic introgressions within the Drosophila yakuba clade. PLoS Genet. 13: e1006971 10.1371/journal.pgen.1006971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini D. A., McGirr J. A., Patel S. S., David J. R., Matute D. R., 2017. The rate of evolution of postmating-prezygotic reproductive isolation in Drosophila. Mol. Biol. Evol. 35: 312–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavre F., Mouton L., Pannebakker B. A., 2009. Chapter 12 Drosophila–parasitoid communities as model systems for host–Wolbachia interactions, pp. 299–331 in Advances in Parasitology. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Wallau G. L., da Rosa M. T., De Re F. C., Loreto E. L., 2016. Wolbachia from Drosophila incompta: just a hitchhiker shared by Drosophila in the new and old world? Insect Mol. Biol. 25: 487–499. 10.1111/imb.12237 [DOI] [PubMed] [Google Scholar]
- Weeks A. R., Turelli M., Harcombe W. R., Reynolds K. T., Hoffmann A. A., 2007. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 5: e114 10.1371/journal.pbio.0050114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert L. A., Araujo-Jnr E. V., Ahmed M. Z., Welch J. J., 2015. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. Biol. Sci. 282: 20150249 10.1098/rspb.2015.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Windsor D. M., 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. Biol. Sci. 267: 1277–1285. 10.1098/rspb.2000.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Zhang W., Guo L. R., 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. Biol. Sci. 261: 55–63. 10.1098/rspb.1995.0117 [DOI] [PubMed] [Google Scholar]
- Wu M., Sun L. V., Vamathevan J., Riegler M., Deboy R., et al. , 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: e69 10.1371/journal.pbio.0020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen J. H., Barr A. R., 1971. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 232: 657–658. 10.1038/232657a0 [DOI] [PubMed] [Google Scholar]
- Yukilevich R., 2012. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 66: 1430–1446. 10.1111/j.1558-5646.2011.01534.x [DOI] [PubMed] [Google Scholar]
- Zabalou S., Charlat S., Nirgianaki A., Lachaise D., Merçot H., et al. , 2004. Natural Wolbachia infections in the Drosophila yakuba species complex do not induce cytoplasmic incompatibility but fully rescue the wRi modification. Genetics 167: 827–834. 10.1534/genetics.103.015990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalou S., Apostolaki A., Pattas S., Veneti Z., Paraskevopoulos C., et al. , 2008. Multiple rescue factors within a Wolbachia strain. Genetics 178: 2145–2160. 10.1534/genetics.107.086488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Wolbachia assemblies (BioProject PRJNA543889, accession numbers GCA_005862115.1, GCA_005862095.1, and GCA_005862135.1) and Sanger sequences (MK950151 and MK950152) are archived in GenBank. All three assemblies were manually corrected using Sanger sequence to accurately represent variation in the cin region. Relevant genetic data and scripts have been uploaded to DRYAD (10.5061/dryad.8n1n677). Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8301035.