SUMMARY
The brain processes information and generates cognitive and motor outputs through functions of spatially organized proteins in different types of neurons. More complete knowledge of proteins and their distributions within neuronal compartments in intact circuits would help in the understanding of brain function. We used unbiased in vivo protein labeling with intravitreal NHS-biotin for discovery and analysis of endogenous axonally transported proteins in the visual system using tandem mass spectrometric proteomics, biochemistry, and both light and electron microscopy. Purification and proteomic analysis of biotinylated peptides identified ~1,000 proteins transported from retinal ganglion cells into the optic nerve and ~575 biotinylated proteins recovered from presynaptic compartments of lateral geniculate nucleus and superior colliculus. Approximately 360 biotinylated proteins were differentially detected in the two retinal targets. This study characterizes axonally transported proteins in the healthy adult visual system by analyzing proteomes from multiple compartments of retinal ganglion cell projections in the intact brain.
In Brief
Axonal protein transport is essential for circuit function. Schiapparelli et al. use unbiased in vivo protein labeling and mass spectrometry to identify ~1,000 proteins in the “RGC axonal transportome.” About 350 retinal proteins are differentially transported to the lateral geniculate nucleus or the superior colliculus, indicating target-specific diversity in presynaptic protein content.
Graphical Abstract
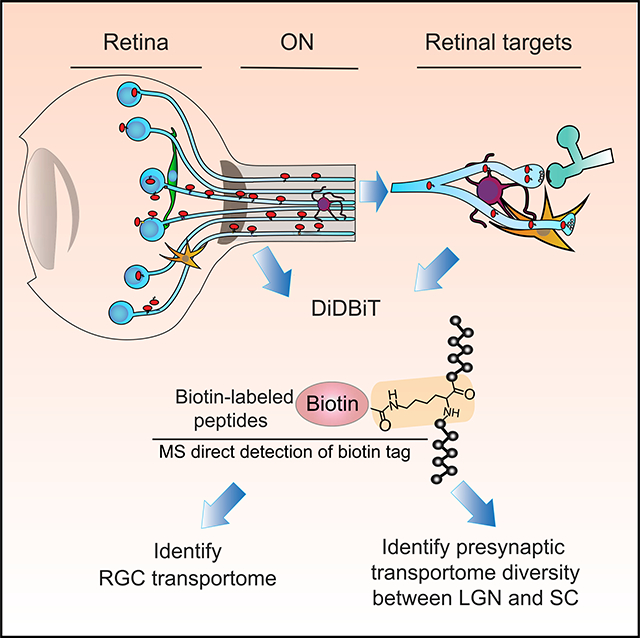
INTRODUCTION
The visual system is composed of a spatially distributed circuit in which diverse retinal ganglion cells (RGCs) project axons, the retinal outputs, to different retinorecipient targets via the optic nerve. Protein localization in cellular compartments is particularly important in the CNS, where neurons are highly polarized, and biochemical processes in dendrites, cell bodies, axons, and presynaptic terminals are specialized. Anatomical and functional studies indicate that visual information processing differs among retinal targets (Dhande et al., 2015; Ellis et al., 2016; Martersteck et al., 2017). This could be accomplished by differences in RGC pre- or postsynaptic protein components or both. Although different target cells, for example in superior colliculus (SC) and lateral geniculate nucleus (LGN), could contain different synaptic proteins that support differential processing of incoming visual information, it’s less clear whether different proteins are transported to different retinal targets. Although understanding of the circuit function depends on knowledge of the component proteins, determining protein distribution within neuronal compartments in the intact CNS has been impeded by the lack of approaches to label populations of endogenously transported proteins in an unbiased manner.
We were interested in dissecting the proteome of RGC projections in the intact brain to identify proteins that are transported from RGC somata into their axons and determining whether RGC proteins are differentially transported to specific retinorecipient targets. To address these points, we combined in vivo RGC protein labeling with N-hydroxysuccinimidobiotin (NHS-biotin) and detection of biotin-modified peptides in visual targets for unbiased proteomic analysis of proteins transported from RGCs into the optic nerve (ON) and two major retinal targets, the SC and the LGN. Mass spectrometry revealed ~1,000 biotinylated proteins in the RGC transportome. Light and electron microscopy demonstrated the distribution of biotinylated proteins within axons in the ON and retinal targets. Remarkably, ~360 proteins were differentially detected in SC or LGN. These data identify axonally transported proteins from RGCs in vivo and demonstrate differential distribution of RGC proteins to distinct retinal targets, suggesting hypotheses for molecular regulation of specificity in axon transport and synaptic function.
RESULTS
In Vivo Intravitreal NHS-Biotin Labels Retinal Proteins Transported into the Visual Pathway
To identify proteins that are transported from RGC somata into axons and synaptic targets in vivo, we labeled proteins in the retina in adult rats with intravitreal injections of NHS-biotin and analyzed biotinylated proteins harvested from the visual system. In vivo protein biotinylation with NHS-biotin has many advantages: it binds covalently to lysines and N-terminal amino acids of proteins, resulting in extensive protein labeling. NHS-biotin cannot be re-incorporated after protein degradation because the succinimide group is quenched after reacting with amino groups and because the resulting biotin-tagged amino acids, such as biotinyl-lysine, cannot be charged onto tRNAs (Watanabe et al., 2007). We harvested tissue from retinas, ONs, SCs, LGNs, and the frontal cortex (FC, a non-visual control area) for analysis by biochemistry, proteomics, and histology (Figures 1A and 1B). Western blots indicated that biotinylated proteins over a wide range of molecular weights were recovered from retina, ON, LGN, and SC, whereas only endogenously biotinylated carboxylases (McKay et al., 2008) were detected in FC and saline-injected control samples (Figure 1C). To test whether we could be detecting diffusion or transport of free biotin, rather than biotinylated protein transport, we injected saline or NHS-biotin intravitreally and collected retinal and LGN tissue. Analysis of biotin revealed substantial protein-bound biotin and free, unbound biotin in the retina after NHS-biotin injections compared with background biotin detected after saline injections. In LGN, only protein-bound biotin was detected, indicating that protein labeling was confined to the retina and that free biotin was not recovered from retinal targets (Figure 1D).
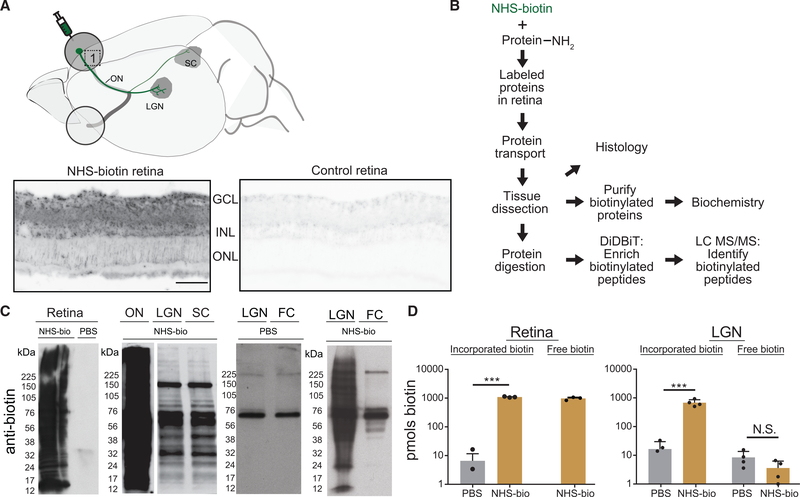
Figure 1. In Vivo Intravitreal NHS-Biotin Injection Labels Retinal Proteins Transported into the Visual Pathway.
(A) Diagram of the rat visual system schematizing the retinal injection of NHS-biotin and retinal ganglion cell projection sites from which transported biotinylated proteins were analyzed. Biotinylated proteins, visualized by tyramide signal amplification in retinal cross sections, were distributed throughout the retina but enriched in the ganglion cell layer (GCL) and inner nuclear layer (INL) in the NHS-biotin-injected tissue only. Images were collected under the same acquisition parameters. Scale bar = 25 μm and applies to both images.
(B) Workflow for in vivo protein labeling strategy and analysis of biotinylated proteins. Adult rats received intravitreal injections once a day for 7 days, and tissue was collected 11 days after the first injection. NHS-biotin binds irreversibly to amino groups of lysine residues and the N-terminals of proteins. Biotinylated proteins are either analyzed in histological sections or purified from brain regions and used for biochemical analysis or tandem MS (MS/MS). Biotin-labeled peptides are identified directly by MS/MS.
(C) Western blot detection of biotinylated proteins from retina, ON, LGN, SC, and the non-visual area, the frontal cortex (FC), following in vivo intravitreal injection of NHS-biotin or saline. Asterisks mark endogenously biotinylated carboxylases at ~60 and 120 kDa, the only labeled proteins in the FC.
(D) Comparison of biotin incorporated into proteins from retina (left) and LGN (right) and free biotin from LGN (right) after intravitreal NHS-biotin or phosphate-buffered saline (PBS) injections. Significantly more biotin was bound to protein in NHS-biotin samples compared with background values in PBS. In LGN samples, significantly more biotin was bound to protein in animals with intravitreal NHS-biotin injections compared with PBS. Comparable low levels of free biotin were detected in LGN from NHS-biotin- and PBS-injected animals, indicating that most biotin in LGN samples was bound to the protein.
Retina, n = 3, ***p < 0.001, two-tailed Student’s t test; LGN, n = 4, ***p < 0.001, two-tailed Holm-Sidak corrected t test. Means ± SEM.
Although we focused on anterograde protein transport in the visual system (Figure 1), we also tested whether NHS-biotin protein labeling could be adapted to different labeling strategies. We exposed the surface of the SC to gelfoam saturated with NHS-biotin and visualized retrograde NHS-biotin-labeled protein transport from SC, through the optic tract and ON, by histology and western blot (Figure S1). Furthermore, unilateral intracortical NHS-biotin injections in postnatal day 3 rat pups produced extensive labeling in callosal axons projecting to the contralateral hemisphere, including cortical layers and the anterior commissure (Figure S2). These observations demonstrate the versatility of NHS-biotin labeling to study axonal cargo in vivo in diverse axonal projections.
We visualized biotinylated proteins in RGC projections in sagittal sections through the optic tract and LGN. Biotin immuno-label was detected in the optic tract and LGN of animals that received intravitreal NHS-biotin injections (Figures 2A and 2B) but not biocytin (biotinyl-lysine) or saline (Figures 2C, 2D, and S3A–S3C). We performed immunoelectron microscopy, visualizing biotin with either 3,3′-diaminobenzidine (DAB) (Figure 2H) or fluoro-nanogold streptavidin (Figures 2F and 2G), which indicated that biotinylated proteins in the ON were confined to RGC axons, and no biotinylated proteins were detected in the surrounding myelinated sheath (Figure 2E). Immunolabeling with 1.4 nm of nanogold particles identified biotinylated proteins inside the axons (Figure 2F). We used fluorescent streptavidin labeling in sections throughout the LGN and imaged a sparsely innervated region at the margin of the LGN to visualize retinogeniculate axons with intensely labeled boutons (Figure 2G), indicative of healthy RGCs. Immunoelectron microscopy also identified biotinylated proteins within presynaptic terminals in the LGN (Figures 2H and 2I). These data indicate that in vivo protein labeling with NHS-biotin allows biochemical analysis of transported proteins and their visualization at the light and ultrastructural levels in sections from the intact visual system.
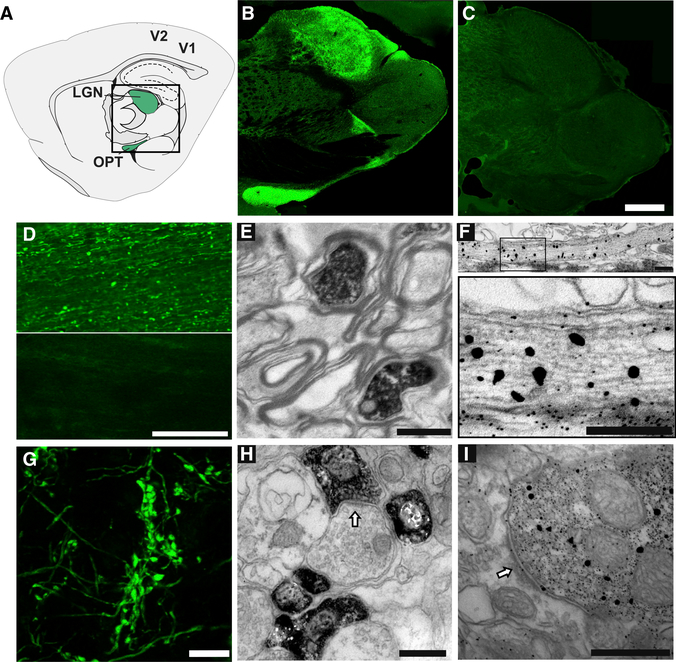
Figure 2. Light and Electron Microscope Detection of Transported Biotinylated Proteins in the Visual Pathway.
(A) Diagram of a sagittal section through the rat brain, adapted from the Paxinos atlas, showing the optic tract (OPT) and LGN (green) in the central boxed region and primary and secondary visual cortex (V1 and V2, respectively).
(B and C) Distribution of biotinylated proteins in the optic tract and LGN, detected with fluorescent streptavidin in sagittal brain sections from animals after intravitreal injection of NHS-biotin (B) or biocytin (C). Images are montages of single optical sections assembled in Photomerge. The montage in (C) was mounted on a black background. Scale bar in (C): 1 mm, applies to (B) and (C).
(D) Immunolabeled biotinylated proteins in the ON after intravitreal injection of NHS-biotin (top) or saline (bottom). Scale bar: 200 μm.
(E) Transmission immuno-electron micrograph of a cross section through the optic tract labeled with streptavidin-horseradish peroxidase (HRP) and DAB/nickel. Biotinylated proteins are contained within the axoplasm of RGC axons. Scale bar: 1 μm.
(F) Immuno-electron micrograph of a transverse section through the optic nerve labeled with 1.4 nm streptavidin-nanogold particles plus gold enhancement showing biotinylated proteins in RGC axons at low (top) and higher (bottom) magnification. The larger gold particles seen with gold enhancement indicate sites of greater biotin signal; individual particles outside of axons are background labeling.
(G-I) Intravitreal NHS-biotin injections label the RGC projections in LGN. Scale bar: 500 nm (G).
Confocal single optical section of fluorescent streptavidin immunolabeled, retinogeniculate axons showing intensely labeled boutons. Scale bar: 10 μm.
(H and I) Transmission immuno-electron micrographs showing the distribution of axonally transported biotinylated proteins labeled with avidin-HRP plus DAB (H) or streptavidin-nanogold (I) in presynaptic profiles in the LGN. Scale bars: 1 μm.
Proteomic Identification of the RGC Axonal Transportome
Although previous analyses helped identify proteins in the retina (Belin et al., 2015; Cagnetta et al., 2018; Harman et al., 2018; Shigeoka et al., 2016), the distribution of the population of RGCs proteins in retinal projections are largely unknown. To study RGC proteins transported into the ON and RGC targets–the “RGC axonal transportome,”–we analyzed biotinylated proteins isolated from the retina, ON, LGN, and SC using tandem mass spectrometry (MS/MS) combined with DiDBiT, a strategy to enrich biotinylated peptides that facilitates direct MS/MS identification of biotinylated proteins (Schiapparelli et al., 2014). We detected 1,765 biotinylated proteins in the retina and 997 biotinylated proteins in the RGC transportome, pooled from the ON, LGN, and SC samples (Figures 3A and 3B; Table S1). By considering only directly detected biotinylated peptides, with an exact mass shift of +226.0776 on lysines, we increased our confidence that identified proteins were NHS-biotin labeled, axonally transported proteins and not unlabeled proteins present from before the labeling protocol or contaminants from the surrounding tissue. Of the 997 proteins in the RGC transportome, 272 were only detected in RGC projections (Figure 3B; Table S1). Our ability to detect those proteins in the RGC transportome was likely due to their relative enrichment in the transportome as they are transported to presynaptic terminals, and the decreased complexity of the transportome samples, in which only RGC axonal proteins are biotinylated, compared with the retinal sample (Schiapparelli et al., 2014; Schirmer et al., 2003). This analysis compared the proteomics data from the retina, which contains the cell bodies of the RGCs and is the source material for the biotinylated proteins in the study, to the RGC transportome, which comprises the RGC proteins that are transported out of the cell body. We compared the retinal proteome with the RGC transportome to contrast the entire set of labeled retinal proteins to the fraction that leaves the cell body and is transported into the axons in the ON. By isolating axonal and presynaptic compartments of RGCs from their somata in vivo, we can now assess RGC protein distribution in different neuronal compartments and explore potential functions of neuronal proteins with proteomic methods.
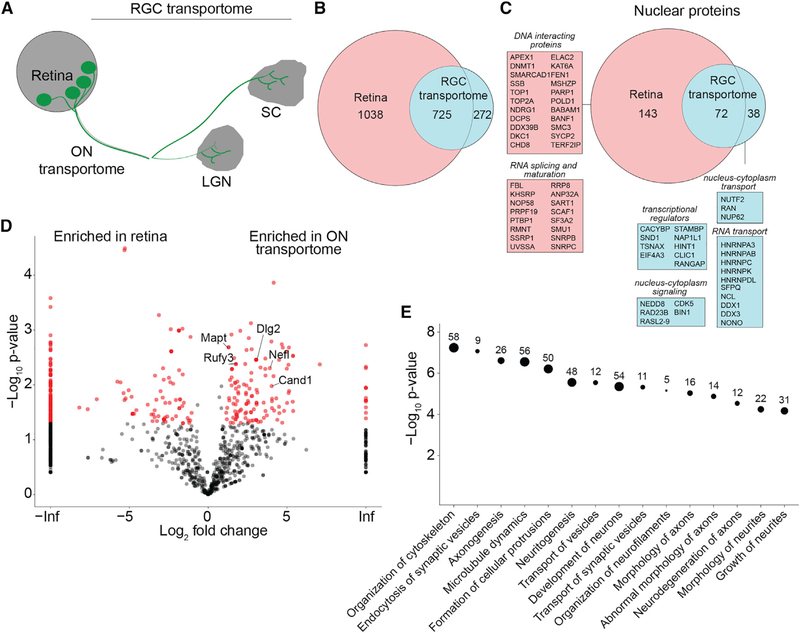
Figure 3. Proteomic Analysis of the RGC and Optic NerveTransportomes.
(A) Diagram of the retinal projection to LGN and SC. The RGC transportome includes the biotinylated proteins transported from retina to ON, SC, and LGN.
(B) Venn diagram of the population of biotinylated proteins in the retina compared with the RGC transportome.
(C) Venn diagram of the populations of biotinylated nuclear proteins in the retina and in the RGC transportome. Nuclear proteins in the RGC transportome are involved in signaling between the nucleus and the cytoplasm, transcriptional regulation, and RNA transport. Nuclear proteins in the retina, but not in the RGC transportome, are DNA-interacting proteins and RNA-splicing and -maturation factors.
(D) Volcano plot comparing biotinylated proteins in the retina and the ON transportome. The plot shows the log2-fold change in enrichment of normalized spectral counts for each protein in the retina and the ON transportome against the –log10 p value, a measure of statistical significance. Proteins marked in red are significantly different between the 2 datasets, n = 3, p < 0.05, t test with Benjamini-Hochberg correction. Data points plotted on the left or right y axes represent proteins that are only detected in one type of dataset.
(E) Ingenuity analysis of the categories of most significantly enriched proteins in the ON transportome. Proteomic data used to generate these figure panels are shown in Table S1. Right-tailed Fisher’s exact test.
Nuclear Proteins in the RGC Transportome
We detected 296 biotinylated nuclear proteins in the retina and 86 in the transportome (Figure 3C; Table S1). Gene ontology analysis indicates that nuclear proteins from the retina were enriched in chromatin- and chromosome-interacting proteins, whereas nuclear proteins in the RGC transportome were enriched in perinuclear, cytoplasmic-nucleus, and endoplasmic reticulum (ER)-nucleus-interacting proteins, categories of proteins that shuttle in and out of the nucleus (Figure S4; Table S1). Examples of proteins annotated to the nucleus that were enriched in the RGC transportome include members of the heterogenous nuclear ribonucleoprotein (HNRNP) family. HNRNPs are a large family of proteins with known functions in splicing, mRNA stabilization, and protein synthesis (Geuens et al., 2016). HNRNP K is required for axon development and regeneration (Liu et al., 2008; Liu et al., 2012), suggesting that other HNRNPs, thus far, not studied in neurons, might function in axons and synapses. In addition, we identified nucleolin and mammalian target of rapamycin (MTOR), which cooperatively regulate RNA localization and translation in axons (Poulopoulos et al., 2019; Terenzio et al., 2018); DDX1, which is involved in RNA granule transport (Kanai et al., 2004); and the small GTPase, RAN, known for its role in nucleocytoplasmic transport in non-neuronal cells, coordinate retrograde axonal-nuclear signaling in response to axon injury (Yudin et al., 2008). We also identified transcriptional regulators, including calcyclin-binding protein CACYBP, which translocates from cytosol to nucleus in a calcium-dependent manner and is involved in extracellular signal-regulated kinase ½ (ERK½)-mediated transcriptional regulation (Filipek et al., 2002; Kilanczyk et al., 2015); the transcriptional regulator SND1 (Cappellari et al., 2014); and TSNAX, which interacts with the axonal protein GAP43 and regulates transcription involved in axonal regeneration (Schröer et al., 2007). Finally, we identify histone proteins in the RGC transportome in vivo, which are also reported to be locally synthesized in growth cones ex vivo (Cagnetta et al., 2018), suggesting diverse mechanisms for regulating histone levels in axons. These findings provide evidence of distributions of nuclear proteins in retinal projections, which may foster discovery of previously unrecognized functions for these proteins in axons and axon terminals.
Dissection of the RGC Transportome into Cellular Compartments: The ON Transportome
The RGC transportome includes the pooled MS/MS samples from ON, LGN, and SC. The LGN and SC samples are the biotinylated proteins in retinal terminals in each target. Here, we consider the constituent cellular compartments separately: the ON transportome; the presynaptic transportome, combined from SC and LGN samples; and finally, the differential LGN transportome versus SC transportome, providing increasing resolution of proteins in different cellular compartments of RGC projections.
The ON transportome is the population of biotinylated proteins detected in the ON. We identified 844 proteins in the ON transportome by MS/MS, providing an inventory of axonally transported proteins from intact CNS tissue. ImmunoEM images (Figure 2) indicate that the biotinylated proteins are confined to axons. We compared the ON transportome with biotinylated retinal proteins in a volcano plot (Figure 3D). Proteins that are enriched in the ON transportome include MAPT (Tau), as well as proteins not previously described in retinal projections: RUFY3, PSMA5, DLG2, and CAND1. Proteins enriched in the retina include Histone H2B and H2A, beta-crystallin, and vimentin. Ingenuity pathway analysis of proteins enriched in the ON transportome indicated that the top five most-significantly enriched categories pertain to diverse axonal functions: organization of cytoskeleton, synaptic vesicle endocytosis, axonogenesis, microtubule dynamics, and formation of cellular protrusions (Figure 3E; Table S1). Even in the adult animal, most of these enriched categories in the ON transportome pertain to growth, development, and morphological dynamics, consistent with the capacity for structural changes in axons in the mature visual system (Hensch and Quinlan, 2018; Stryker and Löwel, 2018).
Previous methods to isolate axonal material for proteomics involved either isolation of myelinated nerves or axoplasm extraction from dissected nerves (Jiménez et al., 2005; Michaelevski et al., 2010). Whole-tissue ON dissection includes the ON sheath with diverse non-neuronal cell types. To determine whether biotinylated proteins identified in the ON transportome are specific to RGC axons without contaminants, we compared the ON transportome to the proteome of the total dissected ON (Figure 4A). The total dissected ON proteome consisted of 2,197 proteins; of which, 690 were present in the ON transportome, and 154 proteins were detected in the ON transportome but not in the total ON proteome (Figure 4B; Table S2). This increased detection of biotinylated proteins in the ON transportome, despite the significantly greater input used for the total ON proteome sample (see Method Details) likely arises because biotin enrichment enhances the MS detection limit of proteins that might be diluted in the more-complex total ON sample (Schiapparelli et al., 2014).
Figure 4. Comparison of the Total Optic Nerve Proteome and Optic Nerve Transportome.
(A) Diagram of the retinal projection to LGN and SC, highlighting the tissue source of the ON transportome.
(B) Venn diagram comparing the MS/MS datasets of the total dissected ON (“Total ON”) from animals without NHS-biotin label and the ON transportome from animals with the intravitreal NHS-biotin label. The Total ON proteome includes proteins from all cell types in the ON (astrocytes, oligodendrocytes, fibroblasts, immune cells, and neurons).
(C) Table of spectral counts for proteins from different cell types in the Total ON proteome and ON transportome. The ON transportome does not contain any proteins that serve as unique identifiers of non-neuronal cell types seen in the Total ON proteome.
(D) Volcano plot comparing the Total ON and ON transportome shows significant enrichment of some neuronal proteins in the ON transportome and a large population of proteins in the Total ON that are not in the ON transportome. n = 3, p < 0.05, t test with Benjamini-Hochberg correction.
(E) Western blot validation of proteins in the ON transportome. Western blots of immunoprecipitation (IP; left) and NeutrAvidin purified proteins (right) comparing immunolabel in ON from animals with intravitreal injection of NHS-biotin and control. TAU, RAS, MTOR, GAPDH, ALIX, CaMKII, HNRNPA2, SNCB, and PSMD2 are enriched in IPs from animals that received NHS-biotin and not in control animals. Experiment were done in triplicate.
Proteomic data used to generate these figures are shown in Table S2.
Analyzing this further, we compared replicates of the ON transportome and dissected ON proteome for markers of oligodendrocytes (MOG, MOBP, and CSPG4), astrocytes (GFAP, GLAST1, GLT-1, and ALDH1L1), immune cells (CD81, CD44, and CD38), fibroblasts (LAMB2 and CD151), and neurons (CAMKII, NEFL, and NCAM). Although neuronal markers and ubiquitously expressed proteins (e.g., glyceraldehyde-3-phosphate dehydrogenase [GADPH]) were detected in both samples, astrocyte, oligodendrocyte, immune cells, and fibroblast components were not detected in the ON transportome (Figures 4C and 4D; Table S2). Interestingly, we detected apolipoprotein E (APOE) in the ON transportome, as well as neuronal receptors for apolipoproteins, LRPAP1 and VLDLR. Although APOE originates in glial, it is released and taken up by neurons as a mechanism of lipid distribution in the brain (Vance and Hayashi, 2010). Some of the most abundant proteins in the ON transportome include α-synuclein (SNCA) and the metabolic proteins GADPH, enolase, and aldolase (Figure 4D; Table S2), which provide ATP to fuel axon transport (Zala et al., 2013). We also detected and validated biotinylated TAU, RAS, MTOR, GAPDH, ALIX, CAMKII, HNRNPA2, SNCB, and PSMD2 in the ON (Figure 4E). Together, these results demonstrate the specificity of in vivo biotinylation to identify the axonal proteome of a specific cell type–RGCs–from intact, complex tissue without requiring isolation of cells of interest.
Distribution of Proteins into Subcellular Compartments: Analysis of the Presynaptic Transportome
Neurons are highly polarized cells with diverse subcellular compartments, such as pre- and postsynaptic elements and the axon initial segment, which are composed of uniquely localized proteins. Understanding neuronal function and the contributions of subcellular specializations to neuronal and circuit function requires an accurate assessment of protein components and their distribution in neurons. Therefore, we next asked whether subsets of proteins in the RGC transportome were specifically enriched in presynaptic terminals. To identify these molecular components, we compared the presynaptic transportome dataset, 576 biotinylated proteins from SC and LGN, to the dataset of 844 proteins in the ON transportome. Half (421) were detected only in the ON transportome, and half (423) overlapped between the ON and presynaptic transportome datasets; 153 proteins in the presynaptic transportome were not detected in the ON transportome, likely because of the relative enrichment of these proteins in the presynaptic compartment (Figure 5A; Table S3). Proteins detected in the presynaptic transportome include components of the core synaptic vesicle fusion machinery and known synaptic proteins, such as vesicle-associated membrane proteins (VAMPs), synaptophysin, RAB3 isoforms, synapsin 1, and syntaxin 1b. We compared the ON and the SC transportome in a volcano plot: in addition to proteins detected uniquely in the ON or SC transportomes, proteins enriched in the SC transportome included mitochondrial proteins and the guanine nucleotidebinding protein, GNAO1, whereas those significantly enriched in the ON transportome included NEDD8, actin, and microtubule-associated proteins (Figure 5B; Table S3).
Figure 5. Analysis of the Presynaptic Transportome.
(A) Venn diagram comparing the ON transportome and presynaptic transportome. The presynaptic transportome contains many classical presynaptic proteins involved in presynaptic machinery for neurotransmitter release.
(B) Volcano plot comparing protein abundance in the ON transportome and the SC transportome. Significantly enriched proteins are in red. Data points plotted on the left or right y axes represent proteins that are only detected in one dataset. n = 3, p < 0.05, t test with Benjamini-Hochberg correction.
(C) Enrichment of presynaptic proteins, identified from the annotated presynaptic protein databases, in the ON, SC, and LGN transportomes normalized to the retina. The NeutrAvidin flow through (LGN fl) is not enriched in presynaptic proteins. Fisher’s exact test, **p < 0.001, †† < 0.001 (Z = −5.35, retina versus ON; Z = −9.9772, retina versus LGN transportome; Z = −11.1862, retina versus SC transportome; Z = −0.9256, p = 0.35238, retina versus total LGN; Z = −4.8628, ON transportome versus LGN transportome; and Z = −5.5885, ON transportome versus SC transportome).
(D) Western blot validation of biotinylated proteins from SC, with NeutrAvidin enrichment of MTOR, MUNC18, and ALIX, but not GFAP, in NHS-biotin treated animals versus control. SC tissue from 3 animals was pooled per sample. Experiments were done in triplicate.
(E) Confocal images of LGN from animals with intravitreal NHS-biotin show biotinylated proteins (green) in RGC presynaptic boutons, with either endogenous synaptotagmin 1 or ALIX (red). Images show colocalization of endogenous proteins with biotin label in presynaptic boutons. Scale bar: 50 μm.
(F) Venn diagram comparing the presynaptic transportome with those of public databases for presynaptic proteins. Proteins detected only in the presynaptic transportome include those that function in vesicle trafficking and those involved in proteostasis.
Proteomic data used to generate these figures are shown in Table S3.
We compared results from our presynaptomes with the public knowledgebases of presynaptic proteins, including SynaptomeDB, compiled and periodically updated from published studies and proteomic resources of synaptosomes, presynaptic proteins, presynaptic active zone, and synaptic vesicles from heterogeneous tissue samples and diverse species (Bayés et al., 2011; Pirooznia et al., 2012). We determined the fraction of biotinylated proteins in the retinal, ON, and presynaptic proteomes that were also present in the public database of presynaptic proteins and calculated their enrichment in the ON and presynaptic transportomes compared with the retina (Figure 5C). About 15% of biotinylated retinal proteins were annotated as presynaptic proteins, increasing significantly in the ON and the presynaptic transportomes in LGN and SC compared with the biotinylated retinal proteome. We demonstrated the presence of biotinylated MTOR, MUNC18, and ALIX, but not GFAP, by western blot in the SC samples after intravitreal NHS-biotin but not after saline injection (Figure 5D). Additionally, synaptotagmin-1, a known presynaptic marker biotin immunofluorescence co-localized with both biotin and ALIX immunofluorescence in the LGN, indicating their presence in RGC boutons (Figure 5E). These data indicate that presynaptic proteins are biotinylated in the retina and become progressively more enriched in the ON and presynaptic transportomes, suggesting specific transport to, and retention in, presynaptic compartments.
We next compared our dataset of 576 proteins in the presynaptic transportome with the 462 proteins in the public presynaptic databases to better understand how our in vivo findings match prior efforts to dissect the presynaptic complex. We found that 192 proteins overlapped between our presynaptic transportome and the public presynaptic database, whereas 384 proteins in the presynaptic transportome were not previously annotated as presynaptic proteins (Figure 5F; Table S3). Many of the proteins present in both datasets have been described as having diverse roles in the presynaptic compartment, such as vesicular transport and sorting, synaptic vesicle exocytosis and endocytosis, and adhesion to postsynaptic compartments (Figure 5F; Table S3). In addition, 270 proteins annotated to the public presynaptic database were not detected in the biotinylated presynaptic transportome of glutamatergic RGCs. These include proteins from non-glutamatergic presynaptic compartments, such as GAD1 and GAD2 (from GABAergic presynaptic neurons), which can be detected in our dataset of the total LGN proteome, a complex mixture of proteins from different cell types (Table S3). Although failure to detect proteins from the presynaptic database could be attributed to their low abundance in our dataset, our methodology nonetheless allows the characterization of specific types of presynaptic compartments, which cannot be otherwise achieved by subcellular fractionation of intact brain tissue.
Finally, we compared our datasets with a postsynaptic density (PSD)-enriched proteomic dataset, consisting of 983 proteins (Bayés et al., 2011; Pirooznia et al., 2012). About 25% of the biotinylated retinal proteins are annotated as postsynaptic proteins, and this representation was enriched ~1.5-fold in the presynaptic transportome (Figures S5B and S5C; Table S3). Some classical excitatory and inhibitory postsynaptic proteins, such as glutamatergic and GABAergic receptor subunits, and postsynaptic scaffolding proteins, including GRIP2, homer, and gephyrin, were only detected in the biotinylated retinal samples but not in the RGC transportome. These data are consistent with the hypothesis that proteins transported from the retina into axons would segregate pre- from postsynaptic proteins. Some of the proteins in the RGC transportome that overlap with the PSD dataset are also annotated as presynaptic proteins, such as SYN1, STX1B, STX1BP, NSF, and SNAP25 (Table S3). These proteins support vesicle fusion in both pre- and postsynaptic compartments (Shimojo et al., 2015; Steinberg et al., 2004), suggesting that the RGC transportome dataset may identify proteins that have more diverse functions in neurons than previously thought. For instance, PSD93 (DLG2), which is part of the postsynaptic scaffold, was present in the ON transportome, consistent with data showing that PSD93 also scaffolds KV1 channels at axon initial segments (Ogawa and Rasband, 2008), which could be captured in the ON transportome.
Analysis of Transportomes to Different Retinal Targets
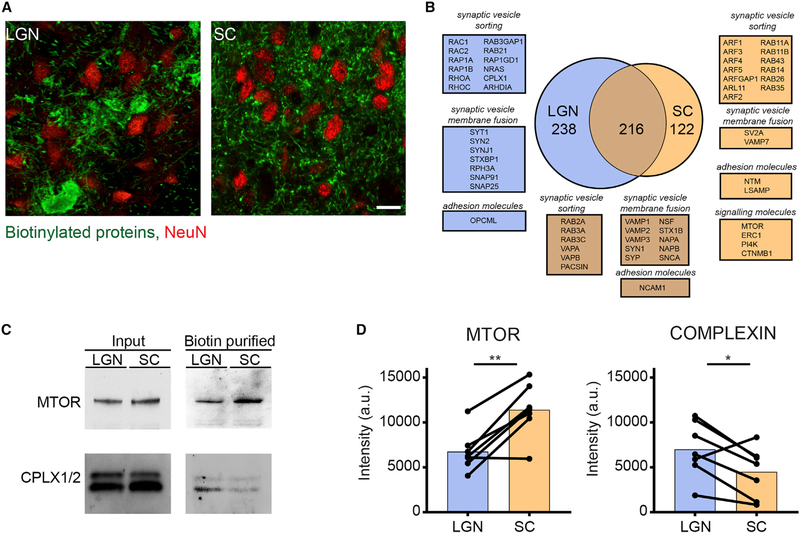
The retina projects to more than 30 distinct targets in the CNS, each thought to have distinct functions (Dhande et al., 2015; Martersteck et al., 2017). The extent to which diverse functions in different retinal targets are subserved by distinct proteins remains unknown. To begin to address that question, we compared proteins targeted to different retinal projections. Immunolabeling transported biotinylated RGC proteins in LGN and SC indicates that retinogeniculate inputs are more densely clustered than retinocollicular inputs (Figure 6A), consistent with previous reports (Hooks and Chen, 2006). Although the mechanisms behind these different synaptic structures is not completely understood, we hypothesized that differential trafficking of proteins from the cell body to specific presynaptic terminals could contribute to their unique features. When we compared the LGN and SC transportomes, we identified 216 proteins that were transported to both LGN and SC, 238 proteins that were preferentially transported to LGN, and 122 proteins that are preferentially transported to SC (Figure 6B; Table S4). Although more than 75% and 86% of proteins in LGN and SC transportomes, respectively, were also detected in ON transportome (Figure 5F), some proteins were identified in the SC and LGN transportomes alone, likely because of their enrichment in the target areas, as mentioned above. The unique members of the retinogeniculate and retinocollicular transportomes may be involved in distinct presynaptic functions in these two retinal targets. For instance, the ARF and RHO GTPase families, which regulate membrane and vesicle fusion at presynaptic sites (Binotti et al., 2016), are selectively enriched in the SC and LGN transportomes, respectively (Figures 6B, S7A, and S7B). These data suggest that presynaptic release and plasticity at SC and LGN retinal projections could be regulated differently, consistent with the distinct functions of LGN and SC in visual information processing (Hong et al., 2014; Kay and Triplett, 2017; Moore et al., 2005; Stein et al., 2014; Suresh et al., 2016). Indeed, validation of differential protein distribution by western blotting confirmed enrichment of transported complexin ½ in the LGN compared with the SC and enrichment of transported MTOR in the SC compared with LGN (Figures 6C and 6D). These results show that target-specific enrichment of transported proteins from a common source may provide information about the diversity of presynaptic inputs and facilitate hypothesis generation regarding circuit function.
Figure 6. Comparison of LGN and SC Transportomes.
(A) Confocal single-optical sections through LGN (left) and SC (right) showing the distribution of biotinylated proteins (green) in characteristic clustered presynaptic profiles in LGN and more evenly distributed presynaptic profiles in superficial layers of SC. Local neuronal somata in LGN and SC were labeled with NeuN antibodies (red). Scale bar, 50 μm.
(B) Venn diagram of LGN and SC transportomes shows that proteins unique to each transportome include different families of GTPases.
(C and D) Western blot validation (C) and quantification (D) of biotinylated proteins in SC and LGN. NeutrAvidin enrichment of both samples showed an enrichment of MTOR in SC and CMPLX½ in LGN. Quantification of samples was performed with normalized optic density of each blot. MTOR, n = 7, **p = 0.005, COMPLEXIN, n = 7, *p = 0.03, two-tailed Student’s t test.
See Table S4 for proteomic data.
DISCUSSION
Here, we combined in vivo, spatially targeted protein labeling with histological, biochemical, and proteomic analysis of labeled proteins to visualize and identify endogenous RGC proteins that are transported into the ON and two functionally distinct retinal targets, the LGN and SC. Axonal protein transport to specific targets is vital to communicate neuronal output required for circuit function, indicated by deficits in transport function in neurodegenerative diseases. Here, we present a cell-type-specific proteomic analysis of axonal projections in the intact brain, using RGC projections to study protein transportomes recovered from two functionally distinct visual targets.
Previous studies investigating axonal transport labeled retinal proteins with radioactive amino acids (Grafstein and Forman, 1980); however, that method did not allow purification of labeled proteins and limited identification of radiolabeled proteins to a candidate-based approach. In addition, radiolabeled amino acids from degraded proteins may have been incorporated into new proteins, confounding interpretation regarding the source of labeled proteins. In our experiments, NHS-biotin labeling generated a covalent tag that could not be reincorporated into other proteins after degradation (Watanabe et al., 2007), supporting the interpretation that biotinylated proteins recovered from the ON and retinal targets originate from RGCs. By taking advantage of axonal transport of biotinylated, endogenous proteins and direct MS/MS detection of biotinylated peptides, we identified hundreds of proteins from glutamatergic RGC axons and presynaptic compartments in intact brain circuits with a depth and specificity not previously reported. In other prior approaches, proteins in synaptic compartments have been analyzed by brain tissue homogenization and subcellular fractionation of synaptosomes, but contamination of the isolated synaptic compartments with other cellular structures, such as astrocyte endfeet (Pielot et al., 2012), or lack of efficient protein extraction from the isolated subcellular compartments limits our understanding of the molecular organization of these specialized neuronal structures. By contrast, our datasets have little to no contamination from complex surrounding tissue, indicated by exclusion of inhibitory synaptic proteins, such as GAD1 and GAD2 in the presynaptic transportomes (Figure 5; Table S3) and by exclusion of non-neuronal proteins in the ON transportome (Figures 3 and 4; Tables S1 and S2). In this manner, we solved a common problem of contaminant proteins in the analysis of compartment-targeted proteomics, which bedevils studies of biotinylated proteins (Alvarez-Castelao et al., 2017; Loh et al., 2016) and subcellular fractionation strategies (Pielot et al., 2012; Poulopoulos et al., 2019).
Several recent studies have demonstrated spatial, temporal, and genetic control over protein labeling strategies. Proximity-based biotin tagging has been invaluable for studying subcellular and molecular phenomena, such as proteomic characterization of organelles or synaptic cleft proteins in reduced experimental systems (Loh et al., 2016; Rhee et al., 2013). Similarly, strategies to label newly synthesized proteins demonstrated proteomic dynamics in response to plasticity paradigms and axon guidance cues (Alvarez-Castelao et al., 2017; Cagnetta et al., 2018; Liu et al., 2018; Schanzenbächer et al., 2016). In vivo NHS-biotin labeling increases coverage of protein biotinylation, partly by removing the dependence on new protein synthesis, and thereby allows the unbiased study of protein transport.
Indeed, the breadth of protein ontologies detected yielded interesting, and in some cases, surprising identifications. For example, the RGC transportome is enriched in proteins known to be present in axons and presynaptic terminals, including neurofilament proteins, tau, kinesins, and proteins that function in vesicular transport and cycling. Recent studies have highlighted the importance of communication between the nucleus and distal processes in neurons (Tasdemir-Yilmaz and Segal, 2016); however, protein components underlying that communication are not fully known (Poulopoulos et al., 2019; Shigeoka et al., 2016). Surprisingly, the RGC transportome is also enriched in proteins not previously annotated to axons, including many nuclear proteins, which are known to shuttle between the nucleus and the cytoplasm in non-neuronal cells (Kanai et al., 2004; Liu et al., 2012; Terenzio et al., 2018; Yudin et al., 2008). It is possible that axonally targeted nuclear proteins acquired distinct functions in neurons based on their distribution into cellular compartments that don’t exist in other cell types. Indeed, more generally, proteins detected in the presynaptic transportome, but not reported in the presynaptic databases, constitute a population of proteins that we have localized to axonal and presynaptic compartments in the retinogeniculate and retinocollicular projections. Exploring this dataset will provide new insights into proteins targeted to axons and/or presynaptic compartments in the visual pathway. The biological functions of many of these proteins have been described based largely on data from non-neuronal cells, as having roles in vesicle sorting, recycling, transport, and secretion; local protein translation; transcriptional regulation; protein transport; membrane adhesion; and RNA transport, all of which may occur in presynaptic compartments. Such findings, including validation of identity and function that could be carried forward into future work, support the premise that such in vivo protein labeling and analysis will be valuable for unbiased discovery of proteins involved in diverse neuronal functions.
MS of the retinocollicular and retinogenicular pre-synaptic transportomes identified several proteins that are differentially enriched in the two targets. The nervous system requires compartmentalized signaling and long-distance axon communication to link complex circuits for information transfer. Given the different roles that the LGN and SC have in visual processing, future studies investigating differentially transported proteins could provide mechanistic insight into the distinct functions of SC and LGN. The retinogeniculate synapse transforms RGC action potentials into LGN firing patterns, yet postsynaptic spike trains do not faithfully resemble the presynaptic signals (Blitz and Regehr, 2003; Usrey et al., 1999). Although α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptor activation, the balance of these components, and short-term plasticity all affect how postsynaptic cells represent RGC input (Blitz and Regehr, 2003), less is known about how presynaptic modulation affects signal transduction. By analyzing RGC transportomics in different retinal targets, we found significantly greater transport of complexin ½ to retinogeniculate synapses than retinocollicular synapses. In cultured cortical neurons, complexin ½ knockdown decreased fast synchronous synaptic vesicle fusion and increased spontaneous fusion (Maximov et al., 2009). Extending this observation to the intact visual system, we hypothesize that enrichment or lack of complexin ½ contributes to differential Ca2+-dependent vesicular release, influencing the divergent postsynaptic responses in LGN and SC. Similarly, we identified different GTPase families in RGC presynaptic terminals in LGN and SC, suggesting different mechanisms may be involved in transport and sorting synaptic vesicles in these retinal targets. This may reflect specialized functions, organization, and morphology of presynaptic compartments in SC and LGN, and these data lay the foundation for future studies of complexin ½ and other presynaptic protein differences among targets.
Dissecting enrichment of different proteins in different retinal targets also informs strategies for regenerating axons after injury. After ON crush, RGCs axons fail to regenerate and die, leading to irreversible blindness. However, after combining MTOR pathway activation and visual stimulation, RGCs can regenerate their axons, reaching multiple targets, including SC and LGN. Nonetheless, behavioral recovery was only seen in the retinocollicular and oculomotor brainstem, not the retinogeniculate pathway (Lim et al., 2016). Avoiding a looming stimulus requires the retinocollicular circuit, but not the visual cortex (Dhande et al., 2015; Lim et al., 2016), whereas the visual cliff test requires retinogeniculate and cortical function (Leamey et al., 2007). Our finding that MTOR is enriched in retinocollicular terminals suggests a hypothesis of biased recovery of RGC connections with greater MTOR content. Significant advances in our understanding of the consequences of impaired protein transport among different neuronal compartments in neurological diseases, including neurogenerative diseases (Devine et al., 2016), highlight the requirement of proper intracellular distribution of proteins for brain function. Indeed, we now further hypothesize that functional recovery of additional visual pathways after injury will require re-establishment of key protein transport to each specific RGC terminal.
Mechanisms underlying differences in proteins transported from the retina to the LGN and SC are not yet known. For instance, they could be due to transport regulation at branch points of individual RGCs, targeted degradation in presynaptic compartments, or differences in projection patterns of RGC subtypes, although a recent study showing that 80% of mouse RGCs projected to both SC and dorsal lateral geniculate nucleus (dLGN) (Ellis et al., 2016) suggests that the likelihood of our findings being because of subtype differences alone is less likely. The distribution of biotin labeling in the ganglion cell layer (Figure 1A) suggests that differential labeling by different RGC types does not account for proteomic differences in SC and LGN. Transportomics of RGC subtypes by genetic labeling (Dhande et al., 2015) and analysis of the presynaptic protein dynamics in SC and LGN will help resolve these questions and advance our understanding of the mechanisms underlying RGC target-specific signal transduction and function. Given the remarkable diversity of chemical synapses in the brain (Nusser, 2018; O’Rourke et al., 2012) and the potentially distinct functions of divergent axon trajectories (Jinno et al., 2007), deeper understanding of pathways regulating differential transport may lead to increased understanding of the wiring in the visual system as well as the potential target-specific regulation of protein transport. Recent studies suggest that proteins can be locally synthesized in axonal compartments (Cagnetta et al., 2018; Poulopoulos et al., 2019; Shigeoka et al., 2016); however, it is highly likely that proteins can be both locally translated and axonally transported in the same cell because these two mechanisms offer different spatial and temporal control over proteostasis. Indeed, we find that the RGC transportome is highly enriched for RNA-binding proteins (Figures 3C, 5F, and S6; Tables S1 and S4). In addition to providing information on the identity of proteins transported from RGC cell bodies into different cellular compartments, these data spark hypothesis-generating insights into protein components underlying the structure and function of neuronal circuits.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to Hollis T. Cline (cline@scripps.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) at the Scripps Research Institute (protocol #08–0082), The Institutional Biosafety Committee of University of California, San Diego (protocol #S13024), and Stanford University (protocol #30550), and complied with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Male Sprague Dawley rats (30–45 day old) from Enviro-Harlan were used for all experiments.
METHODS DETAILS
In vivo biotinylation of retinal proteins
NHS-biotin, N-hydroxysuccinimidobiotin, a membrane permeable biotinylating compound (5mg/300 μL of sterile DMSO, EZ-Link®, Pierce) was prepared immediately before use for eye or brain injections. Intravitreal injections of 5 μl, were given to one or both eyes, depending of the experiment, using a microinjector pressure system (Picosprizer II) with a pulled glass micropipette. The procedure was repeated once a day over 7 days under deep anesthesia with 0.5 mg/kg Medetomidine and 75mg/kg ketamine ip. The eyes were treated with topical antibiotics and analgesics and examined daily. Control animals were injected with biotin or Biocytin (biotinyl-lysine) (Pierce) dissolved in sterile DMSO or with PBS following the same protocol. Rats were processed 11 days after the first injection, as follows: rats were divided into different 2 groups: 1. Rats were euthanized with CO2 and decapitated for brain removal. The tissue was frozen immediately in isopentane in dry ice and stored at −80°C for biochemistry and mass spectrometric studies. 2. Animals were perfused with PBS and then with 4% PFA in 0.1 M phosphate buffer. Tissue was cut into 50 μm sagittal or coronal sections using a vibratome (Leica) and stored in PBS at 4°C for immunofluorescence studies. For electron microscopy, animals were perfused with cold artificial cerebrospinal fluid (ACSF) and then cold 4% PFA plus 0.1% glutaraldehyde in 0.1 M phosphate buffer and sectioned with a vibratome (Leica).
Retrograde labeling from Superior Colliculus
Surgical gelfoam was saturated with NHS-biotin and placed directly on top of the SC bilaterally, as previously described for Fluoro-Gold retrograde labeling (Chiu et al., 2008). Briefly, a midline incision was made on deeply anesthetized rats, exposing the skull. Bilateral holes were drilled 0.5mm from both the sagittal and transverse sutures, and the cortex directly over the dorsal surface of the SC was aspirated. After placement of the gelfoam, the incision was closed with sutures and the animal was treated with antibiotics and analgesics daily.
In vivo biotinylation of motor cortex in rat P3 pups
Postnatal day 3 rat pups were anesthetized on ice and intracranially injected in the motor cortex of the left ventricle using a pulled glass micropipette with 2 μL of 50 mM NHS-biotin dissolved in DMSO. After 5 days the animals were anesthetized with isofluorane and intracardial perfused with PBS and 4% PFA in 0.1 M phosphate buffer (pH = 7.4). The brains were harvested, post-fixed in 4% PFA overnight at 4°C and sectioned with a vibrotome (Leica). Hundred μm coronal sections were incubated with 1:500 goat antibiotin antibody for labeling callosal fibers following protocol described below. Before mounting on glass slides sections were incubated with 1:2000 of 20 mM Hoetch 33342 solution (Invitrogen) for 10 min for counterstaining cell nuclei.
Biotin Measurements
Unincorporated free biotin and biotin incorporated to proteins were measured using a Fluorescence Biotin Quantitation Kit (Thermo) and a fluorescence plate reader (Synergy Mx Microplate Reader, Biotek) by measuring fluorescent excitation/emission at 495/520 according to manufacturer’s instructions. Dissected samples from retina and LGN from 6 animals were weighed and pooled. Protein extracts were generated by homogenizing and sonicating 122 mg of wet tissue from LGN or retina in 1ml of 1 mM EDTA, 50 mM Tris, pH7.5 (TE) buffer and centrifuged at 10,000 g for 15 min at 4°C to remove nuclei and cell debris. The lysates were mixed with 4 mL of cold acetone, incubated at −20°C overnight and centrifuged for 1 h at 4500 g in a swinging bucket rotor to precipitate proteins. Supernatants were collected and evaporated to a volume of ~20 μL in a SpeedVac concentrator (Thermo Scientific) and protein pellets were solubilized in RIPA buffer. As a standard, we used a series of biocytin dilutions (from 0.5 to 10 pmol/μl) according to the manufacturer’s instructions.
Immunohistochemistry in rat brain sections
Sagittal or coronal 50 μm sections containing optic tract, LGN or SC were quenched with 1% Sodium borohydrate for 5 minutes and blocked for 1 h with 3% normal donkey serum and 0.3% Tween 20 in PBS (PBST). Sections were incubated overnight at 4°C with the following primary antibodies: 1:500 Goat anti-biotin (Pierce), 1:200 mouse anti-MAP2 (Millipore), 1:200 mouse anti-NeuN (Millipore), 1:500 rabbit anti-Alix (Millipore), 1:500 mouse anti-Synaptotagmin (MiIlipore). After three washes in PBST, sections were incubated in secondary antibodies: 1:200 dilution of anti-goat alexa 488 or anti-mouse alexa 564 (Invitrogen) for 1 hour at room temperature in blocking buffer. Sections were mounted in Vectashield mounting medium (Vector Labs) and images were collected using a Spinning disc confocal (Ultraview VOX, Perkin Elmer) or a laser scanning confocal (Olympus FV500) microscope. For or obtaining large fields image of rats and pups brains, imaging was performed with Plan Apo 10X objective with 0.45 NA and Plan Apo 20X objective with 0.75 NAon Nikon Alplus confocal system with Andor iXon EMCCD camera system. Large area composite images were made using Nikon NIS elements software using 15% overlap between individual images. All images were background subtracted and processed using Metamorph image processing software.
Electron Microscopy
Detection of biotinylated proteins in the retinogeniculate pathway: Sagittal or coronal 50 μm vibratome sections through LGN were incubated with 100mM glycine in PBS for 2 h and endogenous peroxidase activity was blocked with 0.5% of H2O2 and 1% normal goat serum (NGS) in PBS for 1 hour. Sections were blocked with 10% NGS in PBS for 1 h and incubated overnight with ABC reagent (1 drop of A and 1 drop of B in 5 mL PBS with 1% NGS, Vector Lab). Signal was amplified using tyramide amplification system (TSA kit, Perkin Elmer) and detected by diaminobemzidine reagent (DAB, SigmaFast™) with nickel enhancement. In some sections, biotinylated proteins were labeled by overnight incubation with 1:100 streptavidin-nanoFluorogold 1.4 nm particle size (Nanoprobes) in 1% NGS and 0.001% Triton x (Roche), postfixed for 20 min in Karnovsky fixative, containing 4% paraformaldehyde, 5% glutaraldehyde in PBS, and enhanced for visualization with Goldenhance kit (Nanoprobes) as described (Schikorski, 2010). Sections that contained labeled retinogeniculate axons with DAB or gold particles were then postfixed with 1% OsO4 in PBS for 1 h, dehydrated in ethanol series in water (50%, 70% with 4% uranyl acetate, 90%, 100%), washed 3 times with acetone and flat embedded in epoxy resin (Embed-812, Electron Microscopy Sciences) between two sheets of Aclar plastic (Electron Microscopy Sciences) and polymerized in a vacuum oven overnight at 60°C. Selected sections that contain biotin labeled projections in the optic tract and LGN were cut into ultrathin sections at 50 to 60 nm thickness by using a diamond knife (Diatome, Switzerland) on an ultratome (Powertome XL, RMC products). Ultrathin sections were mounted on a 2×1 mm nickel slot grids (Ted Pella) coated with pioloform film. Sections were examined and imaged in a FEI/Philips CM100 transmision electron microscope operated at 80KV accelerating voltage with 12C-SIA CCD camera.
Western blots
To purify biotinylated proteins for western blot validation, 0.8–2 mg of proteins from ON, LGN, SC or retinal homogenates in a total volume of 1 mL were incubated with 30 μL neutravidin beads (Thermo) at 4°C overnight. The beads were then washed 6 times with 1 mL of RIPA buffer. The bound biotinylated proteins where eluted from the beads with 50 μL Laemmli sample buffer containing 2.5% of 2-mercaptoethanol. Eluates containing purified biotinylated protein were loaded onto 4%–20% SDS/PAGE gradient gel (TGX, Bio-Rad). Proteins were separated by electrophoresis, transferred to nitrocellulose membranes (Bio-Rad) and incubated for 1 h in blocking buffer containing 0.05% Tween 20, 20 mM Tris.HCl, 133 mM NaCl, pH7.4 (TBST) and 5% non-fat milk. The membranes were then incubated in blocking buffer for 24 h with the following antibodies: 1:1000 goat anti-biotin antibody (Thermo); 1:1000 mouse anti-Tau 1 (Chemicon); 1:1000 rabbit anti-Alix (Millipore); 1:1000 rabbit anti-mTOR (Cell Signaling), 1:500 rabbit anti-Ras (Abcam); 1:1000 rabbit anti-Munc18 (Sigma), mouse anti-CaMKII (Novus), 1:10,000 mouse anti-GAPDH (Sigma), 1:1000 rabbit anti-PSMD2 (Cell Signaling), 1:1000 mouse anti-HNRNPA2 (Cell Signaling) and 1:1000 rabbit anti-Complexin ½ (Synaptic Systems). Membranes were washed 3 times for 10 min in TBST and incubated in blocking buffer at room temperature for 1 h with a 1:1600 anti-mouse, antirabbit or anti-goat secondary antibodies conjugated with HRP (Bio-Rad). Bands were detected by chemiluminescence using ECL western blotting substrate® or SuperSignal West femto® (Pierce) and Biomax XAR (KodaK) or Precision™ (BioPioneer, Inc) film. Quantification of optical densities of the bands was made using ImageJ software.
Sample processing for direct identification of biotin labeled proteins using DiDBiT
For mass spectroscopy, tissue samples from 10 to12 brains were pooled for total protein extraction. Retinas, ON, SC, LGN and FC were dissected from fresh brain and homogenized in cold lysis buffer containing 150 mM NaCl, 50 mM TrisHCl (pH7.4), 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor Cocktail (Complete® from Roche). The protein homogenates were briefly sonicated, rotated for 1 h at 4°C and centrifuged for 15 min at 10,000 g at 4°C. Supernatants were collected and protein concentration was measured by DC Protein Assay kit® (Bio-Rad).
For direct identification of biotinylated proteins, we followed the DiDBiT protocol (Schiapparelli et al., 2014). Tissue samples were collected from 10 to 12 animals administrated intravitreal NHS-biotin or saline, pooled and homogenized in RIPA buffer as described above. Protein was quantified and precipitated by adding 3 volumes of methanol, 1 volume of chloroform and 3 volumes of water, vortexed and centrifuged at 15,000 g for 2 min at room temperature. The aqueous and organic phases were removed carefully from the tube without disturbing the protein disc at the interface. Protein pellets were washed once in methanol, air-dried for 10 min and resuspended the in 200 μl of a buffer containing 4 M urea, 50 mM NH4HCO3 and 0.1% ProteaseMax surfactant (Promega) with a brief sonication pulse using a Sonic Dismembrator Model 100 (Fisher Scientific). The protein suspension was reduced by adding of 5 mM Tris(2-carboxyethyl)phosphine (Sigma). The solution was incubated at 55°C with vigorous orbital shaking using a Thermomixer (Eppendorf). Protein alkylation was done by adding 10 mM iodoacetamide (Sigma) and incubating with vigorous shaking in the dark for 20 min. To digest the proteins, we added in the following order: 150 μl of 50 mM NH4HCO3, 2.5 μl of 1% ProteaseMAX dissolved in 50 mM NH4HCO3 and 1:100 (enzyme/protein, w/w) sequencing grade trypsin (Promega) to a final reaction volume of 500 ml. The digestion reactions were incubated for 3 h at 37°C with vigorous orbital shaking. The digestion were stopped by adding 0.1% trifluoroacetic acid, (TFA) (Sigma) and centrifuged at 20,000 g for 20 min at room temperature to remove undigested insoluble material and supernatant containing the peptide mixture was collected and desalted using Sep-Pak tC18 solid-phased extraction cartridges (Waters). Prior to loading the mixture of peptides, the cartridges were washed sequentially with 3 mL of acetonitrile, 3 mL of 0.5% acetic acid, 50% acetonitrile in water, and with 3 mL of 0.1% TFA in water. After loading the peptide mixtures, the cartridges were washed with 3 mL of 0.1% TFA and then with 0.250 mL of 0.5% acetic acid in water. The peptides were eluted into a clean tube with 1 mL of 0.5% acetic acid, 80% acetonitrile in water and dried with a Speed Vac (Thermo). Ten milligrams of dried peptide pellet were solubilized in 1 mL of PBS and incubated with a 200 μl slurry of NeutrAvidin beads (Pierce) for 1 h at room temperature. The beads were precipitated by centrifugation at 1000 g for 5 min and flow through was collected for MS analysis of unbound peptides. Beads were washed 3 times by adding 1 mL of PBS, 3 times with 1 mL of 5% acetonitrile in PBS and with a last wash in ultrapure water. Excess liquid was completely removed from the beads using a micropipette and biotinylated peptides were eluted by adding 0.3 mL of solution containing 0.2% TFA, 0.1% formic acid, 80% acetonitrile in water. The beads were centrifuged at 1000 g and the first elution of biotinylated peptides was transferred to an eppendorf tube. A second elution of 0.3 mL was boiled for 5 min for maximum release of peptides from the beads. A total of 10 elutions were collected and dried separately in a Speed Vac. The enriched biotinylated peptides were resuspended in 0.2 μL PBS and the pH was corrected by adding 20 μl of 1.5 M TrisHCl buffer (pH 7.4). A 10 μl aliquot of the elution was taken to measure biotinylated peptide content using fluorescence biotin quantitation kit as mentioned above.
Mass spectrometry
Soluble peptides were pressure-loaded onto a 250-μm i.d capillary with a kasil frit containing 2 cm of 10 μm Jupiter C18-A material (Phenomenex) followed by 2 cm 5 μm Partisphere strong cation exchanger (Whatman). This column was washed with Buffer A after loading. A100 μm i.d capillary with a 5 μm pulled tip packed with 15 cm 4 μm Jupiter C18 material (Phenomenex) was attached to the loading column with a union and the entire split-column (loading column–union–analytical column) was placed in line with an Agilent 1100 quaternary HPLC (Palo Alto). For transportome analysis, the sample was analyzed using a modified 4-step separation described previously (Washburn et al., 2001). The buffer solutions used were 5% acetonitrile/0.1% formic acid (Buffer A), 80% acetonitrile/0.1% formic acid (Buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (Buffer C). Step 1 consisted of a 35 min gradient from 0%–55% Buffer B, a 5 min gradient from 55%–70% Buffer B, 10 min 100% Buffer Band 27 min 100% Buffer A. Steps 2–3 had the following profile: 5 min of x % Buffer C with (100-x)% buffer A, a 10 min Buffer A, a 5 min gradient from 0%–15% Buffer B, a 70 min gradient from 15%–55% Buffer B, a 5 min gradient from 55%–100% Buffer B, a 5 min 100% Buffer B, and 20 min 100% Buffer A. The Buffer C percentages (X) were 10 and 40 for the steps 2–3, respectively. In the last step, the gradient contained: 5 min of 90% Buffer C with 10% buffer B, a 10 min Buffer A, a 5 min gradient from 0%–15% Buffer B, a 70 min gradient from 15%–55% Buffer B, a 5 min gradient from 55%–100% Buffer B, a 5 min 100% Buffer B, and 20 min 100% Buffer A. For whole proteome analysis, the sample was analyzed using a 11-step separation exactly described as above except the buffer C percentages (X) were 10, 15, 20, 30, 40, 50, 60 and 80% for the steps 2–9, respectively. In the last two steps(I.e., 10 and 11), the gradient contained: 1 min 100% Buffer A, 5 min of 100% Buffer C, a 5 min Buffer A, a 5 min gradient from 0%–15% Buffer B, 70 min gradient from 15%–55% Buffer B, 5 min gradient from 0%–10% Buffer B, 75 min gradient from 10%–45% Buffer B, 10 min 100% Buffer B, and 10 min 100% Buffer A. As peptides eluted from the microcapillary column, they were electrosprayed directly into a Velos mass spectrometer (ThermoFisher) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan FT mass spectrum (300–2,000 m/z) at 60,000 resolution followed by 20 data-dependent IT MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system.
MS Data analysis
MS2 (tandem mass spectra) was extracted from the XCalibur data system format (.RAW) into MS1 and MS2 formats using in house software (RAW_Xtractor) (McDonald et al., 2004). Tandem mass spectra remaining after filtering were searched with Prolucid (Xu et al., 2006) against the UniProt_rat_03–25-2014 concatenated to a decoy database in which the sequence for each entry in the original database was reversed (Peng et al., 2003). All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs (Sadygov et al., 2002). No enzyme specificity was considered for any search. The following modifications were searched for analysis for transportome analyses: a static modification of 57.02146 on cysteine for all analyses, a differential modification of 226.0776 on lysine for modified peptides. For whole proteome analyses, a static modification of 57.02146 on cysteine was searched. Prolucid results were assembled and filtered using the DTASelect (version 2.0) program (Cociorva et al., 2007; Tabb et al., 2002). DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false discovery rate (FDR). In DTASelect, the modified peptides were required to be partially tryptic, less than 10ppm deviation from peptide match, and an FDR at the protein level of 0.01. The FDRs are estimated by the program from the number and quality of spectral matches to the decoy database. For all datasets, the protein FDR was < 1% and the peptide FDR was < 1%.
The Datasets from “ON transportome,” “SC Transportome” and “LGN Transportome” and “Retina biotinylated proteins” are composed of the total detected biotinylated proteins in 6, 4, 3, 4 individual MS runs, respectively. Each sample was prepared by pooling tissue from 10–12 animals. The datasets from “Total ON” are the total detected proteins from two individual MS runs. The starting material of each run was whole ON tissue homogenate in RIPA buffer from 2 animals.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical tests, N values, what N represents, and other details are included in figure legends. Significance is defined as p < 0.05.
The STAT ID function (t test with Benjamini-Hochberg (BH) correction) from Integrated Proteomics Applications, Inc. San Diego, CA. (http://www.integratedproteomics.com) for quantitative proteomic comparisons between: “ON transportome” and “Retina biotinylated proteins,” “ON transportome” and “Total ON” and “SC transportome” and “ON transportome” from Figures 3, 4, and 5, respectively.
Bioinformatic Analysis
Proteins were categorized using open source data bases: Panther.org, Enrichr (http://amp.pharm.mssm.edu/Enrichr/), Synprot (https://www.synprot.de) and Genes to cognition project (G2conline.org). Gene Ontology (GO) analysis was performed using gene symbols to search against the human database in PANTHER (version 11.1) (Mi et al., 2016). To retrieve statistically enriched GO terms and to construct protein interaction networks, we input proteins into STRINGdb (https://string-db.org) (Szklarczyk et al., 2017). We used the high confidence 0.700 required interaction score and included active interaction sources from experiments, and gene function. Data were exported into Cytoscape 3.7 for graphical organization. For the evaluation of enrichment in the transportomes protein list of known presynaptic proteins published in public resourses we used the (http://metamoodics.org/SynaptomeDB/index.php) (Pirooznia et al., 2012). We used the Protein lists: Presynaptic proteins, Presynaptic active zone and presynaptic vesicles. Enrichment was tested using Fisher’s exact test for proportions.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Biotin | Thermo | Cat#31952; RRID:AB_2096845 |
| Anti-Map2 | Millipore | Cat#MAB3418; RRID:AB_94856 |
| Anti-NeuN | Millipore | Cat#MAB377: RRID: AB_2298772 |
| Anti-Alix | Millipore | Cat#ABC40; RRID: AB_10806218 |
| Anti-Synaptotagmin | Millipore | Cat#MAB5200; RRID:AB_2255625 |
| Anti-Tau1 (monoclonal TAU46) | Sigma | Cat#T9450; RRID:AB_477595 |
| Anti-mTOR (monoclonal 7C10) | Cell Signaling | Cat#2983P; RRID:AB_2105622 |
| Anti-GFAP | Dako | Cat#GA52461–2; RRID:AB_10013382 |
| Anti-GAPDH (monoclonal GAPDH-71.1) | Sigma | Cat#G8795; RRID:AB_1078991 |
| Anti-Munc18 (rabbit) | Sigma | Cat#M2694; RRID:AB_477176 |
| ABC-HRP (Vectastain, elite) | Vector labs | Cat#LS-J1001–1 |
| Anti-Ras (monoclonal EP1125Y) | Abcam | Cat#AB52939; RRID:AB_2121042 |
| KIF5A | Abcam | Cat#AB118535 |
| CAMKII (monoclonal 6G9) | Novus | Cat#NB100–1983; RRID:AB_10001339 |
| PSMD2 | Cell Signaling | Cat#1414S; RRID:AB_2797135 |
| Secondary Anti-Mouse Ig conjugated with HRP | Bio-Rad | Cat#172–1011; RRID:AB_11125936 |
| Secondary Anti-rabbit Ig conjugated with HRP | Bio-Rad | Cat#172–1019; RRID:AB_11125143 |
| Secondary Anti-goat Ig conjugated with HRP | Bio-Rad | Cat#172–1034; RRID:AB_11125144 |
| Secondary Anti-mouse Ig conjugated with Alexa568 | Life technologies | Cat#A11004; RRID: AB_2534072 |
| Secondary anti-goat Ig conjugated with Alexa 488 | Life technologies | Cat#A11055; RRID:AB_2534102 |
| Secondary anti-rabbit Ig conjugated with Alexa 568 | Life technologies | CathA10042; RRID:AB_2534017 |
| STREPTAVIDIN ALEXA488 (NANOFLUOROGOLD) | Nanoprobes | Cat#7216–0.5ML; RRID:AB_2315780 |
| STREPTAVIDIN ALEXA488 | Life technologies | Cat#S11223; RRID:AB_2315383 |
| Biological Samples | ||
| Healthy Rat fixed brain and optic nerve tissue. | Envigo (Harlan) | N/A |
| Healthy Rat dissected retinas, Optic nerve, LGN, SC and frontal cortex | Envigo (Harlan) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EZ-Link NHS-biotin | Thermo | Cat#20217 |
| Critical Commercial Assays | ||
| Fluorescence biotin quantitation kit | Thermo | Cat#46610 |
| TSA Biotin system | Perkin Elmer | Cat#NEL700A001KT |
| ABC-HRP (Vectastain, elite) | Vector labs | Cat#LS-J1001–1 |
| Goldenhance EM | Nanoprobes | Cat#2113 |
| Experimental Models: Organisms/Strains | ||
| Rattus novegicus Strain (Sprague Dawley SD) | Envigo (Harlam) | IACUC protocol 08–0082 |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
Highlights.
In vivo protein labeling identifies retinal proteins transported to visual targets
Retinal proteins are differentially transported to presynaptic sites in LGN and SC
Unbiased cell-type-specific proteomic analysis of axon projections in intact brain
Proteomics reveal retinal target-specific diversity in presynaptic protein content
ACKNOWLEDGMENTS
This work was supported by the NIH grants R01EY011261, R01EY027437, P30EY019005, and R01MH103134 and the Hahn Family Foundation to H.T.C.; grants P41GM103533 and R01MH067880 to J.R.Y.; grant P30EY026877 and grants from the Glaucoma Research Foundation and the Research to Prevent Blindness to J.L.G.; grant U01EY027261 to J.L.G., J.R.Y., and H.T.C.; and support from the Helen Dorris Foundation to P.S.
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.037.
REFERENCES
- Alvarez-Castelao B, Schanzenbächer CT, Hanus C, Glock C, Tom Dieck S, Dörrbaum AR, Bartnik I, Nassim-Assir B, Ciirdaeva E, Mueller A, et al. (2017). Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol 35, 1196–1201. [DOI] [PubMed] [Google Scholar]
- Bayés A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, and Grant SG (2011). Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci 14, 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin S, Nawabi H, Wang C,Tang S, Latremoliere A, Warren P, Schorle H, Uncu C, Woolf CJ, He Z, and Steen JA (2015). Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron 86, 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binotti B, Jahn R, and Chua JJ (2016). Functions of Rab proteins at presynaptic sites. Cells 5, E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz DM, and Regehr WG (2003). Retinogeniculate synaptic properties controlling spike number and timing in relay neurons. J. Neurophysiol 90, 2438–2450. [DOI] [PubMed] [Google Scholar]
- Cagnetta R, Frese CK, Shigeoka T, Krijgsveld J, and Holt CE (2018). Rapid cue-specific remodeling of the nascent axonal proteome. Neuron 99, 29–46.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellari M, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O, and Sette C (2014). The transcriptional co-activator SND1 is a novel regulator of alternative splicing in prostate cancer cells. Oncogene 33, 3794–3802. [DOI] [PubMed] [Google Scholar]
- Chiu K, Lau WM, Yeung SC, Chang RC, and So KF (2008). Retrograde labeling of retinal ganglion cells by application of fluoro-gold on the surface of superior colliculus. J. Vis. Exp (16), 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D, Tabb DL, and Yates JR (2007). Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc Bioinformatics Chapter 13, Unit 13.4. [DOI] [PubMed] [Google Scholar]
- Devine MJ, Norkett R, and Kittler JT (2016). DISC1 is a coordinator of intracellular trafficking to shape neuronal development and connectivity. J. Physiol 594, 5459–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Stafford BK, Lim JA, and Huberman AD (2015). Contributions of retinal ganglion cells to subcortical visual processing and behaviors. Annu. Rev. Vis. Sci 1, 291–328. [DOI] [PubMed] [Google Scholar]
- Ellis EM, Gauvain G, Sivyer B, and Murphy GJ (2016). Shared and distinct retinal input to the mouse superior colliculus and dorsal lateral geniculate nucleus. J. Neurophysiol 116, 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek A, Jastrzebska B, Nowotny M, Kwiatkowska K, Hetman M, Surmacz L, Wyroba E, and Kuznicki J (2002). Ca2+-dependent translocation of the calcyclin-binding protein in neurons and neuroblastoma NB-2a cells. J. Biol. Chem 277, 21103–21109. [DOI] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, and Timmerman V (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet 135, 851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstein B, and Forman DS (1980). Intracellular transport in neurons. Physiol. Rev 60, 1167–1283. [DOI] [PubMed] [Google Scholar]
- Harman JC, Guidry JJ, and Gidday JM (2018). Comprehensive characterization of the adult ND4 Swiss Webster mouse retina: Using discovery-based mass spectrometry to decipher the total proteome and phosphoproteome. Mol. Vis 24, 875–889. [PMC free article] [PubMed] [Google Scholar]
- Hensch TK, and Quinlan EM (2018). Critical periods in amblyopia. Vis. Neurosci 35, E014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, Park S, Litvina EY, Morales J, Sanes JR, and Chen C (2014). Refinement of the retinogeniculate synapse by bouton clustering. Neuron 84, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, and Chen C (2006). Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron 52, 281–291. [DOI] [PubMed] [Google Scholar]
- Jiménez CR, Stam FJ, Li KW, Gouwenberg Y, Hornshaw MP, De Winter F, Verhaagen J, and Smit AB (2005). Proteomics of the injured rat sciatic nerve reveals protein expression dynamics during regeneration. Mol. Cell. Proteomics 4, 120–132. [DOI] [PubMed] [Google Scholar]
- Jinno S, Klausberger T, Marton LF, Dalezios Y, Roberts JD, Fuentealba P, Bushong EA, Henze D, Buzsáki G, and Somogyi P (2007). Neuronal diversity in GABAergic long-range projections from the hippocampus. J. Neurosci 27, 8790–8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, and Hirokawa N (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- Kay RB, and Triplett JW (2017). Visual neurons in the superior colliculus innervated by Islet2+ or Islet2− retinal ganglion cells display distinct tuning properties. Front. Neural Circuits 11, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilanczyk E, Filipek A, and Hetman M (2015). Calcyclin-binding protein/Siah-1-interacting protein as a regulator of transcriptional responses in brain cells. J. Neurosci. Res 93, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamey CA, Merlin S, Lattouf P, Sawatari A, Zhou X, Demel N, Glendining KA, Oohashi T, Sur M, and Fässler R (2007). Ten_m3 regulates eye-specific patterning in the mammalian visual pathway and is required for binocular vision. PLoS Biol. 5, e241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Stafford BK, Nguyen PL, Lien BV, Wang C, Zukor K, He Z, and Huberman AD (2016). Neural activity promotes long-distance, target-specific regeneration of adult retinal axons. Nat. Neurosci 19, 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gervasi C, and Szaro BG (2008). A crucial role for hnRNP K in axon development in Xenopus laevis. Development 135, 3125–3135. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yu H, Deaton SK, and Szaro BG (2012). Heterogeneous nuclear ribonucleoprotein K, an RNA-binding protein, is required for optic axon regeneration in Xenopus laevis. J. Neurosci 32, 3563–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, McClatchy DB, Schiapparelli L, Shen W, Yates JR 3rd, and Cline HT (2018). Role of the visual experience-dependent nascent proteome in neuronal plasticity. eLife 7, e33420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. (2016). Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell 166, 1295–1307.e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, et al. (2017). Diverse central projection patterns of retinal ganglion cells. Cell Rep. 18, 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A, Tang J, Yang X, Pang ZP, and Südhof TC (2009). Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science 323, 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, and Yates JR 3rd. (2004). MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom 18, 2162–2168. [DOI] [PubMed] [Google Scholar]
- McKay BE, Molineux ML, and Turner RW (2008). Endogenous biotin in rat brain: implications for false-positive results with avidin-biotin and streptavidinbiotin techniques. Methods Mol. Biol 418, 111–128. [DOI] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, and Thomas PD (2016). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44 (D1), D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelevski I, Medzihradszky KF, Lynn A, Burlingame AL, and Fainzilber M (2010). Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol. Cell. Proteomics 9, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BD 4th, Alitto HJ, and Usrey WM (2005). Orientation tuning, but not direction selectivity, is invariant to temporal frequency in primary visual cortex. J. Neurophysiol 94, 1336–1345. [DOI] [PubMed] [Google Scholar]
- Nusser Z (2018). Creating diverse synapses from the same molecules. Curr. Opin. Neurobiol 51, 8–15. [DOI] [PubMed] [Google Scholar]
- O’Rourke NA, Weiler NC, Micheva KD, and Smith SJ (2012). Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat. Rev. Neurosci 13, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, and Rasband MN (2008). The functional organization and assembly of the axon initial segment. Curr. Opin. Neurobiol 18, 307–313. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, and Gygi SP (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res 2, 43–50. [DOI] [PubMed] [Google Scholar]
- Pielot R, Smalla KH, Müller A, Landgraf P, Lehmann AC, Eisenschmidt E, Haus UU, Weismantel R, Gundelfinger ED, and Dieterich DC (2012). SynProt: a database for proteins of detergent-resistant synaptic protein preparations. Front. Synaptic Neurosci 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Wang T, Avramopoulos D, Valle D, Thomas G, Huganir RL, Goes FS, Potash JB, and Zandi PP (2012). SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics 28, 897–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulopoulos A, Murphy AJ, Ozkan A, Davis P, Hatch J, Kirchner R, and Macklis JD (2019). Subcellular transcriptomes and proteomes of developing axon projections in the cerebral cortex. Nature 565, 356–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, and Ting AY (2013). Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, and Yates JR 3rd. (2002). Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res 1, 211–215. [DOI] [PubMed] [Google Scholar]
- Schanzenbächer CT, Sambandan S, Langer JD, and Schuman EM (2016). Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiapparelli LM, McClatchy DB, Liu HH, Sharma P, Yates JR 3rd, and Cline HT (2014). Direct detection of biotinylated proteins by mass spectrometry. J. Proteome Res 13, 3966–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikorski T (2010). Pre-embedding immunogold localization of antigens in mammalian brain slices. Methods Mol. Biol 657, 133–144. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Yates JR 3rd, and Gerace L (2003). MudPIT: a powerful proteomics tool for discovery. Discov. Med 3, 38–39. [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröer U, Volk GF, Liedtke T, and Thanos S (2007).Translin-associated factor-X (Trax) is a molecular switch of growth-associated protein (GAP)-43 that controls axonal regeneration. Eur. J. Neurosci 26, 2169–2178. [DOI] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, and Holt CE (2016). Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M, Courchet J, Pieraut S, Torabi-Rander N, Sando R 3rd, Polleux F, and Maximov A (2015). SNAREs controlling vesicular release of BDNF and development of callosal axons. Cell Rep. 11, 1054–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein BE, Stanford TR, and Rowland BA (2014). Development of multisensory integration from the perspective of the individual neuron. Nat. Rev. Neurosci 15, 520–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg JP, Huganir RL, and Linden DJ (2004). N-ethylmaleimide-sensitive factor is required for the synaptic incorporation and removal of AMPA receptors during cerebellar long-term depression. Proc. Natl. Acad. Sci. USA 101, 18212–18216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryker MP, and Löwel S (2018). Amblyopia: new molecular/pharmacological and environmental approaches. Vis. Neurosci 35, E018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh V, Çiftçioğlu UM, Wang X, Lala BM, Ding KR, Smith WA, Sommer FT, and Hirsch JA (2016). Synaptic contributions to receptive field structure and response properties in the rodent lateral geniculate nucleus of the thalamus. J. Neurosci 36, 10949–10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, and Yates JR 3rd. (2002). DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz OE, and Segal RA (2016). There and back again: coordinated transcription, translation and transport in axonal survival and regeneration. Curr. Opin. Neurobiol 39, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzio M, Koley S, Samra N, Rishal I, Zhao Q, Sahoo PK, Urisman A, Marvaldi L, Oses-Prieto JA, Forester C, et al. (2018). Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usrey WM, Reppas JB, and Reid RC (1999). Specificity and strength of retinogeniculate connections. J. Neurophysiol 82, 3527–3540. [DOI] [PubMed] [Google Scholar]
- Vance JE, and Hayashi H (2010). Formation and function of apolipoprotein E-containing lipoproteins in the nervous system. Biochim. Biophys. Acta 1801, 806–818. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, and Yates JR 3rd. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Muranaka N, lijima I, and Hohsaka T (2007). Position-specific incorporation of biotinylated non-natural amino acids into a protein in a cell-free translation system. Biochem. Biophys. Res. Commun 361, 794–799. [DOI] [PubMed] [Google Scholar]
- Xu T,Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, and Yates JR (2006). ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics 5, S174–S174. [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, et al. (2008). Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelieres FP, Marco S, and Saudou F (2013). Vesicular glycolysis provides on-board energy for fast axonal transport. Cell 152, 479–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.