Abstract
Fine-mapping of regions linked to the inheritance of hypertension is accomplished by genetic dissection of blood pressure quantitative trait loci (BP QTLs) in rats. The goal of the current study was to further fine-map two genomic regions on rat chromosome 1 with opposing blood pressure effects (BP QTL1b1 and BP QTL1b1a), the homologous region of which on human chromosome 15 harbors BP QTLs. Two new substrains were constructed and studied from the previously reported BP QTL1b1, one having significantly lower systolic BP by 17 mmHg than that of the salt-sensitive (S) rat (P = 0.007). The new limits of BP QTL1b1 were between 134.09 Mb and 135.40 Mb with a 43% improvement from the previous 2.31 Mb to the current 1.31 Mb interval containing 4 protein-coding genes (Rgma, Chd2, Fam174b, and St8sia2), 2 predicted miRNAs, and 4 lncRNAs. One new substrain was constructed and studied from the previously reported BPQTL1b1a having a significantly higher systolic BP by 22 mmHg (P = 0.006) than that of the S rat. The new limits of BPQTL1b1a were between 133.53 Mb and 134.52 Mb with a 32% improvement from the previous1.45 Mb to the current 990.21 Kb interval containing 1 protein-coding gene, Mctp2, and a lncRNA. The congenic segments of these two BP QTLs overlapped between 134.09 Mb and 134.52 Mb. No exonic variants were detected in any of the genes. These findings reiterate complexity of genetic regulation of BP within QTL regions, where elements beyond protein-coding sequences could be factors in controlling BP.
Introduction
Genetic analysis of quantitative traits in rat models has been shown to be a suitable approach to positional cloning of genetic determinants of polygenic traits [1–9]. One such complex polygenic trait is blood pressure (BP), which is extensively studied using rat models. Linkage of human hypertension to a large region on the q-terminus of human chromosome 15 has been detected by four independent studies [10–13]. This region in humans has been mapped to a location on rat chromosome 1 (RNO1) using homology mapping. BP quantitative trait loci (QTLs) in this region on RNO1 have been reported from previous linkage and substitution mapping studies [14–18]. Previously, two BP QTLs were located within 2.31 Mb and 1.45 Mb on rat chromosome 1, referred to as QTL1b1 and QTL1b1a respectively, using the congenic strains S. LEW (Ch1x3x3x3Bx13) and S.LEW (Ch1x3x3x3Bx5), respectively. These strains were developed by introgressing alleles from the normotensive Lewis (LEW) rat onto the genome of the hypertensive Dahl salt-sensitive (S) rat (Fig 1) [16]. These BP QTL regions were further resolved in the current study.
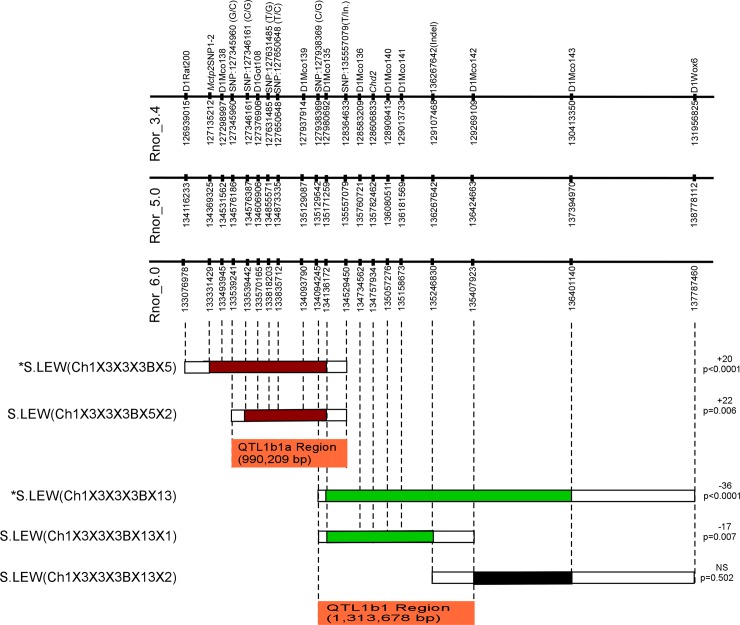
Fig 1. The schematic map of new congenic substrains.
The physical map of RNO1 is shown along with the microsatellite and SNP markers with their locations according to the Ensembl database (www.ensembl.org, RGSC 3.4, 5.0 and 6.0). The horizontal bars below the physical map represent schematics of congenic strains. Introgressed LEW segments are shown with colored bars. A red bar indicates that the congenic strain had a significantly higher BP than S. A green bar indicates that the congenic strain had a significantly lower BP compared with S. A black bar indicates that the congenic strain had no significant change in BP compared with S. The white boxes flanking each of the congenic strains are regions of recombination. The BP effect of each strain compared with S is shown to the right of the schematic of each congenic strain. The locations of the newly identified BP QTLs are indicated as orange bars. *Data from S.LEW (Ch1x3x3x3Bx5) and S.LEW (Ch1x3x3x3Bx13) was previously published and is presented here for completeness [16].
The importance of the current study is that both QTLs were further fine mapped into shorter segments. QTL1b1 was further fine mapped into two shorter segments: (1) S.LEW (Ch1x3x3x3Bx13x1) with <2 Mb, (2) S.LEW (Ch1x3x3x3Bx13x2) within 2 Mb (Fig 1). The first segment on QTL1b1 shows a decreased BP effect, whereas the second segment shows no BP effect. LEW alleles of QTL1b1 had a significant BP lowering effect of the S rat and were mapped within 1.314 Mb containing 10 annotations. QTL1b1a was further fine mapped into one shorter segment: S.LEW (Ch1x3x3x3Bx5x2) within <1 Mb (Fig 1). LEW alleles of QTL1b1a had a significant BP increasing effect of the S rat and were fine-mapped within 990 Kb containing 2 annotations. Genomic DNA sequencing data from these mapped locations has been cataloged. There were no exonic variants within the two newly defined QTL regions but several intronic and intergenic polymorphisms; therefore, these data point to variants outside protein-coding regions as potential candidates for the two BP QTLs with opposing BP effects.
Methods
Animals and diet
All animal research protocols described in this study were approved by the University of Toledo’s Institutional Animal Care and Use Committee. Experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The inbred Dahl salt-sensitive (SS/Jr or S) rat strain was maintained at The University of Toledo College of Medicine and Life Sciences. The Lewis (LEW) rats, originally obtained from Charles River Laboratories (Wilmington, MA), were maintained in our animal facility. In brief, the F1 population was generated by crossing the parental congenic strain with S. The F2 population was generated by intercrossing the F1 rats. The F2 population was genotyped using microsatellite markers throughout the region on rat chromosome 1 from marker D1Rat200 (Chr1:133076978) to D1Wox6 (Chr1:137787460, Rnor 6.0) (Fig 1). The recombinant F2 rats with various introgressed regions of the LEW alleles were backcrossed to S, sorted by genotyping, and then intercrossed to generate homozygous congenic animals on the genomic background of the S rat. S.LEW (Ch1x3x3x3Bx5) is the parental strain for the congenic substrain S.LEW (Ch1x3x3x3Bx5x2). S.LEW (Ch1x3x3x3Bx13) is the parental strain for the congenic substrains S.LEW (Ch1x3x3x3Bx13x1) and S.LEW (Ch1x3x3x3Bx13x2). Rats were maintained on a low salt diet (0.3% NaCl; Harlan Teklad diet 7034, Madison, WI). For experiments involving a high salt regimen (2% NaCl), the Harlan Teklad diet (TD94217) was used.
Microsatellite markers
New microsatellite markers were developed from sequences downloaded from rat genome sequence data available (RGSC 3.4 or RGSC 5.0) through the Ensembl database (http://www.ensemble.org). Marker names and details are deposited with the Rat Genome Database (http://rgd.mcw.edu/). Sequence information on the new polymorphic markers with the prefix D1Mco is provided in the S1 Table. Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) was used to design primers around dinucleotide or trinucleotide repeats. Primer pairs were used to test for polymorphisms between S and LEW genomic DNA. The primer pairs that were polymorphic were used for genotyping F2, backcross, and intercross animals along with DNA from S and LEW as controls.
Genotyping
Congenic substrain DNA was extracted from a tail biopsy using the Promega Wizard SV 96 Genomic DNA Purification System (Promega, Madison, WI, USA). PCR-based genotyping using microsatellite markers was performed using standard techniques as described previously [17].
Blood pressure measurements by radiotelemetry
Rats were weaned at 28–30 days of age and maintained on a low-salt diet (0.3% NaCl). In order to match the blood pressure QTL inference drawn from the previous study of male rats, only male rats were used for the current study [16]. For each congenic strain, about 12–14 male rats were matched by age and weight to an equal number of male S rats. Each cage contained one congenic strain rat and one S rat to minimize environmental effects. At 40–42 days of age, rats were switched to a high-salt diet (2% NaCl) for an additional period of 24 days. After 24 days on high-salt diet, rats were surgically implanted with HD-S10 (S vs. Ch1x3x3x3Bx5x2) or C40 (S vs. Ch1x3x3x3Bx13x1) radiotelemetry transmitters (Data Science International, St Paul, MN) as previously described [14]. Rats were individually housed and allowed to recover from surgery for 3 days before recording blood pressure.
Quantitative real-time PCR
Heart, kidney, brain, and spleen samples were collected from 10 week old S and S.LEW (Ch1x3x3x3Bx13x1) rats that were fed a high-salt (2%) diet. RNA was isolated using the TRIzol method following the recommended procedures (TRIzol, Invitrogen). Concentration of RNA was determined using Nanodrop 2000C. Total RNA was reversed transcribed to cDNA using Superscript III kit (Invitrogen). Then the cDNA was diluted and used as a template for quantitative PCR using SYBR Green. Levels of mRNA expression of Chd2 were analyzed by real-time PCR on a Step One Plus® Real Time PCR Detection System (Applied Biosystems, Foster City, CA), as described previously [15]. By NCBI BLAST of the rat genome, primers were selected for specificity. Using the Step One Plus® Real Time PCR Detection System software, first derivative melting curve analysis verified amplicon specificity. Quantification and normalization of relative gene expression was accomplished using the comparative threshold cycle (CT) method or ΔΔ CT. ΔΔ CT values were converted into ratios by 2- ΔΔ CT and averaged across biological replicates. The expression of the “housekeeping” gene ribosomal protein L36a (GenBank accession no. AA859783) was used for normalization, as this gene did not exhibit differential expression as previously shown [15].
Genomic sequencing and analysis
Genomic variants between SS/Jr and LEW/Crl were obtained from the Rat Genome Database (http://rgd.mcw.edu/). Primers were synthesized by Integrated DNA Technologies (Coralville, IA) and used for manual sequencing. PCR products were sequenced through the Eurofins MWG Operon (Huntsville, AL) sequencing service and sequencing data was analyzed using Sequencher 4.10.1.
Statistical analysis
All statistical analyses of blood pressure studies were conducted using GraphPad Prism 5 (version 5.02). Data was analyzed by independent sample Student’s t-test. The data is presented as the mean ± standard error (Mean ± SEM). A p-value of <0.05 was used as a threshold for statistical significance.
Results
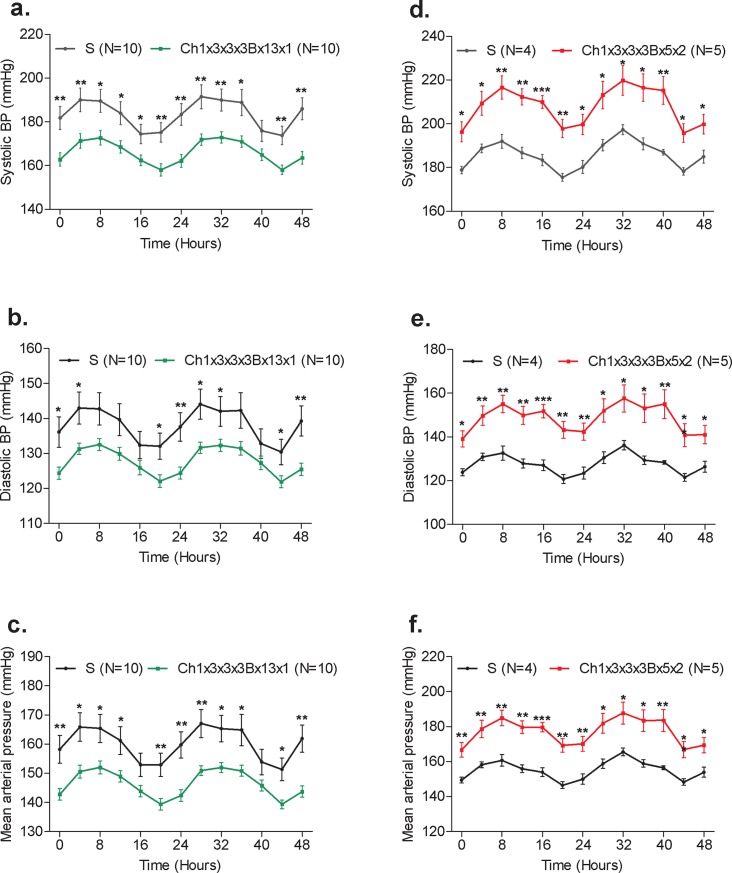
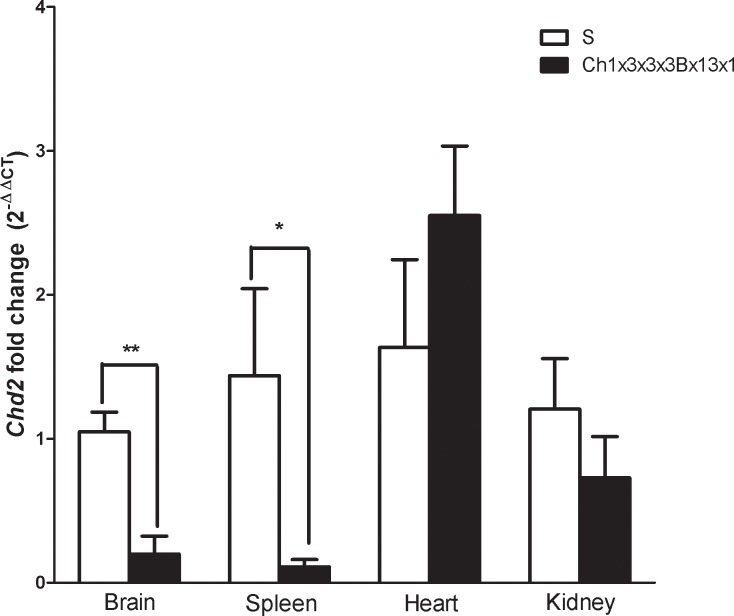
The previously reported congenic substrain S.LEW (Ch1x3x3x3Bx13) had a significant BP lowering effect of -36 mmHg compared with the S rat and spanned the entire 2.31Mb QTL1b1 region from 134.09 to 136.40 Mb [16]. QTL1b1 has been further fine-mapped by constructing and characterizing 2 new congenic substrains S.LEW (Ch1x3x3x3Bx13x1) and S.LEW (Ch1x3x3x3Bx13x2) (Fig 1). These congenic substrains were developed by introgressing LEW rat alleles into the genome of the S rat. One of the newly constructed congenic substrains, S.LEW (Ch1x3x3x3Bx13x1) had a significantly lower systolic BP of 173 ± 7 mmHg compared to the systolic BP of the S rat, 190 ± 4 mmHg (P = 0.007). The systolic, diastolic and mean BP of the congenic strain S.LEW (Ch1x3x3x3Bx13x1) were lower than that of the S rat observed using the telemetry method (Fig 2). The other newly constructed congenic substrain, S.LEW (Ch1x3x3x3Bx13x2), showed no significant difference in BP (190 ± 2 mmHg) compared to the S rat (192 ± 3 mmHg; P = 0.502). Therefore the location of QTL1b1 is further fine-mapped within the congenic segment of S.LEW (Ch1x3x3x3Bx13x1) (Fig 1). The new QTL1b1 location on RNO1 was within a 1.31Mb region from 134.09 to 135.40 Mb. This shows a 43% improvement from the 2.31Mb region to the new 1.31Mb region of QTL1b1. The new 1.31Mb QTL1b1 region contains 4 protein-coding genes (Rgma, Chd2, Fam174b, and St8ia2), 2 predicted miRNAs, and 4 lncRNAs (Table 1). Chd2 (Chromodomain helicase DNA binding protein 2) was prioritized as a candidate gene based on the functional implication that it modifies chromatin structure. Quantitative real-time PCR on heart, kidney, spleen, and brain tissue was performed to determine whether the mRNA expression levels of Chd2 differed between the congenic substrain S.LEW (Ch1x3x3x3Bx13x1) and the S rat. The results indicated a significant down-regulation of Chd2 in brain and spleen in the S.LEW (Ch1x3x3x3Bx13x1) strain compared with S (P = 0.001, P = 0.037, respectively) (Fig 3). No significant difference in expression level was seen in the hearts or kidneys (P = 0.273, P = 0.323, respectively).
Fig 2. Radiotelemetry blood pressure measurements.
Radio-transmitters were implanted into S and congenic strains and their BP was continuously recorded for 48 hours as detailed under the Methods. n = 4–10 rats/group. Systolic BP, systolic blood pressure (a,d); Diastolic BP, diastolic blood pressure (b,e); Mean arterial pressure (c,f). *A p-value of <0.05; **A p-value of <0.01; ***A p-value of <0.001.
Table 1. Annotations within QTL1b1.
| Symbol | Name | Location (Rnor 6.0) | Homologous regions in Human |
|---|---|---|---|
| ENSRNOG00000048799 | Novel miRNA | 134,605,321–134,605,443 | NA |
| Rgma | repulsive guidance molecule family member A | 134,699,299–134,742,514 | Chr 15: 93,042,868–93,088,198 |
| Chd2 | chromodomain helicase DNA binding protein 2 | 134,757,934–134,871,167 | Chr 15: 92,905,067–93,022,512 |
| LOC108349594 | Known lincRNA | 134,873,400–134,887,841 | Chr 15: 92,884,041–92,898,482 |
| ENSRNOG00000051226 | Known lincRNA | 134,921,673–134,922,407 | Chr 15: 92,836,547–92,837,281 |
| ENSRNOG00000041447 | Novel miRNA | 134,946,880–134,946,944 | NA |
| ENSRNOG00000051426 | Novel lincRNA | 134,951,356–134,951,760 | Chr 15: 92,813,676–92,814,080 |
| Fam174b | family with sequence similarity 174, member B | 135,083,974–135,162,685 | Chr 15: 92,580,602–92,659,313 |
| ENSRNOG00000051304 | Known lincRNA | 135,135,833–135,162,693 | Chr 15: 92,593,990–92,620,850 |
| St8sia2 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 | 135,336,374–135,406,700 | Chr 15: 92,356,975–92,427,301 |
Fig 3. Transcript expression analysis of Chd2 in S and S.LEW (Ch1x3x3x3Bx13x1).
Real-time PCR data from S and S.LEW (Ch1x3x3x3Bx13x1) congenic substrain. mRNA obtained from brain, spleen, heart and kidney samples were converted to cDNA then amplified using specific primers. Values are fold change ±s.e.m. **P≤0.01, *P≤0.05.
The previously reported congenic substrain S.LEW (Ch1x3x3x3Bx5) had a significant BP increasing effect of +20 mmHg compared with the S rat and spanned the entire 1.45Mb QTL1b1a region from 133.08 to 134.53 Mb [16]. QTL1b1a was further fine mapped by constructing and characterizing 1 new congenic substrain, S.LEW (Ch1x3x3x3Bx5x2) (Fig 1). S.LEW (Ch1x3x3x3Bx5x2) had a significantly higher systolic BP of 209 ± 5 mmHg compared to the systolic BP of the S rat, 187 ± 3 mmHg (P = 0.006) (Fig 2). Systolic, diastolic, and mean BPs of S.LEW (Ch1x3x3x3Bx5x2) were higher than that of the S rat observed using the telemetry method (Fig 2). Therefore the location of QTL1b1a has been further fine-mapped within the congenic segment of S.LEW (Ch1x3x3x3Bx5x2) (Fig 1). The newly mapped location of QTL1b1a on RNO1 was within a 990Kb region to which QTL1b1a was previously mapped. This represents a 32% improvement from the 1.45Mb region to which QTL1b1a was previously mapped. The newly mapped 990Kb QTL1b1a region contains 1 protein-coding gene, Mctp2, and 1 annotated lncRNA (Table 2). While there were several intronic and intergenic polymorphisms within this region, there were no exonic variants. The exonic variants of Mctp2 lie outside of the QTL region. Gene expression of Mctp2 was previously studied and found to have no significant difference between the previously reported QTL1b1a congenic strain and the S rat [16]. The congenic segments of these two new QTLs overlapped between 134.09Mb and 134.52Mb (Fig 1). The majority of the variants in both QTL regions were located within either intergenic or intronic regions (Table 3). The variants located in highly conserved regions were located within intergenic regions of QTL1b1a. Locations, SNPs and conservation scores are provided in the S2 Table. Genomic variant data within the two QTL regions between SS/Jr and LEW/Crl along with conservation scores were obtained from the Rat Genome Database (http://rgd.mcw.edu/).
Table 2. Annotations within QTL1b1a.
| Symbol | Name | Location (Rnor 6.0) | Homologous regions in Human |
|---|---|---|---|
| Mctp2 | Multiple C2 domains transmembrane 2 | 133,324,301–133,559,975 | Chr 15: 94,231,538–94,483,952 |
| ENSRNOG00000053587 | Novel lincRNA | 134,028,907–134,030,448 | Chr 15: 93,813,356–93,814,897 |
Table 3. Regions of single-nucleotide polymorphisms between SS/Jr and LEW.
| Location | Number of exonic variations (protein coding gene) | Number of intronic variations (#-Gene) |
Number of intergenic SNPs (# of SNPs in highly conserved regions) | Number of 3’ and 5’ UTR SNPs (Gene) |
|---|---|---|---|---|
| QTL1b1a | 0 | • 36-Mctp2 • 1-ENSRNOG00000053587 • 27-pseudogenes |
859 (20) | 36 (Mctp2) |
| QTL1b1 | 0 | 1 -Fam174b | 24 | 0 |
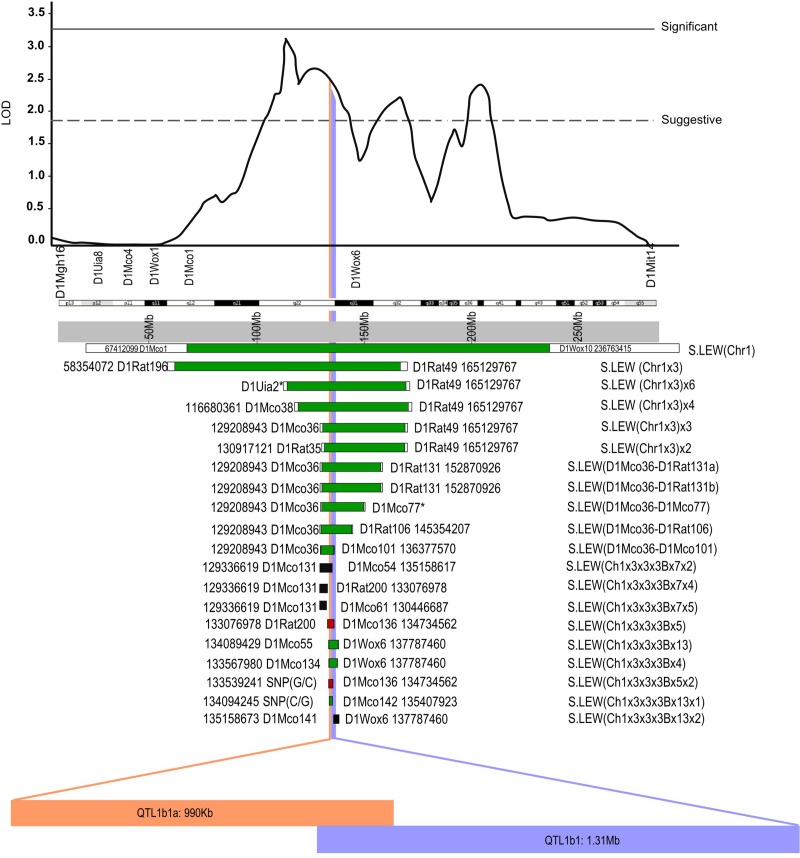
Fig 4 illustrates these two independently functional BP QTLs within the originally detected logarithm of odds (LOD) plot for chromosome 1 by substituting LEW alleles into the S rat genome. QTL1b1 and QTL1b1a are now resolved to regions with candidate variants residing entirely outside the exonic regions of both QTL regions.
Fig 4. Linkage and substitution mapping on RNO1 using S and LEW rats.
The LOD plot for BP was obtained from the study of an F2(S x LEW) population. This is shown at the top followed by the cytogenetic and physical maps of RNO1 [19]. End markers of congenic strains and current physical locations are shown with flanking bars. The physical locations of markers were obtained from the Rat Genome Database (http://www.rgd.mcw.edu) version 6.0. *The location for markers not mapped were determined using nearby markers chosen using the limits of the congenic strains. Congenic strains having a BP lowering effect are shown with a green bar. Congenic strains having no BP effect are shown with a black bar and a red bar indicates that the congenic strain has a BP increasing effect. All congenic strains except the ones currently reported are previously published [14, 16–19]. Vertical blue or orange lines collectively depict the reported QTL regions. The names of QTLs are depicted in the blue or orange horizontal bars along with their respective sizes.
Discussion
While genetic dissection of the human genome is difficult, substitution mapping in rats allows for further dissection of BP QTLs which can be comparatively mapped with human hypertension loci. The subject of this study are two such BP QTLs in the rat which are comparatively mapped within a large region encompassing the q-terminus of human hypertension loci on chromosome 15 [20–22]. This report is a new iteration of substitution mapping of LEW alleles on the S genome on RNO1, wherein two BP QTL regions were previously located within a 2.31Mb and 1.45Mb interval. Data collected from three new congenic substrains showed further mapping of the 2.31Mb BP QTL to a shorter 1.3Mb BP QTL with a BP lowering effect and the 1.45Mb BP QTL to a shorter 990Kb BP QTL with a BP increasing effect. Closely linked BP loci with opposing effects have been shown on other regions of the rat genome [23–25]. This phenomenon shows the complex genetic landscape on BP control in both rats and perhaps in humans.
Further comprehensive mapping of the previously reported 2.31Mb and 1.45Mb intervals has been shown in the current study by three new congenic strains. Genome sequencing of the QTL regions detected a large number of intronic and intergenic sequence variants but no exonic variants. The mapping and sequencing data taken together point to variants beyond protein coding sequences contributing to the BP effects of the two newly mapped genomic intervals on chromosome 1. This data is not surprising because large numbers of intergenic variants associated with several polygenic traits have been cataloged using genome-wide association studies (http://www.genome.gov/gwastudies/). Designing experiments to test the functional relevance of these variants to mechanisms underlying BP control is very difficult when the variants lie outside of protein coding regions. It is possible that these regions encompassing variants within introns and intergenic regions may interact with other chromosomal segments through chromatin remodeling. Therefore, we have prioritized Chd2, a gene located within QTL1b1, as a candidate gene influencing blood pressure. Chd2 has been shown to regulate chromatin and is necessary for appropriate gene expression upon differentiation in mouse embryonic stem cells [26]. Enhancers have been shown to play a role in gene regulation and can regulate a gene from over a million base pairs or more in mammals [27]. Live imaging can be used to learn how enhancers are able to turn a gene on and off over long range distances [28]. Circular chromosome conformation capture (4C) sequencing has been used in recent studies to identify candidate genes for diseases such as atherosclerosis and chronic kidney disease through chromatin interactions with DNA regulatory elements [29, 30]. Therefore, we propose that chromosome conformation capture assay may be an important next step to further prioritize Chd2 as a candidate gene for BP regulation via its effect on the folding of the genome.
Supporting information
(DOC)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported from the University of Toledo and funding from the National Heart Lung and Blood Institute of the National Institutes of Health (HL020176) to BJ. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cheng X, Waghulde H, Mell B, Morgan EE, Pruett-Miller SM, Joe B. Positional cloning of quantitative trait nucleotides for blood pressure and cardiac QT-interval by targeted CRISPR/Cas9 editing of a novel long non-coding RNA. PLoS Genet. 2017;13(8):e1006961 10.1371/journal.pgen.1006961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowley AW Jr, Moreno C, Jacob HJ, Peterson CB, Stingo FC, Ahn KW, et al. Characterization of biological pathways associated with a 1.37 Mbp genomic region protective of hypertension in Dahl S rats. Physiol Genomics. 2014;46(11):398–410. 10.1152/physiolgenomics.00179.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowley AW Jr., Yang C, Kumar V, Lazar J, Jacob H, Geurts AM, et al. Pappa2 is linked to salt-sensitive hypertension in Dahl S rats. Physiol Genomics. 2016;48(1):62–72. 10.1152/physiolgenomics.00097.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng AY. Genetic mechanisms of polygenic hypertension: fundamental insights from experimental models. J Hypertens. 2015;33(4):669–80; discussion 80. 10.1097/HJH.0000000000000479 [DOI] [PubMed] [Google Scholar]
- 5.Deng AY, deBlois D, Laporte SA, Gelinas D, Tardif JC, Thorin E, et al. Novel Pathogenesis of Hypertension and Diastolic Dysfunction Caused by M3R (Muscarinic Cholinergic 3 Receptor) Signaling. Hypertension. 2018;72(3):755–64. 10.1161/HYPERTENSIONAHA.118.11385 [DOI] [PubMed] [Google Scholar]
- 6.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, et al. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci U S A. 2014;111(35):12817–22. 10.1073/pnas.1410745111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AC, Lee JW, Harmon AC, Morris Z, Wang X, Fratkin J, et al. A mutation in the start codon of gamma-crystallin D leads to nuclear cataracts in the Dahl SS/Jr-Ctr strain. Mamm Genome. 2013;24(3–4):95–104. 10.1007/s00335-013-9447-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotchen TA, Cowley AW Jr., Liang M. Ushering Hypertension Into a New Era of Precision Medicine. JAMA. 2016;315(4):343–4. 10.1001/jama.2015.18359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmanabhan S, Joe B. Towards Precision Medicine for Hypertension: A Review of Genomic, Epigenomic, and Microbiomic Effects on Blood Pressure in Experimental Rat Models and Humans. Physiol Rev. 2017;97(4):1469–528. 10.1152/physrev.00035.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang YC, Chiu YF, He CT, Sheu WH, Lin MW, Seto TB, et al. Genome-wide linkage analysis and regional fine mapping identified variants in the RYR3 gene as a novel quantitative trait locus for circulating adiponectin in Chinese population. Medicine (Baltimore). 2016;95(44):e5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraja AT, Hunt SC, Pankow JS, Myers RH, Heiss G, Lewis CE, et al. Quantitative trait loci for metabolic syndrome in the Hypertension Genetic Epidemiology Network study. Obes Res. 2005;13(11):1885–90. 10.1038/oby.2005.231 [DOI] [PubMed] [Google Scholar]
- 12.Montasser ME, Shimmin LC, Hanis CL, Boerwinkle E, Hixson JE. Gene by smoking interaction in hypertension: identification of a major quantitative trait locus on chromosome 15q for systolic blood pressure in Mexican-Americans. J Hypertens. 2009;27(3):491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weder AB, Delgado MC, Zhu X, Gleiberman L, Kan D, Chakravarti A. Erythrocyte sodium-lithium countertransport and blood pressure: a genome-wide linkage study. Hypertension. 2003;41(3 Pt 2):842–6. [DOI] [PubMed] [Google Scholar]
- 14.Joe B, Garrett MR, Dene H, Rapp JP. Substitution mapping of a blood pressure quantitative trait locus to a 2.73 Mb region on rat chromosome 1. J Hypertens. 2003;21(11):2077–84. 10.1097/01.hjh.0000098124.00558.09 [DOI] [PubMed] [Google Scholar]
- 15.Joe B, Letwin NE, Garrett MR, Dhindaw S, Frank B, Sultana R, et al. Transcriptional profiling with a blood pressure QTL interval-specific oligonucleotide array. Physiol Genomics. 2005;23(3):318–26. 10.1152/physiolgenomics.00164.2004 [DOI] [PubMed] [Google Scholar]
- 16.Mell B, Abdul-Majeed S, Kumarasamy S, Waghulde H, Pillai R, Nie Y, et al. Multiple blood pressure loci with opposing blood pressure effects on rat chromosome 1 in a homologous region linked to hypertension on human chromosome 15. Hypertens Res. 2015;38(1):61–7. 10.1038/hr.2014.134 [DOI] [PubMed] [Google Scholar]
- 17.Saad Y, Garrett MR, Rapp JP. Multiple blood pressure QTL on rat chromosome 1 defined by Dahl rat congenic strains. Physiol Genomics. 2001;4(3):201–14. 10.1152/physiolgenomics.2001.4.3.201 [DOI] [PubMed] [Google Scholar]
- 18.Toland EJ, Saad Y, Yerga-Woolwine S, Ummel S, Farms P, Ramdath R, et al. Closely linked non-additive blood pressure quantitative trait loci. Mamm Genome. 2008;19(3):209–18. 10.1007/s00335-008-9093-1 [DOI] [PubMed] [Google Scholar]
- 19.Garrett MR, Dene H, Walder R, Zhang QY, Cicila GT, Assadnia S, et al. Genome scan and congenic strains for blood pressure QTL using Dahl salt-sensitive rats. Genome Res. 1998;8(7):711–23. 10.1101/gr.8.7.711 [DOI] [PubMed] [Google Scholar]
- 20.Shimoyama M, Smith JR, Hayman T, Laulederkind S, Lowry T, Nigam R, et al. RGD: a comparative genomics platform. Hum Genomics. 2011;5(2):124–9. 10.1186/1479-7364-5-2-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoll M, Jacob HJ. Genetic rat models of hypertension: relationship to human hypertension. Curr Hypertens Rep. 2001;3(2):157–64. [DOI] [PubMed] [Google Scholar]
- 22.Stoll M, Kwitek-Black AE, Cowley AW Jr., Harris EL, Harrap SB, Krieger JE, et al. New target regions for human hypertension via comparative genomics. Genome Res. 2000;10(4):473–82. 10.1101/gr.10.4.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopalakrishnan K, Kumarasamy S, Yan Y, Liu J, Kalinoski A, Kothandapani A, et al. Increased Expression of Rififylin in A < 330 Kb Congenic Strain is Linked to Impaired Endosomal Recycling in Proximal Tubules. Front Genet. 2012;3:138 10.3389/fgene.2012.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gopalakrishnan K, Morgan EE, Yerga-Woolwine S, Farms P, Kumarasamy S, Kalinoski A, et al. Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-QT interval and hypertension. Hypertension. 2011;57(4):764–71. 10.1161/HYPERTENSIONAHA.110.165803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saad Y, Yerga-Woolwine S, Saikumar J, Farms P, Manickavasagam E, Toland EJ, et al. Congenic interval mapping of RNO10 reveals a complex cluster of closely-linked genetic determinants of blood pressure. Hypertension. 2007;50(5):891–8. 10.1161/HYPERTENSIONAHA.107.097105 [DOI] [PubMed] [Google Scholar]
- 26.Semba Y, Harada A, Maehara K, Oki S, Meno C, Ueda J, et al. Chd2 regulates chromatin for proper gene expression toward differentiation in mouse embryonic stem cells. Nucleic Acids Res. 2017;45(15):8758–72. 10.1093/nar/gkx475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20(17):R754–63. 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandt MM, Meddens CA, Louzao-Martinez L, van den Dungen NAM, Lansu NR, Nieuwenhuis EES, et al. Chromatin Conformation Links Distal Target Genes to CKD Loci. J Am Soc Nephrol. 2018;29(2):462–76. 10.1681/ASN.2016080875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haitjema S, Meddens CA, van der Laan SW, Kofink D, Harakalova M, Tragante V, et al. Additional Candidate Genes for Human Atherosclerotic Disease Identified Through Annotation Based on Chromatin Organization. Circ Cardiovasc Genet. 2017;10(2). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.