Abstract
Objectives
To assess the methodological quality of prognostic model development studies pertaining to post resection prognosis of pancreatic ductal adenocarcinoma (PDAC).
Design/setting
A narrative systematic review of international peer reviewed journals
Data source
Searches were conducted of: MEDLINE, Embase, PubMed, Cochrane database and Google Scholar for predictive modelling studies applied to the outcome of prognosis for patients with PDAC post resection. Predictive modelling studies in this context included prediction model development studies with and without external validation and external validation studies with model updating. Data was extracted following the Checklist for critical Appraisal and data extraction for systematic Reviews of prediction Modelling Studies (CHARMS) checklist.
Primary and secondary outcome measures
Primary outcomes were all components of the CHARMS checklist. Secondary outcomes included frequency of variables included across predictive models.
Results
263 studies underwent full text review. 15 studies met the inclusion criteria. 3 studies underwent external validation. Multivariable Cox proportional hazard regression was the most commonly employed modelling method (n=13). 10 studies were based on single centre databases. Five used prospective databases, seven used retrospective databases and three used cancer data registry. The mean number of candidate predictors was 19.47 (range 7 to 50). The most commonly included variables were tumour grade (n=9), age (n=8), tumour stage (n=7) and tumour size (n=5). Mean sample size was 1367 (range 50 to 6400). 5 studies reached statistical power. None of the studies reported blinding of outcome measurement for predictor values. The most common form of presentation was nomograms (n=5) and prognostic scores (n=5) followed by prognostic calculators (n=3) and prognostic index (n=2).
Conclusions
Areas for improvement in future predictive model development have been highlighted relating to: general aspects of model development and reporting, applicability of models and sources of bias.
Trial registration number
CRD42018105942
Keywords: hepatobiliary surgery, pancreatic surgery, predictive modeling, risk management
Strengths and limitations of this study.
This is the first systematic review on methodological quality of prognostic models applied to resectable pancreatic cancer.
This review is important and timely, as it is vital that the methodological quality of models designed to support medical decision-making are reviewed at a time when increasing focus and expectation is being placed on personalised predictive medicine.
It highlights limitations in the existing body of research and points towards the direction of future research.
Due to lack of standardisation of reporting of outcomes meta-analysis could not be performed.
Initial title screening was limited to English language.
Introduction
Pancreatic cancer is the fourth and fifth most common cause of cancer deaths in USA and Europe, respectively.1 2 Long-term survival from pancreatic cancer remains poor despite advances in surgical technique and adjuvant treatment,3 yet risk of operative morbidity and mortality remains high with potential benefits of high-risk surgery often nullified by early disease recurrence.4 5 Prognostic models are therefore of great potential benefit in clinical practice. Their application can enhance patient counselling by facilitating the sharing of information. Prognostic models can also support clinical decision-making through risk stratification and support treatment selection by predicting prognostic outcome across competing treatment strategies such as neoadjuvant and surgery-first management pathways.5
Despite a growing interest in prediction research and its methodologies6–12 there is a lack of rigorous application within surgical centres and wider surgical literature of predictive and prognostic models.5 Conversely personalised precision medicine, whereby predictive and prognostic modelling is used to forecast individual patient outcomes, is gaining precedence within contemporary healthcare13 14 and creates an expectation for models to facilitate decision-making and, given the wider socioeconomic context, also guide cost-effective use of resources. Juxtapose these growing expectations with the advent of neoadjuvant therapy making treatment options for resectable pancreatic cancer more complex, and it becomes clear that methods of predictive and prognostic modelling must be assessed if such challenges are to be overcome, as poor methods can result in unreliable and biassed results.12
The aim of this systematic review is to describe and assess the methodological quality of prediction research pertaining to model development studies that predict post resection prognosis of pancreatic ductal adenocarcinoma (PDAC). To our knowledge this is the first such systematic review of its kind. All methodological issues that are considered to be important in prediction research are critically analysed including reporting of: aim, design, study sample, definition and measurement of outcomes and candidate predictors, statistical power and analyses, model validation and results including predictive performance measures.15
Methods
The protocol for this review was published in the PROSPERO online database of systematic reviews (CRD42018105942). This review is reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist.16
A search was undertaken using MEDLINE, Embase, PubMed, Cochrane database and Google Scholar. For each of the five searches, the entire database was included up to and including 31st July 2018 with no further date restrictions or limits applied. Full search strategies and date ranges are detailed in online supplementary appendix S1 (supporting information).
bmjopen-2018-027192supp001.pdf (61.3KB, pdf)
Patient and public involvement
Patients and the public were not involved in the design or conduct of this systematic review.
Study selection
After removal of duplicates, manual screening was carried out by first and second authors, based on the title and abstracts of articles identified in the database searches. Initial title screening was limited to English language. Where this identified relevant studies unavailable in English language translation was sought from colleagues fluent in the language in which the study was published. If this was not possible then language translation software was used.
Articles of probable or possible relevance to this review based on the title and abstract were reviewed in full. This was decided based on the inclusion criteria of prognostic modelling studies applied to the outcome of prognosis for patients with PDAC post resection. Prognostic modelling studies in this context included prognostic model development studies with and without external validation and external validation studies with model updating. We included only prognostic multivariable prediction studies where the aim was to identify a relationship between two or more independent variables and the outcome of interest to predict prognosis. We excluded predictor finding studies and studies that investigated a single predictor, test or marker. Studies investigating only causality between variables and an outcome were excluded. Model impact studies and external validation studies without model updating were excluded as the focus of this systematic review was on assessing the methodological quality of prognostic model development.
Following screening, reference lists and citations of all included papers were manually searched to identify any additional articles. This process was repeated until no new articles were identified.
Data extraction, quality assessment and data analysis
Search design and data extraction was performed by the lead reviewer and with second author performing independent data checking on all studies. Discrepancies were resolved by discussion between the reviewers. Data was extracted to investigate the methodological approach and reporting methods known to affect quality of multivariable predictive modelling studies and followed the Checklist for critical Appraisal and data extraction for systematic Reviews of prediction Modelling Studies (CHARMS) checklist.15 This checklist is designed for appraisal of all types of primary prediction modelling studies including emerging methods of neural network and vector machine learning.15
Data pertaining to the domains outlined in the CHARMS checklist were analysed and presented. These domains include: data sources, sample size, missing data, candidate predictors and model development, performance and evaluation (table 1).15 Risk of bias assessment of included studies was performed according to the Prediction model Risk of Bias Assessment Tool.17
Table 1.
Summary of classification of domains from CHARMS checklist15
| Domain | Key information |
| Data source | Registry data, randomised-controlled-trial data, case-control data, cohort data |
| Participant selection | Participant eligibility (inclusion/ exclusion criteria, description of participants, treatment received) |
| Recruitment methods (setting, location, number of centres, consecutive participants, study dates) | |
| Model outcomes | Definition of outcomes: type (single or combined endpoints), was the same definition used in all participants? |
| Definition of methods for measuring outcomes: same in all participants, blinding, were candidate predictors part of the outcome? | |
| Duration of follow-up or time of outcome occurrence reported | |
| Candidate predictors | Number, type, definition, method and timing of measurement, was assessment blinded, how were candidate predictors handled within the model? |
| Sample size | Number of participants and number of outcomes or events. Event per variable (number of outcomes / number of candidate predictors) |
| Missing data | Number of participants with any missing data, number of participants with missing data for each predictor variable, methods for handling missing data |
| Model development | Modelling methods, methods for selecting predictors to include in multivariable analysis, methods and criteria for selection of predictors during multivariable analysis, shrinking of predictors or regression co-efficients |
| Performance and evaluation | Calibration and discrimination with CIs, classification measures (sensitivity, specificity, predictive value etc), methods for testing performance, comparison of data distribution of predictors for development and validation data sets, in poor validation was model updating performed, alternative presentations of the model (nomogram, calculator, score etc) |
| Presentation of results and discussion | Comparison with other studies, generalisability, strengths and limitations |
CHARMS, Checklist for critical Appraisal and data extraction for systematic Reviews of prediction Modelling Studies.
Results
Initial database searches revealed 23 097 studies that were screened manually by title and abstract. After first round of screening 263 studies underwent full text review with 15 studies identified that satisfied the inclusion criteria (figure 1: PRISMA flowchart).
Figure 1.
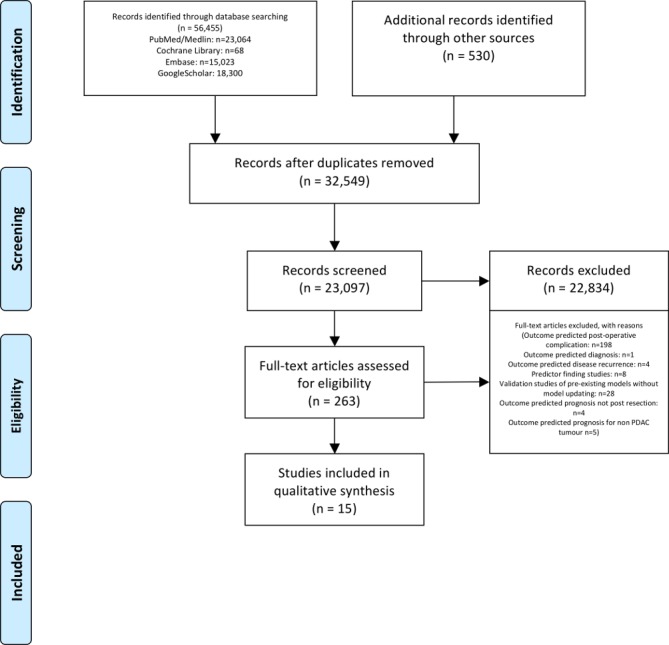
PRISMA 2009 flow diagram. Moher et al.16 PDAC, pancreatic ductal adenocarcinoma; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
This review included a total of 15 model development studies, based on a total of 20 510 patients, published between 2004 and 2018. A full summary of included studies is provided in online supplementary appendix 2 (supporting information) with risk of bias assessment provided in online supplementary appendix 3 (supporting information). The number of model development studies, with (n=3)18–20 and without (n=12) external validation,21–32 increased sharply in recent years (figure 2). Multivariable Cox regression proportional hazard regression was the most commonly employed modelling method (n=13)18–25 27–29 31 32 with two studies employing alternative machine learning techniques (Bayesian model: n=1; Artificial Neural Network (ANN): n=1).26 30 Six models could be applied preoperatively.19 23 24 27 28 31 Five studies focused on predicting poor prognosis (survival time under 7 months n=1, under 9 months n=1, under 12 months n=2, 6, 12 and 18 months survival n=1).18 19 23 26 27 One model predicted prognosis of 3 years or more.32 Seven models predicted prognosis at set time intervals (6 months, 1, 3 and 5 years n=1; 1, 2, 3 years n=2; 1, 3 years n=1 and 1, 3 and 5 years n=3).20–22 24 25 30 31 Two studies did not categories survival time.28 29
Figure 2.
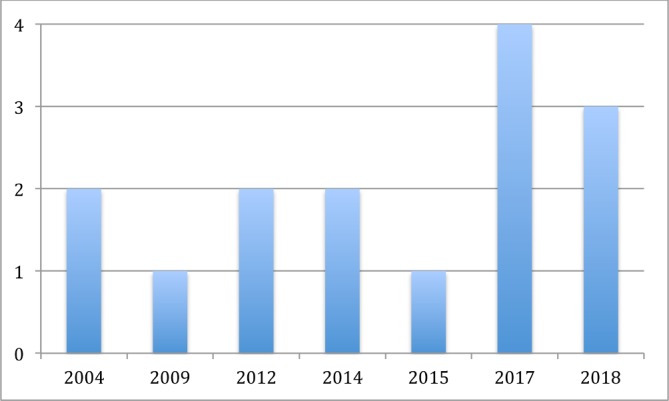
Number of studies published (y-axis) in each year (x-axis).
bmjopen-2018-027192supp002.pdf (68.1KB, pdf)
bmjopen-2018-027192supp003.pdf (67.8KB, pdf)
Source of data, participant selection and follow-up
A cohort design, commonly recommended for prognostic model development,11 was used across all 15 models. Five studies used data from prospectively maintained databases,19 20 22 27 29 with one of these studies collecting data prospectively alongside clinical trials.29 Seven studies used retrospective data.18 23–26 28 31 Three studies used data from the cancer data registry.21 30 32 Prospective cohort designed is recommended as it enables optimal measurement of predictors and outcome.12 Retrospective cohorts are thought to yield poorer quality data11 but do enable longer follow-up time.12
Participant recruitment was well described with inclusion criteria and description of cohort characteristics as well as study dates reported in all 15 studies. Length of follow-up time was clear in 14 studies.18–20 22–32 Consecutive sampling was reported in three studies18 24 26 but whether all consecutive participants were included, or number of participants who refused to participate, could not be evaluated as this was rarely reported across all studies. Non-consecutive sampling can introduce a risk of bias.33–35 The majority of models were developed using single centre databases (n=10)19 20 22–28 31 which can limit the generalisability of the model. This was followed by use of cancer registry database (n=3)21 30 32 and multicentre databases (n=2).18 29
Model outcomes
In all 15 studies outcomes were clearly defined with the same outcome definition and method of measurement applied at all patients. However none of the studies reported blinding the outcome measurement for predictor values. Best practice dictates that assessor of the outcome occurrence should be blinded to ascertainment of the predictor11 36 so as not to bias estimation of predictor effects for the outcome.11 12 Although such a bias would not be a major factor in prediction of all cause mortality,12 15 the majority of studies predicted disease-specific prognosis, whereby bias could come into play in variables requiring subjective interpretation, such as results from imaging.15
Candidate predictors
A variety of candidate predictors were considered across all 15 model development studies (figure 3; table 2). The mean number of candidate predictors was 19.47 (range 7 to 50). The definition, method and timing of measurement of candidate predictors were clear across all 15 studies although, as previously discussed lack of blinding was an issue. Three studies reported categorisation of candidate predictor variables prior to model development.18 20 22 Ten studies specifically detailed how categorical data was analysed as non-binary.18 19 22–25 27 28 30 31 Thirteen studies detailed how time-to-event data was analysed as non-binary.18–21 23–25 27–32 Handling such data as binary is not recommended practice as this can result in less accurate predictions, as with dichotomising predictor variables.37
Figure 3.
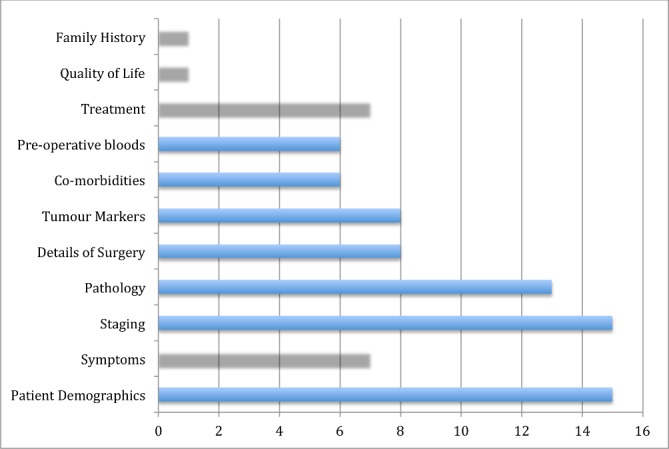
Categories of candidate variables across all studies.
Table 2.
Summary of frequency of included variables in prognostic model development studies
| Variable | Number of models variable is included in | Combined study population of all models in which the variable is included |
| Tumour grade | 9 | 18 815 |
| Age | 8 | 17 565 |
| Tumour stage | 7 | 14 630 |
| Tumour size | 5 | 8154 |
| Gender | 4 | 12 910 |
| Carbohydrate antigen (CA) 19-9 | 4 | 1815 |
| Vascular involvement | 3 | 1059 |
| Tumour location | 3 | 6858 |
| T stage | 3 | 10 433 |
| Margin status | 2 | 774 |
| Lymph node involvement | 2 | 794 |
| Back pain | 2 | 851 |
| Carcinoembryonic antigen (CEA) | 2 | 374 |
| Lymph node ratio | 2 | 6967 |
| Co-morbidities | 2 | 1036 |
| Race | 2 | 12 136 |
| Splenectomy | 1 | 555 |
| Posterior margin positive | 1 | 555 |
| Weight loss | 1 | 555 |
| Platelet count | 1 | 50 |
| Neural Involvement | 1 | 265 |
| Neutrophil to lymphocyte ratio | 1 | 265 |
| Platelet to lymphocyte ratio | 1 | 265 |
| Albumin to globulin ratio | 1 | 265 |
| Quality of life | 1 | 219 |
| Adjuvant therapy | 1 | 219 |
| Radiotherapy | 1 | 12 136 |
| Alkaline phosphate | 1 | 218 |
| Albumin | 1 | 218 |
| Alkaline phosphate to albumin ratio | 1 | 220 |
| Geriatric nutritional index | 1 | 296 |
| Non-metastatic liver disease or insulin resistance | 1 | 296 |
| Marital status | 1 | 6400 |
Statistical power: sample size and missing data
Mean sample size was 1367 (range 50 to 6400). Event per variable (EPV) is the number of predictors assessed compared with the number of events. Statistical power of 10 studies18 19 21 22 24 26 27 29 31 32 could be assessed using the recommended EPV rule of statistical power for Cox regression models of 10 events per candidate predictor, as determined by the smallest group.38–42 Of these studies five did not achieve statistical power according to this rule.18 19 24 26 31 Recently an EPV of 10 has been criticised as being too simplistic for calculating minimum sample size required for models predicting binary and time-to-event outcomes.43 Instead there is a move toward applying the following three criteria to determine the minimum sample size required for such models: (i) predictor effect estimates defined by a global shrinkage factor of ≥0.9, (ii) small absolute difference in the model's apparent and adjusted Nagelkerke's R 2 (≤0.05) and (iii) precise estimation of the overall risk in the population.43 Initial testing of this approach suggests that it will minimise overfitting and ensure precise estimates of overall risk.43
Most studies (n=9) used complete case analysis.19–21 23 24 26–32 This approach results in loss of statistical power and can introduce bias as missing data rarely occurs randomly and often pertains to participant or disease characteristics.12 Two studies reported missing data per candidate variable.22 28 One of these studies handled missing data by predicting input using regression modelling.22 The other study handled missing data by applying the Multivariate Imputation by Chained Equations method assuming data were missing at random.28 Imputation, particularly multiple imputation, of missing data is advocated to reduce bias and maintain statistical power.44–46 Four studies did not give details of missing data.18 19 25 29
Model development
All 15 studies detailed how many candidate predictors were considered but none of the studies detailed how candidate predictors were selected with prior expert knowledge of disease inferred. One study selected predictors on multivariable analysis.22 Most studies (n=12) employed preselection by univariable analysis of predictors for inclusion in multivariable analysis.18–21 23–25 27–29 31 32 Although this method is commonly employed it is not recommended as it carries a greater risk of predictor selection bias, particularly in smaller sample sizes.47 Predictors not significant in univariable analysis may become significantly associated with outcome following adjustment for other predictors.15 Predictors preselected due to large but spurious association with outcome can result in increased risk of overfitting.15 Furthermore multivariable analysis for predictor selection can result in overfitting and unstable models.12 This is a particular risk when outcomes are few but many predictors are analysed.12 None of the studies described shrinkage technique as a method for addressing possible overfitting.15 In the case of low EPV, shrinkage methods could not account for all bias.15
Fourteen studies used backward elimination methods.18–21 23–32 This included an ANN that used single hidden layer back propagation to train the model,26 and a Bayesian model that employed backward step down selection process.30 Of the remaining 12 studies employing this method nominal p value was used as the criteria for predictor inclusion. Ten of these studies used p value <0.05,18 20 21 24 25 27–29 31 32 two of which also reported additionally using Akaike information criteria.28 29 One study used p value <0.1,23 and one study used p value <0.2 for univariate analysis and p value <0.1 for multivariate analysis.19 The use of a small p value has the benefit generating a model from fewer predictors but carries the risk of missing potentially important variables while the use of larger p values potentiates inclusion of predictors of less importance.15 One study reported using multivariable analysis for predictor selection determined by p value but then included non-significant factors in the final model to include all seven candidate variables, therefore effectively employing full model approach.22 While full model approach can avoid selection bias,15 the potential for selection bias still remained in this study, as details were not given on how candidate predictors were decided.
Predictor selection can also incur bias when continuous predictors are categorised.15 Twelve studies reported categorisation.18–25 27 28 30 31 Three studies specifically stated that categorisation was performed prior to modelling.18 20 22 All 12 studies described appropriate statistical techniques for handling continuous variables.
Model performance and evaluation
Eight studies reported calibration of their model,18 19 21 22 25 30–32 most commonly presented as calibration curve. One study reported Hosmer-Lemeshow test,19 a test sometimes criticised for limited statistical power to assess poor calibration and failure to indicate magnitude or direction of miscalibration.15 Twelve studies reported discrimination measured as either c-statistic (n=4)18 22 28 30 or area-under-the-curve (AUC) of the receiver operated curve (n=4)19 20 23 26 or both (n=4).21 25 29 31 Although commonly used, the c-statistic can be influenced by predictor value distribution and be insensitive to inclusion of additional predictors.15 Nine studies reported CIs with discrimination measures.18–22 25 28 30 31 R2 was reported in one study.19 Sensitivity and specificity were also poorly reported (n=2).26 29 Internal validation was rarely performed. Three studies used bootstrapping21 22 28 and four studies used random split method.26 29–31 Three studies included external validation as part of model development.18–20 However, the external validation data sets were small. Shen et al used 17 variables and the external validation data set contained only 61 patients.18 Balzano et al used 56 variables, using univariable analysis to select for multivariable analysis, but the derivation set had only 78 patients and the external validation data set had only 43 patients.19 In one of these studies it was unclear how many events occurred in the external validation cohort.20 None of the studies described external validation of their models separate to the derivation authors and none of the studies described impact analysis of their models.
Presentation of results and discussion
Twelve studies presented both unadjusted and adjusted results of the full model with all candidate predictors considered18–21 23–25 27–29 31 32 and one study presented adjusted results only.22 All 15 studies offered alternative presentation of the model. The most common form of presentation of prognostic models was nomograms (n=5)18 21 22 25 31 and prognostic scores (n=5)19 24 27–29 followed by prognostic calculators (n=3)26 30 32 and prognostic index (n=2).20 23 All 15 studies reported interpretation of models as being for application to clinical practice and all studies discussed comparison, generalisability, strengths and weaknesses of their model as recommended by several guidelines including PRISMA statement.16
Discussion
This systematic review has presented the current state of prognostic model development relating to prognosis following resection of PDAC. By assessing each domain of the CHARMS checklist across the 15 included studies, areas for improvement and direction for future research have been highlighted relating to general aspects of model development and reporting, applicability of models and sources of bias.15
General reporting
General reporting of aspects of model development was found to be clear relating to participant eligibility, recruitment and description as was reporting of follow-up period. Definitions of outcome and number and type of candidate predictors were also generally clearly reported across included studies. The most commonly included variables were tumour grade (n=9), age (n=8), tumour stage (n=7), tumour size (n=5), gender (n=4) and Ca 19–9 (n=4). Vascular involvement, tumour location and tumour stage were each included in three predictive models. Although the number of participants was clearly reported, the number of events at defined times periods of prediction should be more clearly reported to assist assessment of statistical power. Improvement should also be made in the reporting of missing data. The majority of studies used complete case analysis but only two of the remaining studies provided details of missing data per variable.22 28 Across all 15 studies modelling methods were clearly reported. Alternative presentations of models were also offered in all studies to assist application to clinical practice with discussion on strengths, limitations and comparisons also offered.
Applicability
Generalisability of prognostic models is an area for improvement as the majority of models were based in single centre databases. The applicability of these models to patients in neoadjuvant treatment pathways has also not been assessed.
Methods of reporting model performance showed high heterogeneity with only nine studies providing CIs with results,18–22 25 28 30 31 making comment on general applicability difficult. Most models had limited discriminatory performance with AUC below 0.7 and those reporting an AUC nearing 0.9 being based on small sample sizes therefore raising the possibility of overfitting. The two studies employing machine learning methods of ANN26 and Bayesian modelling30 did not report an improved AUC (0.6626 and 0.65,30 respectively). Furthermore calibration, a crucial aspect of model development, was frequently missing or not performed adequately with the calibration curve based on the derivation data set.25 In cases of poor validation whether the model was adjusted or updated was also poorly reported. Only three studies performed external validation18–20 and none of the studies explored impact analysis of their models making comment on the clinical application of the models difficult. Moving forward this could be addressed through access to data sets from meta-analyses of individual participant data, or registry databases containing electronic health records.48 Such big data sets would allow researchers to externally validate, and where needed improve through recalibration, model performance across different settings, populations and subgroups.48
Source of bias
Areas for improvement were also found in limiting sources of bias. As previously mentioned overuse of single centre databases is one area but also the reporting of consecutive sampling, number of participants who refused participation and whether all consecutive participants were included should be more clearly reported. Although handling of candidate predictors, and predictors in modelling, were generally clearly reported including statistical methods for handling categorisation and non-binary variables, their assessment generally did not involve blinding to outcome. Assessment of statistical power of sample size was also not well reported and only two studies used the recommended approach of imputation methods to handle missing data with the majority of studies employing complete case analysis which could both potentiate bias and reduce statistical power.15 None of the included studies gave details on how candidate predictors were identified. In selecting predictors for inclusion in the models the majority of studies employed preselection through univariable analysis followed by multivariable analysis. While such an approach is commonplace it does potentiate overfitting of models, an issue poorly discussed across all studies. Only three studies included external evaluation18–21 and classification measures (sensitivity, specificity, predictive value) were poorly reported, as was comparison of distribution of predictors including missing data.
Future direction of research
The emerging focus on precision medicine means that the future application of predictive modelling will focus on personalised predictive modelling based on patient’s genomic and physiological data.5 The reality however is that such models are not yet in existence and current predictive and prognostic models are limited in scope and value with most only being descriptive in probabilities of adverse events or survival outcomes.5 While this may help to manage patient expectations, existing models fall short in differentiating patients who would, and more importantly would not, benefit from particular treatment options.5 Furthermore some studies have shown that predictive and prognostic models are no better than experience led judgment.49 50 This is reflected in the limited application of predictive and prognostic models within surgical centres and the lack of rigorous application of predictive modelling in surgical literature.5 To integrate fully into clinical practice predictive and prognostic models need to provide predictions beyond length of survival or risk prediction to include fundamentals such as quality of survival time, length of hospital stay, resource utilisation and predicted benefits of competing treatment options available.
The first exciting steps in this path are starting to emerge with the recently published paper by Yamamoto et al demonstrating that a mathematical model can successfully reproduce clinical outcomes using a predictive signature for lower propensity to metastatic disease based on the finding that these primary tumours contain a small fraction of KRAS and CDKN2A, TP53, or SMAD4 genes.51 Although this model requires prospective validation it indicates a future direction of research whereby PDAC treatment can be personalised to the most effective therapeutic modality. The next phase of research will be in integrating breakthroughs in genetic profiling into personalised predictive modelling. In summary, the patient with a favourable genetic profile making metastatic disease from their primary PDAC less likely, but with other pre-existing comorbidities, will still want to know how likely they are to survive an operation and their risk of complications as well as quality adjusted survival predictions for all treatment options available. This is the future personalised predictive medicine supporting cost-effective healthcare.
To conclude, at a time when an increasing focus and expectation is being placed on personalised predictive medicine, this review highlights fundamental aspects of the methodological quality of models that must be improved if future models are to have a clinical impact by supporting decision-making. While many of the models included in this review provided alternative presentations to assist in their clinical application, issues of methodological quality were found that inhibited their clinical impact. These issues included how missing data is handled, the assessment of statistical power, issues of bias associated with candidate predictor selection and a lack of blinding during their assessment. Such issues are augmented by an over reliance on single centre databases which also limits the generalisability of the models. The reporting of model performance is also a key area for improvement. The emerging focus on precision medicine means that the future application of predictive modelling lies in combining each patient’s genomic and clinical data in a meaningful way that will support clinical decision-making at individual patient level. This can only be achieved if future research focuses on improving the methodological quality of model development, regardless of whether they employ traditional or machine learning methods.52
Supplementary Material
Footnotes
Contributors: AB is the first author and undertook data collection, analysis and writing of manuscript. RV-dM was involved in analysis and drafting of manuscript. CMcK was involved in preparing the manuscript.
Funding: Dr Alison Bradley is employed as a Clinical Research Fellow at the University of Strathclyde. This position is funded by NHS Greater Glasgow and Clyde (ref: 160611).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. . Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013;49:1374–403. 10.1016/j.ejca.2012.12.027 [DOI] [PubMed] [Google Scholar]
- 3. Pancreatic Cancer UK Pancreatic cancer UK policy briefing: every life matters: the real cost of pancreatic cancer diagnosis via emergency admission. Available: https://www.pancreaticcancer.org.uk/media/86662/every-im_policybriefing-final.pdf [Accessed 26th Jun 2017].
- 4. Winter JM, Brennan MF, Tang LH, et al. . Survival after resection of pancreatic adenocarcinoma: results from a single institution over three decades. Ann Surg Oncol 2012;19:169–75. 10.1245/s10434-011-1900-3 [DOI] [PubMed] [Google Scholar]
- 5. Lewis RS, Vollmer CM. Risk scores and prognostic models in surgery: pancreas resection as a paradigm. Curr Probl Surg 2012;49:731–95. Dec12 10.1067/j.cpsurg.2012.08.002 [DOI] [PubMed] [Google Scholar]
- 6. Altman DG, Riley RD. Primer: an evidence-based approach to prognostic markers. Nat Clin Pract Oncol 2005;2:466–72. 10.1038/ncponc0287 [DOI] [PubMed] [Google Scholar]
- 7. Altman DG. Prognostic models: a methodological framework and review of models for breast cancer : Lyman GH, Burstein HJ, Breast cancer. translational therapeutic strategies. New York: New York Informa Healthcare, 2007: 11–26. [Google Scholar]
- 8. Altman DG, Lyman GH. Methodological challenges in the evaluation of prognostic factors in breast cancer. Breast Cancer Res Treat 1998;52:289–303. 10.1023/A:1006193704132 [DOI] [PubMed] [Google Scholar]
- 9. McShane LM, Altman DG, Sauerbrei W, et al. . Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005;97:1180–4. 10.1093/jnci/dji237 [DOI] [PubMed] [Google Scholar]
- 10. Rothwell PM. Prognostic models. Pract Neurol 2008;8:242–53. 10.1136/jnnp.2008.153619 [DOI] [PubMed] [Google Scholar]
- 11. Moons KGM, Royston P, Vergouwe Y, et al. . Prognosis and prognostic research: what, why, and how? BMJ 2009;338:b375 10.1136/bmj.b375 [DOI] [PubMed] [Google Scholar]
- 12. Bradley A, van der Meer R, McKay C. Personalized pancreatic cancer management: a systematic review of how machine learning is supporting decision-making. Pancreas 2019;48:598–604. 10.1097/MPA.0000000000001312 [DOI] [PubMed] [Google Scholar]
- 13. Velikova M, van Scheltinga JT, Lucas PJF, et al. . Exploiting causal functional relationships in Bayesian network modelling for personalised healthcare. Int J Approx Reason 2014;55:59–73. 10.1016/j.ijar.2013.03.016 [DOI] [Google Scholar]
- 14. van de Schoot R, Kaplan D, Denissen J, et al. . A gentle introduction to Bayesian analysis: applications to developmental research. Child Dev 2014;85:842–60. 10.1111/cdev.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moons KGM, de Groot JAH, Bouwmeester W, et al. . Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the charms checklist. PLoS Med 2014;11:e1001744 10.1371/journal.pmed.1001744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moher D, Liberati A, Tetzlaff J, et al. . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Open Med 2009;3:e123–30. [PMC free article] [PubMed] [Google Scholar]
- 17. Wolff RF, Moons KGM, Riley RD, et al. . PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med 2019;170:51–8. 10.7326/M18-1376 [DOI] [PubMed] [Google Scholar]
- 18. Shen Y-N, Bai X-L, Gang J, et al. . A preoperative nomogram predicts prognosis of up front resectable patients with pancreatic with pancreatic head cancer and suspected venous invasion. HPB 2018:1–10. [DOI] [PubMed] [Google Scholar]
- 19. Balzano G, Dugnani E, Crippa S, et al. . A preoperative score to predict early death after pancreatic cancer resection. Dig Liver Dis 2017;49:1050–6. 10.1016/j.dld.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 20. Dasari BVM, Roberts KJ, Hodson J, et al. . A model to predict survival following pancreaticoduodenectomy for malignancy based on tumour site, stage and lymph node ratio. HPB 2016;18:332–8. 10.1016/j.hpb.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pu N, Li J, Xu Y, et al. . Comparison of prognostic prediction between nomogram based on lymph node ratio and AJCC 8th staging system for patients with resected pancreatic head carcinoma: a seer analysis. Cancer Manag Res 2018;10:227–38. 10.2147/CMAR.S157940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brennan MF, Kattan MW, Klimstra D, et al. . Prognostic nomogram for patients undergoing resection for adenocarcinoma of the pancreas. Ann Surg 2004;240:293–8. 10.1097/01.sla.0000133125.85489.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanda M, Fujii T, Takami H, et al. . Combination of the serum carbohydrate antigen 19-9 and carcinoembryonic antigen is a simple and accurate predictor of mortality in pancreatic cancer patients. Surg Today 2014;44:1692–701. 10.1007/s00595-013-0752-9 [DOI] [PubMed] [Google Scholar]
- 24. Miura T, Hirano S, Nakamura T, et al. . A new preoperative prognostic scoring system to predict prognosis in patients with locally advanced pancreatic body cancer who undergo distal pancreatectomy with en bloc celiac axis resection: a retrospective cohort study. Surgery 2014;155:457–67. 10.1016/j.surg.2013.10.024 [DOI] [PubMed] [Google Scholar]
- 25. Xu J, Shi K-Q, Chen B-C, et al. . A nomogram based on preoperative inflammatory markers predicting the overall survival of pancreatic ductal adenocarcinoma. Journal of Gastroenterology 2017;32:1394–402. [DOI] [PubMed] [Google Scholar]
- 26. Walczak S, Velanovich V. An evaluation of artificial neural networks in predicting pancreatic cancer survival. J Gastrointest Surg 2017;21:1606–12. 10.1007/s11605-017-3518-7 [DOI] [PubMed] [Google Scholar]
- 27. Hsu CC, Wolfgang CL, Laheru DA, et al. . Early mortality risk score: identification of poor outcomes following upfront surgery for resectable pancreatic cancer. J Gastrointest Surg 2012;16:753–61. 10.1007/s11605-011-1811-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Botsis T, Anagnostou VK, Hartvigsen G, et al. . Modeling prognostic factors in resectable pancreatic adenocarcinomas. Cancer Inform 2009;7:CIN.S3835–291. 10.4137/CIN.S3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L, Xu H-X, He M, et al. . A novel scoring system predicts postsurgical survival and adjuvant chemotherapeutic benefits in patients with pancreatic adenocarcinoma: implications for AJCC-TNM staging. Surgery 2018;163:1280–94. 10.1016/j.surg.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 30. Smith BJ, Mezhir JJ. An interactive Bayesian model for prediction of lymph node ratio and survival in pancreatic cancer patients. J Am Med Inform Assoc 2014;21:e203–11. 10.1136/amiajnl-2013-002171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pu N, Gao S, Xu Y, et al. . Alkaline phosphatase-to-albumin ratio as a prognostic indicator in pancreatic ductal adenocarcinoma after curative resection. J Cancer 2017;8:3362–70. 10.7150/jca.20917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Katz MHG, C-Y H, Fleming JB, et al. . A clinical calculator of conditional survival estimates for resected and unresected pancreatic cancer survivors. Arch Surg 2012;147:6:513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altman DG. Systematic reviews of evaluations of prognostic variables. BMJ 2001;323:224–8. 10.1136/bmj.323.7306.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altman DG, Vergouwe Y, Royston P, et al. . Prognosis and prognostic research: validating a prognostic model. BMJ 2009;338:b605–1435. 10.1136/bmj.b605 [DOI] [PubMed] [Google Scholar]
- 35. Altman DG, Schulz KF, Moher D, et al. . The revised consort statement for reporting randomized trials: explanation and elaboration. Ann Intern Med 2001;134:663–94. 10.7326/0003-4819-134-8-200104170-00012 [DOI] [PubMed] [Google Scholar]
- 36. Laupacis A, Sekar N, Stiell IG. Clinical prediction rules. A review and suggested modifications of methodological standards. JAMA 1997;277:488–94. [PubMed] [Google Scholar]
- 37. Royston P, Altman DG, Sauerbrei W. Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med 2006;25:127–41. 10.1002/sim.2331 [DOI] [PubMed] [Google Scholar]
- 38. Harrell FE. Regression modeling strategies with applications to linear models, logistic regression and survival analysis. New York: Springer Verlag, 2001. [Google Scholar]
- 39. Peduzzi P, Concato J, Kemper E, et al. . A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol 1996;49:1373–9. 10.1016/S0895-4356(96)00236-3 [DOI] [PubMed] [Google Scholar]
- 40. Peduzzi P, Concato J, Feinstein AR, et al. . Importance of events per independent variable in proportional hazards regression analysis. II. accuracy and precision of regression estimates. J Clin Epidemiol 1995;48:1503–10. 10.1016/0895-4356(95)00048-8 [DOI] [PubMed] [Google Scholar]
- 41. Steyerberg EW, Eijkemans MJ, Habbema JD. Stepwise selection in small data sets: a simulation study of bias in logistic regression analysis. J Clin Epidemiol 1999;52:935–42. 10.1016/S0895-4356(99)00103-1 [DOI] [PubMed] [Google Scholar]
- 42. Vittinghoff E, McCulloch CE. Relaxing the rule of ten events per variable in logistic and COX regression. Am J Epidemiol 2007;165:710–8. 10.1093/aje/kwk052 [DOI] [PubMed] [Google Scholar]
- 43. Riley RD, Snell KI, Ensor J, et al. . Minimum sample size for developing a multivariable prediction model: PART II - binary and time-to-event outcomes. Stat Med 2019;38:1276–96. 10.1002/sim.7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361–87. [DOI] [PubMed] [Google Scholar]
- 45. Marshall A, Altman DG, Holder RL. Comparison of imputation methods for handling missing covariate data when fitting a COX proportional hazards model: a resampling study. BMC Med Res Methodol 2010;10:112 10.1186/1471-2288-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Donders ART, van der Heijden GJMG, Stijnen T, et al. . Review: a gentle introduction to imputation of missing values. J Clin Epidemiol 2006;59:1087–91. 10.1016/j.jclinepi.2006.01.014 [DOI] [PubMed] [Google Scholar]
- 47. Collins GS, Reitsma JB, Altman DG, et al. . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 2015;162:55–63. 10.7326/M14-0697 [DOI] [PubMed] [Google Scholar]
- 48. Riley RD, Ensor J, Snell KIE, et al. . External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016;338 10.1136/bmj.i3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Markus PM, Martell J, Leister I, et al. . Predicting postoperative morbidity by clinical assessment. Br J Surg 2005;92:101–6. 10.1002/bjs.4608 [DOI] [PubMed] [Google Scholar]
- 50. Hartley MN, Sagar PM. The surgeon’s “gut feeling” as a predictor of post-operative outcome. Ann R Coll Surg Engl 1994;76(6 Suppl:277–8. [PubMed] [Google Scholar]
- 51. Yamamoto KN, Yachida S, Nakamura A, et al. . Personalized management of pancreatic ductal adenocarcinoma patients through computational modeling. Cancer Res 2017;77:3325–35. 10.1158/0008-5472.CAN-16-1208 [DOI] [PubMed] [Google Scholar]
- 52. Bouwmeester W, Zuithoff NPA, Mallett S, et al. . Reporting and methods in clinical prediction research: a systematic review. PLoS Med 2012;9:e1001221 10.1371/journal.pmed.1001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2018-027192supp001.pdf (61.3KB, pdf)
bmjopen-2018-027192supp002.pdf (68.1KB, pdf)
bmjopen-2018-027192supp003.pdf (67.8KB, pdf)


