Abstract
HIV-1 integration favors active chromatin, which is primarily mediated through interactions between the viral capsid and integrase proteins with host factors cleavage and polyadenylation specificity factor 6 (CPSF6) and lens epithelium-derived growth factor/p75, respectively. Previously published image-based studies had suggested that HIV-1 prefers to integrate into chromatin that associates spatially with the nuclear periphery. Here, we re-evaluated previously reported HIV-1 nuclear distance measures across studies and show that HIV-1 prefers peri-nuclear and mid-nuclear zones similarly, with a common preference between studies mapping to the boundary between these two radial areas. We also discuss emerging roles for the capsid-CPSF6 interaction in facilitating HIV-1 pre-integration complex nuclear import and subsequent intranuclear trafficking to preferred sites of viral DNA integration.
Keywords: HIV nuclear trafficking, CPSF6, HIV integration, HIV capsid, HIV integrase, LEDGF/p75, nuclear import, pre-integration complex
HIV-1, like all retroviruses, reverse transcribes its RNA genome into DNA and integrates the DNA copy into a host cell chromosome. After integration, retroviruses rely on host transcription machinery to produce viral proteins and genomic RNA, both of which co-assemble into nascent viral particles. The HIV-1 ribonucleoprotein complex, which consists of the RNA together with viral nucleocapsid proteins and replication enzymes reverse transcriptase and integrase (IN), is encased by a conical capsid shell composed of the viral capsid (CA) protein (reviewed in [1]).
After HIV-1 enters a susceptible target cell via lipid membrane fusion, reverse transcription happens within a subviral nucleoprotein complex called the Reverse Transcription Complex (RTC) [2]. CA protein within the RTC helps to protect the viral DNA against host defense mechanisms in the cytoplasm [3–5], and the viral capsid shell is gradually shed from the RTC as it is transported towards the nucleus (reviewed in [1, 6, 7]). Once the RTC becomes competent for integration, it is referred to as the pre-integration complex (PIC) [8] and some CA protein remains associated with the HIV-1 PIC after nuclear entry [9–13]. IN functions as part of the intasome nucleoprotein complex composed of an IN multimer and the ends of the linear viral reverse transcript (reviewed in [14]).
HIV-1 integration targeting
HIV-1 integration into cellular DNA is not random, with the virus favoring the interior regions of transcriptionally active genes residing in relatively gene-dense regions of chromatin [15]. HIV-1 integration targeting preference is largely driven by the interaction of two viral proteins, IN and CA, with respective cellular proteins lens epithelium-derived growth factor (LEDGF)/p75 and cleavage and polyadenylation specificity factor (CPSF) 6. Depletion of either of these two host factors results in significant reduction of integration into genes and gene-dense regions [16–21]. Although depleting either LEDGF/p75 or CPSF6 reduces intragenic integration, the two factors influence HIV-1 integration in different ways. LEDGF/p75 depletion shifts intragenic integration towards the 5’ end regions of the genes, whereas CPSF6 depletion results in HIV-l dramatically losing preference for integration near activating epigenetic marks and instead favoring gene-sparse regions. These results suggest that LEDGF/p75 primarily functions to position integration along the genes, whereas CPSF6 predominantly shields HIV-1 from integration into heterochromatin [21]. It was unclear until our recent publication [22] how these contrasting roles of CPSF6 and LEDGF/p75 in HIV-1 integration targeting influenced viral DNA localization inside the nucleus.
HIV-1 nuclear localization during acute infection
Different imaging techniques such as induced double stranded DNA breaks (SCIP for single-cell imaging of HIV-1 provirus) [23, 24], fluorescently labelled IN proteins [25–29], fluorescently labelled cyclophilin A protein that interacts with CA [3], immuno-DNA fluorescent in situ hybridization (FISH) [30], branch-chain DNA FISH [11], click chemistry [9], and stimulated emission depletion (STED) microscopy [10] have been used to track HIV-1 PICs inside the nucleus. Analyses of intranuclear position are facilitated by determining the relative radial distance of the imaged HIV-1 focus from the nuclear envelope (NE), and binning the results into three concentric nuclear zones of equal area [30, 31]. The most peripheral zone, peripheral nuclear (PN), has a width of 0.184 x r (nuclear radius); the mid-nuclear (MN) zone ranges from 0.184 x r to 0.422 x r; and the inner-most central nuclear (CN) zone has a width of 0.422 x r (Fig. 1A). A majority of prior imaging studies had indicated that HIV-1 preferentially integrates into active chromatin within the PN area [3, 23, 25–27, 29] proximal to the nuclear pore [30]. But re-evaluating the reported radial distance measures across studies shows that HIV-1 PICs and proviruses target PN and MN areas similarly, with a preference for the boundary between these two areas [22].
Fig.1. Meta-analysis with correct bin sizes downplays specific targeting of the PN during HIV-1 infection.
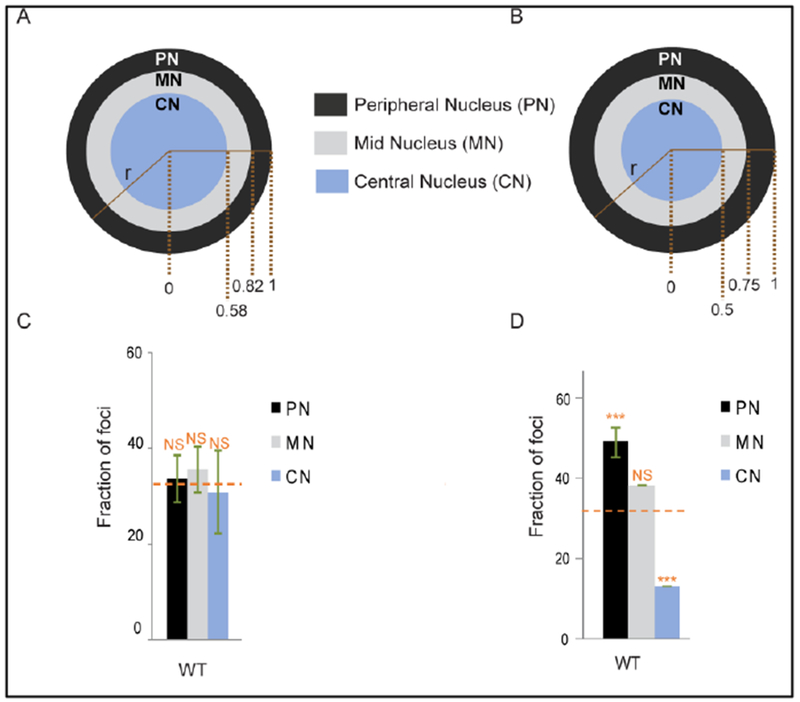
(A) Nucleus is divided into three sections of equal area based on the measured radius (r). (B) The three sectional cutoffs used in [11] that marginally underrepresented the CN and inflated the true PN area. (C) and (D) Bar graphs displaying the proportion of viral DNA from a primary CD4+ T cell sample [11] binned into sectional areas as shown above in respective panels A and B. Orange dashed line indicates the expected random distribution among the PN, MN, and CN areas. *** P < 0.0001, ** P < 0.01 and NS P > 0.05 compared to random using Chi-Square test.
Although HIV-1 normally infects CD4+ cells such as T cells and macrophages, numerous microscopy studies utilized HeLa cells (7.4 μm average nuclear radius) due to their compatibility with imaging technologies. HIV-1 PICs located within 0.4 - 2.0 μm from the NE of HeLa cell nuclei, equating to 0.05 - 0.25 x r, mapping to the PN and MN areas [25]. A separate study reported that the average distance traveled by PICs into HeLa cell nuclei was 1.2-1.6 μm, equating to 0.16-0.22 x r, again representing the PN and MN areas [26]. Tracking the translocation of fluorescently labeled HIV-1 complexes confirmed 1.4 μm (or 0.19 x r) as the average distance travelled into HeLa cell nuclei [29], equating to the interface between the PN and MN areas. Another study tracking the nuclear progression of single HIV-1 particles [3] measured 1.8 μm as the average distance travelled by PICs in HeLa cell nuclei, which is 0.24 x r (MN area). Imaging HIV-1 proviruses using the SCIP technique, Di Primio et al. [23] reported that 55% of HIV-1 proviruses mapped within 1.5 μm from the NE of U2OS cell nuclei, which equates to 0.19 x r and thus is consistent with the PN-MN interface. In this same report, the authors saw that 62% of integrated proviruses mapped within 0.5 μm from the NE of CEMss T cell nuclei at 2 days post infection, placing the virus at 0.09 x r (PN). However, eleven days later, HIV-1 proviruses distributed randomly throughout the T cell nuclei [23]. While the majority of HIV-1 (NL4-3 strain) nuclear foci localized in the PN area of primary CD4+ T cells, the related HIV-1 (BRU) strain localized similarly to the PN and MN areas [30].
Visualizing HIV-1 proviruses by branched-DNA FISH technology, we initially reported preferential localization of HIV-1 PICs and proviruses to the PN area of primary CD4+ T cell nuclei [11]. However, by using radial cutoffs of 0.5 x r for CN and 0.5 x r to 0.75 x r for MN, we since realized our prior bin sizes were modestly unequal (the proper radial cutoffs for respective CN and MN boundaries are 0.58 x r and 0.82 x r; Fig. 1A) [30–32], which underrepresented the CN area by ~25% and inflated PN area by ~30% (Fig. 1A and 1B). The fractional PN localization that we had previously documented became equalized with other nuclear sections when datasets were reanalyzed using the corrected areas (Fig. 1C–D). In summary, imaging experiments do not strongly support preferential localization of HIV-1 in the nuclear periphery. We recently showed, using multiple orthologous approaches, that HIV-1 locates equally to the PN and MN areas, with some penetration into the CN area as well [22]. Using branched-DNA hybridization and the SCIP technique, we failed to observe HIV-1 enrichment at the nuclear periphery in a variety of cell types.
CA-CPSF6 interaction licenses HIV-1 to penetrate cell nuclei
Prior to our work, there was a disagreement regarding the potential role for LEDGF/p75 in the localization of HIV-1 within the nucleus, with two reports indicating that LEDGF/p75 played an important role in peripheral nuclear targeting [27, 30] and two other reports suggesting that LEDGF/p75 does not contribute to intranuclear localization [28, 29]. In terms of CPSF6, we had reported earlier that the interaction with CA was important for HIV-1 to penetrate into the nucleus [11]. To systematically analyze the roles of both cell proteins, we imaged viral DNA foci and mapped sites of HIV-1 integration using an isogenic set of HEK293T cells that were knocked out for LEDGF/p75, CPSF6, or both factors. We additionally visualized the intranuclear localization of preferred integration gene targets in uninfected HEK293T and primary CD4+ T cells. We did not observe a significant role for LEDGF/p75 in determining the localization of HIV-1 inside the nucleus. However, loss of the interaction of CA with CPSF6 dramatically altered virus localization towards the nuclear periphery, with > 60% of HIV-1 foci locating to the PN area in CPSF6 knockout HEK293T cells and in CD4+ T cells infected with a CA mutant virus defective for the interaction with CPSF6. This shift in peripheral localization strongly correlated with integration of HIV-1 into transcriptionally inactive heterochromatin associated with the nuclear lamina [lamin-associated domains (LADs)] located at the nuclear periphery.
The role of CPSF6 in licensing HIV-1 PICs to transcriptionally active genes distal from the nuclear periphery became even more obvious when we analyzed the genes that are repeatedly targeted for HIV-1 integration under normal and CPSF6 depleted conditions. Under normal conditions, HIV-1 targeted comparatively small, transcriptionally active genes that dispersed throughout the nucleus. But when the CA-CPSF6 interaction was disrupted, this preference was lost. A unique set of genes that were larger, transcriptionally less active, and predominantly located in the PN area were enriched for HIV-1 integration upon CPSF6 depletion [22]. Furthermore, evidence for the role of CPSF6 in intranuclear targeting is evident from independent studies that imaged single HIV-1 particles inside the nucleus [3, 10, 11]. Francis and Melikyan [3] reported that while WT HIV-1 penetrated on average 1.8 μm from the NE in HeLa cells, mutant HIV-1 CA virus defective for the interaction with CPSF6 traveled on average only 0.5 μm from the NE. Using two-color STED microscopy, Bejarano et al. [10] reported that CPSF6 is recruited to HIV-1 PICs at the nuclear basket in primary CD4+ macrophages, which is essential for the PICs to release from the NE and travel inside the nucleus. They further observed that the majority of wild–type HIV-1 PICs upon CPSF6 depletion and HIV-1 CA mutant viral PICs defective for CPSF6 binding remained arrested at the NE, unable to enter the nucleus. Thus, the inability for PIC-associated CA to interact with CPSF6 renders HIV-1 unable to penetrate into cell nuclei, redirecting integration into chromatin in association with the nuclear lamina (Fig. 2). Roles for CPSF6 and LEDGF/p75 in the intranuclear localization of other lentiviruses, whether CPSF6 is initially recruited by HIV-1 in the cytoplasm or in the nucleus, and whether CPSF6 accompanies the PIC as it transits beyond the NE are active areas of investigation
Fig.2. Inability to engage CPSF6 shifts HIV-1 localization and integration to the outer region of the nucleus.
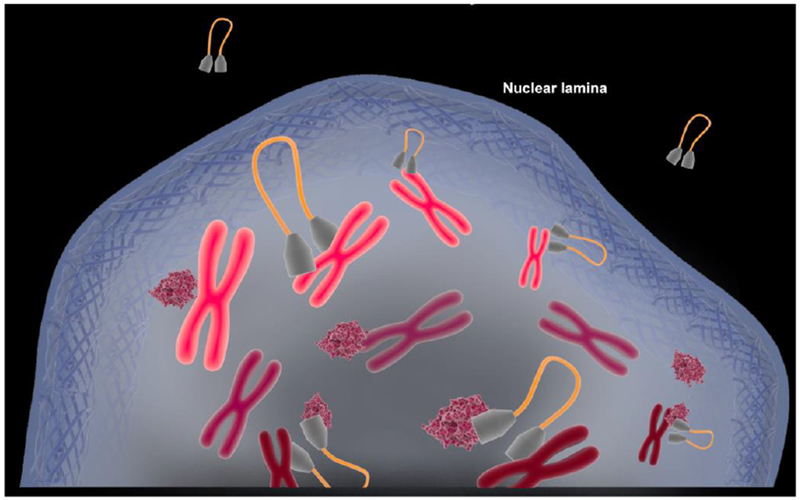
Engagement of CPSF6 (pink globules) by the HIV-1 capsid as part of the PIC (grey polygons with orange DNA loop) is required to bypass the nuclear lamina (peripheral wire mesh) and access interior gene-dense regions of the genome (red chromosomes) for integration. Lack of CPSF6 engagement impedes penetration into the nucleus, resulting in integration into gene-sparse heterochromatin (shown as pink chromosomes) associated with the nuclear lamina.
Acknowledgements
Dr. Engelman acknowledges the support of NIH grants AI039394, AI052014, and AI070042. Dr. Brass acknowledges the funding support from the Bill and Melinda Gates Foundation and Gilead Sciences Inc.
References
- 1.Yamashita M and Engelman AN, Capsid-Dependent Host Factors in HIV-1 Infection. Trends Microbiol, 2017. 25(9): p. 741–755. 10.1016/j.tim.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fassati A and Goff SP, Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol, 2001. 75(8): p. 3626–3635. 10.1128/JVL.75.8.3626-3635.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francis AC and Melikyan GB, Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe, 2018. 23(4): p. 536–548 e6 10.1016/j.chom.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacques DA, et al. , HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature, 2016. 536(7616): p. 349–353. 10.1038/nature19098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasaiyaah J, et al. , HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature, 2013. 503(7476): p. 402–405. 10.1038/nature12769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambrose Z and Aiken C, HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology, 2014. 454–455: p. 371–379. 10.1016/j.virol.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell EM and Hope TJ, HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol, 2015. 13(8): p. 471–483. 10.1038/nrmicro3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowerman B, et al. , A nucleoprotein complex mediates the integration of retroviral DNA. Genes & Development, 1989. 3(4): p. 469–478. 10.1101/gad.3.4.469 [DOI] [PubMed] [Google Scholar]
- 9.Peng K, et al. , Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife, 2014. 3: p. e04114 10.7554/eLife.04114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bejarano DA, et al. , HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. Elife, 2019. 8: p. 8: e41800 10.7554/eLife.41800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin CR, et al. , Direct visualization of HIV-1 replication intermediates shows that capsid and CPSF6 modulate HIV-1 intra-nuclear invasion and integration. Cell Rep, 2015. 13(8): p. 1717–1731. 10.1016/j.celrep.2015.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulme AE, et al. , Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-infected cells. J Virol, 2015. 89(10): p. 5350–5361. 10.1128/JVI.00476-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen NY, et al. , HIV-1 capsid is involved in post-nuclear entry steps. Retrovirology, 2016. 13: p. 28 10.1186/s12977-016-0262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman AN and Cherepanov P, Retroviral intasomes arising. Curr Opin Struct Biol, 2017. 47: p. 23–29. 10.1016/j.sbi.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroder ARW, et al. , HIV-1 integration in the human genome favors active genes and local hotspots. Cell, 2002. 110(4): p. 521–529. 10.1016/S0092-8674(02)00864-4 [DOI] [PubMed] [Google Scholar]
- 16.Ciuffi A, et al. , A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med, 2005. 11(12): p. 1287–1289. 10.1038/nm1329 [DOI] [PubMed] [Google Scholar]
- 17.Shun MC, et al. , LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev, 2007. 21(14): p. 1767–1778. 10.1101/gad.1565107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall HM, et al. , Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One, 2007. 2(12): p. e1340. 10.1371/journal.pone.0001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrijvers R, et al. , HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology, 2012. 9: p. 84 10.1186/1742-4690-9-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh PK, et al. , LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev, 2015. 29(21): p. 2287–2297. 10.1101/gad.267609.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sowd GA, et al. , A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc Natl Acad Sci U S A, 2016. 113(8): p. E1054–E1063. 10.1073/pnas.1524213113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Achuthan V, et al. , Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe, 2018. 24(3): p. 392–404 e8 10.1016/j.chom.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Primio C, et al. , Single-cell imaging of HIV-1 provirus (SCIP). Proc Natl Acad Sci U S A, 2013. 110(14): p. 5636–5641. 10.1073/pnas.1216254110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis AC, et al. , Second generation imaging of nuclear/cytoplasmic HIV-1 complexes. AIDS Res Hum Retroviruses, 2014. 30(7): p. 717–726. 10.1089/aid.2013.0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albanese A, et al. , HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS One, 2008. 3(6): p. e2413 10.1371/journal.pone.0002413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burdick RC, Hu WS, and Pathak VK, Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proc Natl Acad Sci U S A, 2013. 110(49): p. E4780–E4789. 10.1073/pnas.1315996110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vranckx LS, et al. , LEDGIN-mediated Inhibition of Integrase-LEDGF/p75 Interaction Reduces Reactivation of Residual Latent HIV. EBioMedicine, 2016. 8: p. 248–264. 10.1016/j.ebiom.2016.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quercioli V, et al. , Comparative analysis of HIV-1 and murine leukemia virus three-dimensional nuclear distributions. J Virol, 2016. 90(10): p. 5205–5209. 10.1128/JVI.03188-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burdick RC, et al. , Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLoS Pathog, 2017. 13(8): p. e1006570 10.1371/journal.ppat.1006570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marini B, et al. , Nuclear architecture dictates HIV-1 integration site selection. Nature, 2015. 521(7551): p. 227–231. 10.1038/nature14226 [DOI] [PubMed] [Google Scholar]
- 31.Nagai S, et al. , Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science, 2008. 322(5901): p. 597–602. 10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Towbin BD, et al. , Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell, 2012. 150(5): p. 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]


