Abstract
Protein misfolding is linked to a wide array of human diseases1,2. Protective cellular protein quality control (PQC) mechanisms evolved to selectively recognize misfolded proteins and limit their toxic effects3-9. Here we examine how molecular chaperones and the ubiquitin (Ub)-proteasome system (UPS) cooperate to recognize and promote the clearance of soluble misfolded proteins. Using a panel of PQC substrates with distinct characteristics and localization, we define distinct chaperone and ubiquitination circuitries executing quality control in the cytoplasm and nucleus. Proteasomal degradation of cytoplasmic misfolded proteins requires tagging with mixed Lysine-48 (K48) and Lysine-11 (K11) linked ubiquitin chains. A distinct combination of E3 ubiquitin ligases and specific chaperones is required to achieve each type of linkage-specific ubiquitination. Strikingly, proteasomal degradation of nuclear misfolded proteins only requires K48 linked Ub chains and is thus independent of K11 specific E3s and chaperones. The distinct Ub codes for nuclear and cytoplasmic PQC appears linked to the function of the ubiquilin Dsk2, which is specifically required for clearance of nuclear misfolded proteins. Together, our work defines the principles of cytoplasmic and nuclear PQC as distinct, involving combinatorial recognition by defined sets of cooperating chaperones and E3s. A better understanding of how these organelle-specific PQC requirements implement proteome integrity has important implications for our understanding of diseases linked to impaired protein clearance and proteostasis disfunction.
Accumulation of misfolded proteins is toxic to the cell, leading to their accumulation in puncta6-9 and aggregates associated with neurodegenerative diseases1. Therefore, misfolded proteins arising during biogenesis or through proteotoxic damage must be cleared through a poorly understood process involving the cooperation of chaperones and UPS components3-5 (Fig. 1a).
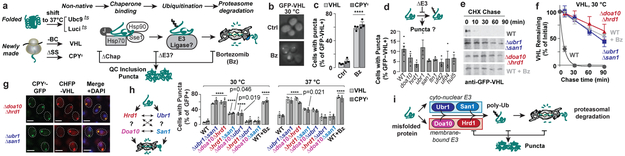
Figure 1 ∣. Combined deletion of certain E3 ligases impairs misfolded protein clearance.
a, Clearance of proteins misfolded for different reasons involves conserved pathways, with initial recognition by Hsp70-Hsp90 family chaperones, ubiquitination by one or more E3 ligases, and targeting for proteasomal degradation. Blocking any step triggers misfolded protein sequestration into puncta.
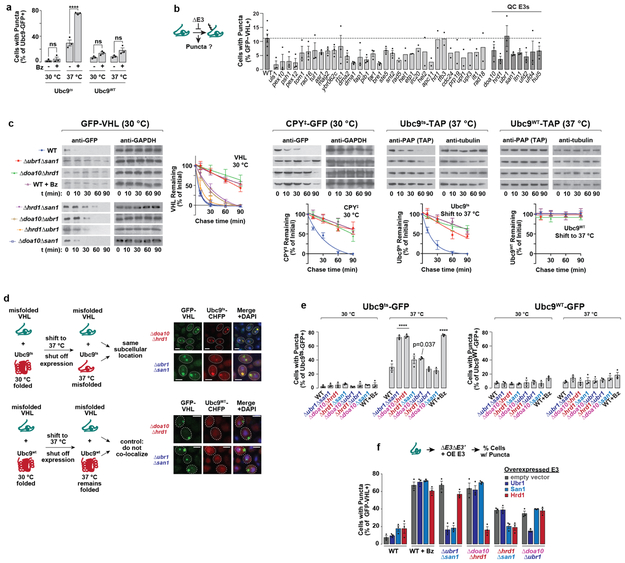
b-c, VHL and CPY‡ form puncta upon proteasome inhibition. WT cells expressing galactose-inducible GFP-VHL or CPY‡-GFP were shifted to glucose media with 50 μM bortezomib (Bz) or vehicle control (Ctrl) for 1 h to shut off expression. Fixed cells were imaged by fluorescence microscopy, c, % cells with GFP-VHL puncta.
d, Deleting individual E3s implicated in PQC does not increase puncta formation. Experiment performed as in b, but in strains with endogenous deletions of E3s.
e-f, Misfolded proteins are stabilized in Δubr1Δsan1 and Δdoa10Δhrd1 strains. Cycloheximide-chase and immunoblot to assess stability of GFP-VHL in E3 double deletion strains, or following 50 μm bortezomib treatment. f, Densitometric quantification of bands relative to t = 0 (mean ± SEM from 3 biologically independent experiments).
g, Multiple misfolded proteins are sequestered in the same subcellular location. Experiment performed as in b, but in strains co-expressing VHL with CPY‡. Images represent over 100 cells from each of 3 biologically independent experiments. Scale bars = 2 μm.
h, Deleting certain E3 pairs increases puncta formation. Experiment performed as in b, but in strains with endogenous deletions of E3 pairs. On the right, cells were shifted to 37 °C for the shut-off.
i, Misfolded protein clearance requires soluble E3s Ubr1 or San1 and membrane-bound E3s Doa10 or Hrd1.
c, d, h, 300 cells were counted per condition. Bars represent mean ± SEM from 3 biologically independent experiments—except for WT in d (7 biologically independent experiments). Statistically significant differences vs. WT by one-way ANOVA + Dunnett’s multiple comparisons test are indicated (adjusted p-value, or ****p < 0.0001).
To better understand PQC of soluble proteins, we used a panel of substrates reflecting different types of misfolding, including two temperature-sensitive proteins (Ubc9ts and Luciferasets), a protein (VHL) that cannot fold without its oligomeric partners Elongin BC, and translocation-defective CPY* lacking its signal sequence (CPY‡)6,7,9,10. Terminally-misfolded GFP-VHL or CPY‡-GFP were cleared within 1 h, with only ~10 % of cells containing GFP-positive puncta (Fig. 1b-c). Blocking proteasomal degradation with bortezomib led to their accumulation in GFP+ puncta. Temperature-sensitive proteins, such as Ubc9ts-GFP, are diffuse when folded at 30 °C but upon misfolding at 37 °C are also degraded through the proteasome or accumulated in puncta (Extended Fig. 1a)6,7. Of note, native Ubc9WT is soluble at either temperature.
To identify the E3 ubiquitin ligases involved in PQC, GFP-VHL was expressed in a library of 37 S. cerevisiae single deletion strains comprising most non-essential E3s (Extended Fig. lb)—including E3s previously implicated in PQC (Fig. 1d). No single deletion caused a significant increase in puncta formation over WT cells, suggesting no single E3 is essential for PQC of this substrate.
Since E3 ligases can have redundant functions11, we next deleted pairs of E3s previously implicated in PQC. The cytoplasmic E3 Ubr1 and the nuclear E3 San1 cooperate in clearance of some cytoplasmic proteins11,12. ER membrane-anchored Hrd1, and Doa10, also localised to the ER and the inner nuclear membrane13, trigger endoplasmic reticulum-associated degradation (ERAD)14. Degradation of all PQC substrates was abrogated in both Δubr1Δsan1 and Δdoa10Δhrd1 strains to the same extent as proteasome inhibition (e.g. cycloheximide (CHX) chase in Fig. 1e-f, Extended Fig. 1c). Notably, all PQC substrates accumulated in the same puncta in these strains (Fig. 1g, Extended Fig. 1d). Therefore, multiple misfolded proteins use the same E3 systems for proteasomal degradation, and are sequestered together in the same PQC compartments in their absence.
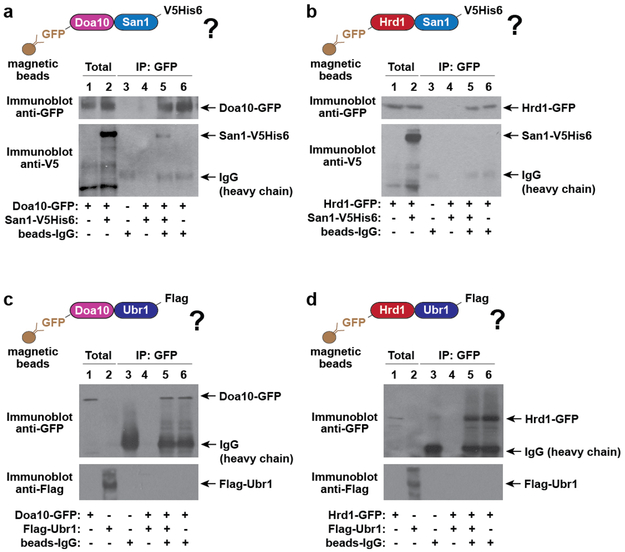
To determine why deletion of either E3 ligase pair similarly stabilized our PQC substrates, we tested the effect of doubly deleting all possible combinations of the four E3 ligases—UBR1, SAN1, DOA10, and HRD1 (Fig. 1h, Extended Fig. 1e). Puncta formation and CHX chase assays showed that only specific combinations of E3 deletions abrogate clearance. Strong stabilization was observed for Δubr1Δsan1 and Δdoa10Δhrd1. A moderate effect was found for Δdoa10Δubr1 and Δhed1Δsan1. Strikingly, Δdoa10Δsan1 and Δhrd1Δubr1 had no effect on PQC, suggesting that these pairs of ligases— Doa10/San1 and Hrd1/Ubr1—provide parallel, optimal combinations for PQC clearance. This E3 circuit logic appeared general, as it operated for all substrates tested at 30 °C and 37 °C. Thus, PQC clearance requires parallel pathways of specific pairs of E3s combining one of the soluble E3s (either Ubr1 or San1, blue) and one of the membrane-bound E3s (either Doa10 or Hrd1, red) (Fig. 1i). Overexpressing an E3 in any of the double deletion strains only rescued clearance in those strains deleted for that particular E3 (Extended Fig. 1f). Therefore, E3 function is not interchangeable, even at higher expression levels, and PQC requires specific E3 combinations. The functional cooperation between E3s may involve a physical complex, as immunoprecipitation experiments show that San1 associates with Doa10 but not with Hrd1 (Extended Fig. 2). We were unable to co-immunoprecipitate Ubr1 with Hrd1 (or Doa10)—these E3s may bind transiently, or cooperate functionally through separate complexes.
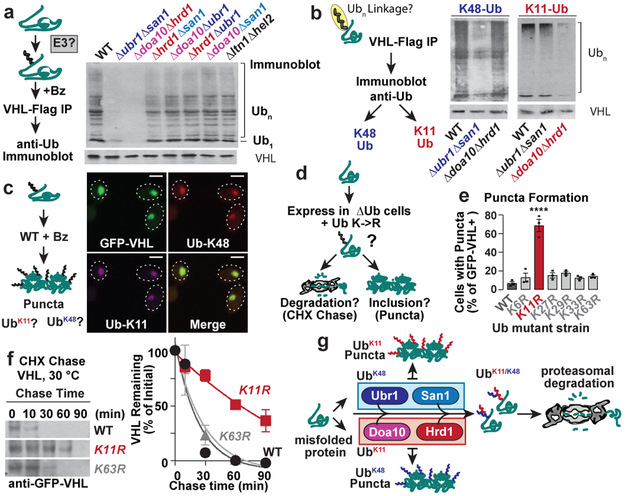
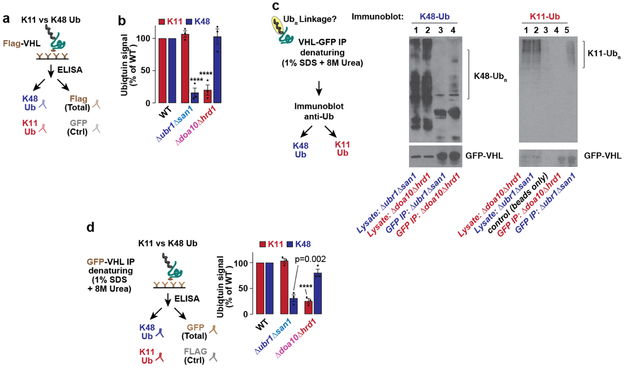
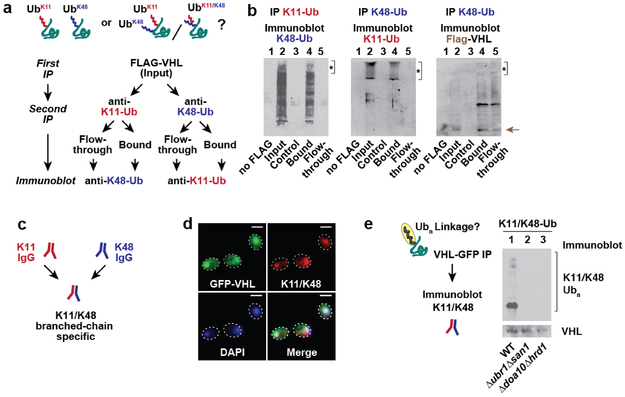
To examine whether the PQC defects observed above were linked to impaired ubiquitination, terminally-misfolded FLAG-tagged VHLL158P (Ref. 10) was expressed in either wild-type or the panel of double E3 deletions. VHL was immunoprecipitated under harsh denaturing conditions, followed by anti-ubiquitin immunoblot detection (Fig. 2a). In WT cells, poly-ubiquitin (Ub) chains were clearly attached to VHL. In the Δubr1Δsan1 and Δdoa10Δhrd1 cells, VHL ubiquitination was significantly reduced, indicating these E3 pairs promote VHL clearance through ubiquitination. No clear differences in VHL ubiquitination were observed in any other strains. A similar strategy examined how E3 deletions specifically affect VHL tagging with the canonical proteasome signal Lysine-48 (K48)-linked Ub (Fig. 2b)5. Surprisingly, K48-linked VHL ubiquitination was strongly reduced in Δubr1Δsan1, but not in Δdoa10Δhrd1 cells. Conversely, the alternative proteasomal targeting signal Lysine-11 (K11)-linked Ub5,15-18 was unaffected in Δubr1Δsan1 cells but was abrogated in Δdoa10Δhrd1 cells. Note that misfolded VHL was ubiquitinated with both K48- and K11-Ub linked chains in WT cells. A quantitative ubiquitin linkage-specific ELISA assay for K11 or K48 linked chains confirmed that Ubr1 and San1 mediate VHL tagging with K48-Ub chains, whereas Doa10 and Hrd1 promote K11-Ub linked ubiquitination (Extended Fig. 3a-b). Upon proteasome inhibition, the VHL puncta co-localised with both K11 and K48 linked Ub (Fig. 2c), supporting the conclusion that VHL itself is modified with distinct K48 and K11 Ub linkages.
Figure 2 ∣. Cytoplasmic misfolded proteins are modified with both K11- and K48-linked ubiquitin chains.
a, VHL ubiquitination is impaired in Δubr1Δsan1 and Δdoa10Δhrd1 strains. Flag-VHL denaturing immunoprecipitation followed by immunoblot for ubiquitin (Ub) in WT or E3 double deletion strains. Deletion of the co-translational E3s LTN1 & HEL2 served as a control.
b, K48 and K11 Ub linkages are reduced on Flag-VHL in Δubr1Δsan1 and Δdoa10Δhrd1 strains, respectively. Experiment performed as in a, but K48 or K11 linkage specific Ub antibodies were used for immunoblot.
c, K48 and K11 Ub co-localise with GFP-VHL puncta. Experiment performed as in Fig. 1b. Fixed cells were spheroplasted and immunostained before imaging by confocal fluorescence microscopy. Images represent >100 cells from each of 3 independent experiments. Scale bars = 2 μm.
d-f, VHL clearance is impaired in the absence of K11-Ub linkages. Cells co-expressing galactose-inducible GFP-VHL, and either WT or Lys-to-Arg mutant Ub (KnR) as their only ubiquitin source, were shifted to glucose media for 1 h to shut off expression. 300 cells were counted per condition. e, % cells with GFP-VHL puncta (mean ± SEM from 3 biologically independent experiments). Only K11R cells significantly altered the percentage of puncta-positive cells vs. WT (one-way ANOVA + Dunnett’s multiple comparisons test, ****p < 0.0001).
f, Cycloheximide-chase and immunoblot to assess GFP-VHL stability in Ub mutant strains. Graphs represent densitometric quantification relative to t = 0 (mean ± SEM from 3 biologically independent experiments).
g, Doa10/Hrd1 and Ubr1/San1 E3 ligases collaborate to ubiquitinate misfolded proteins with branched K11/K48 chains, thereby targeting them for proteasomal clearance. Inhibition of either type of linkage results in the sequestration of the misfolded proteins into puncta.
a, b, f, Immunoblots represent 3 biologically independent experiments.
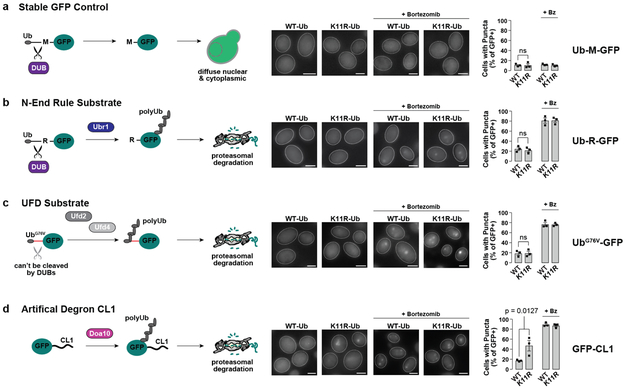
We next examined PQC in cells expressing a single copy of ubiquitin carrying individual K-to-R mutations, unable to form specific linkages (Fig. 2d). Ub-K48R is lethal19, and could not be tested. VHL was degraded in all Ub K-to-R variants except for Ub-K11R cells, where VHL clearance was impaired, leading to accumulation in puncta (Fig. 2e-f). Importantly, K11-Ub linkages were not generally required for proteasomal clearance since non-misfolded cytoplasmic substrates Ub-R-GFP and UbG76V-GFP were degraded normally in Ub-K11R cells (Extended Fig. 4). Consistent with our data implicating ER-resident E3s in K11-Ub tagging of soluble PQC substrates, the Ub-K11R strain was previously shown to be sensitive to ER stress16.
We next examined if misfolded VHL is tagged with mixed linkage K48 and K11 Ub chains. Sequential double immunoprecipitations (IP) isolated FLAG-VHL followed by a subsequent IP for K48 or K11 linked chains (Extended Fig. 5a). The presence of K11 or K48 linked chains in either flow-through or bound fractions was then detected by immunoblot. All the K11 and K48 linked chains were only present in the bound fractions of each IP (Extended Fig. 5b), demonstrating that misfolded VHL is tagged with both K11- and K48- linked ubiquitin chains. Additionally, an antibody that recognises branched K11/K48 linkages with K11 and K48 Ub moieties extending from the same molecule18 reacted with misfolded VHL in both immunofluorescence and IP experiments (Extended Fig. 5c-e). Notably, reactivity with the branched K11/K48 antibody required the presence of both K48- and K11-specific E3s (Extended Fig. 5e). We conclude that a dual Ub code involving K11 and K48 is required for clearance of soluble PQC substrates. K11 ubiquitination is mediated by either of the membrane-bound E3s Doa10 or Hrd1 and K48 ubiquitination is mediated by either of the soluble E3s Ubr1 or San1, thus explaining the requirement for pairs of E3s for PQC clearance. Since deletion of one E3 pair did not abrogate the action of the other, addition of K11 or K48 linked Ub chains does not require a sequential order.
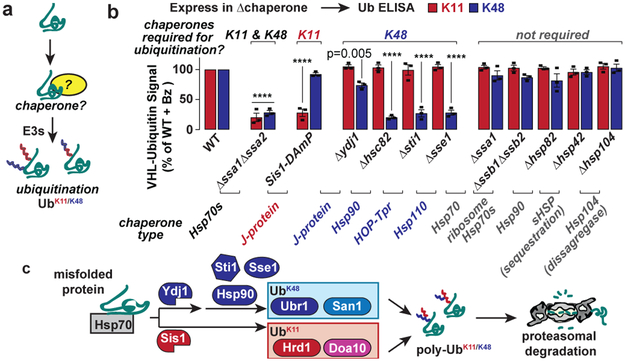
It is unclear how chaperones, key mediators of PQC1,3,4, facilitate ubiquitination and clearance. Chaperones could simply maintain solubility of misfolded proteins or could specifically direct them along an E3 clearance pathway (Fig. 3a). The ubiquitin linkage-specific ELISA assay was used to identify whether chaperones previously implicated in PQC are required to tag misfolded VHL with either K11-Ub or K48-Ub (Fig. 3b). Cells lacking Ssa1 and Ssa2, the major cytosolic Hsp70s required for PQC clearance7,10,20, were strongly impaired in ubiquitination with either linkage. In contrast, ribosome-associated Hsp70s Ssb1 and Ssb2 were dispensable for degradation10 and ubiquitination.
Figure 3 ∣. K11- and K48- linked ubiquitination of misfolded proteins involves different chaperones.
a, Molecular chaperones are involved in the ubiquitination of misfolded proteins by E3s.
b, Relative amounts of K11 and K48 Ub present on Flag-VHL in chaperone deletion strains compared with WT. WT or chaperone deletion strains expressing Flag-VHL were lysed after 1 h bortezomib treatment. Ub linkage ELISA was then performed as described in Extended Fig. 3a. Bars represent normalized values from each strain (mean ± SEM from 3 biologically independent experiments) expressed as a proportion of normalized WT. Strains with statistically significant differences vs. WT by one-way ANOVA + Dunnett’s multiple comparisons test are indicated with the adjusted p-value, or **** for p < 0.0001.
c, K11 and K48 ubiquitination of misfolded proteins proceeds through the action of different chaperone pathways.
Hsp70 substrate binding relies on many J-domain proteins, often themselves chaperones that ferry substrates for Hsp70 binding3,4. We examined VHL ubiquitination in cells lacking two J-domain proteins, Ydj1 and Sis1, implicated in PQC 7-9,11,20-22. Strikingly, each J-domain protein reduced ubiquitination, but in a linkage specific manner. Δydj1 cells specifically reduced K48 ubiquitination, albeit to a modest degree; likely because Ydj1 function is partially redundant with other J-domain cofactor7. Depleting cells of the essential Sis1 left K48 ubiquitination unaffected but caused a dramatic loss of K11 ubiquitination. Of note, three other chaperones important for PQC—the Hsp70 nucleotide exchange factor Sse1 (an Hsp110 chaperone), Hsp90, and Sti1/HOP (which bridges the interaction of Hsp70 with Hsp90)7,10,11,23—were all required for K48 Ub but were dispensable for K11 Ub. We conclude that specific chaperone pathways direct PQC substrates to distinct E3 pathways to promote mixed linkage ubiquitination (Fig. 3c). Sis1 cooperates with Hsp70 in promoting Doa10/Hrd1-mediated K11 ubiquitination, whereas the Ydj1/Sse1/Sti1 chaperones cooperate with Hsp70 and Hsp90 for Ubr1/San1-mediated K48 ubiquitination. Requiring two distinct E3s to communicate with distinct chaperone pathways may provide a checkpoint in the triage decision to refold or degrade.
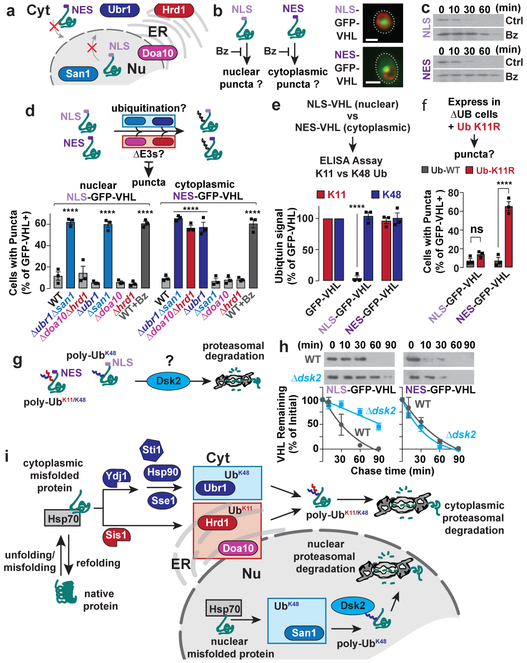
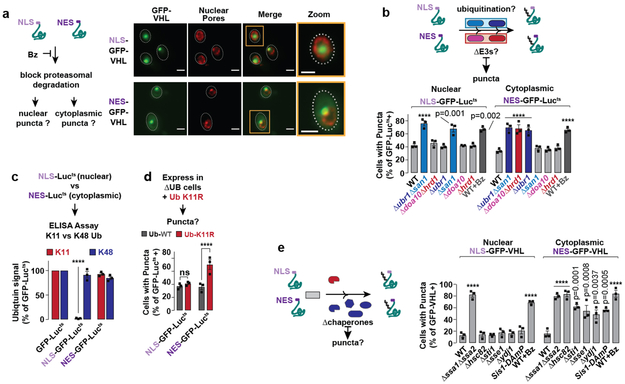
That cytoplasmic misfolded proteins are degraded via a nuclear E3, San1, was puzzling. In principle, misfolded cytoplasmic proteins might be actively imported into the nucleus for degradation 11,12,20,23. Alternatively, they may passively diffuse across the nuclear pores, due to their small size 24. To directly interrogate cytoplasmic-specific and nuclear-specific PQC degradation pathways, we spatially restricted two PQC substrates, VHL and Lucits, to the nucleus, using a nuclear localization signal, or to the cytoplasm, using a nuclear export signal (NLS or NES respectively; Fig. 4a). Similar results were obtained for both substrates, corroborating the generality of our conclusions. Bortezomib treatment showed both nuclear and cytoplasmic variants are degraded by the UPS (Fig. 4b-c, Extended Fig. 6a). NES-GFP-VHL accumulated in cytoplasmic perinuclear puncta, whereas NLS-GFP-VHL accumulated in intranuclear puncta (Fig. 4b). Thus, misfolded proteins are either degraded or form inclusions in the cellular compartment where misfolding occurs.
Figure 4 ∣. Confining misfolded proteins to the nucleus or cytoplasm alters their PQC requirements.
a-b, NLS-GFP-VHL and NES-GFP-VHL accumulate in the nucleus or cytoplasm, respectively, upon proteasome inhibition. Expression of NLS- or NES- GFP-VHL in WT cells was shut off in glucose media with 50 μM bortezomib for 1 h (b). Cells were immunostained for nuclear pores (Nsp1, red) and imaged by fluorescence microscopy. Images represent 3 biologically independent experiments. Scale bars = 2 μm.
c, NLS- and NES- GFP-VHL are cleared by the proteasome. Cycloheximide-chase and immunoblot to assess stability of NLS- and NES- GFP-VHL in WT cells treated with (WT + Bz) or without (WT) 50 μM bortezomib. Immunoblots represent 3 biologically independent experiments.
d, Confining VHL to the nucleus or cytoplasm alters its E3 ligase requirement. % cells with NLS- or NES- GFP-VHL puncta in deletion strains following 1 h shut-off.
e, Nuclear VHL has severely reduced K11-Ub ubiquitin linkages. ELISA performed as in Fig. 2c, but in GFP-multiTrap® instead of α-FLAG-conjugated plates.
f, Nuclear VHL clearance is unaffected by K11-Ub linkages. Experiment performed as in d, but in cells expressing WT or K11R mutant Ub as its only ubiquitin source.
g-h, Clearance of NLS-GFP-VHL requires Dsk2. Cycloheximide-chase performed as in c, but in WT or Δdsk2 cells, g, Densitometric quantification relative to t = 0 (mean ± SEM from 3 biologically independent experiments).
i, Nuclear and cytoplasmic misfolded proteins have distinct clearance requirements. Cytoplasmic misfolded proteins require tagging with both K11- and K48- Ub, by cooperating chaperones and E3s, for proteasomal degradation. In the nucleus, tagging with K48-Ub is sufficient for recognition by Dsk2 and subsequent proteasomal degradation.
d-f, Bars represent mean ± SEM from 3 biologically independent experiments. Statistically significant differences vs. WT (d, f) or GFP-VHL (e) by one-way ANOVA + Dunnett’s multiple comparisons test are indicated (adjusted p-value, or ****p < 0.0001; ns = p > 0.05).
Degradation of cytoplasmic NES-VHL or NES-Lucits required the K11-Ub E3s Doa10 and Hrd1, but only the cytoplasmic K48 E3 Ubr1 (Fig. 4d, Extended Fig. 6b). Therefore, San1 is dispensable for UPS degradation of strictly cytoplasmic PQC substrates. Surprisingly, the nuclear NLS-GFP-VHL or Lucits only required the nuclear K48 E3 San1 for clearance.
Consistent with their E3 requirements, cytoplasm-restricted misfolded NES-proteins were conjugated to both K11 and K48 linked chains at levels similar to their unmodified counterparts. However, nuclear-restricted misfolded NLS-proteins were only ubiquitinated with K48 chains, with the K11 signal reduced to baseline levels (Fig. 4e and Extended Fig. 6c). Notably, K48-Ub tagged nuclear misfolded proteins were efficiently cleared by the proteasome (Fig. 4c-d, Extended Fig. 6a). Confirming the distinct role of K11 ubiquitination in nuclear versus cytoplasmic PQC, clearance of NES-tagged misfolded proteins was impaired K11R-Ub cells, whereas degradation of nuclear-restricted PQC substrates was unaffected (Fig. 4f and Extended Fig. 6d).
To understand the differences between nuclear and cytoplasmic PQC we first focused on ubiquilin Dsk2, a predominantly nuclear25 shuttling factor that physically ferries K48 ubiquitinated proteins to the proteasome.26 Strikingly, cells lacking Dsk2 were impaired in clearance of NLS- but not NES- GFP-VHL (Fig. 4g-h). We propose Dsk2 in the nucleus increases the affinity of K48-only tagged nuclear misfolded proteins for the proteasome.
The role of chaperone circuits in nuclear versus cytoplasmic PQC was examined through functional and proteomic approaches. Depletion of Sis1, required for K11-Ub (Fig. 3b), had no effect on clearance of NLS-tagged proteins; but, as expected, blocked clearance of NES-tagged variants (Extended Fig. 6d). Interestingly, Hsp90, Sti1, Sse1 and Ydj1 were only required for K48 ubiquitination and clearance of cytoplasmic substrates. Only the Hsp70s Ssa1/Ssa2 were required for nuclear PQC.
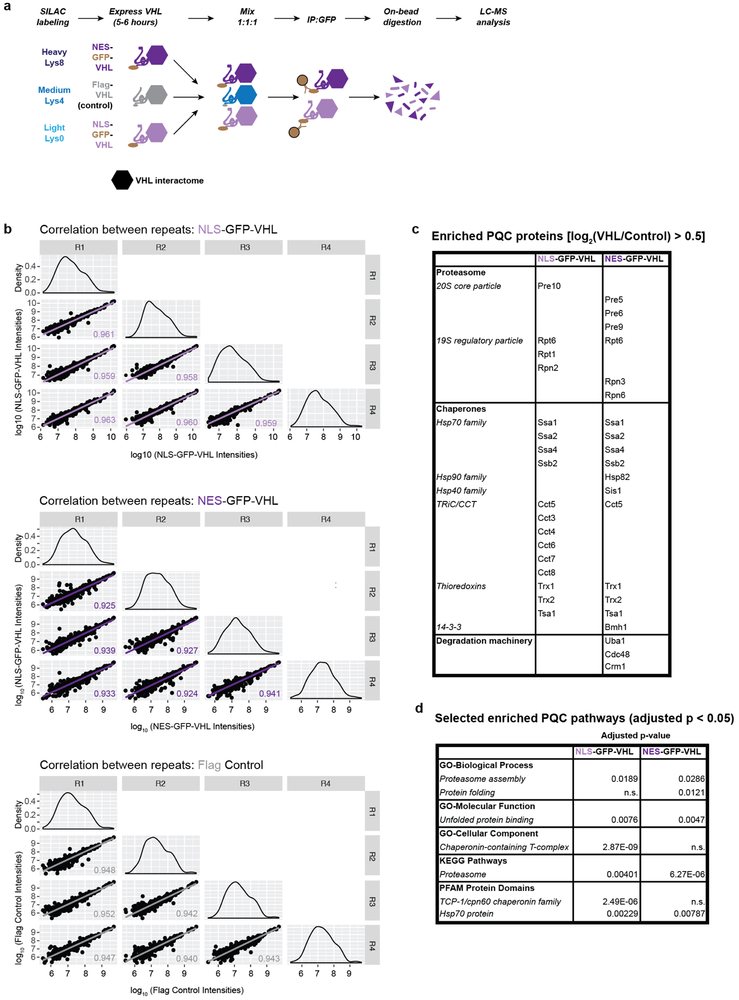
SILAC-based mass spectrometry further indicated PQC of nuclear and cytoplasmic misfolded proteins involves different circuitries. Analyses of IPs for NLS- or NES- GFP-VHL (Extended Fig. 7 and Extended Table 1) revealed localization-dependent proteostasis interactors (log2(VHL/control ratio) > 0.5). Both nuclear and cytoplasmic PQC substrates shared enrichments in proteasomal subunits and several chaperones, including four Hsp70s. Consistent with their selective requirement for cytoplasmic PQC, Hsp82 and Sis1 specifically associated with NES-VHL. Of note, Cdc48/p97 also selectively associated with NES-VHL, whereas the TRiC/CCT chaperonin selectively associated with NLS-VHL (Extended Fig. 7c). The importance of these interactions for PQC will be explored in future studies.
We conclude that clearance of nuclear and cytoplasmic PQC substrates requires distinct Ub codes, distinct E3s, and distinct chaperone sets. Cytoplasmic PQC requires both K11 and K48 linked Ub, while nuclear PQC only requires K48 Ub. Given that the misfolded protein is the same in both compartments, these distinct Ub chain requirements are unlikely to respond to the substrate’s structural properties but instead arise from differences in the proteostasis machineries maintaining nuclear and cytoplasmic proteomes.
Our study opens new perspectives for understanding the circuits and logic of misfolded protein quality control (Fig. 4i). We define a general PQC path where specific non-redundant networks of E3 ligases and chaperones mediate post-translational PQC clearance. Surprisingly, we also uncover significant differences in the pathways of cytoplasmic and nuclear PQC. It is possible that nuclear and cytoplasmic proteasomes differ in composition27 or concentration28, affecting their ability to recognize ubiquitinated substrates effectively. It is also possible that K11-Ub linked chains facilitate recognition by another cytoplasmic PQC factor. Intriguingly, Sis1, required for K11-Ub, can act as a sorting factor9, and is sequestered in protein aggregates7-9,21,29. Nuclear ubiquilin Dsk2 shuttles K48-linked substrates to nuclear proteasomes, whereas in the cytoplasm, mixed K11/K48 chains enhance the affinity of misfolded proteins for cytoplasmic proteasomes, likely by engaging multiple Ub-receptors in the 19S cap30.
The distinct Ub linkage requirements for nuclear and cytoplasmic PQC might respond to regulatory and functional rationales for the triage decision between (re-)folding and targeting to degradation. The importance of the UPS in chromatin regulation31 could sensitize the nuclear proteome to protein misfolding and aggregation32—perhaps leading to relaxed ubiquitination requirements for proteasomal recognition. The cytosol is a site of active protein biogenesis, assembly and targeting, requiring productive folding intermediates to be safeguarded. Requiring a dual Ub code for cytoplasmic PQC, mediated by distinct E3 ligases and chaperones, would provide a checkpoint to ensure that only non-productive, misfolded proteins are degraded. Of interest, mutations in ubiquilin and the PQC E3 ligases identified here are associated with a host of human diseases (Extended Table 2). Future dissection of these circuits in normal and diseased states may provide mechanistic clues and open therapeutic opportunities.
Methods
Yeast media, plasmids, and strains.
Yeast media preparation, growth, transformations and manipulations were performed according to standard protocols. All E3 ubiquitin ligase and chaperone yeast deletion strains used in this study derived from the BY4742 wild-type (WT) strain. Single deletions were generated by homologous recombination using the NAT gene. The Sis1-DAmP strain was also generated in this way. Double deletions were generated with both NAT and hygromycin. All strains were checked by PCR using at least 2 sets of primers.
All ubiquitin lysine-to-arginine mutant strains—expressing a single, galactose-inducible ubiquitin gene—were kind gifts from D. Finley (Harvard University, Massachusetts, USA)23. Yeast strains expressing GFP-tagged Doa10 or Hrd1 from their endogenous loci were from the Yeast-GFP Clone Collection (Thermo Fisher Scientific). The Δdsk2 strain was obtained from the Saccharomyces Genome Deletion Project33. We acknowledge the generous gifts of pGAL-CPY‡-GFP (R. Hampton, University of California San Diego, California, USA), pADH-Flag-Ubr1 and pGAL-San1-V5His6 (D. Wolf, University of Stuttgart, Germany), pFLUC-DM-YFP (F. U. Hartl, Martinsried, Germany), and Ub-M-GFP, Ub-R-GFP, UbG76V-GFP and GFP-CL1 (N. Dantuma, Karolinska Institute, Stockholm, Sweden)34. All other plasmids were constructed using the Gateway cloning technology35, as described previously6.
Gal shut-off protein expression.
Yeast strains transformed with plasmids encoding the galactose-inducible protein of interest were grown overnight in raffinose synthetic media at 30 °C before dilution to an OD600 between 0.05 and 0.1 in galactose synthetic media. The cells were grown for 4-5 h (OD600 0.6-0.8) to induce expression of the galactose-inducible protein. Expression was shut off by switching the cells to glucose synthetic media, and the fate of the existing pool of proteins was assessed according to the appropriate downstream application.
Counting puncta-containing cells.
Cells were grown as described for Gal shut-off protein expression. Following Gal shut-off, cells were allowed to grow at 30 or 37 °C for 1 h in glucose synthetic media. Note that for WT + bortezomib conditions, 50 μM bortezomib (LC Laboratories) was dissolved in the glucose synthetic media prior to addition. Cells were then fixed for 15 min in 4% paraformaldehyde before mounting onto concanavalin A-coated coverslips using ProLong™ Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Fluorescence was visualized using a Zeiss LSM700 confocal microscope with a ×63 oil-immersion lens. Image analysis was performed by ImageJ software (http://rsbwed.nih.gov/ij). 300 cells were counted manually for each blinded sample, and the percentage of cells containing GFP-positive puncta were noted. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparisons test.
Immunofluorescence.
Cells were grown and paraformaldehyde fixed as described above. Fixed cells were spheroplasted by incubating for 20-40 min at 30 °C with Zymolyase 100T (Zymo Research) in potassium phosphate buffer (0.1 M potassium phosphate pH 7.5, 1.2 M sorbitol) supplemented with 25 mM DTT and 5 mM EDTA. The resultant spheroplasts were permeabilized with 0.1% (v/v) Triton X-100 in potassium phosphate buffer for 10 min. For immunostaining, cells were blocked (potassium phosphate buffer with 1 % (w/v) bovine serum albumin (BSA) for 30 min at room temperature) and incubated overnight at 4 °C with antibodies diluted at the appropriate concentration in potassium phosphate buffer with 0.1 % (w/v) BSA. Antibodies used were against K48-Ub covalently linked to Alexa Fluor™ 568 (1:100, Abcam catalogue number ab208136), K11-Ub (1:50, EMD Millipore catalogue number MABS107-I) covalently linked to Alexa Fluor™ 647 NHS Ester (Thermo Fisher Scientific), K11/K48-Ub bispecific antibody (1:500, Genentech) with secondary antibody Cy™5-conjugated donkey anti-human (1:1,000, Jackson ImmunoResearch catalogue number 709-175-149), and Nsp1 (1:500, EnCor catalogue number MCA-32D6) with secondary antibody Alexa Fluor™ 568-conjugated goat anti-mouse (1:1,000, Thermo Fisher Scientific catalogue number A10037) for 1 h at room temperature. Stained cells were mounted onto poly-lysine-coated coverslips using ProLong™ Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Fluorescence was visualized using a Zeiss LSM700 confocal microscope with ×100 NA 1.4 oil-immersion lens. Raw data collected as z-stacks were represented in a single image as maximum intensity projections (ImageJ).
Cycloheximide chase assay.
Cells were grown as described for Gal shut-off protein expression. A first sample of an equivalent of OD600 of 10 (t = 0) was collected prior to shut-off. The rest of the culture was shifted to glucose synthetic media with 50 μg ml1− cycloheximide, and a further 10 OD600 of cells were collected after 10, 30, 60 and 90 min after the shift. Samples from each time-point were pelleted, washed once with 15 mM sodium azide containing 1x Roche cOmplete™ EDTA-free protease inhibitor tablet (Sigma), snap frozen in liquid nitrogen, and stored at −80 °C until all time-points were collected. Proteins were extracted by boiling each sample in an equal volume of 2× SDS Sample Buffer (100 mM Tris-HCl pH 6.8, 4 % (w/v) SDS, 20 % (v/v) glycerol, 200 mM DTT, 0.2 % (w/v) bromophenol blue) for 10 min before detection of protein by SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblotting.
Protein samples from cell lysates or immunoprecipitates were denatured in SDS Sample buffer (95 °C for 5 min) or LDS Sample Buffer (70 °C for 10 min) before separation by SDS-PAGE. Precision Plus Prestained Protein™ Standards (Bio-Rad) were used to estimate protein weight. Proteins were transferred onto PVDF or nitrocellulose membranes (Bio-Rad) and immunoblotted with primary antibodies against GFP (1:1,000, Roche catalogue number 11814460001 or Santa Cruz Biotechnology catalogue number sc-9996), TAP (1:2,000-1:7,500, Sigma catalogue number P1291), GAPDH (1:5,000, Abcam catalogue number ab9485 or 1:10,000 Genetex catalogue number GTX100118), alpha-tubulin (1:2,500, DSHB Hybridoma Product 12G10, deposited by J. Frankel & E.M. Nelson), FLAG (1:1,000, Cell Signaling Technology catalogue number #2368), pan-Ubiquitin (1:1,000, Life Sensors catalogue number VU101), K11-Ub (1:100, EMD Millipore catalogue number MABS107-I), K48-Ub (1:1,000, Cell Signaling Technology catalogue number #12805) or K11/K48-Ub bispecific antibody (1:500, Genentech). Specific primary antibodies used are indicated next to the uncropped immunoblot images in Supplementary Fig. 1. For Immunoblotting of ubiquitin, samples were separated by SDS-PAGE, transferred to a PVDF membrane and denatured by boiling for 10 min at 95 °C prior to antibody incubation. Secondary antibodies used were HRP-conjugated donkey anti-mouse (1:5,000, Jackson ImmunoResearch catalogue number 715-035-150), donkey anti-rabbit (1:5,000, Jackson ImmunoResearch catalogue number 711-035-152) or donkey anti-human (1:5,000, Jackson ImmunoResearch catalogue number 709-035-149). The HRP signal was detected by incubation with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and exposure to GeneMate Blue Ultra Film (BioExpress). Immunoblots shown are representative of 3 independent experiments.
Immunoprecipitation of E3 ligases.
To test for co-immunoprecipitation of E3 ligases, pADH-FLAG-Ubr1 or pGAL-San1-V5His6 plasmids were transfected into yeast strains from the Yeast-GFP collection (expressing GFP-tagged Doa10 or Hrd1 from the endogenous loci). Each strain was grown overnight in raffinose synthetic media at 30 °C before dilution to an OD600 between 0.05 and 0.1 in galactose synthetic media. The cells were grown for 24 h to induce expression of the galactose-inducible San1-V5His6, diluted back to OD600 0.1, and then grown for another 4-6 hours (OD600 0.6-0.8). For consistency between experimental conditions, the same protocol was followed for cells expressing Flag-Ubr1 from the ADH promoter, even though this does not require galactose for expression. Cells were pelleted and washed once with 15 mM sodium azide supplemented with 1x Roche cOmplete™ EDTA-free protease inhibitor tablet (Sigma) and 50 mM 2-chloroacetamide, to inhibit proteases and deubiquitinases, respectively. Pellets were resuspended in an equal volume of Lysis Buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM PMSF, Roche cOmplete™ EDTA-free protease inhibitor tablet (Sigma), 50 mM 2-chloroacetamide, 10 μM PR-619 (Sigma)) and frozen drop-wise in liquid nitrogen by passing through a 20.5 gauge syringe. Frozen samples were lysed by cryogrinding in a Retsch MM-301 (5 cycles, 30 Hz for 3 mins per cycle) and proteins solubilized by adding Triton X-100 (1 % v/v final concentration). Lysates were clarified (16,000xg for 30 min at 4 °C) and quantified for total protein by BCA assay. 2 mg of lysate was incubated with anti-GFP rabbit IgG conjugated to Protein G Dynabeads™ (Thermo Fisher Scientific) for 2 h at 4 °C to immunoprecipitate the GFP-tagged protein complexes, which were then eluted from the beads by heating for 30 min at 65 °C in non-reducing LDS Sample Buffer (Thermo Fisher Scientific). The bead-free samples were reduced with DTT (50 mM final concentration, 10 min at 65 °C) prior to SDS-PAGE analysis.
Immunoprecipitation of ubiquitinated VHL.
For immunoprecipitation of ubiquitinated Flag-VHL, yeast strains were grown as described for Gal shut-off protein expression. Following shut-off in glucose synthetic medium supplemented with 50 μM bortezomib for 1 h at 30 °C, cells were pelleted and snap-frozen in liquid nitrogen. All subsequent steps were performed at 4 °C or on ice. Pellets were resuspended in an equal volume of Urea Lysis Buffer (50 mM Tris-HCl pH 7.5, 8 M urea, 150 mM NaCl, 2 mM EDTA, 1 mM PMSF, Roche cOmplete™ EDTA-free protease inhibitor tablet (Sigma), 50 mM chloroacetamide, 10 μM PR-619 (Sigma)) and lysed by bead beating (5 cycles at 1 min each, with 1 min on ice in between cycles). Following dilution 10-fold in Triton IP buffer (same composition as Urea Lysis Buffer, but with 1 % v/v Triton X-100 instead of 8 M urea), lysates were clarified (16,000xg for 30 min at 4 °C) and quantified for total protein by BCA assay. 2 mg of lysates were incubated with Flag-M2 magnetic beads (Sigma) for 2 h at 4 °C to immunoprecipitate the Flag-tagged protein, which was then eluted from the beads by heating for 30 min at 70 °C in non-reducing LDS Sample Buffer (Thermo Fisher Scientific), to avoid co-elution of the Flag antibody. The bead-free samples were reduced with DTT (50 mM final concentration, 10 min at 65 °C) prior to SDS-PAGE analysis.
The same protocol was used for denaturing immunoprecipitation of ubiquitinated GFP-VHL, but with addition of 1 % w/v SDS in lysis and immunoprecipitation buffers, and incubation with GFP-TRAP_MA magnetic beads (ChromoTek) instead of Flag® M2 magnetic beads.
For double immunoprecipitation experiments, 10 mg of cell lysate was incubated with Flag® M2 magnetic beads for 2 h at 4 °C and eluted from the beads by competition with 3× Flag peptide (Apex Bio). The resultant eluate was subsequently incubated with an antibody against K48-Ub (1:500, Cell Signaling Technologies catalogue number #4289) or K11-Ub (1:50, EMD Millipore catalogue number MABS107-I) covalently conjugated using BS3 to Dynabeads™ Protein G (Thermo Fisher Scientific) overnight at 4 °C. The bead-bound fraction (“eluate”) was eluted by heating for 10 min at 70 °C in non-reducing LDS Sample Buffer, and analyzed alongside the unbound fraction (“flow-through”) by SDS-PAGE.
Ubiquitin linkage ELISA.
For quantification of K11-Ub and K48-Ub linkages on Flag-VHL, cell lysates were prepared as described for immunoprecipitation of ubiquitinated Flag-VHL. 200 μg of lysate protein were added to each well of an ANTI-Flag® M2 coated 96-well plate and incubated for 2 h. All incubation steps were performed at room temperature with gentle shaking. 4 wells were used for each technical replicate (2 replicates per strain per experiment). After washing 4× with Triton IP Buffer to remove unbound protein, each well was incubated for 1 h with rabbit antibodies against one of GFP (1:1,000, Cell Signaling Technology catalogue number #2956), Flag (1:1,000, Cell Signaling Technology catalogue number #2368), K11-Ub (1:50, EMD Millipore catalogue number MABS107-I), or K48-Ub (1:500, Cell Signaling Technology catalogue number #4289) diluted in 100 μL TBS-T with 0.1 % BSA. After another 4× washes, each well was incubated for 1 h with HRP-conjugated donkey anti-rabbit (1:2,000, Jackson ImmunoResearch catalogue number 711-035-152) diluted in 100 μL TBS-T with 0.1 % BSA, washed another 4× with Tween IP Buffer, and incubated for 30 min with 100 μL Pierce™ TMB Substrate (Thermo Fisher Scientific) followed by 100 μL 0.16 M sulfuric acid to stop the reaction. Absorbance was measured at 450 nm.
To calculate the K11-Ub or K48-Ub signal for each strain, the raw absorbance readings were subtracted of the negative control (GFP) signal, and then divided by the Flag signal to account for variations in total Flag-VHL levels. These K11-Ub or K48-Ub signals were then expressed as a proportion of the K11-Ub or K48-Ub signal in the WT strain to allow direct comparison between strains. Bars represent mean ± SEM from 3 individual experiments.
The same protocol was followed for quantification of linkages on GFP-tagged proteins, except with the use of GFP-multiTrap® 96-well plates (ChromoTek) instead of Flag® M2 coated plates, and using the GFP and Flag signals as positive and negative controls, respectively.
SILAC-based mass spectrometry of VHL immunoprecipitates.
WT yeast cells transfected with one of NLS-GFP-VHL, NES-GFP-VHL or Flag-VHL were grown overnight at 30 °C in raffinose-synthetic media supplemented with Light Lys-0 (Cambridge Isotope Laboratories catalogue number ULM-8766-PK), Heavy Lys-8 (Cambridge Isotope Laboratories catalogue number CNLM-291-H-1) or Medium Lys-4 (Cambridge Isotope Laboratories catalogue number DLM-2640-PK), respectively. Cells were diluted to an OD600 between 0.05 and 0.1 in galactose synthetic media supplemented with the appropriate Lysine isotopes and grown for 4-5 h (OD600 0.6-0.8) to induce expression of the galactose-inducible protein. Expression was shut off by switching the cells to glucose synthetic media supplemented with the appropriate Lysine isotope. Pelleted cells were lysed by cryogrinding as described in the ‘Immunoprecipitation of E3 ligases’ section above. 1.5 mg of protein (as quantified by BCA assay) from each of the NLS-GFP-VHL, NES-GFP-VHL and Flag-VHL lysates were mixed prior to immunoprecipitation using GFP-TRAP_MA magnetic beads (ChromoTek) according to the manufacturer’s protocol. After 3 washes with Triton IP Buffer, the beads were washed twice with 50 mM Tris-HCl pH 8 supplemented with 20mM CaCl2. On-bead trypsin digestion and peptide clean-up was performed using the in-StageTip method36. Peptides were analysed on a Q Exactive™ Plus Orbitrap (Thermo Fisher Scientific) connected to a NanoAcquity UPLC system (Waters). An EASY-Spray PepMap® RSLC C18, 3 μm, 75 μm × 15 cm column (Thermo Fisher Scientific) was used to resolve peptides using a binary solvent system (0.1 % formic acid in water as mobile phase A, 0.1 % formic acid in acetonitrile as mobile phase B). The Q Exactive Plus was run on a linear 60-min gradient from 2 % to 30 % B at a flow rate of 300 nL/min. Both precursor and fragment ions were analysed in the FT mode at a mass resolution of 70,000 and 17,500, respectively. After a survey scan, the 10 most intense precursor ions were selected for subsequent fragmentation by HCD.
Raw data from 4 biological replicates were processed using MaxQuant37 (version 1.6.2.3) and searched against the Saccharomyces Genome Database (orf_trans_all.fasta, downloaded in January 2015) with common contaminant entries. The default MaxQuant parameters for a triple SILAC experiment were used, with the exception of ‘Re-quantify’, which was enabled.
The proteinGroups.txt file was filtered to exclude contaminants, reverse hits, hits ‘only identified by site’, and hits for which only 1 peptide was identified. The normalized SILAC ratios were used to generate median fold-change values per protein. Proteins with a log2(Light/Medium) or log2(Heavy/Medium) > 0.5 were counted as ‘enriched’ in NLS- or NES- VHL interactomes, respectively. Enriched proteins from each interactome were subjected to pathway analysis to search for enriched GO Terms, KEGG Pathways and PFAM Protein Domains in either interactome using the STRING database 38 (http://string-db.org. version 10.5).
Data Availability Statement
The data sets generated and/or analysed during this study are available from the corresponding author on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data identifier PXD010660. Uncropped images of all immunoblots shown in this study are in Supplementary Figure 1.
Extended Data
Extended Figure 1 ∣. Puncta formation assay distinguishes between misfolded versus natively-folded proteins.
a, WT cells expressing natively-folded Ubc9WT-GFP or temperature-sensitive Ubc9ts-GFP from a galactose-inducible promoter for 5-6 h at 30 °C were shifted to glucose-containing media for 1 h at 30 °C or 37 °C to shut off expression. Cells were fixed and imaged by fluorescence microscopy. 300 cells were counted per condition, and the percentage of cells with GFP-positive puncta is shown (mean ± SEM from 3 biologically independent experiments). Only cells expressing temperature-sensitive Ubc9ts-GFP showed a statistically significant change in the percentage of puncta-positive cells compared with WT (one-way ANOVA followed by Dunnett’s multiple comparisons test, ****p < 0.0001).
b, Deletion of individual E3 ligases does not increase puncta formation. Experiment performed as in a, but in strains with endogenous deletions of the genes denoted on the x-axis. E3 ligases that are previously implicated in PQC (and shown in Fig. 1d) are grouped to the right hand side. Bars represent mean ± SEM from 3 biologically independent experiments, with the exception of rad5, hel1, etp1, irc20, hel2, apc11, hrt1, tfb3, cdc24, prp19, upf1, upf3, itt1, and rad18 (where bars represent the mean from 2 biologically independent experiments), as well as WT (where bars represent the mean ± SEM from 7 biologically independent experiments). No strains showed statistically significant differences compared with WT by one-way ANOVA followed by Dunnett’s multiple comparisons test.
c, Deleting certain pairs of E3 ligases increases misfolded protein stability. Cyclohexamide-chase followed by immunoblot to assess stability of GFP-VHL, CPY‡-GFP. Ubc9ts-TAP or Ubc9wt-TAP in E3 ligase double deletion strains. For the WT + bortezomib condition, 50 μM bortezomib was added to the glucose-containing media 10 minutes before cyclohexamide treatment. Graphs represent densitometric quantification of bands relative to t = 0 timepoint (mean ± SEM from 3 biologically independent experiments).
d, Multiple misfolded proteins are sequestered in the same subcellular location. Δubr1Δsan1 or Δdoa10Δhrd1 strains co-expressing VHL with temperature-sensitive Ubc9ts (top) or natively-folded Ubc9WT (bottom) from galactose-inducible promoters for 5-6 h at 30 °C were shifted to glucose-containing media for 1 h at 37 °C (for Ubc9ts). Fluorescence microscopy images are representative of at least 100 cells from each of 3 biologically independent experiments.
e, Deletion of certain pairs of E3 ligases increases puncta formation. Experiment performed as in a, but in strains with endogenous deletions of pairs of E3 ligase genes. The right hand panels represents experiments where cells were shifted to 37 °C for the 1 h of galactose shut-off. Bars represent mean ± SEM from 3 biologically independent experiments. Strains for which statistically significant differences were observed by one-way ANOVA followed by Dunnett’s multiple comparisons test compared with WT are indicated with the adjusted p-value, or **** for p < 0.0001.
f, Overexpressing a single E3 ligase does not compensate for loss of others. Ubr1, San1 or Hrd1 were overexpressed alongside GFP-VHL in the indicated strains. The rest of the experiment was performed as in a. Bars represent mean ± SEM from 3 biologically independent experiments.
Extended Figure 2 ∣. San1 forms a complex with Doa10 but not with Hrd1.
a-b, San1-V5His6 co-immunoprecipitates with Doa10-GFP but not with Hrd1-GFP. Yeast cells co-expressing Doa10-GFP (a) or Hrd1-GFP (b) from their endogenous promoters with San1-V5His6 from a galactose-inducible promoter for 16 h were shifted to 37 °C for 1 h, and immediately lysed by cryo-grinding. Native complexes were immunoprecipitated with GFP-Trap®-MA nanobodies before immunoblotting with the indicated antibodies. Immunoblots are representative of 3 biologically independent experiments.
c-d, Flag-Ubr1 does not co-immunoprecipitate with Doa10-GFP or Hrd1-GFP. Experiment was performed as above, but with cells expressing Flag-Ubr1 (from the constitutive ADH promoter) instead of San1-V5His6. Immunoblots are representative of 3 biologically independent experiments.
Extended Figure 3 ∣. K48 and K11 Ub linkages are reduced in Δubr1Δsan1 and Δdoa10Δhrd1 strains, respectively.
a, Schematic illustrating Ub linkage ELISA used to quantify Ub linkages. Flag-VHL from a yeast lysate was immunoprecipitated in an α-Flag-conjugated 96-well plate (4 wells used per sample), and incubated with antibodies against GFP (negative control), Flag, K11-Ub, or K48-Ub. Following incubation with a secondary antibody (α-rabbit-HRP), the strength of each signal was detected by electrochemiluminescence at 450 nm. To quantify the K11 or K48 Ub linkages on Flag-VHL, the α-K11 or α-K48 signal was subtracted of the negative control (α-GFP) and normalized to the total Flag-VHL signal for each sample.
b, Ub linkage ELISA confirms that K48 and K11 Ub linkages are reduced on Flag-VHL in Δubr1Δsan1 and Δdoa10Δhrd1 strains, respectively. WT or E3 double deletion strains expressing Flag-VHL at 30 °C for 5-6 h were lysed after 1 h bortezomib treatment, also at 30 °C. Ub linkage ELISA was then performed as described in c. Bars represent Flag-normalized values from each strain (mean ± SEM from 3 biologically independent experiments) expressed as a proportion of the Flag-normalized WT values. Strains with statistically significant differences compared with WT by one-way ANOVA followed by Dunnett’s multiple comparisons test are indicated (****p < 0.001).
c, GFP-VHL denaturing immunoprecipitation (8M Urea +1% SDS) followed by immunoblot for K48-Ub or K11-Ub in WT or E3 double deletion strains. Immunoblots representative of 3 independent experiments are shown.
d, Relative amounts of K11 and K48 Ub linkages present on GFP-VHL in Δubr1Δsan1 or Δdoa10Δhrd1 strains compared with WT. WT or E3 double deletion strains expressing GFP-VHL at 30 °C for 5-6 h were lysed in denaturing conditions (8M Urea + 1% SDS) after 1 h bortezomib treatment, also at 30 °C. Ub linkage ELISA was then performed using GFP multiTrap® plates. Bars represent GFP-normalized values from each strain (mean ± SEM from 3 biologically independent experiments) expressed as a proportion of the GFP-normalized WT values. Strains for which statistically significant differences were observed by one-way ANOVA followed by Dunnett’s multiple comparisons test compared with WT are indicated with the adjusted p-value, or **** for p < 0.0001.
Extended Figure 4 ∣. K11-Ub linkages are not necessary for proteasomal degradation of all cytoplasmic substrates.
WT or Ub-K11R cells expressing stable Ub-M-GFP (a), the N-End Rule substrate Ub-R-GFP (b), the ubiquitin fusion degradation (UFD) substrate UbG76V-GFP (c) or GFP fused to the artificial degron CL1 (d) from galactose-inducible promoters for 5-6 h at 30 °C were shifted to glucose-containing media for 1 h at 30 °C or 37 °C to shut off expression. Cells were fixed and imaged by fluorescence microscopy. 300 cells were counted per condition, and the percentage of cells with GFP-positive puncta is shown (mean ± SEM from 3 biologically independent experiments). There was a statistically significant increase in puncta when GFP-CL1 (which contains a short amphipathic CL1 helix that could mimic a partially unfolded protein) was expressed in K11R-Ub cells compared with WT cells by one-way ANOVA followed by Dunnett’s multiple comparisons test (p = 0.0127). The differences for all other substrates were not significant (ns, p > 0.05).
Extended Figure 5 ∣. Misfolded VHL is modified with branched K11/K48 ubiquitin chains.
a-b, Both K11 and K48 Ub linkages are present on the same VHL molecule. a, Experiment was designed to determine if both K48 and K11 Ub linkages are present in the same VHL population. Sequential immunoprecipitation, first with a FLAG antibody, followed by a K11 or K48 ubiquitin antibody. The resultant negative control (no Flag: mock Flag + K11 or K48 immunoprecipitation with lysate from cells expressing GFP-VHL instead of Flag-VHL), Eluate and Flow-Through, in addition to samples with just the first Flag immunoprecipitation (Input), were subjected to SDS-PAGE and immunoblotted for the presence of the other Ub linkage (b). Immunoblots representative of 3 biologically independent experiments are shown.
c, Bispecific K11/K48 Ub antibody designed to bind ubiquitin chains with K11 and K48 linkages branching off the same ubiquitin moiety. d, Misfolded VHL co-localises with K11/K48 Ub branched chains. WT cells expressing GFP-VHL from a galactose-inducible promoter for 5-6 h at 30 °C were shifted to glucose-containing media with 50 μM bortezomib for 1 h to shut off expression. Cells were fixed, spheroplasted and detergent-permeabilized before immunostaining with an antibody designed to recognize ubiquitin that had K11 and K48 linkages emanating from the same moiety (K11/K48). Confocal fluorescence microscopy images are representative of at least 100 cells from each of 3 biologically independent experiments. Scale bars = 2 μm. e, VHL is modified with branched K11/K48 Ub chains. GFP-VHL denaturing immunoprecipitation followed by immunoblot for K11/K48 Ub or GFP (VHL) in WT or E3 double deletion strains. Immunoblots representative of 3 biologically independent experiments are shown.
Extended Figure 6 ∣. Nuclear and cytoplasmic proteins require different PQC pathways for clearance.
a, NLS-GFP-VHL and NES-GFP-VHL form a single punctum in the nucleus or cytoplasm, respectively, upon proteasome inhibition. WT cells expressing NLS- or NES- GFP-VHL from a galactose-inducible promoter for 5-6 h at 30 °C were shifted to glucose-containing media with 50 μM bortezomib for 1 h at 30 °C to shut off GFP-VHL expression. Fixed and spheroplasted cells were immunostained for the nuclear pore complex protein Nsp1 prior to imaging by fluorescence microscopy. Representative cells from 3 biologically independent repeats are shown.
b-d, Misfolded Luciferasets confined to the nucleus can be cleared by San1-mediated K48- ubiquitination.
b, The increase in percentage of NLS- and NES- Lucts-GFP puncta-containing cells across the E3 ligase single and double deletion strains is similar to the pattern observed with NLS- and NES- GFP-VHL in Fig. 4. Percentage of cells (mean ± SEM from 3 biologically independent experiments, each with n = 300) containing NLS- or NES- Lucts-GFP puncta in WT, single or double deletion strains after 5-6 h expression of the protein at 30 °C followed by 1 h shut-off at 37 °C. Strains for which statistically significant differences were observed by one-way ANOVA followed by Dunnett’s multiple comparisons test compared with WT are indicated with the adjusted p-value, or **** for p < 0.0001).
c, Misfolded nuclear Luciferasets has severely reduced K11-Ub ubiquitin linkages (****p < 0.0001 by one-way ANOVA followed by Dunnett’s multiple comparisons test). Ubiquitin linkage ELISA was performed on lysates of WT yeast expressing NLS-, NES- or unaltered GFP-Luciferasets at 37 °C as described in Fig. 2c, but in GFP-multiTrap® 96-well plates instead of α-FLAG-conjugated 96-well plates. α-FLAG was used instead of α-GFP as the ELISA negative control. Bars represent mean ± SEM from 3 biologically independent experiments.
d, Misfolded Luciferasets confined to the nucleus does not require K11-Ub linkages for clearance. Experiment was performed as in b, but with yeast strains expressing WT or mutant K11R-Ub as their sole source of ubiquitin. 300 cells were counted per condition, and the percentage of cells with GFP-positive puncta are shown in (mean ± SEM from 3 biologically independent experiments). Only NES-GFP-Luciferasets had a statistically significant change in puncta-positive cells in the K11R strain when compared with WT (one-way ANOVA followed by Dunnett’s multiple comparisons test, ****p < 0.0001; ns = p > 0.05).
e, VHL confined to the nucleus (NLS) or cytoplasm (NES) requires different chaperones for clearance. Experiment was performed as in b, but with the indicated chaperone deletion strains. Bars represent mean ± SEM from 3 biologically independent experiments. Strains for which statistically significant differences were observed compared with WT by one-way ANOVA followed by Dunnett’s multiple comparisons test are indicated with the adjusted p-value, or **** for p < 0.0001.
Extended Figure 7 ∣. Mass spectrometry of VHL interactome identifies distinct PQC circuitries for nuclear and cytoplasmic VHL.
a, Triple SILAC-base mass spectrometry of VHL immunoprecipitates. WT yeast cells transfected with one of NLS-GFP-VHL, NES-GFP-VHL or Flag-VHL were grown overnight at 30 °C in raffinose-synthetic media supplemented with Light Lys-0, Heavy Lys-8, or Medium Lys-4, respectively. Growth of VHL was induced in galactose for 4-5 h before shut off in glucose for 90 min. 1.5 mg of protein from each of the three lysed samples were mixed prior to immunoprecipitation using GFP-TRAP_MA magnetic beads on-bead restriction digestion and peptide clean-up. Peptides were identified using LC-MS analysis prior to analysis using MaxQuant.
b, Strong correlation between the four biological repeats. Raw intensities for Light (NLS-GFP-VHL, top), Heavy (NES-GFP-VHL, middle) and Medium (Flag-VHL Control, bottom) were log10 transformed and plotted as scatter plot matrices. The Pearson correlation coefficient for each pairwise comparison is indicated, and the density distribution of intensities within for repeat is shown in the diagonal axis of the matrices.
c, Enriched PQC proteins in NLS-GFP-VHL and NES-GFP-VHL interactomes. Normalized median Light/Medium (NLS-GFP-VHL) and Heavy/Medium (NES-GFP-VHL) SILAC ratios were log2 transformed. Proteins with log2(SILAC ratio) > 0.5 were considered as enriched, yielding 49 and 56 proteins for the NLS and NES interactomes, respectively. Enriched proteins known to play a role in PQC are shown. Both nuclear and cytoplasmic VHL share enrichments in proteasomal subunits, the Hsp70 chaperones Ssa1, Ssa2, Ssa4 and Ssb2, and the thioredoxins Trx1, Trx2 and Tsa1 (previously implicated in misfolded protein management). All enriched proteins are shown in Extended Table 1.
d, Enriched PQC pathways in NLS-GFP-VHL and NES-GFP-VHL interactomes. The enriched proteins from each interactome (median values from four biologically independent experiments) were subjected to pathway analysis to search for enriched GO Terms, KEGG Pathways and PFAM Protein Domains in either interactome using the STRING database. Selected enriched PQC pathways are shown (p < 0.05 using Fisher’s exact test followed by Benjamini-Hochberg multiple testing correction).
Extended Table 1 ∣. Protein and pathways enriched in nuclear and cytoplasmic interactomes.
Enriched proteins in NLS-GFP-VHL (a) and NES-GFP-VHL interactomes (b). Normalized median Light/Medium (NLS-GFP-VHL) and Heavy/Medium (NES-GFP-VHL) SILAC ratios from 4 biologically independent experiments were log2 transformed. Proteins with log2(SILAC ratio) > 0.5 were considered as enriched, yielding 49 and 56 proteins for the NLS and NES interactomes, respectively.
| Fasta headers | log2(Median NLS/Control) | Fasta headers | log2(Median NES/Control) | |
|---|---|---|---|---|
| URA3 | 2.077 | YNL134C | 3.229 | |
| CCT5 | 1.550 | DBP5 | 2.501 | |
| SSA2 | 1.334 | RKI1 | 2.494 | |
| SSA4 | 1.239 | URA3 | 2.319 | |
| MKT1 | 1.208 | SSA4 | 2.166 | |
| TSA1 | 1.203 | SHM1 | 1.835 | |
| TRX2 | 1.035 | SEC14 | 1.727 | |
| CCT8 | 0.952 | TSA1 | 1.234 | |
| ARO1 | 0.923 | SSA2 | 1.234 | |
| MDN1 | 0.902 | IMD3:IMD2 | 1.213 | |
| RPT6 | 0.879 | SSA1 | 1.199 | |
| CCT7 | 0.873 | TRX2 | 1.054 | |
| TUB1 | 0.867 | GDH1 | 0.984 | |
| CCT3 | 0.841 | BGL2 | 0.964 | |
| GCD6 | 0.837 | TPI1 | 0.880 | |
| TRX1 | 0.808 | RPN3 | 0.876 | |
| BGL2 | 0.790 | CCT5 | 0.820 | |
| VMA2 | 0.775 | CIT1 | 0.803 | |
| AAH1 | 0.773 | TIF4631 | 0.792 | |
| NEW1 | 0.759 | CPA2 | 0.776 | |
| RPA135 | 0.745 | RRP5 | 0.771 | |
| SSA1 | 0.745 | PRE5 | 0.731 | |
| YNL134C | 0.728 | IPP1 | 0.731 | |
| URA7 | 0.710 | MET17 | 0.730 | |
| PRE10 | 0.699 | DYS1 | 0.702 | |
| EFT1;EFT2 | 0.688 | SIS1 | 0.695 | |
| URA2 | 0.684 | TRX1 | 0.675 | |
| RPT1 | 0.670 | MMF1 | 0.672 | |
| NUG1 | 0.657 | ENO2 | 0.672 | |
| HTS1 | 0.649 | PGI1 | 0.670 | |
| SSB2 | 0.642 | RPT6 | 0.665 | |
| SAM1 | 0.630 | PRE9 | 0.658 | |
| RPB2 | 0.615 | HOM6 | 0.645 | |
| KRE33 | 0.605 | HXK2 | 0.628 | |
| RPN2 | 0.578 | ERG10 | 0.625 | |
| FAS2 | 0.561 | BMH1 | 0.625 | |
| ADE6 | 0.552 | UBA1 | 0.616 | |
| MIS1 | 0.550 | PGK1 | 0.612 | |
| YEF3 | 0.545 | HTS1 | 0.608 | |
| CCT6 | 0.540 | YPT52 | 0.607 | |
| CPA2 | 0.524 | SSB2 | 0.607 | |
| CCT4 | 0.520 | URA2 | 0.598 | |
| ADE3 | 0.516 | RPN6 | 0.597 | |
| RRP5 | 0.514 | FPR4 | 0.592 | |
| LEU1 | 0.510 | TKL1 | 0.578 | |
| CRM1 | 0.508 | PRE6 | 0.571 | |
| GCN1 | 0.507 | RNR4 | 0.561 | |
| HIS1 | 0.507 | CDC48 | 0.558 | |
| HIS4 | 0.504 | SDH1 | 0.535 | |
| HSP82 | 0.526 | |||
| TUB1 | 0.520 | |||
| YPL225W | 0.515 | |||
| TAL1 | 0.508 | |||
| LEU1 | 0.506 | |||
| GPM1 | 0.503 |
Extended Table 2 ∣. Human homologues of the ubiquitination machinery characterized in this study are associated with a range of diseases.
Literature-based evidence for disease pathology being directly related to protein quality control is indicated in the last column, where applicable.
| Yeast gene | Human orthologue | Associated Diseases | PQC in disease pathology? |
|---|---|---|---|
| Doa10 | MARCH6/TEB4 | Cri-du-chat syndrome (same chromosomal reqion)39; Lipidoqenesis imbalance 40-42 | - |
| Hrd1 | HRD1 | Alzheimer’s Disease Parkinson’s Disease |
43 (Review) |
| Gp78/AMFR | Cancer Cystic fibrosis ALS Parkinson’s Disease Huntington’s Disease prion disorders |
44 (Review) | |
| Ubr1 | UBR1 | Johanson-Blizzard Syndrome 45 | 45 |
| UBR4 | Episodic Ataxia Type 8 46 | Unclear (yes for related Types 1 & 2) 47,48 | |
| UBR5 | Adult Myoclonal Epilepsy 46 | Unclear (yes for related Lafora Myoclonal Epilepsy) 49 | |
| UBR7 | Autism Spectrum Disorder 46 | 50-56 | |
| San1 | No clear human orthologue | - | - |
| Dsk2 | UBQLN1 | Alzheimer’s Disease Huntington’s Disease |
57,58(Reviews), 59,60 |
| UBQLN2 | ALS Frontotemporal Dementia Huntington’s Disease |
61-65 |
Supplementary Material
Acknowledgements
We thank K. Li, M. Burlingame, and A.L. Burlingame for help with mass spectrometry; R. Andino and F. U. Hartl for critical reading of the manuscript; E.M. Sontag and D.R. Gestaut for sharing the NLS and NES-tagged plasmids, and all members of the Frydman lab for advice. R.S.S. was supported by a Human Frontier Science Program long-term fellowship (LT000695/2015-L). This work was supported by NIH grants to J.F.
References
- 1.Chiti F & Dobson CM Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annual review of biochemistry, doi: 10.1146/annurev-biochem-061516-045115 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Balch WE, Morimoto RI, Dillin A & Kelly JW Adapting proteostasis for disease intervention. Science 319, 916–919, doi: 10.1126/science.1141448319/5865/916 [pii] (2008). [DOI] [PubMed] [Google Scholar]
- 3.Mitchell Sontag E, Samant RS & Frydman J Mechanisms and Functions of Spatial Protein Quality Control. Annual review of biochemistry, doi: 10.1146/annurev-biochem-060815-014616 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Balchin D, Hayer-Hartl M & Hartl FU In vivo aspects of protein folding and quality control. Science 353, aac4354, doi: 10.1126/science.aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kwon YT & Ciechanover A The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends in biochemical sciences 42, 873–886, doi: 10.1016/j.tibs.2017.09.002 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Kaganovich D, Kopito R & Frydman J Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095, doi: 10.1038/nature07195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escusa-Toret S, Vonk WI & Frydman J Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nature cell biology, doi: 10.1038/ncb2838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malinovska L, Kroschwald S, Munder MC, Richter D & Alberti S Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Molecular biology of the cell 23, 3041–3056, doi: 10.1091/mbc.E12-03-0194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park SH et al. PolyQ Proteins Interfere with Nuclear Degradation of Cytosolic Proteins by Sequestering the Sis1p Chaperone. Cell 154, 134–145, doi: 10.1016/j.cell.2013.06.003S0092-8674(13)00704-6 [pii] (2013). [DOI] [PubMed] [Google Scholar]
- 10.McClellan AJ, Scott MD & Frydman J Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748, doi: 10.1016/j.cell.2005.03.024 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Heck JW, Cheung SK & Hampton RY Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proceedings of the National Academy of Sciences of the United States of America 107, 1106–1111, doi: 10.1073/pnas.09105911070910591107 [pii] (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad R, Kawaguchi S & Ng DT A nucleus-based quality control mechanism for cytosolic proteins. Molecular biology of the cell 21, 2117–2127, doi: 10.1091/mbc.E10-02-0111E10-02-0111 [pii] (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng M & Hochstrasser M Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 443, 827–831, doi: 10.1038/nature05170 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Swanson R, Locher M & Hochstrasser M A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes & development 15, 2660–2674, doi: 10.1101/gad.933301 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin L, Williamson A, Banerjee S, Philipp I & Rape M Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665, doi: 10.1016/j.cell.2008.04.012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu P et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145, doi: 10.1016/j.cell.2009.01.041 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yau R & Rape M The increasing complexity of the ubiquitin code. Nature cell biology 18, 579–586, doi: 10.1038/ncb3358 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Yau RG et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell, doi: 10.1016/j.cell.2017.09.040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spence J, Sadis S, Haas AL & Finley D A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Molecular and cellular biology 15, 1265–1273 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad R, Xu C & Ng DTW Hsp40/70/110 chaperones adapt nuclear protein quality control to serve cytosolic clients. The Journal of cell biology, doi: 10.1083/jcb.201706091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers DW, Wolfe KJ, Ren HY & Cyr DM The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PloS one 8, e52099, doi: 10.1371/journal.pone.0052099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiber A, Breuer W, Brandeis M & Ravid T Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Molecular biology of the cell 24, 2076–2087, doi: 10.1091/mbc.E13-01-0010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerriero CJ, Weiberth KF & Brodsky JL Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. The Journal of biological chemistry 288, 18506–18520, doi: 10.1074/jbc.M113.475905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amm I & Wolf DH Molecular mass as a determinant for nuclear San1-dependent targeting of misfolded cytosolic proteins to proteasomal degradation. FEES letters 590, 1765–1775, doi: 10.1002/1873-3468.12213 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Biggins S, Ivanovska I & Rose MD Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. The Journal of cell biology 133, 1331–1346 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchiya H et al. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol Cell 66, 488–502 e487, doi: 10.1016/j.molcel.2017.04.024 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Fabre B et al. Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics. Mol Cell Proteomics 12, 687–699, doi: 10.1074/mcp.M112.023317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.RuSse1l SJ, Steger KA & Johnston SA Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. The Journal of biological chemistry 274, 21943–21952 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Miller SB et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. The EMBO journal 34, 778–797, doi: 10.15252/embj.201489524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X et al. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure 24, 1257–1270, doi: 10.1016/j.str.2016.05.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben Yehuda A et al. Ubiquitin Accumulation on Disease Associated Protein Aggregates Is Correlated with Nuclear Ubiquitin Depletion, Histone De-Ubiquitination and Impaired DNA Damage Response. PloS one 12, e0169054, doi: 10.1371/journal.pone.0169054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Y et al. Nuclear export of misfolded SOD1 mediated by a normally buried NES-like sequence reduces proteotoxicity in the nucleus. Elife 6, doi: 10.7554/eLife.23759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winzeler EA et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Dantuma NP, Lindsten K, Glas R, Jellne M & Masucci MG Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18, 538–543, doi: 10.1038/75406 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Alberti S, Gitler AD & Lindquist S A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919, doi: 10.1002/yea.1502 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulak NA, Pichler G, Paron I, Nagaraj N & Mann M Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nature methods 11, 319–324, doi: 10.1038/nmeth.2834 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372, doi: 10.1038/nbt.1511 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368, doi: 10.1093/nar/gkw937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassink G et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J 388, 647–655, doi: 10.1042/BJ20041241 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loregger A et al. A MARCH6 and IDOL E3 Ubiquitin Ligase Circuit Uncouples Cholesterol Synthesis from Lipoprotein Uptake in Hepatocytes. Molecular and cellular biology 36, 285–294, doi: 10.1128/MCB.00890-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson I, Luu W, Kristiana I & Brown AJ Squalene mono-oxygenase, a key enzyme in cholesterol synthesis, is stabilized by unsaturated fatty acids. Biochem J 461, 435–442, doi: 10.1042/BJ20131404 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Zelcer N et al. The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Molecular and cellular biology 34, 1262–1270, doi: 10.1128/MCB.01140-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nomura J et al. Neuroprotection by Endoplasmic Reticulum Stress-Induced HRD1 and Chaperones: Possible Therapeutic Targets for Alzheimer's and Parkinson's Disease. Med Sci (Basel) 4, doi: 10.3390/medsci4030014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joshi V, Upadhyay A, Kumar A & Mishra A Gp78 E3 Ubiquitin Ligase: Essential Functions and Contributions in Proteostasis. Front Cell Neurosci 11, 259, doi: 10.3389/fncel.2017.00259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zenker M et al. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome). Nat Genet, 1345–1350, doi: 10.1038/ng1681 (2005). [DOI] [PubMed] [Google Scholar]
- 46.George AJ, Hoffiz YC, Charles AJ, Zhu Y & Mabb AM A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders. Front Genet 9, 29, doi: 10.3389/fgene.2018.00029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mezghrani A et al. A destructive interaction mechanism accounts for dominant-negative effects of misfolded mutants of voltage-gated calcium channels. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 4501–4511, doi: 10.1523/JNEUROSCI.2844-07.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manganas LN et al. Episodic ataxia type-1 mutations in the Kv1.1 potassium channel display distinct folding and intracellular trafficking properties. The Journal of biological chemistry 276, 49427–49434, doi: 10.1074/jbc.M109325200 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Mittal S, Dubey D, Yamakawa K & Ganesh S Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum Mol Genet 16, 753–762, doi: 10.1093/hmg/ddm006 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Atkin TA, Brandon NJ & Kittler JT Disrupted in Schizophrenia 1 forms pathological aggresomes that disrupt its function in intracellular transport. Hum Mol Genet 21, 2017–2028, doi: 10.1093/hmg/dds018 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Crider A, Ahmed AO & Pillai A Altered Expression of Endoplasmic Reticulum Stress-Related Genes in the Middle Frontal Cortex of Subjects with Autism Spectrum Disorder. Mol Neuropsychiatry 3, 85–91, doi: 10.1159/000477212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Jaco A, Comoletti D, King CC & Taylor P Trafficking of cholinesterases and neuroligins mutant proteins. An association with autism. Chem Biol Interact 175, 349–351, doi: 10.1016/j.cbi.2008.04.023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Jaco A et al. A mutation linked with autism reveals a common mechanism of endoplasmic reticulum retention for the alpha,beta-hydrolase fold protein family. The Journal of biological chemistry 281, 9667–9676, doi: 10.1074/jbc.M510262200 (2006). [DOI] [PubMed] [Google Scholar]
- 54.De Jaco A et al. Neuroligin trafficking deficiencies arising from mutations in the alpha/beta-hydrolase fold protein family. The Journal of biological chemistry 285, 28674–28682, doi: 10.1074/jbc.M110.139519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujita E et al. Autism spectrum disorder is related to endoplasmic reticulum stress induced by mutations in the synaptic cell adhesion molecule, CADM1. Cell Death Dis 1, e47, doi: 10.1038/cddis.2010.23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulbrich L et al. Autism-associated R451C mutation in neuroligin3 leads to activation of the unfolded protein response in a PC12 Tet-On inducible system. Biochem J 473, 423–434, doi: 10.1042/BJ20150274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.El Ayadi A, Stieren ES, Barral JM & Boehning D Ubiquilin-1 and protein quality control in Alzheimer disease. Prion 7, 164–169, doi: 10.4161/pri.23711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marin I The ubiquilin gene family: evolutionary patterns and functional insights. BMC Evol Biol 14, 63, doi: 10.1186/1471-2148-14-63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Safren N et al. Ubiquilin-1 overexpression increases the lifespan and delays accumulation of Huntingtin aggregates in the R6/2 mouse model of Huntington's disease. PloS one 9, e87513, doi: 10.1371/journal.pone.0087513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Natunen T et al. Relationship between ubiquilin-1 and BACE1 in human Alzheimer's disease and APdE9 transgenic mouse brain and cell-based models. Neurobiology of disease 85, 187–205, doi: 10.1016/j.nbd.2015.11.005 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Deng HX et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215, doi: 10.1038/nature10353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osaka M, Ito D & Suzuki N Disturbance of proteasomal and autophagic protein degradation pathways by amyotrophic lateral sclerosis-linked mutations in ubiquilin 2. Biochem Biophys Res Commun 472, 324–331, doi: 10.1016/j.bbrc.2016.02.107 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Teyssou E et al. Novel UBQLN2 mutations linked to amyotrophic lateral sclerosis and atypical hereditary spastic paraplegia phenotype through defective HSP70-mediated proteolysis. Neurobiol Aging 58, 239 e211–239 e220, doi: 10.1016/j.neurobiolaging.2017.06.018 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Zeng L et al. Differential recruitment of UBQLN2 to nuclear inclusions in the polyglutamine diseases HD and SCA3. Neurobiology of disease 82, 281–288, doi: 10.1016/j.nbd.2015.06.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hjerpe R et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 166, 935–949, doi: 10.1016/j.cell.2016.07.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and/or analysed during this study are available from the corresponding author on reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data identifier PXD010660. Uncropped images of all immunoblots shown in this study are in Supplementary Figure 1.