Abstract
Genetic robustness, or the ability of an organism to maintain fitness in the presence of mutations, can be achieved via protein feedback loops. Recent evidence suggests that organisms may also respond to mutations by upregulating related gene(s) independently of protein feedback loops, a phenomenon called transcriptional adaptation. However, the prevalence of transcriptional adaptation and its underlying molecular mechanisms are unknown. Here, by analyzing several models of transcriptional adaptation in zebrafish and mouse, we show a requirement for mRNA degradation. Alleles that fail to transcribe the mutated gene do not display transcriptional adaptation and exhibit more severe phenotypes than alleles displaying mutant mRNA decay. Transcriptome analysis reveals the upregulation of a substantial proportion of the genes that exhibit sequence similarity with the mutated gene’s mRNA, suggesting a sequence dependent mechanism. Besides implications for our understanding of disease-causing mutations, these findings will help design mutant alleles with minimal transcriptional adaptation-derived compensation.
Recent advances in reverse genetic tools have greatly enhanced our ability to analyze gene function in a wide range of organisms. These studies have reinforced prior observations that many engineered mutants do not exhibit an obvious phenotype, reviving interest in the concept of genetic robustness. Several mechanisms have been proposed to explain genetic robustness, including functional redundancy1, rewiring of genetic networks2, and the acquisition of adaptive mutations in the case of rapidly proliferating organisms such as yeast3. In a previous report4, we proposed genetic compensation, and specifically transcriptional adaptation5, as another underlying mechanism, whereby a deleterious mutation can lead to the increased expression of related genes which themselves can assume the function of the mutated gene. Importantly, we provided evidence that this increased expression can be induced by a process lying upstream of the loss of protein function, implying the existence of an unknown trigger. Here, in order to investigate the underlying molecular machinery, we developed and investigated several models of transcriptional adaptation in zebrafish and mouse cell lines.
Transcriptional adaptation in zebrafish and mouse
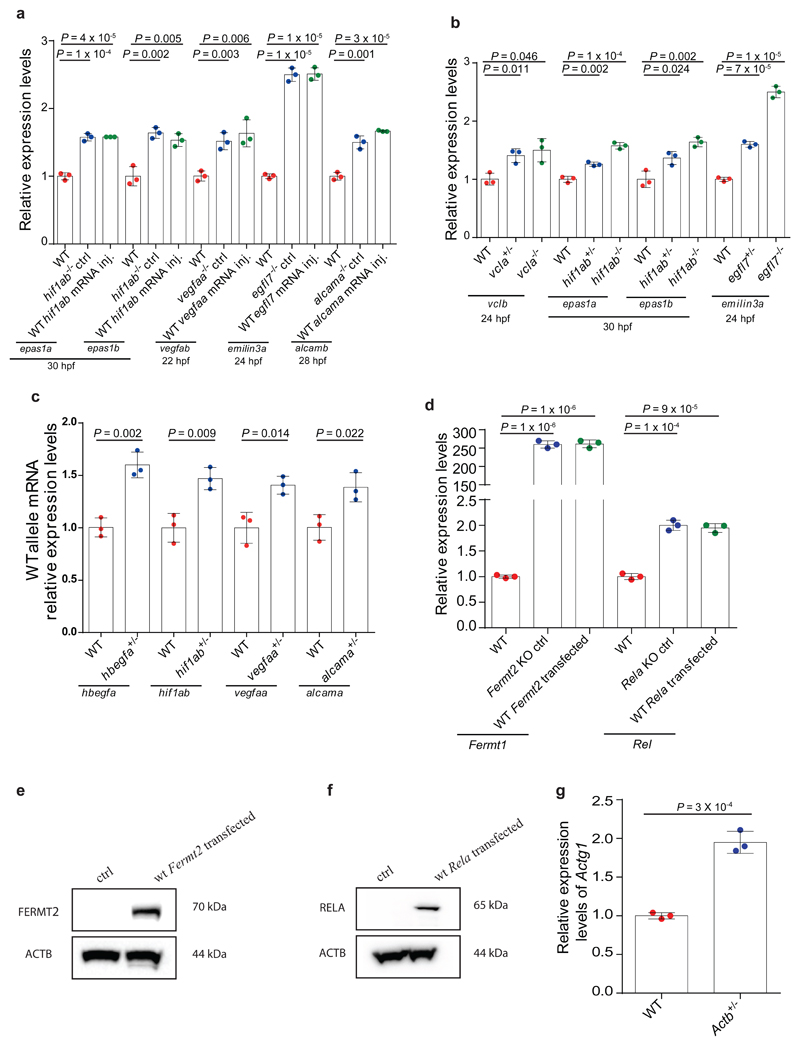
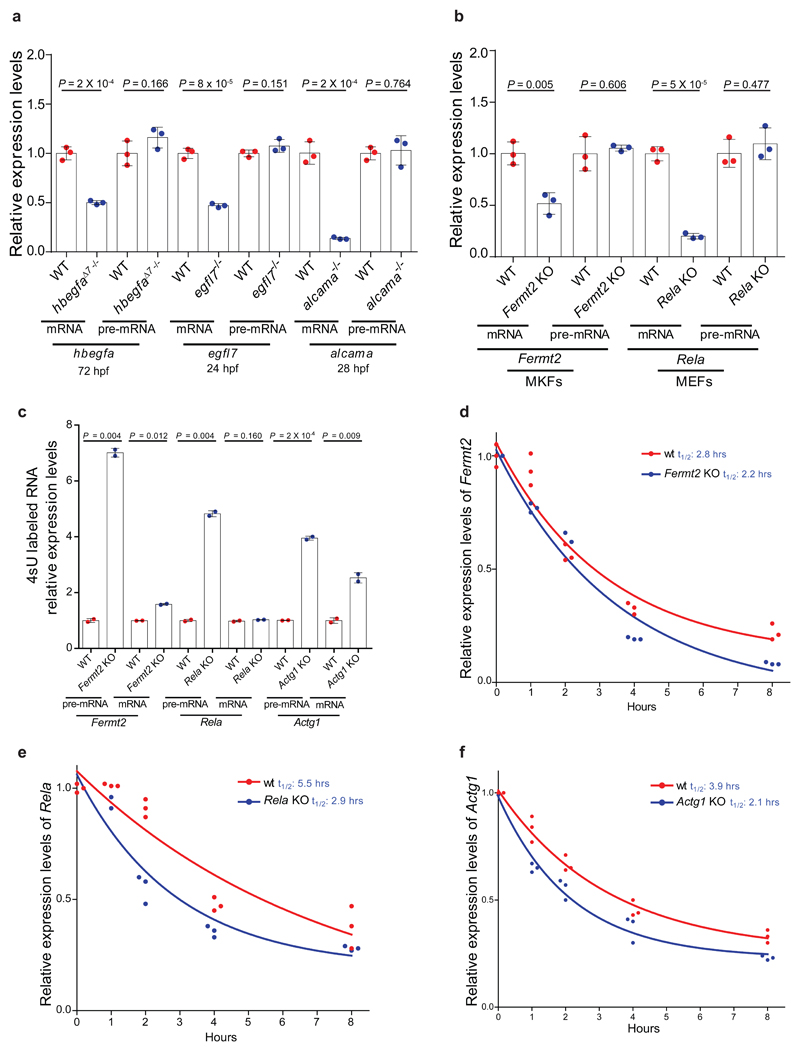
We started by analyzing different zebrafish and mouse mutants that harbor a premature termination codon (PTC) or have their last exon deleted (Extended Data Fig. 1). hbegfa, vcla, hif1ab, vegfaa, egfl7 and alcama zebrafish mutants exhibit increased mRNA levels of a paralogue or family member (hereafter referred to as ‘adapting gene’), namely hbegfb, vclb, epas1a and epas1b, vegfab, emilin3a and alcamb, respectively (Fig. 1a). Injection of wild-type (wt) hif1ab, vegfaa, egfl7 and alcama mRNA into the respective mutants did not have any effect on this transcriptional adaptation response (Extended Data Fig. 2a), suggesting that it is triggered upstream of the loss of protein function. Moreover, we found that vcla, hif1ab and egfl7 heterozygous animals also display transcriptional adaptation, albeit less pronounced than that observed in the homozygous mutants (Extended Data Fig. 2b), indicating that transcriptional adaptation is a dominant phenomenon. Notably, we also observed upregulation of the wt allele in hbegfa, hif1ab, vegfaa and alcama heterozygous embryos (Extended Data Fig. 2c). Similarly, we found that Fermt2 mutant mouse kidney fibroblasts (MKFs), Rela and Actg1 mutant mouse embryonic fibroblasts (MEFs), and Actb mutant mouse embryonic stem cells (mESCs) (hereafter referred to as the knockout (K.O.) alleles) exhibit increased mRNA levels of Fermt1, Rel, Actg2 and Actg1, respectively (Fig. 1b), that transfection of wt Fermt2 and Rela in the respective mutant cells did not dampen the transcriptional adaptation response (Extended Data Fig. 2d-f), and that Actb heterozygous mESCs upregulate Actg1 (Extended Data Fig. 2g). Altogether, these data strongly indicate that the loss of protein function is not the trigger for the transcriptional adaptation response observed in these models.
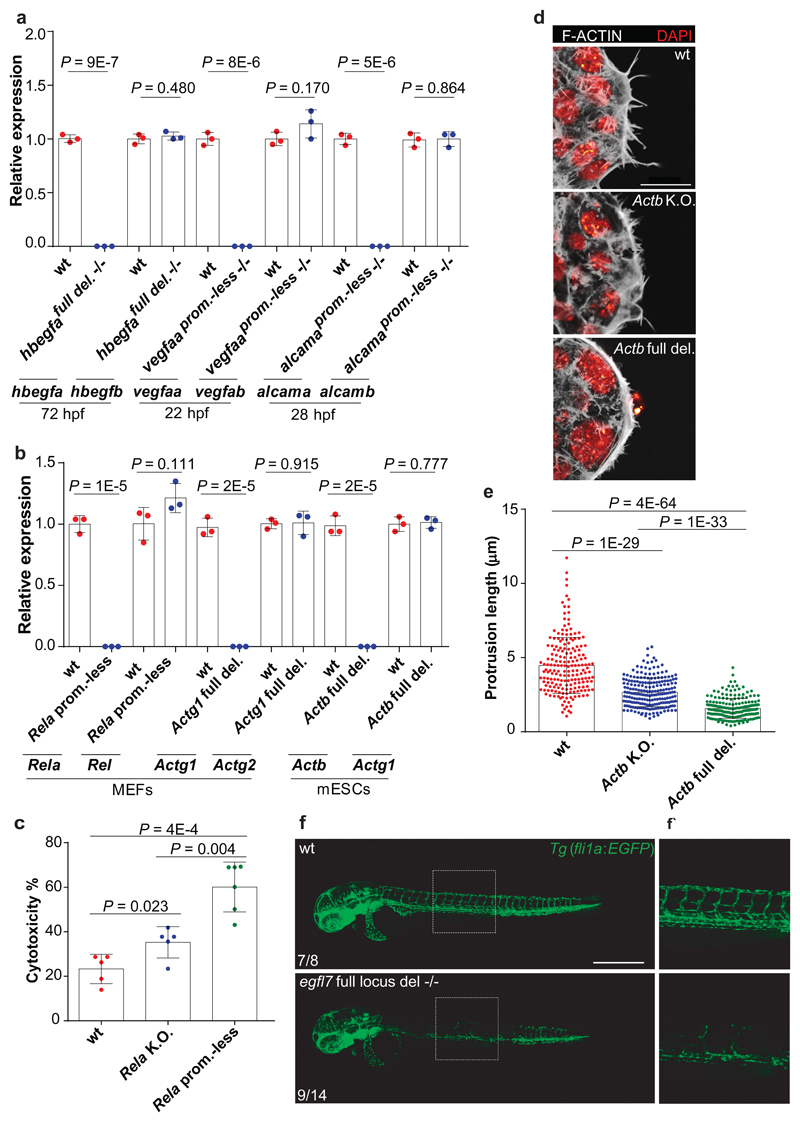
Figure 1. Transcriptional adaptation in zebrafish and mouse correlates with mutant mRNA decay.
a, qPCR analysis of hbegfb, vclb, epas1a and epas1b, vegfab, emilin3a and alcamb mRNA levels in hbegfa, vcla, hif1ab, vegfaa, egfl7 and alcama wt and mutant zebrafish. b, qPCR analysis of Fermt1, Rel, Actg2 and Actg1 mRNA levels in Fermt-2, Rela, Actg1 and Actb wt and K.O. cells. c, d, qPCR analysis of hbegfb and vclb (c) and hbegfa and vcla (d) mRNA levels in the indicated hbegfa and vcla mutant alleles. e, qPCR analysis of hif1ab, vegfaa, egfl7 and alcama mRNA levels in hif1ab, vegfaa, egfl7 and alcama wt and mutant zebrafish. f, qPCR analysis of Fermt-2, Rela, Actg1 and Actb mRNA levels in Kindlin-2, Rela, Actg1 and Actb wt and K.O. cells. a-f, n = 3 biologically independent samples. wt expression set at 1. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
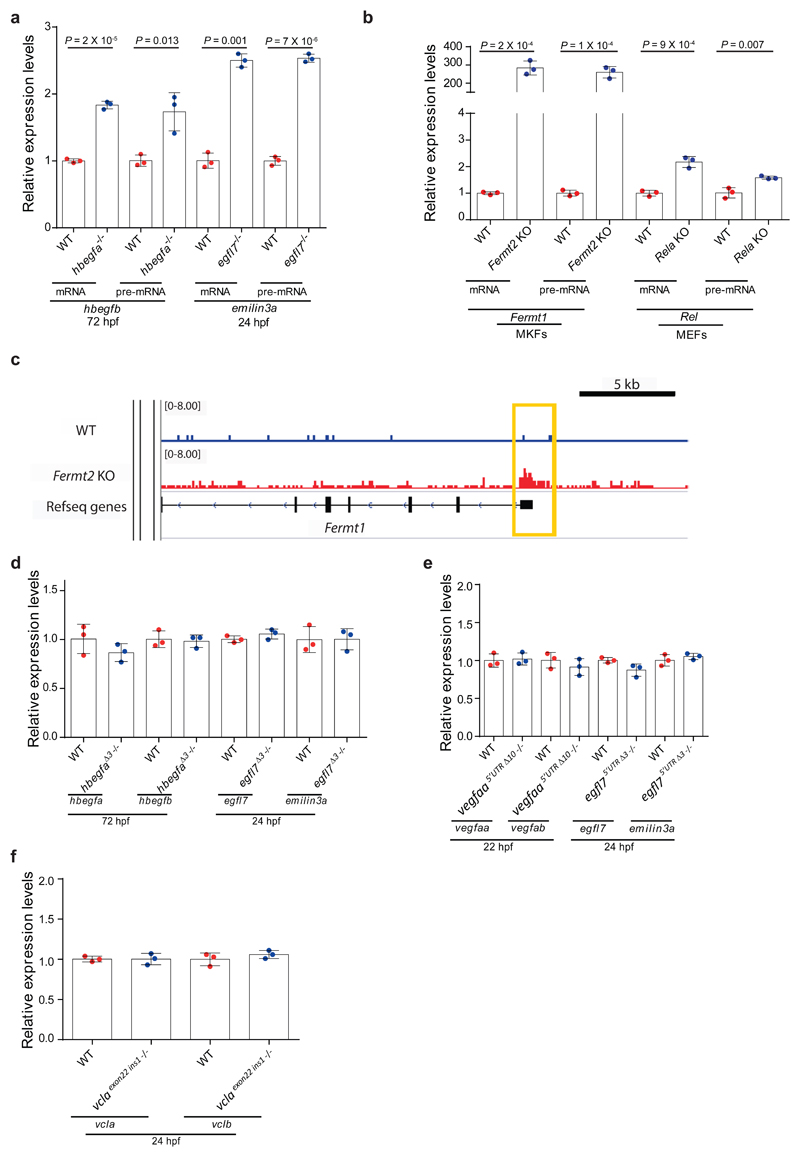
To determine whether the increased mRNA levels were due to increased transcription of the adapting gene or increased mRNA stability, we measured pre-mRNA levels of hbegfb and emilin3a in hbegfa and egfl7 mutants and found that they were also upregulated (Extended Data Fig. 3a). Similar findings were obtained for Fermt1 and Rel pre-mRNA levels in Fermt2 and Rela K.O. cells (Extended Data Fig. 3b). Together, these data indicate an increase in transcription of the adapting genes. In addition, Fermt2 K.O. MKFs display increased chromatin accessibility at the Fermt1 transcription start site (TSS) as observed by ATAC-seq (Extended Data Fig. 3c).
RNA decay triggers transcriptional adaptation
Since the loss of protein function does not appear to be the trigger for the transcriptional adaptation response, we investigated two other possibilities, the DNA lesion and the mutant mRNA. In the context of the zebrafish studies, we identified several mutant alleles which do not display transcriptional adaptation (Extended Data Fig. 3d-f), indicating that a DNA lesion by itself is not sufficient to trigger transcriptional adaptation, or that specific DNA lesions are required. Notably, while analyzing various mutant alleles, we found that unlike hbegfaΔ7 and vclaΔ13, two other PTC bearing alleles, hbegfasa18135 and vclasa14599, do not display transcriptional adaptation (Fig. 1c, Extended Data Fig. 1). To investigate the reason for this difference, we examined mutant mRNA levels and observed limited or no decrease in the hbegfasa18135 and vclasa14599 alleles (Fig. 1d). This correlation between mutant mRNA decrease and transcriptional adaptation was observed in all alleles examined (Fig. 1e, f, Extended Data Fig. 3-f). To determine whether the decreased mRNA levels were due to decreased transcription or reduced stability, we analyzed pre-mRNA levels of hbegfa, egfl7 and alcama in hbegfaΔ7, egfl7 and alcama mutant zebrafish, and found that unlike the mRNA levels, the pre-mRNA levels remained unchanged, or were slightly upregulated, compared to wt (Extended Data Fig. 4a). Similar findings were observed in Fermt2 and Rela K.O. cells (Extended Data Fig. 4b). Moreover, metabolic labeling of newly synthesized transcripts revealed similar or increased levels of Fermt2, Rela and Actg1 mutant transcripts compared to wt (Extended Data Fig. 4c), while transcription inhibition assays revealed shorter half-lives (Extended Data Fig. 4d-f). Together, these data indicate that mutants which exhibit transcriptional adaptation have reduced mutant mRNA levels due to mRNA decay.
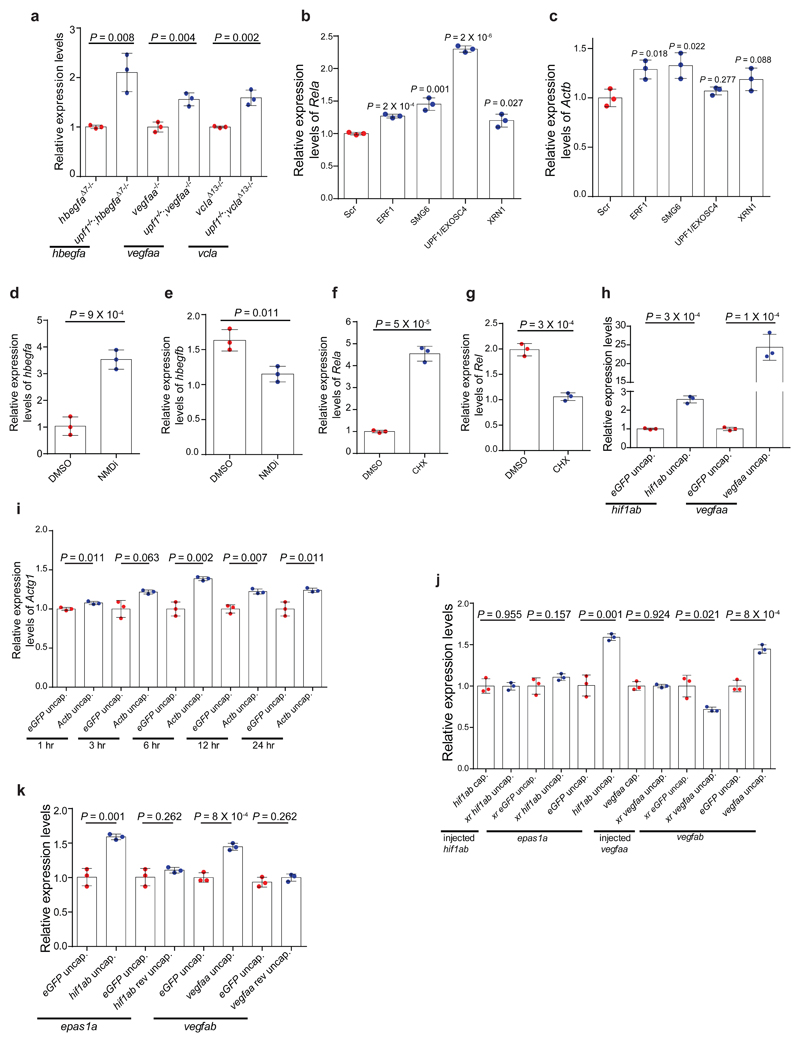
To investigate the role of the mRNA surveillance machinery in transcriptional adaptation, we genetically inactivated Upf1, a key non-sense mediated decay (NMD) factor6, in hbegfaΔ7, vegfaa and vclaΔ13 zebrafish mutants. Inactivating upf1 in these mutants led to decreased levels of mutant mRNA decay (Extended Data Fig. 5a) and loss of transcriptional adaptation (Fig. 2a). Similarly, we observed a decrease, or loss, of transcriptional adaptation in Rela and Actb K.O. cells when knocking down proteins involved in the mRNA surveillance machinery (Figure 2b, c, Extended Data Fig. 5b, c). Pharmacological inhibition of NMD in hbegfaΔ7 mutants (Extended Data Fig. 5d, e) and blocking translation, as an alternative approach to inhibit mRNA decay, in Rela K.O. cells (Extended Data Fig. 5f, g) led to similar observations. We next asked whether inducing mRNA degradation in wt zebrafish or mouse cells by using uncapped RNAs, which are known to be rapidly degraded by 5’ to 3’ exonucleases7, could trigger transcriptional adaptation. Indeed, injection of uncapped hif1ab or vegfaa RNAs into wt embryos induced transcriptional adaptation (Fig. 2d) including an increase in endogenous hif1ab and vegfaa expression (Extended Data Fig. 5h). Similarly, transfection of uncapped Actb RNA into wt mESCs led to Actg1 upregulation (Extended Data Fig. 5i). Moreover, injection of uncapped hif1ab or vegfaa RNAs with an upstream sequence which renders them resistant to 5’ to 3’ exonuclease-mediated decay8 did not induce transcriptional adaptation (Extended Data Fig. 5j). Taken together, these data indicate a major role for mRNA degradation in triggering transcriptional adaptation. Notably, injection of uncapped RNA synthesized from the non-coding strand of hif1ab or vegfaa did not lead to an increase in epas1a or vegfab mRNA levels (Extended Data Fig. 5k), suggesting a possible role for the RNA sequence itself in transcriptional adaptation.
Figure 2. Mutant mRNA decay is required for transcriptional adaptation.
a, qPCR analysis of hbegfb, vegfab, and vclb mRNA levels in upf1;hbegfa, upf1;vegfaa and upf1;vcla double mutant zebrafish. b, qPCR analysis of Rel mRNA levels after siRNA mediated knockdown of indicated proteins in Rela K.O. cells. c, qPCR analysis of Actg1 mRNA levels after siRNA mediated knockdown of indicated proteins in Actb K.O. cells. d, qPCR analysis of injected hif1ab, epas1a, injected vegfaa, and vegfab RNA levels in 6 hpf wt embryos injected with the indicated RNA. b, c, Scr: Scrambled siRNA control. d, wt or control expression set at 1. a-d, n = 3 biologically independent samples. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
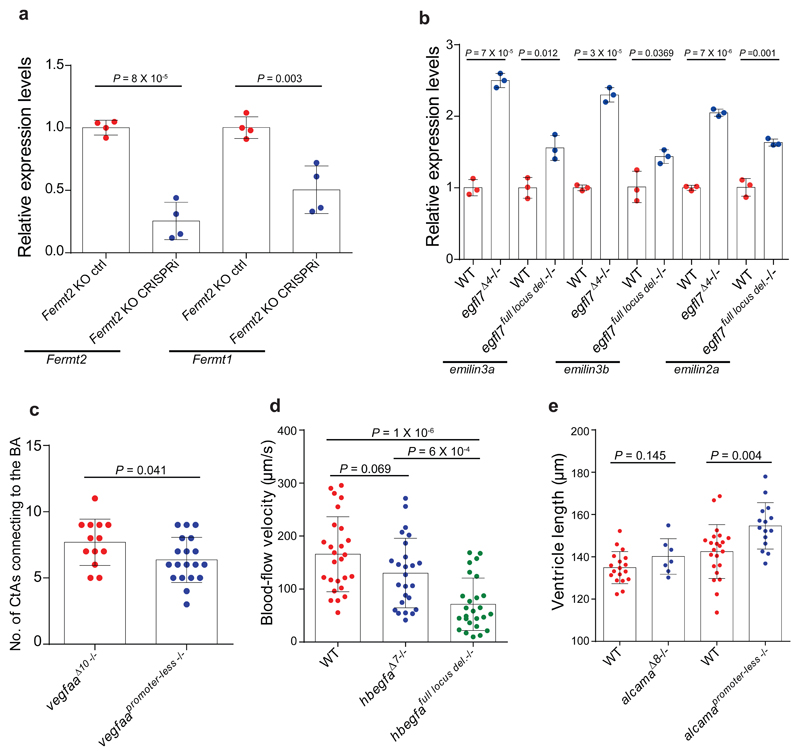
If mutant mRNA degradation is required for transcriptional adaptation, alleles that fail to transcribe the mutated gene should not display this response. To this end, we used the CRISPR/Cas9 technology to generate such alleles (hereafter referred to as RNA-less alleles) by deleting promoter regions or the entire gene locus. Indeed, RNA-less alleles of hbegfa, vegfaa and alcama fail to upregulate hbegfb, vegfab and alcamb, respectively (Fig. 3a). Similarly, in mouse cells, RNA-less alleles of Rela, Actg1 and Actb fail to upregulate the adapting gene (Fig. 3b). We also attempted to generate a promoter-less allele for Fermt2 in MKFs; however, the obtained clones exhibited proliferation defects that prevented their expansion. As an alternative, we used CRISPR interference and found that reducing transcription of the mutant Fermt2 gene in Fermt2 K.O. cells led to a decrease in Fermt1 mRNA levels (Extended Data Fig. 6a). Notably, we observed that the promoter-less Rela MEFs were more sensitive to TNFα-induced apoptosis9 than the Rela K.O. MEFs (Fig. 3c). Similarly, mESCs with an Actb full locus deletion displayed less protrusive activity and more severe growth defects than Actb K.O. cells (Fig. 3d, e). We also generated an egfl7 RNA-less allele and observed that, compared to the phenotypically wt egfl7Δ4 allele4, the mutants displayed pronounced vascular defects (Fig. 3f) as well as milder upregulation of the emilin genes (Extended Data Fig. 6b). Similarly, vegfaa promoter-less mutants displayed a stronger central artery (CtA) sprouting phenotype than vegfaaΔ10 mutants (Extended data Fig. 6c); hbegfa RNA-less mutants displayed slow blood circulation, a phenotype not observed in hbegfaΔ7 mutants (Extended data Fig. 6d); and alcama promoter-less mutants but not alcamaΔ8 mutants exhibited an elongated cardiac ventricle (Extended Data Fig. 6e). Therefore, use of RNA-less alleles can uncover phenotypes not observed in alleles exhibiting mutant mRNA degradation.
Figure 3. Alleles that fail to transcribe the mutated gene do not display transcriptional adaptation.
a, qPCR analysis of hbegfa, hbegfb, vegfaa, vegfab, alcama and alcamb mRNA levels in zebrafish lacking the full hbegfa locus or the vegfaa or alcama promoter compared to wt siblings. b, qPCR analysis of Rela, Rel, Actb, Actg1, Actg1 and Actg2 mRNA levels in MEFs and mESCs lacking the Rela promoter or the full Actg1 or Actb locus compared to wt cells. c, Cytotoxicity assay following treatment with TNFα of wt, Rela K.O. and Rela promoter-less MEFs. Percentages normalized relative to DMSO-treated cells. d, Confocal micrographs of wt, Actb K.O. and Actb full locus deletion mESCs. Actin filaments are depicted in white, nuclei in red. e, Actin filament protrusion length in wt, Actb K.O. and Actb full locus deletion mESCs. f, Confocal micrographs of 48 hpf Tg(fli1a:eGFP) wt and egfl7 full locus deletion mutant siblings; lateral views, anterior to the left. Higher magnifications of dashed boxes shown in f’. Scale bars: e, 20 µm; f, 500 µm. a, b, wt expression set at 1. a-c, e, n = 3 (a, b); 5 (c); 189, 219 and 205 (e) independent experiments. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values. d, f, These experiments were repeated twice independently with similar results.
Sequence similarity and transcriptional adaptation
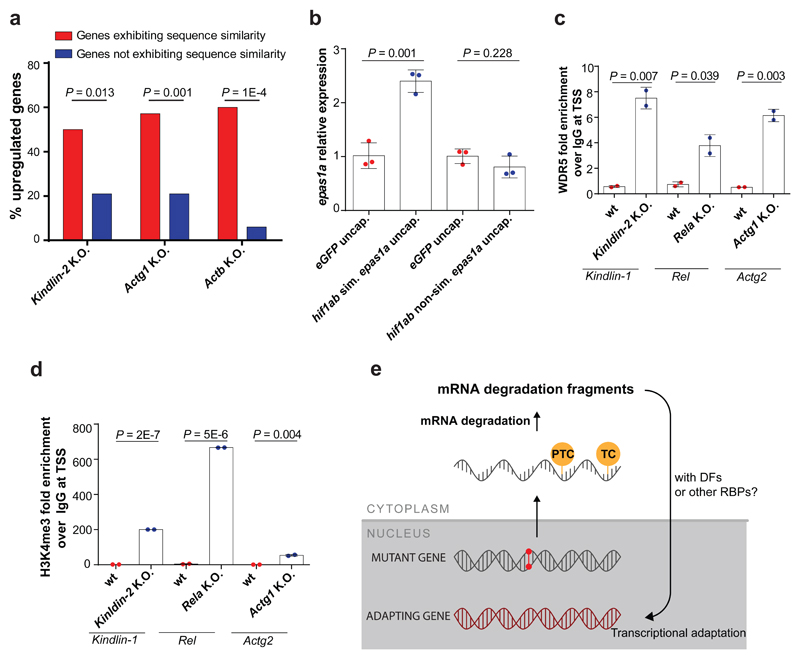
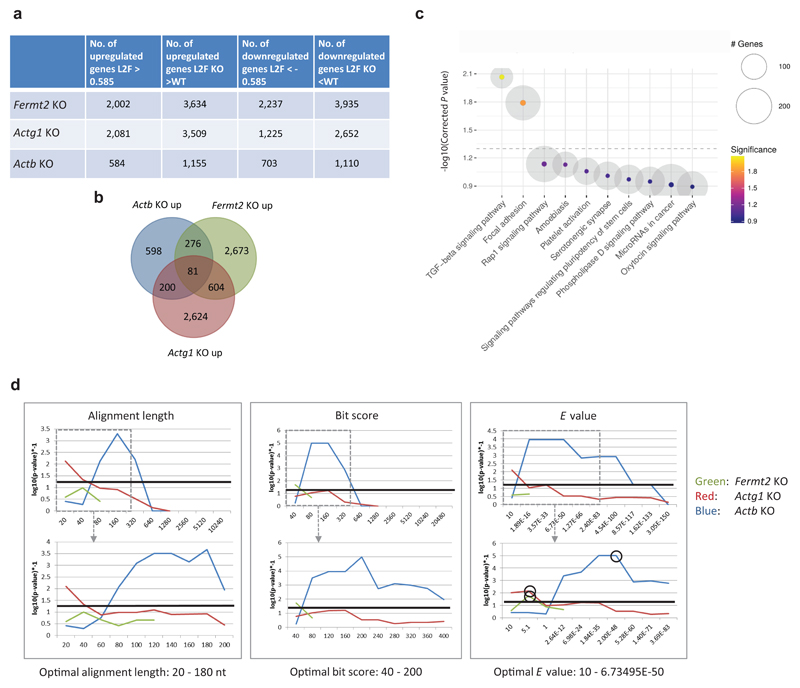
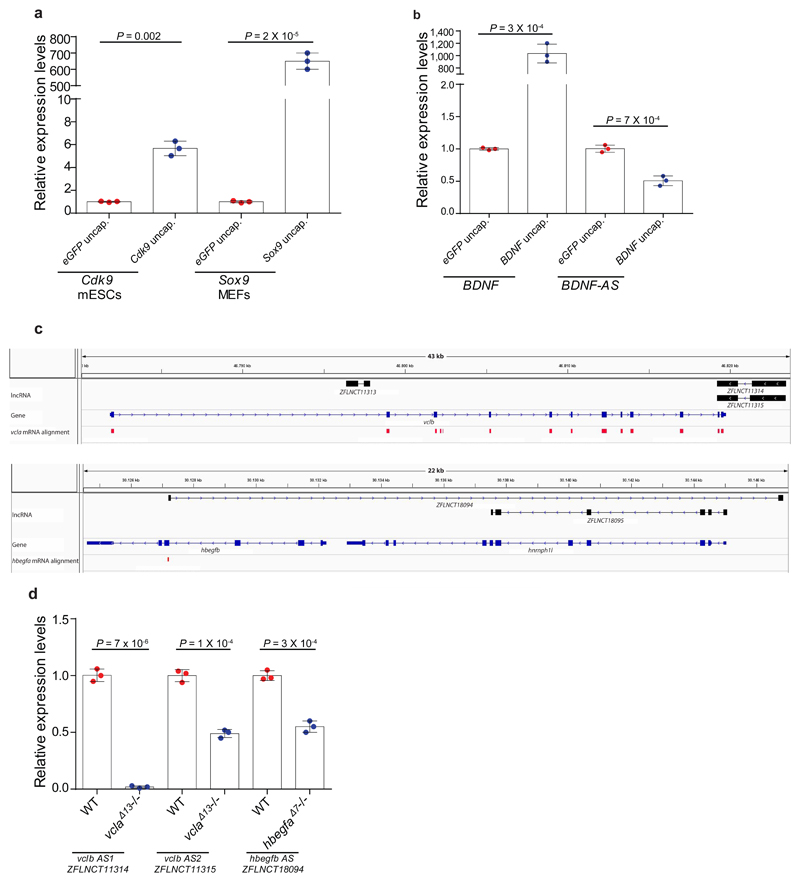
Next, we performed transcriptome analysis of Fermt2, Actg1 and Actb K.O. cells and observed the upregulation of hundreds of genes in K.O. cells compared to wt (Extended Data Fig. 7a, b). Notably, only 81 genes were upregulated in all three models, with no signs of a stress-induced response (Extended Data Fig. 7a-c, Supplementary Table 1). A first-pass analysis of the upregulated genes in each model showed that a disproportionate number of them exhibited sequence similarity to the mutated gene. We thus explored the correlation between upregulation and sequence similarity based on several similarity thresholds (see Methods). Using the optimal E-values to identify ‘similar’ genes, we observed that at least 50% of them were significantly upregulated in the different K.O. cells, compared to a maximum of 21% of randomly selected non-similar genes (Fig. 4a, Extended Data Fig. 8a-c). Notably, 7 of the 12 upregulated ‘similar’ genes in Actg1 K.O. cells were not upregulated in Actg1 RNA-less cells, and 4 of the 6 upregulated ‘similar’ genes in Actb K.O. cells were not upregulated in Actb RNA-less cells (Extended Data Fig. 8a-c). Interestingly, we also observed that 4 of the 9 ‘similar’ genes that were not upregulated in Actg1 K.O. cells at the mRNA level were upregulated at the pre-mRNA level (Extended data Fig. 8d). Additional studies showed that injection of uncapped mouse Actb RNA into zebrafish embryos led to an increase in zebrafish actb1 mRNA levels (Extended Data Fig. 8e, Supplementary Data), in line with the sequence similarity analyses mentioned above.
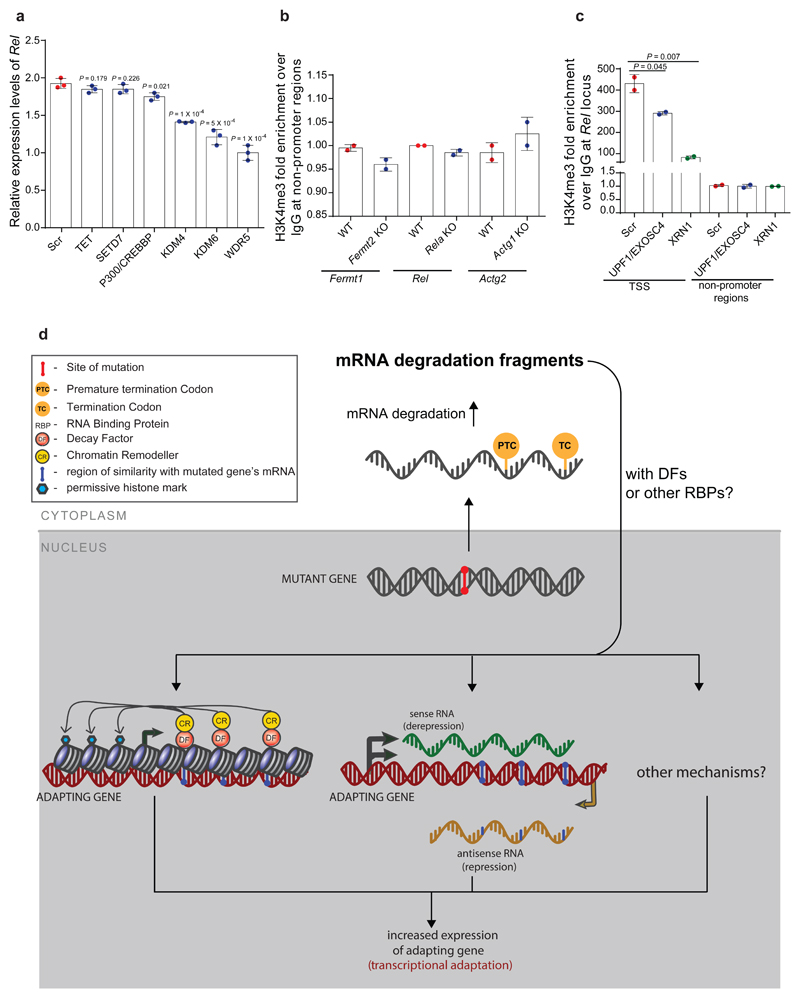
Figure 4. Transcriptional adaptation favors genes exhibiting sequence similarity to the mutated gene’s mRNA and is associated with permissive histone marks.
a, Percentage of significantly upregulated (Log2 Fold Change K.O. > wt and P ≤ 0.05) protein-coding genes exhibiting sequence similarity with Fermt-2, Actg1 or Actb and those not exhibiting sequence similarity. b, qPCR analysis of epas1a mRNA levels in 6 hpf wt zebrafish injected with uncapped RNA composed solely of the hif1ab mRNA sequences similar to epas1a or uncapped RNA composed solely of the hif1ab mRNA sequences not similar to epas1a. c, d, ChIP-qPCR analysis of Wdr5 (c) and H3K4me3 (d) occupancy near the TSS of Fermt1, Rel and Actg2 in Fermt-2, Rela and Actg1 K.O. cells, respectively, compared to wt. Quantification of enrichment shown as fold-enrichment over IgG control. e, Current putative simplified model of transcriptional adaptation to mutations. TC: termination codon; DFs: decay factors; RBPs: RNA binding proteins. b, Control expression set at 1. b-d, n = 3 (b); 2 (c, d); biologically independent samples. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
To begin to investigate how sequence similarity could play a role in transcriptional adaptation, we first injected uncapped transcripts containing only similar or non-similar sequences into zebrafish embryos. Using the hif1ab model, we observed that only transcripts containing sequences similar to epas1a led to an increase in epas1a mRNA levels (Fig. 4b, Extended Data Fig. 8f, Supplementary Data). We also generated synthetic transcripts containing hif1ab sequences similar to the promoter, exons, introns, or 3’UTR of epas1a. Injection of uncapped versions of these transcripts revealed that those exhibiting sequence similarity to exons or introns induced transcriptional adaptation while those exhibiting sequence similarity to the 3’UTR did not; and the transcripts exhibiting sequence similarity to the promoter induced only a mild response (Extended Data Fig. 8g). These data are consistent with the transcriptome analyses of Fermt2, Actg1 and Actb K.O. cells (Extended Data Fig. 8a-c): genes exhibiting sequence similarity to the mutated gene’s mRNA in their 3’UTR were not up-regulated while those exhibiting sequence similarity to promoters exhibited mostly a mild up-regulation. Altogether, these data suggest that, at least in some cases, sequence similarity plays a role in transcriptional adaptation.
Epigenetic remodeling in adapting genes
In the past decade, it has become evident that the control of mRNA stability plays an important role in gene expression10–13. Indeed, several studies reported that mRNA decay factors can translocate to the nucleus and interact with histone modifiers and chromatin remodelers to modulate gene expression14–17. We thus performed a targeted siRNA screen in Rela K.O. cells to identify epigenetic modulators involved in transcriptional adaptation (Supplementary Table 2). Knockdown of JMJD2 or KDM6, which remove the inhibitory H3K9me3 and H3K27me3 histone marks, respectively, dampened the transcriptional adaptation response; however, the strongest effect was observed after WDR5 knockdown (Extended Data Fig. 9a). WDR5 is part of the COMPASS complex which generates the permissive histone mark H3K4me3. Chromatin immunoprecipitation (ChIP) revealed enrichment of WDR5 and H3K4me3 at the TSS of Fermt1, Rel and Actg2 in Fermt2, Rela and Actg1 K.O. cells (Fig. 4c, d and Extended Data Fig. 9b). Moreover, knockdown of UPF1/EXOSC4 or XRN1 in Rela K.O. cells led to the depletion of H3K4me3 at the Rel TSS (Extended Data Fig. 9c). Altogether, these data suggest a model whereby following mutant mRNA degradation, decay factors translocate to the nucleus and bind to specific loci, possibly guided by decay intermediates, and recruit histone modifiers and/or chromatin remodelers to upregulate transcription (Fig. 4e and Extended Data Fig. 9d).
While investigating this model, we noted a study reporting that transfection of short fragments of Cdk9 or Sox9 mRNA could lead to an increase in expression of these genes18. Mechanistically, these RNA fragments were found to downregulate native antisense transcripts which normally function as negative regulators of Cdk9 and Sox9 expression. Notably, we found that transfection of uncapped Cdk9 or Sox9 RNA also led to upregulation of these genes (Extended Data Fig. 10a). Furthermore, knockdown of a BDNF antisense transcript was reported to cause the upregulation of the sense transcript, a response that involved a decrease in the inhibitory H3K27me3 histone mark19. We observed that transfection of uncapped BDNF sense RNA led to a downregulation of the antisense transcript and a concomitant upregulation of the sense one (Extended Data Fig. 10b). Notably, we also observed a downregulation of anti-sense transcripts at the hbegfb and vclb loci in hbegfaΔ7 and vclaΔ13 mutants, respectively (Extended Data Fig. 10c, d). These data indicate that acting on antisense transcripts is one possible mechanism through which mRNA decay intermediates could induce transcriptional adaptation in a sequence specific manner (Fig. 4e and Extended Data Fig. 9c).
Discussion
Despite its potential importance5, transcriptional adaptation to mutations and its underlying molecular mechanisms remain poorly understood. Here we show that the mRNA surveillance machinery is important not only to prevent the translation of defective transcripts but also to buffer against mutations by triggering the transcriptional upregulation of related genes including the mutated gene itself (See Supplementary Discussion).
For a number of human genetic diseases, missense or in frame indels, which are less likely to lead to mutant mRNA degradation, are reportedly more common in affected individuals than potentially RNA-destabilizing nonsense mutations or out-of-frame indels20–25. Interestingly, a study on Marfan syndrome patients reported that when compared to individuals with FBN1 missense mutations, the mildest form of the disease was observed in an individual displaying very low mutant FBN1 transcript levels due to an out-of-frame indel leading to a PTC in the FBN1 coding sequence26. Similar observations were reported for mutations in the HBB gene27. While the current dogma is that pathogenic missense mutations tend to be more common in affected individuals as they might lead to constitutively active or dominant negative proteins, we propose that nonsense mutations might be less common as they might lead to mRNA decay-triggered upregulation of related genes and therefore not cause significant symptoms. Detailed transcriptomic analyses of relevant individuals will help test this hypothesis. Moreover, recent studies28,29 of healthy individuals have reported homozygous loss-of-function mutations in several genes (including EGFL7 and RELA, which were studied here), and it will be interesting to investigate whether degradation of the mutant transcripts is associated with a transcriptional adaptation response that protects them. Such analyses may help us understand why some mutations cause disease while others do not. They may also help identify new modifier genes whose expression levels could be further modulated for therapeutic purposes.
Methods
Statistics and reproducibility
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. All experiments were performed at least twice unless otherwise noted. P< 0.05 was accepted as statistically significant.
Zebrafish husbandry
All zebrafish (Danio rerio, strain: Tüb/AB) husbandry was performed under standard conditions in accordance with institutional (Max Planck Gesellschaft) and national ethical and animal welfare guidelines approved by the ethics committee for animal experiments at the Regierungspräsidium Darmstadt, Germany (permit number: B2/1017). All experiments were performed on zebrafish embryos or larvae between 6 hours post fertilization (hpf) and 6 days post fertilization (dpf). We used the following previously published mutant and transgenic lines: hif1abbns90 30, vegfaabns1 31, egfl7s980 4, egfl7s981 4, hbegfasa18135 and vclasa14599 (Sanger institute zebrafish mutation project; http://www.sanger.ac.uk/resources/zebrafish/zmp/), Tg(fli1a:EGFP)y1 32, TgBAC(etsrp:eGFP)ci1 33.
Cell culture
Wt and Fermt2 knockout MKFs were a generous gift from R. Fässler (MPI for biochemistry, Martinsried, Germany). Wt and Rela knockout MEFs were a generous gift from A. Hoffmann (UCLA, USA). mESCs from the C57BL/6 mouse strain were a generous gift from J. Kim (MPI for heart and lung research, Bad Nauheim, Germany). None of the cell lines were authenticated by the authors. All cell lines tested negative for mycoplasma contamination.
Undifferentiated mESCs were maintained in PluriQ-ES-DMEM (MTI-GlobalStem) consisting of high glucose Dulbecco's minimal essential medium supplemented with 15% ES cell qualified fetal bovine serum (Millipore), 2 mM glutamine, 1% non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.1 mM β-mercaptoethanol, 1000 U/ml ESGRO (LIF, Chemicon international) and 2i (3 μM CHIR99021 and 1 μM PD0325901, Sigma). mESCs were grown in 0.1% gelatin-coated plates and passaged every second day. MEFs and MKFs were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo) supplemented with 10% bovine calf serum (HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin. All cells were grown at 37°C, 95% humidity with 5% CO2 and experiments were performed on cells passaged less than 20 times.
Genotyping
Heterozygous fish were incrossed; DNA and RNA were extracted from at least 24 individual embryos or larvae using TRIzol (Life Technologies) followed by phenol-chloroform extraction. In brief, single embryos or larvae were lysed and homogenized in TRIzol using the NextAdvance Bullet Blender® Homogenizer (Scientific instrument services, inc.). Chloroform was then added and phase separation was obtained following vortexing and centrifugation. The top aqueous phase (containing RNA) was isolated and stored at -80°C and the bottom organic phase (containing DNA) was subjected to ethanol purification to precipitate the DNA. Purified DNA was then dissolved in water and genotyping was performed using high resolution melt analysis (HRMA) or PCR. RNA from genotyped +/+ and -/- zebrafish was then pooled for further purification and cDNA synthesis. For each experiment, this process was performed on embryos from at least 3 different crosses/parents.
HRMA was used to genotype all mutant zebrafish and mouse cell lines with the exception of the RNA-less alleles which were genotyped by PCR. Primer sequences used for genotyping are listed in Supplementary Table 3.
qPCR expression analysis
qPCR was performed in a CFX Connect Real-Time System (Biorad). RNA was isolated using TRIzol and at least 500 ng RNA was used for reverse transcription using the Maxima First Strand cDNA synthesis kit (Thermo). All reactions were performed in at least technical duplicates and the results represent biological triplicates. qPCR was performed at the embryonic or larval stage when the wild-type version of the mutated gene exhibits its highest expression level. For hbegfa mutants treated with NMDi, data were analyzed at 6 dpf and not at 72 hpf as the drug seemed to be effective only when used for 3 days between 72 hpf and 6 dpf; treatment of earlier stage embryos was not possible due to toxicity. Primers were designed using Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). Primers to detect pre-mRNA were designed to bind around intron-exon boundaries. Allele-specific primers were designed such that they amplify the wt but not the mutant allele. To determine the injected capped and uncapped RNA levels, a universal reverse primer corresponding to the adaptor sequence at the 3’ end of the injected RNAs was used along with forward primers designed in close proximity; in this way we were able to distinguish between endogenous and injected RNAs. Several primer pairs along the gene’s cDNA were used to assess transcription in the candidate promoter-less alleles. Only mutant alleles that exhibited less than 10% of wt mRNA levels with all the above mentioned primer pairs were used in the study. For the upf1 double mutant data, the figures show expression levels of the adapting gene in the double mutants relative to its expression levels in the upf1 single mutants. Expression levels of the mutated gene in the upf1 double mutants are relative to those in hbegfa, vegfaa, or vcla single mutants. Equal numbers of cells were used for the mouse cell line qPCR experiments. rpl13, gapdh and Actb, 18srRNA, Gapdh were used to normalize zebrafish and mouse experiments, respectively. rpl13 was chosen as a reference gene for zebrafish experiments as its expression levels were not changed between wild-type and egfl74, hbegfa, vcla, vegfaa and alcama mutant embryos (unpublished microarray data). Primer sequences used for the qPCR experiments are listed in Supplementary Table 4. Fold changes were calculated using the ΔΔCt method. All Ct and dCt values are listed in the source data files. Ct values for the reference genes ranged between 11 and 18 except for 18srRNA which ranged between 6 and 8.
In vitro transcription and RNA microinjections
cDNAs encoding hif1ab, vegfaa, egfl7 and alcama full length mRNAs were amplified using whole embryo cDNA as template. PCR fragments were ligated into pCS2+ between BamHI and XbaI (NEB) sites. All constructs were verified by sequencing. Plasmids were linearized using NotI (NEB) and in vitro transcribed using the mMESSAGE mMACHINE T7 kit (Life Technologies). RNA was then purified using an RNA Clean and Concentrator kit (Zymo Research). 10 to 100 pg of each mRNA was injected into embryos from heterozygous incrosses at the one-cell stage. At 22-30 hpf, embryos were collected in TRIzol for qPCR analysis.
To transcribe uncapped RNAs, cDNA was used to amplify zebrafish hif1ab and vegfaa and mouse Actb, while genomic DNA was used to amplify mouse Cdk9 and Sox9 (single exon for Cdk9 and the entire genomic locus (exons + introns) for Sox9) since their expression levels were too low to amplify from cDNA, all using a reverse primer that contains the adapter sequence 5’ GCCAAGCTATTTAGGTGACACTATAG 3’, subsequently used for qPCR detection of the injected transcripts as described above. To generate uncapped xrFRAG transcripts, the xrFRAG sequence8 was cloned upstream of the hif1ab and vegfaa coding sequences in pCS2+ which was then linearized using Not1 for in vitro transcription. In vitro transcription of uncapped transcripts was performed using T7 RNA polymerase (Promega) and the RNA was purified using an RNA Clean and Concentrator kit. The RNAs were injected into one-cell stage zebrafish embryos (50 pg) or transfected into cells (1 μg) in 12 well plates.
In vitro transcription of synthetic transcripts containing different sequences of hif1ab mRNA
Oligos containing hif1ab sequences similar to epas1a were ligated together along with a 5’ T7 promoter sequence. Similar sequences were identified using a highly sensitive BLASTn analysis with a word size of 7 and an E-value of up to 1,000,000. The alignment was visualized using Kablammo at the less sensitive E-value of 25 due to display constraints (Extended Data Fig. 8f). The same approach was used to generate transcripts of hif1ab sequences not similar to epas1a using the non-alignable sequences. In vitro transcription was performed using the mMESSAGE mMACHINE T7 kit.
gRNA design
gRNAs were designed using the CRISPR design tool CHOPCHOP (http://chopchop.cbu.uib.no/)34. To generate RNA-less alleles, double gRNAs were designed to flank the region to be deleted. gRNAs used to delete promoter regions were designed at least 500 bp upstream and downstream from the transcription start site of the respective gene. Sequences of the gRNAs used in this study and their genomic binding sites are listed in Supplementary Table 5.
Generation of zebrafish mutants
alcama mutant fish were generated using the TALEN technology35,36. The other mutants were generated using the CRISPR/Cas9 system as previously described37,38. To generate mutants, Cas9 mRNA (100 pg) and a gRNA targeting the gene of interest (50 pg) were co-injected into zebrafish embryos at the one-cell stage. To generate RNA-less alleles, two gRNAs were co-injected with Cas9 mRNA. The following mutants were generated for this study: hbegfabns189 (hbegfaΔ7), hbegfabns203 (hbegfaΔ3), hbegfabns243 (hbegfafull locus del.), vclabns241 (vclaΔ13), vclabns300 (vclaexon22_ins1), vegfaabns301 (vegfaa5’UTRΔ10), vegfaabns242 (vegfaapromoter-less), egfl7bns303 (egfl75’UTRΔ3), egfl7bns302 (egfl7full locus del.), alcamabns201 (alcamaΔ10), alcamabns244 (alcamapromoter-less), upf1ya3319 (upf1Δ3ins1).
hbegfaΔ7, hbegfasa18135, hbegfafull locus del., vclaΔ13, vclasa14599, vclaexon22_ins1, alcamaΔ10, and alcamapromoter-less mutants do not exhibit any obvious defects under a dissecting microscope. upf1 mutants exhibit pericardial edema by 5 dpf and die by 10 dpf.
Transfections and isolation of mutant mouse cell clones
Fermt2 MKFs and Rela K.O. MEFs were previously published39,40. The other mutants were generated using the CRISPR/Cas9 system. gRNAs targeting Rela, Actg1 and Actb were cloned into the PX458 or PX459 vectors (Addgene, #48138 and #62988) as previously described41. The final bicistronic vector (hereafter referred to as ‘nuclease plasmid’) encoded the gRNA and the Cas9 nuclease. mESCs were transfected with nuclease plasmids in antibiotic-free medium in a 24 multi-well plate using Xfect (Takara) according to manufacturer's protocol and split into a 10 cm dish the next day. Transfected cells were selected with 0.5 µg/ml puromycin (Sigma) for 4 days and clones were isolated and genotyped using HRMA and sequencing. MKFs and MEFs were electroporated using Nucleofection™ (Lonza) with 5 ug nuclease plasmid DNA according to manufacturer’s protocol. Two days following transfection, cells expressing eGFP were subjected to single cell sorting into 96-well plates using a FACSAria III (BD Biosciences). Three to four weeks following single-cell sorting, clones were isolated and genotyped by PCR and sequencing. For generation of RNA-less alleles, cells were co-transfected with two nuclease plasmids.
Actg1NSD (Actg1 K.O.) and Actg1full locus del. mutant MEFs do not exhibit any gross morphological defects.
Overexpression plasmids
Fermt2 and Rela cDNAs were amplified using MEF cDNA as template. PCR fragments were ligated into the pcDNA3.1 mammalian expression vector (Thermo) between the BamHI and XbaI sites. Plasmids were transfected into the respective mutant cells using FuGene 6 (Promega) according to manufacturer’s protocol. Two days post-transfection, cells were selected with 0.5 mg/ml and 2 mg/ml G418 (Sigma) for Fermt2 and Rela K.O. cells, respectively. A week later, cells were lysed in modified RIPA buffer for western blot analysis. This experiment was performed once.
Uncapped RNA transfections
Uncapped eGFP, Cdk9 and Sox9 RNAs were transfected into mESCs or MEFs using Lipofectamine Messenger Max (Thermo) transfection reagent according to manufacturer’s protocol. 6 hours post transfection, cells were trypsinized and collected in TRIzol for RNA extraction.
CRISPRi
3 gRNAs targeting the Fermt2 promoter and transcription start site were cloned into a plasmid encoding a fusion protein of catalytically-dead Cas9 and Krüppel-associated box (KRAB) repressor (Addgene, #71237) as previously described42. Fermt2 K.O. cells were transfected with the three plasmids and 48 hours post-transfection, cells positive for eGFP were sorted into TRIzol for RNA extraction.
RNA interference
siRNAs are listed in Supplementary Table 2. siRNA transfections were performed using the RNAimax transfection reagent (Thermo) according to manufacturer’s protocol. 48 hours post-transfection, cells were collected in TRIzol for RNA extraction except for knockdowns of SMG6, ERF1 and XRN1 in which case cells were collected 24 hours post-transfection. All siRNAs were used at 10 nM and gave a knockdown efficiency of 70 to 90% (as assessed by mRNA levels); to knock down XRN1 in mESCs, a lower concentration of siRNAs was used (2.5 nM; which led to a knockdown efficiency of 20%) as stronger knockdown affected housekeeping genes as well (data not shown). A scrambled (Scr) siRNA (Sigma, #SIC00), which does not bind to any of the mouse transcripts, was used as a negative control.
For each experiment, the figures show expression levels of the adapting gene in K.O. cells relative to its expression levels in wt cells treated with the same siRNA. Expression levels of the mutant gene in the K.O. cells treated with a given siRNA are relative to those in K.O. cells treated with Scr siRNA. All Ct and dCt values are shown in the source data files.
Pharmacological treatments
To inhibit NMD, 72 hpf wild-type and hbegfa mutant larvae were treated with 10 uM NMDi1443 or DMSO, and 3 days later, they were collected in TRIzol for qPCR analysis. No gross alterations were observed in the drug treated larvae. To inhibit RNA degradation through translation blockade, wild-type and Rela K.O. cells were treated with 200 µg/ml cycloheximide (sigma) or DMSO for 5 hours then collected in TRIzol for RNA extraction.
For each experiment, the figures show expression levels of the adapting gene in mutant larvae, or K.O. cells, relative to its expression levels in wt larvae, or cells treated with the same drug. Expression levels of the mutated gene in the mutant larvae, or K.O. cells treated with a given drug, are relative to those in untreated mutant larvae, or K.O. cells treated with DMSO. All Ct and dCt values are shown in the source data files.
RNA metabolic labeling
Metabolic labeling was performed as previously described44,45. Briefly, cells were treated with 200 µM 4-thiouridine (4sU, Sigma) for 1 hour followed by phenol-chloroform RNA extraction. 80 µg of RNA was incubated with biotin-HPDP (Thermo) to specifically biotinylate the newly transcribed 4sU labeled RNAs. Biotinylated RNAs were then pulled down using the μMacs Streptavidin Kit (Miltenyi) and at least 100 ng of pulled RNA was used for reverse transcription and downstream qPCR analysis. This experiment was performed once with two biological replicates.
Quantifying mRNA half-lives by transcription inhibition
Wild-type, Fermt2 K.O., Rela K.O., and Actg1 K.O. cells were treated with 10 µg/ml Actinomycin D (Sigma) to block transcription. Cells were then collected in TRIzol at 0, 1, 2, 4 and 8 hours post treatment for RNA extraction. 18srRNA was used as a housekeeping gene as its expression level was not affected over the time course of treatment (data not shown). Half-lives were then quantified from fitted nonlinear exponential decay curves.
Cytotoxicity assay
7,000 cells were seeded per well in a 96-well plate and incubated with media containing 25 ng/ml of mouse TNFα. 24 hours later, cells were washed with PBS and incubated for 5 hours with media containing 3 mg/ml MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide). The formed formazan (product of MTT’s metabolism by viable cells) was resuspended in 50% DMSO: 50% ethanol solution and optical densities (O.D.) were measured at 572 nm using a FLUOstar Omega spectrophotometer (BMGH Labtech). % cytotoxicity was measured as the ratio of O.D. difference between DMSO and TNFα treated cells to that of DMSO treated cells. This experiment was performed once.
mESC staining
Cells were fixed with 4% PFA for 15 min at room temperature (RT), permeabilized with 0,3% Triton X-100 (in PBS) for 10 min, and then incubated with 1:1000 phalloidin-568 (Thermo) in 3% BSA for 1 hr at RT. After 3x 15 min washes with PBT (0,1% Triton X-100 in PBS), samples were counterstained for 5 min at RT with 1:5000 DAPI (Sigma) and mounted for imaging with Dako Fluorescent mounting medium. Images were obtained on a Zeiss LSM700 confocal microscope using a LD C-Apochromat 63x/1.15 W Corr M27 objective. Protrusion length was measured using ImageJ.
Immunostaining
100 hpf larvae from alcamapromoter-less +/- and alcama Δ8 +/- incrosses were collected and fixed in 4% paraformaldehyde. After removing the fixative with PBS/0.1% Tween washes, larvae were incubated with 3 µg/ml proteinase K for 1 hour then washed with PBS/1% BSA/1%DMSO/0.5% Triton-X (PBDT) before being incubated for 2 hours with phalloidin Alexa-568 (Invitrogen) at RT to label F-Actin. They were then washed with PBS/0.1% Tween and mounted for imaging the heart. Images of the hearts were acquired using a Zeiss LSM880 Axio Examiner confocal microscope with a W Plan-Apochromat 20x/1.0 objective. The ventricle length, or long axis of the ventricle, was measured from the apex of the ventricle to the junction of the ventricle with the bulbus arteriosus using Zen Black. In the first experiment, larvae were genotyped after imaging.
alcamapromoter-less larvae were obtained from F2 heterozygous parents generated by outcrossing the founder and F1 fish. alcama Δ8 larvae were obtained from an incross of F3 heterozygous parents generated by consecutive outcrosses.
Confocal microscopy
A Zeiss LSM 700 confocal microscope was used for live imaging of the trunk and brain vasculature. Embryos were anaesthetized with a low dose of tricaine, placed in a glass-bottomed Petri dish (MatTek) on a layer of 1.2% low melt agarose and imaged using Plan-Apochromat 10×/0.45 and LCI Plan-Neofluar 25×/0.8 objectives.
In wt embryos, at 60 hpf, an average of 13 CtAs have connected to the basilar artery (BA)46.
vegfaapromoter-less larvae were obtained from F2 heterozygous parents generated by outcrossing the founder and F1 fish.
To measure blood flow velocity, we performed time-lapse imaging of the trunk region of 78 hpf wild-type, maternal zygotic hbegfaΔ7 -/- and maternal zygotic hbegfafull locus del. -/- larvae using a Zeiss Spinning disc CSU-X1 confocal microscope with a high-speed camera, and quantified blood flow velocity in the dorsal aorta by measuring the time needed by erythrocytes to move 200 μm at the level of the fifth and sixth somites using Zen Blue as previously described47.
hbegfafull locus del. larvae were obtained from F2 homozygous parents generated by outcrossing the founder.
hbegfaΔ7 and vegfaaΔ10 larvae were obtained from an incross of F3 heterozygous parents that were generated by consecutive outcrosses.
Western blot analysis and antibodies
MKFs and MEFs were lysed in modified RIPA buffer (150 mM NaCl, 50 mM TrisHCl pH 7.4, 1% IGEPAL, 0.1% sodium deoxycholate, 1 mM EDTA) supplemented with protease inhibitors (cOmplete ULTRA Mini, Roche) and PMSF. 35 µg of protein samples were separated using SDS-PAGE on precast TGX gradient gels (Biorad) (β-ACTIN control was run on the same gel as a loading control) and then electrophoretically transferred to polyvinylidine fluoride membranes (Biorad) using a Transblot Turbo Transfer System (Biorad). Membranes for RELA and Fermt2 were blocked in 5% BSA (sigma) or 5% Non-fat milk (Sigma), respectively, for 1 hr then probed with primary antibodies overnight at 4°C. The following day, membranes were probed with peroxidase conjugated secondary antibodies for 1 hr at RT. For analysis, membranes were incubated with ECL (Clarity Western ECL Substrate, Biorad) and imaged using a ChemiDoc MP system (Biorad). The following antibodies were used: Fermt2 (Millipore, MAB2617; clone 3A3, 1:1,000), RELA (Cell Signaling Technology, #6956, 1:1,000), β-ACTIN (Cell Signaling Technology, #8457, 1:1000), anti-mouse IgG-HRP (Thermo, 31430, 1:10,000), and anti-rabbit IgG-HRP (Thermo, 31460, 1:10,000).
Chromatin immunoprecipitation
ChIP was performed using the truChIP Chromatin Shearing Reagent kit (Covaris) using 30 million cells/IP according to manufacturer’s protocol. Chromatin was sheared using Bioruptor (Diagenode) to generate fragments between 200-400 bp. Immunoprecipitation was then performed as previously described48. The following antibodies were used: rabbit IgG (4μg/IP, # 026102, Thermo Fischer), WDR5 (4μg/IP, #13105, Cell signaling) and H3K4me3 (4μg/IP, #9751, Cell signaling). Following immunoprecipitation and reverse cross-linking, samples were purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) following manufacturer’s protocol for samples containing SDS.
ATAC-seq material extraction and library preparation
Cells were trypsinized and washed with PBS. Counting of cells was performed with MOXI Z Mini Automated Cell Counter Kit (Orflo) and 50,000 cells were used for ATAC library preparation using Tn5 Transposase from Nextera DNA Sample Preparation Kit (Illumina). The cell pellet was resuspended in 50 µl PBS and mixed with 25 µl TD-Buffer, 2.5 µl Tn5, 0.5 µl 10% NP-40 and 22 µl water. This cell/Tn5 mixture was incubated at 37°C for 30 min with occasional snap mixing. Transposase treatment was followed by 30 min incubation at 50°C together with 500 mM EDTA pH8.0 for optimal recovery of digested DNA fragments. For neutralization of EDTA, 100 µl of 50 mM MgCl2 was added followed by purification of the DNA fragments by MinElute PCR Purification Kit (Qiagen). Amplification of the library together with indexing was performed as previously described49. Libraries were mixed in equimolar ratios and sequenced on a NextSeq500 platform using V2 chemistry.
ATAC-seq analysis
The samples were assessed for quality using FastQC (Andrews S. 2010, FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Trimmomatic version 0.3350 was employed to trim reads after a quality drop below a mean of Q20 in a window of 5 nucleotides. Only reads above 30 nucleotides were cleared for further analyses. In order to normalize all samples to the same sequencing depth, 27 million reads per sample were randomly selected for further analysis. These reads were mapped versus the mm10 version of the mouse genome with STAR 2.4.2a51 using only unique alignments to exclude reads with unclear placing. The reads were further deduplicated using Picard 1.136 (Picard: A set of tools (in Java) for working with next generation sequencing data in the BAM format; http://broadinstitute.github.io/picard/) to avoid PCR artefacts leading to multiple copies of the same original fragment. The Macs2 peak caller version 2.1.0 was employed to identify peaks. The minimum qvalue was set to -1.5 and FDR was changed to 0.01. In order to determine thresholds for significant peaks, the data were manually inspected in IGV 2.3.5252. Peaks overlapping ENCODE blacklisted regions (known mis-assemblies, satellite repeats) were excluded. In order to be able to compare peaks between samples, the resulting lists of significant peaks were overlapped and unified to represent identical regions. After conversion of BAM files to BigWig format with deep Tools bamCoverage53, the counts per unified peak per sample were computed with BigWigAverageOverBed (UCSC Genome Browser Utilities, http://hgdownload.cse.ucsc.edu/downloads.html). Raw counts for unified peaks were submitted to DESeq2 for normalization54. Spearman correlations were produced to identify the degree of reproducibility between samples using R. To allow a normalized display of samples in IGV, the raw BAM files were normalized for sequencing depth (number of mapped deduplicated reads per sample) and noise level (number of reads inside peaks). Two factors were computed and applied to the original BAM files using bedtools genomecov55 resulting in normalized BigWig files for IGV.
RNA-seq
RNA was isolated using the miRNeasy micro Kit (Qiagen). To avoid contamination by genomic DNA, samples were treated by on-column DNase digestion (DNase-Free DNase Set, Qiagen). Total RNA and library integrity were verified on LabChip Gx Touch 24 (Perkin Elmer). 1µg of total RNA was used as input for SMARTer Stranded Total RNA Sample Prep Kit - HI Mammalian (Clontech). Sequencing was performed on a NextSeq500 instrument (Illumina) using v2 chemistry, resulting in an average of 25-30M reads per library with 1x75bp single end setup.
RNA-seq analysis
The resulting raw reads were assessed for quality, adapter content and duplication rates with FastQC. Reaper version 13-100 was employed to trim reads after a quality drop below a mean of Q20 in a window of 10 nucleotides56. Only reads of at least 15 nucleotides were cleared for subsequent analyses. Trimmed and filtered reads were aligned versus the Ensembl mouse genome version mm10 (GRCm38) using STAR 2.5.3a with the parameters “--outFilterMismatchNoverLmax 0.1 --alignIntronMax 200000”51. The number of reads aligning to genes was counted with featureCounts 1.6.0 from the Subread package57. Only reads mapping at least partially inside exons were admitted and aggregated per gene. Reads overlapping multiple genes or aligning to multiple regions were excluded. Differentially expressed genes were identified using DESeq2 version 1.14.158. Genes were classified as significantly differentially expressed with P< 0.05 without assigning a specific minimum or maximum regulation fold change as transcriptional adaptation might not necessarily lead to strong upregulation levels. The Ensemble annotation was enriched with UniProt data (release 24.03.2017) based on Ensembl gene identifiers (Activities at the Universal Protein Resource (UniProt)).
Sequence similarity and subsampling analyses
In order to identify similarity to one of the three query nucleotide sequences (Fermt2, Actg1 and Actb), the longest respective transcript was selected (ENSMUST00000071555, ENSMUST00000045905, ENSMUST00000100497) and compared to the whole genome using BLASTn59. Genes were defined to be similar to the mutated gene when a partial match was identified inside the target gene body or its promoter region (ie, 2 kb upstream of the TSS). Several alignment parameters were surveyed to identify the optimal degree of similarity. Alignment length, bit score, and E-value were queried to identify the dynamic range and optimal values using subsampling analyses (see below). E-value denotes the probability for the match to have resulted by chance when considering the whole target database (the genome in this case).
A subsampling approach was used to calculate a ranked P value for the significance of the percentage of upregulated genes in subsamples of a specific size (equivalent to the number of ‘similar’ protein coding genes) for each mouse cell line model. Briefly, the following algorithm was repeated 10,000 times: 1) get X random protein coding genes, 2) count the percentage of upregulated genes in this subsample. The resulting list was filtered for subsamples with an equal or higher than expected number of upregulated genes according to a previous comparison (e.g., for Fermt2, 18 protein coding genes exhibit sequence similarity to its mRNA [=subsample size], 9 of which were also upregulated [=expectation]). The number of subsamples showing at least as many upregulated genes as the expectation represents the rank of the comparison. The ranked P value was computed by dividing that rank by the total number of iterations (=10,000). Optimal thresholds varied for the different cell line models and ranged between 1) 20 and 180 nucleotides alignment length, 2) 40 and 200 bit score (combination of alignment quality and length), and 3) 10 to 6.73E-50 maximum E-value. We selected the following maximum E-values from the optimal range for the follow up similarity analyses: 5.1 for Fermt2 and Actg1, and 2E-48 for Actb. The much stricter E-value for Actb was necessary due to its repetitive 3’UTR which results in misleading “noisy” matches. These E-value thresholds translate into local nucleotide sequence alignments ranging from 24 to 1901 nucleotides in length with 75 to 96% identity.
Sequence alignments of hif1ab, Actb and epas1a
The BLASTn alignments of the longest transcript of hif1ab (ENSDART00000018500) with the epas1a gene body including its promoter (2 kb upstream of TSS) were visualized using Kablammo60 at word size 7 and E-value of 25 (Extended Data Fig. 8f). Two additional alignments were performed to show homologous regions of the synthetic hif1ab transcript compared to 1) the original source transcript ENSDART00000018500 using MUSCLE61, and 2) epas1a including its promoter (TSS-2000) using BLASTn. The uncapped RNA composed solely of the hif1ab sequences similar to epas1a was 1277 nucleotides in length, and that composed solely of the hif1ab sequences not similar to epas1a was 1929 nucleotides in length. The similarity of the coding sequences of zebrafish actb1 transcript ENSDART00000054987 (Query) to the mouse Actb transcript ENSMUST00000100497 (Subject) was assessed with a MUSCLE alignment. All these alignments can be found in Supplementary Data.
Gene set enrichment analyses
Genes strongly upregulated in all three K.O. samples versus the respective wild-type samples (P< 0.05, L2F > 0.585) were used for gene set enrichment analyses using KOBAS62.
Extended Data
Extended Data Figure 1. Schematic illustration of the mutant alleles generated for this study.
Partial DNA sequences of the different mutant alleles generated for this study, and images of gels providing evidence for deletions in RNA-less alleles. Red: mutation; green: stop codon in alleles with a PTC; arrows: genotyping primers.
Extended Data Figure 2. Transcriptional adaptation is independent of the loss of protein function.
a, qPCR analysis of epas1a and epas1b, vegfab, emilin3a and alcamb mRNA levels in wt and hif1ab, vegfaa, egfl7 and alcama mutant embryos injected with eGFP mRNA (control) or wt hif1ab, vegfaa, egfl7 or alcama mRNA. b, qPCR analysis of vclb, epas1a and epas1b and emilin3a mRNA levels in vcla, hif1ab and egfl7 wt, heterozygous and mutant zebrafish. c, qPCR analysis of hbegfa, hif1ab, vegfaa and alcama mRNA levels in hbegfa, hif1ab, vegfaa and alcama wt and heterozygous zebrafish using primers specific for the wt allele. d, qPCR analysis of Fermt1 and Rel mRNA levels in wt and Fermt2 and Rela K.O. cells transfected with empty vectors (control) or plasmids encoding wt Fermt2 or RELA. e, Western blot analysis of Fermt2 and ACTB levels in Fermt2 K.O. cells transfected with empty vectors (control) or plasmids encoding wt Fermt2. f, Western blot analysis of RELA and ACTB levels in Rela K.O. cells transfected with empty vectors (control) or plasmids encoding wt RELA. g, qPCR analysis of Actg1 mRNA levels in wt and heterozygous Actb mESCs. a-d, g, n = 3 biologically independent samples. wt or control expression set at 1 for each assay. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values. e, f, These experiments were repeated twice independently with similar results. For western blots’ source data, see Supplementary Figure 1.
Extended Data Figure 3. Transcriptional adaptation involves enhanced transcription and is independent of the DNA lesion itself.
a, qPCR analysis of hbegfb and emilin3a mRNA and pre-mRNA levels in hbegfa and egfl7 wt and mutant zebrafish. b, qPCR analysis of Fermt1 and Rel mRNA and pre-mRNA levels in Fermt2 and Rela wt and K.O. cells. c, Integrated genome viewer tracks of Fermt1 (Fermt1) locus showing ATAC-seq signals in wt and Fermt2 K.O. cells. d, qPCR analysis of hbegfa, hbegfb and egfl7, emilin3a mRNA levels in hbegfa and egfl7 wt and Δ3 mutant zebrafish. e, qPCR analysis of vegfaa, vegfab and egfl7, emilin3a mRNA levels in vegfaa and egfl7 wt and 5’UTR mutant zebrafish. f, qPCR analysis of vcla and vclb mRNA levels in vcla wt and last exon (exon 22) mutant zebrafish. a, b, d-f, n = 3 biologically independent samples. wt expression set at 1 for each assay. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Extended Data Figure 4. Reduction in mutant transcript levels is due to mRNA decay.
a, qPCR analysis of hbegfa, egfl7 and alcama mRNA and pre-mRNA levels in hbegfa, egfl7 and alcama wt and mutant zebrafish. b, qPCR analysis of Fermt2 and Rela mRNA and pre-mRNA levels in Fermt2 and Rela wt and K.O. cells. c, qPCR analysis of 4sU labeled Fermt-2, Rela and Actg1 mRNA and pre-mRNA levels in Fermt-2, Rela and Actg1 wt and K.O. cells. d, Fitted exponential decay curves of Fermt2 mRNA levels in wt and Fermt2 K.O. cells. e, Fitted exponential decay curves of Rela mRNA levels in wt and Rela K.O. cells. f, Fitted exponential decay curves of Actg1 mRNA levels in wt and Actg1 K.O. cells. a-f, n = 3 (a, b, d-f); 2 (c) biologically independent samples. a-c, wt expression set at 1 for each assay. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Extended Data Figure 5. RNA decay induces transcriptional adaptation.
a, qPCR analysis of hbegfa, vegfaa, and vcla mRNA levels in upf1;hbegfa, upf1;vegfaa and upf1;vcla double mutant zebrafish. b, qPCR analysis of Rela mRNA levels after siRNA mediated knockdown of indicated proteins in Rela K.O. cells. c, qPCR analysis of Actb mRNA levels after siRNA mediated knockdown of indicated proteins in Actb K.O. cells. d, qPCR analysis of hbegfa mRNA levels in 6 dpf hbegfa mutants treated with NMD inhibitor (NMDi). e, qPCR analysis of hbegfb mRNA levels in 6 dpf hbegfa mutants treated with NMDi. f, qPCR analysis of Rela mRNA levels in Rela K.O. cells treated with cycloheximide (CHX). g, qPCR analysis of Rel mRNA levels in Rela K.O. cells treated with CHX. h, qPCR analysis of endogenous hif1ab and vegfaa mRNA levels in 6 hpf wt embryos injected with uncapped hif1ab or vegfaa RNA. i, qPCR analysis of Actg1 mRNA levels in mESCs transfected with uncapped Actb RNA at different times post-transfection. j, qPCR analysis of injected hif1ab, epas1a and injected vegfaa, vegfab RNA levels in 6 hpf wt embryos injected with uncapped hif1ab or vegfaa transcripts with or without a 5’ xrFRAG sequence. hr, hour. k, qPCR analysis of epas1a and vegfab mRNA levels in 6 hpf wt zebrafish embryos injected with uncapped sense or antisense hif1ab or vegfaa RNA. b, c, Scr: Scrambled siRNA control. a-d, f, h-k, wt or control expression set at 1 for each assay. a-k, n = 3 biologically independent samples. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Extended Data Figure 6. Mutant mRNA decay helps confer genetic robustness.
a, qPCR analysis of Fermt2 and Fermt1 mRNA levels following CRISPRi mediated knockdown of Fermt2 transcription in Fermt2 K.O. cells. b, qPCR analysis of emilin3a, emilin3b and emilin2a mRNA levels in wt, egfl7Δ4 mutant and egfl7full locus del. mutant 20 hpf zebrafish. c, Number of CtAs connecting to the basilar artery (BA) in 58 hpf vegfaaΔ10 and vegfaapromoter-less mutants. d, Blood flow velocity in 78 hpf wt, hbegfaΔ7 and hbegfafull locus del. mutants. e, Quantification of the cardiac ventricle length in 100 hpf wt, alcama Δ8 and alcamapromoter-less mutant larvae. a, b, wt or control expression set at 1 for each assay. a-e, n = 3 (a, b); 13 and 19 (c); 25 (d); 18, 7, 22 and 15 (e) animals. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Extended Data Figure 7. Analysis of sequence similarity parameters in models of transcriptional adaptation.
a, Numbers of differentially expressed genes in the different K.O. cell line models; P ≤ 0.05; these genes are distributed throughout the genome (data not shown). b, Venn diagram of genes upregulated in the three different cell line models with Log2 Fold (L2F) K.O. > wt and P ≤ 0.05. c, KEGG pathway enrichment analysis for genes commonly upregulated in Fermt2, Actg1 and Actb K.O. cells compared to wt. The top 10 pathways based on P value are displayed. The dashed line marks a P value of 0.05. Circle sizes aim to provide scale; outer gray circles represent the total number of genes in the pathway while centered colored circles represent the number of genes in the pathway that are commonly upregulated. d, Impact of various values of three different BLASTn alignment quality parameters (alignment length, Bit score, E-value) on the significance of the observed correlation between up-regulation and sequence similarity and thereby the identification/predication of putative adapting genes. E-value describes the probability of the match resulting from chance (lower = better), while Bit score evaluates the combination of alignment quality and length (higher = better). Each diagram shows the negative log10 of the significance P value (higher = better) on the Y-axis, and the respective parameter value on the X-axis. A P value of 0.05 is marked with a black horizontal line. The E-value thresholds used in our analyses are highlighted with a circle. Lines ending preliminarily indicate a lack of any remaining alignments after that point. The first row of diagrams explores large variations of thresholds in an attempt to identify the total range, while the second row focuses on the most relevant window for the three genes investigated. The optimal thresholds differ considerably depending on the gene analyzed. a-d, n = 2 biologically independent samples. DESeq2 tests for significance of coefficients in a negative binomial GLM (Generalized Linear Model) with the Wald test. P values were not multiple testing corrected.
Extended Data Figure 8. Expression level of genes exhibiting sequence similarity in the different mouse cell line models.
a-c, RNA-seq analysis of genes exhibiting sequence similarity with Fermt2 (a), Actg1 (b) or Actb (c) in K.O. cells compared to wt. L2F: Log2 Fold change. Bold: Significantly upregulated in K.O. cells relative to wt. Red: L2F>0, blue: L2F<0. Green: P value or P adjusted value ≤ 0.05. Purple: Genes that exhibit sequence similarity with the mutated gene’s mRNA in their promoter region. Yellow: Genes that exhibit sequence similarity with the mutated gene’s mRNA in their 3’UTR region. Other non-colored genes exhibit sequence similarity with the mutated gene’s mRNA in their exons or introns. Boxed: upregulated in K.O. but not RNA-less cells; no Fermt2 RNA-less allele was analyzed. d, qPCR analysis of Ubapl, Fmnl2, Cdk12 and Actr1a pre-mRNA levels in Actg1 K.O. cells relative to wt. e, qPCR analysis of actb1 mRNA levels in 6 hpf wt zebrafish injected with uncapped mouse Actb RNA. f, Schematic representation of regions of sequence similarity between hif1ab mRNA and epas1a locus. TSS: transcription start site. Grey shaded triangles represent the alignments; intensity represents alignment quality and width at the base represents length of the similarity region. g, qPCR analysis of epas1a mRNA levels in 6 hpf wt zebrafish embryos injected with uncapped RNA composed solely of the hif1ab sequences similar to epas1a promoter, exons, introns, or 3’UTR. a-e, g, n = 2 (a-c); 3 (d, e, g) biologically independent samples. a-c, DESeq2 tests for significance of coefficients in a negative binomial GLM (Generalized Linear Model) with the Wald test. P values were not multiple testing corrected. d, e, g, wt or control expression set at 1 for each assay. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Extended Data Figure 9. Transcriptional adaptation involves chromatin remodeling dependent on decay factor activity.
a, qPCR analysis of Rel mRNA levels after siRNA mediated knockdown of the indicated proteins in Rela K.O. cells. b, ChIP-qPCR analysis of H3K4me3 occupancy at non-promoter regions (as a control) of Fermt1, Rel and Actg2 in Fermt-2, Rela and Actg1 K.O. cells, respectively, compared to wt. c, ChIP-qPCR analysis of H3K4me3 occupancy near the Rel TSS and a non-promoter region (as a control) after siRNA mediated knockdown of the indicated proteins in Rela K.O. cells. Scr: Scrambled siRNA control. d, Current expanded model of transcriptional adaptation to mutations. RNA decay fragments may act as intermediates to bring decay factors, and chromatin remodelers, to adapting gene loci, thereby triggering increased gene expression. Alternatively, RNA decay fragments may function to repress antisense RNAs at the adapting gene loci allowing for increased sense mRNA expression. It is, however, likely that additional mechanisms are involved in transcriptional adaptation, and possibly in a gene-dependent manner. a-c, n = 3 (a); 2 (b, c) biologically independent samples. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Extended Data Figure 10. Antisense transcripts as potential players in the transcriptional adaptation response.
a, qPCR analysis of Cdk9 and Sox9 mRNA levels in cells transfected with uncapped Cdk9 or Sox9 RNA. b, qPCR analysis of BDNF and BDNF-AS mRNA levels in HEK293T cells transfected with uncapped BDNF RNA. c, Integrated genome viewer tracks of vclb and hbegfb loci showing the location of the annotated antisense transcripts. Two alignments of 105 and 147 bp were observed between vcla mRNA and vclb AS RNAs, and an alignment of 39 bp was observed between hbegfa mRNA and hbegfb AS RNA. d, qPCR analysis of vclb and hbegfb antisense RNA levels in vcla and hbegfa wt and mutant zebrafish. a, b, d, Control expression set at 1 for each assay. n = 3 biologically independent samples. Error bars, mean, s.d. Two-tailed student’s t-test used to assess P values.
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgments
We thank V. Serobyan, F. Mueller (U. Birmingham, UK), Z. Jiang, A. Beisaw and F. Gunawan for discussion and comments on the manuscript and J. Pestel for the alcama mutant. We also thank A. Atzberger for support with cell sorting and N. Gehring and V. Böhm for providing the xrFRAG plasmid. M.A.E.B. was supported by a Boehringer Ingelheim Fonds PhD fellowship. Research in the Stainier lab is supported by the Max Planck Society, EU, DFG and Leducq Foundation.
Footnotes
Data availability
ATAC-seq and RNA-seq data were deposited to Gene Expression Omnibus under accession codes GSE107075 and GSE114212.
Author contributions M.A.E.B. designed and performed most of the experiments, analyzed data and wrote the manuscript; Z.K. and A.R. designed and performed mESC experiments, some imaging and edited the manuscript; C.K. performed bioinformatics analyses; S.G. performed ATAC- and RNA-seq; N.F. and K.K. performed some qPCR experiments; G.L.M.B. performed some imaging; C.T. generated the upf1 mutant under the supervision of A.J.G; S.L., R.F., and C.G. provided unpublished mutants; D.Y.R.S. helped design the experiments and analyze data, supervised the work, and wrote the manuscript; all authors commented on the manuscript.
Author information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing interests.
References
- 1.Tautz D. Redundancies, development and the flow of information. BioEssays : news and reviews in molecular, cellular and developmental biology. 1992;14:263–266. doi: 10.1002/bies.950140410. [DOI] [PubMed] [Google Scholar]
- 2.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 3.Teng X, et al. Genome-wide consequences of deleting any single gene. Molecular cell. 2013;52:485–494. doi: 10.1016/j.molcel.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 5.El-Brolosy MA, Stainier DYR. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes & development. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee C, et al. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell reports. 2012;2:674–684. doi: 10.1016/j.celrep.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm V, Gerbracht JV, Marx MC, Gehring NH. Interrogating the degradation pathways of unstable mRNAs with XRN1-resistant sequences. Nature communications. 2016;7 doi: 10.1038/ncomms13691. 13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doi TS, et al. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkon R, Zlotorynski E, Zeller KI, Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotech. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun M, et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome research. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 15.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes & development. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. The EMBO journal. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haimovich G, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Ghanbarian H, et al. Small RNA-directed epigenetic programming of embryonic stem cell cardiac differentiation. Scientific reports. 2017;7 doi: 10.1038/srep41799. 41799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature biotechnology. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisensmith RC, Woo SL. Molecular basis of phenylketonuria and related hyperphenylalaninemias: mutations and polymorphisms in the human phenylalanine hydroxylase gene. Human mutation. 1992;1:13–23. doi: 10.1002/humu.1380010104. [DOI] [PubMed] [Google Scholar]
- 21.Myerowitz R. Tay-Sachs disease-causing mutations and neutral polymorphisms in the Hex A gene. Human mutation. 1997;9:195–208. doi: 10.1002/(sici)1098-1004(1997)9:3<195::aid-humu1>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Genschel J, Schmidt HH. Mutations in the LMNA gene encoding lamin A/C. Human mutation. 2000;16:451–459. doi: 10.1002/1098-1004(200012)16:6<451::aid-humu1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Chuzhanova NA, Anassis EJ, Ball EV, Krawczak M, Cooper DN. Meta-analysis of indels causing human genetic disease: mechanisms of mutagenesis and the role of local DNA sequence complexity. Human mutation. 2003;21:28–44. doi: 10.1002/humu.10146. [DOI] [PubMed] [Google Scholar]
- 24.Ferec C, Cutting GR. Assessing the Disease-Liability of Mutations in CFTR. Cold Spring Harbor Perspectives in Medicine. 2012;2 doi: 10.1101/cshperspect.a009480. a009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Q, et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. The New England journal of medicine. 2014;370:911–920. doi: 10.1056/NEJMoa1307361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietz HC, et al. Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics. 1993;17:468–475. doi: 10.1006/geno.1993.1349. [DOI] [PubMed] [Google Scholar]
- 27.Hall GW, Thein S. Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood. 1994;83:2031–2037. [PubMed] [Google Scholar]
- 28.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nature genetics. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 29.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerri C, et al. Hif-1alpha regulates macrophage-endothelial interactions during blood vessel development in zebrafish. Nature communications. 2017;8 doi: 10.1038/ncomms15492. 15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi A, et al. Regulation of Vegf signaling by natural and synthetic ligands. Blood. 2016;128:2359–2366. doi: 10.1182/blood-2016-04-711192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Developmental biology. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 33.Proulx K, Lu A, Sumanas S. Cranial vasculature in zebrafish forms by angioblast cluster-derived angiogenesis. Developmental biology. 2010;348:34–46. doi: 10.1016/j.ydbio.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 34.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic acids research. 2016;44:W272–276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic acids research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle EL, et al. TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat -1. PloS one. 2013;8:e82120. doi: 10.1371/journal.pone.0082120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PloS one. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vejnar CE, Moreno-Mateos MA, Cifuentes D, Bazzini AA, Giraldez AJ. Optimized CRISPR-Cas9 System for Genome Editing in Zebrafish. Cold Spring Harbor protocols. 2016;2016 doi: 10.1101/pdb.prot086850. pdb.prot086850. [DOI] [PubMed] [Google Scholar]
- 39.Theodosiou M, et al. Fermt2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 2016;5:e10130. doi: 10.7554/eLife.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gapuzan ME, Yufit PV, Gilmore TD. Immortalized embryonic mouse fibroblasts lacking the RelA subunit of transcription factor NF-kappaB have a malignantly transformed phenotype. Oncogene. 2002;21:2484–2492. doi: 10.1038/sj.onc.1205333. [DOI] [PubMed] [Google Scholar]
- 41.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin L, et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.can-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radle B, et al. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. Journal of visualized experiments : JoVE. 2013 doi: 10.3791/50195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun W, Chen W. Metabolic Labeling of Newly Synthesized RNA with 4sU to in Parallel Assess RNA Transcription and Decay. Methods in molecular biology (Clifton, N.J.) 2018;1720:25–34. doi: 10.1007/978-1-4939-7540-2_3. [DOI] [PubMed] [Google Scholar]
- 46.Vanhollebeke B, et al. Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/beta-catenin pathway during brain angiogenesis. eLife. 2015;4 doi: 10.7554/eLife.06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon HB, et al. In vivo modulation of endothelial polarization by Apelin receptor signalling. Nature communications. 2016;7 doi: 10.1038/ncomms11805. 11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blecher-Gonen R, et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nature protocols. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 49.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson JT, et al. Integrative Genomics Viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic acids research. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis MP, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods (San Diego, Calif.) 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 60.Wintersinger JA, Wasmuth JD. Kablammo: an interactive, web-based BLAST results visualizer. Bioinformatics. 2015;31:1305–1306. doi: 10.1093/bioinformatics/btu808. [DOI] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic acids research. 2011;39:W316–322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.