Synopsis
Overall survival rates for pediatric patients with high-risk or relapsed rhabdomyosarcoma (RMS) have not improved significantly since the 1980s. Recent studies have identified a number of targetable vulnerabilities in RMS, but these discoveries have infrequently translated into clinical trials. We propose streamlining the process by which agents are selected for clinical evaluation in RMS. We believe that strong consideration should be given to the development of combination therapies that add biologically targeted agents to conventional cytotoxic drugs. One example of this type of combination is the addition of the WEE1 inhibitor, AZD1775, to the conventional cytotoxic chemotherapeutics, vincristine and irinotecan.
Keywords: rhabdomyosarcoma, early phase clinical trials, cancer biology, genomics
Introduction
Rhabdomyosarcoma (RMS) is the third most common extracranial solid tumor of childhood, accounting for 3 percent of childhood cancers and comprising approximately 350 cases in the US annually. RMS is also found in adult patients, which accounts for approximately 100 additional cases annually1. RMS tumor cells morphologically resemble cells arrested in the early stages of skeletal muscle development 2. However, a large percentage of RMS tumors occur in locations normally lacking skeletal muscle such as the head and neck, genitourinary tract, and retroperitoneum 3. Childhood RMS is subdivided into two major subtypes, PAX-fusion negative (previously called embryonal RMS) and PAX-fusion positive (previously called alveolar RMS), which have distinct histological features and genetic alterations. Spindle cell/sclerosing RMS has recently emerged as a third pediatric RMS subtype (Figure 1), while a fourth RMS subtype, pleiomorphic RMS, is seen exclusively in adults. Current treatment regimens use PAX-fusion status for risk assignment as the PAX-fusion positive group denotes a high-risk subtype with a less favorable prognosis than those who are PAX-fusion negative 4,5. Spindle cell/sclerosing RMS also has a poor prognosis 6.
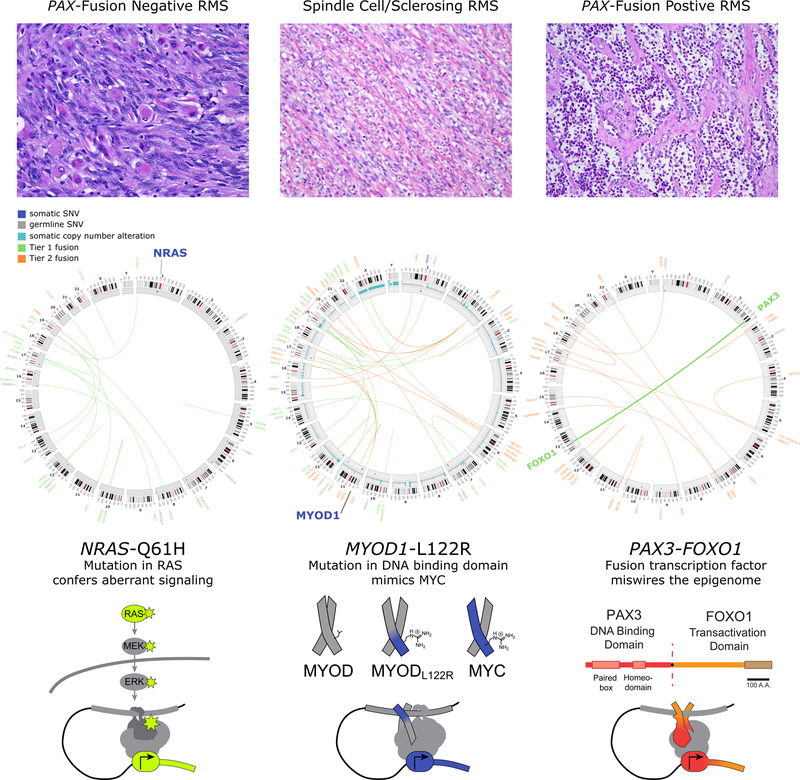
Figure 1.
Histopathology and Genomics of the RMS subtypes
Representative H&E staining (top), Circos plots (middle), and schematics of the genomic drivers (bottom) of each of the three RMS subtypes, PAX-fusion negative RMS, spindle cell/sclerosing RMS and PAX-fusion positive RMS. Histology courtesy of C.R. Antonescu. Circos plots adapted from 22,25. In these plots, chromosomes 1–22 as well as the sex chromosomes are arrayed in a circle and arranged clockwise. The length of the chromosome in Mb is depicted outside each representation. Mutations for each tumor are indicated outside the chromosome number. Somatic single nucleotide variants (SNVs, blue), germline SNVs (gray), Tier 1 chromosomal translocations (translocations with strong clinical significance, green) and Tier 2 chromosomal translocations (translocations with potential clinical significance, orange) are shown. The outermost track of the Circos plot is a representation of the cytogenetic banding pattern of each chromosome, with the centromere colored red. For the spindle cell/sclerosing RMS tumor, somatic copy number alterations (aqua) are shown in the center track. In the innermost track, dots represent a somatic (blue) or germline (gray) SNV. The position of the dot within the track is representative of the variant allele frequency (VAF) for that SNV: SNVs with a higher VAF are positioned closer to the center of the Circos plot. In the center of the plot, lines link genes that are partners in Tier 1 (green) or Tier 2 (orange) translocations. In this figure, driver mutations for each RMS subtype are bolded (NRAS mutation, MYOD1 mutation and PAX3-FOXO1 translocation) are highlighted.
Treatment for RMS is multidisciplinary, including chemotherapy plus local control via surgical resection and/or radiation therapy. Relapse-free survival rates with this aggressive treatment regimen approach 90% for patients with low-risk disease and 70–80% for patients with localized disease, with significant treatment-associated morbidity 7. However, the 5-year event-free survival rate for patients with metastatic disease at diagnosis continues to be less than 30% 8, and patients with relapsed disease have a similarly dismal prognosis 9. Neither the survival rates nor the side effects of treatment for high-risk RMS have changed appreciably in the last 30 years. Improvement in these survival rates is dependent upon identification of clinically effective agents that target RMS-specific vulnerabilities.
In this consensus manuscript generated from input from clinicians, scientists, patients, and advocacy groups, we summarize the recent progress that has been made in the understanding of RMS biology and advances in RMS treatment. The genetically-engineered Drosophila, zebrafish, or mouse models; human cell lines; or patient-derived xenografts available for basic or translational research are reveiwed (Table 1 and 2). Recent preclinical successes, such as the combination of the WEE1 inhibitor, AZD1775, with irinotecan and vincristine10, will be discussed. We also highlight other drugs and drug combinations that are currently under preclinical study in RMS, including MEK inhibitors in combination with PI3 kinase/mTOR, IGF-1R or CDK4/6 inhibitors 11–13, HDAC inhibitors 14,15, DNA methyltransferase inhibitors 16, PARP inhibitors in combination with temozolomide 17, SMO inhibitors 18, asparaginase 19, and Aurora kinase inhibitors (J Shipley and B Schäfer, personal communication) (Table 2). In addition, we highlight biological and clinical questions that remain unanswered for RMS, as well as new questions that have been identified. We conclude with our recommendations to improve the efficiency of translation of scientific findings into clinical trials.
Table 1:
Available animal models for pediatric RMS. Models highlighted here were either discussed at the Summer’s Way RMS workshop or identified through a comprehensive review of the literature.
| PAX-fusion negative: | ||||
|---|---|---|---|---|
| Organism | Type | Model | Reference | |
| Mouse | Genetically engineered | HGF/SF;Ink4a/Arf−/− M-Cre-Trp53−/− M-Cre-Trp53−/−;Ptch1+/− Myf5-Cre-Trp53−/−;Ptch1+/− Pax7-CreER-Trp53−/−;Ptch1+/− Pax7CE/+, LSL-KrasG12D/+;Trp53Fl/Fl aP2-Cre;SmoM2 aP2-Cre;SmoM2;Cdkn2aFl/Fl |
70 44,48,71 45,47 |
|
| Syngeneic | Myoblast Trp53−/− ;KRASG12D Myoblast Trp53 −/− ;FGFR4V550E |
35,72 | ||
| Xenograft | SkMC/HSMM + T/t-Ag + hTERT + HRASG12V Human cell lines Patient-derived xenografts |
10,50,73 | ||
| Zebrafish | Genetically engineered | rag2-KRASG12D cdh15-KRASG12D mylz2-KRASG12D |
74–76 | |
| Xenograft | Human cell lines Patient-derived xenografts |
|||
| PAX-fusion positive: | ||||
| Organism | Type | Model | Reference | |
| Mouse | Genetically engineered | M-Cre-Pax3-Foxo1;Trp53−/− Myf6-Cre-Pax3-Foxo1;Trp53−/− Stk3F/F;Stk4F/F;Pax3PF/PF;Cdkn2aF/F;Myf6ICN/+ |
49,77–79 | |
| Xenograft | Dbt myoblast + Pax3-Foxo1 + MYCN HSMM + PAX3-FOXO1 + hTERT + MYCN Human cell lines Patient-derived xenografts |
79 51 10,73 |
||
| Zebrafish | Genetically engineered | CMV-GFP2A-PAX3FOXO1;tp53M214K/M214K |
74,80 | |
| Xenograft | Human cell lines Patient-derived xenografts |
|||
| Drosophila | Genetically engineered | Mhc-Gal4;UAS-Pax7-Foxo1 | 81 | |
Table 2:
Preclinical targets in pediatric RMS
| Target | Potential Agent |
|---|---|
| Asparagine metabolism | PEG-asparaginase |
| Aurora kinases | alisertib |
| BRD4 inhibitor | OTX015 |
| CDK4/6 | palbociclib |
| DNA methyltransferases | 5-azacytidine |
| Histone deacetylases | entinostat |
| IGF-1R | ganitumab |
| MEK 1/2 | trametinib |
| NOTCH | RO4929097 |
| PARP | olaparib |
| PI3 kinase/ mTOR | buparlisib |
| SMO | vismodegib |
| VANGL | N/A |
| WEE1 | AZD1775 |
Critical Biological Problems
Significant progress has been made in the last decade in the understanding of the molecular basis for RMS development. These advances in knowledge are the result of several large-scale next generation sequencing studies of primary RMS tumors and extensive mechanistic studies facilitated in part by a variety of animal and xenograft models of RMS (Table 1). Through these genomic and mechanistic studies, RMS biologists have comprehensively characterized the landscape of mutations, copy number changes, genomic rearrangements, DNA methylation and histone modification changes, and defined a number of molecular mechanisms that drive RMS subtypes. The major conclusion of these studies is that there are two molecularly distinct subtypes of childhood RMS, defined by the presence (PAX-fusion positive RMS) or absence (PAX-fusion negative RMS) of a PAX gene rearrangement. Genomic characterization has further revealed several targetable vulnerabilities in these tumor subtypes. Due to this finding, the presence or absence of a PAX-fusion has been incorporated as part of the diagnostic criteria for RMS 4,5.
PAX-fusion positive RMS is associated with one of several balanced chromosomal translocations resulting in the creation of an aberrant transcription factor. In most cases, the N-terminal DNA binding domain of PAX3 is fused to the C-terminal transactivation domain of FOXO1. Less commonly, the DNA binding domain of PAX7 is fused to FOXO1, or rarely, PAX3 is fused to the nuclear receptor coactivator, NCOA1, or the chromatin remodeler, INO80D. The gene expression patterns of tumors driven by any of these aberrant transcription factors are similar, although the PAX3-FOXO1 fusion carries a worse prognosis than other fusions 4,20,21. PAX-fusion positive RMS tumors have a low mutation rate, but commonly exhibit whole genome duplication and focal amplification of MYCN or CDK4 22. PAX3-FOXO1 binds to and seeds the formation of super enhancers found at the loci for MYCN and the myogenic master transcription factors 23. There are areas of DNA hyper-and hypomethylation that can distinguish between the PAX-fusion positive and negative subtypes24. These recurrent epigenetic changes suggest that the use of epigenetic modulators might have therapeutic benefit for patients with PAX-fusion positive RMS.
PAX-fusion negative RMS tumors, in contrast, have a higher rate of single nucleotide variations. Recurrent mutations in known cancer genes such as HRAS, NRAS, KRAS, ALK, FGFR4, PIK3CA, FBXW7, NF1, TP53, CTNNB1, or BCOR are found in this subtype, yet some sequenced tumors do not have an identifiable driver mutation 22,25,26. Importantly, we now appreciate that the majority of fusion negative RMS tumors are driven by RAS pathway activation. However, the prognostic implications of the mutations in these known cancer genes have yet to be defined. Loss of imprinting at chromosome 11p15.5, leading to paternal isodisomy and resulting overexpression of IGF2 is nearly universal among fusion negative RMS tumors. These tumors also have complex karyotypes owing to chromosome and chromosome-arm level gains and losses 22. In addition to loss of function TP53 mutations, PAX-fusion negative tumors have focal amplification of the TP53 negative regulator, MDM2 27, such that altered TP53-dependent transcription is a common feature of these tumors. The DNA methylation pattern in PAX-fusion negative tumors is similar to that of normal tissues 24, but super enhancers are observed in these tumors at MYC, negative regulators of MAP kinase signaling, and the myogenic master transcription factors 13. While PAX-fusion negative RMS is associated with several familial cancer predisposition syndromes, including Li-Fraumeni syndrome and the RASopathies, particularly neurofibromatosis type 1 and Costello syndrome, most children diagnosed with PAX-fusion negative RMS do not have a family history of cancer28. A better understanding of the risk factors for RMS development in these patients is needed.
Sclerosing/spindle cell RMS has recently been characterized as a separate RMS subtype. This aggressive sarcoma is frequently driven by mutations in the myogenic master transcription factor, MYOD1 29. Mutations in the RAS or PI3 kinase pathways frequently co-occur with the MYOD1 mutations 6. In addition, a majority of sclerosing/spindle cell RMS tumors also harbor VGLL2-related fusions, with a subset harboring NCOA2 rearrangements 30. Epigenetic and transcriptomic characterization, functional studies, and animal model development is needed to better understand the biology of this RMS subtype.
Ongoing biological questions
Despite the progress made with genomic characterization of PAX-fusion positive and PAX-fusion negative RMS, many important biological questions remain including:
PAX-fusion positive RMS tumors show a higher propensity to metastasize compared to PAX-fusion negative tumors, and loss of TP53 increases the invasive potential of PAX-fusion negative RMS tumors in a zebrafish model 31, but what are the mechanisms that govern invasion and metastasis in PAX-fusion positive and negative RMS?
The YAP/TAZ 32,33, RAS/RAF/MEK/ERK 13,34, PI3 kinase/mTOR 35, MYOD/MYF5 36,37, Notch 38–40, WNT 34,41, Hedgehog 42, and EZH2 43 pathways have been implicated in PAX-fusion negative RMS as blocking muscle differentiation; can this knowledge be leveraged diagnostically or therapeutically?
Animal modeling studies have shown that PAX-fusion negative RMS can be initiated from myogenic and non-myogenic (endothelial) precursors, while differentiating fetal myoblasts are most poised to develop PAX-fusion positive RMS 14,44–51. Since the RMS cell of origin influences not only histological identity but also site of disease and response to therapy 14, what are the RMS cell(s) of origin in human disease?
What is the role of immune and other cells in the tumor microenvironment in driving RMS progression, metastasis, and therapy resistance?
What are the risk factors and germline mutations associated with an increased risk of RMS development?
What are the most predictive preclinical models for RMS and can these be exploited for rapid and better prioritization of pre-clinical therapy testing?
How can we best leverage new technologies, such as CRISPR-Cas9 screening of protein domains 52, to identify new drug targets for RMS?
What are the mechanisms by which MYOD1L122R drives spindle cell/sclerosing RMS tumorigenesis?
Critical Clinical Problems
Despite the surge in our understanding of the molecular mechanisms underlying RMS in recent years, the clinical translation of such discoveries has lagged behind. Since 2014, there have been only two interventional trials opened specifically for patients with RMS: one for the upfront treatment of a subgroup of newly diagnosed patients (NCT02567435) and one for the treatment of patients with relapsed or refractory disease (NCT03041701). The problem with the lack of newly opened trials is two-fold; questions about which new scientific findings may have clinical benefit or applicability are left unanswered, and patients have limited access to experimental treatment options, which they are in dire need of after standard therapies have been exhausted.
Notably, the rationale for each of the two aforementioned trials is based on substantial preclinical data implicating the importance of the relevant pathways in RMS. The upfront trial is a Children’s Oncology Group (COG) study for patients with newly diagnosed intermediate risk RMS comparing vincristine, actinomycin D and cyclophosphamide alternating with vincristine and irinotecan (VAC/VI) with VAC/VI plus temsirolimus for this subgroup (NCT02567435). This study was initiated following the outcome of a prior COG study showing superiority of a temsirolimus containing regimen for RMS patients at relapse 53, as well as abundant preclinical data showing the importance of the mTOR pathway for RMS survival and growth 54,55. The trial for patients with relapsed or refractory RMS is a phase I/II study investigating the safety and efficacy of the combination of the IGF-1R monoclonal antibody, ganitumab with the SRC family kinase inhibitor, dasatinib (NCT03041701). Earlier preclinical work described the efficacy of small molecule and antibody-based inhbitors of IGF-1R in RMS 56, and previous early phase clinical studies demonstrated that IGF-1R antibodies yielded meaningful but short-lived responses in patients with relapsed RMS 57. However, the addition of an IGF-1R antibody to upfront intensive multiagent chemotherapy did not improve outcomes for unselected patients with metastatic disease 58. The current trial is based on further preclinical work showing that inhibition of IGF-1R activates a SRC family kinase bypass resistance pathway. Cotargeting IGF-1R with a monoclonal antibody such as ganitumab and SRC family kinases with dasatinib provided therapeutic enhancement in animal models 59, which supported clinical translation of this combination.
Despite the lack of RMS-specific clinical trials that have been initiated in recent years, several early phase clinical trials have been initiated for patients with solid tumors or sarcomas that include patients with RMS among the eligible participants. These include studies of new cytotoxic agents or new cytotoxic combinations; targeted agents; immunotherapeutic agents or modalities and allogeneic cellular transplants; or new applications of local control methods such as hyperthermic intraperitoneal chemotherapy60. Additional details for these trials can be found in Table 3. For several of these trials, promising preclinical data exist to support pursuit of these therapeutic targets and agents in RMS 11,14,34,35,54,61–65. However, for many, minimal or no published preclinical data exist, and there is a limitation with currently available RMS models to adequately evaluate some of these therapies (e.g. the need to evaluate immunotherapeutics in immune competent animal models). In addition, since these types of clinical studies typically enroll a small number of patients with each tumor subtype, they rarely provide sufficient information about activity in a given tumor type. Furthermore, patients treated on these smaller early phase studies typically are heavily pre-treated with a high burden of disease, which may make interpretation of outcomes difficult.
Table 3:
Early Phase Clinical Trials for which Patients with RMS are Eligible
| Agent | Class/ Molecular Target | NCT Number | Date of Initiation |
|---|---|---|---|
| eribulin | Cytotoxic |
NCT03441360 NCT03245450 |
2/21/18 8/10/17 |
| Nab-paclitaxel | Cytotoxic |
NCT02945800 NCT03507491 |
10/26/16 4/25/18 |
| PEN-866 | Cytotoxic | NCT03221400 | 7/8/17 |
| afatinib | Targeted - ErbB | NCT02372006 | 2/26/15 |
| cabozantinib | Targeted/ multi-kinase | NCT02867592 | 8/16/16 |
| erlotinib | Targeted - EGFR | NCT02689336 | 2/23/16 |
| copanlisib | Targeted - PI3 kinase | NCT03458728 | 3/8/18 |
| lenvatinib | Targeted - multi-kinase | NCT03245151 | 8/10/17 |
| entinostat | Targeted - HDAC | NCT02780804 | 5/24/18 |
| Pediatric MATCH | Targeted – multiple | NCT03155620 | 5/16/17 |
| HER2 CAR T cell | Immunotherapy - HER2 | NCT00902044 | 5/14/09 |
| nivolumab/ipilimumab | Immunotherapy | NCT02304458 | 12/2/14 |
| enoblituzumab | Immunotherapy | NCT02982941 | 12/6/16 |
| Allogeneic HSCT | Transplant |
NCT02890758 NCT02508038 |
9/7/16 3/24/15 |
| High intensity focus ultrasound | Local control |
NCT02557854 NCT02536183 |
9/23/15 8/31/15 |
| Stereotactic body radiation therapy | Local control | NCT02581384 | 10/21/15 |
| Hyperfractionated radiation therapy | Local control | NCT03651375 | 8/29/18 |
Ongoing clinical questions
Given that there have been so few RMS-specific clinical trials despite the advances that have been made in understanding the biology of this disease, a number of important questions remain regarding how best to move agents from the bench to the bedside and design informative trials. These include:
What is the threshold for preclinical data that is sufficient to initiate a clinical trial?
Which new drugs/pathways should be prioritized?
How should we address the disease-free period that high-risk patients experience between end of therapy and relapse? Should we be giving maintenance therapy 66, and if so, with what?
How can we better provide local control in sites such as the abdomen and pelvis where high local failure rates continue?
Since RMS has a relatively low mutational burden and is unlikely to be immunogenic, how can we best leverage immunotherapeutic treatment options against RMS? Can we improve our modeling of these agents with development of humanized animal models?
How can we best design rational combinations of targeted agents, conventional chemotherapeutics, and/or immunotherapeutics?
How can we better engage the adult sarcoma centers to participate in RMS-specific trials?
Can we specifically target the clinically aggressive, MYOD1-mutant, spindle cell/sclerosing RMS subtype 67?
Future Directions and Consensus Goals
The primary concerns of both investigators and patient advocates centers on eliminating the deleterious side effects of the available treatment options and optimizing translation of preclinical findings into clinical trials. To best address this, the criteria by which drugs are selected for inclusion in clinical trials must be standardized. In addition, clinical trials must be designed such that our ability to build upon our knowledge of RMS biology to inform future trials is maximized. Finally, improved access to information about clinical trials should be provided to patients with known poor prognosis and for patients who are expected to achieve remission with severe long-term sequelae. Specific recommendations to achieve these goals are outlined below.
Initiate RMS-specific clinical trials based upon robust preclinical work
Our consensus opinion, based on the currently available preclinical data, is that the combination of the WEE1 inhibitor, AZD1775, with the chemotherapeutic agents vincristine and irinotecan should be prioritized for evaluation in a clinical trial for patients with RMS. WEE1 is a tyrosine kinase that is activated in response to DNA damage. WEE1 phosphorylates and inactivates CDK1, which halts progression through the G2/M checkpoint and allows for DNA repair prior to initiation of mitosis. WEE1 inhibition in the setting of chemotherapy-induced DNA damage leads to mitotic catastrophe. AZD1775 has been studied preclinically in RMS 64 as well as in a COG Phase I trial in combination with irinotecan (NCT02095132), but the number of patients with RMS enrolled on that study was small, and the results have not yet been reported.
We as a community feel comfortable proposing AZD1775 in combination with irinotecan and vincristine as the next clinical trial for patients with initially metastatic or relapsed/refractory RMS. However, we encourage investigators to establish pharmacodynamic markers, such as assays for DNA damage, in animal models of RMS treated with this combination in preparation for potential use as early response markers for patients receiving AZD1775/vincristine/irinotecan on study. As outlined above, additional therapies that warrant further preclinical testing include bromodomain inhibitors 23 and HDAC inhibitors in PAX-fusion positive RMS and MEK inhibitors in RAS-driven PAX-fusion negative RMS. We would encourage investigators that are engaged in preclinical research to be mindful of the criteria needed to support the clinical translation of novel drugs (see Table 4) and to design experiments that attempt to address these questions. Finally, we encourage investigators to make use of novel clinical trial designs, such as basket trials, so that the clinical efficacy of new drugs and combinations can be assessed rapidly with a minimum number of enrolled patients.
Table 4:
Criteria for Prioritization of Drugs for Translation to Clinical Trials in Pediatric RMS. We propose that at least 6 of these criteria be met for an agent to be prioritized for clinical translation, and meeting criteria 4 and 5 should be required.
| 1. | Is there a biomarker identified in human subjects that predicts sensitivity to targeting the pathway (i.e. SNV, amplification, deletion, etc)? |
| 2. | Does the tumor depend on this target in vitro? |
| 3. | Does the tumor depend on this target in vivo? |
| 4. | Is the proposed drug efficacious in vitro (5 – 8 independent cell lines, if available)? |
| 5. | Is the proposed drug efficacious in vivo (at least 3 independent models, including genetically engineered, cell line xenograft and patient derived xenograft models)? |
| 6. | Does the presence of the biomarker from question 1 predict response to the proposed drug in vitro and in vivo? |
| 7. | Are the concentrations of proposed drug required for efficacy in vitro achieved in vivo? Achievable in patients? |
| 8. | Are resistance mechanisms to the proposed drug known? |
| 9. | Are there drugs with which the proposed drug synergizes in vitro and in vivo? |
| 10. | Is the proposed drug formulated in such a way that it can be delivered to pediatric patients? |
Maximize information learned from each patient diagnosed with RMS
We recognize that our best resource for understanding RMS biology and for developing therapies that improve survival while minimizing side effects of treatment are the patients with RMS themselves. Since RMS is a rare disease, each patient who is diagnosed with RMS in North America be offered enrollment on the COG study, Project:EveryChild (NCT02402244). This project aims to create both a database of clinical data as well as a biorepository of disease-specific specimens. In addition, for patients with relapsed RMS who have somatic mutational analysis indicating an actionable finding, clinicians should offer enrollment on a clinical trial, such as the Pediatric MATCH in North America (NCT03155620) or ESMART (NCT02813135) in Europe. In this way, RMS-specific responses to these agents can be prospectively evaluated. Furthermore, efforts should be made to incorporate on-treatment tumor and liquid biopsies into treatment trials for newly diagnosed and relapsed patients. Tumor tissue and circulating tumor cells can be used to evaluate target-specific pharmacodynamic markers, while circulating tumor DNA and exosomes can be used as early markers of response to therapy 68. These studies are essential for determining that the intended target is engaged by the drug administered, as well as for improving our understanding of intrinsic and acquired resistance to therapies. This knowledge will, in turn, inform future clinical trials. As well, establishing liquid biopsies and newer nuclear medicine imaging techniques such as FLT-PET as early markers of disease response will facilitate completion of trials in a timely manner, such that follow-up trials that build upon knowledge gained from our current trials can begin.
Establish international, multi-disciplinary research teams to facilitate discovery
Several of the critical biological problems described above are currently being investigated by more than one of the members of the RMS community. For example, many investigators are interested in targeting the myogenic transcription factor, MYOD1, while several investigators are interested in targeting oncogenic RAS in RMS. We suggest that investigators continue to assemble into collaborative groups aimed at efficiently translating understanding of these sub-topics of RMS biology into clinical trials and encourage these groups to collectively pursue funding opportunities to support this type of research. To facilitate data sharing, the COG is working to establish a centralized database to provide all investigators access to the genomic and clinical outcome data that has already been generated.
In conclusion, we predict that the research described above, conducted by our collaborative community of investigators, has the potential to produce additional RMS-specific clinical trials in the near future. In the next phase of RMS research, we aim to improve upon those trials with the ultimate goal of understanding RMS biology and identifying treatments for RMS that provide meaningful clinical benefit and minimize toxicity.
Acknowledgements
We thank the clinicians, basic scientists, and translational researchers as well as patients, their families, and representatives from five RMS patient advocacy groups (Summer’s Way, Focus on Rhabdo, Friends of TJ, Super Sam Foundation, Rein in Sarcoma, and Sarcoma Patient Alliance Coalition) that assembled for the Summer’s Way RMS workshop in May 2018. Many are authors on this consensus manuscript. The authors of this manuscript certify they have no conflicts of interest. We are also grateful to S.X. Skapek for attendance at the Summer’s Way Rhabdomyosarcoma workshop and critical reading of the manuscript. This was the third such international workshop to follow those prevsiously held at the Banbury Center at Cold Spring Harbor Laboratory in May of 2014 69and the National Cancer Institute in April of 2015. This work was supported by R24OD016761 (D.M.L.), R01CA154923 (D.M.L.), R01CA215118, (D.M.L.), R01 R01CA226926 (D.M.L), and the Liddy Shriver Sarcoma Initiative.
Abbreviations:
- COG
Children’s Oncology Group
- HDAC
histone deacetylase
- PARP
poly (ADP-ribose) polymerase
- RMS
rhabdomyosarcoma
- VAC
vincristine, actinomycin D, cyclophosphamide
- VI
vincristine, irinotecan
References
- 1.Shern JF, Yohe ME, Khan J. Pediatric Rhabdomyosarcoma. Crit Rev Oncog 2015;20(3–4):227–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer 2015;15(7):426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuchi K, Keller C. The not-so-skinny on muscle cancer. Cancer Cell 2012;22(4):421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Missiaglia E, Williamson D, Chisholm J, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol 2012;30(14):1670–1677. [DOI] [PubMed] [Google Scholar]
- 5.Skapek SX, Anderson J, Barr FG, et al. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children’s oncology group report. Pediatr Blood Cancer 2013;60(9):1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agaram NP, LaQuaglia MP, Alaggio R, et al. MYOD1-mutant spindle cell and sclerosing rhabdomyosarcoma: an aggressive subtype irrespective of age. A reappraisal for molecular classification and risk stratification. Mod Pathol 2019;32(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malempati S, Hawkins DS. Rhabdomyosarcoma: review of the Children’s Oncology Group (COG) Soft-Tissue Sarcoma Committee experience and rationale for current COG studies. Pediatr Blood Cancer 2012;59(1):5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudzinski ER, Anderson JR, Chi YY, et al. Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: A report from the Children’s Oncology Group. Pediatr Blood Cancer 2017;64(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pappo AS, Anderson JR, Crist WM, et al. Survival after relapse in children and adolescents with rhabdomyosarcoma: A report from the Intergroup Rhabdomyosarcoma Study Group. J Clin Oncol 1999;17(11):3487–3493. [DOI] [PubMed] [Google Scholar]
- 10.Stewart E, Federico SM, Chen X, et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017;549(7670):96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renshaw J, Taylor KR, Bishop R, et al. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res 2013;19(21):5940–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart E, McEvoy J, Wang H, et al. Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell 2018. [DOI] [PMC free article] [PubMed]
- 13.Yohe ME, Gryder BE, Shern JF, et al. MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci Transl Med 2018;10(448). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abraham J, Nunez-Alvarez Y, Hettmer S, et al. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes Dev 2014;28(14):1578–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bharathy N, Berlow NE, Wang E, et al. The HDAC3-SMARCA4-miR-27a axis promotes expression of the PAX3:FOXO1 fusion oncogene in rhabdomyosarcoma. Sci Signal 2018;11(557). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ecke I, Petry F, Rosenberger A, et al. Antitumor effects of a combined 5-aza-2’deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res 2009;69(3):887–895. [DOI] [PubMed] [Google Scholar]
- 17.Yan CB, Brunson DC, Tang Q, et al. Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 2019;epub. [DOI] [PMC free article] [PubMed]
- 18.Geyer N, Ridzewski R, Bauer J, et al. Different Response of Ptch Mutant and Ptch Wildtype Rhabdomyosarcoma Toward SMO and PI3K Inhibitors. Front Oncol 2018;8:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hettmer S, Schinzel AC, Tchessalova D, et al. Functional genomic screening reveals asparagine dependence as a metabolic vulnerability in sarcoma. Elife 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res 2006;66(14):6936–6946. [DOI] [PubMed] [Google Scholar]
- 21.Williamson D, Missiaglia E, de Reynies A, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol 2010;28(13):2151–2158. [DOI] [PubMed] [Google Scholar]
- 22.Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4(2):216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gryder BE, Yohe ME, Chou HC, et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 2017;7(8):884–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W, Chatterjee B, Wang Y, et al. Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma. Mod Pathol 2015;28(9):1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Shern JF, Wei JS, et al. Clonality and evolutionary history of rhabdomyosarcoma. PLoS Genet 2015;11(3):e1005075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Stewart E, Shelat AA, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 2013;24(6):710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L, Zheng Y, Liu J, et al. Integrative Bayesian Analysis Identifies Rhabdomyosarcoma Disease Genes. Cell Rep 2018;24(1):238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupo PJ, Danysh HE, Plon SE, et al. Family history of cancer and childhood rhabdomyosarcoma: a report from the Children’s Oncology Group and the Utah Population Database. Cancer Med 2015;4(5):781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohsaka S, Shukla N, Ameur N, et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet 2014;46(6):595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alaggio R, Zhang L, Sung YS, et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am J Surg Pathol 2016;40(2):224–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ignatius MS, Hayes MN, Moore FE, et al. tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. Elife 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deel MD, Slemmons KK, Hinson AR, et al. The Transcriptional Coactivator TAZ Is a Potent Mediator of Alveolar Rhabdomyosarcoma Tumorigenesis. Clin Cancer Res 2018;24(11):2616–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tremblay AM, Missiaglia E, Galli GG, et al. The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell 2014;26(2):273–287. [DOI] [PubMed] [Google Scholar]
- 34.Chen EY, DeRan MT, Ignatius MS, et al. Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc Natl Acad Sci U S A 2014;111(14):5349–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinnon T, Venier R, Yohe M, et al. Functional screening of FGFR4-driven tumorigenesis identifies PI3K/mTOR inhibition as a therapeutic strategy in rhabdomyosarcoma. Oncogene 2018;37(20):2630–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenente IM, Hayes MN, Ignatius MS, et al. Myogenic regulatory transcription factors regulate growth in rhabdomyosarcoma. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, MacQuarrie KL, Analau E, et al. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev 2009;23(6):694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ignatius MS, Hayes MN, Lobbardi R, et al. The NOTCH1/SNAIL1/MEF2C Pathway Regulates Growth and Self-Renewal in Embryonal Rhabdomyosarcoma. Cell Rep 2017;19(11):2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raimondi L, Ciarapica R, De Salvo M, et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21(Cip1) expression and hampers tumour cell growth in vitro and in vivo. Cell Death Differ 2012;19(5):871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slemmons KK, Crose LES, Riedel S, Sushnitha M, Belyea B, Linardic CM. A Novel Notch-YAP Circuit Drives Stemness and Tumorigenesis in Embryonal Rhabdomyosarcoma. Mol Cancer Res 2017;15(12):1777–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes MN, McCarthy K, Jin A, et al. Vangl2/RhoA Signaling Pathway Regulates Stem Cell Self-Renewal Programs and Growth in Rhabdomyosarcoma. Cell Stem Cell 2018;22(3):414–427 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teot LA, Schneider M, Thorner AR, et al. Clinical and mutational spectrum of highly differentiated, paired box 3:forkhead box protein o1 fusion-negative rhabdomyosarcoma: A report from the Children’s Oncology Group. Cancer 2018;124(9):1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciarapica R, Carcarino E, Adesso L, et al. Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS. BMC Cancer 2014;14:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blum JM, Ano L, Li Z, et al. Distinct and overlapping sarcoma subtypes initiated from muscle stem and progenitor cells. Cell Rep 2013;5(4):933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond CJ, Hanna JA, Garcia MR, et al. Hedgehog Pathway Drives Fusion-Negative Rhabdomyosarcoma Initiated From Non-myogenic Endothelial Progenitors. Cancer Cell 2018;33(1):108–124 e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geltzeiler M, Li G, Abraham J, Keller C. The case for primary salivary rhabdomyosarcoma. Front Oncol 2015;5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatley ME, Tang W, Garcia MR, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell 2012;22(4):536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hettmer S, Liu J, Miller CM, et al. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc Natl Acad Sci U S A 2011;108(50):20002–20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev 2004;18(21):2608–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer Res 2005;65(11):4490–4495. [DOI] [PubMed] [Google Scholar]
- 51.Naini S, Etheridge KT, Adam SJ, et al. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer Res 2008;68(23):9583–9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 2015;33(6):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mascarenhas L, Meyer WH, Lyden E, et al. Randomized phase II trial of bevacizumab and temsirolimus in combination with vinorelbine (V) and cyclophosphamide (C) for first relapse/disease progression of rhabdomyosarcoma (RMS): A report from the Children’s Oncology Group (COG). Journal of Clinical Oncology 2014;32(15_suppl):10003–10003. [Google Scholar]
- 54.Hawkins DS, Spunt SL, Skapek SX, Committee COGSTS. Children’s Oncology Group’s 2013 blueprint for research: Soft tissue sarcomas. Pediatr Blood Cancer 2013;60(6):1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le X, Pugach EK, Hettmer S, et al. A novel chemical screening strategy in zebrafish identifies common pathways in embryogenesis and rhabdomyosarcoma development. Development 2013;140(11):2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan X, Helman LJ. Levels of PTEN protein modulate Akt phosphorylation on serine 473, but not on threonine 308, in IGF-II-overexpressing rhabdomyosarcomas cells. Oncogene 2003;22(50):8205–8211. [DOI] [PubMed] [Google Scholar]
- 57.Pappo AS, Vassal G, Crowley JJ, et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research Through Collaboration study. Cancer 2014;120(16):2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malempati S, Weigel BJ, Chi YY, et al. The addition of cixutumumab or temozolomide to intensive multiagent chemotherapy is feasible but does not improve outcome for patients with metastatic rhabdomyosarcoma: A report from the Children’s Oncology Group. Cancer 2018. [DOI] [PMC free article] [PubMed]
- 59.Wan X, Yeung C, Heske C, Mendoza A, Helman LJ. IGF-1R Inhibition Activates a YES/SFK Bypass Resistance Pathway: Rational Basis for Co-Targeting IGF-1R and Yes/SFK Kinase in Rhabdomyosarcoma. Neoplasia 2015;17(4):358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zmora O, Hayes-Jordan A, Nissan A, et al. Cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC) for disseminated intra-abdominal malignancies in children-a single-institution experience. J Pediatr Surg 2018;53(7):1381–1386. [DOI] [PubMed] [Google Scholar]
- 61.Chen EY, Dobrinski KP, Brown KH, et al. Cross-species array comparative genomic hybridization identifies novel oncogenic events in zebrafish and human embryonal rhabdomyosarcoma. PLoS Genet 2013;9(8):e1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gee MF, Tsuchida R, Eichler-Jonsson C, Das B, Baruchel S, Malkin D. Vascular endothelial growth factor acts in an autocrine manner in rhabdomyosarcoma cell lines and can be inhibited with all-trans-retinoic acid. Oncogene 2005;24(54):8025–8037. [DOI] [PubMed] [Google Scholar]
- 63.Heske CM, Mendoza A, Edessa LD, et al. STA-8666, a novel HSP90 inhibitor/SN-38 drug conjugate, causes complete tumor regression in preclinical mouse models of pediatric sarcoma. Oncotarget 2016;7(40):65540–65552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kahen E, Yu D, Harrison DJ, et al. Identification of clinically achievable combination therapies in childhood rhabdomyosarcoma. Cancer Chemother Pharmacol 2016;78(2):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolb EA, Gorlick R, Reynolds CP, et al. Initial testing (stage 1) of eribulin, a novel tubulin binding agent, by the pediatric preclinical testing program. Pediatr Blood Cancer 2013;60(8):1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klingebiel T, Boos J, Beske F, et al. Treatment of children with metastatic soft tissue sarcoma with oral maintenance compared to high dose chemotherapy: report of the HD CWS-96 trial. Pediatr Blood Cancer 2008;50(4):739–745. [DOI] [PubMed] [Google Scholar]
- 67.Agaram NP, LaQuaglia MP, Alaggio R, et al. MYOD1-mutant spindle cell and sclerosing rhabdomyosarcoma: an aggressive subtype irrespective of age. A reappraisal for molecular classification and risk stratification. Mod Pathol 2018. [DOI] [PMC free article] [PubMed]
- 68.Shulman DS, Klega K, Imamovic-Tuco A, et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children’s Oncology Group. Br J Cancer 2018;119(5):615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hettmer S, Li Z, Billin AN, et al. Rhabdomyosarcoma: current challenges and their implications for developing therapies. Cold Spring Harb Perspect Med 2014;4(11):a025650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharp R, Recio JA, Jhappan C, et al. Synergism between INK4a/ARF inactivation and aberrant HGF/SF signaling in rhabdomyosarcomagenesis. Nat Med 2002;8(11):1276–1280. [DOI] [PubMed] [Google Scholar]
- 71.Rubin BP, Nishijo K, Chen HI, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell 2011;19(2):177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKinnon T, Venier R, Dickson BC, et al. Kras activation in p53-deficient myoblasts results in high-grade sarcoma formation with impaired myogenic differentiation. Oncotarget 2015;6(16):14220–14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol 2013;3:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayes MN, Langenau DM. Discovering novel oncogenic pathways and new therapies using zebrafish models of sarcoma. Methods Cell Biol 2017;138:525–561. [DOI] [PubMed] [Google Scholar]
- 75.Langenau DM, Keefe MD, Storer NY, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev 2007;21(11):1382–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Storer NY, White RM, Uong A, et al. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development 2013;140(14):3040–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keller C, Capecchi MR. New genetic tactics to model alveolar rhabdomyosarcoma in the mouse. Cancer Res 2005;65(17):7530–7532. [DOI] [PubMed] [Google Scholar]
- 78.Oristian KM, Crose LES, Kuprasertkul N, et al. Loss of MST/Hippo Signaling in a Genetically Engineered Mouse Model of Fusion-Positive Rhabdomyosarcoma Accelerates Tumorigenesis. Cancer Res 2018;78(19):5513–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandey PR, Chatterjee B, Olanich ME, et al. PAX3-FOXO1 is essential for tumour initiation and maintenance but not recurrence in a human myoblast model of rhabdomyosarcoma. J Pathol 2017;241(5):626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kendall GC, Watson S, Xu L, et al. PAX3-FOXO1 transgenic zebrafish models identify HES3 as a mediator of rhabdomyosarcoma tumorigenesis. Elife 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galindo RL, Allport JA, Olson EN. A Drosophila model of the rhabdomyosarcoma initiator PAX7-FKHR. Proc Natl Acad Sci U S A 2006;103(36):13439–13444. [DOI] [PMC free article] [PubMed] [Google Scholar]