Summary
Regulatory T cells (Treg cells) deficient in the transcription factor Foxp3 lack suppressor function and manifest a T effector (Teff) cell-like phenotype. We demonstrate that Foxp3 deficiency dysregulates metabolic checkpoint kinase mTORC2 signaling and gives rise to augmented aerobic glycolysis and oxidative phosphorylation. Specific deletion of the mTORC2 adaptor gene Rictor in Foxp3-deficient Treg cells ameliorated disease in a Foxo1 transcription factor-dependent manner. Rictor deficiency reestablished a subset of Treg cell genetic circuits and suppressed the Teff cell-like glycolytic and respiratory programs, which contributed to immune dysregulation. Treatment of Treg cells from patients with FOXP3 deficiency with mTOR inhibitors similarly antagonized their Teff cell-like program and restored suppressive function. Thus, regulatory function can be reestablished in Foxp3-deficient Treg cells by targeting their metabolic pathways, providing opportunities to restore tolerance in Treg cell disorders.
Introduction
Regulatory T cells (Treg cells) enforce peripheral immunological tolerance by suppressing immune responses to self-antigens and innocuous environmental antigens 1, 2. The Forkhead transcription factor Foxp3 is essential for Treg cell differentiation and function 1, 3, 4. While Foxp3 is not essential for thymic Treg cell development, Foxp3-deficient Treg cells (ΔTreg cells) lack regulatory function 3, 4. The core Treg cell transcriptome and epigenome are largely preserved in ΔTreg cells, although the expression of individual genes is frequently decreased 3, 4, 5. Nevertheless, ΔTreg cells acquire additional phenotypic and transcriptional attributes akin to those of effector T (Teff) cell 3, 4, 6. These findings led to the concept that Foxp3 stabilizes the transcriptome of Treg cells and prevents their degeneration into Teff cells 7.
Teff cells undergo a profound change in their bioenergetic profile in favor of augmented aerobic glycolysis, oxidative phosphorylation (OXPHOS) and glutaminolysis as well as the de novo acquisition of biosynthetic pathways such as fatty acid synthesis 8. Treg cell metabolism is biased towards fatty acid oxidation (FAO) and pyruvate-dependent OXPHOS 9, 10, 11, 12, whereas glycolysis is kept under strict control. Enforced expression of Foxp3 promotes OXPHOS and suppresses glycolysis 13. Reciprocally, upregulation of the glucose transporter Glut1 by toll-like receptor signaling, acting via the mammalian Target of Rapamycin Complex 1 (mTORC1), or by its enforced expression promoted Treg cell proliferation but dampened Foxp3 expression and Treg cell suppressive function 14. We reasoned that interventions that deprive the Teff cell-like programs of ΔTreg cells of metabolic support may allow recovery of regulatory function by virtue of the preservation in those cells of a core regulatory transcriptome.
Results
Lineage-specific, Cre-mediated recombination in Foxp3-deficient Treg cells
To enable genetic manipulations in ΔTreg cells, we derived a bicistronic loss of function Foxp3 allele, Foxp3ΔEGFPiCre, that contained a Frt element inserted between exons 9 and 10 that created an aberrant 3’ splice junction site, leading to a premature stop codon in the Foxp3 transcript. A ribosomal entry sequence inserted in the 3’ untranslated region of Foxp3 directed the expression of a humanized Cre recombinase (iCre) fused with the enhanced green fluorescent protein (EGFP) (Supplementary Fig. 1a–d). Treg cells of Foxp3ΔEGFPiCre mice expressed EGFP but not Foxp3 (Supplementary Fig. 1e, f). Analysis of Foxp3ΔEGFPiCre mice harboring a Cre-inducible yellow fluorescent protein (YFP) transgene in the Rosa26 locus (R26YFP) revealed that all EGFP+ cells were YFP+ and virtually all within the CD4+ T cell population (Supplementary Figure 1e). Comparison with wild-type mice carrying a bacterial artificial chromosome (BAC) harboring a Foxp3 promoter-driven EGFP and Cre recombinase and crossed with R26YFP (Foxp3EGFPCreR26YFP) showed that the EGFP+ cells of Foxp3ΔEGFPiCre mice were expanded, in agreement with previous studies on Foxp3-deficient mice (Supplementary Fig. 1e) 3, 4. Within the YFP+ cell population, the frequency of cells that have downregulated their Foxp3 locus activity (EGFP–YFP+) was modestly increased in the Foxp3ΔEGFPiCreR26YFP males, pointing to incremental ΔTreg cell instability.
Analysis of Foxp3EGFPCre and Foxp3ΔEGFPiCre EGFP– and EGFP+ CD4+ T cells revealed that Foxp3 transcripts were highly enriched in EGFP+ cells (Supplementary Fig. 1g). Foxp3 transcripts were 20 fold higher in Foxp3EGFPCre EGFP+ compared to Foxp3ΔEGFPiCre EGFP+ cells, reflecting a positive feed-forward regulation by Foxp3 of its own gene expression (Supplementary Fig. 1g) 4. EGFP+ ΔTreg cells exhibited similar demethylation of the Foxp3 CNS2 promoter region as EGFP+ control Treg cells from Foxp3EGFP knockin reporter allele mice, indicating that the ΔTreg cells were epigenetically of Treg cell lineage (Supplementary Fig. 1h).
Analysis of Foxp3ΔEGFPiCre/+ heterozygous females revealed that similar to the indigenous wild-type Foxp3 allele, expression of Foxp3ΔEGFPiCre in thymocytes was overwhelmingly restricted to the CD4+ lineage, starting at the CD4 single positive thymocyte stage (Supplementary Fig. 1i). However, the ΔTreg cells were dramatically decreased in the periphery compared to wild-type Treg cells, indicative of their reduced fitness (Supplementary Fig. 1i) 3, 4. Overall, these results established that Foxp3ΔEGFPiCre allele was specifically expressed in the Treg cell lineage, and rendered these cells totally deficient in Foxp3 while enabling EGFP and Cre recombinase expression.
Inactivation of mTORC2 in ΔTreg cells ameliorates disease
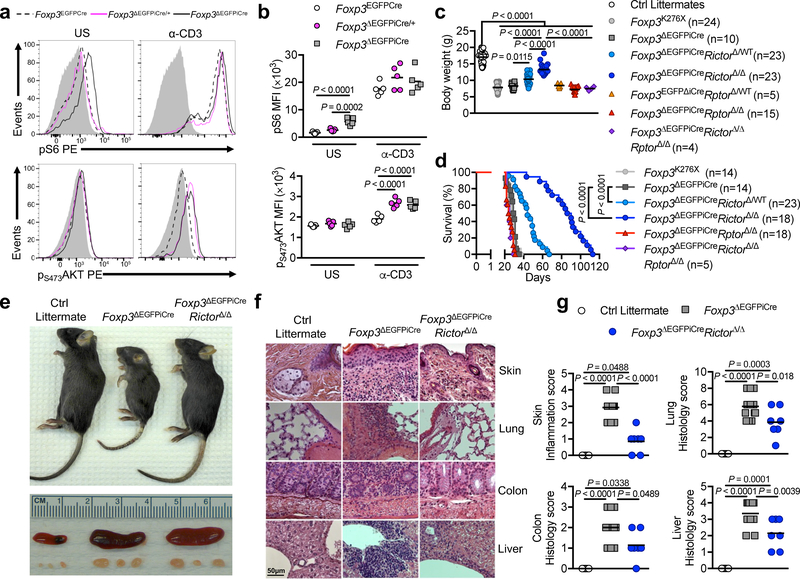
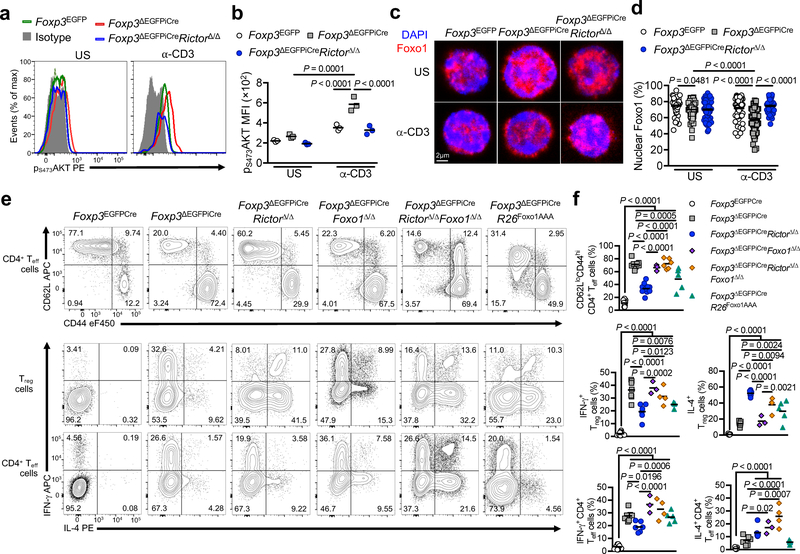
The mTOR pathways play a key role in promoting Teff cell function 15, 16. The mTOR kinase exists as part of two complexes: mTORC1 and mTORC2, that differentially contribute to TH cell effector functions, while mTOR-deficient T cells differentiate in vitro into iTreg cells 17, 18, 19. mTORC1 acts as a finely-tuned Treg cell metabolic checkpoint essential for Treg cell homeostasis and function 20. Activated mTORC1 phosphorylates the downstream substrates S6 and 4EBP, while activated mTORC2 phosphorylates the serine/threonine kinase AKT at serine residue 473 (pS473AKT) 20, 21. ΔTreg cells from Foxp3ΔEGFPiCre hemizygous mutant males, but not from heterozygous Foxp3ΔEGFPiCre/+ females, had increased pS6 and p4EBP at steady state compared to Foxp3-sufficient Treg cells from Foxp3EGFPCre mice (Fig. 1a,b and Supplementary Fig. 2a,b). However, both ΔTreg cell populations upregulated their pS6 phosphorylation in response to anti-CD3 mAb stimulation to the same extent as Foxp3-sufficient Treg cells, indicating that increased basal mTORC1 activity in male ΔTreg cells was cell-extrinsic. Induction of pT308AKT, a target of upstream Phosphoinositide-dependent kinase 22, was also increased to a similar extent in Treg and ΔTreg cells of Foxp3EGFPCre and Foxp3ΔEGFPiCre mice, respectively, upon anti-CD3 mAb stimulation (Supplementary Fig. 2a,b).
Figure 1. Inactivation of mTORC2 but not mTORC1 in ΔTreg cells mitigates Foxp3 deficiency.
(a,b) Representative flow cytometric analysis (a) and mean fluorescence intensity (MFI) (b) of phosphorylated S6 (pS6) and phosphorylated AKT at Ser473 residu (pS473AKT) expression of unstimulated (US) or anti-CD3 stimulated (α-CD3) Treg cells from Foxp3EGFPCre, Foxp3ΔEGFPiCre/+ and Foxp3ΔEGFPiCre mice (n=5 per group). Results represent 1 of 3 independent experiments. (c,d) Body weight at 25–28 days of age (c) and survival (d) of Foxp3K276X, Foxp3ΔEGFPiCre, Foxp3ΔEGFPiCreRictorΔ/WT, Foxp3ΔEGFPiCreRictorΔ/Δ, Foxp3ΔEGFPiCreRptorΔ/WT, Foxp3ΔEGFPiCreRptorΔ/Δ, Foxp3ΔEGFPiCreRictorΔ/ΔRptorΔ/Δ mice and control littermates. Results represent a pool of 5 independent experiments. (e) Gross appearance of Foxp3ΔEGFPiCre, Foxp3ΔEGFPiCreRictorΔ/Δ and control littermate and their respective spleens and peripheral lymph nodes. (f,g) Representative microscopic pictures of H&E staining (original magnification ×200) (f) and histological scores (g) of skin, lung, colon and liver of Foxp3ΔEGFPiCre (n=14), Foxp3ΔEGFPiCreRictorΔ/Δ (n=7) and control littermates (n=5). Results represent a pool of 3 independent experiments. Statistical significance was determined by a one-way ANOVA with Tukey’s multiple comparisons (c, g), two-way ANOVA with Sidak’s multiple comparisons (b) and log rank test (d) (P values as indicated).
pS473AKT was selectively increased in ΔTreg cells of both hemizygous males and heterozygous females following anti-CD3 mAb stimulation, indicating that Foxp3 deficiency dysregulated mTORC2 activation in a cell intrinsic manner irrespective of the inflammatory environment (Fig. 1a,b). The increased mTORC2 activity in ΔTreg cells was associated with decreased protein expression of phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase phosphatase and tensin homolog (PTEN) and PH-domain leucine-rich-repeat protein phosphatase 1 (PHLPP1), two phosphatases implicated in mTORC2 regulation 23, 24 (Supplementary Fig. 2c). We also evaluated proximal TCR signaling in ΔTreg cells of Foxp3ΔEGFPiCre mice by measuring Zeta-chain-associated protein kinase 70 (ZAP70) phosphorylation at tyrosine 319 residue upon anti-CD3 mAb stimulation, an event critical to downstream calcium mobilization, Ras activation and NFAT-dependent transcription 25. ΔTreg cells exhibited attenuated ZAP70 phosphorylation in response to anti-CD3 mAb stimulation compared to Foxp3-sufficient Treg cells (Supplementary Fig. 2d). These results suggested that the enhanced mTORC2 activation in ΔTreg cells of Foxp3ΔEGFPiCre mice reflected impaired regulation by phosphatases.
Male mice hemizygous for the Foxp3ΔEGFPiCre allele were runted and died early due to autoimmune lymphoproliferation, similar to another Foxp3-deficient mouse strain, Foxp3K276X, that we previously generated 26 (Fig. 1c–f). We examined the role of dysregulated mTOR signaling in ΔTreg cells of Foxp3ΔEGFPiCre mice in their disease by cell-specific deletion of Rptor and Rictor, encoding the mTORC1 and mTORC2 regulators Raptor and Rictor, respectively. Expression of Rptor and Rictor transcripts was specifically abrogated in EGFP+ cells of Foxp3ΔEGFPiCreRptorΔ/Δ and Foxp3ΔEGFPiCreRictorΔ/Δ mice, respectively (Supplementary Fig. 2e,f). Rictor protein expression was increased in EGFP+ cells of Foxp3ΔEGFPiCre mice, mirroring the increase in its transcripts, and was also abrogated in the ΔTreg cells of Foxp3ΔEGFPiCreRictorΔ/Δ mice, as confirmed by immunoblotting (Supplementary Fig. 2g). ΔTreg cell-specific Rictor deletion markedly increased the body weight and survival of Foxp3ΔEGFPiCre mice in a gene dose-dependent manner, whereas Rptor deletion did not (Fig. 1c,d). Furthermore, combined deletion of Rictor and Rptor in ΔTreg cells of Foxp3ΔEGFPiCreRictorΔ/ΔRptorΔ/Δ mice did not increase survival or body weight compared to Foxp3ΔEGFPiCre or Foxp3ΔEGFPiCreRptorΔ/Δ mice (Fig. 1c,d). Foxp3ΔEGFPiCreRictorΔ/Δ mice had decreased tissue inflammation compared to Foxp3ΔEGFPiCre mice but persistent lymphoproliferation (Fig. 1e–g).
The marked increase in ΔTreg cells in Foxp3ΔEGFPiCre mice was reversed in Foxp3ΔEGFPiCreRptorΔ/Δ but not Foxp3ΔEGFPiCreRictorΔ/Δ mice, indicating its dependence on mTORC1 activation (Supplementary Fig. 2h). The phenotype of EGFP+ ΔTreg cells of Foxp3ΔEGFPiCre mice closely resembled that of ΔTreg cells analyzed in earlier models 3, 4. In addition to absent Foxp3 expression, ΔTreg cells had lower expression of CD25 and CD73 and higher expression of ICOS, Helios, GITR, CD127, IL-2 and Granzyme B compared to WT Treg cells (Supplementary Fig. 2i,j and L-M.C., data not shown). ΔTreg cells of Foxp3ΔEGFPiCreRictorΔ/Δ mice had upregulated expression of ICOS, Nrp1, GITR and CD73 compared to ΔTreg cells from Foxp3ΔEGFPiCre mice (Supplementary Fig. 2i,j). These results indicated that mTORC2 was specifically dysregulated in ΔTreg cells in a cell-intrinsic manner, and promoted disease in Foxp3-deficient mice.
Rictor deficiency restores ΔTreg cell suppressor function
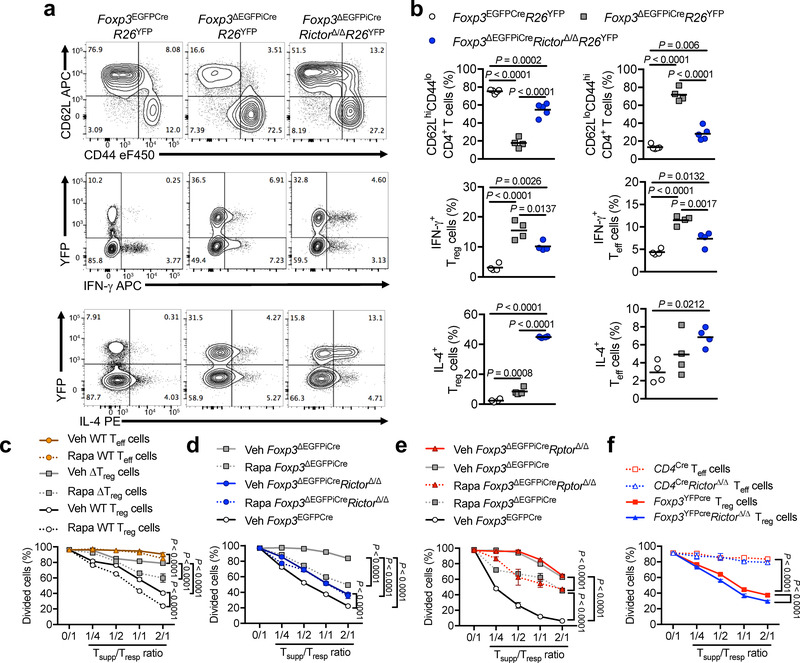
Analysis of Foxp3ΔEGFPiCreRictorΔ/Δ mice revealed that the frequencies of CD4 and CD8 effector memory cells were decreased by about 50% compared to Foxp3ΔEGFPiCre mice, whereas those of naive and central memory T cells were increased (Fig. 2a,b and Supplementary Fig. 3a,b). Expression of the TH1-associated transcription factor T-bet and IFN-γ was markedly increased in ΔTreg and Teff cells of Foxp3ΔEGFPiCre compared to control Foxp3EGFPCre mice, but was down-regulated by Rictor deficiency (Foxp3ΔEGFPiCreRictorΔ/Δ) By contrast, expression of the TH2-associated transcription factor Gata-3 and IL-4, which was also increased in Foxp3ΔEGFPiCre ΔTreg and Teff cells, was further upregulated in ΔTreg cells by Rictor deficiency, thus confirming a critical role for mTORC2 in TH1 but not TH2 programming of ΔTreg cells (Fig. 2a, b and Supplementary Fig. 3a,b). In contrast, expression of ROR-γt and IL-17 was unchanged (Supplementary Fig. 3a,b).
Figure 2. mTORC2 blockade endows ΔTreg cells with regulatory function.
(a,b) Representative flow cytometric analysis (a) and frequencies (scatter plots with mean) (b) of CD62LhiCD44lo and CD62LloCD44hi CD4+ T cells or IFN-γ and IL-4 expression by CD4+ Treg (YFP+) and Teff (YFP–) cells from spleen of Foxp3EGFPCreR26YFP (n=5 for CD62LhiCD44lo and CD62LloCD44hi CD4+ T cells, n=4 for IFN-γ and IL-4 expression), Foxp3ΔEGFPiCreR26YFP (n=4) and Foxp3ΔEGFPiCreRictorΔ/ΔR26YFP mice (n=5 for CD62LhiCD44lo and CD62LloCD44hi CD4+ T cells, n=4 for IFN-γ and IL-4 expression). Results represent 1 of 4 independent experiments (c) In vitro suppression of the proliferation of WT CD4+ Teff cells (denoted as T responder or Tresp) by Foxp3EGFPCre Teff cells pre-treated for 1h with DMSO (Veh WT Teff cells) or 1μM Rapamycin (Rapa WT Teff cells), Foxp3EGFPCre Treg cells pre-treated for 1h with DMSO (Veh WT Treg cells) or 1μM Rapamycin (Rapa WT Treg cells) or Foxp3ΔEGFPiCre Treg cells pre-treated for 1h with DMSO (Veh ΔTreg cells) or 1μM Rapamycin (Rapa ΔTreg cells) (denoted as T suppressor or Tsupp) (n=3 per group). Results represent 1 of 3 independent experiments (d, e) In vitro suppression of Tresp cells by Foxp3EGFPCre Treg cells (Foxp3EGFPCre), Foxp3ΔEGFPiCre Treg cells pre-treated for 1h with DMSO (Veh Foxp3ΔEGFPiCre) or 1μM Rapamycin (Rapa Foxp3ΔEGFPiCre), Foxp3ΔEGFPiCreRictorΔ/Δ Treg cells pre-treated for 1h with DMSO (Veh Foxp3ΔEGFPiCreRictorΔ/Δ) or 1μM Rapamycin (Rapa Foxp3ΔEGFPiCreRictorΔ/Δ) (n=3 per group) (Results represent 1 of 3 independent experiments) (d), and Foxp3ΔEGFPiCreRptorΔ/Δ Treg cells pre-treated for 1h with DMSO (Veh Foxp3ΔEGFPiCreRptorΔ/Δ) or 1μM Rapamycin (Rapa Foxp3ΔEGFPiCreRptorΔ/Δ) (n=3 per group) (Results represent 1 of 3 independent experiments) (e). (f) In vitro suppression of Tresp cells by Rictor-sufficient or -deficient Teff (CD4+CD25− cells from CD4cre and CD4creRictorΔ/Δ) and Foxp3-sufficient Treg (Foxp3YFPcre and Foxp3YFPcreRictorΔ/Δ) cells (n=3 per group) (Results represent 1 of 2 independent experiments). Results are expressed as mean ± SEM in panels c-f. Statistical significance was determined by a one-way ANOVA (b) or two-way ANOVA with Tukey’s multiple comparisons (c-f) (P values as indicated).
We next analyzed the impact of mTOR inhibition on Treg, ΔTreg and Teff cell in vitro suppressive capacities. Pre-treatment of Teff cells with the mTOR inhibitor Rapamycin did not confer them suppressive capacity. Foxp3ΔEGFPiCre ΔTreg cells had a small but detectable suppressive activity as compared to Teff cells, which was augmented by Rapamycin pretreatment. Rapamycin pretreatment also enhanced the suppressive function of wild-type Treg from Foxp3EGFPCre (Fig. 2c).
The positive impact of rapamycin on ΔTreg cell suppressive capacity mapped to mTORC2. Rictor deletion substantially upregulated ΔTreg cell-mediated suppression (Fig. 2c), an effect that was not related to decreased ΔTreg cell proliferation, as evidenced by Ki-67 staining (Supplementary Fig. 3a,b). Furthermore, Rictor but not Raptor deficiency abrogated the enhanced ΔTreg cell suppressive activity mediated by Rapamycin, indicating that Rapamycin promoted ΔTreg cell suppression by inhibiting mTORC2 (Fig. 2d,e).
We further compared the suppressive capacity of CD4+CD25– Teff cells isolated from Cd4Cre versus Cd4CreRictorΔ/Δ mice. Results showed that Rictor deficiency did not confer any suppressive capacity to Teff cells. In contrast, Rictor deficiency in otherwise Foxp3-sufficient Treg cells (Foxp3YFPCreRictorΔ/Δ) resulted in a small but significant improvement in their suppressive capacity (Fig. 2f), consistent with the results of treating wild-type Treg cells with Rapamycin (Fig. 2c).
The significance of improved wild-type Treg cell function upon Rictor deficiency was evaluated in vivo using the lymphopenia colitis model 27. Foxp3YFPCreRictorΔ/Δ Treg cells proved superior in their capacity to control the intestinal inflammation in Rag1-deficient mice reconstituted with naive CD4+CD45RBhigh Teff cells. Under limiting Treg cell availability (1:10 Treg:Teff cell ratio), the Foxp3YFPCreRictorΔ/Δ Treg cells prevented weight loss, improved the intestinal inflammation and the inflammation-induced colon shortening, and suppressed the infiltration of the intestines by Teff cells, including IFN-γ+, IL-17+ and IFN-γ+IL-17+ cells (Supplementary Fig. 4a–g).
We next analyzed the impact of Rictor deletion in ΔTreg cells on their stability. Whereas ex-Treg cells, corresponding to the EGFP– fraction of total YFP+ CD4 T cells, represented approximately 7% in Foxp3-sufficent control mice (Foxp3EGFPcreR26YFP) and 15% in Foxp3-deficient Foxp3ΔEGPiCreR26YFP mice, their frequency was significantly decreased to 9–10% in Foxp3ΔEGFPiCreRictorΔ/ΔR26YFP mice (Supplementary Fig. 3a,b), indicating that mTORC2 destabilized ΔTreg cells, possibly by affecting their epigenetic demethylation 28.
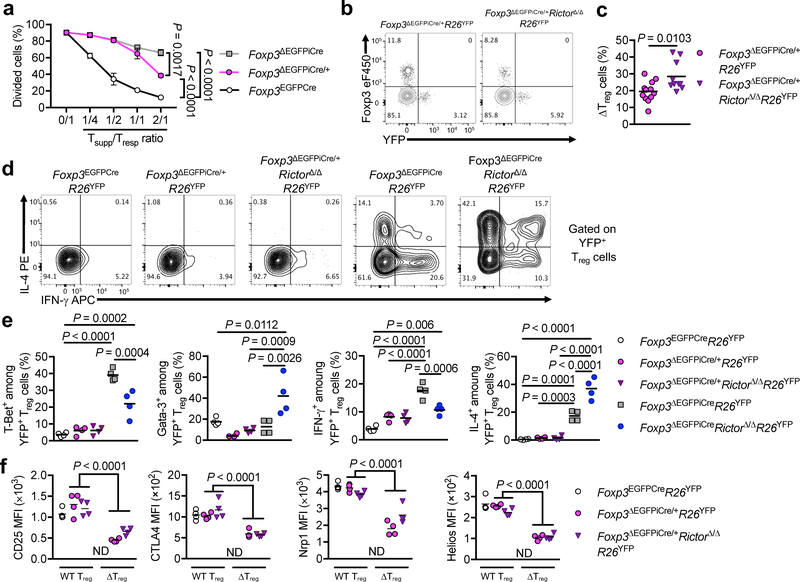
We employed heterozygous Foxp3ΔEGFPiCre/+ female mice, which are phenotypically normal, to examine the impact of Rictor deficiency on ΔTreg cells in the absence of systemic inflammation. ΔTreg cells isolated from Foxp3ΔEGFPiCre/+ females had more suppressive activity at high ΔTreg/Teff cell ratios than those of Foxp3ΔEGFPiCre males, indicative of a detrimental role for inflammation in the loss of ΔTreg suppressive function (Fig. 3a). To assess Treg cell fitness, we examined the frequencies of wild-type and ΔTreg cells in Foxp3ΔEGFPiCre/+R26YFP versus Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP heterozygous female mice. Whereas ΔTreg cells were profoundly under-represented in Foxp3ΔEGFPiCre/+ mice compared to Treg cells carrying the wild-type Foxp3 allele, their frequency increased in Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP mice, indicative of their improved fitness (Fig. 3b,c). ΔTreg cells from Foxp3ΔEGFPiCre/+R26YFP and Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP heterozygous females minimally expressed T-bet/IFN-γ and Gata-3/IL-4 compared to ΔTreg cells of hemizygous males Fig. 3d,e). These results indicate that the acquisition by ΔTreg cells of TH cell-like phenotypes was augmented by the intense inflammatory environment in Foxp3-deficient males. Further characterization of ΔTreg cells of Foxp3ΔEGFPiCre/+R26YFP and Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP heterozygous females revealed increased expression of CD25 and Nrp1 in Rictor-deficient ΔTreg cells (Fig. 3f). Overall, the results in Fig. 2 and Fig. 3 show that mTORC2 was cell intrinsically dysregulated in ΔTreg cells, and that its inhibition substantially restored ΔTreg cell function.
Figure 3: Cell intrinsic and extrinsic determinants of the Teff cell-like phenotype of ΔTreg cells.
(a) In vitro suppression of CD4+ Teff cell (Tresp) proliferation by Foxp3-sufficient Treg cells from Foxp3EGFPcre mice and ΔTreg cells from Foxp3ΔEGFPiCre/+ and Foxp3ΔEGFPiCre mice (Tsupp) (n=3 per group). Results are expressed as mean ± SEM and represent 1 of 2 independent experiments. (b, c) Representative flow cytometric analysis of Foxp3 and YFP among CD4+ T cells (b) and frequencies of ΔTreg cells (c) among total Treg cells from Foxp3ΔEGFPiCre/+R26YFP (n=12) and Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP (n=10) mice. Results are expressed as scatter plot and mean and represent a pool of 3 independent experiments. (d, e) Representative flow cytometric analysis of IL-4 and IFN-γ among YFP+ Treg cells (d) and frequencies (Scatter plot and mean) of T-Bet+, Gata-3+, IFN-γ+ and IL-4+ YFP+ Treg cells (e) from Foxp3EGFPCreR26YFP, Foxp3ΔEGFPiCre/+R26YFP, Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP, Foxp3ΔEGFPiCreR26YFP and Foxp3ΔEGFPiCreRictorΔ/ΔR26YFP mice (n=4 per group). Results represent 1 of 2 independent experiments. (f) CD25, CTLA4, Nrp1 and Helios MFI in WT Treg and ΔTreg cells from Foxp3EGFPCreR26YFP, Foxp3ΔEGFPiCre/+R26YFP, Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP female mice (n=4 per group). ND, Not Determined. Results represent 1 of 2 independent experiments. Statistical significance was determined by unpaired t-test (c), one-way ANOVA (e) or two-way ANOVA (a, f) with Tukey’s multiple comparisons (P values as indicated).
mTORC2 dependent and independent transcriptional programs in ΔTreg cells
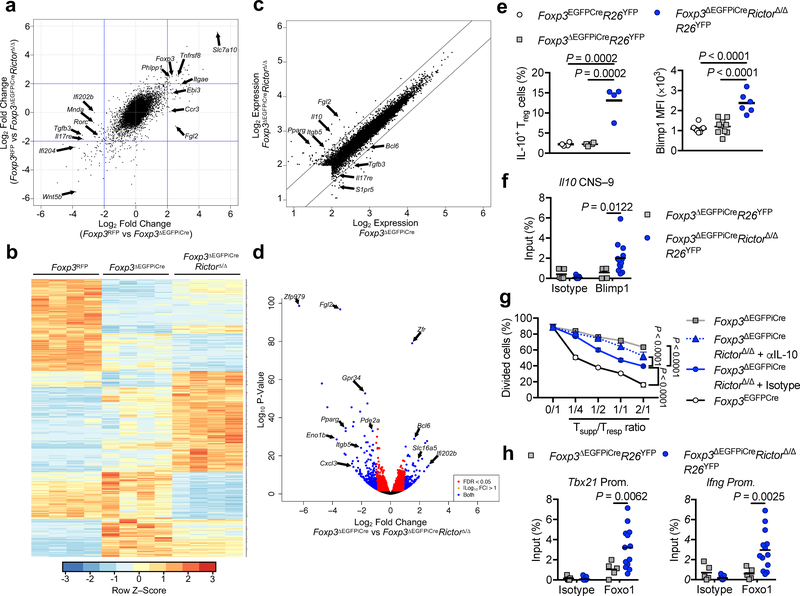
We compared the transcriptomes of ΔTreg and Treg isolated from female mice that carried one mutant Foxp3ΔEGFPiCre allele and a second competent Foxp3 allele (Foxp3RFP) that also directed the expression of the red fluorescent protein (RFP), thus allowing for color sorting of the respective populations 29. We extended these studies to examine the transcriptome of ΔTreg cells of Foxp3ΔEGFPiCre/+RictorΔ/Δ females to identify transcriptional pathways altered by Rictor. Results revealed global changes in the transcriptional landscape induced by Foxp3 deficiency. Concurrent Rictor deficiency normalized some of these changes, while also inducing de novo Foxp3-independent transcriptional alterations of its own Fig. 4a,b and Data Set 1). ΔTreg cells exhibited decreased expression of some genes associated with the Treg cell transcriptome (e.g. Itgae, Fgl2, Nrp1, Ebi3a, Il2ra, Ikzf2, Tigit) and the acquisition of several others associated with Teff cells, including TH17 cells (e.g. Rorc, Tgfb3, Il17a, Nr1d1, Sgk1), cytokines and cellular activators (Il2, Il21, Cd40lg).
Figure 4. mTORC2-dependent and -independent gene expression profiles in ΔTreg cells.
Gene expression profiles of Foxp3RFP (WT), Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ Treg cells (n=4 per group) isolated from spleen of Foxp3RFP/ΔEGFPiCreR26YFP and Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP female mice. (a), Gene expression profiles represented as Fold change (Foxp3RFP vs Foxp3ΔEGFPiCre) versus Fold change (Foxp3RFP vs Foxp3ΔEGFPiCreRictorΔ/Δ). (b), Heatmap representation, (c), Log2 expression and (d) volcano plot of differential gene expression in Foxp3ΔEGFPiCre versus Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells. FDR, false discovery rate; log2FC, log2(fold change). (e), Flow cytometric analysis of IL-10 and Blimp1 expression in Treg cells of Foxp3EGFPCre (n=4 and n=8 respectively) and ΔTreg cells of Foxp3ΔEGFPiCre (n=4 and n=10 respectively) and Foxp3ΔEGFPiCreRictorΔ/Δ (n=4 and n=6 respectively) mice. Results are expressed as scatter plot and mean and represent 1 of 3 independent experiments. (f), Chromatin immunoprecipitation (ChIP) assay for the binding of Blimp1 and control isotype to the Il10 conserved non-coding sequence located 9kb in proximal of il10 promoter (il10 CNS–9) in Foxp3ΔEGFPiCre (n=5) and Foxp3ΔEGFPiCreRictorΔ/Δ (n=13) ΔTreg cells. Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. (g), In vitro suppression of Tresp cells by Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells carried out in the presence of either an isotype control or anti-IL-10 mAb (n=3 per group). Results are expressed as mean ± SEM and represent 1 of 2 independent experiments. (h) ChIP assays for the binding of Foxo1 and control isotype to Tbx21 and Ifng promoters in Foxp3ΔEGFPiCre (n=5) and Foxp3ΔEGFPiCreRictorΔ/Δ (n=13) ΔTreg cells. Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. Statistical significance was determined one-way ANOVA (e) or two-way ANOVA (f-h) with Tukey’s or Sidak’s multiple comparisons (P values as indicated).
Relevant to the mTOR pathway, ΔTreg cells had increased expression of Prr5l, encoding an mTORC2 regulator that directs its substrate specificity 30, and decreased expression of Phlpp1, encoding the phosphatase PHLPP1 23. Concurrent Rictor deficiency upregulated a subset of the core Treg transcriptome genes, including Fgl2, Il10, Lag3, Tnfrsf18 (Gitr), Ebi3, Nrp1, Il2ra, Ctla4 and Nt5e. It also down-regulated the expression of genes associated with the TH17 (Rorc, Tgfb3) and T follicular helper cell (Cxcr5 and Bcl6) lineages, while upregulating the Bcl6 antagonist Prdm1 Fig. 4c,d and Data Set 2). These results suggested that Rictor deficiency altered the ΔTreg cell transcriptome in favor of improved regulatory function.
Of particular interest was the role of increased IL-10 and Blimp1 expression in Rictor-deficient ΔTreg cells (Fig. 4e). Blimp1 binds to the regulatory CNS –9 element located proximal to the Il10 gene to promote Il0 transcription 31. Chromatin immunoprecipitation (ChIP) studies demonstrated increased binding of Blimp1 to the Il10 CNS–9 element of Rictor-deficient ΔTreg cells (Fig. 4f). IL-10 neutralization largely abrogated the improved in vitro suppressive capacity of Rictor deficient ΔTreg cells, indicating a mechanistic role for this pathway in their augmented regulator function (Fig. 4g).
mTORC2 signaling in ΔTreg cells activates the AKT-FOXO1 axis
We examined mTORC2 activity, as monitored by pS473AKT staining, in wild-type Treg cells from Foxp3EGFP mice, and in ΔTreg cells from Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ mice. The increase in pS473AKT staining in Foxp3ΔEGFPiCre ΔTreg cells following their stimulation with anti-CD3+anti-CD28 mAbs was totally reversed in the Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells Fig. 5a,b). AKT phosphorylates the transcription factor Foxo1, resulting in its retention in the cytosol and its ubiquitination and degradation 32. Foxo1 in turn negatively regulates TH1 differentiation of Treg cells by suppressing the transcription of Ifng and other TH1 genes 33, 34. Expression of several Foxo1-regulated genes, including Il7r, Cd55, Cd83, Emb, Etv5 and Mafg, was affected in Foxp3ΔEGFPiCre ΔTreg cells and normalized in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells (Data Set 2). Foxo1 was decreased in the nucleus in anti-CD3 mAb-stimulated ΔTreg cells of Foxp3ΔEGFPiCre compared to control Foxp3EGFP Treg cells, a deficit that was reversed in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells Fig. 5c,d). Also, Rictor deletion increased Foxo1 binding at the Ifng and Tbx21 promoters (Fig. 4g).
Figure 5. Contribution of AKT/Foxo1 axis to the Rictor-dependent ΔTreg cell phenotype.
(a) Representative flow cytometric analysis and (b) MFI of p473-AKT expression in unstimulated (US) and anti-CD3/CD28 mAb-stimulated (α-CD3+α-CD28) Treg cells from Foxp3EGFPCre, Foxp3ΔEGFPiCre, Foxp3ΔEGFPiCreRictorΔ/Δ mice (n=3 per group). Results are expressed as scatter plot and mean and represent 1 of 3 independent experiments. (c,d) Representative confocal microscopic merge image of Foxo1 (Red) and DAPI (Blue) (c), and percent of nuclear Foxo1 (d) in unstimulated (US) or 0.1μg/mL anti-CD3 (α-CD3) stimulated Treg cells from Foxp3EGFPCre (n=53 for US, n=71 for α-CD3), Foxp3ΔEGFPiCre (n=74 for US, n=95 for α-CD3), Foxp3ΔEGFPiCreRictorΔ/Δ (n=78 for US, n=46 for α-CD3) mice. Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. (e) Representative flow cytometric analysis and (f) frequencies (scatter plot and mean) of CD62LloCD44hi CD4+ Teff cells, and IFN-γ and IL-4 expression by CD4+ Treg and Teff cells in spleens of Foxp3EGFPCre (n=10), Foxp3ΔEGFPiCre (n=7), Foxp3ΔEGFPiCreRictorΔ/Δ (n=6), Foxp3ΔEGFPiCreFoxo1Δ/Δ (n=3), Foxp3ΔEGFPiCreRictorΔ/ΔFoxo1Δ/Δ (n=4) and Foxp3ΔEGFPiCreR26Foxo1AAA (n=5) mice. Results represent a pool of 3 independent experiments. Statistical significance was determined one-way ANOVA with Tukey’s multiple comparisons (f) or two-way ANOVA with Sidak’s multiple comparisons (b, d) (P values as indicated).
We examined the consequences of deleting a floxed Foxo1 gene in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells. The triple mutant Foxp3ΔEGFPiCreRictorΔ/ΔFoxo1Δ/Δ mice were similar to single mutant Foxp3ΔEGFPiCre and the double mutant Foxp3ΔEGFPiCreFoxo1Δ/Δ mice in terms of growth and survival, dysregulated Teff cells and IFN-γ expression in both ΔTreg and Teff cells, whereas the IL-4 response was unaffected Fig. 5e,f and Supplementary Fig. 5a–e). Foxo1 deletion reversed the upregulation of Il10 expression in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg (Supplementary Fig. 5f). Reciprocally, ΔTreg cell-specific expression of Cre-regulated AKT-insensitive Foxo1 transgene (R26Foxo1AAA) partially recapitulated the effects of Rictor deficiency in improving weight gain, prolonging survival and augmenting ΔTreg cell suppressive capacity (Supplementary Fig. 5a–d) 33. It also down-regulated Teff cell activation and the TH1 programming of ΔTreg cells Fig. 5e,f and Supplementary Fig. 5e,g). These results indicated that the effects of Rictor deletion in promoting ΔTreg cell function largely proceeded by Foxo1-dependent mechanisms.
Rictor deficiency resets the metabolism of ΔTreg cells
Dysregulation of the mTORC2-Foxo1 axis upregulates glycolysis and OXPHOS 35, 36. Treg cells and naive T cells exhibit low levels of glycolysis in favor of increased OXPHOS, whereas Teff cells manifest increased aerobic glycolysis, products of which feed into DNA and protein synthesis 37. Foxp3ΔEGFPiCre ΔTreg cells had increased expression of several enzymes in the glycolytic and pentose phosphate shunt pathways, several of which were reset back to baseline upon concurrent Rictor deficiency (e.g. Gpi, Aldoa, Eno1, Pkm2 and the pentose phosphate shunt enzymes) (Supplementary Fig. 6a). Some glycolytic enzymes were down-regulated by Rictor deficiency but remained increased above their levels in wild-type Treg cells (e.g. Hk2, Gapdh), while others such as Tpi and the glycolytic regulator Pfkfb3, which were upregulated in Foxp3-deficient Treg cells, were either unaffected or further upregulated by Rictor deficiency.
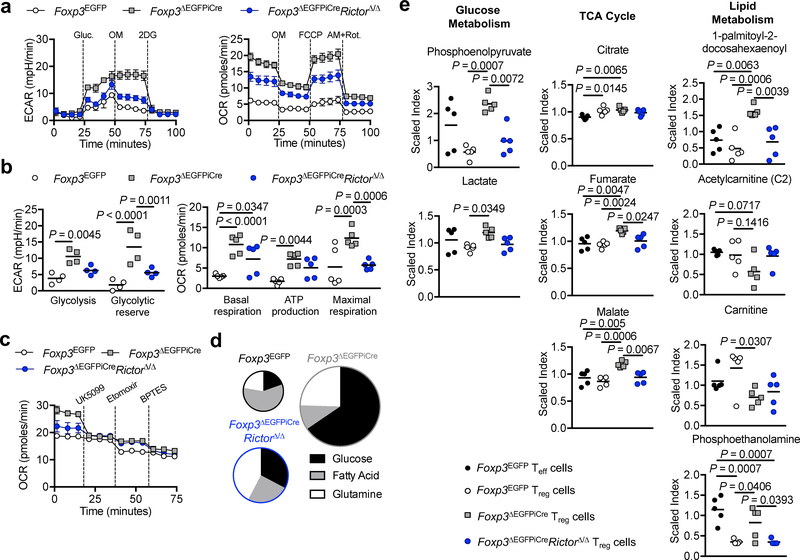
Unlike wild-type Treg cells, Foxp3ΔEGFPiCre ΔTreg cells stimulated with anti-CD3 mAb and IL-2 exhibited exaggerated lactate production similar to activated Teff cells, which was markedly reduced in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells (Supplementary Fig. 6b). Glycolysis and OXPHOS were further evaluated in Foxp3EGFP (wild-type) Treg cells and Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells isolated from hemizygous males using an extracellular metabolic flux analyzer. Of the three Treg cell populations, Foxp3ΔEGFPiCre ΔTreg cells exhibited the largest increase in extracellular acidification rate (ECAR), a measure of glycolysis, upon D-glucose supplementation, while Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg had an intermediate phenotype Fig. 6a,b). All three Treg cells populations exhibited a modest drop in ECAR upon Oligomycin supplementation, indicating that glycolysis had peaked following glucose addition 38.
Figure 6. mTORC2 promotes aerobic glycolysis and OXPHOS in ΔTreg cells.
(a, b) Extracellular acidification rate (ECAR) under glycolysis stress test conditions (n=4 per group) and Oxygen consumption rate (OCR) under mitochondrial stress test conditions (n=5 per group) of Treg/ΔTreg cells isolated from Foxp3EGFP, Foxp3ΔEGFPiCre or Foxp3ΔEGFPiCreRictorΔ/Δ males (n=4 per group). Results are expressed as mean ± SEM and represent a pool of 2 independent experiments. (c) OCR under mitochondrial fuel test conditions (n=4 per group). Results are expressed as mean ± SEM and represent a pool of 2 independent experiments (d) Pie chart representation of the contribution of glucose, fatty acids and glutamine to the OXPHOS capacities in Foxp3EGFP, Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells. (e) Quantification of metabolites (expressed as scaled index) of glucose metabolism (Phosphoenolpyruvate and lactate), tricarboxylic acid (TCA) cycle (Citrate, fumarate and Malate) and lipid metabolism (Acetylcarnitine (C2), Carnitine, 1-palmitoyl-2-decosahexaenoyl and Phospho-ethanolamine) isolated from Foxp3EGFP, Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells (n=5 per group). Results are expressed as mean ± SEM and represent 1 experiment. Statistical significance was determined one-way ANOVA with Tukey’s multiple comparisons (e) or two-way ANOVA with Sidak’s multiple comparisons (b) (P values as indicated).
OXPHOS, as assessed by the oxygen consumption rate (OCR), was also markedly increased in Foxp3ΔEGFPiCre ΔTreg cells. Both basal respiration, ATP-coupled respiration and respiratory reserve were increased in Foxp3ΔEGFPiCre ΔTreg compared to control Treg cells whereas Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg had an intermediate phenotype.. Fig. 6a,b).
The ECAR and OCR of ΔTreg cells of Foxp3ΔEGFPiCre/+ females were also highly increased, indicative of cell-intrinsic metabolic changes (Supplementary Fig. 6c,d). Rictor deficiency completely reversed the increase in ECAR in ΔTreg cells of Foxp3ΔEGFPiCre/+ females, but only partially reversed the increase in OCR, indicating that additional pathways contributed to the latter’s dysregulation (Supplementary Fig. 6c,d).
The contribution of inputs from glycolysis, glutaminolysis and FAO to the OCR was measured upon the sequential addition of specific metabolic inhibitors: UK5099 for glycolysis, Etomoxir for FAO and BTPES for glutaminolysis (Fig. 6c). Whereas the OCR in wild-type Treg cells is fatty acid-dependent, it becomes glucose- and to a lesser extent glutamate-dependent in Foxp3ΔEGFPiCre ΔTreg cells, pointing to a shift from FAO to aerobic glycolysis and, secondarily, glutaminolysis (Fig. 6c). Rictor deficiency reduced the OCR and decreased aerobic glycolysis Fig. 6c). Transcripts of key factors associated with aerobic glycolysis, including Myc and Hif1a 12, 39, were markedly increased in Foxp3ΔEGFPiCre ΔTreg cells, and partially corrected in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells (Supplementary Fig. 6e).
Metabolomic analysis on Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells, and control Treg and Teff cells from Foxp3EGFP mice revealed increased glycolytic (phosphoenolpyruvate, lactate) and citric acid cycle (fumarate, malate) metabolites in Foxp3ΔEGFPiCre ΔTreg cells, which were reversed in Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells Fig. 6d and Supplementary Fig. 6f). Foxp3ΔEGFPiCre ΔTreg cells were deficient in carnitine, necessary for the transport of fatty acids to the mitochondria for beta oxidation, which was not corrected by Rictor deficiency Fig. 6d and Supplementary Fig. 6f) 40. These results established that Foxp3ΔEGFPiCre ΔTreg cells exhibited robust mTORC2-dependent aerobic glycolysis and OXPHOS.
Metabolic blockade inhibits the Teff-like phenotype of ΔTreg cells
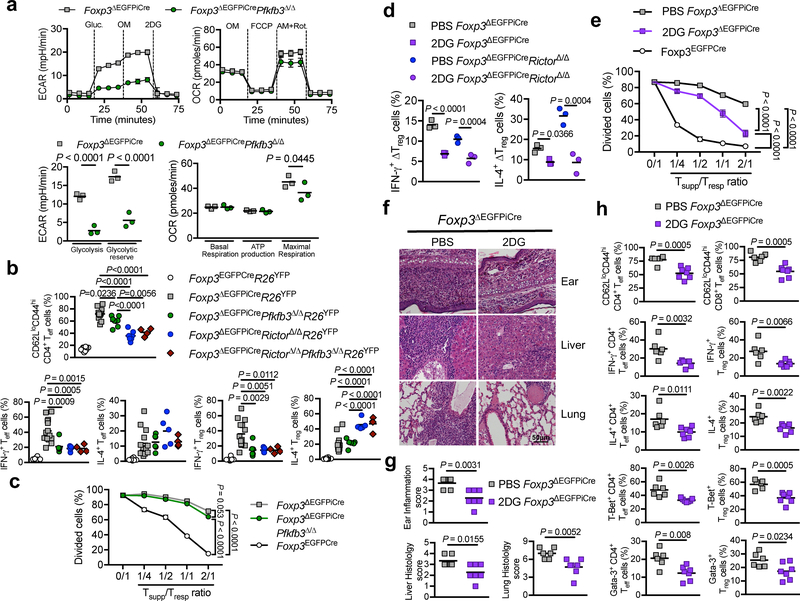
The role of dysregulated glycolysis in ΔTreg cell dysfunction was probed by cell-specific deletion of Pfkfb3, encoding 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3. This enzyme synthesizes fructose-2,6-bisphosphate, an allosteric activator of phosphofructokinase-1 and a powerful stimulator of glycolysis 41. ΔTreg cell-specific deletion of Pfkfb3 (Foxp3ΔEGFPiCrePfkfb3Δ/Δ) normalized their ECAR, while minimally impacting the OCR (Fig. 7a). Foxp3ΔEGFPiCrePfkfb3Δ/Δ mice showed decreased memory Teff cells, at values intermediate between Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ mice (Fig. 7b). ΔTreg cell-specific PFKFB3 deficiency suppressed IFN-γ production by ΔTreg and Teff cells, whereas IL-4 production was not affected. Concurrent deletion of Pfkfb3 and Rictor were not additive, indicating that effects of PFKFB3 deficiency on glycolysis were subsumed under mTORC2 deficiency (Fig. 7b). PFKFB3 deficiency marginally improved in vitro ΔTreg cells suppression of Teff cell proliferation (Fig. 7c), Thus, the inhibition of dysregulated glycolysis by Rictor or Pfkfb3 deletion restored ΔTreg cells regulation of TH1 cell responses.
Figure 7. Blockade of glycolysis improves the scurfy phenotype of Foxp3ΔEGFPiCre mice.
(a) ECAR under glycolysis stress test conditions and OCR under mitochondrial stress test conditions of ΔTreg cells isolated from Foxp3ΔEGFPiCre or Foxp3ΔEGFPiCrePfkfb3Δ/Δ males (n=3 per group). Results are expressed as mean ± SEM and represent 1 of 2 independent experiments. (b) Frequencies of CD62LloCD44hi CD4+ Teff cells, and IFN-γ and IL-4 expression by CD4+ Treg and Teff cells in spleens of Foxp3EGFPCreR26YFP, Foxp3ΔEGFPiCreR26YFP, Foxp3ΔEGFPiCrePfkfb3Δ/ΔR26YFP, Foxp3ΔEGFPiCreRictorΔ/ΔR26YFP, Foxp3ΔEGFPiCreRictorΔ/ΔPfkfb3Δ/ΔR26YFP mice. Results are expressed as scatter plot and mean and represent a pool of 3 independent experiments. (c) In vitro suppression of CD4+ Teff cell (Tresp) proliferation by Foxp3EGFPCre, Foxp3ΔEGFPiCre or Foxp3ΔEGFPiCrePfkfb3Δ/Δ Treg cells (Tsupp) (n=3 per group). Results are expressed as mean ± SEM and represent 1 of 2 independent experiments. (d) Frequencies of IFN-γ+ and IL-4+ Foxp3ΔEGFPiCreR26YFP and Foxp3ΔEGFPiCreRictorΔ/ΔR26YFP sorted Treg cells that were either sham treated or treated ex-vivo with 2-deoxyglucose (2DG) (n=3 per group). Results are expressed as scatter plot and mean and represent 1 of 2 independent experiments. (e) In vitro suppression of CD4+ Teff cell (Tresp) proliferation by Foxp3ΔEGFPiCre ΔTreg (Tsupp) cells that were pre-treated for 12h with either PBS (Veh ΔTreg cells) or 2DG (2DG ΔTreg cells) (n=3 per group). Results are expressed as mean ± SEM and represent 1 of 2 independent experiments. (f, g) Representative hematoxylin and eosin-stained tissue histological sections (f) and tissue histological scores (g) of ears, livers and lungs of Foxp3ΔEGFPiCreR26YFP mice treated with PBS (n=) or 2 μg/g 2DG (n=) every other day from day 14 to day 28. Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. (h) Frequencies of CD62LloCD44hi CD4+ and CD8+ Teff cells, IFN-γ+ and IL-4+ Treg and CD4+ Teff cells and T-Bet+ and Gata-3+ Treg and CD4+ Teff cells in Foxp3ΔEGFPiCreR26YFP mice treated with PBS (n=6) or 2DG (n=7). Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. Statistical significance was determined unpaired t-test (g, h), one-way ANOVA with Tukey’s multiple comparisons (b) or two-way ANOVA with Sidak’s multiple comparisons (a, d) or with Tukey’s multiple comparisons (c, e) (P values as indicated).
We further investigated the dysregulated metabolism of ΔTreg cell using 2-deoxyglucose (2DG), a competitive inhibitor of glycolysis. While Pfkfb3 deletion attenuated the ECAR in ΔTreg cells, 2DG more profoundly suppressed it (Fig. 6a). 2DG strongly inhibited the production by Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells of both of IFN-γ and IL-4 (Fig. 7d), and at high concentrations partially restored Foxp3ΔEGFPiCre ΔTreg cell suppression, suggesting a role for aerobic glycolysis-dependent TH cytokine production in poor ΔTreg cell suppression (Fig. 7e).
In vivo, treatment of Foxp3ΔEGFiCreR26YFP mice with 2DG resulted in decreased tissue inflammation Fig. 7f–h). 2DG reduced the frequencies of memory CD62LloCD44hi CD4+ and CD8+ T cells, and the production of IL-4 and IFN-γ and the expression of Gata-3 and T-bet expression by both CD4+ Teff and Treg cells. 2DG treatment of Foxp3ΔEGFPiCreRictorΔ/Δ mice also reduced the memory frequencies and cytokine production by CD4+ and CD8+ T cells (Supplementary Fig. 7).
mTOR inhibition upregulates human mutant FOXP3 Treg cell suppressive function
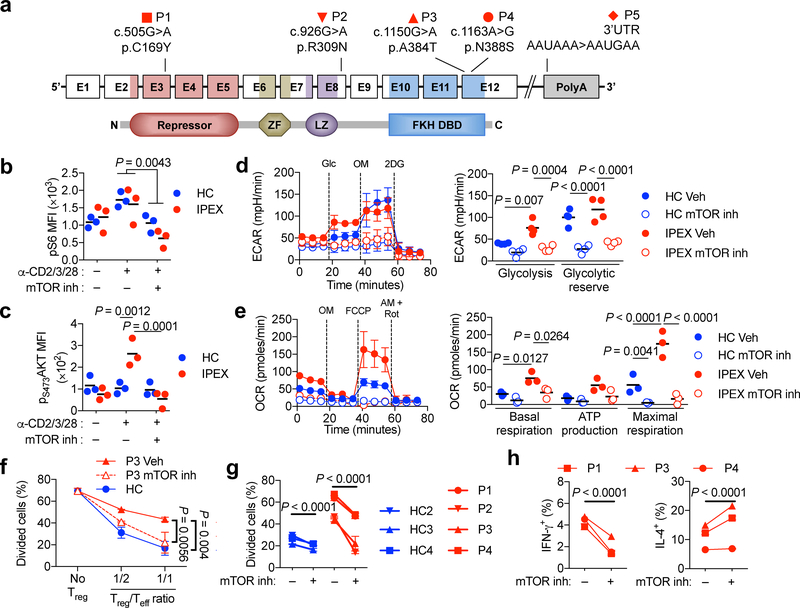
FOXP3 deficiency causes a human autoimmune lymphoproliferative disease, Immune dysregulation, Enteropathy, Polyendocrinopathy X-Linked or IPEX 42. Similar to ΔTreg cells, IPEX Treg cells are present in the periphery but are deficient in suppressive functions 43, 44. We analyzed Treg cells of five IPEX patients with different FOXP3 mutations, including N-terminal (C169Y), linker region (R309N) and forkhead domain (A384T and N388S) missense mutations and an inactivating mutation in the 3’ polyadenylation signal (Fig. 8a). The C169Y, R309N and N388S are novel mutations, while the A384T and the 3’ polyadenylation signal mutation have been reported 45. All five IPEX patients exhibited detectable but lower expression of FOXP3 in their mutant Treg cells, defined as CD4+CD25+CD127lo 46, 47, compared to control subject Treg cells (Supplementary Figure 8). There was a similar increase in pS6 phosphorylation in control and IPEX Treg cells in response to cell activation, which was abrogated by treatment with the dual mTOR inhibitor Ku-0063794 (Fig 8b). By contrast, increased pS473AKT phosphorylation was exclusively observed in stimulated IPEX Treg cells, which was also abrogated by the inhibitor (Fig 8c). Thus, mTORC2 activity was also dysregulated in IPEX Treg cells.
Figure 8. mTOR inhibition augments the suppressive function of human FOXP3 mutant Treg cells.
(a) Schematic representation of FOXP3 illustrating the exons, the protein domains and mapped mutations of five patients. Amino acid changes are referred to by their single letter code. The N-terminal proline rich repressor domain (Repressor), zinc finger (ZF) motif, leucine zipper domain (LZ) and the forkhead DNA-binding domain (FKH DBD) are indicated. (b,c) Mean Fluorescence Intensity (MFI) of pS6 and pS473AKT in Treg cells of healthy control subjects (HC) (n=3) and IPEX patients (n=3; P1, P2, P3) stimulated with anti-CD2/CD3/CD28 mAbs (α-CD2/3/28) in the absence or presence of a competitive dual mTOR inhibitor (mTOR inh). Results are expressed as scatter plot and mean and represent 1 of 2 independent experiments. (c) Representative ECAR tracings (HC1 and P1), expressed as mean ± SEM. (d) Evaluation of glycolysis and glycolytic reserve (n=5 each for HC and IPEX groups). Results are expressed as mean ± SEM and represent a pool of 5 independent experiments. (e) Representative OCR tracings (HC2 and P2). Basal respiration, ATP production and maximal respiration in DMSO (Vehicle) or mTOR inhibitor (mTOR inh) treated HC and IPEX Treg cells (HC n=3 and IPEX: P1, P2, P4). (f) In vitro suppression of third party CD4+ Teff cell (Tresp) proliferation by Treg (Tsupp) cells of a HC or an IPEX subject (P3) that were pre-treated with vehicle or mTOR inhibitor. (g) Compilation of in vitro suppression assay results for HC and IPEX subjects at the ratio of 1:1 Treg:Teff cells without or with Treg cell mTOR inhibitor pretreatment. (h) IFN-γ and IL-4 secretion by vehicle or mTOR inhibitor pre-treated IPEX Treg cells. Statistical significance was one-way ANOVA with Tukey’s multiple comparisons (h) or two-way ANOVA with Sidak’s multiple comparisons (b-d) or with Tukey’s multiple comparisons (e, f, g) (P values as indicated).
Examination of metabolic activities revealed that IPEX Treg cells exhibited increased ECAR in response to D-glucose, indicative of heightened glycolytic function, while showing equivalent glycolytic reserves following oligomycin addition (Fig. 8d). Both glycolytic components were inhibited by treatment of Treg cells with the dual mTOR inhibitor. IPEX Treg cells also exhibited exaggerated OCR responses, which were also inhibited by treatment with the dual mTOR inhibitor (Fig. 8e).
While control Treg cells effectively suppressed Teff cell proliferation, IPEX Treg cells did not Fig. 8f,g). Pretreatment of the latter with the dual mTOR inhibitor uniformly improved their suppressor function irrespective of the underlying mutation involved Fig. 8f,g). mTOR inhibition also upregulated the suppressive function of Treg cells of healthy subjects Fig. 8g), similar to Foxp3-sufficient but Rictor-deficient mouse Treg cells (Fig. 2f and Supplementary Fig. 4). IPEX Treg cells expressed both IFN-γ and IL-4, indicative of their Teff-like phenotype. mTOR inhibition suppressed the TH1 but not the TH2 response, reflecting the role of mTORC2 in driving the TH1 response in ΔTreg cells (Fig. 8h). These results support the targeting of mTOR pathways to restore Treg cell function in IPEX, as well as augmenting Foxp3-sufficient Treg cell function in immune dysregulatory diseases.
Discussion
We demonstrate that the skewing of Treg cells towards a Teff cell-like phenotype upon Foxp3 deficiency is critically dependent on a limited set of molecular pathways, including mTORC2 and glycolysis. Furthermore, inhibition of these pathways specifically in Foxp3-deficient Treg cells partially restored regulatory function and attenuated disease. Targeting the same pathways may also improve the stability and function of Foxp3-sufficient Treg cells in inflammatory and autoimmune diseases.
Our studies implicated impaired regulation by phosphatases, including PTEN and PHLPP1, in mTORC2 dysregulation in ΔTreg cells. Pten deletion dysregulates mTORC2 and results in the loss of Treg cell regulatory function 38. PTEN and AKT phosphatase PHLPP1 were markedly decreased in ΔTreg cells, while transcripts encoding both Rictor and the Rictor-associated protein PRR5L, which regulates mTORC2 activity in a substrate-specific manner 30, were increased. Collectively, these abnormalities may favor heightened mTORC2 activity.
The mechanisms by which mTORC2 dysregulation impair ΔTreg cell regulatory functions included disruption of Blimp1 and Foxo1-dependent induction of IL-10 31, 48. The salutary effects of Rictor deficiency on ΔTreg cells were were reversed by Foxo1 deficiency. Reciprocally expression of a Foxo1 gain-of-function mutant transgene recapitulated the effects of Rictor deficiency, rendering ΔTreg cells less TH1 cell-like, consistent with the role of Foxo1 in suppressing Treg cells TH1 reprogramming 33.
The relationship of the metabolic resetting induced by Rictor deletion with enhanced ΔTreg cell function was further established using pharmacological and genetic approaches, including inhibition of aerobic glycolysis with 2-DG and ΔTreg cell-specific deletion of Pfkfb3. Rictor deficiency did not attenuate the TH2 program in ΔTreg cells and in fact exacerbated it, an effect not observed upon glycolytic pathway inhibition with 2DG treatment or Pfkfb3 deletion. These results suggest that the TH2 cell program in ΔTreg cells is regulated by mTORC2 by distinct mechanisms(s)
Unlike ΔTreg cells of Foxp3ΔEGFPiCre hemizygous male mice, those of Foxp3ΔEGFPiCre/+ heterozygous females manifested residual suppressive activity and did not express TH cell cytokines, indicating that the intense inflammatory environment of mutant male mice further contributed to the Teff cell-like phenotype of ΔTreg cells. These findings suggested that suppression of systemic immune activation may act in an adjunct manner with mTORC2 inhibition to further antagonize the Teff cell-like phenotype of ΔTreg cells and to promote their regulatory function.
IPEX-causing FOXP3 mutations may completely inactivate FOXP3 or more selectively impair its functions 45. Studies on mice harboring IPEX-causing FOXP3 mutations, including I363V, A384T and R397W, have identified divergent mechanisms by which these mutations impair Foxp3 function 49, 50. Common to these mutations is the acquisition by the Treg cells of Teff cell-like attributes, with the impaired regulatory function 49. IPEX Treg cells also responded favorably to mTOR inhibitors, with decreased cytokine expression and improved suppression, indicating that targeting mTORC2 is a viable therapeutic strategy in this disorder. More broadly, our studies open up the possibility of combinatorial interventions that target distinct metabolic pathways in Treg cells to restore immune tolerance in a variety of immune dysregulatory diseases.
Methods
Human subjects
Male subjects with IPEX-causing FOXP3 mutations and healthy controls were recruited under protocols approved by the local Institutional Review Boards at the respective referring centers. Patient P1, P2, P3 and P4 each harbored a missense mutation at c.505G>A (p.C169Y), c.926G>A (p.R309N), c.1150G>A (p.A384T), c.1163A>G (p.N388S) respectively (reference sequence NM_014009.3). Patient P5 suffered from a previously described inactivating mutation in the FOXP3 3’ polyadenylation signal 51.
Generation of Foxp3ΔEGFPiCre mice
Foxp3 genomic DNA was isolated from a bacterial artificial chromosome clone (Genome Systems) and subcloned into the plasmid vector pKO (Lexicon). A DNA cassette encoding for an improved Cre recombinase (iCre) was isolated from the paavCAG-iCre plasmid (Addgene) and inserted at the BSGRI site of a pIRES2-EGFP (Addgene) vector containing a DNA cassette composed of an internal ribosomal entry sequence (IRES) linked to downstream EGFP and SV40 poly-A sequences. The IRES-EGFP-iCre sequence was inserted by blunt-end ligation at the SSPI restriction site immediately downstream of the Foxp3 translational stop codon and upstream of the endogenous polyadenylation signal. A PGK-neo cassette was also inserted at the EcoRI site in intron 9 of Foxp3 in the opposite orientation of Foxp3 and was flanked by two FRT sites to allow excision by FLP-mediated recombination. The targeting construct also included a diphtheria toxin gene (DT) for negative selection against randomly inserted targeting constructs.
Targeting plasmids were introduced by electroporation into SCC10 embryonic stem cells and subjected to G418 selection. Resistant clones were screened by Southern blot. Successfully targeted clones were injected into C57BL/6 blastocysts, and chimeric males were mated with Wild-type (WT) C57/BL6/J females to derive N1 females that have transmitted the bicistronic allele together with the PGK-neo cassette insert (Foxp3EGFPiCre-neo) in the germline. The PGK-neo cassette was removed by mating founder males with Flp-deleter female mice that harbor a constitutive, ubiquitously expressed FLP recombinase allele 52. Male offspring hemizygous for Foxp3ΔEGFPiCre allele (minus the PGK-neo cassette) presented Scurfy phenotype. The PCR products of Exon9/Exon10 obtained using complementary and genomic DNA (cDNA and gDNA, respectively) were amplified by the following primer sequences: Foxp3 Exon9 Forward 5’-CTTCCACAACATGGACTACTTCAA-3’ and Foxp3 Exon10 Reverse 5’-AAGTAGGCGAACATGCGAGT-3’. The PCR product obtained from gDNA was purified and sequenced by Sanger sequencing using Foxp3 Exon9 Forward primer.
Mice
Foxp3EGFPCre, Foxp3YFPCre, CD4Cre, Rptorfl/fl, Foxo1fl/fl, R26YFP and Rag1–/– mice were obtained from the Jackson Laboratory. Rictorfl/fl mice were obtained from the Mutant Mouse Regional Resource Center. R26Foxo1AAA and Pfkfb3fl/fl mice were generated as described 33, 41. Foxp3K276X, Foxp3EGFP, CD45.1 Foxp3EGFP, Foxp3ΔEGFPiCre and their respective crosses were backcrossed 8–10 generations on C57/BL6/J, excepted Foxp3ΔEGFPiCreFoxo1Δ/Δ and Foxp3ΔEGFPiCreRictorΔ/ΔFoxo1Δ/Δ strain. A list of all mouse strains used is reported in Supplementary Table 1. Excepted when it was specified, 25–28 days old mice were used in this study. The mice were housed under specific pathogen-free conditions and used according to the guidelines of the institutional Animal Research Committees at the Boston Children’s Hospital.
Real-time PCR
Total RNA was isolated from sorted cells with RNeasy kit (Qiagen). Reverse transcription was performed with the SuperScript II RT-PCR system (Invitrogen) and quantitative real-time reverse transcription (RT)-PCR with Taqman® Fast Universal PCR master mix, internal house-keeping gene mouse (Actin VIC-MGB dye) and specific target gene primers (FAM Dye) (Applied Biosystems) on Step-One-Plus machine. Relative expression was normalized to Actin for genes encoding for the enzymes of the glycolysis cascade (Scl2a3, Hk1, Hk2, Gpi, Pfkfb3, Aldoa, Gapdh, Tpi, Pdk1, Pgam1, Pgam5, Eno1, Pkm2, Ldha, Scl16a1), the pentose phosphate cycle (G6pdx, Pgd, Rpia, Rpe), Foxp3, Rictor, Rptor, HIF1a and Myc and calculated as fold change compared to WT CD4+GFP– Teff cells or WT CD4+GFP– Treg cells isolated from Foxp3EGFP mice.
Flow cytometry
Viability dye and antibodies against mouse CD4, CD8, CD16/CD32, CD90.2, CTLA4, ICOS, CD73, Blimp-1 (biolegend), CD25, CD44, CD45.1, CD45.2, CD62L, Foxp3, T-Bet, Gata-3, Helios, IFN-γ, IL-4, IL-17A, IL-2, IL-10, OX40, Nrp1, GITR, pS6, p4EBP (eBioscience), ROR-γt, pT308AKT (BD biosciences), pS473AKT (Cell signaling) were used. Cell suspensions were incubated for 10 min with CD16/CD32 then stained for surface markers and viability dye for 20 min on PBS/0.5%FCS. Foxp3, Helios, T-Bet, Gata-3, ROR-γt and CTLA4 staining was performed overnight using the BD Cytofix/Cytoperm™ kit. For cytokine detection, cell suspensions were pre-incubated with 50 ng/mL PMA, 500 ng/mL ionomycin and 10 μg/mL brefeldin A for 4h in complete medium. Following CD16/32 blocking with specific mAbs, the cells were surface stained for the indicated markers then permeabilized and stained intracellularly overnight with mAbs against IL-2, IL-4, IL-10 IFN-γ, or IL-17 using the BD Cytofix/Cytoperm™ kit. For pS473AKT, pT308AKT, pS6 and p4EBP staining, spleen cells were stimulated for 30 min with soluble anti-CD3 mAb (1μg/mL), then fixed with PBS/2% paraformaldehyde for 20min, permeabilized in 90% methanol for 30 min on ice and stained for CD4, pS473AKT and pT308AKT or CD4, pS6 and p4EBP in PBS. For ex-vivo Treg cell stimulation, isolated Treg cells were cultured for 2 days with plate-bound anti-CD3 (1μg/mL) and IL-2 (100U/mL) in presence or absence of 2DG (Sigma; 2mg/mL) and PMA/ionomycin/BrefeldinA the last 4 hours before staining for IFN-γ and IL-4. All flow cytometry acquisitions were performed on a BD Fortessa cytometer using DIVA software (BD Biosystems) and analyzed using FlowJo Version 10 (Tree Star). All mouse and human antibodies used are listed in Supplementary Table 2 and 3, respectively.
Suppression assays
For mouse studies, CD4+YFP– Teff cells from Foxp3EGFPcreR26YFP mice were isolated by cell sorting on FACSAria (Becton Dickinson), labeled with CellTrace™ Violet Cell Proliferation dye (Life Technologies) according to the manufacturer’s instructions and used as responder cells. Treg (CD4+YFP+) cells were similarly isolated by cell sorting and used as suppressor cells. Responder cells were cultured in triplicates at a fixed number of 105 cells/well in 96-well round-bottom plates and stimulated for 3 days with 1 μg/mL of soluble anti-CD3 mAb in the presence of 4 × 105 feeder spleen cells from Rag1−/− mice. In some experiments, Teff and Treg cells were pretreated for 1h with Rapamycin (Sigma; 1μM) or vehicle (DMSO), or 2DG (40mg/mL) or vehicle (PBS) overnight before being extensively washed and used as suppressor cells. For human studies, CD4+CD127hiCD25lo T cells from control subjects were by cell sorter, labeled with CellTrace™ Violet Cell Proliferation dye (Life technologies) according to the manufacturer’s instructions and used as responder cells. Treg (CD4+CD127loCD25hi) cells were isolated on FACSAria from control or IPEX subjects and used as suppressor cells. Responder cells were cultured in triplicates at a fixed number of 5 × 104 cells per well and stimulated for 3 days with T cell activation and expansion beads (Miltenyi) in 96-well, round-bottom plates.
Adoptive transfer induced Colitis
Naïve (CD45.1 CD4+CD45RBhighGFP–) and Treg (CD4+YFP+) cells are respectively isolated from the spleen of CD45.1 Foxp3EGFP and CD45.2 Foxp3YFPCre or Foxp3YFPCreRictorΔ/Δ mice. Colitis was induced in Rag1–/– males by i.p. injection of 5.105 CD45.1 naïve ± 5.104 Treg cells. Mice were weighed and monitored for signs of disease twice weekly. Large intestines were dissected from the mice and the fecal contents were flushed out using PBS containing 2% FCS. The intestines were cut into 1cm pieces and treated with PBS containing 2% FCS, 1.5 mM dithiothreitol, and 10mM EDTA at 37 °C for 30 min with constant stirring to remove mucous and epithelial cells. The tissues were then minced and the cells were dissociated in RPMI containing collagenase (2 mg/mL collagenase II; Worthington), DNase I (100 μg/mL; Sigma), 5mM MgCl2, 5mM CaCl2, 5mM HEPES, and 10% FBS with constant stirring at 37 °C for 45 min. Leukocytes were collected at the interface of a 40%/70% Percoll gradient (GE Healthcare). The cells were washed with PBS containing 2% FCS and used for experiments.
Immunoblotting
Cell extracts were prepared by using RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with a complete protease inhibitor cocktail (Roche), a Phos STOP phosphatase inhibitor cocktail (Roche). The lysates were mixed with 4x loading buffer (Biorad) and denatured by heating for 5 minutes in 100°C. Samples were subjected to SDS-PAGE. The resolved proteins were then electrically transferred to a PVDF membrane (Millipore). Immunoblotting was probed with indicated antibodies followed by anti-rabbit IgG, HRP-linked antibody. The protein bands were visualized by using a SuperSignal West Pico chemiluminescence ECL kit (Pierce). Signal intensities of immunoblot bands were quantified by Image J software. For pY319-ZAP70 and ZAP70 immunoblotting, cell-sorted Treg cells were stimulated with plate-bound anti-CD3 (145–2C11, 1 μg/mL) for 0, 2, 5 or 10 min prior protein extraction. A list of all antibodies used for immunoblotting is reported in Supplementary Table 4.
Metabolic profiling
Mouse splenocyte suspensions were enriched for CD4+ T cells by positive selection with magnetic beads (Miltenyi). Treg cells were cell sorted based on GFP expression and stimulated with plate-bound anti-CD3 (145–2C11, 1 μg/mL) and 100U recombinant mouse IL-2 (Peprotech) overnight. Sorted human Treg (CD4+CD127loCD25hi) cells were isolated from blood after PBMCs enrichment by Ficoll-Paque (Sigma-Aldrich) density gradient centrifugation and treated with the competitive dual mTOR inhibitor KU 0063794 (Tocris) at 8 μg/mL or with vehicle (DMSO) in presence of T cell activation and expansion beads (Miltenyi) and 100U/mL of recombinant human IL-2 (Peprotech). Cells were washed and seeded in Seahorse 8 wells plate at 105 cells per well. ECAR and OCR were measured for both mouse and human Treg cells using an XFp Extracellular Flux Analyzer respectively under glycolysis, mitochondrial stress and mitochondrial fuel test conditions (Seahorse Bioscience-Agilent). For glycolysis stress test, assay buffer was made of non-buffered DMEM medium supplemented with 2 mM glutamine andD-glucose, Oligomycin and 2-DG were sequentially injected at a final concentration of 10mM, 1μM and 50mM, respectively. For Mitochondrial stress test, assay buffer was made of non-buffered DMEM medium supplemented with 2.5 mM D-glucose, 2 mM glutamine and 1 mM sodium pyruvate and Oligomycin, Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) and Rotenone/Antimycin A were sequentially injected at a final concentration of 1μM, 1μM and 500nM, respectively. For Mitochondrial fuel test, assay buffer was made of non-buffered DMEM medium supplemented with 2.5 mM D-glucose, 2 mM glutamine and 1 mM sodium pyruvate and UK5099, Etomoxir and Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) were sequentially injected at a final concentration of 2μM, 4μM and 3μM, respectively. Baseline ECAR (for glycolysis stress test) and OCR (for mitochondrial stress and mitochondrial fuel tests) values were averaged between technical replicates for these first three successive time intervals. For lactate production by ELISA, 105 purified mouse naïve Teff cells (CD4+CD62LhiCD44lo) from Foxp3EGFP mice or Treg cells from Foxp3EGFP, Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ mice were seeded in flat bottom 96 well plate in complete RPMI medium in absence or presence of plate-bound anti-CD3 (1μg/mL) and 100U/mL of recombinant IL-2. Supernatants were collected after 48h and extracellular lactate production was measured using L-Lactate Assay Kit (Abcam). For metabolomics profiling, 2.5×106 purified Treg cells from Foxp3EGFP, Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ mice were seeded in flat bottom 48 well plate in complete RPMI medium in presence of plate-bound anti-CD3 (1μg/mL) and 100U/mL of recombinant IL-2. Supernatants and cell pellets were collected after 48h and analyzed for metabolomics studies (Metabolom ®, Morrisville, North Carolina, USA).
Transcriptome profiling
Splenic Treg (CD4+RFP+ or CD4+YFP+) cells were double-sorted from 4 weeks old female Foxp3ΔEGFPiCre/RFPR26YFP and Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP mice. (n=4 per group). Cells were collected directly into TRIzol (Invitrogen). Total RNA was extracted and converted into double-stranded DNA (dsDNA), using SMART-Seq v4 Ultra Low Input RNA kit (Clontech). dsDNA was then fragmented to 200- to 300-bp-sized fragments, using M220 Focused-ultrasonicator (Covaris), and these were used for the construction of libraries for Illumina sequencing, using the KAPA Hyper Prep Kit (Kapa Biosystems). Libraries were then quantified using Qubit dsDNA HS (high-sensitivity) Assay Kit on Agilent High Sensitivity DNA Bioanalyzer. RNA sequencing data was demultiplexed by using perfect matches to indices and was quality-inspected using FastQC. The sequencing data was aligned to the mm10 build (Gencode annotation) of the mouse genome using STAR62, and counts were quantified using HTSeq63. Raw counts were filtered for non-mitochondrial protein-coding genes with at least three counts in one sample, and were normalized using the DESeq2 package in R64. Pairwise comparisons of differential gene expression were computed using DESeq2.
Histology
Large intestine, lung, Liver and ear sections were stained with hematoxylin and eosin. Histopathological scoring of tissue was done by a blinded observer, and the final scores reflected averages of scores from 5 different ×200 fields per tissue per mouse. Large intestinal sections were scored as follows 53: 0, no inflammation; 1, mild, scattered infiltrates; 2, moderate infiltrates without loss of epithelium integrity; 3, moderate and diffuse or severe inflammation; 4, Severe inflammation associated with loss of the epithelial barrier integrity. Lung inflammation was scored separately for cellular infiltration around blood vessels and airways, as follows: 0, no infiltrates; 1, few inflammatory cells; 2, a ring of inflammatory cells 1 cell layer deep; 3, a ring of inflammatory cells 2–4 cells deep; 4, a ring of inflammatory cells >4 cells deep. A composite score was determined by adding the inflammatory scores for both vessels and airways. Liver inflammation was scored at portal areas, as follows: 0, no inflammatory cells; 1, mild, scattered infiltrates; 2, moderate infiltrates occupying less than 50% of the portal areas; 3, extensive infiltrates in the portal areas; 4, severe, with infiltrates completely packing the portal area and spilling over into the parenchyma. Ear inflammation was scored as followed: 0, no inflammation, no infiltration; Mild inflammation associated with few cells infiltration; 2, moderately severe inflammation associated with mild infiltration; 3, severe inflammation associated with large infiltration of cells and mild skin dryness; 4, very severe inflammation associated with skin dryness and cartilage erosion.
Methylation analysis
The methylation status of Foxp3 Treg-specific demethylation region (TSDR or CNS2) in splenic Treg cells of 25 d old male mice was assessed by bisulfite sequence analysis, as described 54. The TSDR of converted DNA was amplified by methylation-specific primer sequences: Foxp3 CNS2 Forward 5’-TATTTTTTTGGGTTTTGGGATATTA-3’ (forward) and Foxp3 CNS2 Reverse 5’-AACCAACCAACTTCCTACACTATCTAT-3’. The PCR product was subcloned and sequenced 54. Blast analyses were done by comparing the resulting sequences with converted Foxp3 gene sequences.
Confocal microscopy
Confocal microscopic analysis of Foxo1 nuclear and cytoplasmic distribution was carried out as described 28. Treg cells were purified and incubated on pre-coated coverslip (poly-L-lysin 50 μg/mL, ± anti-CD3 0.1 μg/mL) at 37°C for 30 min in RPMI/10%FCS. After fixation with PBS/4% paraformaldehyde, cells were permeabilized with PBS/0.1% saponin and blocked on PBS/4% bovine serum albumin (BSA). Cells were incubated with 1:100 diluted rabbit anti-Foxo1 (C29H4, Cell Signaling) followed by 1:500 diluted Alexa fluor 555-anti rabbit secondary antibody (Life technologies) in PBS/1%BSA. Slides were mounted with gold anti-fade reagent with DAPI (Invitrogen). Images were acquired with a Zeiss LSM700 confocal microscopy and ZEN imaging software. Five to 10 fields were selected randomly and total cells in the field were analyzed for percentage of Foxo1 nuclear localization using ImageJ software. Percentage of nuclear Foxo1 localization was obtained by the formula: 100 X corrected nuclear fluorescence/corrected total cell fluorescence and corrected fluorescence was obtained by the formula: Integrated Density – (Area of selected cell or nucleus X Mean fluorescence of background).
Chromatin Immunoprecipitation
ΔTreg cells were isolated from spleen of Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ mice. Cells were first cross-linked with 10% paraformaldehyde (PFA) for 8 minutes at room temperature (RT). Chromatin was centrifuged and the pellet was resuspended in lysis buffer I for 20 minutes at RT. The chromatin was pelleted again at 8000 rpm for 5 min at 4°C. The chromatin was then resuspended in 75 μl lysis buffer II containing 4% SDS. The chromatin was incubated for 5 min at RT to ensure the disruption of the nucleus and the release of the chromatin. The supernatant was then diluted to 1% SDS content using lysis buffer II without any SDS. Afterwards, the chromatin was sonicated using the bioruptor (Diagenode, Denville, NJ, USA) toward a proper size of 200–400 bp per DNA fragment for further immunoprecipitation. For the chromatin immunoprecipitation (ChIP) 3 antibodies were used to pull down the DNA fragments. Blimp1 (clone 3H2-E8, Invitrogen, USA), Foxo1 (clone 3B6, Invitrogen, USA) and mock IgG1 isotype control (Invitrogen, USA). Quantitative RT-PCR with the precipitated chromatin was performed to calculate the percentage of input and a fold change Blimp1/Isotype control or Foxo1/Isotype control was performed for each sample. All amplifications were performed in triplicate with SYBR Green PCR Master Mix (Qiagen) and specific primers for the different locations (5’-CTTGAGGAAAAGCCAGCATC-3’ Forward and 5’-TTTGCGTGTTCACCTGTGTT-3’ Reverse primers for il10 CNS–9, 5’-AGGCGAGATCTGAAGTGCAT-3’ Forward and 5’-CCTGCCGGTTGTAATCAACT-3’ Reverse primers for Tbx21 Promoter and 5’-TACTTCCTGCTCAGACCTGC-3’ Forward and 5’-TTCCCATCTCCTTCCTGTGG-3’ Reverse primers for Ifng Promoter). Control ChIP was performed with a respective isotype control antibody to ensure specificity. After normalization of the data according to the isotype control, the specific pulldown (percentage of input chromatin) was calculated.
Statistical analysis
Data were analyzed by paired and unpaired two-tailed Student’s t-test, one- and two-way ANOVA with post-test analyses and log-rank test, as indicated. Differences in mean values were considered significant at a P < 0.05.
Data Availability
Data presented in the manuscript, including de-identified patient results, will be made available to investigators following request to the corresponding author. Any data and materials to be shared will be released via a Material Transfer Agreement. RNAseq datasets have been deposited in the GEO with the accession code GSE129472.
Supplementary Material
Acknowledgements
This work was supported by a National Institutes of Health grants 2R01 AI065617 (to T.A.C.) and RO1 AI102888-01A1 (to M.L).
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Josefowicz SZ, Lu LF & Rudensky AY Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nutsch KM & Hsieh CS T cell tolerance and immunity to commensal bacteria. Curr Opin Immunol 24, 385–391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin W et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol 8, 359–368 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Gavin MA et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature 445, 771–775 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Ohkura N et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37, 785–799 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Kaech SM & Cui W Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudensky AY Regulatory T cells and Foxp3. Immunol Rev 241, 260–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck MD, Sowell RT, Kaech SM & Pearce EL Metabolic Instruction of Immunity. Cell 169, 570–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beier UH et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. FASEB J 29, 2315–2326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerriets VA et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest 125, 194–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalek RD et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 186, 3299–3303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi LZ et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med 208, 1367–1376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angelin A et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab 25, 1282–1293 e1287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerriets VA et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol 17, 1459–1466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollizzi KN & Powell JD Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol 36, 13–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman NM & Chi H mTOR Links Environmental Signals to T Cell Fate Decisions. Frontiers in immunology 5, 686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valmori D et al. Rapamycin-mediated enrichment of T cells with regulatory activity in stimulated CD4+ T cell cultures is not due to the selective expansion of naturally occurring regulatory T cells but to the induction of regulatory functions in conventional CD4+ T cells. J Immunol 177, 944–949 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Chapman NM, Karmaus PW, Zeng H & Chi H mTOR and metabolic regulation of conventional and regulatory T cells. J Leukoc Biol (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delgoffe GM et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng H et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarbassov DD, Guertin DA, Ali SM & Sabatini DM Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson SJ et al. Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regulatory T cells. J Immunol 186, 5533–5537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha S et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol 16, 178–187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams BL et al. Phosphorylation of Tyr319 in ZAP-70 is required for T-cell antigen receptor-dependent phospholipase C-gamma1 and Ras activation. EMBO J 18, 1832–1844 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin W et al. Allergic dysregulation and hyperimmunoglobulinemia E in Foxp3 mutant mice. J Allergy Clin Immunol 116, 1106–1115 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Leach MW, Bean AG, Mauze S, Coffman RL & Powrie F Inflammatory bowel disease in C.B-17 scid mice reconstituted with the CD45RBhigh subset of CD4+ T cells. Am J Pathol 148, 1503–1515 (1996). [PMC free article] [PubMed] [Google Scholar]
- 28.Charbonnier LM, Wang S, Georgiev P, Sefik E & Chatila TA Control of peripheral tolerance by regulatory T cell-intrinsic Notch signaling. Nat Immunol 16, 1162–1173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan YY & Flavell RA Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A 102, 5126–5131 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan X et al. PRR5L degradation promotes mTORC2-mediated PKC-delta phosphorylation and cell migration downstream of Galpha12. Nature cell biology 14, 686–696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann C et al. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J Exp Med 211, 1807–1819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plas DR & Thompson CB Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem 278, 12361–12366 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Ouyang W et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature 491, 554–559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerdiles YM et al. Foxo transcription factors control regulatory T cell development and function. Immunity 33, 890–904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagiwara A et al. Hepatic mTORC2 Activates Glycolysis and Lipogenesis through Akt, Glucokinase, and SREBP1c. Cell Metabolism 15, 725–738 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Wilhelm K et al. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature 529, 216–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearce EL, Poffenberger MC, Chang CH & Jones RG Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huynh A et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol 16, 188–196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longo N, Frigeni M & Pasquali M Carnitine transport and fatty acid oxidation. Biochim Biophys Acta 1863, 2422–2435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Bock K et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 154, 651–663 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Chatila TA et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest 106, R75–81 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacchetta R et al. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest 116, 1713–1722 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gavin MA et al. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A 103, 6659–6664 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bacchetta R, Barzaghi F & Roncarolo MG From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Annals of the New York Academy of Sciences (2016). [DOI] [PubMed] [Google Scholar]
- 46.Liu W et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med 203, 1701–1711 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seddiki N et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 203, 1693–1700 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L et al. Mammalian Target of Rapamycin Complex 2 Controls CD8 T Cell Memory Differentiation in a Foxo1-Dependent Manner. Cell reports 14, 1206–1217 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Hayatsu N et al. Analyses of a Mutant Foxp3 Allele Reveal BATF as a Critical Transcription Factor in the Differentiation and Accumulation of Tissue Regulatory T Cells. Immunity (2017). [DOI] [PubMed] [Google Scholar]
- 50.Bin Dhuban K et al. Suppression by human FOXP3+ regulatory T cells requires FOXP3-TIP60 interactions. Sci Immunol 2 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Bennett CL et al. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA-->AAUGAA) leads to the IPEX syndrome. Immunogenetics 53, 435–439 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Raymond CS & Soriano P High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One 2, e162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivas MN et al. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J Clin Invest 122, 1933–1947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitt EG et al. IL-10 produced by induced regulatory T cells (iTregs) controls colitis and pathogenic ex-iTregs during immunotherapy. J Immunol 189, 5638–5648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in the manuscript, including de-identified patient results, will be made available to investigators following request to the corresponding author. Any data and materials to be shared will be released via a Material Transfer Agreement. RNAseq datasets have been deposited in the GEO with the accession code GSE129472.