Mature biofilms are known for their high-level tolerance to antibiotics; however, antibiotic susceptibility of sessile cells during early-stage biofilm formation is not well understood. In this study, we aim to fill this knowledge gap by following bacterial antibiotic susceptibility during early-stage biofilm formation. We found that the attached cells have a dynamic change in antibiotic susceptibility, and during certain phases, they can be more sensitive to antibiotics than planktonic counterparts in the same culture. Using surface chemistry-controlled patterned biofilm formation, cell-surface and cell-cell interactions were found to affect the antibiotic susceptibility of attached cells. Collectively, these findings provide new insights into biofilm physiology and reveal how adaptation to the attached life form may influence antibiotic susceptibility of bacterial cells.
KEYWORDS: antibiotic tolerance, biofilm, cell-cell interaction, cell-surface interaction, patterned biofilm
ABSTRACT
Bacteria form complex multicellular structures on solid surfaces known as biofilms, which allow them to survive in harsh environments. A hallmark characteristic of mature biofilms is the high-level antibiotic tolerance (up to 1,000 times) compared with that of planktonic cells. Here, we report our new findings that biofilm cells are not always more tolerant to antibiotics than planktonic cells in the same culture. Specifically, Escherichia coli RP437 exhibited a dynamic change in antibiotic susceptibility during its early-stage biofilm formation. This phenomenon was not strain specific. Upon initial attachment, surface-associated cells became more sensitive to antibiotics than planktonic cells. By controlling the cell adhesion and cluster size using patterned E. coli biofilms, cells involved in the interaction between cell clusters during microcolony formation were found to be more susceptible to ampicillin than cells within clusters, suggesting a role of cell-cell interactions in biofilm-associated antibiotic tolerance. After this stage, biofilm cells became less susceptible to ampicillin and ofloxacin than planktonic cells. However, when the cells were detached by sonication, both antibiotics were more effective in killing the detached biofilm cells than the planktonic cells. Collectively, these results indicate that biofilm formation involves active cellular activities in adaption to the attached life form and interactions between cell clusters to build the complex structure of a biofilm, which can render these cells more susceptible to antibiotics. These findings shed new light on bacterial antibiotic susceptibility during biofilm formation and can guide the design of better antifouling surfaces, e.g., those with micron-scale topographic structures to interrupt cell-cell interactions.
IMPORTANCE Mature biofilms are known for their high-level tolerance to antibiotics; however, antibiotic susceptibility of sessile cells during early-stage biofilm formation is not well understood. In this study, we aim to fill this knowledge gap by following bacterial antibiotic susceptibility during early-stage biofilm formation. We found that the attached cells have a dynamic change in antibiotic susceptibility, and during certain phases, they can be more sensitive to antibiotics than planktonic counterparts in the same culture. Using surface chemistry-controlled patterned biofilm formation, cell-surface and cell-cell interactions were found to affect the antibiotic susceptibility of attached cells. Collectively, these findings provide new insights into biofilm physiology and reveal how adaptation to the attached life form may influence antibiotic susceptibility of bacterial cells.
INTRODUCTION
As the earliest form of life on earth, bacteria have developed remarkable strategies to survive in harsh environments, one being the formation of multicellular structures of biofilms with cells attached to a surface and embedded in a self-produced extracellular matrix (1). Biofilms are well-documented to provide bacterial cells protection from a variety of stresses, such as heavy metals, organic solvents, and antimicrobials (2–4). Biofilms of pathogenic bacteria allow these cells to tolerate higher concentrations of antibiotics (up to 1,000 times) than planktonic cells of the same species, leading to chronic infections and facilitating the development of multidrug-resistant strains, such as “superbugs” (2, 5). High-level antibiotic tolerance has been a major focus of previous biofilm research and is viewed as a hallmark characteristic of biofilms (2, 5). It is commonly stated that biofilm cells are more tolerant to antibiotics than their planktonic counterparts (6), except for few scarcely reported findings that chemical or mechanical biofilm removal can sensitize dispersed cells to antibiotics (7–10). How antibiotic susceptibility changes during biofilm formation is not well understood. The lack of a holistic view and some seeming discrepanc-ies among literature emphasize the need for a more in-depth understanding of antibiotic susceptibility during biofilm formation.
Biofilm cells obtain high-level tolerance to antibiotics through multiple mechanisms (2, 11–13). First, the polymeric matrix can protect biofilm cells from antibiotics by blocking or retarding the penetration of certain antibiotic molecules (2, 13). It is well documented that, for most antibiotics, the killing of biofilm cells is limited to the outer layers (14). However, for the molecules that do penetrate the extracellular matrix and reach biofilm cells, the killing effects still can be significantly reduced due to the slow growth of biofilm cells and the need for growth-associated targets required for the action of most antibiotics (2, 13). It is worth noticing that these results are largely based on mature biofilms. How antibiotic susceptibility changes during early events in the formation of a biofilm has not been well explored.
The early-stage biofilm formation includes initial adhesion and microcolony formation; both are important to the subsequent development of biofilm architecture and the physiology of biofilm cells (15–19). The generally accepted model is that motile bacteria use cell surface appendages, such as flagella and pili, to explore a surface and make the decision of switching from planktonic growth to biofilm formation (20–23). The attached cells also interact to form small colonies before further biofilm growth, matrix production, and maturation can take place (24, 25). These are rather active processes and, thus, are expected to be energy expensive and require coordination of cellular activities (24, 25). Thus, we hypothesize that bacterial antibiotic tolerance does not increase to a high level right after attachment but rather changes gradually during early-stage biofilm formation. We further hypothesize that cells involved in the interaction between cell clusters have a higher susceptibility to antimicrobials than those embedded in cell clusters, contributing to the well-known spatial variation in biofilm antibiotic susceptibility. In this study, we tested these hypotheses by following antibiotic susceptibility during early-stage biofilm formation of Escherichia coli and comparing the levels of susceptibility between cells in cell clusters and those involved in the interaction between cell clusters. We present evidence that biofilm cells are not always more tolerant to antibiotics than planktonic cells in the same culture. While the cells in mature biofilms have reduced metabolic activities, early events in the formation of a biofilm require active interaction between cells, which has a profound impact on the structure of biofilm formation and also leads to a window of elevated cellular activities and, thus, higher antibiotic susceptibility.
RESULTS
Antibiotic susceptibility of E. coli RP437 changed dynamically during early-stage biofilm formation.
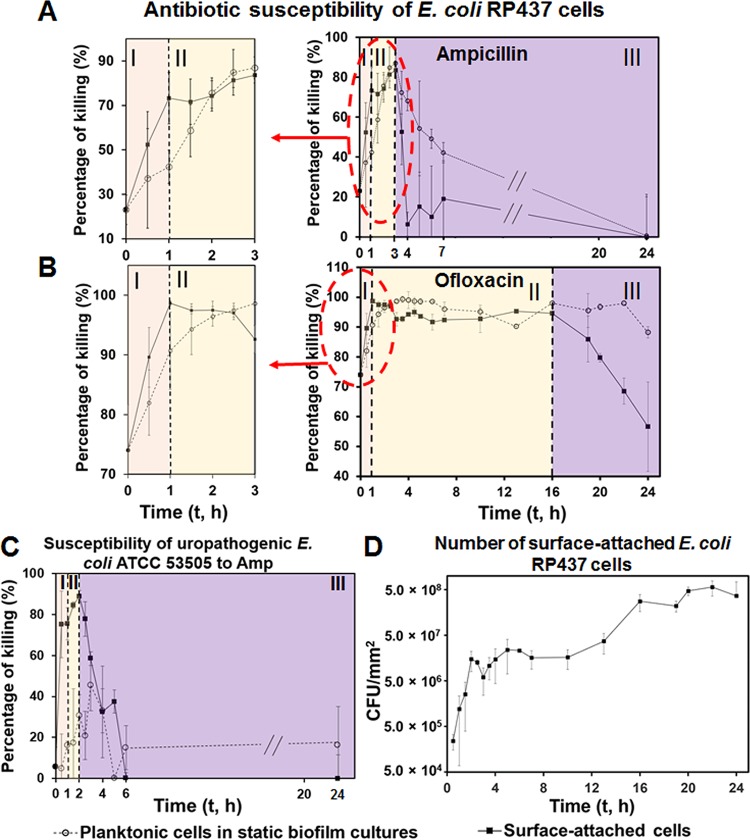
To follow the change in antibiotic susceptibility during early events in biofilm formation, E. coli RP437 biofilms were harvested at different time points during biofilm formation, followed with antibiotic treatment (200 μg/ml ampicillin [Amp] or 10 μg/ml ofloxacin [Ofx]) for 1 h in 0.85% NaCl. We chose these two concentrations because we were studying biofilm cells, which are known to have high-level tolerance to antibiotics (13). These two concentrations are 20 times greater than the MICs of E. coli RP437 (5 and 0.5 μg/ml for Amp and Ofx, respectively) and have been used to study E. coli persisters in biofilms (26, 27). Amp is effective only against active cells, while Ofx is known to also kill cells in the stationary phase (28). Although Amp showed lower killing effects in 0.85% NaCl than in a nutrient-abundant medium (LB) (see Fig. S1 in the supplemental material), we chose 0.85% NaCl solutions because this choice allows us to characterize the killing activity in the absence of growth (a confounding factor) and specifically compare the susceptibilities of bacterial cells at their native stage during early-stage biofilm formation. This condition has been widely used for biofilm research, including some of our previous studies (29–33). As expected (2, 12), E. coli RP437 cells in mature biofilms (24 h) are not susceptible to antibiotics (Fig. 1A and B and Fig. S2 in the supplemental material). Treatments with 200-μg/ml Amp did not show significant killing effects on 24-h biofilm cells, and 10-μg/ml Ofx only killed 24-h biofilm cells by 56.6% ± 15.0% (values are means ± standard deviations throughout; n ≥ 5). However, before entering this stage, the antibiotic susceptibility of attached E. coli RP437 cells exhibited a dynamic change over time (Fig. 1A and B and Fig. S2), during which antibiotic susceptibility first increased and reached a peak level before decreasing as the biofilm matured.
FIG 1.
E. coli antibiotic susceptibility during early stages of biofilm formation. (A and B) Reduction of surface-attached E. coli RP437 cells by 1-h treatment with 200-μg/ml Amp (A) or 10-μg/ml Ofx (B). (C) Reduction of surface-attached uropathogenic E. coli ATCC 53505 cells by 1-h treatment with 200-μg/ml Amp. (D) Number of surface-attached E. coli RP437 cells on glass surfaces. Dotted lines indicate the time points when major changes in antibiotic susceptibility occurred, which correspond to the three phases (I, II, and III) marked in panels A, B, and C. E. coli biofilms were formed on glass surfaces. The antibiotic susceptibility was tested in 0.85% NaCl solution (no carbon source) to minimize the effects of cell growth. At least five biological replicates were tested for each data point.
In this study, the biofilm cultures were inoculated with stationary-phase planktonic cells, which are known to be less susceptible to antibiotics than cells in the exponential phase (34). After planktonic cells were inoculated in fresh medium, their susceptibility to Amp and Ofx increased rapidly (Fig. 1A and B). After reaching a peak level at around 3 h after inoculation (86.9% ± 4.9% and 98.6% ± 0.5% killing by Amp and Ofx, respectively; n ≥ 5), the growth of planktonic E. coli RP437 cells in static cultures started entering the late exponential phase (see Fig. S3 in the supplemental material), leading to the decrease in susceptibility to Amp. In contrast, Ofx, an antibiotic that is known to be also effective against stationary-phase cells (28), remained effective in killing planktonic cells after 3 h of incubation.
Similar to that of planktonic cells in the same cultures, the susceptibility of initially attached E. coli RP437 cells to both antibiotics increased first upon inoculation. After reaching a peak level at 1 h after inoculation (68.8% ± 1.7% and 98.7% ± 0.4% of killing by Amp and Ofx, respectively; n ≥ 5), susceptibility remained rather unchanged and then decreased rapidly, e.g., 3 h after inoculation for Amp and 16 h after inoculation for Ofx (Fig. 1A and B). A similar change in susceptibility was also observed during the early-stage biofilm formation of uropathogenic E. coli ATCC 53505 (to 200-μg/ml Amp) (Fig. 1C) and Pseudomonas aeruginosa PAO1 (to 10-μg/ml tobramycin [Tob]) (see Fig. S4 in the supplemental material). Thus, the observed change is not strain specific. The data indicate that antibiotic susceptibility during early-stage biofilm formation can be divided into three phases, namely, phase I, II, and III (Fig. 1). In phase I, initially attached cells were more sensitive to antibiotics than planktonic cells, while surface-attached cells became comparably susceptible or less susceptible to antibiotics than planktonic cells in phases II and III. For example, 200-μg/ml Amp killed 73.3% ± 11.5%, 81.3% ± 3.3%, and 6.3% ± 6.0% of attached cells and 42.4% ± 7.5%, 84.9% ± 4.9%, and 68.1% ± 4.9% of planktonic cells in the same cultures at 1, 2.5, and 4 h after inoculation, respectively (n ≥ 5; P = 0.009, 0.7698, and < 0.0001, respectively; two-way analysis of variance [ANOVA] adjusted by Tukey test) (Fig. 1A and B). Although the changes in susceptibility to Amp and Ofx are similar during early-stage E. coli RP437 biofilm formation under our experimental conditions, the transition time from phase II to III differed between these two antibiotics (t = 3 h for Amp and t = 16 h for Ofx).
To understand how biofilm growth affects antibiotic susceptibility, we followed the attachment and growth of E. coli RP437 cells during early-stage biofilm formation. Instead of having two continuous phases (exponential and stationary phases) as planktonic cells in the same cultures (Fig. S3), the change in the number of attached cells appeared to have more phases. Upon inoculation, the number of attached cells increased quickly during the first 2 h. According to the tracking of E. coli RP437 on a glass surface (see Fig. S5 in the supplemental material), this increase in cell number was due to the attachment of planktonic cells instead of the division of attached cells. Between 2 and 10 h, the cell number remained rather stable. Then, it increased again between 10 and 16 h, followed by another plateau (Fig. 1D). The change in Amp and Ofx susceptibility from phase I to II (t = 1 h) occurred during a rapid increase in the number of initially attached cells, which explains the increase in antibiotic susceptibility. The transition from phase II to III (3 h for Amp and 16 h for Ofx) correlated well with changes in the number of attached cells (Fig. 1D), indicating the possible role of biofilm structure in antibiotic susceptibility.
Bacterial initial adhesion enhanced antibiotic susceptibility.
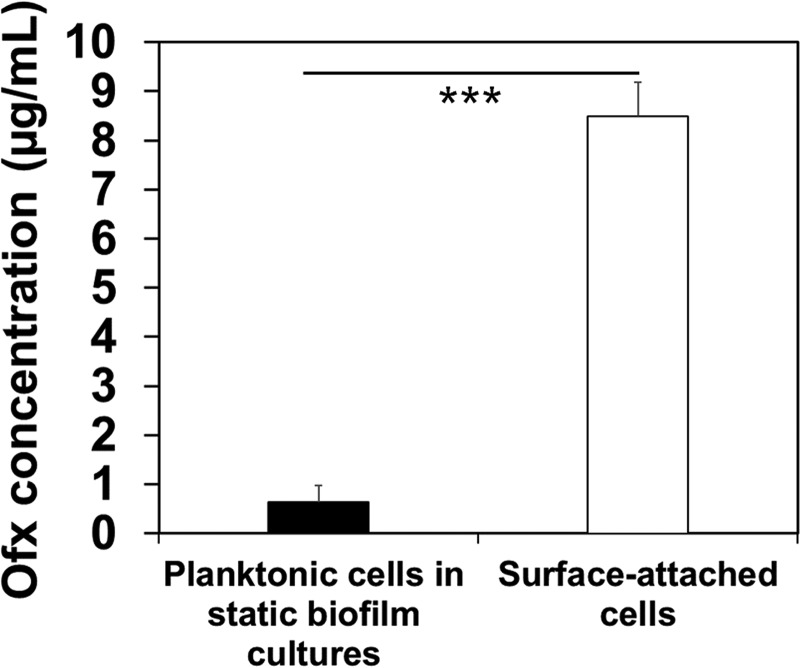
In the first hour after biofilm inoculation (Fig. 1A to C, phase I of antibiotic susceptibility graphs), initially attached cells became sensitive to antibiotics faster than planktonic cells (e.g., 2.6 and 1.5 times faster for Amp and Ofx, respectively; P < 0.01 for both, two-way ANOVA adjusted by Tukey test). This phenomenon was also observed for uropathogenic E. coli ATCC 53505 and P. aeruginosa PAO1 (Fig. 1C and Fig. S4), indicating that it is not specific to E. coli RP437. To corroborate the results and understand the cause of such changes, we compared the antibiotic concentrations in the sessile and planktonic E. coli RP437 cells. The liquid chromatography-mass spectrometry (LC-MS) results showed that after 15 min of incubation with 10 μg/ml Ofx, the intracellular drug concentration was 0.6 ± 0.3 μg/ml and 8.5 ± 0.7 μg/ml in planktonic and attached cells, respectively (n ≥ 3; P ≤ 0.0001, one-way ANOVA) (Fig. 2). This finding indicates that cell-surface interaction during the very early-stage biofilm formation can promote antibiotic penetration, consistent with the higher killing activities against early attached cells.
FIG 2.
The concentration of Ofx in surface-attached and planktonic cells at 15 min after inoculation (***, P < 0.0005). The antibiotic treatment was conducted in 0.85% NaCl solution. E. coli RP437 biofilms were formed on glass surfaces in LB medium, and each condition was tested with at least three biological replicates.
To understand if efflux plays a role in the difference in Ofx concentration, we tested Amp susceptibility of E. coli ΔacrA, a mutant of AcrAB-TolC efflux pump system, and compared it with that of its isogenic wild type (E. coli BW25113). If the change in efflux activity is responsible for the difference in intracellular concentration of an antibiotic between attached and planktonic cells, we would expect that mutation in efflux would abolish/significantly reduce such difference. The results show that, although the ΔacrA mutant is more sensitive to Amp than the wild-type strain in general, there is still a similar difference in Amp susceptibility between its surface-attached cells and planktonic cells (see Fig. S6 in the supplemental material). Because Amp is a substrate of this efflux system (35, 36), this result suggests that other factors are also involved.
According to Perozo et al. (37), the contact between bacteria and a substratum surface during initial attachment can deform the bacterial cell wall and, thus, causes the opening of mechanosensitive channels (MSC) on the membrane of E. coli cells, which triggers the secretion of extracellular polymeric substrates (EPSs) for subsequent biofilm formation (37–39). We hypothesize that the increase in pore size and other unknown changes in bacterial membranes upon initial attachment may also cause the increased penetration of antibiotics in sessile cells and, consequently, elevated antibiotic susceptibility among these cells, as observed in our study. Further study is needed to identify the exact factors affecting antibiotic penetration/accumulation during early-stage biofilm formation.
Cellular activities and biofilm-associated antibiotic susceptibility.
Under our experimental conditions, initially attached cells started active growth/elongation (see Fig. S7A in the supplemental material) with high-level cellular activities at around 1 h after inoculation (Fig. S7B). Consistently, surface-attached cells at this stage (late phase I and phase II) (Fig. 1A to C) showed a similar high-level susceptibility as planktonic cells did to Amp in 0.85% NaCl, an antibiotic that is known to be effective against metabolically active cells (40).
In addition to growth, biofilm formation also involves cell-cell interactions. In our earlier study, we observed that E. coli RP437 cells from adjacent clusters actively interact and form connections between them (24). Since such activities need energy, we hypothesized that such interactions may also play a role in biofilm antibiotic susceptibility. To test this hypothesis, we followed the antibiotic susceptibility of E. coli RP437 cells in patterned biofilms during early-stage biofilm formation. Patterned biofilms were formed on gold-coated glass surfaces with square-shaped CH3-SAM modified areas (20 μm by 20 μm) and various interpattern distances (D = 2, 5, or 10 μm) as we described previously (24). After 2 h of biofilm formation, patterned biofilms were harvested and treated with 200-μg/ml Amp for 1 h. Cell viability was determined using live/dead staining (Fig. 3A), and fluorescence signals were quantified using COMSTAT (41) (Fig. 3B). The results in Fig. 3A showed that the cells involved in interaction between clusters (94.4% ± 6.2% killing; n = 3) (Fig. 3B) were more susceptible to Amp (200 μg/ml) than those in the middle of clusters (14.2% ± 3.5% killing; n = 3; P < 0.0001, two-way ANOVA adjusted by Tukey test) (Fig. 3B). This finding indicates that cells involved in intercluster interactions were more sensitive to Amp.
FIG 3.
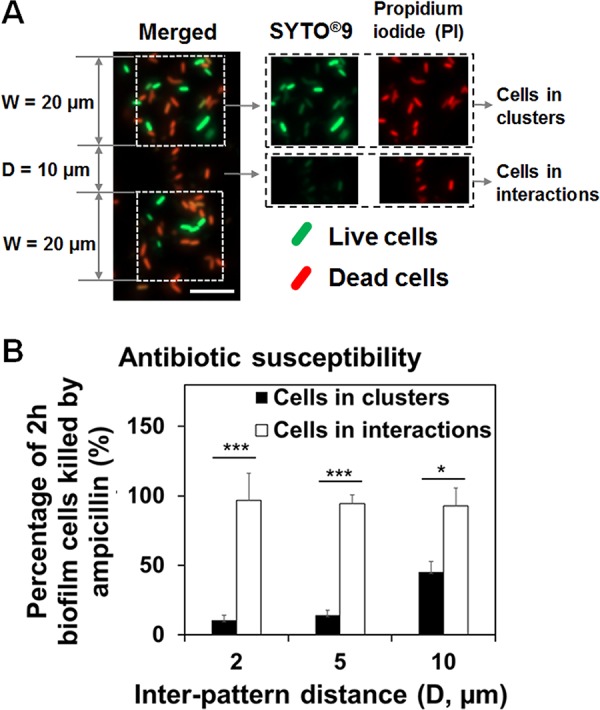
Cell-cell interactions affected the Amp susceptibility of attached cells. (A) Representative fluorescence images of 2-h patterned biofilms treated with 200-μg/ml Amp and labeled with live/dead staining (bar, 10 μm). The antibiotic treatment was conducted in 0.85% NaCl solution. (B) Percentage of 2-h patterned biofilm cells (W = 20 μm and D = 2, 5, or 10 μm) killed by 200-μg/ml Amp (*, P < 0.05; **, P < 0.005; and ***, P < 0.0005). E. coli RP437 biofilms were formed on gold-coated glass surfaces in LB medium, and each condition was tested with three biological replicates.
Cells embedded in early-stage biofilms remained active and sensitive to antibiotics if dispersed prior to antibiotic treatment.
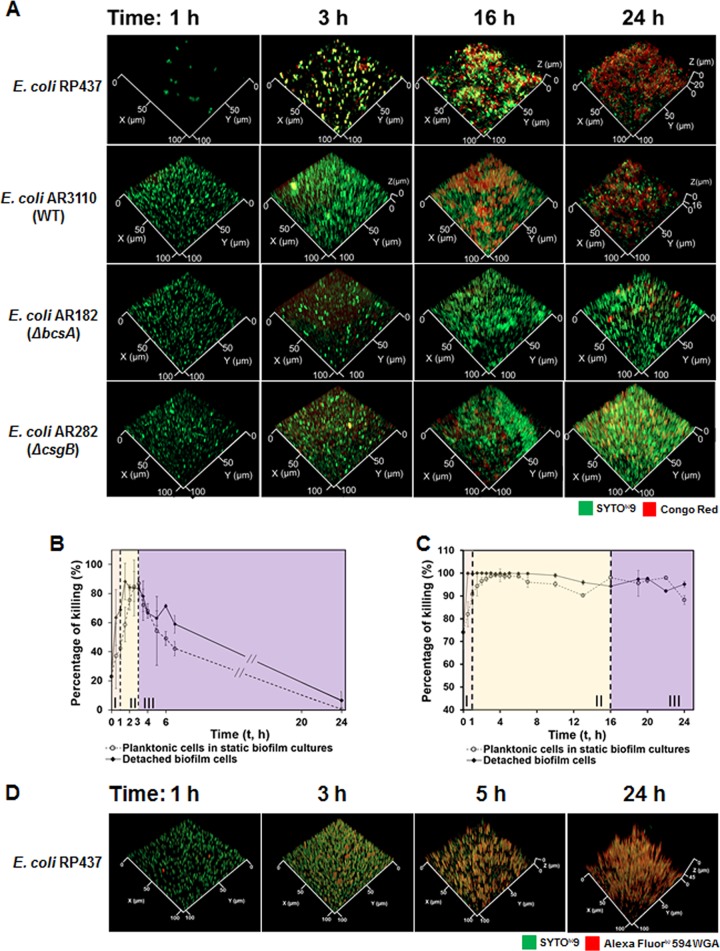
Susceptibility of E. coli RP437 biofilm cells to Amp and Ofx entered phase III at different time points (3 h for Amp and 16 h for Ofx), during which antibiotic susceptibility started decreasing. In addition, the change in biofilm susceptibility to both Amp and Ofx decreased faster than that of planktonic cells (Fig. 1A to C). When treated with 200-μg/ml Amp for 1 h, the killing of initially attached cells dropped 4.1 times faster than planktonic cells (77.4% versus 18.9%; P = 0.0012, two-way ANOVA adjusted by Tukey test). It is well known that planktonic cells become more tolerant to Amp upon entering stationary phase (42, 43). Biofilm cells have other mechanisms for antibiotic tolerance (2, 16, 44). During biofilm formation, bacteria produce extracellular polymeric substrates (EPSs) to facilitate three-dimensional (3D) biofilm formation when the surface coverage of initially attached cells reached saturation (∼10%) (39, 45), which is one of the well-known mechanisms that protect biofilm cells from antibiotics (39, 45, 46). In our system, we observed that the coverage of glass surfaces reached a plateau (12.0% ± 0.3%; n = 3) at 3 h after inoculation (see Fig. S8 in the supplemental material). To understand if the EPS produced at this critical time point had contributed to the different antibiotic susceptibility between biofilm and planktonic cells observed in this study, we followed the EPS production during early-stage biofilm formation using both Congo red and Alexa Fluor 594-WGA (Fig. 4). Congo red binds to cellulose and curli, and wheat germ agglutinin (WGA) targets sialic acid and N-acetyl-d-glucosamine, all of which are major components of E. coli EPS (25, 47). From 2.5 to 3 h, the biomass labeled with Congo red increased by 17.4 times based on the fluorescence signal (Fig. 4A and Fig. S9A in the supplemental material), indicating that E. coli RP437 cells start producing EPS at 3 h after inoculation. It coincides with the Alexa Fluor 594-WGA staining results, which also showed an increase in signal at this time point (Fig. 4D). Since the biofilm matrix of E. coli is known to impede β-lactam penetration significantly (48), this change correlates well with the significant decrease in Amp susceptibility in 0.85% NaCl among attached cells (77.4% decrease between 3- and 4-h biofilms; P < 0.0001, two-way ANOVA adjusted by Tukey test) (Fig. 1A). Consistently, the decrease in Amp susceptibility at 4 h after inoculation was also abolished by the deletion of bscA for cellulose synthesis or csgB for curli production compared to the wild-type strain (Fig. S9B and C). Collectively, these results indicate that cellulose and curli may influence the antibiotic susceptibility of surface-attached E. coli cells. Unlike β-lactam, the E. coli biofilm matrix cannot prevent fluoroquinolone penetration effectively (48), which explains why the susceptibility of biofilm cells to Ofx (fluoroquinolone family) did not show significant change at the same time point (P = 0.9257, two-way ANOVA adjusted by Tukey test) (Fig. 1B). A significant decrease in biofilm susceptibility to Ofx was observed around 16 h after inoculation (P < 0.05, two-way ANOVA adjusted by Tukey test), which could be the result of other mechanisms, such as the increase in biofilm thickness, and subsequent change in cellular activities and physiology (Fig. 4A).
FIG 4.
Cells embedded in early-stage biofilms remained active and sensitive to antibiotics if dispersed prior to antibiotic treatment. (A) Representative fluorescence images of E. coli RP437, E. coli AR3110 (wild type), E. coli AR182 (ΔbscA), and E. coli AR282 (ΔcsgB) biofilms at different time points (1, 3, 16, and 24 h) during biofilm formation. The embedded cells were labeled with SYTO9, and the cellulose in the biofilm matrix was labeled with Congo red. (B and C) Susceptibility of different cell populations (detached biofilm cells and planktonic cells in the same static biofilm cultures) to Amp (B) or Ofx (C) during early events of biofilm formation. The susceptibility of planktonic cells was repeated (from Fig. 1A and B) in these two panels as references for comparison. The antibiotic treatment was conducted in 0.85% NaCl solutions. (D) Representative fluorescence images of E. coli RP437 biofilms. Alexa Fluor 594-WGA stains sialic acid and N-acetyl-d-glucosamine in biofilm matrix, while SYTO9 stains surface-attached E. coli RP437 cells. E. coli RP437 biofilms were formed on glass surfaces in LB medium, and each condition was tested with three biological replicates (n = 3).
To further investigate if the change in antibiotic susceptibility was due to biofilm matrix or physiological changes in these cells, we followed the antibiotic susceptibility of biofilm cells after detaching them by gentle sonication. The mechanical force itself did not affect antibiotic susceptibility of E. coli RP437 cells (see Fig. S10 in the supplemental material). Interestingly, cells in young microcolonies were found more susceptible to antibiotics than planktonic cells if they were dispersed prior to antibiotic treatment (Fig. 4B and C and Fig. S11 in the supplemental material). For instance, in 6-h biofilms, 200-μg/ml Amp can kill only 10.1% ± 25.3% (n ≥ 5) surface-attached biofilm cells. However, this percentage increased to 71.4% ± 1.2% (n = 3) when biofilm cells were released by sonication prior to Amp treatment, which was higher than that of planktonic cells in the same culture (49.1% ± 4.7% killing; n ≥ 5; P = 0.0196, two-way ANOVA adjusted by Tukey test) (Fig. 4B). A similar result was obtained when cells were exposed to 10-μg/ml Ofx (Fig. 4C). When the antibiotic was used to treat 24-h biofilms, Ofx killed 96.9% ± 1.3% (n = 3) detached biofilm cells which is higher than the killing of both surface-attached biofilm cells (56.6% ± 15%; n ≥ 5; P < 0.0001, two-way ANOVA adjusted by Tukey test) and planktonic cells in the same biofilm cultures (88.2% ± 1.9%; n ≥ 5; P = 0.0008, two-way ANOVA adjusted by Tukey test). Thus, without the protection of the biofilm matrix, cells in early-stage biofilms can be more susceptible to antibiotics than planktonic cells, possibly due to the activities of cell cluster interactions and biofilm EPS synthesis.
Since Amp is not effective against metabolically inactive cells (49), the high-level Amp susceptibility of detached biofilm cells indicates that cells in early-stage biofilms are not dormant. To understand if these cells are indeed active, we used a reporter strain, E. coli ASV, with a chromosomally tagged unstable variant of green fluorescent protein (GFP) under the promoter rrnBP1, which allows one to track cellular activities using fluorescence microscopy (50). The result (Fig. S7B) demonstrates that the GFP signal from initially attached cells (1 h after inoculation) is >20 times stronger than that from planktonic cells in static biofilm cultures or shaken planktonic cultures in LB medium (P < 0.0001, one-way ANOVA). This finding indicates an increase in cellular activities in these newly attached cells and is consistent with our observation that initially attached cells are more sensitive to Amp and Ofx (Fig. 1A and B).
DISCUSSION
Biofilm formation is a dynamic process that includes initial adhesion, microcolony formation, maturation, and dispersion (51). Once a mature biofilm is formed, bacterial cells are extremely difficult to eradicate due to high-level tolerance to antimicrobials. However, how antibiotic susceptibility changes during biofilm formation is still not fully understood, which motivated this study.
It is interesting to us that bacterial activities during biofilm formation can render these attached cells even more susceptible to antibiotics than planktonic cells in the same culture during the early stages of biofilm formation. To our best knowledge, this has not been reported before. The killing of biofilm cells in Fig. 1 was determined using the standard assay by counting the viable cells attached on glass surfaces before and after treatment. Amp did not cause notable detachment, while Ofx caused moderate detachment. The detached cells were found to be highly sensitive to antibiotics (LIVE/DEAD images not shown), indicating that the killing of early-stage biofilms by Ofx in Fig. 1 is even underestimated. To more specifically study the susceptibility of biofilm cells in the absence of EPS, we detached the cells before antibiotic treatment. The results in Fig. 4 indicate the higher susceptibility of detached cells than of planktonic cells to Amp and Ofx during early-stage biofilm formation. Before EPS production started, we observed that cell-surface interaction led to an increase in antibiotic accumulation in attached cells, which helps explain why surface-attached cells can be more sensitive to antibiotics than planktonic cells during this stage of biofilm formation. With the production of EPS started at 3 h after inoculation, the susceptibility of biofilm to Amp decreased presumably due to the decrease in antibiotic penetration. As for Ofx, an antibiotic that has better permeability through E. coli biofilm matrix and killing effects on cells with lower metabolic activities than Amp (28, 48), the decrease in Ofx susceptibility of biofilm cells occurred later (16 h after inoculation) when the cellular activities started decreasing. Consistently, we demonstrate that without the protection of EPS, antibiotic susceptibility of dispersed early-stage biofilm cells was higher than planktonic cells due to active cellular activities and cell-cell interactions. These results inform us that antibiotics are more effective against initially attached cells than microcolonies, and the treatment on early-stage biofilms with biofilm matrix can be more effective by inducing a chemical or mechanical dispersion first, which are consistent with previous observations (7–10). The new finding is that biofilm susceptibility to antibiotics can be altered by influencing the level of interactions between cell clusters. Future studies using cell patterning and flow control may reveal further details about biofilm physiology and help develop better biofilm controls (52–55). This is part of our ongoing effort.
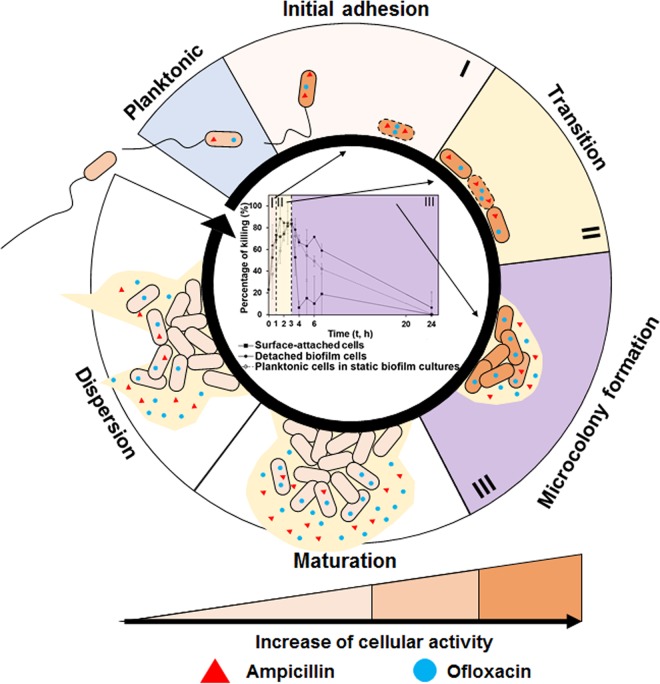
In summary, this study provides missing information about antibiotic susceptibility during early-stage biofilm formation. The main findings are summarized in Fig. 5. During reversible attachment, bacterial cells need to overcome repulsive forces from the surface to attach. During irreversible attachment, the change in mechanical forces sensed by flagella/pili of motile bacteria and membrane receptors of nonmotile bacteria can trigger cellular responses, leading to the switch from free-swimming to surface-attached phase (56). The upregulated cellular activities in initially attached cells render these cells more prone to antibiotic attack, and our data show that under certain conditions are even more susceptible than planktonic cells in the same culture (phase I). Antibiotic susceptibility of permanently attached cells can further increase due to the adhesion-triggered antibiotic penetration into these cells (phase I). After the initial adhesion, cells that are close to each other will initiate active cell-cell interaction (phase II). During this process, cells will divide, grow, rearrange their position, and consequently become highly sensitive to antibiotics. Under our experimental conditions, we define biofilm formation during this stage as a transition between initial attachment and microcolony formation, during which surface-attached cells are highly sensitive to antibiotics (phase II). After this stage, biofilm susceptibility to the antibiotics that have high affinity to biofilm matrix will start decreasing due to the production of EPS (phase III). However, the embedded cells are still susceptible to antibiotics if dispersed from the EPS matrix. As for the antibiotics with low affinity to biofilm matrix (better penetration), the level of susceptibility will remain for a longer period of time until the cells become rather dormant in mature biofilms.
FIG 5.
A hypothetic model of antibiotic susceptibility during early-stage biofilm formation. Phases I, II, and III in this diagram are the main findings from this study. Cells with dotted lines in the cell membrane represent these initially attached cells or cells in interactions with increased membrane permeability. The mechanisms in biofilm maturation and dispersion are based on literature. They are included here to show the whole process of biofilm formation and associated antibiotic tolerance.
The results of this study shed new light on the understanding of early-stage biofilm formation and indicate that attached cells are not always highly tolerant to antibiotics. There is a time window that surface-attached cells are more sensitive than their planktonic counterparts, suggesting an opportunity to maximize the antifouling effects by combining other physical/chemical factors with conventional antibiotics. The results of this study complement current knowledge on biofilm-associated antibiotic resistance and may guide the design of better strategies for biofilm control, e.g., the design of smart biomaterials and medical devices that can interrupt bacteria cell-cell interactions.
MATERIALS AND METHODS
Bacterial strains and media.
The strains used in this study are summarized in Table S1 in the supplemental material. Bacterial cells were routinely grown at 37°C with shaking at 200 rpm in lysogeny broth (LB) (57) containing 10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter sodium chloride overnight. The overnight cultures were used to inoculate 20-ml fresh LB solution with an optical density at 600 nm (OD600) of 0.05 for biofilm formation.
Surface preparation.
Chemically modified gold surfaces (with 5-nm Ti and 10-nm Au) were prepared using microcontact printing as described previously (24). Briefly, the polydimethylsiloxane (PDMS) surfaces with square-shaped microtopographic patterns (10 μm tall and 20 μm wide with 2-, 5-, 10-, 15-, or 20-μm interpattern distance) were used to pattern the gold surfaces with HS(CH2)14CH3 (10 mM in 95% ethanol; Sigma-Aldrich Co., St. Louis, MO). Then, patterned gold surfaces were soaked in HS(CH2)11(OCH2CH2)3OH (10 mM in 95% ethanol; Sigma-Aldrich Co.) for 24 h to modify the remaining area with self-assembled monolayer presenting tri(ethylene glycol) groups. After 24 h of incubation, the gold surfaces were dried using filtered air (sterilized using 0.22-μm filters) and transferred into a clean petri dish for biofilm formation.
Biofilms were also formed on glass slides with a dimension of 2.5 cm by 2.0 cm. Before biofilm inoculation, glass slides were sterilized by soaking in 95% ethanol for 30 min and then transferred to clean petri dishes for drying at 50°C for 40 min. To test biofilm formation on glass slides, clean glass slides were placed at the bottom of clean petri dishes. Biofilm formation was conducted at 37°C without shaking.
Antibiotic susceptibility.
To evaluate the antibiotic susceptibility of bacterial cells during biofilm formation, different cells (planktonic cells in static biofilm cultures, detached biofilm cells, and surface-attached cells) were collected at different time points. For Amp, samples were taken every 30 min during the first 7 h of incubation and then at 24 and 48 h after inoculation. For Ofx, samples were taken at every 30 min during the first 7 h and then at 10, 13, 16, 19, 20, 22, and 24 h. For the Amp and Tob treatment during the early-stage biofilm formation of uropathogenic E. coli ATCC 53505 and P. aeruginosa PAO1, samples were taken every 30 min during the first 3 h of incubation and then at 4, 5, 6, and 24 h after inoculation.
Planktonic cells were harvested at the same time points from static biofilm cultures and washed three times before being treated with an antibiotic for 1 h in 0.85% NaCl solution at 37°C with shaking at 200 rpm. Planktonic cells grown with shaking at 200 rpm were also harvested at 2 h after inoculation and treated with Amp for 1 h in LB medium at 37°C with shaking at 200 rpm to confirm the killing effects of Amp. Glass slides with biofilms were washed three times with clean 0.85% NaCl solution before antibiotic treatment at 37°C without shaking. To physically detach biofilm cells, each glass surface with biofilm was washed three times and then transferred to 3-ml clean 0.85% NaCl solution in a test tube. The surface was sonicated for 30 s followed by pipetting for 1 min to detach biofilm cells. The detached biofilm cells were collected by centrifugation at 13,200 × g for 5 min and resuspended in appropriate antibiotic solution for 1-h treatment at 37°C. To test the effect of sonication on bacterial antibiotic susceptibility, planktonic cells from static biofilm cultures were also sonicated for 30 s and pipetted for 1 min before antibitoic treatment. Since the biofilm cells were detached into clean 0.85% NaCl solutions in test tubes after the wash, biofilm cells would not contaminate the planktonic population, which was harvested before biofilms were taken for washing.
To evaluate antibiotic susceptibility, the number of cells with and without antibiotic treatment was determined using drop plate method to count CFUs, as described previously (58). Antibiotic susceptibility was also evaluated using LIVE/DEAD BacLight bacterial viability kit (Life Technologies Inc., Carlsbad, CA). Using this method, all cells were labeled with SYTO9 in green fluorescence, and the dead cells were labeled with propidium iodide (PI) in red fluorescence. Fluorescence signals were quantified using MATLAB-based COMSTAT (41). The percentage of cells killed by antibiotics was calculated by dividing the surface coverage of dead cells (red fluorescence signals) with the surface coverage of all cells (both red and green fluorescence signal).
Bacterial cellular activity during early attachment.
E. coli ASV (rrnBP1-gfp) (both planktonic cells in static biofilm cultures and detached biofilm cells) was taken at 1 h after inoculation and resuspended in 0.85% NaCl solution. The intensity of GFP signal was measured using a Synergy2 microplate reader (BioTek, Winooski, VT) and normalized by cell number. Stationary and exponential phases of E. coli ASV cells and 0.85% NaCl were used as controls. Each condition was tested with three biological replicates.
Biofilm matrix production.
To track biofilm EPS production during biofilm formation, 40-μg/ml Congo red (Sigma-Aldrich, St. Louis, MO) was added into biofilm culture during inoculation. After biofilm formation, the samples were washed three times with 0.85% NaCl solution before the biofilm cells were labeled with SYTO9 in 0.85% NaCl solution (vol/vol, 1.5:1) for 15 min. The labeled samples were imaged using an Axio Imager M1 fluorescence microscope (Carl Zeiss Inc., Berlin, Germany).
To corroborate the results, Alexa Fluor 594-WGA (Thermo Fisher Scientific, Waltham, MA) was also used to stain E. coli RP437 biofilms taken at 1, 3, 5, and 24 h after inoculation. The biofilm samples were stained by incubating in phosphate-buffered saline (PBS) supplemented with 100-μg/ml Alexa Fluor 594-WGA for 30 min after three times of washing in PBS. The biofilm cells were labeled with SYTO9 before they were imaged using an Axio Imager M1 fluorescence microscope.
Microscopy and data analysis.
Labeled cells were transferred onto microscopic slides and imaged using an Axio Imager M1 fluorescence microscope with a camera (Orca-Flash 4.0 LT; Hamamatsu Photonics, Hamamatsu City, Japan).
For labeled planktonic cells, two-dimensional (2D) pictures were taken. Cells were vortexed before imaging to ensure even distribution. For biofilm cells, especially mature biofilms, three-dimensional (3D) images were taken. At least 5 random spots were imaged for each sample. The ratio between different signals was analyzed using MATLAB-based COMSTAT (41) and Zen Pro 10.1 (Carl Zeiss Inc., Berlin, Germany).
Ofx penetration into 1-h biofilm cells and planktonic cells.
To quantify the amount of Ofx that penetrated biofilm and planktonic cells, cells were harvested and then treated with 10-μg/ml Ofx for 15 min. We shortened the Ofx treatment time from 1 h (in killing tests) to 15 min to avoid the artifact in quantifying antibiotic concentration due to cell death/cell lysis, since treatment for 15 min resulted in no killing and, thus, ensured the integrity of cells (see Fig. S12 in the supplemental material). After being washed once with 0.85% NaCl solution, cells were detached and then lysed with chloroform to release the accumulated antibiotic into the solvent. Cell debris was removed by centrifugation (5,000 × g for 5 min), and the solvent with released antibiotic was collected. The solvents were evaporated in a vacuum desiccator. Antibiotics were resuspended in 80% methanol solution for quantification using Thermo LTQ Orbitrap mass spectrometer at State of New York, Upstate Medical University (Syracuse, NY).
Statistics.
SAS 9.1.3, Windows version (Cary, NC), was used for all statistical analyses. Either one-way or two-way ANOVA followed by Tukey test was used depending on the nature of the test. Results with P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the U.S. National Science Foundation (CBET-1706061 and DMR-1836723) and U.S. National Institutes of Health (1R21AI142424-01) for supporting parts of this study.
We are grateful to Arne Heydorn at the Technical University of Denmark for providing the COMSTAT software. We also thank the Cornell NanoScale Science & Technology Facility for the access to photolithography facilities and Ebbing de Jong from the Core Facility for Proteomics & Mass Spectrometry for the analysis of Ofx concentrations using LC-MS (NIH grant 1S10OD023617-01A1).
D.R. and H.G. conceived the concept and designed the experiments. H.G., S.W.L., Z.J., and J.C. carried out the experiments. D.R., H.G., and Z.J. cowrote the paper. All authors discussed the results and comments on the manuscript.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00034-19.
REFERENCES
- 1.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 2.Hall CW, Mah TF. 2017. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 3.Azizi S, Kamika I, Tekere M. 2016. Evaluation of heavy metal removal from Wastewater in a modified packed bed biofilm reactor. PLoS One 11:e0155462. doi: 10.1371/journal.pone.0155462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein AK, Pokroy B, Seminara A, Aizenberg J. 2011. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci U S A 108:995–1000. doi: 10.1073/pnas.1011033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood TK. 2017. Strategies for combating persister cell and biofilm infections. Microb Biotechnol 10:1054–1056. doi: 10.1111/1751-7915.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollet C, Gal L, Guzzo J. 2009. Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: a comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa. FEMS Microbiol Lett 290:135–142. doi: 10.1111/j.1574-6968.2008.01415.x. [DOI] [PubMed] [Google Scholar]
- 7.Sultana ST, Call DR, Beyenal H. 2016. Eradication of Pseudomonas aeruginosa biofilms and persister cells using an electrochemical scaffold and enhanced antibiotic susceptibility. NPJ Biofilms Microbiomes 2:2. doi: 10.1038/s41522-016-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart PS, Rayner J, Roe F, Rees WM. 2001. Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J Appl Microbiol 91:525–532. doi: 10.1046/j.1365-2672.2001.01413.x. [DOI] [PubMed] [Google Scholar]
- 9.Berlanga M, Gomez-Perez L, Guerrero R. 2017. Biofilm formation and antibiotic susceptibility in dispersed cells versus planktonic cells from clinical, industry and environmental origins. Antonie Van Leeuwenhoek 110:1691–1704. doi: 10.1007/s10482-017-0919-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee SW, Gu H, Kilberg JB, Ren D. 2018. Sensitizing bacterial cells to antibiotics by shape recovery triggered biofilm dispersion. Acta Biomater 81:93–102. doi: 10.1016/j.actbio.2018.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penesyan A, Gillings M, Paulsen IT. 2015. Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20:5286–5298. doi: 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen I. 2015. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis 34:877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 13.Lewis K. 2001. Riddle of biofilm resistance. Antimicrob Agents Chemother 45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciofu O, Rojo-Molinero E, Macià MD, Oliver A. 2017. Antibiotic treatment of biofilm infections. APMIS 125:304–319. doi: 10.1111/apm.12673. [DOI] [PubMed] [Google Scholar]
- 15.Crouzet M, Le Senechal C, Brözel VS, Costaglioli P, Barthe C, Bonneu M, Garbay B, Vilain S. 2014. Exploring early steps in biofilm formation: set-up of an experimental system for molecular studies. BMC Microbiol 14:253. doi: 10.1186/s12866-014-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. 2017. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Htwe Mon Y-R, Ritter AL, Falkinham JO III, Ducker WA. 2018. Effects of colloidal crystals, antibiotics, and surface-bound antimicrobials on Pseudomonas aeruginosa surface density. ACS Biomater Sci Eng 4:257–265. doi: 10.1021/acsbiomaterials.7b00799. [DOI] [PubMed] [Google Scholar]
- 18.Chang YR, Weeks ER, Ducker WA. 2018. Surface topography hinders bacterial surface motility. ACS Appl Mater Interfaces 10:9225–9234. doi: 10.1021/acsami.7b16715. [DOI] [PubMed] [Google Scholar]
- 19.Lagree K, Mon HH, Mitchell AP, Ducker WA. 2018. Impact of surface topography on biofilm formation by Candida albicans. PLoS One 13:e0197925. doi: 10.1371/journal.pone.0197925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song F, Brasch ME, Wang H, Henderson JH, Sauer K, Ren D. 2017. How bacteria respond to material stiffness during attachment: a role of Escherichia coli flagellar motility. ACS Appl Mater Interfaces 9:22176–22184. doi: 10.1021/acsami.7b04757. [DOI] [PubMed] [Google Scholar]
- 21.Gu H, Chen A, Song XR, Brasch ME, Henderson JH, Ren DC. 2016. How Escherichia coli lands and forms cell clusters on a surface: a new role of surface topography. Sci Rep 6:29516. doi: 10.1038/srep29516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lele PP, Hosu BG, Berg HC. 2013. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A 110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berne C, Ducret A, Hardy GG, Brun YV. 2015. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu H, Hou S, Yongyat C, De Tore S, Ren D. 2013. Patterned biofilm formation reveals a mechanism for structural heterogeneity in bacterial biofilms. Langmuir 29:11145–11153. doi: 10.1021/la402608z. [DOI] [PubMed] [Google Scholar]
- 25.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Wu N, Zhang S, Yuan Y, Zhang W, Zhang Y. 2017. Variable persister gene interactions with (p)ppGpp for persister formation in Escherichia coli. Front Microbiol 8:1795. doi: 10.3389/fmicb.2017.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amato SM, Brynildsen MP. 2014. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One 9:e93110. doi: 10.1371/journal.pone.0093110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Völzing KG, Brynildsen MP. 2015. Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. mBio 6:e00731. doi: 10.1128/mBio.00731-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niepa TH, Gilbert JL, Ren D. 2012. Controlling Pseudomonas aeruginosa persister cells by weak electrochemical currents and synergistic effects with tobramycin. Biomaterials 33:7356–7365. doi: 10.1016/j.biomaterials.2012.06.092. [DOI] [PubMed] [Google Scholar]
- 30.Pan JC, Bahar AA, Syed H, Ren D. 2012. Reverting antibiotic tolerance of Pseudomonas aeruginosa PAO1 persister cells by (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one. PLoS One 7:e45778. doi: 10.1371/journal.pone.0045778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu H, Lee SW, Buffington SL, Henderson JH, Ren D. 2016. On-demand removal of bacterial biofilms via shape memory activation. ACS Appl Mater Interfaces 8:21140–21144. doi: 10.1021/acsami.6b06900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou S, Gu H, Smith C, Ren D. 2011. Microtopographic patterns affect Escherichia coli biofilm formation on poly(dimethylsiloxane) surfaces. Langmuir 27:2686–2691. doi: 10.1021/la1046194. [DOI] [PubMed] [Google Scholar]
- 33.Wood TL, Gong T, Zhu L, Miller J, Miller DS, Yin B, Wood TK. 2018. Rhamnolipids from Pseudomonas aeruginosa disperse the biofilms of sulfate-reducing bacteria. NPJ Biofilms Microbiomes 4:22. doi: 10.1038/s41522-018-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pletnev P, Osterman I, Sergiev P, Bogdanov A, Dontsova O. 2015. Survival guide: Escherichia coli in the stationary phase. Acta Naturae 7:22–33. doi: 10.32607/20758251-2015-7-4-22-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anes J, McCusker MP, Fanning S, Martins M. 2015. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol 6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perozo E, Kloda A, Cortes DM, Martinac B. 2002. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat Struct Biol 9:696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 38.Harapanahalli AK, Younes JA, Allan E, van der Mei HC, Busscher HJ. 2015. Chemical signals and mechanosensing in bacterial responses to their environment. PLoS Pathog 11:e1005057. doi: 10.1371/journal.ppat.1005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carniello V, Peterson BW, van der Mei HC, Busscher HJ. 2018. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv Colloid Interface Sci 261:1. doi: 10.1016/j.cis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 41.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 42.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 44.Gu H, Kolewe KW, Ren D. 2017. Conjugation in Escherichia coli biofilms on poly(dimethylsiloxane) surfaces with microtopographic patterns. Langmuir 33:3142–3150. doi: 10.1021/acs.langmuir.6b04679. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Busscher HJ, van der Mei HC, Norde W, Krom BP, Sjollema J. 2011. Analysis of the contribution of sedimentation to bacterial mass transport in a parallel plate flow chamber: part II: use of fluorescence imaging. Colloids Surf B Biointerfaces 87:427–432. doi: 10.1016/j.colsurfb.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Yan J, Sharo AG, Stone HA, Wingreen NS, Bassler BL. 2016. Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc Natl Acad Sci U S A 113:E5337-43. doi: 10.1073/pnas.1611494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hufnagel DA, Depas WH, Chapman MR. 2015. The biology of the Escherichia coli extracellular matrix. Microbiol Spectr 3(3):MB-0014-2014. doi: 10.1128/microbiolspec.MB-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh R, Sahore S, Kaur P, Rani A, Ray P. 2016. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog Dis 74:ftw056. doi: 10.1093/femspd/ftw056. [DOI] [PubMed] [Google Scholar]
- 49.Kaldalu N, Mei R, Lewis K. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob Agents Chemother 48:890–896. doi: 10.1128/AAC.48.3.890-896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song F, Koo H, Ren D. 2015. Effects of material properties on bacterial adhesion and biofilm formation. J Dent Res 94:1027–1034. doi: 10.1177/0022034515587690. [DOI] [PubMed] [Google Scholar]
- 52.Collins DJ, Morahan B, Garcia-Bustos J, Doerig C, Plebanski M, Neild A. 2015. Two-dimensional single-cell patterning with one cell per well driven by surface acoustic waves. Nat Commun 6:8686. doi: 10.1038/ncomms9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren D, Xia Y, You Z. 2013. Multiplexed living cells stained with quantum dot bioprobes for multiplexed detection of single-cell array. J Biomed Opt 18:096005. doi: 10.1117/1.JBO.18.9.096005. [DOI] [PubMed] [Google Scholar]
- 54.Ren D, Xia Y, Wang J, You Z. 2013. Micropatterning of single cell arrays using the PEG-silane and biotin–(strept)avidin system with photolithography and chemical vapor deposition. Sens Actuator B Chem 188:340–346. doi: 10.1016/j.snb.2013.07.037. [DOI] [Google Scholar]
- 55.Ren D, Cui M, Xia Y, You Z. 2012. Micropatterning and its applications in biomedical research. Pro Biochem Biophys 39:931–944. doi: 10.3724/SP.J.1206.2011.00228. [DOI] [Google Scholar]
- 56.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 58.Naghili H, Tajik H, Mardani K, Razavi Rouhani SM, Ehsani A, Zare P. 2013. Validation of drop plate technique for bacterial enumeration by parametric and nonparametric tests. Vet Res Forum 4:179–183. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.