Bacterial type 4 pili (T4P) have been termed the “Swiss Army knives” of bacteria because they perform numerous functions, including host cell interaction, twitching motility, colony formation, DNA uptake, protein secretion, and surface sensing. The pilus fiber continuously elongates or retracts, and these dynamics are functionally important. Curiously, only a subset of T4P systems employ T4P retraction ATPases to power T4P retraction. Here, we show that one of the strongest T4P machines, the gonococcal T4P, retracts without a retraction ATPase. Biophysical characterization reveals strongly reduced force and speed compared to retraction with ATPase. We propose that bacteria encode retraction ATPases when T4P have to generate high-force-supporting functions like twitching motility, triggering host cell response, or fluidizing colonies.
KEYWORDS: Neisseria gonorrhoeae, molecular motor, pilus, twitching motility
ABSTRACT
Bacterial type 4 pili (T4P) belong to the strongest molecular machines. The gonococcal T4P retraction ATPase PilT supports forces exceeding 100 pN during T4P retraction. Here, we address the question of whether gonococcal T4P retract in the absence of PilT. We show that pilT deletion strains indeed retract their T4P, but the maximum force is reduced to 5 pN. Similarly, the speed of T4P retraction is lower by orders of magnitude compared to that of T4P retraction driven by PilT. Deleting the pilT paralogue pilT2 further reduces the speed of T4P retraction, yet T4P retraction is detectable in the absence of all three pilT paralogues. Furthermore, we show that depletion of proton motive force (PMF) slows but does not inhibit pilT-independent T4P retraction. We conclude that the retraction ATPase is not essential for gonococcal T4P retraction. However, the force generated in the absence of PilT is too low to support important functions of T4P, including twitching motility, fluidization of colonies, and induction of host cell response.
IMPORTANCE Bacterial type 4 pili (T4P) have been termed the “Swiss Army knives” of bacteria because they perform numerous functions, including host cell interaction, twitching motility, colony formation, DNA uptake, protein secretion, and surface sensing. The pilus fiber continuously elongates or retracts, and these dynamics are functionally important. Curiously, only a subset of T4P systems employ T4P retraction ATPases to power T4P retraction. Here, we show that one of the strongest T4P machines, the gonococcal T4P, retracts without a retraction ATPase. Biophysical characterization reveals strongly reduced force and speed compared to retraction with ATPase. We propose that bacteria encode retraction ATPases when T4P have to generate high-force-supporting functions like twitching motility, triggering host cell response, or fluidizing colonies.
INTRODUCTION
Bacterial type 4 pili (T4P) are among the strongest molecular machines known to date. In some species, they generate forces exceeding 100 pN (1–3), i.e., 20-fold higher than the force generated by muscle myosin. Force generation has been linked to diverse functions, including twitching motility (4–7), host cell interaction (8–11), and regulation of biofilm structure and dynamics (12–17). For all of these functions, the retraction ATPase PilT is required. Interestingly, some T4P systems involved in protein secretion, DNA uptake during transformation, or surface sensing bear no pilT-like gene. Very recently, it has been shown that T4P can retract in the absence of a retraction ATPase (18–20). The forces generated by these pili, however, are an order of magnitude lower than the force observed for Neisseria gonorrhoeae T4P retraction. It remains unclear whether gonococci can retract T4P in the absence of PilT.
The T4P filament is a helical structure built from thousands of major pilin subunits and various minor pilins (21, 22). Recent advances in cryo-electron microscopy together with high-resolution structures of the individual components have given insight into the structure of the complex machinery that shuttles pilin subunits from the cytoplasmic membrane into the growing pilus (23–27) (see Fig. S1 in the supplemental material). Two ATPases power elongation (PilF) (28) and retraction (PilT) (29) of the pilus, respectively. Biophysical, electron microscopy, and crystallographic studies suggest that these motors form oblong hexameric rings (30, 31). It was proposed that sequential ATP binding leads to functionally relevant deformations that propagate around the ring in opposite directions for the elongation and retraction ATPase (24). These sequential deformations of the ATPases would couple to the pilus fiber through a platform complex. (23, 24). Since the T4P fiber is helical, opposite rotations driven by PilF and PilT would then power elongation and retraction of the T4P fiber, respectively. The exact coupling mechanism remains unclear.
While all bacteria generating T4P encode elongation ATPases, not all encode retraction ATPases. For example, DNA uptake during transformation has been reported to require a retraction ATPase in N. gonorrhoeae and Vibrio cholerae (29, 32). However, Bacillus subtilis and Streptococcus pneumoniae do not carry a clear pilT homologue, but they still employ T4P for DNA uptake (33). The first T4P system shown to retract T4P in the absence of a retraction ATPase was the toxin-coregulated pilus of V. cholerae (18). The maximum force generated by these T4P was in the range of 4 pN. Furthermore, the Tad pilus of Caulobacter crescentus generates somewhat higher force, in the range of 12 pN (19). We note that it is unknown how force generation depends on experimental conditions. For two T4P systems that naturally encode a retraction ATPase, force generation was observed when pilT was deleted. First, in Myxococcus xanthus, deletion of pilT leads to strong reduction of T4P retraction frequency and almost complete loss of twitching motility (2). The force was reduced 2-fold. M. xanthus carries four pilT paralogues, and it is therefore unclear whether any of them encodes a functional retraction ATPase. Second, the competence pilus of V. cholerae showed T4P retraction when pilT was deleted (20), while the transformation rate was severely reduced. Forces generated by the competence pilus were on the lower side (8 pN), even in the presence of functional PilT, and dropped by 2-fold in a pilT deletion strain. Therefore, it was interesting to find out how severely deletion of pilT and its paralogues affected force generation in N. gonorrhoeae, one of the strongest T4P machines.
Here, we set out to address this question by characterizing velocity and force generation in gonococcal pilT deletion strains. We show that gonococcal T4P indeed retract in the absence of pilT and its paralogues. Interestingly, both force and speed of PilT-independent T4P retraction are lower by orders of magnitude compared to retraction in wild-type (wt) T4P, explaining why T4P retraction has been overlooked so far. We investigate putative energy sources of PilT-independent T4P retraction by characterizing the effects of proton motive force (PMF).
RESULTS
T4P retract at low speed in a pilT deletion mutant.
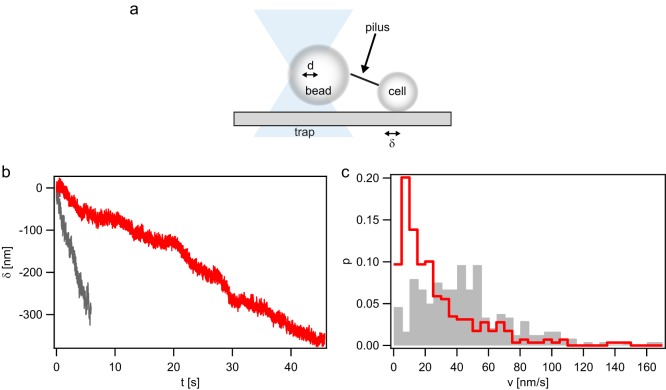
Deleting the gene encoding the T4P retraction ATPase PilT was long believed to be in accord with generating strains that are incapable of T4P retraction (4). However, recent experiments reported T4P retraction in the absence of pilT or homologues (20). We set out to investigate whether T4P retraction occurred in gonococcal pilT deletion strains. We used a laser tweezers assay to probe for T4P retraction (Fig. 1a). A bacterium was immobilized with a glass cover slide. Using a laser trap, a polystyrene bead was placed adjacent to the bacterium. When a T4P bound to the bead and retracted, the bead was deflected by distance d from the center of the laser trap. The force acting on the bead is proportional to the optical restoring force (F). In force clamp mode, d was kept constant by moving the microscope stage by distance δ with respect to the center of the laser trap. Thus, by measuring δ, we can determine the velocity of T4P retraction at constant force.
FIG 1.
pilT-independent T4P retraction in a ΔpilT strain (Ng178). (a) Sketch of the experimental setup. A gonococcal cell is attached to a glass surface, and a polystyrene bead trapped in a laser trap is placed in close proximity. When a T4P binds to the bead, retraction deflects the bead from the center of the trap by distance d. d is proportional to the optical restoring force (F). In force clamp mode, F is held constant by moving the surface-bound cell by distance δ. Thus, δ is a measure of the T4P length change. (b and c) Typical examples of T4P length change δ as a function of time (b) and velocity distribution at forces clamped at F = 4 pN (gray, n = 239) and F = 8 pN (red, n = 298), respectively (c).
To start with, we clamped the force at F = 8 pN. This is the lowest force that we typically used for characterizing T4P retraction in the wt strain with functional PilT. We found that the ΔpilT strain indeed deflected the bead from the center of the laser trap, indicating T4P retraction (Fig. 1b). The distribution of velocities showed a maximum around a velocity (v) of 5 nm · s−1 and a pronounced tail toward higher velocities (Fig. 1c). We assessed whether similar retractile behavior occurred in a different trapping geometry. A dual trap (17) was used to trap a single spherical bacterium in each trap in position clamp mode. This setup was not influenced by microscope drift. Again, T4P retraction was observed (see Fig. S2a in the supplemental material). Furthermore, we used a configuration where we trapped a spherical bacterium in one trap and a bead in the second trap. Likewise, T4P retraction was observed (Fig. S2b). We conclude that gonococcal T4P retract in the absence of the retraction ATPase PilT.
Next, the effect of force on the speed of PilT-independent T4P retraction was investigated. We measured the velocity when the force was clamped to F = 4 pN (Fig. 1b). As expected, the distribution of velocities shifted to higher values compared to those at F = 8 pN. The distribution showed a maximum around v = 40 nm · s−1 and, again, a tail at higher velocities (Fig. 1c). No clear T4P elongation events were observed.
In summary, gonococcal T4P retract in the absence of the retraction ATPase PilT, but the speed is two orders of magnitude lower than the speed measured in the PilT-producing strain.
PilT-independent T4P retraction generates lower force than PilT-driven retraction.
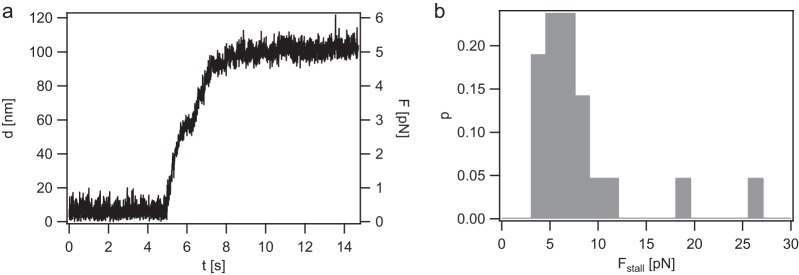
Gonococcal T4P are among the strongest molecular machines described so far, generating 150 pN during retraction (5, 34). We addressed force generation in the absence of the T4P retraction ATPase PilT. To this end, T4P retraction was probed using the laser trap in the position clamp mode. As the T4P pulled on the bead, the deflection increased and, concomitantly, the force increased. Eventually, the speed leveled off (Fig. 2a). A stalling event was defined as an event where the force did not change for at least one second, i.e., dF/dt = 0 for 1 s or longer. The stalling forces were distributed around a mean (±standard deviation [SD]) of F = 4.7 ± 0.7 pN (Fig. 2b). We note that there were outliers at considerably higher forces. They were disregarded when calculating the average stalling force. Most likely, these stalling events were caused by multiple T4P pulling on the bead simultaneously.
FIG 2.
Stalling of pilT-independent T4P retraction in the ΔpilT (Ng178) strain. (a) Typical stalling event in position clamp mode. Deflection of the bead from the center of the laser trap (d) and force (F) are plotted as functions of time. (b) Distribution of stalling forces (Fstall). n = 21.
We conclude that PilT-independent T4P retraction generates force in the range of 5 pN, i.e., 20-to 30-fold lower than that generated by PilT-powered T4P retraction.
Deletion of pilT2 in a ΔpilT background reduces the speed of T4P retraction.
N. gonorrhoeae bears two pilT paralogues on its genome, namely, pilU and pilT2. pilU resides in the same operon as pilT, and its deletion shows little effect on T4P retraction in wt gonococci (35). Deletion of pilT2 in the wt background causes a reduction of T4P retraction speed by a factor of ∼2 (35). It was conceivable that one of the pilT paralogues coded for proteins that powered PilT-independent T4P retraction. To find out whether deletion of the paralogues affected the velocity of T4P retraction, we used the laser tweezers with force clamped to 4 pN. Interestingly, the speed was strongly reduced in the ΔpilT2 ΔpilT strain (Fig. 3). The ΔpilU ΔpilT strain showed no significant change in T4P retraction compared to the ΔpilT strain. As described before, the force clamp assay may be subject to drift of the bacterium with respect to the laser trap. To make sure that the residual bead deflection was not caused by drift, we used the dual-trap assay and probed the following strains for T4P retraction: ΔpilT2 ΔpilT, ΔpilU ΔpilT, and ΔpilT2 ΔpilU ΔpilT strains. All three strains showed T4P retraction (Table S1), indicating that even in a strain that had all three pilT paralogues deleted, T4P retract. Examples for retraction of the ΔpilT2 ΔpilT strain are shown in Fig. S3, where the bacteria approached each other by >500 nm within 30 s. We conclude that deletion of pilT2 slows but does not inhibit T4P retraction.
FIG 3.
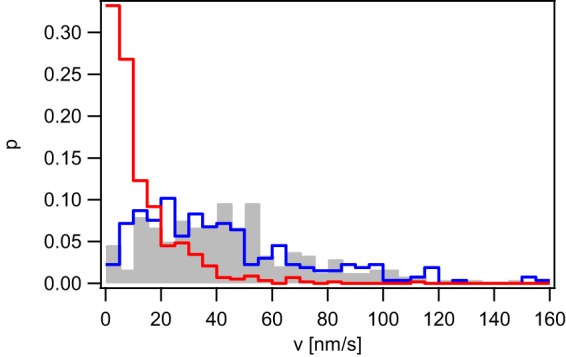
T4P retraction velocity depends on pilT2. Velocity distribution of the ΔpilT (gray, Ng178, n = 239), ΔpilT ΔpilU (blue, Ng185, n = 265), and ΔpilT ΔpilT2 (red, Ng184, n = 587) strains at F = 4 pN.
Depletion of proton motive force slows PilT-independent T4P retraction.
Depletion of proton motive force reduces the speed of gonococcal T4P retraction by 2-fold (36, 37). To test whether proton motive force powers pilT-independent T4P retraction, ΔpilT cells were treated with the uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP). CCCP shuttles protons across the membrane in the direction of the proton gradient and depletes PMF. Cells were incubated with 50 μM CCCP for 15 min prior to usage in dual laser tweezers. Notably, ΔpilT cells were able to generate forces between 15 and 50 min post CCCP treatment (Table S1). However, the speed of T4P retraction was strongly reduced (Fig. 4). We conclude that PilT-independent retraction is slowed but not fully inhibited by depleting PMF.
FIG 4.
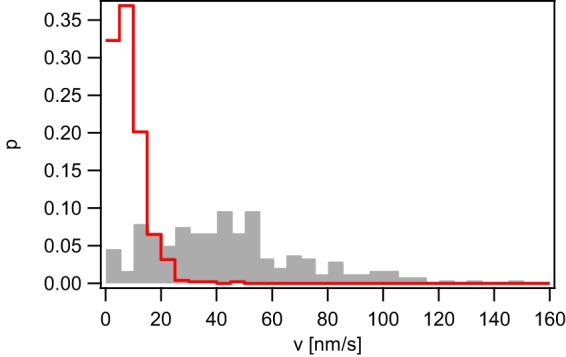
T4P retraction velocity depends on proton motive force. Velocity distribution of the ΔpilT strain (Ng178) in the absence (gray, n = 239) and in the presence (red, n = 477) of 50 μM CCCP at F = 4 pN.
DISCUSSION
Putative effects of PilT2.
When pilT2 is deleted in a strain generating PilT, the speed of T4P retraction is reduced by ∼2-fold through an unknown mechanism (35). Here, we found that in a ΔpilT background, deleting pilT2 again reduces the speed of T4P retraction by ∼4-fold. A straightforward interpretation of this result would be that PilT2 can power T4P retraction at a low speed in the absence of PilT. The finding that application of CCCP reduces the speed by a similar factor is in agreement with this result. Depleting the proton motive force reduces the ATP levels considerably within 5 to 10 min (37) and would thus inhibit the ATPase-driven T4P retraction. Other mechanisms explaining the effect of PilT2 are possible. For example, we cannot rule out the possibility that the elongation ATPase PilF is involved in T4P retraction. Interaction of PilT2 with PilF may tune PilF activity.
Comparing T4P retraction in the presence and absence of the T4P retraction ATPase.
It is important to note that thus far a gonococcal pilT deletion strain was considered to be incapable of T4P retraction (4, 9, 17, 38, 39). In our own studies characterizing single T4P retraction (1, 34) we have overlooked retraction in the absence of pilT because the velocity is close to zero in the force range in which we have worked so far. The lowest force was 8 pN, i.e., above the mean stalling force of PilT-independent retraction. Most other studies have probed for T4P retraction by measuring phenotypic consequences of T4P retraction such as twitching motility or DNA transformation. Indeed, deleting pilT inhibits gonococcal twitching motility and transformation (4, 29, 40).
T4P retraction in the absence of the retraction ATPase PilT has been recently reported for various bacterial species. First, T4P retraction was reported in T4P systems that naturally lack the gene encoding the T4P retraction ATPase (18, 19). Second, in T4P systems that naturally bear genes encoding the T4P retraction ATPase, retraction ATPases were deleted and yet T4P retraction was still observed (2, 20). Here, we show that deletion of pilT in the gonococcal T4P system has a strong effect force generation. While wt T4P generate force in the range of 150 pN (5), the stalling forces measured for the ΔpilT strain are in the range of 5 pN. Moreover, the retraction velocity is strongly reduced. It is interesting to compare the forces generated by various T4P systems in the presence and absence of the retraction ATPase. In the absence of the retraction ATPase, the forces generated by T4P retraction are fairly low, in the range of 3 to 12 pN (18, 19). The exception is T4P retraction in an M. xanthus ΔpilT strain, in which forces in the range of 70 pN are generated (2). However, it is likely that one of the pilT paralogues powers retraction for this specific strain. On the other hand, T4P retraction powered by the retraction ATPase shows a wide range of forces, from 8 pN up to 150 pN (2, 5, 20, 41). Taken together, we conclude that the retraction ATPase PilT consistently increases the force generated by T4P retraction in different bacterial species.
Which biological functions related to T4P retraction require retraction ATPases? T4P systems lacking retraction ATPases are associated with protein secretion (18), surface sensing (19), and DNA in uptake systems in Gram-positive bacteria (33). We propose that high force generation may not be required for these functions. DNA uptake in Gram-negative species is strongly impaired in the absence of the retraction ATPases (32, 40), suggesting a role of high force generation during threading into the periplasm. Twitching motility is another function of T4P (4). T4P elongate, adhere to a surface, and subsequently pull the cell body forward by retraction. The rupture forces of T4P from abiotic surfaces (5, 42) and from other T4P (17) are in the range of 50 pN. During cellular movement, T4P must detach from the surface; otherwise, movement stalls (5, 43). Therefore, the motor force must exceed 50 pN, consistent with PilT-powered retraction. Similarly, T4P retraction regulates the kinetics of T4P-T4P attachment and detachment within bacterial colonies (17). Colonies formed by N. gonorrhoeae and Neisseria meningiditis generating functional PilT behave like liquids, whereas pilT deletion strains behave in a glasslike manner (16, 17). Liquidlike behavior facilitates colonization of blood vessels during meningococcal infection (16) and cell sorting with respect to differential T4P-T4P adhesion forces (14). Another function of T4P retraction that requires high forces is signaling to host cells. When epithelial cells are infected with gonococci or mock infected with T4P-coated beads, cytoskeletal proteins accumulate to the site of infection (8–10, 38). This accumulation depends on PilT and force application for gonococcal and mock infection, respectively (9, 10). We conclude that T4P retraction in the absence of a retraction ATPase is sufficient for some T4P-related functions, but for those functions that call for high forces, retraction ATPases are required.
Implications of retraction ATPase-independent T4P retraction for chemomechanical coupling in the T4P machine.
Recently, progress has been made in understanding chemomechanical coupling in the T4P machine (see Fig. S1 in the supplemental material). Most likely, ATP binding and/or hydrolysis induces conformational changes of the retraction ATPase hexamer parallel to the membrane (23, 24, 31). Structural data are consistent with a wave of conformational changes around the ring leading to rotational motion of the platform complex. T4P retraction force, however, is generated perpendicular to the membrane by collapse of pilins from the fiber into the membrane, and it remains to be determined how putative rotation of the platform complex shuffles pilins from the fiber into the cytoplasmic membrane. The fact that T4P retraction occurs in the absence of the retraction ATPase evokes speculations about the energetics of the T4P machine. Deleting the gene encoding the elongation ATPase leads to nonpiliated bacteria (28). So far, to our knowledge, filament assembly has not been reported in the absence of an elongation ATPase. These two facts strongly suggest that the energy provided by ATP hydrolysis in the elongation ATPase is required for T4P polymerization. Part of this energy may be stored in the T4P fiber and power T4P depolymerization when the elongation ATPase has dissociated from the complex.
Interestingly, little T4P elongation was observed in this study. If the retraction and elongation ATPases bound alternatively and stochastically, then we would expect to see occasional switching from slow T4P retraction to fast elongation (indicative of binding of the elongation ATPase). The lack of T4P elongation in our study may suggest that T4P have to retract fully prior to binding of the elongation ATPase. This finding is consistent with processive retraction of toxin-coregulated pili in V. cholerae, in which it was proposed that insertion of minor pilins blocked elongation and triggered retraction (18). Previously, we observed that T4P retraction frequently switched to elongation in gonococcal strains that had strongly reduced concentrations of PilT (34, 44). However, the frequency of these elongation events increased strongly with external force, and elongation events were rarely observed at forces as low as 8 pN (34), in agreement with results of the present study.
Conclusion.
We have shown that gonococcal T4P retract in the absence of the retraction ATPase PilT. Both the speed and the maximum force of pilT-independent retraction are orders of magnitude lower than those in PilT-powered retraction, explaining why pilT-independent T4P retraction has been overlooked so far. Our findings, together with recent results for other species, strongly suggest that T4P retraction without PilT is a general phenomenon. We thus propose that the T4P elongation ATPase is necessary to provide energy for fiber formation, while retraction occurs spontaneously. Figuring out the chemomechanical coupling within the T4P machine, especially the dynamics of the platform complex in the presence and absence of the retraction ATPase, will be a future challenge.
MATERIALS AND METHODS
Growth conditions.
Gonococcal base agar was made from 10 g/liter Bacto agar (BD Biosciences, Bedford, MA), 5 g/liter NaCl (Roth, Darmstadt, Germany), 4 g/liter K2HPO4 (Roth), 1 g/liter KH2PO4 (Roth), 15 g/liter Bacto proteose peptone no. 3 (BD), and 0.5 g/liter soluble starch (Sigma-Aldrich, St. Louis, MO), and supplemented with 1% IsoVitaleX: 1 g/liter d-glucose (Roth), 0.1 g/liter l-glutamine (Roth), 0.289 g/liter l-cysteine–HCl · H20 (Roth), 1 mg/liter thiamine pyrophosphate (Sigma-Aldrich), 0.2 mg/liter Fe(NO3)3 (Sigma-Aldrich), 0.03 mg/liter thiamine HCl (Roth), 0.13 mg/liter 4-aminobenzoic acid (Sigma-Aldrich), 2.5 mg/liter β-NAD (Roth), and 0.1 mg/liter vitamin B12 (Sigma-Aldrich). GC medium is identical to the base agar composition but lacks agar and starch.
Bacterial strains.
All bacterial strains were derived from the gonococcal strain MS11 (VD300). In all strains, we deleted the G4 motif by replacing it with the aac gene conferring resistance against apramycin. The G4 motif is essential for antigenic variation of the major pilin subunit (45). Pilin antigenic variation modifies the primary sequence of the pilin gene. To generate the ΔpilT2 ΔpilT strain (Ng184), Ng150 (46) was transformed with genomic DNA from ΔpilT2 (35) and subsequently with DNA from the ΔpilT strain (Ng178). To generate the ΔpilU ΔpilT strain (Ng185), Ng150 (46) was transformed with genomic DNA from GU2 (47) and subsequently with DNA from the ΔpilT strain (Ng178). To generate the ΔpilT2 ΔpilU ΔpilT strain (Ng186), Ng150 (46) was transformed with genomic DNA from GU2 (47) and the ΔpilT2 strain (35), and subsequently with DNA from the ΔpilT strain (Ng178).
Transformants were selected on agar plates containing the respective antibiotics (Table S1).
Characterization of T4P retraction.
Retraction velocities and stalling forces were measured using an optical tweezers setup assembled on a Zeiss Axiovert 200 microscope (34). In short, all measurements were carried out in retraction assay medium consisting of phenol red-free Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY) with 2 mM l-glutamine (Gibco), 8 mM sodium pyruvate (Gibco) and 30 mM HEPES (Roth). A suspension of bacteria and carboxylated latex beads with a diameter of 2 μm (Polysciences, Warrington, PA) was applied to a microscope slide and sealed. All measurements were performed at 37°C. The trap stiffness was determined by power spectrum analysis of the Brownian motion of a trapped bead to be 0.5 pN/nm ± 10%. The retraction velocities were measured in force clamp mode. During the experiment, a bead was trapped and held close to an immobilized bacterium at the surface. Eventually, a pilus attached to the bead, and its retraction lead to a deflection out of the equilibrium position. As soon as the deflection of the bead reached the threshold deflection corresponding to a force of 4 or 8 pN, a force feedback algorithm held the displacement constant by moving the sample in the x-y plane using a piezo stage. Stalling forces were measured in position clamp mode.
Double laser trap.
In order to investigate single cell interactions, we followed a previously developed protocol (17). Two optical traps were installed in an inverted microscope (Nikon TE2000 C1). The trapping laser (20I-BL-106C, 1,064 nm, 5.4 W; Spectra Physics) was directed into a water immersion objective (Plan Apochromate VC; 60×; numerical aperture [NA], 1.20; Nikon). Manipulation of the laser was done with a 2-axis acousto-optical deflector (DTD-274HD6 colinear deflector; IntraAction Corp., USA). Bacterial interaction was recorded with a charge-coupled device (CCD) camera (Sensicam qe; PCO, Kelheim, Germany). The optical trap was calibrated via the equipartition and drag force methods. At 100% laser power, the average stiffness is 0.11 ± 0.01 pN/nm. The linear regime extends up to 80 pN.
Depletion of proton motive force.
Cells were incubated with 50 μM CCCP for 15 min prior to usage in dual laser tweezers. To check that 15 min is sufficient to affect cells, twitching motility of pilT-expressing ΔG4 cells was checked by bright field microscopy. Consistent with previous results (37), cells showed low-speed twitching motility after 15 min of treatment with 50 μM CCCP and high-speed twitching motility without CCCP.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Katrina Forest, Lisa Craig, and the members of the Maier lab for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft through grant MA3898.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00778-18.
REFERENCES
- 1.Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. 2002. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci U S A 99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clausen M, Jakovljevic V, Sogaard-Andersen L, Maier B. 2009. High-force generation is a conserved property of type IV pilus systems. J Bacteriol 191:4633–4638. doi: 10.1128/JB.00396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biais N, Ladoux B, Higashi D, So M, Sheetz M. 2008. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol 6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 5.Marathe R, Meel C, Schmidt NC, Dewenter L, Kurre R, Greune L, Schmidt MA, Muller MJ, Lipowsky R, Maier B, Klumpp S. 2014. Bacterial twitching motility is coordinated by a two-dimensional tug-of-war with directional memory. Nat Commun 5:3759. doi: 10.1038/ncomms4759. [DOI] [PubMed] [Google Scholar]
- 6.Zaburdaev V, Biais N, Schmiedeberg M, Eriksson J, Jonsson AB, Sheetz MP, Weitz DA. 2014. Uncovering the mechanism of trapping and cell orientation during Neisseria gonorrhoeae twitching motility. Biophys J 107:1523–1531. doi: 10.1016/j.bpj.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabass B, Koch MD, Liu G, Stone HA, Shaevitz JW. 2017. Force generation by groups of migrating bacteria. Proc Natl Acad Sci U S A 114:7266–7271. doi: 10.1073/pnas.1621469114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merz AJ, Enns CA, So M. 1999. Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol Microbiol 32:1316–1332. doi: 10.1046/j.1365-2958.1999.01459.x. [DOI] [PubMed] [Google Scholar]
- 9.Howie HL, Glogauer M, So M. 2005. The N. gonorrhoeae type IV pilus stimulates mechanosensitive pathways and cytoprotection through a pilT-dependent mechanism. PLoS Biol 3:e100. doi: 10.1371/journal.pbio.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Opitz D, Maier B. 2011. Rapid cytoskeletal response of epithelial cells to force generation by type IV pili. PLoS One 6:e17088. doi: 10.1371/journal.pone.0017088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biais N, Higashi DL, Brujic J, So M, Sheetz MP. 2010. Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc Natl Acad Sci U S A 107:11358–11363. doi: 10.1073/pnas.0911328107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 13.Anyan ME, Amiri A, Harvey CW, Tierra G, Morales-Soto N, Driscoll CM, Alber MS, Shrout JD. 2014. Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 111:18013–18018. doi: 10.1073/pnas.1414661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldewurtel ER, Kouzel N, Dewenter L, Henseler K, Maier B. 2015. Differential interaction forces govern bacterial sorting in early biofilms. Elife 4:e10811. doi: 10.7554/eLife.10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hockenberry AM, Hutchens DM, Agellon A, So M. 2016. Attenuation of the type IV pilus retraction motor influences Neisseria gonorrhoeae social and infection behavior. mBio 7:e01994-16. doi: 10.1128/mBio.01994-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonazzi D, Lo Schiavo V, Machata S, Djafer-Cherif I, Nivoit P, Manriquez V, Tanimoto H, Husson J, Henry N, Chaté H, Voituriez R, Duménil G. 2018. Intermittent pili-mediated forces fluidize Neisseria meningitidis aggregates promoting vascular colonization. Cell 174:143–155.E116. doi: 10.1016/j.cell.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Welker A, Cronenberg T, Zollner R, Meel C, Siewering K, Bender N, Hennes M, Oldewurtel ER, Maier B. 2018. Molecular motors govern liquidlike ordering and fusion dynamics of bacterial colonies. Phys Rev Lett 121:118102. doi: 10.1103/PhysRevLett.121.118102. [DOI] [PubMed] [Google Scholar]
- 18.Ng D, Harn T, Altindal T, Kolappan S, Marles JM, Lala R, Spielman I, Gao Y, Hauke CA, Kovacikova G, Verjee Z, Taylor RK, Biais N, Craig L. 2016. The Vibrio cholerae minor pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLoS Pathog 12:e1006109. doi: 10.1371/journal.ppat.1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison CK, Dalia TN, Vidal Ceballos A, Wang JC, Biais N, Brun YV, Dalia AB. 2018. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat Microbiol 3:773–780. doi: 10.1038/s41564-018-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Hospenthal MK, Costa TRD, Waksman G. 2017. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol 15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 23.Chang Y-W, Rettberg LA, Treuner-Lange A, Iwasa J, Søgaard-Andersen L, Jensen GJ. 2016. Architecture of the type IVa pilus machine. Science 351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCallum M, Tammam S, Khan A, Burrows LL, Howell PL. 2017. The molecular mechanism of the type IVa pilus motors. Nat Commun 8:15091. doi: 10.1038/ncomms15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, Craig L. 2016. Structure of the Neisseria meningitidis type IV pilus. Nat Commun 7:13015. doi: 10.1038/ncomms13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancl JM, Black WP, Robinson H, Yang Z, Schubot FD. 2016. Crystal structure of a type IV pilus assembly ATPase: insights into the molecular mechanism of PilB from Thermus thermophilus. Structure 24:1886–1897. doi: 10.1016/j.str.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Chang Y-W, Kjær A, Ortega DR, Kovacikova G, Sutherland JA, Rettberg LA, Taylor RK, Jensen GJ. 2017. Architecture of the Vibrio cholerae toxin-coregulated pilus machine revealed by electron cryotomography. Nat Microbiol 2:16269. doi: 10.1038/nmicrobiol.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freitag NE, Seifert HS, Koomey M. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol 16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 29.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 30.Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. 2007. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misic AM, Satyshur KA, Forest KT. 2010. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J Mol Biol 400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci U S A 110:17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 34.Clausen M, Koomey M, Maier B. 2009. Dynamics of type IV pili is controlled by switching between multiple states. Biophys J 96:1169–1177. doi: 10.1016/j.bpj.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurre R, Hone A, Clausen M, Meel C, Maier B. 2012. PilT2 enhances the speed of gonococcal type IV pilus retraction and of twitching motility. Mol Microbiol 86:857–865. doi: 10.1111/mmi.12022. [DOI] [PubMed] [Google Scholar]
- 36.Kurre R, Maier B. 2012. Oxygen depletion triggers switching between discrete speed modes of gonococcal type IV pili. Biophys J 102:2556–2563. doi: 10.1016/j.bpj.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurre R, Kouzel N, Ramakrishnan K, Oldewurtel ER, Maier B. 2013. Speed switching of gonococcal surface motility correlates with proton motive force. PLoS One 8:e67718. doi: 10.1371/journal.pone.0067718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higashi DL, Lee SW, Snyder A, Weyand NJ, Bakke A, So M. 2007. Dynamics of Neisseria gonorrhoeae attachment: microcolony development, cortical plaque formation, and cytoprotection. Infect Immun 75:4743–4753. doi: 10.1128/IAI.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hepp C, Maier B. 2016. Kinetics of DNA uptake during transformation provide evidence for a translocation ratchet mechanism. Proc Natl Acad Sci U S A 113:12467–12472. doi: 10.1073/pnas.1608110113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gangel H, Hepp C, Muller S, Oldewurtel ER, Aas FE, Koomey M, Maier B. 2014. Concerted spatio-temporal dynamics of imported DNA and ComE DNA uptake protein during gonococcal transformation. PLoS Pathog 10:e1004043. doi: 10.1371/journal.ppat.1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribbe J, Baker AE, Euler S, O’Toole GA, Maier B. 2017. Role of cyclic di-GMP and exopolysaccharide in type IV pilus dynamics. J Bacteriol 199 doi: 10.1128/JB.00859-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holz C, Opitz D, Greune L, Kurre R, Koomey M, Schmidt MA, Maier B. 2010. Multiple pilus motors cooperate for persistent bacterial movement in two dimensions. Phys Rev Lett 104:178104. doi: 10.1103/PhysRevLett.104.178104. [DOI] [PubMed] [Google Scholar]
- 43.Ponisch W, Weber CA, Juckeland G, Biais N, Zaburdaev V. 2017. Multiscale modeling of bacterial colonies: how pili mediate the dynamics of single cells and cellular aggregates. New J Physics 19:015003. doi: 10.1088/1367-2630/aa5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maier B, Koomey M, Sheetz MP. 2004. A force-dependent switch reverses type IV pilus retraction. Proc Natl Acad Sci U S A 101:10961–10966. doi: 10.1073/pnas.0402305101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rotman E, Seifert HS. 2014. The genetics of Neisseria species. Annu Rev Genet 48:405–431. doi: 10.1146/annurev-genet-120213-092007. [DOI] [PubMed] [Google Scholar]
- 46.Zollner R, Oldewurtel ER, Kouzel N, Maier B. 2017. Phase and antigenic variation govern competition dynamics through positioning in bacterial colonies. Sci Rep 7:12151. doi: 10.1038/s41598-017-12472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park HS, Wolfgang M, Koomey M. 2002. Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect Immun 70:3891–3903. doi: 10.1128/IAI.70.7.3891-3903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.