Although the gut anaerobe Bacteroides thetaiotaomicron is a prominent member of the healthy human gut microbiota, little is known about its capacity to adhere to surfaces and form biofilms. Here, we identify that alteration of a surface-exposed protein corresponding to a type of pili found in many Bacteroidetes increases B. thetaiotaomicron biofilm formation. This study lays the ground for establishing this bacterium as a model organism for in vitro and in vivo studies of biofilm-related phenotypes in gut anaerobes.
KEYWORDS: adhesion, Bacteroides thetaiotaomicron, biofilm, type V pili, commensal, gut, symbiont
ABSTRACT
Bacteroides thetaiotaomicron is a prominent anaerobic member of the healthy human gut microbiota. While the majority of functional studies on B. thetaiotaomicron addressed its impact on the immune system and the utilization of diet polysaccharides, B. thetaiotaomicron biofilm capacity and its contribution to intestinal colonization are still poorly characterized. We tested the natural adhesion of 34 B. thetaiotaomicron isolates and showed that although biofilm capacity is widespread among B. thetaiotaomicron strains, this phenotype is masked or repressed in the widely used reference strain VPI 5482. Using transposon mutagenesis followed by a biofilm positive-selection procedure, we identified VPI 5482 mutants with increased biofilm capacity corresponding to an alteration in the C-terminal region of BT3147, encoded by the BT3148-BT3147 locus, which displays homology with Mfa-like type V pili found in many Bacteroidetes. We show that BT3147 is exposed on the B. thetaiotaomicron surface and that BT3147-dependent adhesion also requires BT3148, suggesting that BT3148 and BT3147 correspond to the anchor and stalk subunits of a new type V pilus involved in B. thetaiotaomicron adhesion. This study therefore introduces B. thetaiotaomicron as a model to study proteinaceous adhesins and biofilm-related phenotypes in this important intestinal symbiont.
IMPORTANCE Although the gut anaerobe Bacteroides thetaiotaomicron is a prominent member of the healthy human gut microbiota, little is known about its capacity to adhere to surfaces and form biofilms. Here, we identify that alteration of a surface-exposed protein corresponding to a type of pili found in many Bacteroidetes increases B. thetaiotaomicron biofilm formation. This study lays the ground for establishing this bacterium as a model organism for in vitro and in vivo studies of biofilm-related phenotypes in gut anaerobes.
INTRODUCTION
Bacterial biofilms are widespread surface-attached communities in which reduced diffusion and a heterogeneous structure favor physical and metabolic contacts between bacteria and induce novel behaviors compared to individual planktonic microorganisms (1, 2). In the past decades, biofilms were often investigated under aerobic conditions using genetically amenable aerobic bacterial models, and anaerobic biofilm studies mainly focused on oral anaerobes. Although these studies considerably advanced our understanding of this important bacterial lifestyle, many clinically and ecologically important processes take place under anaerobic conditions and are carried out by strict or facultative commensal and pathogenic anaerobes (3). This is particularly the case for the vertebrate gut microbiota, which is vastly dominated by anaerobic Firmicutes and Bacteroidetes (4, 5). However, although the intestinal microbiota provides protection from pathogen colonization and impacts host intestinal physiology and health (6–11), the contributions of its structural organization and of the biofilm formation capacities of gut anaerobes are still poorly understood.
One of the most abundant and best-studied members of the healthy human gut microbiota is the strictly anaerobic bacterium Bacteroides thetaiotaomicron (12). B. thetaiotaomicron was shown to contribute to the development of the intestinal mucosal layer and modulate the function of the intestinal immune system and is considered to play a key role in the breakdown of diet- and host-derived polysaccharides (13–16). Whereas biofilm formation could be involved in several important aspects of B. thetaiotaomicron biology, including intestinal colonization, inter- and intraspecies interactions, and virulence in extraintestinal sites (17, 18), studies investigating B. thetaiotaomicron adhesion and biofilm formation are still scarce. A transcriptomic analysis performed on biofilm formed on a carbon paper surface in chemostats showed an upregulation of several polysaccharide utilization loci (PUL) involved in polysaccharide foraging, mucin degradation enzymes, as well as of two out of eight B. thetaiotaomicron capsular systems (19). This study, however, did not reveal significant differences in the expression levels of genes encoding extracellular appendages usually associated with bacterial surface adhesion, leaving open the question of whether adhesins contribute to B. thetaiotaomicron biofilm formation.
In the present study, we show that although in vitro biofilm development is widespread among natural B. thetaiotaomicron isolates, the reference B. thetaiotaomicron strain VPI 5482 (ATCC 29148) commonly used in human gut microbiota studies forms poor biofilm. We hypothesized that functions involved in VPI 5482 in vitro biofilm formation could be repressed or masked, and the use of transposon mutagenesis and a positive-selection procedure allowed us to identify a mutation in the BT3147 gene, altering the C-terminal region of BT3147 and increasing VPI 5482 biofilm formation. We show that the BT3148-BT3147 locus displays homology with the Mfa-like type V pili found in many Bacteroidetes species (20) and could encode a surface-exposed type V pilus involved in B. thetaiotaomicron adhesion. We expect this work to lay the groundwork for further characterization of B. thetaiotaomicron adhesins in VPI 5482 and other biofilm-forming isolates, establishing this species as a model organism for in vitro and in vivo studies of biofilm-related phenotypes in gut anaerobes.
RESULTS
The reference strain B. thetaiotaomicron VPI 5482 forms poor in vitro biofilm.
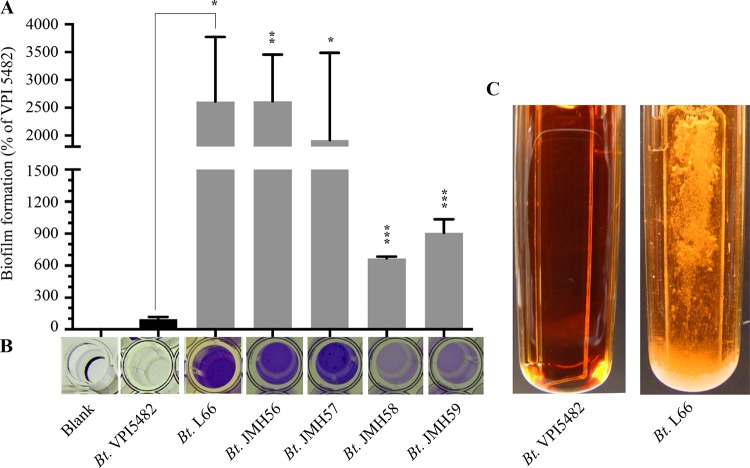
In order to analyze the ability of B. thetaiotaomicron VPI 5482 (ATCC 29148) (12) to form biofilm, we performed in vitro biofilm assays, using either a static 96-well-plate model followed by crystal violet staining or dynamic continuous-flow biofilm microfermentors. Although B. thetaiotaomicron VPI 5482 adhesion and biofilm formation were previously reported in a chemostat on carbon paper (19), we observed poor or no biofilm formation in both biofilm models (Fig. 1A and C). We then tested the biofilm capacities of 34 B. thetaiotaomicron isolates (see Table S1 in the supplemental material) and found that 41% of them (14 out of 34) produce more biofilm than VPI 5482 (Fig. 1A and B and Fig. S1). Taken together, these results indicate that whereas the ability to form biofilm is widespread among natural B. thetaiotaomicron isolates, the widely used reference strain VPI 5482 is a poor biofilm former under the tested growth conditions.
FIG 1.
The B. thetaiotaomicron VPI 5482 strain forms poor biofilms compared to different isolates. (A and B) Biofilm formation in a 96-well-plate biofilm assay followed by crystal violet staining of B. thetaiotaomicron (Bt) VPI 5482 and various biofilm-forming B. thetaiotaomicron isolates (see also Fig. S1 in the supplemental material). Error bars indicate standard deviations from 3 technical replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Biofilm formation on glass slides inserted into continuous-flow biofilm microfermentors. The images were taken 48 h after inoculation.
Selection of B. thetaiotaomicron VPI 5482 mutants with increased biofilm capacity.
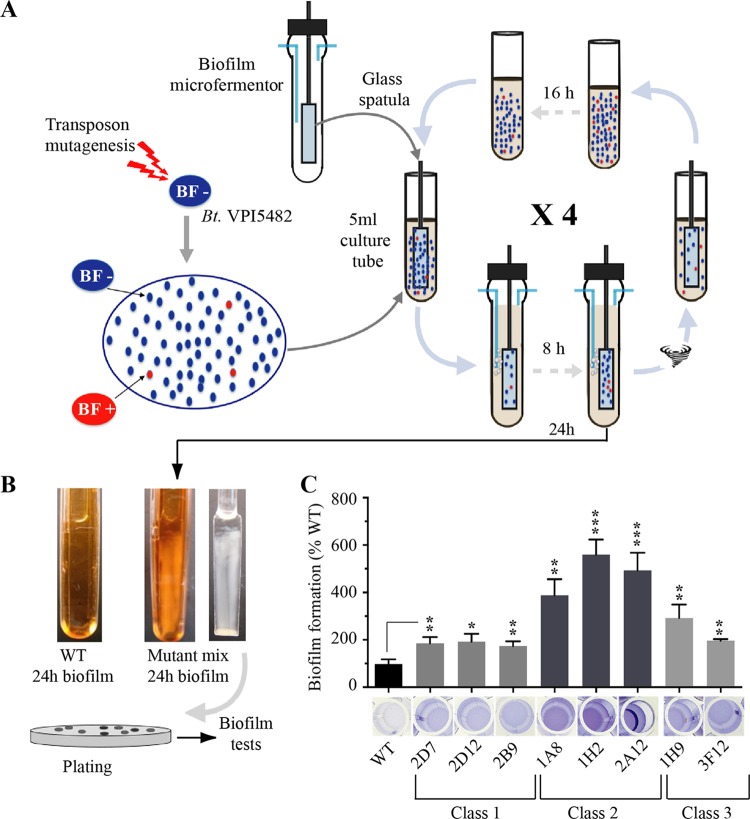
Considering the ability of several B. thetaiotaomicron strains to form biofilm, we hypothesized that functions involved in biofilm formation could be repressed or masked in the B. thetaiotaomicron VPI 5482 strain. To test this hypothesis and identify B. thetaiotaomicron VPI 5482 mutants with increased biofilm formation ability, we generated a library of over 10,000 mariner transposon mutants and subjected them to a positive-selection procedure for biofilm formation. Briefly, we resuspended a mix of B. thetaiotaomicron VPI 5482 transposon mutants in a tube with 15 ml of brain heart infusion salt (BHIS) medium (adjusted to an optical density at 600 nm [OD600] of 1), and we placed a microfermentor glass spatula into this solution for 10 min, to allow initial adhesion (Fig. 2A). The spatula was then washed in BHIS medium to remove nonadherent bacteria. Subsequently, the spatula potentially carrying the most adherent mutants was inserted into a continuous-flow biofilm microfermentor to allow surface growth and favor positive selection of B. thetaiotaomicron mutants with increased adhesion capacity. After 8 h, the spatula-associated bacterial population was resuspended in fresh BHIS medium, and the resulting culture grown overnight was used as the inoculum for another cycle of positive selection (Fig. 2A). After 4 cycles of positive selection, the biofilm microfermentor inoculated with a culture potentially enriched in biofilm-positive B. thetaiotaomicron VPI 5482 mutants displayed, after 24 h, strong biofilm formation compared to a microfermentor inoculated with the original wild-type (WT) VPI 5482 strain (Fig. 2B). After resuspension of the corresponding biofilm, B. thetaiotaomicron VPI 5482 mutants were subjected to serial dilution in BHIS medium and plated on selective agar plates. We reisolated the resulting colonies and individually tested mutants using a 96-well-plate biofilm assay. This led us to identify several mutants with an improved biofilm-forming capacity compared to WT VPI 5482.
FIG 2.
Selection of B. thetaiotaomicron VPI 5482 mutants with increased biofilm capacities. (A) Schematic representation of the positive-selection strategy used to identify transposon mutants with increased biofilm production. BF, biofilm formation. (B) Biofilm biomass formation on the internal spatulas of continuous-flow biofilm microfermentors after 24 h. (Left) Wild-type B. thetaiotaomicron VPI 5482; (middle and right) microfermentors and spatulas inoculated with a culture enriched for biofilm production after 4 cycles of positive selection. (C) Comparison of the biofilm formation capacities of WT B. thetaiotaomicron and eight identified mutants with an insertion in the BT3147 gene. (Bottom) Crystal violet staining in 96-well microtiter plates; (top) corresponding biomass quantification after resuspension in an acetone-ethanol mix and absorbance measured at 575 nm. Biofilm formation capacities of the WT (VPI 5482) have been set to 100%. Error bars indicate standard deviations from 3 technical replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Transposon insertion in the 3′ end of the Mfa/type V pilin homolog gene BT3147 increases biofilm formation.
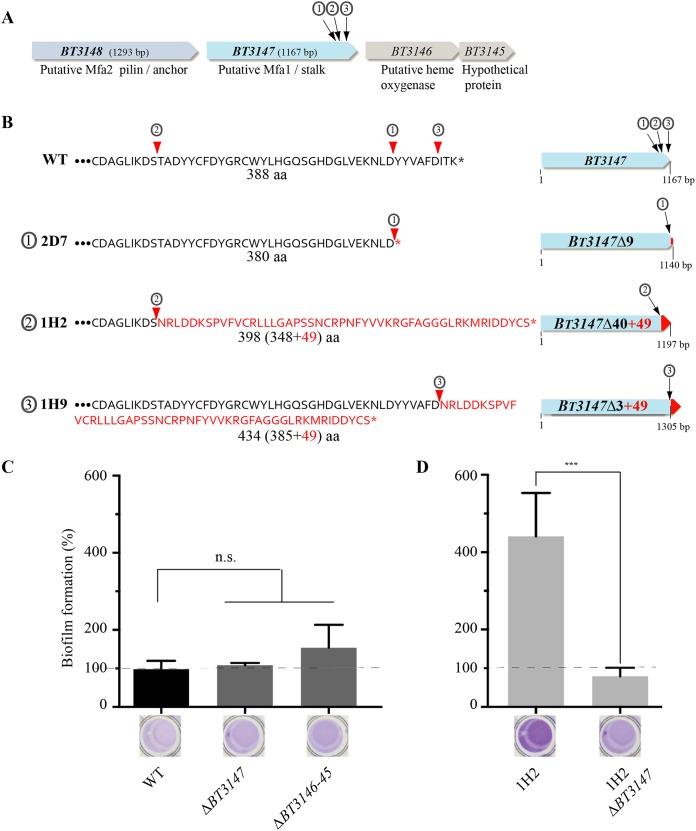
We determined that 8 of the identified biofilm-positive mutants (Fig. 2C) had a transposon inserted in BT3147, a gene located downstream of BT3148 and upstream of BT3146-BT3145 (Fig. 3A). Although protein sequence analysis showed only weak homologies, a search of structure databases revealed that the N-terminal region (residues 49 to 192) and the central region (residues 171 to 388) of BT3147 contain a Pfam 08842 (Mfa2) domain corresponding to the Mfa2/Mfa1 pilin of recently defined type V pili that are ubiquitous in Porphyromonas gingivalis and other gut Bacteroidetes (20). All eight transposon insertions were located in the last 40 codons of BT3147 and produced three different deletion-insertion events in the BT3147 coding sequence (Fig. 3B). Class 1 mutants (3 mutants, 2D7, 2D12, and 2B9) (Fig. 2C) correspond to an out-of-frame transposon insertion at bp 1138, deleting the last 9 amino acids from BT3147 without adding any transposon-derived coding sequence (here named BT3147Δ9). One representative of class 1, 2D7, was chosen for further analysis. Class 2 mutants (3 mutants, 1A8, 1H2, and 2A12) (Fig. 2C) correspond to an in-frame transposon insertion at position 1047, deleting the last 40 amino acids of BT3147 and replacing them with 49 amino acids from the transposon sequence, leading to a chimeric 398-amino-acid-long sequence (BT3147Δ40+49). One representative of class 2, 1H2, was chosen for further analysis. Finally, class 3 mutants (2 mutants, 1H9 and 3F12) (Fig. 2C) correspond to a 434-amino-acid fusion protein arising from the deletion of the last 3 BT3147 codons replaced by 49 codons from the transposon sequence (BT3147Δ3+49) (Fig. 3B).
FIG 3.
Insertions of a transposon in the 3′ end of B. thetaiotaomicron BT3147 increase biofilm formation. (A) Genetic organization of the BT3148-BT3145 B. thetaiotaomicron locus with indication of the three transposon insertion points in the 3′ end of BT3147. 2D7 corresponds to class 1 (①) mutants. 1H2 corresponds to class 2 (②) mutants. 1H9 corresponds to class 3 (③) mutants. (B) Schematic representation of the genetic consequences of transposon insertion in BT3147. aa, amino acids. (C) Comparison of the biofilm formation capacity of WT B. thetaiotaomicron (VPI 5482) with those of the class 2 transposon mutant 1H2 and different deletion strains lacking either BT3147 (ΔBT3147) or BT3146 and BT3145 (ΔBT3146-BT3145). (Bottom) Crystal violet staining in 96-well microtiter plates; (top) corresponding quantification after resuspension in acetone-ethanol and absorbance measured at 575 nm. (D) Comparison of biofilm formation capacities of the B. thetaiotaomicron 1H2 mutant and the 1H2 ΔBT3147 strain, derived from the biofilm-forming 1H2 transposon mutant after deletion of 1,046 bp of BT3147 located upstream of the transposon insertion. Biofilm formation capacities of the WT (VPI 5482) have been set to 100%. Error bars indicate standard deviations from 3 biological replicates. ***, P < 0.001; n.s., not significant.
To confirm the link between biofilm production and transposon insertion in BT3147, we deleted the BT3147 gene in the wild-type VPI 5482 strain; however, this approach did not lead to an increase in biofilm formation (Fig. 3C). We then hypothesized that insertion of the transposon in the 3′ extremity of BT3147 could prevent (by a polar effect) or enhance the expression of the downstream genes BT3146 and BT3145 (Fig. 3A). However, deletion of BT3146-BT3145 did not lead to a significant alteration of biofilm formation compared to the WT VPI 5482 strain (Fig. 3C). In addition, the comparison of VPI 5482 and 1H2 transcription using transcriptome sequencing (RNAseq) analysis showed that the insertion of the transposon did not alleviate potential premature transcription termination in the wild-type strain. Moreover, we did not detect any antisense transcript in the BT3147-BT3145 region that could be interfering with BT3147 transcription (Fig. S2). Nevertheless, deletion of the 1,046 bp of BT3147 (1,167 bp) located upstream of the transposon insertion point in the class 2 1H2 mutant, the strongest biofilm former among our 8 mutants (Fig. 2C), abolished biofilm formation in this strain (Fig. 3D). This result demonstrated that the presence of the mariner transposon per se does not promote biofilm formation and that the observed biofilm phenotype requires at least part of the BT3147 gene.
C-terminal truncation of BT3147 promotes BT3148-dependent B. thetaiotaomicron biofilm formation.
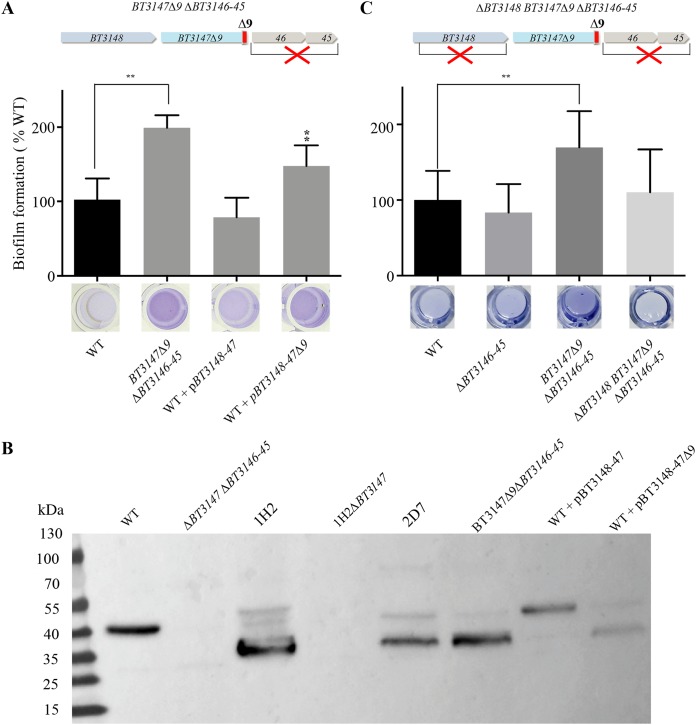
We then further investigated the consequences of transposon insertions on the BT3147 protein by reproducing one of the genetic configurations created by transposon insertion. We chose class 1 biofilm-forming mutants, as they correspond to the truncation of the last 9 C-terminal amino acids of BT3147 (BT3147Δ9) without an added transposon-derived coding sequence. Since the downstream genes BT3146 and BT3145 are not involved in the studied phenotype (Fig. 3C and Fig. S2), we constructed chromosomal BT3147Δ9 de novo by removing the VPI 5482 region encompassing the last 9 codons of BT3147 and the downstream genes BT3146 and BT3145, replacing it with a stop codon (Fig. 4A). We then compared the resulting BT3147Δ9 ΔBT3146-BT3145 strain with the wild-type strain VPI 5482 and showed that it displayed an increased biofilm formation capacity (Fig. 4A and Fig. S3). Consistently, whereas chromosome-based expression of BT3148-BT3147 displayed a wild-type biofilm phenotype, the expression of BT3148-BT3147Δ9 led to increased biofilm formation compared to WT VPI 5482 (Fig. 4A).
FIG 4.
C-terminal truncation of the BT3147 protein promotes B. thetaiotaomicron biofilm formation. (A, top) Schematic genetic organization of the BT3147Δ9 ΔBT3146-BT3145 mutant; (middle) comparison of the biofilm formation capacities of WT B. thetaiotaomicron VPI 5482 and the indicated mutant or plasmid-containing strains; (bottom) crystal violet staining in 96-well microtiter plates. Shown is the corresponding quantification after resuspension in acetone-ethanol and the absorbance measured at 575 nm. Biofilm formation capacities of the WT (VPI 5482) have been set to 100%. Error bars indicate standard deviations from 3 biological replicates. **, P < 0.01. (B) Immunodetection of BT3147 in WT B. thetaiotaomicron VPI 5482 and the indicated mutants or complemented strains expressing either full-length BT3147 (pBT3148-47) or a truncated version of BT3147.
The use of an antiserum raised against BT3147 enabled the immunodetection of a truncated form of BT3147 in whole-cell extracts of the BT3147Δ9 ΔBT3146-45 strain, class 1 (2D7) and class 2 (1H2) transposon mutants, and the wild-type strain constitutively expressing BT3147Δ9 (Fig. 4B). However, while WT (40.6 kDa) and truncated (1H2, 41.6 kDa; 2D7 and de novo BT3147Δ9, 39.6 kDa) versions of BT3147 have very similar theoretical sizes, the truncated forms of BT3147 migrated at an apparently lower molecular mass of ∼30 to 35 kDa, suggesting additional cleavage of these forms (see Discussion).
Finally, considering that BT3148, encoded by the gene located immediately upstream of BT3147 (Fig. 3A), displays homology with the B. thetaiotaomicron Fim3B/Mfa2 homolog BT2657 (PDB code 4QDG) and Fim2B/Mfa2 homolog BT1062 (PDB code 3GF8), we tested the contribution of BT3148 to BT3147Δ9-dependent biofilm formation. Deletion of BT3148 in B. thetaiotaomicron BT3147Δ9 ΔBT3146-45 resulted in a significant decrease in biofilm formation (Fig. 4C). Taken together, these results demonstrated that a C-terminal truncation of BT3147 promotes biofilm formation in B. thetaiotaomicron.
Localization of a putative BT3148-BT3147 type V pilus on the B. thetaiotaomicron surface.
BT3148 and BT3147 display structural similarities with Mfa1/type V pili identified in the anaerobic oral pathogen P. gingivalis and other gut Bacteroidetes (20). Two types of pili were described in P. gingivalis: the long FimA and the short Mfa1. FimA filaments are more than 1 μm long and fragile and promote autoaggregation and biofilm formation (21). FimA corresponds to the major structural component of the FimA filaments, FimB corresponds to the anchor protein, and FimC to FimE correspond to accessory proteins (22). The shorter Mfa1-type pili are 0.1- to 0.5-μm-long filaments essentially composed of the major stalk protein Mfa1 bound to the cell surface via the membrane-located anchor protein Mfa2 (Fig. 5A). In addition to mfa1 and mfa2, the P. gingivalis gene cluster often includes the genes mfa3 to mfa5, which encode accessory proteins. Since BT3148 displays homology with the B. thetaiotaomicron Fim3B/Mfa2 homolog BT2657 (PDB code 4QDG) and Fim2B/Mfa2 homolog BT1062 (PDB code 3GF8), we propose that it could be a B. thetaiotaomicron VPI 5482 type V pilus anchor (Fig. 5A and Fig. S3). BT3147, on the other hand, displays structural homology with Bacteroides eggerthii DSM 20697 BegFim1A-like proteins belonging to the FimA/Mfa1 protein family (PDB code 4GPV) and could correspond to the stalk component of Bacteroidia type V pili. We could not identify candidates for accessory proteins to the putative BT3148-BT3147 pilus (Fig. 5A and Fig. S4).
FIG 5.
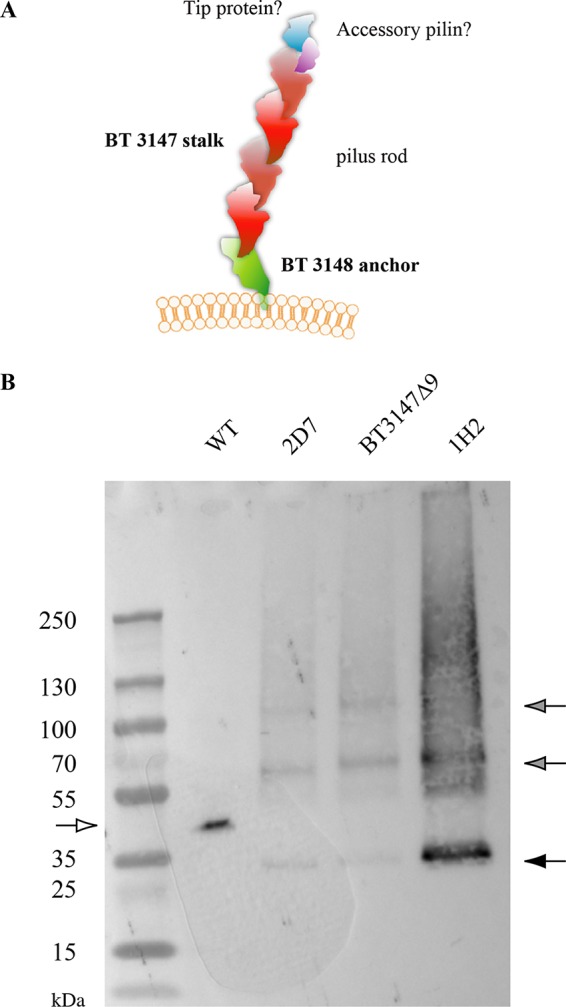
BT3147 is exposed on the B. thetaiotaomicron surface as part of a potential type V pilus. (A) Model of the putative BT3148-BT3147 pilus. (B) Western blot analysis of BT3147-containing structures released after surface shaving of the indicated B. thetaiotaomicron strains and mutants, followed by immunodetection using anti-BT3147 antibodies. The white arrow indicates the WT, and the filled arrows indicate the BT3147Δ9 variant of BT3147 and its potential multimers.
Based on these analyses, we hypothesized that BT3147 could be the major pilin of a structure exposed at the B. thetaiotaomicron cell surface. However, attempts to visualize the BT3147-based pilus using immunofluorescence as well as immunostaining coupled with transmission electron microscopy analyses were unsuccessful, as is sometimes the case for thin or masked fimbrial surface structures. Nevertheless, the proteinase K sensitivity of BT3147 on treated whole cells suggested that this protein is surface exposed (Fig. S5). We therefore tried to detect BT3147-containing structures by releasing B. thetaiotaomicron protruding surface appendages in the supernatant using a blender-based shaving procedure that does not lead to detectable bacterial lysis. We concentrated the protein obtained using this surface shaving procedure with WT strain VPI 5482, the BT3147Δ9 ΔBT3146-BT3145 strain, the class 1 transposon mutant strain (2D7), and the class 2 transposon mutant strain (1H2), and we performed immunodetection using anti-BT3147 antibodies.
We detected BT3147 in all samples, with truncated BT3147 forms migrating at a lower molecular mass than the WT, as described above (Fig. 4B and Fig. 5B). However, while the WT displayed a full-length and monomeric protein, the biofilm-forming BT3147Δ9 ΔBT3146-BT3145 and the 2D7 and 1H2 transposon mutant strains showed multiple specific products, potentially indicative of protein polymerization during pilus formation (Fig. 5B).
In vivo relevance of BT3147-mediated biofilm formation.
The demonstration that BT3147 is exposed on the B. thetaiotaomicron surface as part of a potential type V pilus structure contributing to B. thetaiotaomicron biofilm formation prompted us to investigate the in vivo role of BT3148-BT3147 in B. thetaiotaomicron adhesion or colonization. To this end, we used a germfree mouse model of intestinal colonization and performed in vivo mixed-culture competition experiments, comparing the respective colonization capacities of the competing strains administered intragastrically at a 1:1 mixed ratio (2 × 107 cells/ml). We first verified that B. thetaiotaomicron ΔBT3146-BT3145 showed a similar colonization capacity as the wild-type VPI 5482 strain, indicating that deletion of BT3146-BT3145 does not affect colonization (Fig. 6A). We then found that ΔBT3148-BT3145 has the same colonization capacity as the wild type (Fig. 6B) and that the biofilm-forming BT3147Δ9 ΔBT3146-BT3145 strain shows a similar colonization capacity as the wild-type VPI 5482 strain (Fig. 6C). This analysis therefore showed that the putative BT3148-BT3147 pilus or its biofilm-promoting truncated form does not significantly contribute to the B. thetaiotaomicron intestinal colonization capacity under the tested conditions.
FIG 6.
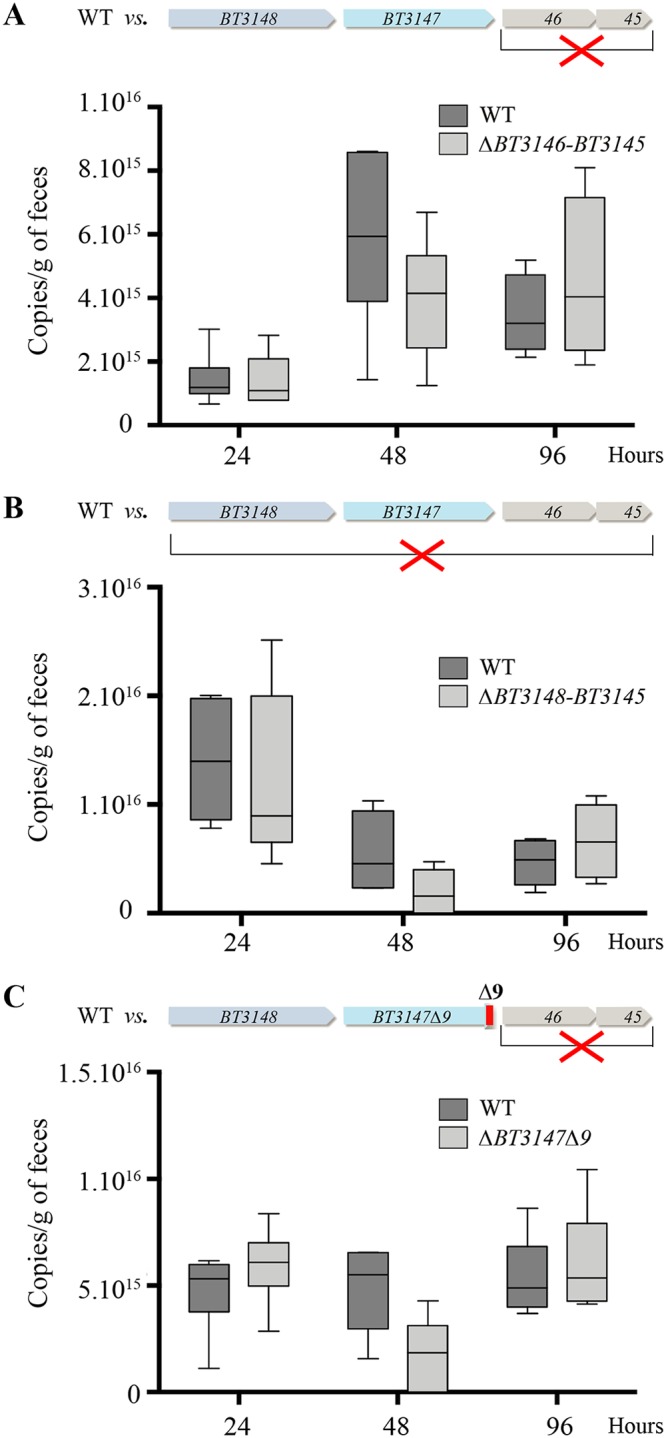
Contribution of the BT3148-BT3145 genes to B. thetaiotaomicron in vivo intestinal colonization in germfree mice. (A) In vivo mixed-culture competition experiments comparing the respective colonization capacities of the WT and ΔBT3146-BT3145 strains. (B) In vivo mixed-culture competition experiments comparing the respective colonization capacities of the WT and ΔBT3148-BT3145 strains. (C) In vivo mixed-culture competition experiments comparing the respective colonization capacities of the WT and the biofilm-forming BT3147Δ9 ΔBT3146-BT3145 strain. All strains were administered intragastrically at a 1:1 ratio (2 × 107 cells/ml). Comparisons were performed after collection of fecal samples 24 h, 48 h, and 96 h after the initial gavage and strain-specific qPCR on frozen fecal material samples and did not show any significant statistical difference between the tested strains.
DISCUSSION
In this study, we investigated the biofilm formation capacity of the prominent gut symbiont B. thetaiotaomicron. We found that many B. thetaiotaomicron isolates displayed an increased biofilm formation capacity compared to the reference B. thetaiotaomicron VPI 5482 strain that is used in most gut microbiota studies. This widespread but highly variable capacity to form biofilm is observed in many aerobic bacteria, including the facultative anaerobe Escherichia coli (23) and Staphylococcus epidermidis (24), and is consistent with a study performed in Bacteroides fragilis, a closely related intestinal anaerobe which also displays a variable biofilm formation capacity (25). Biofilm formation by pathogenic bacteria is a major cause of chronic and recurring infections (2), and pathogenicity and abscess formation by B. fragilis are associated with biofilm formation and adhesion to the peritoneal epithelium (26, 27). Whereas B. thetaiotaomicron usually behaves as an intestinal nonpathogenic bacterium, it is also found associated with abdominal and deep-wound infections, representing up to 14% of the bacteria found at these sites (28). Since 5 out of 14 of the best biofilm-forming strains identified in our study were isolated from different infection sites (bone biopsy specimens, blood samples, or abscesses) (see Table S1 in the supplemental material), this suggests that biofilm formation by B. thetaiotaomicron could be correlated with opportunistic extraintestinal infections, and future investigations could provide insights into this aspect of B. thetaiotaomicron biology.
Our static and dynamic biofilm assays show that B. thetaiotaomicron strain VPI 5482 forms poor biofilms in vitro, which is consistent with a study performed by TerAvest and colleagues, who obtained a VPI 5482 biofilm only after 8 days of continuous incubation in a chemostat (19). Whereas we did not observe significant biofilm accumulation in VPI 5482 even after 48 h, this result might reflect the fact that VPI 5482 biofilm formation is repressed under laboratory conditions. For example, uropathogenic E. coli is known to cause infections by adhering to the bladder epithelium using fimbriae but displays poor biofilm formation in vitro (29, 30). Similarly, Huang and colleagues demonstrated that poor adherence of B. fragilis is enhanced on mucin-covered plates, which mimics the intestinal mucosa, the natural habitat of B. fragilis (31).
Our approach to uncover the biofilm potential and biofilm-related functions of B. thetaiotaomicron VPI 5482 resulted in the identification of two putative Mfa1-like proteins (BT3148 and BT3147) potentially involved in biofilm formation in B. thetaiotaomicron. BT3147 and BT3148 have N-terminal domain homologies with Mfa1-type pili of the closely related anaerobic oral pathogen P. gingivalis, in which two types of pili were described, the long FimA and the short Mfa1, which represent the prototype of a recently defined type V pilus that is ubiquitous in gut Bacteroidetes (20). In VPI 5482, the BT3148-BT3147 genes appear to encode a minimal Mfa1-type operon encoding only the potential pilus anchor (BT3148) and stalk pilin (BT3147) (Fig. 5A). Intriguingly, studies of Mfa1-type pili in P. gingivalis showed that the presence of minor pilins is necessary for proper pilus function and that deletion of the minor pilus component mfa4 results in a complete loss of function (32, 33). The apparent lack of the mfa3 to mfa5 genes encoding accessory pilins near putative mfa1- and mfa2-like genes (BT3147 and BT3148, respectively) suggests that BT3148-BT3147 might correspond to a new minimal type V pilus operon that does not contain or has lost its accessory pilins during evolution.
The contribution of the putative Mfa-like BT3148-BT3147-based pilus to biofilm formation is revealed only upon removal of the last 9 C-terminal amino acids of BT3147. The potential polymerization observed in our immunodetection experiments (Fig. 5B) suggested that C-terminal truncation of the putative stalk pilin BT3147 could lead to the synthesis of an extended pilus, thus enhancing in vitro adhesion. This is surprising, as it has been suggested that the C terminus plays a critical role in type V pilus assembly (34), and the observed polymerization could correspond to an artifact introduced upon C-terminal truncation of BT3147. However, a similar phenotype had been described for P. gingivalis, in which deletion and truncation of the anchor resulted in the production of an unusually long pilus that promoted aggregation and biofilm formation (35, 36). The exact mechanism of synthesis of Mfa1- and FimA-like pili has not yet been resolved; however, recent structural studies of crystal structures and cysteine-based cross-linking showed an interaction between beta strands of Mfa1 N and C termini (20, 34), indicating that polymerization could include strand exchange between the C-terminal and N-terminal pilin domains (37). In P. gingivalis, maturation of Mfa1 pilins involves an N-terminal proteolytic cleavage step performed by the RgpA/B arginine protease of P. gingivalis. This proteolysis takes place in a loop connecting two N-terminal β-strands, enabling the removal of one strand and freeing a groove for the insertion of a C-terminal β-strand coming from another Mfa1 pilin subunit. It is speculated that in this processive assembly process, Mfa1/FimA stalk pilin subunits need to undergo conformational changes to allow them to connect to the N terminus of the next pilin unit (20). The critical role of the Mfa1/FimA C terminus during type V pilus assembly was recently confirmed by Hall and colleagues, as they showed that the presence of the last 6 C-terminal amino acids is necessary for polymerization (34). The reason why truncation of the putative Mfa1-like B. thetaiotaomicron pilin BT3147 seems to stimulate rather than impair polymerization and biofilm formation is not clear but could reflect a structural difference in the C terminus of BT3147 compared to P. gingivalis Mfa1. We indeed could not find any homology associated with the BT3147 C terminus and the Mfa1-type structural model (PDB code 4GPV) (Fig. S3). In contrast, Phyre structural model prediction of BT3147 suggests the presence of a long and unstructured C terminus in BT3147 (Fig. S4). This suggests that at least some of the B. thetaiotaomicron type V pilin structures might differ from those of P. gingivalis and could potentially require additional or different posttranslational pilus modifications during the assembly process. In our study, removal of the last 9 C-terminal amino acids of BT3147 might have exposed the A1 C-terminal strand, which was shown to be involved in the interaction with the N terminus (34), allowing for pilus polymerization, as suggested by the polymerization pattern observed in the BT3147Δ9 ΔBT3146-BT3145 strain (Fig. 5B). In support of this possibility, we showed that BT3147Δ9 detected using anti-BT3147 antibodies migrates at a size about 10 kDa lower than that of the WT protein. We therefore hypothesize that the truncation in the BT3147Δ9 protein could unmask a protease cleavage site and undergo an extra proteolytic cleavage step. Preliminary results using mass spectrometry analysis of the shorter band detected in the 1H2 mutant indicated a protein size of 27 kDa, in accordance with the protein size observed by immunodetection and consistent with a further maturation of the protein.
Alternatively, considering that all type V pilins (major, minor, and accessory) share domain homology in their N termini, we cannot completely rule out that BT3147 belongs to the group of accessory pilins (Mfa3-Mfa5-like) whose C-terminal structure does not allow further pilus elongation (20). In this scenario, deletion of the last 9 C-terminal amino acids of BT3147 could remove a peptide normally blocking polymerization, thus resulting in the observed polymerization in the BT3147Δ9 ΔBT3146-BT3145 strain.
We also show that BT3147 is constitutively expressed in the wild-type strain, suggesting that the BT3148-BT3147 genes are not cryptic. However, as we were unable to see differences in the colonization of the intestinal tract of germfree mice using strains with the WT or the mutated form of BT3147, the in vivo function of this locus is yet to be determined. In P. gingivalis, the Mfa1-type filaments promote heterologous cell-to-cell interactions, including adhesion to an oral pathogen often found in dental plaque, Streptococcus gordonii (46), and future work will determine whether a similar function is carried out by B. thetaiotaomicron BT3148-BT3147.
Our study demonstrates the widespread ability of B. thetaiotaomicron to form biofilm and brings the first evidence for the existence of functional type V pilus structures as mediators of adhesion in B. thetaiotaomicron in vitro. We expect this work to contribute to the introduction of B. thetaiotaomicron proteinaceous adhesins and biofilm-related functions in intestinal colonization and microbiota-host interactions in this anaerobic gut symbiont.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table S2 in the supplemental material. B. thetaiotaomicron was grown in BHIS broth (38) supplemented with erythromycin (20 μg/ml), tetracycline (2.5 μg/ml), gentamicin (200 μg/ml), or 5′-fluoro-2′-deoxyruidine (FUdR) (200 μg/ml) when required and incubated at 37°C under anaerobic conditions using anaerobic jars with anaerobic atmosphere generators or in a C400M Ruskinn anaerobic-microaerophilic station. E. coli S17λpir was grown in lysogeny broth (LB) supplemented with ampicillin (100 μg/ml) when required and incubated at 37°C. All media and chemicals were purchased from Sigma-Aldrich.
Biofilm formation in microfermentors.
Continuous-flow biofilm microfermentors containing a removable glass spatula were used as described previously (39) (see also https://research.pasteur.fr/en/tool/biofilm-microfermenters/), with or without internal bubbling of a filter-sterilized compressed mix of 90% nitrogen–5% hydrogen–5% carbon dioxide (anaerobic biofilm conditions). Biofilm microfermentors were inoculated by placing the spatula in a culture solution adjusted to an OD600 of 0.5 (containing 2.5 × 108 bacteria/ml) for 10 min. The spatula was then reintroduced into the microfermentor. The flow rate was then adjusted (30 ml/h) so that the total time for renewal of microfermentor medium was shorter than the bacterial generation time, thus minimizing planktonic growth by constant dilution of nonbiofilm bacteria.
Transposon mutagenesis and positive selection.
We used pSAMbt, a plasmid previously used for random mariner-based transposon mutagenesis (40), to generate a library of approximately 10,000 B. thetaiotaomicron mutants carrying erythromycin-resistant transposons. The mutants were pooled and subjected to a positive-selection procedure using biofilm microfermentors. The mutant pool was put into contact with the glass microfermentor spatula for 10 min, and the spatula was then introduced into the microfermentor and incubated with internal bubbling agitation to select for strongly adherent bacteria during 8 h. Following incubation, the bacteria attached to the spatula were resuspended in 15 ml of fresh BHIS medium supplemented with erythromycin and incubated anaerobically overnight. On the next day, the culture grown overnight was used to start another round of positive selection. This procedure was repeated four times, with the last microfermentor incubation lasting 24 h instead of 8 h. The biofilm composed of adherent transposon mutants developing on the spatula was then recovered and plated out on BHIS-erythromycin agar to obtain single colonies that were then individually tested for biofilm formation using 96-well crystal violet staining.
Biofilm microtiter plate assay.
Cultures grown overnight were diluted to an OD600 of 0.01 (5 × 106 cells/ml) and inoculated in technical triplicates into a polystyrene Greiner 96-well plate. Incubation was done at 37°C under anaerobic conditions for 24 h. The supernatant with free-floating bacteria was removed by careful pipetting, and 100 μl of Bouin’s solution (0.9% picric acid, 9% formaldehyde, and 5% acetic acid) was applied during 15 min to fix the biofilm attached to the well. Next, the fixation solution was washed 3 times with water, and biofilm was stained with a 1% crystal violet solution during 15 min. After removal of the crystal violet solution, biofilms were washed 3 times with water. For quantification of biofilm formation, dried stained biofilms were resuspended in a 1:5 acetone-ethanol mixture, and the absorbance was measured at 575 nm using a infinite M200 Pro plate reader. Biofilm formation of different B. thetaiotaomicron strains is represented in values normalized to the average biofilm formation of the wild-type strain VPI 5482.
Construction of B. thetaiotaomicron mutants.
All experiments and genetic constructions of B. thetaiotaomicron were made in the VPI 5482_Δtdk strain, which was developed for a 2-step selection procedure for unmarked gene deletion by allelic exchange, as previously described (41). Therefore, the VPI 5482_Δtdk strain is referred to as the wild type in this study. All primers used in this study are described in Table S3 in the supplemental material.
For the generation of deletion strains by allelic exchange in B. thetaiotaomicron, we used the pExchange-tdk vector (41). Briefly, 1-kb upstream and downstream regions of the target sequence were cloned into the pExchange-tdk vector and transformed into the E. coli S17λpir strain, which was used to deliver the vector to B. thetaiotaomicron by conjugation. Conjugation was carried out by mixing exponentially grown cultures of the donor and the recipient strains at a 2:1 ratio, and mating was done by placing the mixture on a Millipore 0.45-μm hydrophilic cellulose ester filter disc (catalog no. HAWP29325) on top of BHIS agar plates at 37°C under aerobic conditions overnight. After this, bacteria were recovered by resuspending and vortexing the filters in fresh BHIS medium and then plated on selective BHIS agar supplemented with erythromycin for selection of B. thetaiotaomicron transconjugants that underwent the first recombination event and gentamicin to ensure the exclusion of any E. coli growth. Erythromycin- and gentamicin-resistant colonies of B. thetaiotaomicron were then subjected to a second round of selection on BHIS agar plates supplemented with FUdR for selection of double-recombination colonies. The resulting deletion mutants were confirmed by PCR and sequencing.
For constitutive gene expression, we used the pNBU2-bla-erm vector (42), which inserts itself into the 5′ untranslated region of tRNASer, thus providing chromosome-based expression. In order to provide equal expression levels of different target genes, we used the promoter of the BT1311 gene (coding for the sigma factor RpoD), which had been cloned from a PCR fragment into the multiple-cloning site of pNBU2-bla-erm to yield pNBU1311. Target genes were cloned (start to stop codons) using the NdeI/NotI sites of pNBU1311, and the resulting vector was then transformed into the E. coli S17λpir strain. Transfer of the expression vector to B. thetaiotaomicron was done by conjugation as described above, this time with only one selection step. The transformed B. thetaiotaomicron strains were selected on erythromycin, and vector insertions were confirmed by PCR using pNBU1311-specific primers.
RNAseq analysis.
Total RNA was extracted from exponentially grown cultures of wild-type B. thetaiotaomicron using an MP Biomedicals FastRNA Pro blue kit according to the provider’s manual and treated with an Ambion Turbo DNA-free kit to remove possible DNA contamination. All samples were checked for residual genomic DNA contamination with the primer pair DNAPol3-F and DNAPol3-R (Table S3) and were considered DNA-free if no amplification was detected at <30 cycles. rRNA was removed using Illumina Ribo-Zero rRNA removal kits, the purified RNA concentration was measured with a NanoDrop instrument, and RNA quality was checked by using an Agilent 2100 bioanalyzer. The obtained RNA samples were fragmented and used for cDNA library preparation using TruSeq stranded mRNA library preparation kit set A. Samples were sequenced with an Illumina HiSeq2500 sequencer. Sequence quality was analyzed by using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and reads were aligned against the reference VPI 5482 genome using the Bowtie2 R package (43).
Structural analyses.
Conserved domain analyses were performed using the CDSEARCH/cdd v3.15 database (44). A three-dimensional (3D) homology search was performed using the Phyre2 program in the intensive mode (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
In vivo mixed-culture competition experiments in germfree mice.
Animal experiments were done at the Animalerie Axénique de MICALIS (ANAXEM) platform (Microbiologie de l’Alimentation au Service de la Santé [MICALIS], Jouy-en-Josas, France) according to official authorization no. 3441-2016010614307552 delivered by a French ministry (Education Nationale, Enseignement Supérieur, et Recherche). The protocol was approved by a local ethic committee on animal experimentation (committee no. 45). All animals were housed in flexible-film isolators (Getinge-La Calhène, Vendôme, France) under controlled environmental conditions (12-light/12-h dark cycle, temperature of between 20°C and 22°C, and humidity of between 45 and 55%). Mice were provided with sterile tap water and a gamma-irradiated standard diet (catalog no. R03-40; SAFE, Augy, France), ad libitum. Their bedding was composed of wood shavings, and they were also given cellulose sheets as enrichment.
Methods.
Six male C3H/HeN germfree mice were gavaged with 200-μl bacterial suspensions containing 2 × 107 cells/ml consisting of a 1:1 mix of wild-type B. thetaiotaomicron and one of the tested strains: ΔBT3146-BT3145, ΔBT3148-BT3145, or BT3147Δ9 ΔBT3146-BT3145. The wild-type B. thetaiotaomicron strain carried an erythromycin resistance gene (VPI 5482/pNBU2-bla-erm) (Table S1) for later distinction from the tested erythromycin-sensitive strains. Six mice were used under each condition (wild type with ΔBT3146-BT3145, wild type with ΔBT3148-BT3145, and wild type with BT3147Δ9 ΔBT3146-BT3145). At 24, 48, and 96 h postinoculation, feces were collected and homogenized in 1 ml of 1× phosphate-buffered saline (PBS), and serial dilutions were plated onto BHIS agar plates with and without erythromycin (20 μg/ml). The intestinal colonization capacity of the wild type and different mutants was estimated by assessing their abundance in feces using quantitative PCR (qPCR). Total bacterial DNA was isolated from the fecal samples with a QIAamp DNA stool kit, and 10 ng was used as the template for qPCR amplification of gyrA and ermG using Brilliant II SYBR green qPCR master mix according to the manufacturer’s manual. The reaction was performed in a Bio-Rad thermal cycler, and absolute quantities of target genes were determined using the standard-curve quantification method. The gene copy number was calculated from the absolute quantities using the following formula: (starting quantity [nanograms] × 60,221 × 1023 molecules/mol)/(amplicon length [base pairs] × 660 g/mol × 1 × 109 ng/g). The abundances of the wild-type and the competing mutant strains were estimated by quantifying the copy number of genes coding for the gyrase A subunit (gyrA-BT0899) and ermG, conferring resistance to erythromycin. While gyrA is present in the wild-type as well as in the mutant strains, ermG is present only in the wild-type strain. Therefore, the abundance of the wild-type strain corresponds to the copy number of ermG, and the mutant strain abundance is deduced as the difference in the copy numbers between gyrA (total population) and ermG (wild type).
Statistical analysis.
Unpaired two-tailed Mann-Whitney nonparametric tests were performed using Prism 6.0 for Mac OS X (GraphPad Software, Inc.). Each experiment was performed at least 3 times.
Generation of an antiserum against BT3147.
Polyclonal antibodies against BT3147 were raised in rabbits injected with a mixture of two peptides: peptide 1 (C-VYDETGKTEVKQETTF-coNH2 [corresponding to BT3147 amino acids 54 to 69]) and peptide 2 (C-VADGKTTGTESDTKT-coNH2 [corresponding to BT3147 amino acids 169 to 183]). The design and synthesis of the peptides, as well as the production and purification of the antibodies, were performed at the Covalab Research Institute, Villeurbane, France. A mix of peptides 1 and 2 with Freund adjuvant was used for immunization of rabbits by 3 intradermal injections at day 0, day 21, and day 42 of the immunization protocol, followed by a subcutaneous injection at day 74. At day 88 of the protocol, anti-BT3147 antibodies were purified from rabbit serum using affinity chromatography with a mix of peptides 1 and 2 as column ligands.
Preparation of protein extracts from shaved bacteria.
Cultures grown overnight in BHIS medium were washed and resuspended in 1× PBS. “Shaving” of surface pili was performed as previously described (45). Briefly, 100 ml of the culture was grown in BHIS medium for 48 h at 37°C without agitation under anaerobic conditions. The culture was centrifuged (6,000 rpm for 10 min), the pellet was resuspended in 1× PBS, and potential surface appendages were “shaved” on ice using a mini-hand blender using 5 1-min pulses (19,600 rpm). Next, the samples were centrifuged at 6,000 rpm for 1 h at 4°C, and proteins from the supernatant were precipitated using 10% trichloroacetic acid (TCA) for 48 h at 4°C, followed by centrifugation at 20,000 rpm for 1 h at 4°C. The pellet was washed with 100% acetone and resuspended in 100 μl of 8 M urea. Eight micrograms of the shaved surface protein was analyzed by Western blotting using anti-BT3147 antibodies.
Western blot analysis.
Cultures were grown in BHIS medium overnight, and 1 ml at an OD600 of 2 was centrifuged (7,000 rpm for 5 min), resuspended in 100 μl of 1× Laemmli buffer with 250 U of Benzonase nuclease (catalog no. E0114; Sigma), and incubated for 5 min at 95°C. Aliquots of these raw cell extracts or protein extracts from shaved bacteria were run on Mini-Protean TGX stain-free precast gels (Bio-Rad) in 1× TGX buffer and then transferred to a nitrocellulose membrane using a Trans-Blot Turbo transfer system (Bio-Rad). Blocking was performed during 1 h in a 5% solution of dry milk and 0.05% Tween–1× PBS (1× PBST). The membranes were then incubated overnight in 1× PBST with anti-BT3147 antiserum at a 1:1,000 dilution, at 4°C with agitation. Membranes were washed in 1× PBST and then incubated with the secondary antibody (anti-rabbit IgG conjugated with horseradish peroxidase at a 1:3,000 dilution; Promega). After a washing the excess second antibody, specific bands were visualized using the ECL prime detection method (GE Healthcare).
Supplementary Material
ACKNOWLEDGMENTS
We thank Andy Goodman and Justin Sonnenburg for generously providing plasmids and strains used as genetic tools in this study, Marie-Cécile Ploy and Luc Dubreuil for providing B. thetaiotaomicron strains, and Bruno Dupuy and Isabelle Martin-Verstraete for their help and advice regarding the handling of anaerobic bacteria.
This work was supported by an Institut Pasteur grant; by the French government’s Investissement d’Avenir Program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant no. ANR-10-LABX-62-IBEID); and by the Fondation pour la Recherche Médicale (grant no. DEQ20180339185 Equipe FRM 2018). J. Mihajlovic was supported by the Pasteur Paris University International Doctoral Program and the Fondation pour la Recherche Médicale (grant no. FDT20160435523).
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00650-18.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Lebeaux D, Ghigo J-M, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes M, Borras L, Seco A, Ferrer J. 2015. Identification and quantification of microbial populations in activated sludge and anaerobic digestion processes. Environ Technol 36:45–53. doi: 10.1080/09593330.2014.934745. [DOI] [PubMed] [Google Scholar]
- 4.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajilić-Stojanović M, de Vos WM. 2014. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenburg JL, Angenent LT, Gordon JI. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol 5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 7.Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR. 2004. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol 28:23–27. doi: 10.1080/01913120490275196. [DOI] [PubMed] [Google Scholar]
- 8.Swidsinski A, Loening-Baucke V, Lochs H, Hale L-P. 2005. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol 11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macfarlane S, Dillon JF. 2007. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol 102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 10.Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, Stein E, Vadivelu J, Roslani AC, Malik AA, Wanyiri JW, Goh KL, Thevambiga I, Fu K, Wan F, Llosa N, Housseau F, Romans K, Wu X, McAllister FM, Wu S, Vogelstein B, Kinzler KW, Pardoll DM, Sears CL. 2014. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A 111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, Cho K, Wick EC, Hechenbleikner EM, Uritboonthai W, Goetz L, Casero RA Jr, Pardoll DM, White JR, Patti GJ, Sears CL, Siuzdak G. 2015. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab 21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 13.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 14.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol 4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 15.Zocco MA, Ainora ME, Gasbarrini G, Gasbarrini A. 2007. Bacteroides thetaiotaomicron in the gut: molecular aspects of their interaction. Dig Liver Dis 39:707–712. doi: 10.1016/j.dld.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viljanen MK, Linko L, Lehtonen OP. 1988. Detection of Bacteroides fragilis, Bacteroides thetaiotaomicron, and Bacteroides ovatus in clinical specimens by immunofluorescence with a monoclonal antibody to B. fragilis lipopolysaccharide. J Clin Microbiol 26:448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feuillet L, Carvajal J, Sudre I, Pelletier J, Thomassin JM, Drancourt M, Cherif AA. 2005. First isolation of Bacteroides thetaiotaomicron from a patient with a cholesteatoma and experiencing meningitis. J Clin Microbiol 43:1467–1469. doi: 10.1128/JCM.43.3.1467-1469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.TerAvest MA, He Z, Rosenbaum MA, Martens EC, Cotta MA, Gordon JI, Angenent LT. 2014. Regulated expression of polysaccharide utilization and capsular biosynthesis loci in biofilm and planktonic Bacteroides thetaiotaomicron during growth in chemostats. Biotechnol Bioeng 111:165–173. doi: 10.1002/bit.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Q, Shoji M, Shibata S, Naito M, Sato K, Elsliger M-A, Grant JC, Axelrod HL, Chiu H-J, Farr CL, Jaroszewski L, Knuth MW, Deacon AM, Godzik A, Lesley SA, Curtis MA, Nakayama K, Wilson IA. 2016. A distinct type of pilus from the human microbiome. Cell 165:690–703. doi: 10.1016/j.cell.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. 2009. Surface components of Porphyromonas gingivalis. J Periodontal Res 44:1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 22.Nagano K, Hasegawa Y, Abiko Y, Yoshida Y, Murakami Y, Yoshimura F. 2012. Porphyromonas gingivalis FimA fimbriae: fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA genotypes. PLoS One 7:e43722. doi: 10.1371/journal.pone.0043722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansan-Almeida R, Pereira AL, Giugliano LG. 2013. Diffusely adherent Escherichia coli strains isolated from children and adults constitute two different populations. BMC Microbiol 13:22. doi: 10.1186/1471-2180-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begovic J, Jovcic B, Papic-Obradovic M, Veljovic K, Lukic J, Kojic M, Topisirovic L. 2013. Genotypic diversity and virulent factors of Staphylococcus epidermidis isolated from human breast milk. Microbiol Res 168:77–83. doi: 10.1016/j.micres.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Reis ACM, Silva JO, Laranjeira BJ, Pinheiro AQ, Carvalho CBM. 2014. Virulence factors and biofilm production by isolates of Bacteroides fragilis recovered from dog intestinal tracts. Braz J Microbiol 45:647–650. doi: 10.1590/S1517-83822014000200037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung DR, Kasper DL, Panzo RJ, Chitnis T, Grusby MJ, Sayegh MH, Tzianabos AO, Chtinis T. 2003. CD4+ T cells mediate abscess formation in intra-abdominal sepsis by an IL-17-dependent mechanism. J Immunol 170:1958–1963. doi: 10.4049/jimmunol.170.4.1958. [DOI] [PubMed] [Google Scholar]
- 27.Gibson FC III, Onderdonk AB, Kasper DL, Tzianabos AO. 1998. Cellular mechanism of intraabdominal abscess formation by Bacteroides fragilis. J Immunol 160:5000–5006. [PubMed] [Google Scholar]
- 28.Brook I. 1989. Pathogenicity of the Bacteroides fragilis group. Ann Clin Lab Sci 19:360–376. [PubMed] [Google Scholar]
- 29.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. 2007. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog 3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valle J, Da Re S, Henry N, Fontaine T, Balestrino D, Latour-Lambert P, Ghigo JM. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc Natl Acad Sci U S A 103:12558–12563. doi: 10.1073/pnas.0605399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang JY, Lee SM, Mazmanian SK. 2011. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 17:137–141. doi: 10.1016/j.anaerobe.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikai R, Hasegawa Y, Izumigawa M, Nagano K, Yoshida Y, Kitai N, Lamont RJ, Yoshimura F, Murakami Y. 2015. Mfa4, an accessory protein of Mfa1 fimbriae, modulates fimbrial biogenesis, cell auto-aggregation, and biofilm formation in Porphyromonas gingivalis. PLoS One 10:e0139454. doi: 10.1371/journal.pone.0139454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng C, Wu J, Xie H. 2011. Differential expression and adherence of Porphyromonas gingivalis FimA genotypes. Mol Oral Microbiol 26:388–395. doi: 10.1111/j.2041-1014.2011.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall M, Hasegawa Y, Yoshimura F, Persson K. 2018. Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis. Sci Rep 8:1793. doi: 10.1038/s41598-018-20067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa Y, Iwami J, Sato K, Park Y, Nishikawa K, Atsumi T, Moriguchi K, Murakami Y, Lamont RJ, Nakamura H, Ohno N, Yoshimura F. 2009. Anchoring and length regulation of Porphyromonas gingivalis Mfa1 fimbriae by the downstream gene product Mfa2. Microbiology 155:3333–3347. doi: 10.1099/mic.0.028928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagano K, Hasegawa Y, Murakami Y, Nishiyama S, Yoshimura F. 2010. FimB regulates FimA fimbriation in Porphyromonas gingivalis. J Dent Res 89:903–908. doi: 10.1177/0022034510370089. [DOI] [PubMed] [Google Scholar]
- 37.Lee JY, Miller DP, Wu L, Casella CR, Hasegawa Y, Lamont RJ. 2018. Maturation of the Mfa1 fimbriae in the oral pathogen Porphyromonas gingivalis. Front Cell Infect Microbiol 8:137. doi: 10.3389/fcimb.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacic MK, Smith CJ. 2008. Laboratory maintenance and cultivation of Bacteroides species. Curr Protoc Microbiol Chapter 13:Unit 13C.1. doi: 10.1002/9780471729259.mc13c01s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 40.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Shoemaker NB, Wang G-R, Salyers AA. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol 182:3559–3571. doi: 10.1128/JB.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsonneur F, Martin FA, Mallet A, Martinez-Gil M, Semetey V, Ghigo J-M, Beloin C. 2016. Functional analysis of Escherichia coli Yad fimbriae reveals their potential role in environmental persistence. Environ Microbiol 18:5228–5248. doi: 10.1111/1462-2920.13559. [DOI] [PubMed] [Google Scholar]
- 46.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627–1636. https://www.ncbi.nlm.nih.gov/pubmed/12055284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.