The finding that exposure to DNase I impairs biofilm formation or leads to the dispersal of early stage biofilms has led to the realization of extracellular genomic DNA (eDNA) as a structural component of the biofilm matrix. However, little is known about the contribution of intrinsic DNases to the weakening of the biofilm matrix and dispersion of established biofilms. Here, we demonstrate for the first time that nucleases are induced in dispersed Pseudomonas aeruginosa cells and are essential to the dispersion response and that degradation of matrix eDNA by endogenously produced/secreted EndA is required for P. aeruginosa biofilm dispersion. Our findings suggest that dispersing cells mediate their active release from the biofilm matrix via the induction of nucleases.
KEYWORDS: DNA-specific endonuclease I, EddB, EndA, biofilm, dispersion, endonuclease, nuclease
ABSTRACT
The dispersion of biofilms is an active process resulting in the release of planktonic cells from the biofilm structure. While much is known about the process of dispersion cue perception and the subsequent modulation of the c-di-GMP pool, little is known about subsequent events resulting in the release of cells from the biofilm. Given that dispersion coincides with void formation and an overall erosion of the biofilm structure, we asked whether dispersion involves degradation of the biofilm matrix. Here, we focused on extracellular genomic DNA (eDNA) due to its almost universal presence in the matrix of biofilm-forming species. We identified two probable nucleases, endA and eddB, and eddA encoding a phosphatase that were significantly increased in transcript abundance in dispersed cells. However, only inactivation of endA but not eddA or eddB impaired dispersion by Pseudomonas aeruginosa biofilms in response to glutamate and nitric oxide (NO). Heterologously produced EndA was found to be secreted and active in degrading genomic DNA. While endA inactivation had little effect on biofilm formation and the presence of eDNA in biofilms, eDNA degradation upon induction of dispersion was impaired. In contrast, induction of endA expression coincided with eDNA degradation and resulted in biofilm dispersion. Thus, released cells demonstrated a hyperattaching phenotype but remained as resistant to tobramycin as biofilm cells from which they egress, indicating EndA-dispersed cells adopted some but not all of the phenotypes associated with dispersed cells. Our findings indicate for the first time a role of DNase EndA in dispersion and suggest weakening of the biofilm matrix is a requisite for biofilm dispersion.
IMPORTANCE The finding that exposure to DNase I impairs biofilm formation or leads to the dispersal of early stage biofilms has led to the realization of extracellular genomic DNA (eDNA) as a structural component of the biofilm matrix. However, little is known about the contribution of intrinsic DNases to the weakening of the biofilm matrix and dispersion of established biofilms. Here, we demonstrate for the first time that nucleases are induced in dispersed Pseudomonas aeruginosa cells and are essential to the dispersion response and that degradation of matrix eDNA by endogenously produced/secreted EndA is required for P. aeruginosa biofilm dispersion. Our findings suggest that dispersing cells mediate their active release from the biofilm matrix via the induction of nucleases.
INTRODUCTION
Surface-associated communities of bacteria, encased in a polymeric matrix of their own matrix, are referred to as biofilms. The surface association of biofilms can be to inert or living surfaces (1). Biofilms form as part of a developmental life cycle that is initiated by planktonic cells attaching to surfaces and turns full circle when biofilm cells liberate themselves from the biofilm structure and disperse. Dispersion is apparent by the hollowing of biofilm microcolonies, with central voids generated by single cells breaking free from the biofilm, or by an overall erosion of the three-dimensional biofilm structure (2–12). Dispersion likely occurs in response to the accumulation of waste products or oxygen depletion in the interior of biofilms, making dispersion a process by which bacteria avoid stress associated with biofilm growth and seek nutrient rich environments better suited for survival at new locales (3, 5, 8, 12). Dispersion, furthermore, can be induced in response to heavy metals and nitric oxide (NO), upon exposure to elevated concentrations of carbon sources, amino acids, and ammonium chloride, or by the sensing of cis-2-decenoic acid, an interkingdom fatty acid signaling molecule belonging to the (B)DSF family (5, 13).
While dispersed cells revert to the planktonic mode of growth and phenotypically resemble planktonic cells, it is now apparent that while dispersed cells actively escape as single cells from the biofilm, dispersed cells differ from planktonic cells. Dispersed cells were found to exhibit protein production and gene expression profiles that are distinct from those of planktonic cells and biofilms from which they escaped (3, 14–17). Moreover, dispersed cells appear to be more primed to reattach following egress from the biofilm (14, 18), to produce more matrix-degrading enzymes (16), to be more virulent than planktonic cells when tested using various acute and chronic virulence models (15, 16, 19), and depending on the antibiotic used, to be as susceptible as planktonic cells or resistant to antimicrobial agent (14, 20). These differences have led to dispersed cells being referred to as a third phenotype that is distinct from the phenotypes of both planktonic and biofilm cells (3, 15, 16). The distinct phenotype of dispersed cells, however, was found to be reversible and somewhat short lived. Using quantitative reverse transcriptase PCR (qRT-PCR) and antimicrobial susceptibility assays, Chambers et al. (14) demonstrated that in Pseudomonas aeruginosa, differences between planktonic and dispersed cells remained for 2 h postdispersion, with additional time being required for dispersed cells to express genes serving as signs of exponential growth.
Levels of the intracellular signaling molecule c-di-GMP have important implications for the bacterial mode of growth, with low c-di-GMP levels fostering the motile lifestyle and high intracellular levels promoting aggregative behavior, including attachment and biofilm formation (21, 22). It is thus not surprising that the induction of dispersion by P. aeruginosa biofilms has been linked to increased expression of fliC (encoding flagellin type B) (23), with cells actively leaving the biofilms, the activation of phosphodiesterases, and reduced levels of c-di-GMP present in dispersed cells relative to that in biofilm cells from which they egressed (6, 7, 9, 24). Additional support for c-di-GMP playing an important role in dispersion stems from the finding that planktonic cells expressing yhjH (also known as pdeH) encoding an Escherichia coli phosphodiesterase are similar with respect to gene expression, virulence, and susceptibility to antibiotics to dispersed cells obtained in response to NO (19, 25). The findings further underscored that dispersed cells are characterized by c-di-GMP levels that are significantly reduced relative to that in biofilm or even planktonic cells (26, 27).
Low c-di-GMP levels present in dispersed cells have led to the notion of dispersion being a consequence of reduced intracellular c-di-GMP levels, likely through the activation of phosphodiesterases (6, 7, 19). However, little is known about the subsequent events leading to biofilm dispersion. Considering dispersion coincides with a reduction in c-di-GMP levels, it is likely that dispersion coincides with phenotypes associated with low c-di-GMP levels, such as motility and the planktonic mode of growth, and/or a reversion of high c-di-GMP-associated phenotypes, such as reduced adhesiveness and decreased production of biofilm matrix components (21, 22, 28–33). The latter include the Psl and Pel polysaccharides by P. aeruginosa. In addition, localization of the large adhesin LapA has been linked to c-di-GMP. LapA is localized at the outer membrane at high c-di-GMP levels but released from the cell surface upon proteolytic cleavage by the protease LapG at low c-di-GMP (34). Not surprisingly, the release of the large adhesin LapA from the cell surface via cleavage by protease LapG has been shown to coincide in a dispersion response by Pseudomonas putida and Pseudomonas fluorescens biofilms (24).
Given the impact of c-di-GMP modulation on biofilm matrix components and considering that dispersion coincides with biofilm erosion and single cells escaping the biofilm structure, with dispersed cells producing more matrix-degrading enzymes than planktonic or biofilm cells (16), it is likely that dispersion relies on factors that degrade or at least compromise the integrity of the biofilm matrix. Matrix degradation playing a role in dispersion is supported by the plethora of matrix-degrading factors such as proteases, deoxyribonucleases, and glycoside hydrolases having been linked to biofilm dispersal (15, 16, 35–38). However, most studies have relied on inducing dispersion by the exogenous addition of these factors, and specific matrix-degrading factors that are intrinsic to the biofilm dispersion response remain uncharacterized.
The goal of this study was to identify specific matrix-degrading factors that contribute to dispersion. The biofilm matrix is composed of a variety of polysaccharides, proteins, adhesins, and extracellular genomic DNA (eDNA) (39, 40). While biofilm dispersion is common among biofilm-forming species, the compositions of the biofilm matrix differ significantly among biofilm-forming species. More specifically, the polysaccharides and adhesins present in the biofilm matrix appear to be species or even strain specific. However, one matrix component that appears to be common to all biofilm matrices is eDNA. We therefore asked whether dispersion relies on eDNA degradation to weaken or compromise the matrix encasing biofilms for bacteria to escape.
RESULTS
Identification of probable or predicted nucleases.
To determine whether the degradation of eDNA plays a role in the dispersion response, we surmised that eDNA degradation would involve nucleases that are active in the periplasmic or extracellular space and that the respective genes are induced upon induction of the dispersion response. To identify potential extracellular enzymes with nucleolytic activity, we made use of the biofilm model organism Pseudomonas aeruginosa PAO1 and screened the genome sequence for the presence of genes encoding nucleases using the search term “nuclease” (41). The screen revealed 15 genes encoding probable or predicted nucleases (Table 1). Of the 15 “nucleases,” according to https://www.pseudomonas.com/, 10 were predicted to be localized in the cytoplasm, while EndA (PA2749) and EddA (PA3910) were predicted to be localized in the extracellular space (Table 1). The respective localizations were confirmed by Lewenza et al. (42) using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. While subcellular prediction tools place EddB (PA3909) with equal likelihood in the cytoplasmic membrane, periplasm, outer membrane, or extracellular space, https://www.pseudomonas.com/ indicates EddB is localized in the extracellular space (Table 1). This subcellular localization is based on EddB harboring cleavable type I signal peptides (42) and having been experimentally confirmed to be secreted (43). An additional two “nucleases,” PA4424 and PA5048, were listed as unknown (Table 1). The amino acid sequences of PA5048 and PA4424 lack N-terminal signaling peptides enabling translocation across membranes (42), likely suggesting a cytoplasmic location.
TABLE 1.
Genes encoding potential and confirmed nucleases in the genome of P. aeruginosa PAO1a
| PA number | Gene name | Localization | N-terminal signal | Description |
|---|---|---|---|---|
| PA0965 | ruvC | Cytoplasmic | None | Holliday junction resolvase |
| PA2545 | xthA | Cytoplasmic | None | Exonuclease III |
| PA2585 | uvrC | Cytoplasmic | None | Endonuclease |
| PA2749 | endA | Extracellular | Type I | Endonuclease, DNA-specific endonuclease I |
| PA3232 | PA3232 | Cytoplasmic | None | Probable nuclease |
| PA3495 | Nth | Cytoplasmic | None | Endonuclease III |
| PA3725 | recJ | Cytoplasmic | None | Single-stranded-DNA-specific exonuclease |
| PA3909 | eddB | Extracellular | Type I | Extracellular DNA degradation protein |
| PA3910 | eddA | Extracellular | Type I | Alkaline phosphatase, PhoD-type phosphataseb |
| PA4172 | PA4172 | Cytoplasmic | None | Probable nuclease, endonuclease III-like |
| PA4281 | sbcD | Cytoplasmic | None | Exonuclease |
| PA4316 | scbB | Cytoplasmic | None | Exodeoxyribonuclease I, exonuclease I |
| PA4424 | PA4424 | Unknown | None | Endonuclease II-like (yraN homolog) |
| PA5048 | PA5048 | Unknown | None | Probable nuclease, SNase-like |
| PA5147 | mutY | Cytoplasmic | Endonuclease, endonuclease III-like |
As we anticipated nucleases to act on eDNA to weaken the biofilm matrix, with eDNA degradation likely occurring extracellularly, all candidates predicted to be located in the cytoplasm were excluded. We likewise excluded PA4424 and PA5048 due to localization. This reduced the number of potential candidates to three: EndA, EddA, and EddB.
We furthermore asked whether there are indications that the three remaining candidates indeed encoded potential nucleases. Based on sequence analysis, endA likely encodes a probable DNA-specific endonuclease I. EddB, encoded by PA3909, was previously reported to be required for DNA degradation and to promote P. aeruginosa growth when DNA was supplied as the sole phosphate source (43, 44). eddB is downstream of eddA (PA3910), with eddA encoding a PhoD-like alkaline phosphatase that possesses both monoesterase and phosphodiesterase activities (45). Despite EddA having been shown to harbor alkaline phosphatase activity, we included EddA in our further investigation, as EndA has been speculated to be required for phosphorus acquisition from DNA (46) and EddA homologs, such as the Zymomonas mobilis CP4 PhoD alkaline phosphatase, have been shown to hydrolyze nucleotides (47).
Dispersion coincides with increased transcript abundance of genes in the endA and eddA operons.
Having identified three potential candidates, we next determined their transcript abundance in response to dispersion cues. We reasoned that if EndA, EddA, or EddB plays a role in the dispersion response, then induction of dispersion would coincide with increased transcript abundance. To address this question, we determined the transcript abundance by qRT-PCR using RNA isolated from biofilms by two mutants, the ΔbdlA and PAO1/bdlA_G31A strains. By doing so, we took advantage of the finding that BdlA is central to the dispersion response to obtain homogenous populations of dispersing and nondispersing cells. This is supported by the fact that ΔbdlA biofilms are impaired in the dispersion response to a large variety of dispersion cues (9, 10), while biofilms overproducing BdlA_G31A, which transmits a constant signal-on bias for dispersion, are hyperdispersive (48). Previous findings indicated the hyperdispersive phenotype correlates with reduced biofilm biomass accumulation, but with a 2- to 3-fold increase of bacteria present in biofilm effluents compared to that in wild-type biofilms over the course of 5 days of biofilm growth (48). Moreover, we determined the transcript abundance of only eddA, not eddB, as the genes eddA and eddB were previously reported to be part of a 2-gene operon and to be coregulated (43, 45).
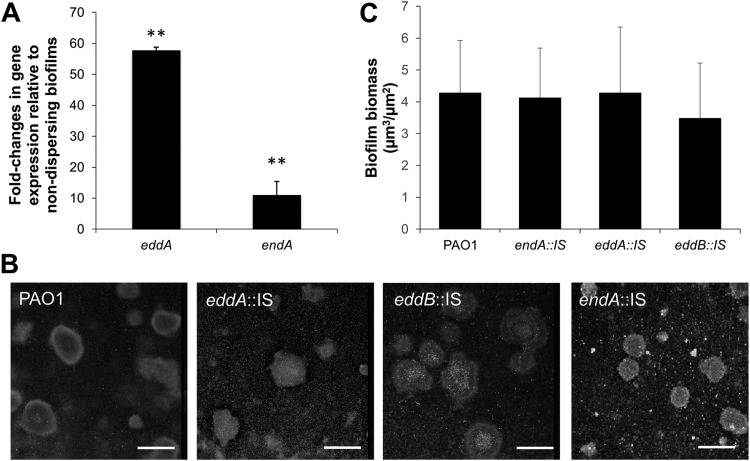
Relative to nondispersing biofilms, hyperdispersive biofilms were characterized by a significant 11-fold increase in endA transcript abundance and a 58-fold increase in the transcript abundance of eddA (Fig. 1A). The findings suggested the expression of endA and eddA was induced under dispersion conditions. It is of interest to note that based on quantification cycle (Cq) values, endA and eddA were transcribed in nondispersing ΔbdlA biofilm cells.
FIG 1.
Transcript abundance of endA and eddA in hyperdispersive biofilms relative to that in nondispersing biofilms cells, and contribution of endA, eddA, and eddB to biofilm formation. (A) Fold changes of endA and eddA transcript abundance in hyperdispersive biofilms relative to that in nondispersing biofilms, as determined using qRT-PCR. Biofilms by the hyperdispersive strain PAO1/pJN-bdlA-G31A and the nondispersive ΔbdlA cells were grown for 3 days under flowing conditions in tube reactors using 5-fold diluted VBMM. cysD was used as a housekeeping control. Error bars indicate standard deviations. **, P < 0.05 relative to ΔbdlA biofilms. (B) Representative confocal images of the biofilm architecture by P. aeruginosa PAO1 and endA::IS, eddA::IS, and eddB::IS mutant strains. Biofilms were grown for 5 days under flowing conditions in flow cells and stained prior to microscopy using the Live/Dead BacLight viability stain. Bars, 100 μm. (C) COMSTAT analysis of the biofilm biomass. All experiments were performed in triplicate. Error bars indicate standard deviations.
Insertional inactivation of endA, eddA, and eddB has no apparent effect on the biofilm architecture under flowing conditions.
Previous findings indicated that nucleases contribute to biofilm formation and dynamics (43, 44, 49). As our qRT-PCR analysis indicated that endA and eddA (and indirectly, eddB) are transcribed in biofilms, we next determined whether inactivation of endA, eddA, and eddB affected biofilm formation. We therefore made use of mutant strains harboring transposon insertions in endA, eddA, and eddB. The respective mutant strains are referred to as endA::IS, eddA::IS, and eddB::IS strains. Biofilms of the strains were grown for 5 days under flowing conditions in flow cells after which, the biofilm architecture was viewed by confocal microscopy. Visual inspection of biofilms by endA::IS, eddA::IS, and eddB::IS strains revealed an overall architecture characterized by the presence of large microcolonies that resembled biofilms formed by P. aeruginosa PAO1 (Fig. 1B). Quantitative analysis of the biofilm biomass using COMSTAT furthermore indicated insertional inactivation of endA, eddA, and eddB had apparent effect on the biofilm biomass accumulation under flowing conditions (Fig. 1C).
Inactivation of endA impairs biofilm dispersion in response to multiple dispersion cues.
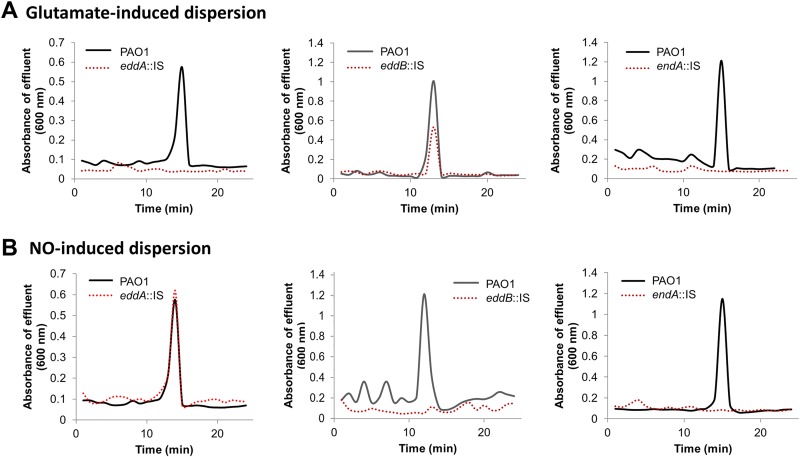
Considering the increased expression of endA, eddA, and eddB under dispersion-inducing conditions, we next asked whether the nucleases EndA and EddB or phosphatase EddA contributes to the dispersion response by P. aeruginosa biofilms. Previous findings indicated that dispersion can be induced by exposure to various exogenous dispersion cues (5, 13), including nitric oxide (NO) and glutamate (6, 9, 18, 23, 50). Furthermore, it has been demonstrated that dispersion can be detected by a decrease in the biofilm biomass and void formation, as determined using flow-cell-grown biofilms in conjunction with microscopy, as well as by a sharp increase in the absorbance (600 nm) in the effluent within 15 to 20 min upon induction of dispersion, as determined using tube reactors (6, 9, 10, 23, 48, 51). We made use of the latter approach to ease the collection of dispersed cells. Biofilms by the P. aeruginosa mutant (endA::IS, eddA::IS, and eddB::IS) strains were first grown for 5 days under flowing conditions in a tube reactor and then exposed to two different dispersion cues, glutamate or NO, to induce dispersion. While exposure of wild-type PAO1 biofilms to the dispersion cue glutamate coincided with increased turbidity of the medium effluent of biofilm tube reactors, no such increase was noted upon challenging biofilms by endA::IS and eddA::IS cells with glutamate (Fig. 2A). However, biofilms by eddB::IS cells dispersed in response to glutamate (Fig. 2A). Likewise, challenging biofilms by endA::IS and eddB::IS cells with NO failed to induce a dispersion response (Fig. 2B). In contrast, biofilms by eddA::IS cells dispersed in response to NO in a manner similar to that of biofilms by wild-type P. aeruginosa PAO1 (Fig. 2B).
FIG 2.
Insertional inactivation of endA impairs the dispersion response by P. aeruginosa when exposed to glutamate or nitric oxide (NO), while insertional inactivation of eddA or eddB impairs dispersion in a manner dependent on the dispersion cue used. Biofilms were grown for 5 days in 5-fold diluted VBMM in tube reactors. Dispersion was induced by the addition of glutamate (A) or nitric oxide (NO) (B) to the growth medium. Sodium nitroprusside served as a source of NO. Absorbance of biofilm tube reactor effluents after induction of dispersion is shown. Dispersion assays were performed at least three times, with each biological replicate consisting of 4 technical replicates. Representative dispersion profiles are shown.
Given that eddA and eddB were previously reported to be part of a 2-gene operon and to be coregulated (43, 45), our finding of EddA only contributing to dispersion to glutamate while EddB only contributed to dispersion to NO was surprising. To exclude polar effects of the transposon inserts, we determined whether complementation restored the dispersion response. Interestingly, multicopy expression of eddB restored dispersion by the eddB::IS biofilm in response to NO, while multicopy expression of eddA restored dispersion by the eddA::IS biofilm in response to glutamate (see Fig. S1 in the supplemental material). The findings indicated that the difference in dispersion phenotypes was not due to polar effects and that EddA and EddB likely have to work in concert to induce dispersion to multiple dispersion cues. In contrast, EndA alone contributed to dispersion in response to both glutamate and NO.
endA encodes a secreted protein capable of degrading genomic DNA derived from P. aeruginosa.
Given that, of the three candidates, only EndA contributed to biofilm dispersion in response to two dispersion cues, we selected EndA for further investigation. As indicated above, endA likely encodes a probable DNA-specific endonuclease I. Moreover, the deduced amino acid sequence of EndA shares ∼50% identity to the periplasmic nucleases EndA from E. coli and Vvn from Vibrio vulnificus, as well as to the extracellular nucleases Dns from Vibrio cholerae and Aeromonas hydrophila, and EndA from Shewanella oneidensis MR-1 (52–56). In addition, Gnanadhas et al. (44) reported that exposure of P. aeruginosa biofilms to l-methionine results in increased DNase activity in culture supernatants, with l-Met treatment coinciding with upregulated expression of four DNase genes (sbcB, endA, eddB, and recJ). While the findings suggested EndA is an active nuclease and likely to be secreted, the DNase activity of EndA or its location was not confirmed in the study.
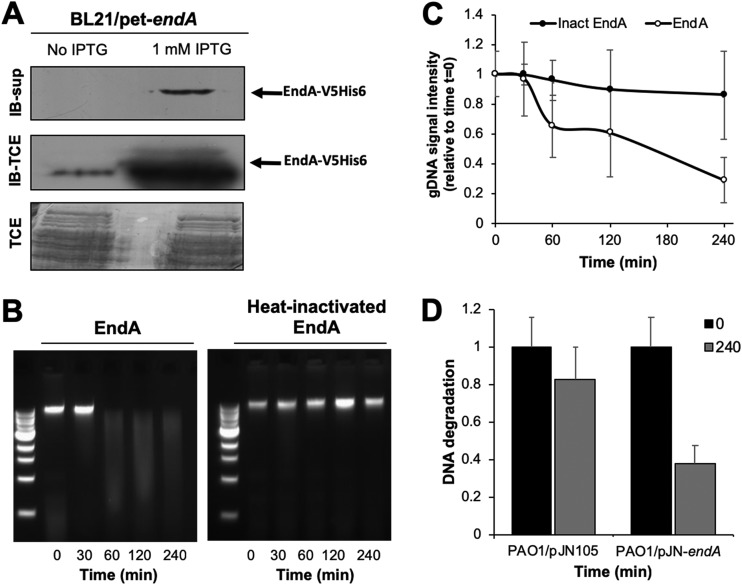
To determine whether EndA is secreted, we overproduced the protein in E. coli (BL21/pET-endA_V5/6×His) under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. The gene endA was modified to harbor C-terminal V5 and 6×His tags, resulting in a calculated molecular mass of EndA of 29.5 kDa. Uninduced cells were used as a control. Relative to uninduced cells, a protein having an apparent mass of ∼28 kDa was detected in total cell extracts after IPTG induction, as determined using immunoblot analysis and anti-V5 antibodies (Fig. 3A). More importantly, however, a protein of similar size was detected in culture supernatants of induced but not uninduced cells (Fig. 3A; see Fig. S2), suggesting that EndA is indeed secreted.
FIG 3.
EndA is an extracellular nuclease. (A) Immunoblot analysis of EndA produced by E. coli BL21/pet-endA_V5/His in the absence and presence of IPTG in culture supernatants (IB-sup) and total extracts (IB-TCE). A total of 20 μg of total cell extracts and 1.5 μg of BL21/pet-endA supernatants were loaded. Coomassie-stained SDS gels showing total cell extracts (TCE) after transfer were used as loading controls. (B) Representative agarose gel images demonstrating EndA-dependent degradation of genomic DNA over a period of 240 min. EndA purified from E. coli culture supernatants was used. (C) Quantitative analysis of the nuclease activity of purified EndA (EndA) and purified EndA that was inactivated by heat (Inact EndA). (D) Quantitative analysis of the nuclease activity of supernatants obtained from P. aeruginosa PAO1 overexpressing endA relative to that in PAO1 harboring the empty plasmid pJN105. Experiments were performed in triplicate. Error bars indicate standard deviations.
To characterize the activity of the V5/6×His-tagged EndA protein, we determined the level of DNA degradation using genomic DNA (gDNA) obtained from P. aeruginosa upon the addition of purified EndA. EndA was purified from culture supernatants to apparent homogeneity (Fig. S2). To this end, we measured gDNA degradation by visualizing the residual DNA by separation on agarose gels after incubation with EndA. As a negative control, we used heat-inactivated EndA. The purified enzyme was capable of degrading gDNA within less than 60 min (Fig. 3B and C). In contrast, heat-inactivated EndA was unable to do so (Fig. 3B and C). These assays demonstrated that gDNA was readily degraded by EndA, while no DNA degradation occurred when heat-inactivated EndA was added.
Dispersion coincides with EndA-dependent reduction in the eDNA content present in biofilms.
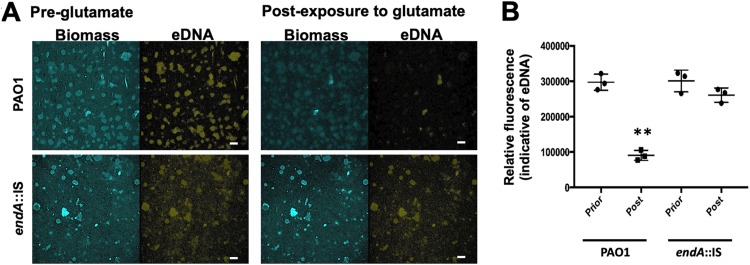
Similar to the E. coli-produced EndA, EndA produced by P. aeruginosa was found to be secreted (Fig. S2) and capable of degrading gDNA (Fig. 3D). Therefore, we next asked whether EndA is capable of degrading eDNA present in P. aeruginosa biofilms. To do so, we first determined whether we can visualize eDNA by confocal microscopy, by staining eDNA with propidium iodide as previously reported (57). We made use of strain PAO1 carrying plasmid pMRP9-1, in which the green fluorescent protein (GFP) is constitutively expressed, and established biofilms in 24-well plates under static conditions. Established biofilms were either left untreated or exposed to salmon sperm DNA and then subsequently stained with propidium iodide. While untreated biofilms were stained by propidium iodide throughout (see Fig. S3A), biofilms that were exposed to exogenously added DNA primarily stained at the periphery of biofilm microcolonies (Fig. S3B). COMSTAT analysis furthermore suggested that while addition of exogenous DNA had no effect on the biofilm biomass (visualized via GFP), the propidium iodide-stained eDNA significantly increased relative to that in untreated control biofilms (Fig. S3C and D). The same settings used for eDNA detection were used to visualize eDNA prior to and after induction of the dispersion response. To do so, biofilms by PAO1 and endA::IS cells were grown in flow cells for 5 days. Prior to the induction of dispersion by glutamate, biofilms were stained with propidium iodide and images were acquired (Fig. 4A, “pre-glutamate”). No significant difference in the biofilm architecture was noted, with biofilms by PAO1 and endA::IS cells characterized by the presence of microcolonies. Moreover, staining of eDNA by propidium iodide revealed no differences between biofilms formed by the wild type and the endA::IS mutant (Fig. 4A). The lack of a significant biofilm phenotype under hydrodynamic conditions upon insertional inactivation of endA strongly indicated that this nuclease has only a minor role, if any, in biofilm formation.
FIG 4.
EndA contributes to eDNA degradation in biofilm upon induction of dispersion. (A) Representative confocal images of 5-day-old biofilms by PAO1 and endA::IS strains prior to and after addition of the dispersion cue glutamate. eDNA was visualized using propidium iodide. Bars, 100 μm. (B) Quantitative analysis of the relative fluorescence associated with eDNA. Experiments were performed in triplicate, with each biological replicate consisting of 4 technical replicates. **, P < 0.05 relative to biofilms prior to exposure to glutamate (pre-glutamate).
Then, biofilms were exposed to the dispersion cue glutamate, and 30 min after exposure to glutamate, additional images of the same biofilm were acquired (Fig. 4A, “post-exposure to glutamate”). A comparison of the propidium iodide-stained eDNA detectable in biofilms prior to and after addition of glutamate suggests a reduction in eDNA in biofilms by P. aeruginosa PAO1. Quantitation of the fluorescence indicative of eDNA confirmed a significant reduction in eDNA after induction of dispersion in response to glutamate (Fig. 4B). In contrast, little to no difference in eDNA was noticed in endA::IS biofilms prior to and after addition of glutamate (Fig. 4A). The finding was confirmed by quantitative analysis of propidium iodide-based fluorescence (Fig. 4B). Overall, our findings suggested that while dispersion by the P. aeruginosa wild type results in a significant reduction in the overall detectable eDNA content, impaired dispersion coincides with a lack of eDNA modulation. The findings furthermore suggested that EndA likely contributes to the degradation of eDNA present in biofilms after induction under dispersion-inducing conditions.
Induction of endA coincides with a reduction of eDNA present in biofilms.
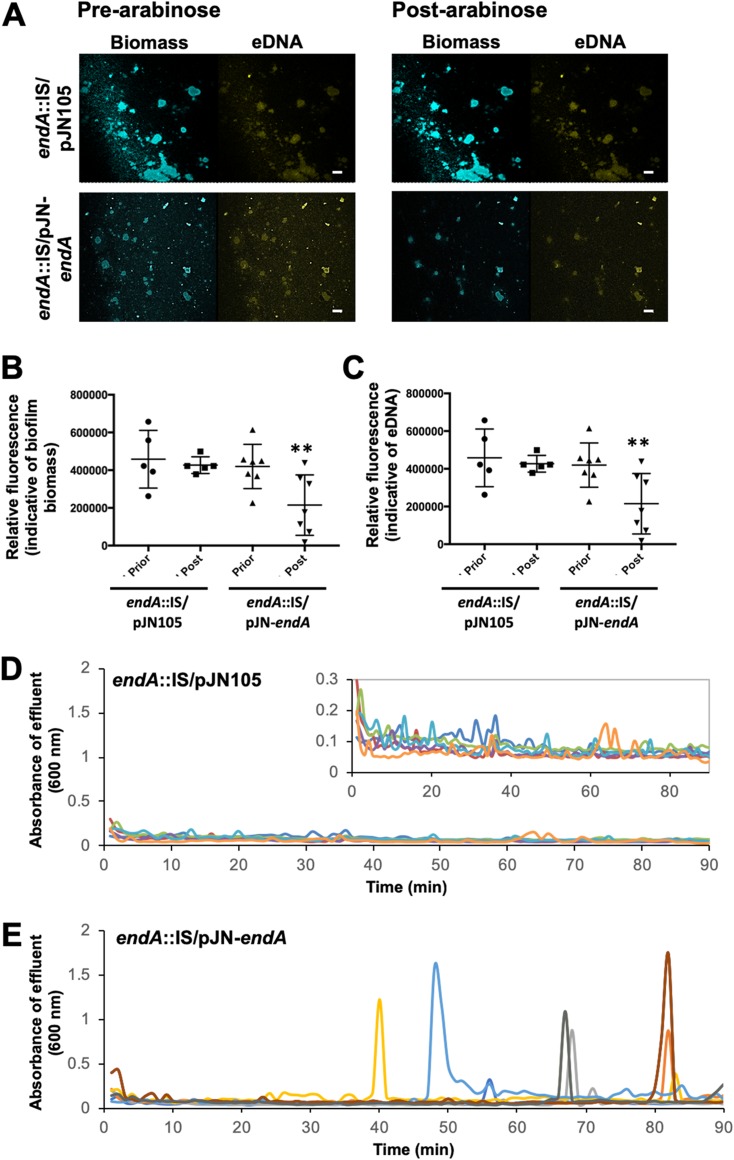
To further confirm a role of EndA in eDNA degradation, we next determined the fate of eDNA upon overexpression of endA. We therefore made use of a complemented endA::IS/pJN-endA strain which harbored endA under the control of the arabinose-inducible PBAD promoter. The endA::IS/pJN105 strain was used as a control. Biofilms by the mutant strains were grown for 5 days in the absence of arabinose to ensure the establishment of biofilms. Following 5 days of growth, arabinose was added to the growth medium for a period of 90 min to induce the transcription of endA. Prior to and following arabinose supplementation, the biofilm architecture and eDNA were monitored by confocal microscopy (Fig. 5A). Relative to uninduced endA::IS/pJN105 biofilms, exposure to arabinose appeared to have no effect on the propidium stainable eDNA content of endA::IS/pJN105 biofilms (Fig. 5A). Moreover, no apparent change in the biofilm biomass was noted (Fig. 5A) The findings were supported by the quantitative analysis of fluorescence indicative of the biofilm biomass (Fig. 5B) and eDNA (Fig. 5C) prior to and after addition of arabinose. In contrast, induction of gene expression, upon exposure to arabinose, resulted in a reduction of the biofilm biomass and detectable eDNA present in biofilms by the endA::IS/pJN-endA strain (Fig. 5A). Analysis of the detectable fluorescence obtained following image analysis confirmed the induction of endA gene expression results in a significant reduction of both the biofilm biomass (Fig. 5B) and eDNA (Fig. 5C) relative to that in untreated endA::IS/pJN-endA biofilms.
FIG 5.
Induction of endA gene expression coincides with eDNA degradation and dispersion by P. aeruginosa biofilms. (A) Representative confocal images of 5-day-old biofilms by endA::IS/pJN105 and endA::IS/pJN-endA strains prior to and after addition of 0.8% arabinose. eDNA was visualized using propidium iodide. Bars, 100 μm. (B) Quantitative analysis of the relative fluorescence associated with the biofilm biomass. (C) Quantitative analysis of the relative fluorescence associated with eDNA. Experiments were performed in triplicate, with each biological replicate consisting of 4 technical replicates. **, P < 0.05 versus uninduced (prior) biofilms. Dispersion upon induction of endA gene expression. Graphs shown are representative of three independent biofilm replicates. (D) Dispersion profiles of biofilms by P. aeruginosa endA::IS harboring the empty plasmid pJN105 upon exposure to arabinose. Inset, zoomed-in graph to show biological replicates. (E) Detection of dispersion following induction of endA gene expression, with gene expression induced upon addition of 0.8% arabinose to the growth medium. Different colors represent biological replicates.
Induction of endA results in biofilm dispersion.
Our findings suggested not only that EndA contributes to eDNA degradation but also that induction of endA gene expression coincides with a significant reduction in the biofilm biomass. As biofilm biomass loss is indicative of dispersion, we wanted to confirm whether induction of endA gene expression indeed results in biofilm dispersion. We therefore made use of tube-reactor-grown endA::IS/pJN-endA biofilms harboring endA under the control of the arabinose-inducible PBAD promoter. The endA::IS/pJN105 strain was used as a control. Mutant biofilms were allowed to form biofilms for 5 days in the absence of arabinose. Then, biofilms were exposed to arabinose to induce endA gene expression, and biofilm effluents were collected for a period of 90 min after addition of arabinose to the growth medium. Effluents by endA::IS/pJN105 biofilms appeared to have an average absorbance of ∼0.1 over the entire 90 min period (Fig. 5D). The addition of arabinose to the growth medium resulted in repeated dispersion events of various intensities by endA::IS/pJN-endA biofilms over the course of 90 min (Fig. 5E), a response that was absent in ΔendA::IS/pJN105 biofilms used as a control (Fig. 5D). Our findings strongly suggested that induction of endA gene expression results in dispersion. It is of interest to note that the dispersion events noted upon induction of endA gene expression were similar to those previously observed upon induction of bdlA_G31A gene expression by P. aeruginosa biofilms (48). Moreover, the findings are in agreement with the reduction of the biofilm biomass noted upon induction of endA gene expression in flow-cell-grown biofilms (Fig. 5A and B).
Cells generated upon induction of endA gene expression share characteristics with dispersed cells.
Chambers et al. (14) reported dispersed cells are more primed to reattach following egress from the biofilm and are more susceptible to tobramycin than biofilm cells. We therefore asked if dispersed cells obtained in response to endA gene expression adopt phenotypes generally associated with dispersed cells. As the induction of endA gene expression to induce dispersion resulted in cells dispersing at various time points (Fig. 5E) that makes it difficult to collect a sufficient number of dispersed cells, we instead made use of biofilms continuously overexpressing endA, to mimic hyperdispersive biofilms. The system is similar to the one making use of the overexpression of bdlA_G31A to generate hyperdispersive biofilms (Fig. 1A).
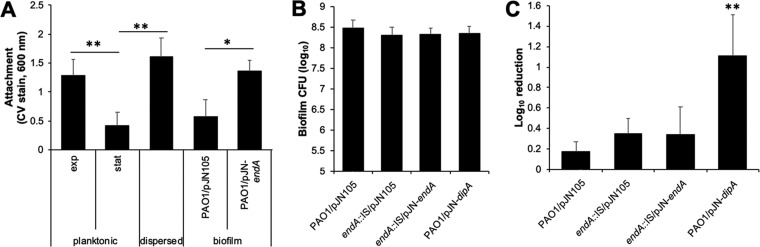
To assess attachment, biofilms (PAO1/pJN105 and PAO1/pJN-endA) were grown for 3 days in the presence of arabinose (0.1%) to ensure endA gene expression. Then, biofilms were harvested, homogenized to disrupt cell aggregates, diluted to an optical density at 600 nm (OD600) of 0.2, and used as inoculum in attachment assays. Attachment was assessed by crystal violet (CV) staining following 2 h of incubation. Planktonic cells grown to exponential and stationary phases, and dispersed cells obtained in response to glutamate, were used as controls. In agreement with previous findings (14), dispersed cells attached more efficiently than cells grown to stationary phase (Fig. 6A). Attachment by biofilm cells by PAO1/pJN105 was comparable to that noted for planktonic cells grown to stationary phase (Fig. 6A). In contrast, biofilm cells overexpressing endA (PAO1/pJN-endA) attached as efficiently as dispersed cells (Fig. 6A).
FIG 6.
Induction of endA gene expression results in cells that more readily attach but are as susceptible to tobramycin as biofilms. For all experiments, biofilms were grown under flowing conditions in tube reactors in the presence of 0.1% arabinose. Following 3 days of growth, biofilms were then harvested and the resulting cells were subjected to attachment and susceptibility assays. (A) Attachment assays were carried out in 96-well plates, with each well inoculated with 200 μl of bacterial culture adjusted to an OD of 0.2. The adhering biomass was determined 2 h postinoculation using crystal violet (CV) staining. Absorbance was determined at 600 nm. Experiments were carried out at least in triplicate, with each repeat comprising 12 technical replicates. Error bars denote standard deviations. *, significantly different from PAO1/pJN105 biofilms; **, significantly different from P. aeruginosa grown planktonically to stationary phase; P < 0.01 as determined by ANOVA and SigmaStat. exp, exponential phase; stat, stationary phase. (B) Average numbers of viable cells present in biofilms, tube reactor grown, as determined using CFU count prior to exposure to tobramycin. Biofilm CFU is expressed as log10. Error bars represent standard deviations. Experiments were performed in triplicate, with each repeat comprising a technical duplicate. (C) Susceptibility phenotype of indicated strains to tobramycin (50 μg/ml) was determined using viability plate assays, calculated as CFU counts. Susceptibility is expressed as log10 reduction in viability. Experiments were performed in triplicate, with each repeat comprising technical duplicates. Error bars represent standard deviations. **, P < 0.005 versus PAO1/pJN105 by ANOVA.
We furthermore assessed the susceptibility phenotype. Biofilms (PAO1/pJN105, PAO1/pJN-endA, and endA::IS/pJN105) were grown as described above, harvested, homogenized to disrupt cell aggregates, and diluted to an OD600 of 0.2. The obtained cells were subsequently exposed to tobramycin (50 μg/ml) for 1 h. Homogenized biofilm cells not treated with tobramycin were used as a control. Under the conditions tested, no significant difference in the biofilm biomass was noted (Fig. 6B), apparent by biofilms being composed on average of ∼2.5 × 108 CFU/biofilm. Moreover, no difference in susceptibility to tobramycin, as determined using log10 reduction, was noted (Fig. 6C). Using a similar approach, we furthermore determined the susceptibility of biofilms overexpressing dipA, encoding the phosphodiesterase DipA (PAO1/pJN-dipA). This strain was chosen as DipA was previously demonstrated to be required for dispersion (7) by contributing to the relay of dispersion cue sensing into the modulation of c-di-GMP levels (6). The exposure of biofilms by PAO1/pJN-dipA to tobramycin coincided with significantly increased susceptibility (Fig. 6C). Taken together, our findings suggested that induction of endA gene expression coincides with P. aeruginosa adopting some but not all phenotypes generally associated with dispersed cells.
DISCUSSION
The goal of this study was to identify specific matrix-degrading factors that contribute to dispersion. Specifically, we were interested in determining whether P. aeruginosa employs nucleases to weaken the biofilm matrix to ultimately disperse from the biofilm. But why focus on nucleases and why eDNA? eDNA has long been known as a nutrient and a source of phosphorus and nitrogen (43). In 2002, however, Whitchurch and coworkers demonstrated an additional function of eDNA, that of a structural element, a factor mediating cell-cell as well as cell-surface interactions in Pseudomonas aeruginosa biofilms (58). Since then, eDNA has been shown to be deposited on the stalk of biofilms, enabling the formation of mushroom-shaped microcolonies (59), to enhance biofilm formation by various bacterial species (60–63), likely by enabling direct or indirect interactions with the bacterial cell surface (64, 65), and to cross-link matrix components such as the Pel polysaccharide present in the P. aeruginosa biofilm matrix (57). eDNA serving a structural role has been confirmed for several biofilm-forming species, as DNase I treatment strongly decreased the ability of various bacteria to attach to a surface and the subsequent formation of 3-dimensional structures was severely negatively affected (39, 40, 49, 66). Given that eDNA is a structural element of the biofilm matrix, eDNA degradation has likewise been shown to weaken the biofilm matrix. In fact, several studies have reported that treatment with DNase I results in the release of large amounts of biomass in a large number of other biofilm-forming species, including P. putida, Staphylococcus aureus, S. oneidensis, and Bacillus licheniformis (40, 49, 52, 66–69). However, DNase treatment leading to the detachment of biofilms was found to be limited to young, but not mature, flow chamber-grown P. aeruginosa biofilms (58), probably due to mature biofilms harboring increasing amounts of matrix material other than extracellular DNA, and speculated to occur to improve nutrient supply.
Here, we identified a phosphatase, EddA, and two nucleases, EndA and EddB, as being induced upon induction of dispersion. Both EndA and EddB were found to be required for P. aeruginosa biofilm dispersion, with the role of EddB limited to dispersion in response to NO. The previously characterized phosphatase EddA likewise contributed to dispersion, but its role appeared to be limited to dispersion in response to glutamate. Our findings suggested the roles of EddA and EddB are complementary, with EddA and EddB likely working in concert to induce dispersion to multiple dispersion cues.
In contrast, EndA alone contributed to multiple dispersion cues. Further characterization indicated EndA is highly homologous to other bacterial nucleases, including EndA from E. coli, Vvn from Vibrio vulnificus, Dns from Vibrio cholerae, and EndA from S. oneidensis MR-1 (52–56). All of these enzymes are exported from the cytoplasm and either remain in the periplasm, as do Vvn and E. coli EndA, or are released into the medium, as are Dns and the S. oneidensis EndA (52–56). Our findings here strongly suggest that the P. aeruginosa EndA, like Dns and the S. oneidensis EndA, is an extracellular enzyme. Similar to other EndA orthologs, the P. aeruginosa EndA belongs to the family of ββα-metal endonucleases, also known as His-Me finger endonucleases, that form a large and diverse protein superfamily (70). The classification is based on a conserved structural motif, a β-hairpin followed by an α-helix, with a single metal ion being crucial for function (71). Based on sequence alignments (not shown), P. aeruginosa EndA harbors all the highly conserved amino acid residues that were previously identified as critical for activity, including the eight cysteine residues that form four disulfide bonds required for proper folding. The findings are in agreement with P. aeruginosa EndA having DNase activity.
Insertional inactivation of endA had no significant effect on biofilm development or eDNA accumulation in the matrix. Likewise, endA deletion had no effect on growth in liquid. This is in contrast to previously described nucleases belonging to the ββα-metal endonucleases that affected attachment and/or biofilm formation or displayed an aggregative phenotype in liquid. The difference in phenotypes indicated EndA by P. aeruginosa contributes to eDNA degradation in a manner independent of eDNA being a nutrient. Instead, our findings suggested EndA is involved in eDNA degradation to weaken the biofilm matrix and thus enables the escape from the biofilm matrix. This is supported by inactivation of endA resulting in impaired dispersion response, while induction of endA gene expression coincided with dispersion events. The noted dispersion events were similar to those previously observed upon induction of bdlA_G31A gene expression by P. aeruginosa biofilms (48). In addition, while dispersion by wild-type biofilms coincided with a reduction in the stainable eDNA content, endA::IS biofilms failed to demonstrate a reduction in eDNA. In contrast, induction of endA coincided with a significant reduction in eDNA present in biofilms. The findings furthermore demonstrated that EndA is sufficient to disperse established biofilms.
To our knowledge, this is the first report of DNase EndA being required for dispersion, with induction of endA gene expression resulting in biofilm dispersion and reduction in eDNA. Our findings raise several questions. First, if eDNA degradation results in weakening of the biofilm matrix to enable dispersion, is eDNA the only matrix component that is being degraded to enable dispersion? Second, are additional matrix components degraded? The biofilm matrix by the nonmucoid lab strain P. aeruginosa PAO1 is primarily composed of eDNA, protein, and the two polysaccharides Pel and Psl. However, four classes of nonmucoid P. aeruginosa matrix-producing strains have been identified, with the classification based on the type and amount of exopolysaccharides present in the biofilm matrix. Class I and II strains primarily utilize Pel and Psl, respectively, as the biofilm matrix polysaccharides, with PAO1 belonging to class II. Class III strains utilize both Pel and Psl, while class IV strains overproduce both polysaccharides (72). The different classes of matrix producers in P. aeruginosa alone suggest that if polysaccharide degradation is involved in the dispersion response, each matrix producer is expected to have a unique set of matrix-degrading enzymes. These questions are subject to future investigations. An additional line of inquiry will address the question of how endogenously produced DNases differ from exogenously added DNases when it comes to inducing dispersion. This is based on DNase treatment only leading to the detachment of young biofilms (58), while endA induction coincided with dispersion of established biofilms. One explanation could be that the access to eDNA becomes more limited to exogenously added DNases as the biofilm, and the extent of the biofilm matrix, matures.
We also asked how similar EndA induction-derived dispersed cells are to dispersed cells obtained in response to the dispersion cues glutamate or NO. In agreement with previous findings (14), EndA induction-derived cells were as hyperadhesive as dispersed cells and attached more readily than biofilm cells. In contrast, EndA induction-derived cells were as susceptible as biofilm cells. The findings suggested that while induction of endA coincided with dispersion, endA induction was not sufficient to mimic all phenotypes ascribed to dispersed cells. Recent findings suggested a link between c-di-GMP in drug susceptibility. Gupta et al. (73) demonstrated that P. aeruginosa planktonic cells were rendered more resistant to antimicrobial agents by increasing intracellular c-di-GMP levels to those more commonly found in biofilm cells, while biofilm cells were rendered susceptible upon decreasing c-di-GMP levels. It is thus likely that EndA-induced dispersion is not affecting c-di-GMP. Considering that dispersion has been reported to be the result of an overall reduction in the intracellular c-di-GMP content (6, 7, 19), our finding furthermore suggests endA induction is likely a response of c-di-GMP modulation. But how can this be? While direct evidence is lacking, indirect evidence suggests a role of AmrZ. Originally described to inversely regulate alginate production and swimming motility in P. aeruginosa, AmrZ is now recognized as a global regulator of multiple virulence factors, including c-di-GMP, extracellular polysaccharide production, including of Pel and Psl polysaccharides, and flagella (74). Support for AmrZ playing a role in dispersion stems from AmrZ affecting gcbA expression and inversely regulating exopolysaccharide production and motility (74). Additionally, Chua et al. (15) demonstrated that amrZ is differentially expressed in dispersed cells relative to that in planktonic cells by using whole-transcriptome shotgun sequencing (RNA-seq). However, to address this question, future experiments will be required to elucidate c-di-GMP-dependent pathways leading to dispersion and, more specifically, endA induction.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 2. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic cultures were grown in flasks at 220 rpm at 37°C using Vogel and Bonner citrate minimal medium (VBMM) (75). For plasmid maintenance, antibiotics were used at the following concentrations: 250 μg/ml carbenicillin and 50 μg/ml gentamicin for P. aeruginosa and 50 μg/ml ampicillin for E. coli. Unless indicated otherwise, arabinose was used at 1% to induce gene expression.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 tonA | Life Technologies |
| BL21 | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Life Technologies |
| P. aeruginosa | ||
| PAO1 | Wild-type strain PAO1 | B. H. Holloway |
| ΔbdlA strain | ΔbdlA in PAO1, Kmr | 9 |
| endA::IS strain | PAO1 PA2749::ISlacZ Tetr | 82 |
| eddA::IS strain | PAO1 PA3910::ISlacZ Tetr | 82 |
| eddB::IS strain | PAO1 PA3910::ISlacZ Tetr | 82 |
| Plasmids | ||
| pET101D | Vector for directional cloning and V5/6×His fusion protein expression, Ampr | Life Technologies |
| pET-endA | IPTG-inducible expression of C-terminal V5/6×His-tagged endA cloned into pET101D, Ampr | This study |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD Gmr | 83 |
| pJN-bdlA-G31A | Arabinose-inducible expression of C-terminal 6×His-tagged bdlA with G31A mutation cloned into pJN105, Gmr | 48 |
| pJN-dipA_V5 | Arabinose-inducible expression of C-terminal V5/6×His-tagged dipA (PA5017) cloned in pJN105, Gmr | 7 |
| pJN-endA_V5 | Arabinose-inducible expression of C-terminal V5/6×His-tagged endA (PA2749) cloned in pJN105, Gmr | This study |
| pMJT1 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD Gmr | 84 |
| pMJT-eddA_V5 | C-terminal V5-tagged PA3910 cloned into pMJT-1 XbaI and SacI; araC-PBAD Carbr | This study |
| pMJT-eddB_V5 | C-terminal V5-tagged PA3909 cloned into pMJT-1 XbaI and SacI; araC-PBAD Carbr | This study |
| pMRP9-1 | Enables continuous expression of gfp, Carbr | 85 |
Strain construction.
C-terminal V5/6×His tagging of EndA was accomplished by subcloning into pET101D (Life Technologies, Carlsbad, CA). The tagged constructs were then cloned into pJN105. The identity of all vector inserts was confirmed by PCR and sequencing. Plasmids were introduced into P. aeruginosa via conjugation. Additionally, transposon insertional inactivation of endA and eddA was confirmed by PCR and sequencing. Primers used for strain construction and confirmation are listed in Table 3.
TABLE 3.
Primers used
| Oligonucleotide | Sequence |
|---|---|
| qRT-PCR | |
| endA_FqPCR | GCTTTCCCGTTTGTTTGT |
| endA_RqPCR | TAGAGCTTCCAGCCGATT |
| cysD_FOR qPCR | CTGGACATCTGGCAATACAT |
| cysD_REV qPCR | TCTCTTCGTCAGAGAGATGC |
| eddA_FORqPCR | CCGACCAGTCGATCTTCTA |
| eddA_REVqPCR | TCCAGACGAAACGGATATT |
| Transposon insertional inactivation check primers | |
| endA_FOR | TTTCCCGTTTGTTTGTAGGC |
| endA_REV | CAGGGTATGTCCGCAGGT |
| eddA_FOR | ATGAGTGGGATGGACCTCAAGCGCCGC |
| eddA_REV | TCAGGCGCCGTCGGGCTG |
| eddB_FOR | AAGACCTTCGTCCTCGCCAAC |
| eddB_REV | ATAGATCAGCCCCACGGCAAT |
| Cloninga | |
| pJN105 MCS_FOR | TAGCGGATCCTACCTGACGC |
| pJN105 MCS_REV | CCATTCGCCATTCAGGCTG |
| endA_FpET | CACCCGCATGCTTTCCCGTTTGTTT |
| endA_RpET | GCGTCGACGCGAGGATAG |
| endA_FpETNheI | gctacgCGCATGCTTTCCCGTTTGTTT |
| pETHis_XbaI_rev | GCGCGCtctagaTCAATGGTGATGGTGATG |
| eddB_XbaI_FOR | GCGCGCGCtctagaATGCACCCCTTGCGTAACGCC |
| eddB_SacI-V5_REV | GCGCGCGCgagctcTCACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCCCTGCGGTGCTTCTT CATCGC |
| eddA_XbaI_FOR_FOR | GCGCGCGCtctagaATGAGTGGGATGGACCTCAAG |
| eddA_SacI-V5_REV | GCGCGCGCgagctcTCACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCGGCGCCGTCGGGCTG |
Restriction sites are in lowercase letters. The sequence of the V5 tag is underlined.
Biofilm growth.
Biofilms were grown for 5 days under continuous flow conditions in biofilm tube reactors or flow cells. The flow rate was 0.2 ml/min using 5-fold diluted VBMM medium. For plasmid maintenance, 10 μg/ml carbenicillin and 2 μg/ml gentamicin were added. Where indicated, the growth medium was supplemented with 0.1% arabinose to induce expression of genes of interest. Flow-cell-grown biofilms were stained using propidium iodide or a Live/Dead BacLight viability stain kit (Invitrogen, Carlsbad, CA) to visualize eDNA. The biofilm architecture was visualized via confocal laser scanning microscopy (CLSM) using a Leica TCS SP5 confocal microscope. The CLSM images were processed using LAS AF software. Quantitative analysis of biofilm architecture was accomplished using MATLAB with the COMSTAT software package (76).
RNA extraction and quantitative reverse transcriptase PCR.
To obtain RNA from dispersing and nondispersing biofilms, ΔbdlA and PAO1/bdlA_G31A mutant biofilms were grown in biofilm tube reactors in 5-fold diluted VBMM medium supplemented with 0.1% arabinose to induce bdlA_G31A gene expression. Following 5 days of growth, biofilm cells were collected directly into equal volumes of RNA Protect (Qiagen, Hilden, Germany). Isolation of mRNA and cDNA synthesis were carried out as previously described (77–79). qRT-PCR was performed using the Bio-Rad CFX Connect Real-Time PCR detection system and SsoAdvanced SYBR green supermix (Bio-Rad, Hercules, CA) with oligonucleotides listed in Table 3. cysD was used as a control. Relative transcript quantitation was accomplished using the CFX Manager software (Bio-Rad, Hercules, CA), by first normalizing transcript abundance (based on the threshold cycle [CT] value) to cysD followed by determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Biofilm dispersion.
Dispersion assays were performed using biofilms grown for 5 days in flow cells or tube reactors. Image acquisition using flow-cell-grown biofilms was performed so that the same biofilm microcolonies were observed prior to and after addition of dispersion cues or 0.8% arabinose. For tube-reactor-grown biofilms, dispersion was induced by the sudden addition of l-glutamate (18 mM) or (500 μM) sodium nitroprusside to the growth medium, as previously described (9, 50). Sodium nitroprusside was used as a source of NO. In addition, biofilms were exposed to 0.8% arabinose to induce endA gene expression to determine whether induction of gene expression resulted in dispersion events. Regardless of the dispersion cue used, dispersed cells were collected from the tube reactor effluents at 1-min intervals for a total of 24 min or 90 min, using 96-well microtiter plates. The absorbance of the biofilm effluents was assessed by spectrophotometry at 600 nm. Dispersion events were characterized by an increase in the effluent optical density (OD), with the OD being at least two times greater than the baseline.
Fluorescence analysis of the biofilm biomass and eDNA prior to and after induction of dispersion.
To quantify the amount of biomass and eDNA of biofilms prior to and after dispersion, the relative fluorescence intensities of confocal images that were acquired prior to and after addition of dispersion cues or arabinose were determined. Each image was analyzed for the relative fluorescence intensity indicative of biofilm biomass (green channel) and eDNA (red channel) using the Intensity Luminance V1 software (80).
Immunoblot analysis.
Confirmation of V5/6×His-tagged EndA production was assessed by SDS-PAGE and immunoblot analysis. E. coli BL21 (BL21/pET-endA_V5/6×His) was grown planktonically in LB medium containing ampicillin for plasmid maintenance to exponential phase. Then, endA gene expression was induced by IPTG for 2 h. Uninduced cells were used as controls. Cells were subsequently harvested by centrifugation for 5 min at 16,000 × g, resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 0.3 mg phenylmethanesulfonyl fluoride, sonicated on ice with six 10-s bursts at 4 W, and then centrifuged for 5 min at 21,200 × g to pellet cell debris and unbroken cells. Furthermore, culture supernatants were collected. The protein concentrations were determined using a modified Lowry assay (Thermo Scientific, Waltham, MA) and bovine serum albumin as a standard. The samples (1.5 μg) were resolved on an 11% polyacrylamide gel and subsequently transferred onto polyvinylidene difluoride (PVDF) membrane using a Turbo Trans-Blot apparatus (Bio-Rad, Hercules, CA). Western blots were first probed with anti-V5 antibody followed by a secondary anti-mouse IgG antibody (Cell Signaling Technology, Danvers, MA). The blots were subsequently developed using Immun-Star WesternC chemiluminescence reagents (Bio-Rad, Hercules, CA). Following transfer, SDS-PAGE gels were Coomassie stained to ensure equal loading. Likewise, the presence of V5/6×His-tagged EndA production in culture supernatants of PAO1/pJN-endA_V5/6×His, grown planktonically in LB medium containing gentamicin for plasmid maintenance to exponential phase, was assessed as described above. Culture supernatants by strain PAO1/pJN105 harboring the empty pJN105 vector were used as a control.
Purification of 6×His-tagged proteins.
EndA V5/6×His-tagged protein was purified from culture supernatants. First, culture supernatants were concentrated using Vivaspin 2 columns (Sartorius Stedim Biotech, Göttingen, Germany) according to the manufacturer’s protocol, and the concentrated supernatants were loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) affinity resin (Qiagen, Hilden, Germany). After washing to remove unbound protein, the resin was then washed with buffer, and EndA was eluted using an imidazole gradient according to the manufacturer’s instructions for native protein purification. Fractions containing EndA were confirmed by SDS-PAGE. EndA-containing fractions were pooled and then desalted and concentrated using Vivaspin 2 columns.
DNA degradation assays.
The DNA degradation assays were adapted from those described by Heun et al. (52) in order to confirm the nuclease activity of EndA. Briefly, purified EndA, obtained from E. coli culture supernatants, at a concentration of 1.5 μg was used and incubated with 250 ng of P. aeruginosa gDNA in 10 mM Tris-HCl (pH 7.6) buffer supplemented with dithiothreitol (DTT) (10 mM). Samples were incubated at 37°C, and aliquots (20 μl) were removed at 0, 30, 60, 120, and 240 min. Samples were stored at −20°C until analyzed by DNA gel electrophoresis using a 1% agarose gel. The integrity and quantity of the gDNA were analyzed by ImageJ (81). We likewise carried out DNA degradation assays using culture supernatants (1.5 μg) obtained from P. aeruginosa PAO1/pJN-endA. Culture supernatants by strain PAO1/pJN105 harboring an empty vector were used as a control.
Assessment of attachment.
Initial attachment to a polystyrene surface was measured using the polystyrene microtiter dish assay system (96-well) as previously described (14) with the following modifications. Biofilms by endA::IS and PAO1 strains harboring empty vectors or overexpressing endA or dipA were grown for 3 days in 5-fold-diluted VBMM in the presence of 0.1% arabinose to induce gene expression. This was performed to obtain populations with homogenous phenotypes. Biofilms were harvested and homogenized using a Tissue-Tearor. Each well was inoculated with 200 μl of biofilms, adjusted to an optical density at 600 nm of ∼0.2. In addition, OD-adjusted planktonic cells grown to exponential and stationary phases, as well as dispersed cells obtained in response to glutamate, were used. The 96-well plates were then incubated for 2 h at 37°C with shaking at 220 rpm to ensure proper aeration. All experiments were carried out at least in triplicate, with each repeat comprising six technical replicates.
Biofilm antibiotic susceptibility testing.
To determine whether induction of endA gene expression mimicking dispersion events rendered biofilm cells more susceptible to antimicrobial agents, biofilms were grown for 3 days with arabinose (0.1%). Then, biofilms were harvested, homogenized using a Tissue-Tearor, and centrifuged at 16,000 × g for 1 min at 22°C, and cell pellets were resuspended in fresh VBMM to an OD600 of 0.2. The resulting suspension was exposed tobramycin (50 μg/ml) for 1 h at 37°C. The suspension was subsequently serially diluted and plated on LB agar. Viability was determined via CFU counts, and susceptibility was expressed as log reduction.
Statistical analysis.
All experiments were carried out at least in triplicate. Student’s t test was performed for pairwise comparisons of groups, and multivariate analyses were performed with a one-way analysis of variance (ANOVA), followed by a Tukey’s test to compare the means of all treatment groups. All statistical analyses were performed using the Prism 5 software (GraphPad Software, La Jolla, CA).
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00059-19.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Davies DG. 1999. Regulation of matrix polymer in biofilm formation and dispersion, p 93–112. In Wingender J, Neu TR, Flemming H-C (ed), Microbial extrapolymeric substances, characterization, structure and function. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies DG, Marques C. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrova OE, Sauer K. 2016. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr Opin Microbiol 30:67–78. doi: 10.1016/j.mib.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basu Roy A, Sauer K. 2014. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol 94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu Roy A, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 9.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies DG. 2011. Biofilm dispersion, p 1–28. Biofilm highlights. Springer, Berlin, Germany. [Google Scholar]
- 13.Marques CN, Davies DG, Sauer K. 2015. Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals (Basel) 8:816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambers JR, Cherny KE, Sauer K. 2017. Susceptibility of Pseudomonas aeruginosa dispersed cells to antimicrobial agents is dependent on the dispersion cue and class of the antimicrobial agent used. Antimicrob Agents Chemother 61:e00846-17. doi: 10.1128/AAC.00846-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyle. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, Hassett DJ, Davies DG, Sauer K. 2014. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog 10:e1004168. doi: 10.1371/journal.ppat.1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming D, Rumbaugh K. 2018. The consequences of biofilm dispersal on the host. Sci Rep 8:10738. doi: 10.1038/s41598-018-29121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barraud N, Kelso MJ, Rice SA, Kjelleberg S. 2015. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr pharm Des 21:31–42. doi: 10.2174/1381612820666140905112822. [DOI] [PubMed] [Google Scholar]
- 19.Chua SL, Hultqvist LD, Yuan M, Rybtke M, Nielsen TE, Givskov M, Tolker-Nielsen T, Yang L. 2015. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat Protoc 10:1165–1180. doi: 10.1038/nprot.2015.067. [DOI] [PubMed] [Google Scholar]
- 20.Chua SL, Tan S-Y, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. 2013. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:2066–2075. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 23.Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 25.Christensen LD, van Gennip M, Rybtke MT, Wu H, Chiang W-C, Alhede M, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-GMP level in the bacteria. Infect Immun 81:2705–2713. doi: 10.1128/IAI.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 28.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a c-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 189:8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol 188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 32.Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol 54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- 33.Poudyal B, Sauer K. 2018. The PA3177 gene encodes an active diguanylate cyclase that contributes to the biofilm antimicrobial tolerance but not biofilm formation by P. aeruginosa. Antimicrob Agents Chemother 62:e01049-18. doi: 10.1128/AAC.01049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical Implications, and potential therapeutic uses. J Dent Res 89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J Bacteriol 185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S, Su T, Wu H, Liu S, Wang D, Zhao T, Jin Z, Du W, Zhu M-J, Chua SL, Yang L, Zhu D, Gu L, Ma LZ. 2015. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res 25:1352–1367. doi: 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleming D, Rumbaugh K. 2017. Approaches to dispersing medical biofilms. Microorganisms 5:E15. doi: 10.3390/microorganisms5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flemming H-C. 2016. EPS-then and now. Microorganisms 4:E41. doi: 10.3390/microorganisms4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 41.Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman F. 2009. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res 37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewenza S, Gardy JL, Brinkman FSL, Hancock R. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res 15:321–329. doi: 10.1101/gr.3257305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulcahy H, Charron-Mazenod L, Lewenza S. 2010. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol 12:1621–1629. doi: 10.1111/j.1462-2920.2010.02208.x. [DOI] [PubMed] [Google Scholar]
- 44.Gnanadhas DP, Elango M, Datey A, Chakravortty D. 2015. Chronic lung infection by Pseudomonas aeruginosa biofilm is cured by l-methionine in combination with antibiotic therapy. Sci Rep 5:16043. doi: 10.1038/srep16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilton M, Halverson TWR, Charron-Mazenod L, Parkins MD, Lewenza S. 2018. Secreted phosphatase and deoxyribonuclease are required by Pseudomonas aeruginosa to defend against neutrophil extracellular traps. Infect Immun 86:e00403-18. doi: 10.1128/IAI.00403-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewenza S. 2013. Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front Microbiol 4:21. doi: 10.3389/fmicb.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez PF, Ingram L. 1995. Cloning, sequencing and characterization of the alkaline phosphatase gene (phoD) from Zymomonas mobilis. FEMS Microbiol Lett 125:237–245. doi: 10.1111/j.1574-6968.1995.tb07364.x. [DOI] [PubMed] [Google Scholar]
- 48.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gödeke J, Heun M, Bubendorfer S, Paul K, Thormann KM. 2011. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Appl Environ Microbiol 77:5342–5351. doi: 10.1128/AEM.00643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heun M, Binnenkade L, Kreienbaum M, Thormann KM. 2012. Functional specificity of extracellular nucleases of Shewanella oneidensis MR-1. Appl Environ Microbiol 78:4400–4411. doi: 10.1128/AEM.07895-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang MC, Chang SY, Chen SL, Chuang SM. 1992. Cloning and expression in Escherichia coli of the gene encoding an extracellular deoxyribonuclease (DNase) from Aeromonas hydrophila. Gene 122:175–180. doi: 10.1016/0378-1119(92)90046-R. [DOI] [PubMed] [Google Scholar]
- 54.Focareta T, Manning PA. 1987. Extracellular proteins of Vibrio cholerae: molecular cloning, nucleotide sequence and characterization of the deoxyribonuclease (DNase) together with its periplasmic localization in Escherichia coli K-12. Gene 53:31–40. doi: 10.1016/0378-1119(87)90090-4. [DOI] [PubMed] [Google Scholar]
- 55.Jekel M, Wackernagel W. 1995. The periplasmic endonuclease I of Escherichia coli has amino-acid sequence homology to the extracellular DNases of Vibrio cholerae and Aeromonas hydrophila. Gene 154:55–59. doi: 10.1016/0378-1119(94)00835-G. [DOI] [PubMed] [Google Scholar]
- 56.Li CL, Hor LI, Chang ZF, Tsai LC, Yang WZ, Yuan HS. 2003. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J 22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 59.Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 60.Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol 72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2011. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol 81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinberger RE, Holden PA. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol 71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol 76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vilain S, Pretorius JM, Theron J, Brözel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75:2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nijland R, Hall MJ, Burgess JG. 2010. Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS One 5:e15668. doi: 10.1371/journal.pone.0015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boles BR, Horswill AR. 2011. Staphylococcal biofilm disassembly. Trends Microbiol 19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 69.Dominiak DM, Nielsen JL, Nielsen PH. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol 13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 70.Jablonska J, Matelska D, Steczkiewicz K, Ginalski K. 2017. Systematic classification of the His-Me finger superfamily. Nucleic Acids Res 45:11479–11494. doi: 10.1093/nar/gkx924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W. 2011. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys 44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. 2012. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. 2014. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol 92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schweizer HP. 1991. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol 173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 77.Allegrucci M, Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol 189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allegrucci M, Sauer K. 2008. Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol 190:6330–6339. doi: 10.1128/JB.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marques CNH, Craver SA. 2015. Quantification of respiratory activity in biofilms. Bio Protoc 5:e1591. [Google Scholar]
- 81.Rasband W. 1997. ImageJ. U.S. National Institutes of Health, Bethesda, MD. [Google Scholar]
- 82.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 84.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest 117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.