Amyloid proteins are the major proteinaceous components of biofilms, which are associated with up to 65% of human bacterial infections. Amyloids produced by human cells are also associated with diseases such as Alzheimer’s. The amyloid monomeric subunits self-associate to form oligomers, protofibrils, and finally mature fibrils. Amyloid β oligomers are more cytotoxic to cells than the mature amyloid fibrils. Here we detected oligomeric intermediates of curli for the first time. Like the oligomers of amyloid β, intermediate curli fibrils were more cytotoxic than the mature curli fibrillar aggregates when incubated with bone marrow-derived macrophages. The discovery of cytotoxic curli intermediates will enable research into the roles of amyloid intermediates in the pathogenesis of Salmonella and other bacteria that cause enteric infections.
KEYWORDS: Escherichia coli, Salmonella, amyloid, biofilm, curli
ABSTRACT
Enterobacteriaceae produce amyloid proteins called curli that are the major proteinaceous component of biofilms. Amyloids are also produced by humans and are associated with diseases such as Alzheimer’s. During the multistep process of amyloid formation, monomeric subunits form oligomers, protofibrils, and finally mature fibrils. Amyloid β oligomers are more cytotoxic to cells than the mature amyloid fibrils. Oligomeric intermediates of curli had not been previously detected. We determined that turbulence inhibited biofilm formation and that, intriguingly, curli aggregates purified from cultures grown under high-turbulence conditions were structurally smaller and contained less DNA than curli preparations from cultures grown with less turbulence. Using flow cytometry analysis, we demonstrated that CsgA was expressed in cultures exposed to higher turbulence but that these cultures had lower levels of cell death than less-turbulent cultures. Our data suggest that the DNA released during cell death drives the formation of larger fibrillar structures. Consistent with this idea, addition of exogenous genomic DNA increased the size of the curli intermediates and led to binding to thioflavin T at levels observed with mature aggregates. Similar to the intermediate oligomers of amyloid β, intermediate curli aggregates were more cytotoxic than the mature curli fibrils when incubated with bone marrow-derived macrophages. The discovery of cytotoxic curli intermediates will enable research into the roles of amyloid intermediates in the pathogenesis of Salmonella and other bacteria that cause enteric infections.
IMPORTANCE Amyloid proteins are the major proteinaceous components of biofilms, which are associated with up to 65% of human bacterial infections. Amyloids produced by human cells are also associated with diseases such as Alzheimer’s. The amyloid monomeric subunits self-associate to form oligomers, protofibrils, and finally mature fibrils. Amyloid β oligomers are more cytotoxic to cells than the mature amyloid fibrils. Here we detected oligomeric intermediates of curli for the first time. Like the oligomers of amyloid β, intermediate curli fibrils were more cytotoxic than the mature curli fibrillar aggregates when incubated with bone marrow-derived macrophages. The discovery of cytotoxic curli intermediates will enable research into the roles of amyloid intermediates in the pathogenesis of Salmonella and other bacteria that cause enteric infections.
INTRODUCTION
Bacteria thrive in numerous, vastly different environments. Biofilms, which protect bacteria from environmental insults, immune defense mechanisms, and antimicrobial agents, are associated with up to 65% of human infections (1). The extracellular matrix (ECM) of a biofilm is composed of proteins, DNA, and polysaccharides (2, 3) and is primarily responsible for the recalcitrance of bacterial infections to antimicrobial substances and the immune system (4).
Amyloid proteins are characterized by their conserved cross-β-sheet quaternary structure (5). It is estimated that amyloids decorate the biofilms of more than 40% of bacteria (6). Amyloid curli, the best-characterized bacterial amyloids, are produced by members of the Enterobacteriaceae family. Curli are encoded by the csgBAC and csgDEFG operons and assembled via a nucleation precipitation pathway encoded by a type VIII secretion system (7, 8). The csgA gene encodes the major subunit of the fibril, CsgA, and the csgB gene encodes a minor subunit, CsgB, a nucleator protein (9–12). Together these proteins transport and assemble curli fibrils outside of the bacterial cell. Curli fibrils are critical for the structural integrity and stability of the biofilm. Previous work from our lab has described the inclusion of extracellular DNA (eDNA) into curli fibrils from Salmonella enterica serotype Typhimurium and Escherichia coli during biofilm formation. DNA incorporation increases the amyloid fibrillization rate of synthetic curli monomers (13). Like curli, the bacterial amyloids produced by Staphylococcus aureus, phenol-soluble modulins (PSMs), also associate with eDNA (14, 15).
Amyloids not only are found in bacteria but also have been observed in humans. More than 60 amyloidogenic proteins are expressed under homeostatic conditions, but their physiological roles remain unclear. These amyloids can accumulate and form deposits that are associated with diseases such as Alzheimer’s, type II diabetes, Huntington’s, and secondary amyloidosis (16–18). In each of these diseases, a unique amyloid accumulates in an organ associated with the disease. For instance in Alzheimer’s disease, amyloid β accumulates in the brain, forming plaques. The formation of fibrillar amyloid β has been extensively studied, and intermediate forms of amyloid β such as oligomers and protofibrils have been described. Briefly, the monomeric amyloid β first forms oligomeric units that have been shown to be cytotoxic to immune cells (19–21). Oligomeric units then form protofibrils, which then form mature fibrillar amyloid β. Once the amyloid β fibrils have fully formed, they are less cytotoxic. Similar to bacterial amyloid curli and PSMs, amyloid β also binds to DNA (22). Although bacterial and human amyloids do not have homology in their amino acid sequences, amyloids are characterized by their conserved β-sheet fibrillar structure and common functional properties (2, 7, 9, 23, 24). This conserved structure is recognized by the innate immune receptors such as the Toll-like receptor 2 (TLR2)/TLR1 heterocomplex and Nod-like receptor protein 3 (NLRP3) inflammasome (25, 26). It is thought that bacteria produce amyloids as part of their biofilm, introducing a common molecular pattern for host recognition (23). Intriguingly, recent studies suggested a connection between bacterial amyloid curli and two complex human diseases, systemic lupus erythematosus and Parkinson’s disease (13, 27). However, the exact mechanism by which curli contribute to these human diseases in vivo remains unclear.
Curli assembly involves fibrillization, which has been monitored using approaches similar to those used to characterize fibrillization of amyloid β (e.g., binding to thioflavin T [ThT]), but structural intermediates in the curli fibrillization process had not been identified (28, 29). We found that we could interfere with the formation of mature curli fibrils by increasing turbulence during the biofilm formation in liquid culture. For the first time, we were able to purify cytotoxic intermediate forms of curli that resemble protofibrillar structures formed by amyloid β.
RESULTS
High turbulence leads to decreased biofilm formation.
In the laboratory setting, curli expression is induced in S. Typhimurium cells grown under stress such as low temperature or low osmolarity (30–32). If these conditions are applied to liquid static cultures, biofilm formation is observed at the air-liquid interface. Recently, it was also reported that the addition of dimethyl sulfoxide (DMSO) triggers curli expression and leads to the formation of biofilms even in shaken cultures (33). We use DMSO to trigger curli expression in order to purify curli from S. Typhimurium. Briefly, cultures of S. Typhimurium are grown in yeast extract supplemented with Casamino Acids (YESCA) broth containing 4% DMSO. Cultures are shaken at 200 rpm for 72 h at 26°C. At this time point, biofilm (also termed the pellicle) is observed forming at the air-liquid-flask interface. Eight 150-ml cultures are pooled, and curli are purified as described previously (34). In an attempt to increase production, we increased the culture volume from 150 ml to 500 ml and used a 1-liter flask. Although the starting inoculum ratio and all other variables were the same, we did not observe the production of a robust pellicle biofilm at the air-liquid interface in the 1-liter flask. In addition, we observed a clear difference in turbulence in the 150-ml and 500-ml culture volumes. The larger volume culture was more turbulent than the smaller volume culture, and foamy bubbles were observed in the 500-ml culture (see Movie S1 in the supplemental material). Finally, by decreasing the shaking speed of the 500-ml culture in the 1-liter flask, we were able to visibly decrease the turbulence (see Movie S2 in the supplemental material) and restore the biofilm formation (see Fig. S1 in the supplemental material). Additionally, we examined the effect of different speeds that influence the turbulence seen in the batch culture flasks. In the 1-liter flask, decreasing turbulence was able to increase the biofilm pellicle formed on the air-liquid-flask interface. In contrast, increasing turbulence in the 150-ml flask disrupted pellicle formation, causing a decrease in pellicle size (Fig. S1).
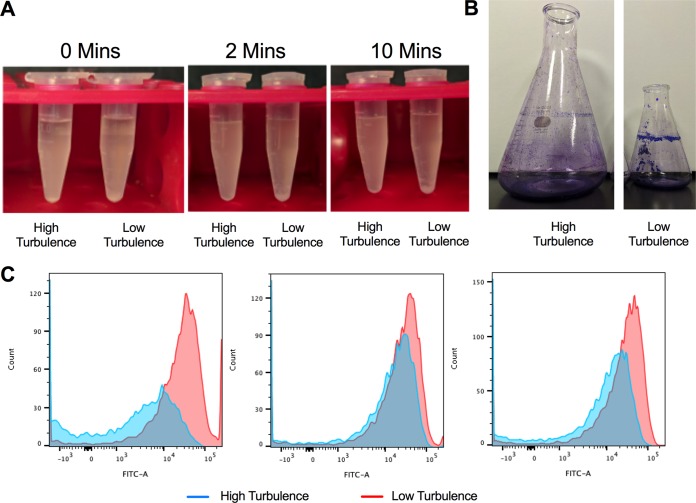
We also observed that cultures with higher turbulence had less visible aggregation of bacteria. Visible aggregation and sedimentation were observed after only 2 min in an aliquot taken from the 150-ml culture (at 72 h of incubation), whereas there was no visible aggregation or sedimentation in the sample taken from the 500-ml culture after 10 min (Fig. 1A) or 72 h (see Fig. S2 in the supplemental material). In the 150-ml culture, the biofilm pellicle was clearly visible and stained brightly with crystal violet (Fig. 1B). In the 500-ml culture, which had higher turbulence, the pellicle stained poorly with crystal violet (Fig. 1B). This difference in biofilm was not due to bacterial growth rate, as shown by optical density determination, but potentially was due to a difference in the ratios of planktonic and biofilm-forming bacteria (Fig. S2).
FIG 1.
Higher turbulence leads to decreased biofilm formation. (A) Sedimentation assay of aliquots of cultures from high-turbulence (500-ml) and low-turbulence (150-ml) cultures allowed to sediment for 0 min, 2 min, and 10 min. (B) Crystal violet staining of the pellicle-associated biofilm from high-turbulence and low-turbulence growth conditions. (C) Flow cytometry histograms of curli induction as measured by GFP expressed from a reporter plasmid conjugated to the csgBA promoter at 24 h (left), 48 h (middle), and 72 h (right) under high-turbulence and low-turbulence growth conditions.
As the major proteinaceous component of the enteric bacterial biofilm, curli fibrils are important for cell-to-cell interactions and surface attachment (35). As the biofilm pellicle and aggregation differed depending on the turbulence of culture, we first investigated whether curli expression differed under the two conditions. To evaluate curli expression, we used a reporter strain of S. Typhimurium that contains the PcsgBA::gfp reporter plasmid. When the csgBA gene is expressed, green fluorescent protein (GFP) is produced and can be detected by flow cytometry. GFP expression was detected in both cultures, although in the 500-ml high-turbulence cultures, expression was delayed and decreased relative to that in the 150-ml low-turbulence cultures (Fig. 1C). These data suggest that although the curli structural components are expressed, pellicle formation is not optimal under high turbulence.
Intermediate forms of curli aggregates are detectable under high-turbulence conditions.
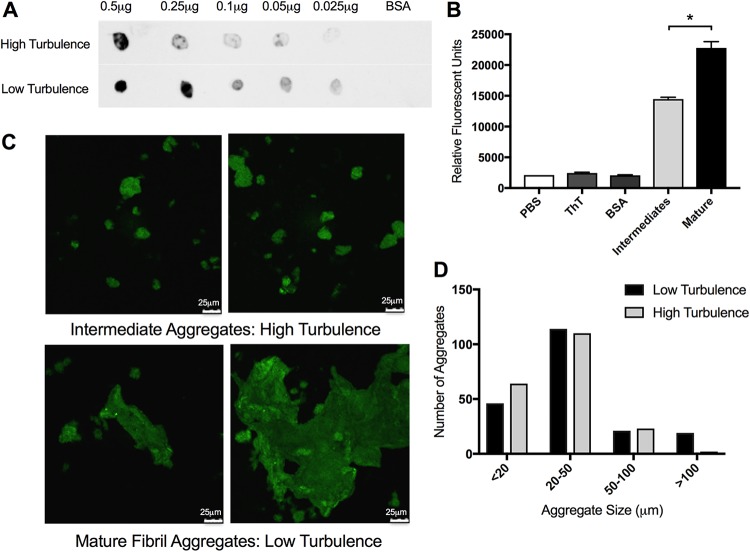
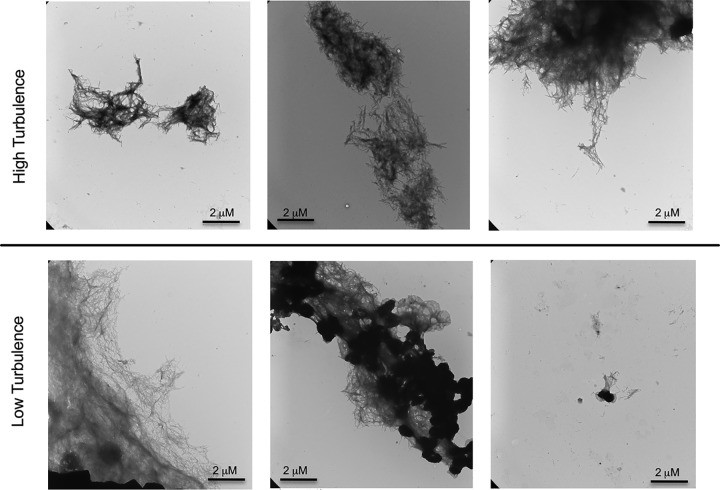
Although there was no pellicle in the 500-ml culture grown with shaking at 200 rpm, we were able to purify curli from both the 150-ml and the 500-ml culture conditions. When we performed a dot blot using anti-CsgA antibodies, CsgA was detected in both protein preparations, although the amount of curli recognized by the antibody was smaller in the sample from the 500-ml culture (Fig. 2A). We next stained samples with the amyloid-specific stain thioflavin T (ThT). Fluorescence due to ThT binding to curli increases as fibrillization increases (36). The protein preparations from both conditions stained with ThT; however, the curli purified under high-turbulence conditions showed lower levels of ThT fluorescence compared to the curli purified from the low-turbulence conditions (Fig. 2B). Consistent with this, the curli purified under high-turbulence conditions displayed smaller curli aggregates under confocal microscopy compared to the curli purified from the low-turbulence conditions (Fig. 2C). Using the LAS AF software, we measured the size of the curli aggregates across multiple fields of view. After counting 200 aggregates from each condition, we found the average size of curli masses purified under high-turbulence conditions to be smaller than that of the mature aggregates purified from the low-turbulence conditions (Fig. 2D). Therefore, we termed these smaller curli masses curli intermediates. To confirm the physical differences in these aggregates, we also used transmission electron microscopy (TEM). Fibrillar structures were visible in both curli preparations purified from high-turbulence and low-turbulence conditions (Fig. 3). However, the curli purified from low-turbulence conditions displayed larger aggregates with spots showing greater electron density (Fig. 3, middle lower panel). Even small aggregates from the low-turbulence conditions showed electron-dense regions (Fig. 3, right lower panel), supporting the idea that these aggregates are highly fibrillized and aggregated into a compact structure compared to the intermediates that do not show this dense staining. Additionally, the density of the staining in the mature aggregates may be attributable to greater DNA incorporation and therefore structural support for these larger aggregations.
FIG 2.
Curli intermediates are detected under high-turbulence growth conditions. (A) Dot blot analysis of decreasing concentrations of curli purified from high-turbulence and low-turbulence cultures was completed on PVDF membrane. BSA was used as a negative control. Blots were imaged on the Li-Cor Odyssey imaging system. (B) Aliquots of purified curli aggregates (400 μg/ml) from high-turbulence and low-turbulence growth conditions were stained with 10 μM ThT and quantified after a 10-min incubation at room temperature on a fluorescent plate reader at excitation 440 nm/emission 500 nm. Data are means (±standard error of the mean [SEM]) from three replicates. *, P < 0.05. (C) Curli purified from high-turbulence (left panel) and low-turbulence (right panel) growth conditions stained with 10 μM thioflavin T were imaged at ×60 magnification by confocal microscopy. Scale bars are 25 μm. (D) Enumeration of difference in size of aggregates calculated by confocal microscopy images. Aggregates were measured within the LAS AF confocal system and grouped into one of four groups. Two hundred aggregates were counted for each condition.
FIG 3.
Transmission electron microscopy of high- and low-turbulence curli aggregates showing electron density differences. Purified curli from high-turbulence and low-turbulence conditions stained by negative staining were imaged at ×16,500 magnification by transmission electron microscopy, showing electron density differences.
Overall, these experiments showed that although biofilm formation is inhibited by the increased turbulence in the bacterial culture, CsgA is expressed under both high- and low-turbulence conditions; however, larger mature fibrillar aggregates were observed only under lower turbulence, where the biofilm was visible at the air-liquid-flask interface.
DNA induces formation of mature curli fibrillar aggregates.
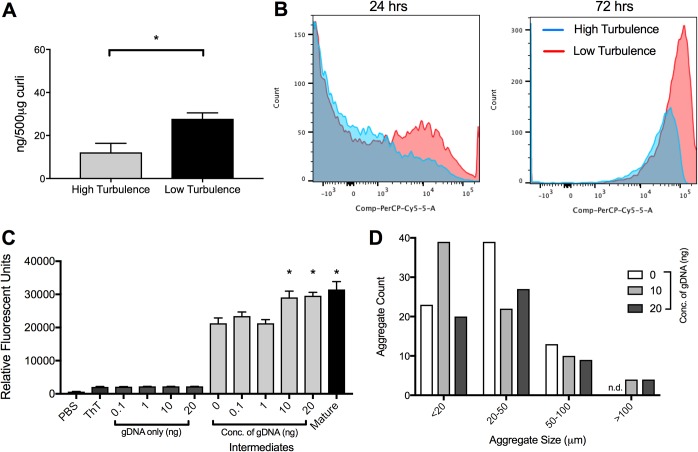
Previous work from our lab has shown that eDNA associates with curli fibrils during the development of the mature biofilm and purified curli contains eDNA (13). Moreover, eDNA accelerates the fibrillization of curli fibrils (13). The reduced recognition of curli purified from 500-ml highly turbulent cultures by the anticurli antibodies could be due to either a reduction of curli per μg of protein preparation or reduced binding to the curli intermediates by anticurli antibodies. To investigate whether there are differences in the association of eDNA with curli under the two culture conditions, we first examined the DNA content in purified curli aggregates. We performed a phenol-chloroform DNA extraction from 500 μg of the two curli preparations. We isolated more DNA from the curli purified under low-turbulence conditions than from those purified from the high-turbulence culture (Fig. 4A). This suggests that DNA contributes to the formation of the larger mature fibrillar aggregates observed in the low-turbulence preparation.
FIG 4.
DNA is necessary for formation of mature curli fibrillar aggregates. (A) DNA content per 500 μg of curli preparations from high-turbulence and low-turbulence conditions. Plotted are means (±SEM) of data. One representative experiment of three independent experiments is shown. (B) Flow cytometry histograms of aliquots of cultures stained with propidium iodide after 24 h and 72 h in high-turbulence and low-turbulence cultures. (C) A purified curli preparation from a highly turbulent culture was treated with or without S. Typhimurium genomic DNA (gDNA) at the indicated concentrations. Fluorescence of samples incubated at 37°C for 24 h and stained with ThT was determined. Plotted are means (±SEM) from three replicates. The asterisks indicate significant increases (P < 0.05) relative to intermediate aggregates without DNA. (D) Enumeration of difference in size of aggregates calculated by confocal microscopy images. Aggregates were measured within the LAS AF confocal system and grouped into one of four groups. At least 60 aggregates were counted for each condition (0 to 75, 10 to 75, and 20 to 60 counts).
Cell death is a major source of eDNA during biofilm formation. We hypothesized that there is increased cell death under low-turbulence conditions where the mature fibrillar aggregates and biofilm are forming. To test this, we stained bacterial cells under both culture conditions with a live/dead stain and evaluated cell death via flow cytometry. At 24 h, few dead cells were observed. However, at 72 h, the time that we see a large amount of mature biofilm in low-turbulence cultures, there was clearly more cell death in the low-turbulence culture than in the high-turbulence culture (Fig. 4B). We reason that the low-turbulence cultures are poorly aerated due to the biofilm pellicle that forms on the air-liquid-flask interface, and this causes cell death and DNA release into culture media. It may be necessary for curli to complex with DNA to form the mature fibril aggregates necessary for biofilm formation.
As there is an association between DNA content and mature matrix formation, we sought to determine whether supplementing the smaller curli intermediates with DNA would lead to the formation of larger curli masses. To do this, we incubated the curli intermediates obtained from the high-turbulence culture with increasing concentration of genomic DNA (gDNA) purified from S. Typhimurium and then stained with ThT. As the amount of DNA increased, the relative fluorescence levels increased (Fig. 4C). This behavior was also observed with purified curli upon addition of exogenous DNA in a previous study (13). When adding exogenous DNA to mature curli aggregates, we observed no increase in relative fluorescence intensity (see Fig. S3A in the supplemental material). Finally, using confocal microscopy, we observed that the addition of gDNA to intermediates increased the size of the aggregates. While there were no aggregates above 100 μm observed in untreated intermediate, addition of genomic DNA resulted in aggregates above 100 μm. There were no differences in the number of aggregates smaller than 20 μm or 20 to 100 μm (Fig. 4D). The addition of gDNA to the mature aggregates also did not show an increase in aggregate size, similar to the ThT results (Fig. S3B). Interestingly, when we isolated curli intermediates at earlier time points, we observed an increase in the number of fibril aggregates smaller than 20 μm (Fig. S3C).
Curli intermediates are more cytotoxic to macrophages than the mature fibrillar aggregates.
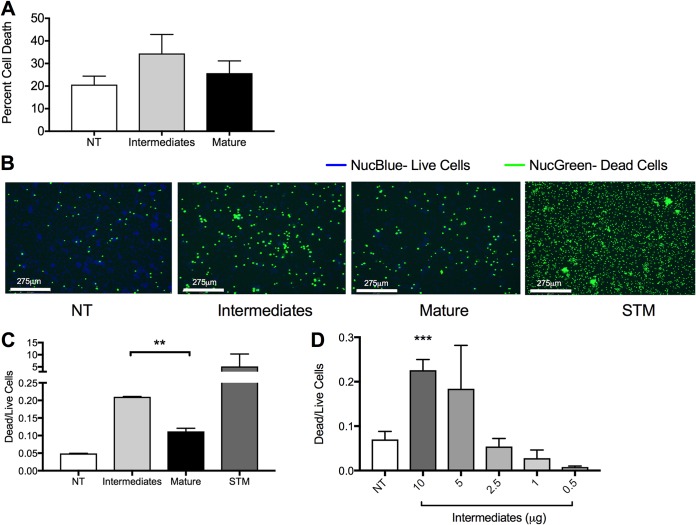
The early intermediate forms of human amyloid β are more cytotoxic to immune cells than mature amyloid β fibrils (19–21). As curli fibrils are structurally similar to amyloid β, we evaluated the toxicities of curli prepared from high- and low-turbulence culture conditions. We treated bone marrow-derived macrophages (BMDMs) with the two different curli preparations for 24 h and performed a lactose dehydrogenase (LDH) assay to evaluate cell death. The percentage of dead cells was higher in the BMDMs treated with the preparation from the turbulent culture, which contains curli intermediates, than in cell samples treated with the mature curli aggregates from the low-turbulence culture (Fig. 5A). To confirm these findings, BMDMs were treated with the two preparations, stained with the Ready Probe cell viability kit, and imaged on the EVOS FL Auto2 fluorescence microscope. S. Typhimurium was used as a positive control. More dead cells were detected in the BMDMs treated with the preparation from the high-turbulence culture containing the curli intermediates than the preparation from the low-turbulence culture (Fig. 5B). The dead and live cells were quantified, demonstrating that the ratio of dead to live cells was significantly higher in the wells treated with the curli intermediates than mature curli fibrillar aggregates (Fig. 5C). Furthermore, decreasing the concentration of curli intermediates decreased the cytotoxicity observed on BMDMs (Fig. 5D).
FIG 5.
Curli intermediates are more cytotoxic than mature aggregates. (A) An LDH assay was used to determine the percentage of cytotoxicity in BMDMs treated with curli preparations from high-turbulence and low-turbulence cultures. Shown are means and a representative experiment from three independent experiments. (B) Images of BMDMs seeded at 2.5 × 105 cells/well and not treated (NT) or treated with curli preparations from high-turbulence or low-turbulence cultures or with S. Typhimurium (STM). Cells were stained with NucBlue, which stains nuclei of live cells, and NucGreen, which stains nuclei of dead cells. Each image is a small portion of the full well. Representative images are shown for one of three independent experiments. (C) The ratios of dead to live cells obtained by analysis of multiple images accounting for the central 20% of well per condition in duplicate. Plotted are means (±SEM) from all images per condition. The asterisks indicate significant difference between the effects of preparations from high- and low-turbulence cultures. (D) The ratios of dead to live cells obtained by analysis of multiple images accounting for the central 20% of the well per condition in duplicate. Plotted are means (±SEM) from all images per condition. **, P < 0.01; ***, P < 0.001.
DISCUSSION
Amyloid deposits appear to underlie symptoms of many debilitating human diseases, including Alzheimer’s disease (5). Studies on human amyloids have revealed that various intermediate structures form during amyloid polymerization. Soluble monomeric amyloid units first form oligomers, which then polymerize into protofibrillar structures. These protofibrils cross-assemble to form thicker mature fibrils (37). Curli fibrillar assembly in Enterobacteriaceae is finely regulated via a type VIII section system, which results in a highly efficient nucleation precipitation process (7, 8). The CsgA subunit efficiently fibrillizes in the presence of CsgB nucleator protein, which seeds the fibrillization process. At high concentrations without the CsgB protein, CsgA self-assembles into fibrils (9–12). Intermediate oligomeric structures of curli, or any other bacterial amyloid, had not been isolated prior to this study. Here we describe for the first time intermediate protofibrillar structures of the bacterial amyloid curli. We observed that increasing the volume in our S. Typhimurium culture led to a more turbulent culture, which prevented biofilm formation at the air-liquid-flask interface. Curli intermediate structures purified from the highly turbulent culture were smaller and displayed decreased fibrillization kinetics and decreased density of aggregation. The bacteria in highly turbulent cultures produced curli accompanied by smaller amounts of dead cells and therefore eDNA. DNA stabilized the intermediate forms of curli into larger aggregates. Similar to amyloid β, curli intermediates were more cytotoxic to immune cells than its mature fibrillar aggregates (Fig. 6).
FIG 6.
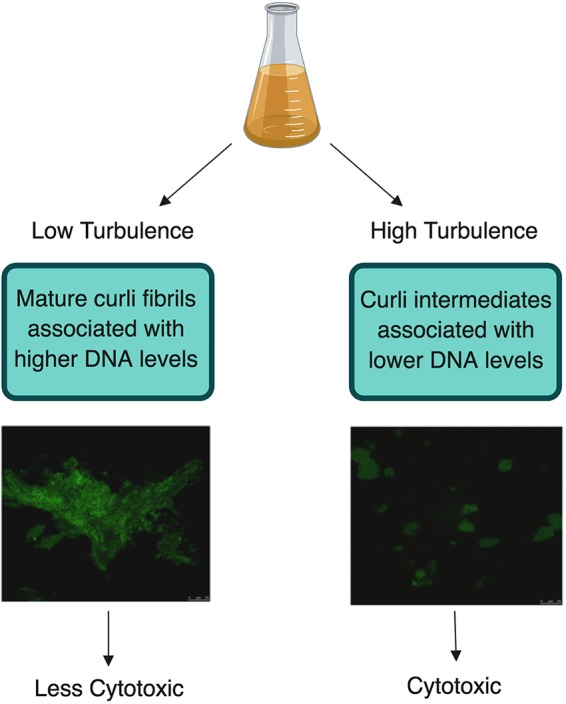
Working model. This figure is a comparison of the two different curli preparations used throughout our experiments as well as the difference involved in their processing. High-turbulence growth conditions produced smaller curli intermediates, which incorporate less DNA and are cytotoxic to murine macrophages. Alternatively, low-turbulence growth conditions led to larger fibrillar aggregates, which incorporate more DNA and are not cytotoxic to murine macrophages.
Oxygen tension is a major signal that regulates the csg gene cluster expression in S. Typhimurium (30). It was previously reported that oxygen-deprived biofilms produce larger amounts of extracellular polymers (38, 39). Consistent with this, we observed more curli expression and increased aggregation under the low-turbulence culture conditions where the biofilm pellicle potentially limited oxygen penetration into the culture. In contrast, in high-turbulence cultures, the bubbles observed could be indicative of better oxygenation. In the high-turbulence cultures, decreased curli expression and mature fibril aggregate formation were observed compared to levels in low-turbulence cultures. Although we were not able to detect any differences in the growth using the two conditions, flow cytometry analysis revealed a higher percentage of dead cells in the low-turbulence culture. We reason that the formation of the biofilm pellicle may limit the oxygen penetration into the forming biofilm and increase the cell death. Similar observations were made for Pseudomonas aeruginosa, in which cell death and lysis occur during the development of biofilms (40).
It was previously demonstrated that curli associates with DNA during biofilm formation (13). Both prokaryotic DNA and eukaryotic DNA accelerate the fibrillization of curli (13), although it is unclear whether DNA is needed for the formation of mature fibrillar structures. Here, we showed that reduced cell death was associated with smaller aggregate structures of curli, suggesting that the DNA released from dying cells is important for the assembly of larger, mature curli fibrillar aggregates. Consistent with this idea, addition of bacterial DNA to intermediates isolated from highly turbulent cultures was sufficient to trigger the assembly of larger curli masses. The turbulence may also prevent formation of large fibril aggregates due to physical disruption. Previous work on β2-macroglobulin, another human amyloid, showed that turbulent conditions resulted in intermediate forms of the fibrils (21). Intriguingly, the TEM analysis revealed that curli fibrils purified from the low-turbulence culture that contains a pellicle biofilm displayed highly aggregated electron-dense spots in the images. This level of aggregation could be due to highly organized curli and DNA networks as several recent studies showed that amyloid and DNA form into highly ordered complexes, which also affect their immunomodulatory capacity (41–43).
Studies of amyloid β have shown that oligomeric structures and protofibrils are more cytotoxic and cause a greater degree of membrane disruption than their mature fibrillar counterparts (21). This cytotoxicity was attributed to an enhanced ability of the oligomeric structures to permeabilize membranes, a mechanism shared by many amyloidogenic proteins (44, 45). By permeabilizing the membrane, it is hypothesized that oligomers or protofibrils cause a chemical imbalance within the cell that leads to deregulated calcium-dependent signal transduction pathways. The immune system recognizes the mature fibrillar amyloids as a conserved molecular pattern via Toll-like receptor 2 (TLR2)/TLR1 heterocomplex (26, 43, 46–50) and the NLRP3 inflammasome (25, 51–54). NLRP3 inflammasome activation leads to the activation of caspase 1 and has been associated with an inflammatory type of cell death. In the case of amyloid β, although NLRP3 is activated, which then leads to the activation of caspase 1, no cell death was observed (55), suggesting that this pathway is not associated with the cytotoxic events observed for the intermediate amyloid structures. Similarly, although curli activate the NLRP3 inflammasome, this activation leads only to caspase-1 activation, not cell death (25). Therefore, the cytotoxicity associated with the intermediate protofibrillar structures of curli may result from membrane disruption; however, this needs to be confirmed.
Amyloid proteins are produced by up to 40% of bacteria. Both commensal and pathogenic organisms produce amyloids that are secreted to the extracellular environment (2, 6, 23). Our study provides insight into the fibrillization kinetics of curli as well as the role of mature curli fibrillar aggregates in the assembly of bacterial extracellular matrix and its stability as we showed that intermediate structures of bacterial amyloids form and that the presence of bacterial DNA accelerates mature fibrillar aggregate formation, limiting cytotoxic effects. Further work is needed to identify their role in host-microbe interactions and their immunological role within the host.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. Typhimurium strain IR715, a fully virulent, spontaneous nalidixic acid-resistant derivative of strain ATCC 14028, was grown in Luria-Bertani broth (LB) supplemented with 50 μg/ml nalidixic acid at 37°C (56). The S. Typhimurium msbB mutant was previously described (57). This strain was grown in 100 μg/ml kanamycin at 37°C.
Purification of curli.
Curli aggregates were purified using a previously described protocol with some modifications (34). Briefly, an overnight culture of S. Typhimurium IR715 msbB was grown in LB with appropriate antibiotic selection with shaking (200 rpm) at 37°C. Overnight cultures were then diluted 1:100 in yeast extract supplemented with Casamino Acids (YESCA) broth with 4% DMSO to enhance curli production (33). Bacterial cultures were grown in the either 150 ml liquid YESCA medium containing 4% DMSO in a 250-ml flask or in 500 ml liquid YESCA medium in a 1-liter flask. These cultures were grown at 26°C for 72 h with shaking (200 rpm). Bacterial pellets were collected by centrifugation, resuspended in 10 mM Tris-HCl at pH 8.0, and treated with a mixture of 0.1 mg/ml RNase A (Sigma, R5502) from bovine pancreas, 0.1 mg/ml DNase I (Sigma, DN25), and 1 mM MgCl2 for 20 min at 37°C. Bacterial cells were then broken by sonication (30% amplification for 30 s twice). Next, lysozyme was added (1 mg/ml; Sigma, L6876), and samples were incubated at 37°C. After 40 min, 1% SDS was added, and the samples were incubated for 20 min at 37°C with shaking (200 rpm). After this incubation, curli were pelleted by centrifugation (10,000 rpm in a J2-HS Beckman centrifuge with rotor JA-14 for 10 min at 4°C) and then resuspended in 10 ml Tris-HCl (pH 8.0) and boiled for 10 min. A second round of enzyme digestion was then performed as described above. Curli were then pelleted, washed in Tris-HCl at pH 8.0, and resuspended in 2× SDS-PAGE buffer and boiled for 10 min. The samples were then electrophoresed on a 12% separating/3 to 5% stacking gel run for 5 h at 20 mA (or overnight at 100 V). Fibrillar aggregates are too large to pass into the gel and therefore remain within the well of the gel and can be collected. Once collected, the curli aggregates were washed three times with sterile water and then extracted by being washed twice with 95% ethanol. Purified curli were then resuspended in sterile water. Concentrations of curli aggregates were determined using the bicinchoninic acid (BCA) assay according to the manufacturer’s instructions (Novagen, 71285-3).
Sedimentation assay.
To determine the differences in aggregation and planktonic cell populations between high-turbulence and low-turbulence conditions, 1 ml of culture was collected in a 1.5-ml tube at 72 h. These tubes were allowed to sit at room temperature to allow aggregates to precipitate over time. Images were taken at time zero and at multiple time points until no further sedimentation was observed (∼30 min).
Crystal violet staining.
To image the pellicle-associated biofilm ring on the flask of shaken batch cultures, biofilms were stained with 1% crystal violet. Once the liquid culture was removed from the flasks—either for centrifugation or to be discarded—1% crystal violet was carefully added to the flask as to not displace the biofilm material. The staining solution was then gently rolled around the flask until the pellicle ring was fully stained, and the remaining crystal violet solution was removed. The crystal violet-stained pellicle-associated biofilm rings were then imaged using a Google Pixel 2.
Flow cytometry.
All flow cytometry experiments were completed using the FACSCanto II fluorescence-activated cell sorter (FACS). Plasmid pDW6, which encodes a promoterless green fluorescent protein (GFP), and pDW5, which contains GFP under the control of a tetracycline promoter, were kindly provided by Brad Cookson from the University of Washington (58). S. Typhimurium expressing GFP under the control of the curli promoter was obtained by cloning the csgBA promoter from S. Typhimurium IR715 using primer 5′-GGAATTCGAGACGTGGCATTAACCTGGACAGCACAA-3′ and reverse primer 5′-GGGATCCGCTGTCACCCTGGACCTGGTCGTACATAGC-3′. The resulting PCR product was cloned upstream of the gene encoding GFP on plasmid pDW6, yielding plasmid pCT125 (PcsgBA::gfp), which was then electroporated into S. Typhimurium IR715.
To investigate the expression of csgBA under different batch culture conditions, we used S. Typhimurium IR715, which contains pCT125 or pDW6 as a negative control. The cultures were examined once every 24 h for expression of csgBA via GFP expression. Briefly, 1 ml was sampled from shaken batch cultures and centrifuged at 10,000 rpm for 5 min. These pellets were then resuspended in 500 μl phosphate-buffered saline (PBS), and GFP was detected in the fluorescein isothiocyanate (FITC) channel by FACS.
To investigate cell death in batch cultures, 1 ml was sampled from each of the growth conditions and pelleted. This pellet was washed once with PBS, resuspended in propidium iodide (2 μl/ml), and incubated for 15 min at room temperature protected from light. These cells were then washed once with PBS and resuspended in 500 μl PBS. Propidium iodide staining was evaluated in the peridinin chlorophyll protein (PerCP)-Cy5.5 channel by FACS.
Thioflavin T assay.
A 1:1 mixture of 50 μl of curli (400 μg/ml) and 50 μl 10 μM thioflavin T was added to wells of a black 96-well plate and incubated for 10 min. After incubation, the relative fluorescence intensity was determined by a BMG Labtech POLARstar Omega plate reader using an excitation of 440 nm and an emission of 500 nm. In some experiments, genomic DNA (0.1, 1, 10, 20, 100, and 200 ng) extracted from S. Typhimurium cells and dissolved in PBS was added to curli preparations. These mixtures were then incubated at 37°C for 24 h.
Confocal laser scanning microscopy.
For confocal images of purified curli, preparations from each of the two conditions were stained with a 1:1 ratio of 10 μM thioflavin T to curli (400 μg/ml), and 5 μl was spotted onto a microscopy slide. Spots were allowed to dry completely, and then images were analyzed on the Leica SP5 microscope with a TCS confocal system at magnifications as noted. For enumeration of aggregates by size, the LAS AF confocal system was used to draw scale bars on aggregates from multiple fields, and aggregates’ sizes were recorded up to total numbers of 60 to 200 aggregates.
Transmission electron microscopy.
Purified curli were prepared as described above, but were diluted to 1 mg/ml and frozen in 25% sucrose solution. Samples were absorbed to 200-mesh copper grids coated with Formvar/carbon (Electron Microscopy Sciences, Hatfield, PA) and stained with 2% uranyl acetate. Samples were visualized with an FEI Tecnai T12 transmission electron microscope equipped with 2K × 2K Megaplus camera model ES 4.0 (Roper Scientific MASD, San Diego, CA).
DNA extraction.
Beginning with 500 μg of curli fibril complexes based on the BCA assay, complexes were centrifuged at 10,000 rpm for 3 min and resuspended in 550 μl of TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA), 30 μl of 10% sodium dodecyl sulfate, and 20 μl of 20 mg/ml proteinase K and mixed without vortexing. This mixture was incubated at 37°C for 1 h. After incubation, 100 μl of 5 M NaCl and 80 μl of cetyltrimethylammonium bromide (CTAB) were mixed into the solution. After incubation at 37°C for 10 min, 300 to 400 μl of phenol-chloroform–isoamyl alcohol (Fisher BP1754-400) was added and mixed well without vortexing. The mixture was then centrifuged at 13,000 rpm for 5 min at 4°C. The supernatant was transferred to a 1.5-ml tube, and 700 μl of chloroform was added. The solution was mixed by pipetting and centrifuged again. Once again, the supernatant was transferred to a clean 1.5-ml tube, and an equal volume of isopropanol was added and shaken by hand to mix. This mixture was then incubated at −20°C for at least 30 min to precipitate DNA. The DNA was pelleted by centrifugation at 12,000 rpm for 5 min at 4°C. The pellet was then washed with 1 ml of 70% ethanol and spun down once more at 7,500 rpm for 5 min. The final DNA pellet was resuspended in 30 μl TE buffer, and DNA content was measured using a NanoDrop2000 spectrophotometer.
Lactate dehydrogenase assay.
Bone marrow-derived macrophages were generated from the leg bones of C57BL/6 wild-type mice. Macrophages were differentiated as previously described (49). Cells were plated at 5 × 105 cells/well and stimulated for 24 h with curli fibrillar aggregates (10 μg) prepared with or without shaking and a 1:20 multiplicity of infection with fully virulent S. Typhimurium IR715. S. Typhimurium IR715 was diluted 1:40 from an overnight culture in LB supplemented with 5 mM NaCl and grown statically at 37°C for 2 h. Supernatants and cell lysates were analyzed using the lactate dehydrogenase (LDH) assay. The LDH assay was performed using the Cytotox 96 nonradioactive cytotoxicity assay (Promega, G1780) according to the manufacturer’s protocol. Briefly, supernatants were collected from stimulated cells. The 10× lysis buffer supplied with the kit was diluted to 1× and added to cells in medium or PBS, and samples were incubated for 30 min at 37°C. Cell lysates were then collected for total LDH determination. One vial of substrate was resuspended in 12 ml of assay buffer, and 50 μl was added to each well of either supernatant or lysate (50 μl) in a 96-well plate. The plate was then incubated in the dark at room temperature for 15 min. Following incubation, 50 μl of stop solution supplied with the kit was added to each well. Absorbance was read on the BMG Labtech POLARstar Omega plate reader at 490 nm. Cell death was calculated as a ratio of LDH activity in supernatant to LDH activity of lysed cells.
Live/dead staining.
Cells were stained using the ReadyProbes cell viability imaging kit (Thermo Fisher, R37609) following the manufacturer’s instructions. Briefly, 2 drops of each color stain was added per 1 ml of medium, and stain was aliquoted into each well and incubated at room temperature for 15 min protected from light. Cells were then imaged on the EVOS FL Auto 2 microscope. Analysis of images to determine the percentage of dead cells to live cells was done using the HCS Studio software system.
Dot blots.
Dot blotting to detect curli within curli aggregates was completed by first carefully spotting 2 μl of serially diluted complexes beginning with a 500-μg/ml solution on a piece of polyvinylidene difluoride (PVDF) membrane. The spots were given 1 to 2 h to fully dry. Next, the membrane was blocked in Tris-buffered saline (TBS [pH 7.5]) with 5% nonfat dry milk for 1 h at room temperature with rocking. The membrane was then incubated in 1:500 primary anti-CsgA antibody (generated in rabbit) diluted in TBS blocking buffer with 0.05% Tween 20 for 1 h with rocking. The membrane was then washed three times with TBS blocking buffer for 10 min each wash. Finally, the membrane was incubated in a 1:5,000 Li-Cor secondary antibody (anti-rabbit antibody) and blocking buffer with Tween for 1 h at room temperature with rocking. The membrane was then washed three times in TBS blocking buffer and then once with PBS and imaged on the Odyssey imaging system (Li-Cor).
Statistical analysis.
Data were analyzed using Prism software (GraphPad, San Diego, CA). Student's t test was used as appropriate. Error was determined by standard error of the mean. P values of <0.05 were considered significant and are noted on the figures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joice Kanefsky for useful discussions and Ronald Lucarelli for the generous donation of bone marrow-derived macrophages.
Work in Ç.T.’s lab was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases, grants AI125429, AI132996, and AI126133. A.E. was supported by CCSG P30CA006927.
The funders had no role in study design, data collection and interpretation, or decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00095-19.
REFERENCES
- 1.Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, Hussain T, Ali M, Rafiq M, Kamil MA. 2018. Bacterial biofilm and associated infections. J Chin Med Assoc 81:7–11. doi: 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Hufnagel DA, Tukel C, Chapman MR. 2013. Disease to dirt: the biology of microbial amyloids. PLoS Pathog 9:e1003740. doi: 10.1371/journal.ppat.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. 1987. Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 4.Hathroubi S, Mekni MA, Domenico P, Nguyen D, Jacques M. 2017. Biofilms: microbial shelters against antibiotics. Microb Drug Resist 23:147–156. doi: 10.1089/mdr.2016.0087. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. 2006. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. 2007. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol 9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 7.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desvaux M, Hebraud M, Talon R, Henderson IR. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol 17:139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Barnhart M, Chapman M. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor J, Zhou Y, Salgado P, Patwardhan A, McGuffie M, Pape T, Grabe G, Ashman E, Constable S, Simpson P, Lee W-C, Cota E, Chapman M, Matthews S. 2011. Atomic resolution insights into curli fiber biogenesis. Structure 19:1307–1316. doi: 10.1016/j.str.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammer ND, Schmidt JC, Chapman MR. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Smith DR, Jones JW, Chapman MR. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem 282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo PM, Rapsinski GJ, Wilson RP, Oppong GO, Sriram U, Goulian M, Buttaro B, Caricchio R, Gallucci S, Tukel C. 2015. Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 42:1171–1184. doi: 10.1016/j.immuni.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. 2016. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol 99:123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherny I, Rockah L, Levy-Nissenbaum O, Gophna U, Ron EZ, Gazit E. 2005. The formation of Escherichia coli curli amyloid fibrils is mediated by prion-like peptide repeats. J Mol Biol 352:245–252. doi: 10.1016/j.jmb.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 17.Hull RL, Westermark GT, Westermark P, Kahn SE. 2004. Islet amyloid: a critical entity in the pathogenesis of type 2 diabetes. J Clin Endocrinol Metab 89:3629–3643. doi: 10.1210/jc.2004-0405. [DOI] [PubMed] [Google Scholar]
- 18.Ross CA, Poirier MA. 2004. Protein aggregation and neurodegenerative disease. Nat Med 10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 19.Balducci C, Beeg M, Stravalaci M, Bastone A, Sclip A, Biasini E, Tapella L, Colombo L, Manzoni C, Borsello T, Chiesa R, Gobbi M, Salmona M, Forloni G. 2010. Synthetic amyloid-beta oligomers impair long-term memory independently of cellular prion protein. Proc Natl Acad Sci U S A 107:2295–2300. doi: 10.1073/pnas.0911829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 21.Xue WF, Hellewell AL, Hewitt EW, Radford SE. 2010. Fibril fragmentation in amyloid assembly and cytotoxicity: when size matters. Prion 4:20–25. doi: 10.4161/pri.4.1.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloney B, Lahiri DK. 2011. The Alzheimer's amyloid beta-peptide (Abeta) binds a specific DNA Abeta-interacting domain (AbetaID) in the APP, BACE1, and APOE promoters in a sequence-specific manner: characterizing a new regulatory motif. Gene 488:1–12. doi: 10.1016/j.gene.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tursi SA, Tukel C. 2018. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications. Microbiol Mol Biol Rev 82:e00028-18. doi: 10.1128/MMBR.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyama BH, Weissman JS. 2011. Amyloid structure: conformational diversity and consequences. Annu Rev Biochem 80:557–585. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapsinski GJ, Wynosky-Dolfi MA, Oppong GO, Tursi SA, Wilson RP, Brodsky IE, Tükel Ç. 2015. Toll-like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect Immun 83:693–701. doi: 10.1128/IAI.02370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tükel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JPM, Bäumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen SG, Stribinskis V, Rane MJ, Demuth DR, Gozal E, Roberts AM, Jagadapillai R, Liu R, Choe K, Shivakumar B, Son F, Jin S, Kerber R, Adame A, Masliah E, Friedland RP. 2016. Exposure to the functional bacterial amyloid protein curli enhances alpha-synuclein aggregation in aged Fischer 344 rats and Caenorhabditis elegans. Sci Rep 6:34477. doi: 10.1038/srep34477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dueholm MS, Nielsen SB, Hein KL, Nissen P, Chapman M, Christiansen G, Nielsen PH, Otzen DE. 2011. Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 50:8281–8290. doi: 10.1021/bi200967c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sleutel M, Van den Broeck I, Van Gerven N, Feuillie C, Jonckheere W, Valotteau C, Dufrêne YF, Remaut H. 2017. Nucleation and growth of a bacterial functional amyloid at single-fiber resolution. Nat Chem Biol 13:902–908. doi: 10.1038/nchembio.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstel U, Romling U. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ Microbiol 3:638–648. doi: 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 31.Olsen A, Arnqvist A, Hammar M, Normark S. 1993. Environmental regulation of curli production in Escherichia coli. Infect Agents Dis 2:272–274. [PubMed] [Google Scholar]
- 32.Romling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- 33.Lim JY, May JM, Cegelski L. 2012. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Appl Environ Microbiol 78:3369–3378. doi: 10.1128/AEM.07743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol 173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. 2014. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio 4:e00645-13. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue C, Lin TY, Chang D, Guo Z. 2017. Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R Soc Open Sci 4:160696. doi: 10.1098/rsos.160696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnabel J. 2010. Protein folding: the dark side of proteins. Nature 464:828–829. doi: 10.1038/464828a. [DOI] [PubMed] [Google Scholar]
- 38.Applegate DH, Bryers JD. 1991. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol Bioeng 37:17–25. doi: 10.1002/bit.260370105. [DOI] [PubMed] [Google Scholar]
- 39.Thormann KM, Saville RM, Shukla S, Spormann AM. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185:4585–4592. doi: 10.1128/jb.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee EY, Zhang C, Di Domizio J, Jin F, Connell W, Hung M, Malkoff N, Veksler V, Gilliet M, Ren P, Wong G. 2019. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat Commun 10:1012. doi: 10.1038/s41467-019-08868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt NW, Jin F, Lande R, Curk T, Xian W, Lee C, Frasca L, Frenkel D, Dobnikar J, Gilliet M, Wong GC. 2015. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat Mater 14:696–700. doi: 10.1038/nmat4298. [DOI] [PubMed] [Google Scholar]
- 43.Tursi SA, Lee EY, Medeiros NJ, Lee MH, Nicastro LK, Buttaro B, Gallucci S, Wilson RP, Wong GCL, Tukel C. 2017. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog 13:e1006315. doi: 10.1371/journal.ppat.1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bucciantini M, Calloni G, Chiti F, Formigli L, Nosi D, Dobson CM, Stefani M. 2004. Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J Biol Chem 279:31374–31382. doi: 10.1074/jbc.M400348200. [DOI] [PubMed] [Google Scholar]
- 45.Demuro A, Mina E, Kayed R, Milton SC, Parker I, Glabe CG. 2005. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J Biol Chem 280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 46.Cheng N, He R, Tian J, Ye PP, Ye RD. 2008. Cutting edge: TLR2 is a functional receptor for acute-phase serum amyloid A. J Immunol 181:22–26. doi: 10.4049/jimmunol.181.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jana M, Palencia CA, Pahan K. 2008. Fibrillar amyloid-beta peptides activate microglia via TLR2: implications for Alzheimer's disease. J Immunol 181:7254–7262. doi: 10.4049/jimmunol.181.10.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S, Liu Y, Hao W, Wolf L, Kiliaan AJ, Penke B, Rube CE, Walter J, Heneka MT, Hartmann T, Menger MD, Fassbender K. 2012. TLR2 is a primary receptor for Alzheimer's amyloid beta peptide to trigger neuroinflammatory activation. J Immunol 188:1098–1107. doi: 10.4049/jimmunol.1101121. [DOI] [PubMed] [Google Scholar]
- 49.Tükel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akçelik M, Adams LG, Bäumler AJ. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 50.Tukel C, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Baumler AJ. 2009. Responses to amyloids of microbial and host origin are mediated through Toll-like receptor 2. Cell Host Microbe 6:45–53. doi: 10.1016/j.chom.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ather JL, Ckless K, Martin R, Foley KL, Suratt BT, Boyson JE, Fitzgerald KA, Flavell RA, Eisenbarth SC, Poynter ME. 2011. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J Immunol 187:64–73. doi: 10.4049/jimmunol.1100500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, Gelpi E, Halle A, Korte M, Latz E, Golenbock DT. 2013. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niemi K, Teirila L, Lappalainen J, Rajamaki K, Baumann MH, Oorni K, Wolff H, Kovanen PT, Matikainen S, Eklund KK. 2011. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J Immunol 186:6119–6128. doi: 10.4049/jimmunol.1002843. [DOI] [PubMed] [Google Scholar]
- 54.Westwell-Roper C, Nackiewicz D, Dan M, Ehses JA. 2014. Toll-like receptors and NLRP3 as central regulators of pancreatic islet inflammation in type 2 diabetes. Immunol Cell Biol 92:314–323. doi: 10.1038/icb.2014.4. [DOI] [PubMed] [Google Scholar]
- 55.Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stojiljkovic I, Baumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Baumler AJ. 2005. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun 73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol 61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.