Typhoid fever is caused by Salmonella enterica serovar Typhi, and 3% to 5% of patients become chronic gallbladder (GB) carriers (also known as “Typhoid Marys”). We have previously demonstrated a role for Salmonella biofilm formation on gallstones as a primary mechanism of carriage. In this study, we found that the important biofilm extracellular matrix component curli fimbria is induced in an in vitro human GB model system. This induction is specific to human bile and increases as the biofilm matures. We also found that the biofilm and curli regulator CsgD play a key role in this observed induction. This work further enhances our understanding biofilm-mediated chronic carriage and provides a potential target for eliminating persistent GB infection by S. Typhi.
KEYWORDS: curli fimbriae, Salmonella, bile, biofilm, chronic carriage
ABSTRACT
Typhoid fever is caused primarily by Salmonella enterica serovar Typhi. Approximately 3% to 5% of individuals infected with S. Typhi become chronic carriers with the gallbladder (GB) as the site of persistence, as gallstones within the GB are a platform on which the bacteria form a biofilm. S. Typhi is a human-restricted pathogen; therefore, asymptomatic carriers represent a critical reservoir for further spread of disease. To examine the dynamics of the Salmonella biofilm during chronic carriage, the human gallstone (GS) environment was simulated by growing biofilms on cholesterol-coated surfaces in the presence of bile, and the transcriptional profile was determined. Some of the most highly activated genes corresponded to the curli fimbria operon, with the major structural component csgA upregulated >80-fold. The curli protein polymer is a major component of the extracellular matrix (ECM) in Salmonella biofilms. The upregulation of curli fimbriae by human bile was validated through reverse transcription-quantitative PCR (qRT-PCR), microscopy, and Western blotting. Interestingly, this activation appears human specific, as qRT-PCR showed repression of csgA in biofilms grown in mouse or ox bile. Comparative transcriptional studies of the two divergent csg operons suggest an early activation of both operons in minimal medium complemented with glucose that quickly diminishes as the biofilm matures. However, in the presence of human bile, there is a modest activation of both operons that steadily increases as the biofilm matures. Understanding the effect of the GB environment on key biofilm-associated factors can help target antibiofilm therapeutics or other preventative strategies to eradicate chronic carriage.
IMPORTANCE Typhoid fever is caused by Salmonella enterica serovar Typhi, and 3% to 5% of patients become chronic gallbladder (GB) carriers (also known as “Typhoid Marys”). We have previously demonstrated a role for Salmonella biofilm formation on gallstones as a primary mechanism of carriage. In this study, we found that the important biofilm extracellular matrix component curli fimbria is induced in an in vitro human GB model system. This induction is specific to human bile and increases as the biofilm matures. We also found that the biofilm and curli regulator CsgD play a key role in this observed induction. This work further enhances our understanding biofilm-mediated chronic carriage and provides a potential target for eliminating persistent GB infection by S. Typhi.
INTRODUCTION
Typhoid fever, caused primarily by Salmonella enterica serovar Typhi, is a life-threatening systemic disease that is responsible for significant morbidity and mortality worldwide, causing more than 12 million infections and 129,000 deaths annually (1). Common symptoms include high fever, malaise, and abdominal pain. If left untreated, up to 10% of convalescent patients excrete S. Typhi in their feces for up to 3 months. Furthermore, 3% to 5% of individuals infected with S. Typhi become chronic carriers, shedding bacteria for more than one year (2). The gallbladder (GB) is the most common site of persistence. S. Typhi is a human-restricted pathogen; therefore, asymptomatic carriers represent a critical reservoir for further spread of disease. We have demonstrated that gallstones aid in the development and maintenance of GB carriage in a mouse model and in humans, serving as a substrate to which Salmonella attaches and forms a protective biofilm (3, 4). However, the molecular basis of chronic carriage of Salmonella in the GB, both from the host and bacterial perspectives, is poorly understood.
Biofilms are bacterial communities that attach to a biological or nonbiological surface and are encased by a bacterium-initiated matrix (5). This structure allows bacteria to survive in harsh environments such as the GB. The main components of the Salmonella biofilm matrix include curli fimbriae, the O-antigen capsule, cellulose, colanic acid, Vi-antigen, and biofilm-associated proteins (6–8). The major protein component is curli fimbria, which is known to be important for biofilm formation and plays a role in virulence and alteration of the immune response (9–11). Curli fibers are similar to eukaryotic amyloid fibers and are involved in cell aggregation and cell adhesion. Curli genes are arranged in two divergent curli-specific gene (csg) operons with independent promoters: one contains the structural components CsgA and CsgB (csgBAC), and the second contains the regulator CsgD and other structural proteins (csgDEFG). CsgD is a transcriptional regulator that is central to biofilm production. It directly regulates the csgBAC operon, cellulose, and the BapA protein (7, 12, 13).
To examine the dynamics of a Salmonella enterica serovar Typhimurium (a model for S. Typhi infection) biofilm during chronic carriage, the human GB environment was simulated in vitro by growing biofilms on a cholesterol surface in the presence of human bile, followed by determination of the biofilm transcriptional profile. This study describes the observed induction of curli fimbria genes under in vitro GB conditions. This upregulation was human specific and continued to increase as the biofilm matured.
RESULTS
Salmonella differential gene expression analysis using an in vitro simulated GB environment.
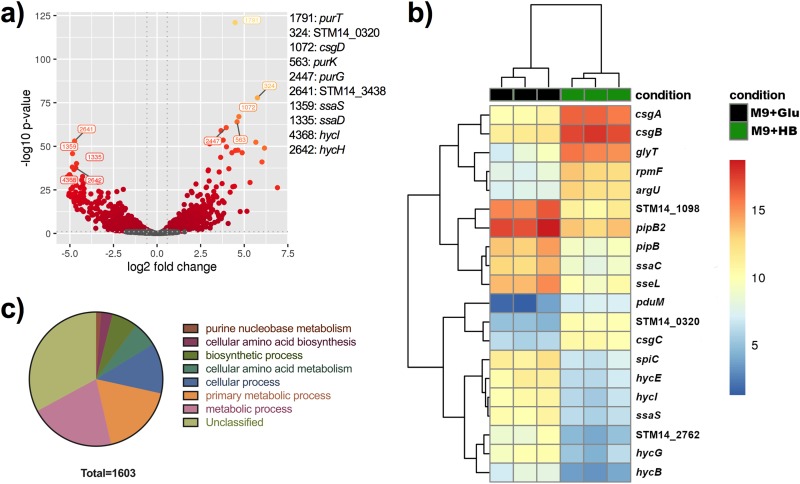
We used transcriptome sequencing (RNA-Seq) analysis to study the gene expression profile of S. Typhimurium under human GB-like conditions. This environment (biofilms grown on cholesterol-coated surfaces and 10% human bile) induced a global shift in the transcriptional profile. RNA-Seq identified a total of 1,603 genes that were differentially expressed with a fold change (FC) equal to or greater than 1.5 and a P value of 0.05 or lower (FC ≥ 1.5, P ≤ 0.05) as a cutoff (Fig. 1 and Table 1; see also Table S1 in the supplemental material). There was a balance between up- and downregulated genes (Fig. 1a), and biological replicates were observed to cluster closely together (Fig. 1b). The top induced gene was the tRNA glyT (FC = 182.9), followed by csgA and csgB (FC = 82.9 and 76.33, respectively). csgA and csgB are part of one of the two curli fimbria operons and code for the main protein polymer (CsgA) and a nucleator protein that is crucial for fimbral assembly (CsgB). As expected, RNA sequencing detected various known bile resistance genes (14), including marA (FC = 3), which we had identified in a previous study (15), the tol genes tolQ (FC = 2.2) and tolR (FC = 2.1), efflux pump genes emrR (FC = 2.6) and emrA (FC = 2.7), ompF (FC = 1.9), and the bile transporter yfeH (FC = 5.1). To gain a better understanding of how GB conditions affect gene expression, a functional analysis was performed using the Panther classification system tool (16). This biological process overrepresentation analysis identifies whether there is a statistical overrepresentation or underrepresentation of the genes and/or proteins in the test list relative to the reference list. A global look showed that the majority of the differentially expressed genes fell in the unclassified category, followed by metabolic and cellular processes as well as amino acid biosynthesis and metabolism (Fig. 1c). The top overrepresented biological processes were O-antigen metabolic/biosynthetic process with a 4.64-fold overrepresentation, cobalamin (vitamin B12) metabolic/biosynthetic process with a 3.88-fold increase, and purine metabolism with a 3.04-fold increase (see Table S2). Other interesting operons or gene families that were differentially expressed are listed in Table S3. Those demonstrated to be upregulated include purine metabolism (pur), curli fimbriae (csg), and propanediol utilization (pdu). Those that were downregulated were fimbriae (fim), SPI2 (ssr, ssa, and sse), and hydrogenases (hyp and hyc). Interestingly, both curli operons were highly upregulated under human GB conditions (Table 1). Curli genes are arranged in two operons with independent promoters, one contains the main structural components CsgA and CsgB (csgBAC) and the second (csgDEFG) contains the regulator CsgD and other structural proteins (Fig. 2a). Curli fimbriae are a fundamental component of Salmonella biofilms, and so we decided to further investigate their role under human GB conditions.
FIG 1.
Differentially expressed genes between biofilms grown in a simulated human GS environment versus that in minimal medium plus glucose. (a) Volcano plot. The x axis specifies the fold change (FC) and the y axis specifies the negative logarithm to the base 10 of the t test P values. Dashed lines represent the filtering criteria (FC ≥ 1.5, P ≤ 0.05). Red and orange dots represent probe sets for transcripts expressed at significantly higher or lower levels. (b) Dendrogram and unsupervised hierarchical clustering heat map (using Euclidean distance) of gene expression based on the log ratio fragments per kilobase per million mapped reads (FPKM) data. Each column represents a biological replicate from each experimental condition; rows represent genes. Red, upregulation; blue, downregulation. The vertical distances on each branch of the dendrogram represent the degrees of similarity between gene expression profiles of various groups. (c) Biological function of enriched genes in HB biofilms.
TABLE 1.
Top differentially expressed genes in biofilms grown under in vitro human GS conditions by RNA-Seqa
| Gene | Fold change | Function |
|---|---|---|
| glyT | 182.9 | tRNA-Gly |
| csgA | 82.93 | Major curlin subunit |
| csgB | 76.33 | Minor curlin subunit |
| STM14_0320 | 57.86 | Type VI secretion system contractile sheath small subunit |
| rpmF | 56.75 | 50S ribosomal protein L32 |
| pduM | 51.58 | Propanediol utilization protein |
| argU | 47.68 | tRNA-Arg |
| proM | 37.09 | tRNA-Pro |
| csgC | 31.74 | Curli assembly protein |
| cysT | 27.59 | tRNA-Cys |
| csgD | 27.14 | csgAB operon transcriptional regulatory protein |
| csgE | 26.57 | Curli production assembly/transport component |
| purK | 25.24 | N5-Carboxyaminoimidazole ribonucleotide synthase |
| hycA | –24.88 | Formate hydrogenlyase regulatory protein |
| ygaF | –24.91 | Predicted dehydrogenase |
| ssaG | –25.24 | Type III secretion system, needle protein |
| STM14_1971 | –25.25 | Membrane protein |
| sseE | –26.24 | Type III secretion system regulator LcrR/chaperone CesD2 |
| ssaD | –26.6 | Secretion system apparatus protein |
| hycH | –27.25 | Protein for processing of HycE, formate hydrogenlyase maturation |
| sseJ | –27.72 | Secreted effector protein |
| ssaI | –27.82 | Secretion system apparatus |
| STM14_1694 | –28.11 | SPI-2 type III secretion system translocon protein SseB |
| STM14_3438 | –28.12 | Hypothetical protein |
| hydN | –28.56 | Electron transport protein (FeS center) from formate to hydrogen |
| ssaK | –28.58 | Secretion system apparatus protein |
| ssaJ | –29.28 | Secretion system apparatus lipoprotein |
| ssaT | –29.77 | Secretion system apparatus protein |
| STM14_2760 | –29.99 | Tail fiber assembly protein |
| hycD | –30.15 | Hydrogenase 3, membrane subunit |
| STM14_3365 | –30.32 | Hypothetical protein |
| STM14_2763 | –30.43 | Hypothetical protein |
| ssaS | –31.12 | Secretion system apparatus |
| hycI | –31.77 | Protease involved in processing C-terminal end of HycE |
| STM14_3192 | –31.83 | Hypothetical protein |
| STM14_3222 | –32 | Hypothetical protein |
| STM14_2759 | –32.27 | Hypothetical protein |
| STM14_2761 | –32.74 | Hypothetical protein |
| ssaR | –32.78 | Secretion system apparatus |
| STM14_1693 | –32.8 | Hypothetical protein |
| hycG | –33.57 | Hydrogenase activity, HycG, ortholog |
| sseL | –33.69 | Deubiquitinase |
| ssaC | –35.23 | Secretion system apparatus protein |
| pipB2 | –35.71 | Homolog of pipB, putative pentapeptide repeats |
| hycB | –36.07 | Hydrogenase-3, iron-sulfur subunit |
| hycE | –36.23 | Hydrogenase 3, large subunit |
| pipB | –37.41 | Pathogenicity island encoded protein, SPI3 |
| STM14_1098 | –39.54 | Hypothetical protein |
| STM14_2762 | –39.8 | Hypothetical protein |
| spiC | –40.97 | SPI-2 type III secretion system protein |
Curli fimbria genes are in boldface font.
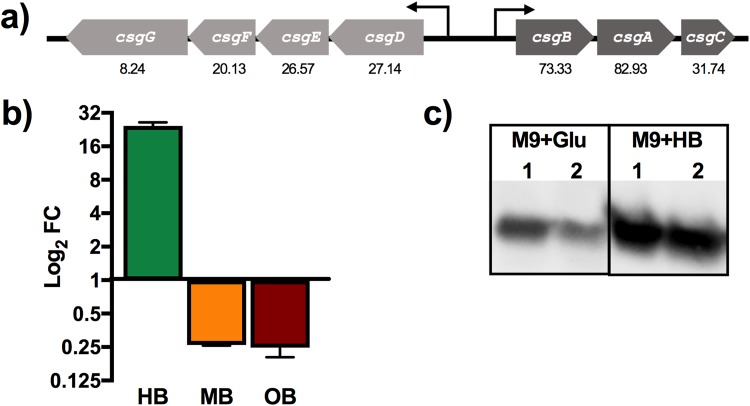
FIG 2.
(a) Graphical representation of curli operons. RNA-Seq fold changes are presented under each gene. (b) qRT-PCR log2 fold change in csgA expression from biofilms grown in M9 supplemented with human bile (HB; 10%), mouse bile (MB; 2.5%), or ox bile (OB; 2.5%) with respect to that in M9 supplemented with glucose (10 mM); rpoB was used as a reference. (c) Western blot analysis of the CsgA protein expressed in biofilms grown in M9 minimal medium with glucose (Glu) or HB (lanes 1 and 2 correspond to biological replicates under the same growth conditions).
qRT-PCR analysis demonstrates human bile-specific regulation of csgA.
To validate the RNA-Seq results at the transcriptional level, reverse transcription-quantitative PCR (qRT-PCR) was performed. Figure 2b shows the fold changes for csgA in a 7-day biofilm grown in minimal medium (M9) with human (10%), mouse (2.5%), or ox (2.5%) bile. The control was csgA in a biofilm grown in minimal medium with glucose (10 mM). rpoB was used as the reference gene. The fold changes for csgA in a biofilm in human bile showed the same trend as for the RNA-Seq, with a 28.38 FC with respect to that in M9 plus glucose. Interestingly, the effect was human bile specific, as csgA was downregulated in both ox and mouse bile.
Human bile induces production of CsgA in Salmonella biofilms.
The data described above only provided evidence of curli overexpression in the presence of human bile at the transcriptional level. To verify that the human GB environment induces production of curli fimbriae at a translational level, total protein from 7-day-old biofilms was collected and analyzed by Western blotting for CsgA. As observed in Fig. 2c, the addition of human bile visibly increased CsgA production. Image Lab quantification showed a 2.76-fold increase in CsgA in the biofilms grown with human bile versus that in the control with M9 plus glucose.
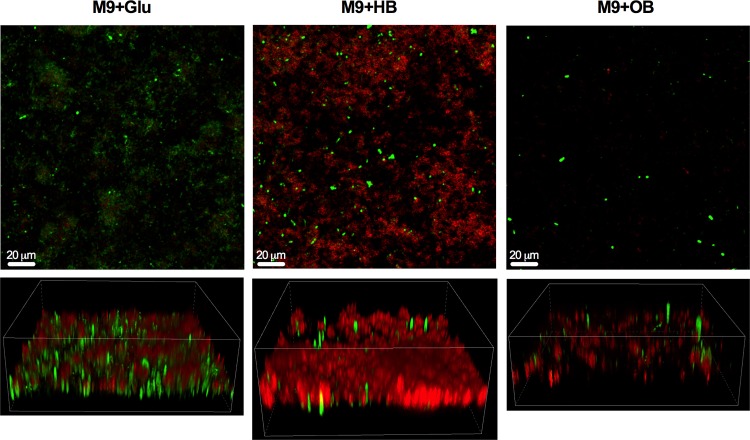
Confocal imaging confirms increased production of curli fimbriae in the presence of human bile.
Like all amyloids, curli bind specifically to the dye Congo red and show a strong signal in the Texas Red channel on a fluorescence microscope. When biofilms were grown in M9 supplemented with glucose, as expected, curli fimbriae were detected. However, curli fimbria production was significantly increased when the minimal medium was supplemented with human bile. In general, biofilms grew less well in the presence of ox bile, but low levels of curli were still detected by confocal scanning laser microscopy (Fig. 3).
FIG 3.
Curli fimbria activation in human bile (HB). Representative images of 7-day biofilms of S. Typhimurium grown in M9 plus glucose (Glu) (control), M9 plus human bile (HB), and M9 plus ox bile (OB). Biofilms were incubated with Congo red for 5 min to stain curli fimbriae, rinsed twice with KPi buffer, and subsequently fixed in 2% paraformaldehyde (PFA). Green represents GFP-expressing Salmonella. All images are at ×20 magnification. The upper panels are top-down views of the biofilm, while the bottom panels are three-dimensional (3-D) Z-stacks.
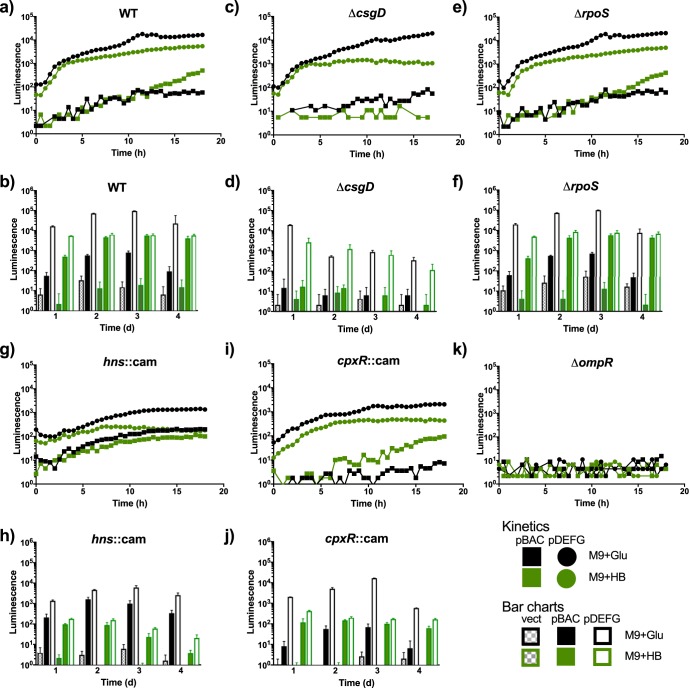
Curli gene expression is sustained and increases over time only in the presence of human bile.
Reporter plasmids with the promoter for each of the divergent curli operons cloned upstream of the lux genes were used to analyze expression in different strains and under different conditions. In the wild type (WT) strain, there was very high expression of the csgDEFG operon in minimal medium supplemented with glucose over the 16-h measurement period. The expression in the presence of human bile was also high but showed slightly lower values than that in M9 with glucose. The csgBAC operon, on the other hand, was expressed at lower levels, took approximately 10 h to reach detectable levels, and was expressed at higher levels when grown in M9 with human bile (Fig. 4a). Since the RNA-Seq had been performed on a mature biofilm, curli expression was measured over a longer period of time (4 days). Interestingly, the csgDEFG operon continued to maintain very high expression levels in M9 plus glucose, while there was modest expression of csgBAC at days 2 and 3 that returned to low levels at day 4. When grown in human bile, on the other hand, there was a high and steady expression of csgDEFG at all time points and an increase of csgBAC expression at day two, which was maintained through day 4 (Fig. 4b).
FIG 4.
Temporal expression of curli genes under different growth conditions. Gene expression measured as a function of light production (log10 luminescence [counts per second]) versus time for the csgBAC and csgDEFG::lux promoter fusions. Luminescence measurements in each indicated background strain for the first 18 h of growth (a, c, e, g, i, and k) and daily measurements (b, d, f, h, and j). Representative expression curves and bar charts from one experiment performed in triplicates and repeated three times with similar results are shown. M9+Glu, M9 plus glucose (control); M9+HB, M9 plus human bile.
We examined the role of the biofilm and curli operon regulator CsgD and other upstream transcriptional regulators in the presence of human bile. CsgD did not regulate its own expression, in accordance with previous reports (17), as high expression of the csgDEFG genes was observed in M9 plus glucose in a csgD mutant background (Fig. 4c and d). Additionally, in a ΔcsgD background, the csgDEFG expression in minimal medium with glucose was considerably greater than that with human bile after 16 h (approximately 1 log). However, the csgBAC operon was expressed at very low levels with both glucose and human bile in the csgD mutant background (Fig. 4c), and these expression patterns were similarly low and did not increase during biofilm maturation (Fig. 4d). The sigma factor RpoS was not involved in curli regulation under the conditions tested, as the promoter activity of the curli operons look nearly identical to that for the WT strain (Fig. 4e and f). The histone-like nucleoid structuring protein (hns product), on the other hand, affected expression. In an hns mutant background, both operons were active at approximately 1 log lower than the WT at early time points (Fig. 4g). Interestingly, at later time points in the hns mutant, the expression of csgBAC in human bile was greatly diminished (>2 logs), while in M9 plus glucose, it was slightly higher than the WT (Fig. 4h). The response regulator CpxR has been shown to repress curli expression in Escherichia coli (18). Under our conditions, at early time points in the cpxR mutant, csgDEFG activity was approximately 1 log lower than in the WT, while csgBAC was only slightly reduced (Fig. 4i). At later time points, the csgBAC operon expression was reduced approximately 2 logs in the presence of human bile and was even lower in glucose (Fig. 4j). Finally, the regulator OmpR was crucial for the expression of both operons (Fig. 4k).
DISCUSSION
The GB stores and concentrates bile, secreting it into the intestine after a meal to aid digestion. Bile not only plays an important role in the emulsification and absorption of fats but also is a strong antimicrobial agent (19). The high concentrations of bile in the GB make it an extremely hostile environment for microbial life; nonetheless, enterobacteria such as Salmonella have developed mechanisms to survive and establish themselves in this niche (14). Salmonella must carefully regulate gene expression in order to persist in the GB environment, and our data reported here reflect this. We observed a global shift in the transcriptional profile of Salmonella grown in vitro under human GB/Salmonella chronic carriage conditions (cholesterol-coated plates and 10% human bile), with 1,603 genes differentially expressed, among them, known bile resistance genes (14), including marA (FC = 3), which we had previously identified in a microarray study as upregulated in medium complemented with sodium cholate (Ox bile) (15), the tol genes tolQ and tolR, efflux pump genes emrR and emrA, ompF (FC = 1.9), and a putative bile transporter yfeH (FC = 5.1). As expected with bacterial genomes, a majority of the differentially expressed genes fell in the unclassified category, as most prokaryotic genomes have nearly 30% uncharacterized genes (Table 1; see also Table S1 in the supplemental matieral) (20).
Repressed genes include those for fimbriae (fim) and hydrogenases (hyp and hyc), which interestingly, were upregulated in our previous ox bile study (21). Many genes in Salmonella pathogenicity island 2 (SPI2) (ssr, ssa, and sse) were downregulated, which was previously observed in biofilms grown in a flow-through system (22).
Panther analysis, which groups affected genes into overrepresented biological processes, identified (Table S2) O-antigen capsule, which we previously showed to be upregulated in the presence of ox bile (23), further highlighting the importance of biofilm formation in the GB environment, as well as cobalamin (vitamin B12) metabolism, which has been implicated in stress resistance in natural biofilms (24), and purine metabolism, which was reported to be involved in persistence in Staphylococcus aureus (25) and biofilm formation in Streptococcus sanguinis (26) and Burkholderia (27).
Many of the top activated genes belonged to the two curli fimbria operons (Table 1). Curli fimbriae are the major protein component of the Salmonella biofilm extracellular matrix, and curli is known to be crucial for biofilm formation, virulence, and immune alteration (9–11, 28). Interestingly, we had not observed differential regulation of the structural genes csgAB in previous studies using ox bile (21). The strong induction of the csgD promoter in human bile is also quite significant, as it is a central biofilm transcriptional regulator, influencing the expression of many other biofilm components (7, 29, 30).
Another interesting finding was the apparent host specificity of bile-mediated curli induction. Salmonella biofilms grown in medium containing mouse or ox bile demonstrated repression of csgA, opposite to the dramatic activation in the presence of human bile (Fig. 2b). Differential expression of genes between mouse and ox bile was previously observed (31). Importantly, this activation was maintained at the protein level as observed by Western blot analysis (Fig. 2c). Congo red staining of curli visualized by confocal microscopy clearly confirms the increased abundance of curli fimbriae by exposure to human bile (Fig. 3).
The observed curli gene expression patterns were examined using reporter strains to help explain the activation observed in human bile. As we see in Fig. 4a and b, the csgDEFG operon was highly activated in both M9 plus glucose and M9 plus human bile early and through 4 days. Contrarily, expression of the csgBAC operon started at much lower levels but was still somewhat enhanced in human bile. In M9 plus glucose, expression peaked at around 3 days and then started to drop off, while in M9 plus human bile, expression steadily increased and was maintained through the duration of the assay. As expected, the transcriptional regulator CsgD plays a crucial role. In a csgD mutant background, Fig. 4c and d show a decrease in the expression of csgBAC to low/undetectable levels, while the csgDEFG operon was expressed at quantities similar to that of the WT. This is perhaps unsurprising given that it is well established that CsgD regulates the csgBAC (32) promoter but not its own expression (12). Remarkably, csgDEFG, while highly expressed, declines in expression in the csgD mutant over time in both types of media (human bile and glucose). Interestingly, the sigma factor RpoS, which regulates various stationary-phase stress-induced genes, was not involved in curli regulation in either of the conditions tested (Fig. 4e and f). RpoS plays a key role in curli expression in E. coli (33), but its role in Salmonella has not been completely elucidated (29). Conversely, the transcriptional regulators Hns and CpxR did affect our observed phenotype. Hns is a global regulator that has been shown to positively regulate csgDEFG (13, 29). Our results reflect this, as the activity of both promoters dropped approximately 1 log in this mutant background at early time points (Fig. 4g). Importantly, at later time points, csgBAC activity only diminished in human bile (2 logs or more) but stayed at WT levels in M9 plus glucose, suggesting that this transcriptional regulator could play an important role during exposure to human bile. CpxR, on the other hand, has been proposed to be a curli repressor (18, 34), but under our conditions, it had the opposite effect. Both curli promoters showed decreased activity in a cpxR mutant background at early time points (Fig. 4i). As the biofilm matured, the csgDEFG operon showed increased activity in glucose, reaching WT levels at day 3. The csgBAC promoter, on the other hand, showed a decrease activity under both conditions, especially in glucose (Fig. 4j).
How does human bile then activate curli gene expression, especially since the csgDEFG operon is still highly expressed in human bile in the absence of CsgD? The data obtained from the reporter plasmids in different mutant backgrounds could indicate that the transcriptional regulator Hns is involved in this phenotype. This of course could be the effect of decreased csgD expression and not necessarily direct involvement of Hns itself, although the fact that it affects the expression of the csgBAC promoter only in human bile could suggest otherwise. This work provides some clues as to what is causing this phenotype, be we do not yet have a definitive answer. We speculate that an unknown factor present in human bile activates expression through the action of an unknown curli transcriptional regulator. Future work will involve the testing of bile salts and other human bile components, fractionation of human bile, as well as heat/protease treatment of bile to begin to identify this activating substance.
In addition to fostering biofilm development, curli activation has been demonstrated to dampen the inflammatory response (35) and thus may aid in establishing chronic GB infection by helping to provide an environment conducive to bacterial growth and biofilm formation. Although curli expression in Salmonella and E. coli has been detected in the host (10, 11, 36–38), the subject is still hotly debated (8), as it had been thought to be expressed only at 28°C to 30°C (29, 39, 40). Our work could potentially point to a new avenue of study regarding the role of curli fimbriae in vivo, as past studies in Salmonella have not examined bile and biofilms as factors contributing to curli activation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. Typhimurium strains are listed in Table S4 in the supplemental material. Strains were streaked on Luria-Bertani (LB) agar plates and incubated at 37°C overnight. Single colonies were used to start overnight (O/N) liquid cultures. Planktonic cells were grown at 37°C on a rotating drum in LB or tryptic soy broth (TSB). Strains containing the green fluorescent protein (GFP) plasmid (pFPV25.1) or the luciferase reporter plasmid (pCS26) were supplemented with ampicillin (Amp; 100 μg ml−1) or kanamycin (Kan; 100 μg ml−1), respectively, to keep selective pressure on the plasmids.
Biofilm growth.
S. Typhimurium biofilms were grown on nontreated polystyrene 96-well plates (Corning, Kennebunkport, ME) by normalizing overnight (O/N) cultures grown in TSB to an optical density at 600 nm (OD600) of 0.8 and then diluting 1:10 into biofilm growth medium (M9 supplemented with glucose [10 mM], human bile [10%; collected under an IRB protocol approved by the University of Texas Health Science Center at San Antonio], mouse bile [2.5%], or ox bile [2.5%]) and dispensing 0.1 ml per well. The plates were incubated at 30°C in a GyroMini nutating mixer (LabNet International, Inc., NJ) at 24 rpm. To simulate gallstone (GS) conditions, wells containing human, mouse, and ox bile were precoated with cholesterol (Sigma-Aldrich, St. Louis, MO) by adding 5 mg/ml in a 1:1 solution of isopropanol-ethanol and air drying overnight (23). The medium was changed every other day. Salmonella biofilms were grown at different concentrations of human bile in order to choose the optimal conditions for RNA extractions (see Fig. S1). Biofilm growth was quantified using the crystal violet assay as previously described (23). A concentration of 10% was chosen because it resulted in similar growth to the control (M9 plus glucose). Mouse bile was more limited, and so a lower concentration was used. Ox bile concentrations for biofilm growth were previously optimized in our lab (data not shown).
RNA isolation.
Biofilms were grown under the above-outlined conditions for 7 days. The biofilm was washed with 100 μl phosphate-buffered saline (PBS) twice to remove planktonic cells, scraped from the surface of the plates with a 200-μl pipette tip, transferred to a microcentrifuge tube, and centrifuged at 10,000 × g for 3 min. RNA was extracted using the hot phenol method. Briefly, the supernatant was removed, and pellets were resuspended in 475 μl AE buffer (50 mM sodium acetate [NaOAc], 10 mM EDTA, pH 5.2). Forty microliters of 20% SDS and 475 μl of phenol were added and incubated for 10 min at 65°C, shaking every minute. Samples were put on ice for 5 min and centrifuged for 15 min at 10,000 rpm at 4°C. Supernatants were transferred to new microcentrifuge tubes and 475 μl of chloroform was added, mixed, and centrifuged for 10 min at 2,000 rpm at 4°C. The top aqueous layer was placed in a new microcentrifuge tube, and RNA was precipitated by adding 500 μl isopropanol and 50 μl sodium acetate (2 M), centrifuging for 20 min (12,000 rpm, 4°C), and washing with 250 μl 70% cold ethanol. Finally, samples were treated with DNase (12.5% of total reaction volume, catalog number M0303L; New England BioLabs, Ipswich, MA, USA) for 45 min.
RNA-Seq library construction and sequencing.
Total RNA quality was assessed using an Agilent 2100 bioanalyzer and RNA Nano Chip kit (Agilent Technologies, CA) to ensure that the RNA integrity number (RIN) was ≥7 and that there were no RIN value outliers. rRNA was removed from 1.5 μg of RNA with a Ribo-Zero rRNA removal kit for bacteria (Epicentre Biotechnologies, WI). To generate a directional signal in the RNA-Seq data, libraries were constructed from first-strand cDNA using the ScriptSeq v2 RNA-Seq library preparation kit (Epicentre Biotechnologies, WI). Briefly, 50 ng of rRNA-depleted RNA was fragmented and reverse transcribed using random primers containing a 5ʹ tagging sequence, followed by 3ʹ-end tagging with a terminus-tagging oligonucleotide to yield ditagged single-stranded cDNA. Following purification by a magnetic bead-based approach, the ditagged cDNA was amplified by limit-cycle PCR using primer pairs that anneal to tagging sequences and add adaptor sequences required for sequencing cluster generation. Amplified RNA-Seq libraries were purified using an AMPure XP System (Beckman Coulter). The quality of libraries was determined via an Agilent 2200 TapeStation using high sensitivity D1000 tape and quantified using a Kapa SYBR Fast qPCR kit (KAPA Biosystems, Inc, MA). One hundred fifty-base pair sequence reads were generated using the Illumina HiSeq 4000 platform.
RNA-Seq data analysis.
Each sample was aligned to the GCF_000022165.1 assembly of the Salmonella Typhimurium strain 14028S reference from NCBI (NC_016856.1) using version 0.7.5a of the BWAMEM aligner (41). Features were identified from the GFF file that came with the assembly from NCBI. Feature coverage counts were calculated using HTSeq (42). Differentially expressed features were calculated using DESeq2 (43) and custom scripts developed in-house to perform RNA-Seq analysis.
Statistical analysis.
Significant differentially expressed features between the two groups are those that have an absolute value of fold change of ≥1.5 and an adjusted P value of ≤0.10 (10% false discovery rate [FDR]).
qRT-PCR.
RNA was extracted as described above. Two hundred fifty nanograms of RNA was reversed transcribed into cDNA using a cDNA synthesis kit (Quantabio, Beverly, MA). Twenty nanograms of cDNA was added to IQ SYBR green PCR master mix (Bio-Rad, Hercules) containing 250 nM primers specific for csgA or rpoB (housekeeping gene) (see Table S5). Samples were run in triplicates and experiments were repeated three times using the Bio-Rad CFX96 iCycler apparatus. Relative copy numbers were calculated according to the threshold cycle (2−ΔΔCT) method (44) using Bio-Rad CFX Maestro software.
Western blot analysis.
CsgA was detected in whole-cell lysates using a polyclonal antisera generated against E. coli CsgA (gift of Matthew Chapman, University of Michigan). Western blotting was conducted as previously described (45). In brief, biofilms were grown for 1 week as outlined above, cells were scraped from plates and resuspended in 1 ml PBS (pH 7.4), and total protein was normalized by Bradford assay (Bio-Rad, Hercules, CA). Twenty micrograms of protein was pelleted, the supernatant was removed, and the pellet was resuspended in 70 μl of 88% formic acid (Sigma-Aldrich, St. Louis, MO). Samples were dried using a SpeedVac and resuspended in 20 μl H2O. Laemmli loading buffer was added to a final volume of 40 μl, and 10 μg of protein was loaded onto a 15% SDS-PAGE gel along with Bio-Rad Western C chemiluminescent molecular weight standard. Proteins were electrophoresed at 60 V until the dye front reached the bottom of the gel. Proteins in the gel were transferred for 2 h at 150 mA to a methanol (MeOH)-activated polyvinylidene difluoride (PVDF) membrane and blocked for 1 h in 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS) at room temperature. The membrane was incubated with anti-CsgA antibody (1:10,000 in 5% BSA-TBS with Tween 20 [TBST] overnight and 4°C), washed in TBST (5 washes, 10 min each), and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (1:12,500 in 5% BSA-TBST for 1 h, room temperature). The membrane was washed in TBST (5 washes, 10 min each) and visualized using the Bio-Rad (Hercules, CA) ChemiDoc system. Relative protein quantities were quantified using Image Lab software (Bio-Rad, Hercules, CA).
Confocal microscopy.
GFP-expressing strains (Table S2) were grown as biofilms as described above. After 7 days, the medium was removed, and biofilms were washed twice with KPi buffer (28.9 mM KH2PO4, 21.1 mM K2HPO4, pH 7.2), stained with Congo red (CR; 0.5 g/liter solution in KPi buffer) for 5 min, and then rinsed twice with KPi buffer (45). Biofilms were subsequently fixed in 2% paraformaldehyde (Affymetrix, Cleveland, OH) in PBS. Biofilms were later imaged in a Nikon A1R live cell inverted confocal microscope.
Generation of mutants and cloning procedures.
Mutations were performed by using the lambda Red mutagenesis method (46). Oligonucleotide primers used to perform gene deletion, cloning, and site-directed mutagenesis are listed in Table S5.
Luciferase reporter strain assays.
For bioluminescence assays, reporter plasmids were introduced into different S. Typhimurium strains (Table S1). The plasmids (kindly provided by Aaron White) are based on pCS26 and have the promoter regions of either the csgBAC or csgDEFG curli operon cloned upstream of the lux genes (47). Overnight cultures grown in TSB were diluted 1:100 in M9 supplemented with either 10 mM glucose (control) or 5% human bile to a final volume of 200 μl in 96-well clear-bottom black plates (9520, Costar; Corning, Tewksbury, MA). The plate was covered with a Breathe-Easy sealing membrane (Sigma-Aldrich, St. Louis, MO) prior to starting the assays. Cultures were assayed for luminescence (0.1 s) and absorbance (620 nm, 0.1 s) every 30 min during growth at 30°C in a Spectramax M3 (Molecular Devices, Sunnyvale, CA). Each condition had 4 replicate wells and each experiment was repeated at least 3 times.
Supplementary Material
ACKNOWLEDGMENTS
The work presented here was funded by grants AI109002 and AI116917 from the National Institutes of Health to J.S.G.
We thank Sara Cole at the Campus Microscopy and Imaging Facility (CMIF) in The Ohio State University for her help with microscopy experiments, Wayne Schwesinger at University of Texas HSC at San Antonio for the human bile samples, Aaron White at the University of Saskatchewan for the reporter plasmids, Matthew Chapman at the University of Michigan for the anti-CsgA antibody, and Yixin Shi at the Arizona State University for the cpxR and hns mutants.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00055-19.
REFERENCES
- 1.Crump JA, Mintz ED. 2010. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis 50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. 2002. Typhoid fever. N Engl J Med 347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 3.Prouty AM, Schwesinger WH, Gunn JS. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun 70:2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford RW, Rosales-Reyes R, Ramírez-Aguilar Mde L, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. 2010. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci U S A 107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Escobedo G, Marshall JM, Gunn JS. 2011. Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 9:9–14. doi: 10.1038/nrmicro2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Niu H, Wu S, Huang R. 2014. CsgD regulatory network in a bacterial trait-altering biofilm formation. Emerg Microbes Infect 3:1. doi: 10.1038/emi.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tursi SA, Tükel Ç. 2018. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications. Microbiol Mol Biol Rev 82:e00028-18. doi: 10.1128/MMBR.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adcox HE, Vasicek EM, Dwivedi V, Hoang KV, Turner J, Gunn JS. 2016. Salmonella extracellular matrix components influence biofilm formation and gallbladder colonization. Infect Immun 84:3243–3251. doi: 10.1128/IAI.00532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White AP, Gibson DL, Grassl GA, Kay WW, Finlay BB, Vallance BA, Surette MG. 2008. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect Immun 76:1048–1058. doi: 10.1128/IAI.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphries A, Deridder S, Bäumler AJ. 2005. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect Immun 73:5329–5338. doi: 10.1128/IAI.73.9.5329-5338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Römling U, Rohde M, Olsén A, Normark S, Reinköster J. 2000. AgfD the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol 36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 13.Gerstel U, Park C, Römling U. 2004. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol 49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- 14.Gunn JS. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect 2:907–913. doi: 10.1016/S1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 15.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 16.Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Römling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella Typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- 18.Jubelin G, Vianney A, Beloin C, Ghigo J-M, Lazzaroni J-C, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki T, Moschetta A, Lee Y-K, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. 2006. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaoz U, Murali TM, Letovsky S, Zheng Y, Ding C, Cantor CR, Kasif S. 2004. Whole-genome annotation by using evidence integration in functional-linkage networks. Proc Natl Acad Sci U S A 101:2888–2893. doi: 10.1073/pnas.0307326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Escobedo G, Gunn JS. 2013. Identification of Salmonella enterica serovar Typhimurium genes regulated during biofilm formation on cholesterol gallstone surfaces. Infect Immun 81:3770–3780. doi: 10.1128/IAI.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton S, Bongaerts RJ, Mulholland F, Cochrane B, Porter J, Lucchini S, Lappin-Scott HM, Hinton JC. 2009. The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genomics 10:599. doi: 10.1186/1471-2164-10-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford RW, Gibson DL, Kay WW, Gunn JS. 2008. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun 76:5341–5349. doi: 10.1128/IAI.00786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 25.Yee R, Cui P, Shi W, Feng J, Zhang Y. 2015. Genetic screen reveals the role of purine metabolism in Staphylococcus aureus persistence to rifampicin. Antibiotics (Basel) 4:627–642. doi: 10.3390/antibiotics4040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge X, Kitten T, Chen Z, Lee SP, Munro CL, Xu P. 2008. Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence. Infect Immun 76:2551–2559. doi: 10.1128/IAI.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JK, Jang HA, Won YJ, Kikuchi Y, Heum Han S, Kim CH, Nikoh N, Fukatsu T, Lee BL. 2014. Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J 8:552–563. doi: 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biesecker S, Nicastro L, Wilson R, Tükel Ç. 2018. The functional amyloid curli protects Escherichia coli against complement-mediated bactericidal activity. Biomolecules 8:5. doi: 10.3390/biom8010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerstel U, Römling U. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella Typhimurium. Res Microbiol 154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antunes LCM, Wang M, Andersen SK, Ferreira RBR, Kappelhoff R, Han J, Borchers CH, Finlay BB. 2012. Repression of Salmonella enterica phoP expression by small molecules from physiological bile. J Bacteriol 194:2286–2296. doi: 10.1128/JB.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol 18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x. [DOI] [PubMed] [Google Scholar]
- 33.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol 7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 34.Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol Lett 178:169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x. [DOI] [PubMed] [Google Scholar]
- 35.Oppong GO, Rapsinski GJ, Tursi SA, Biesecker SG, Klein-Szanto AJP, Goulian M, McCauley C, Healy C, Wilson RP, Tükel C. 2015. Biofilm-associated bacterial amyloids dampen inflammation in the gut: oral treatment with curli fibres reduces the severity of hapten-induced colitis in mice. NPJ Biofilms Microbiomes 1:15019. doi: 10.1038/npjbiofilms.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natanson C, Hoffman WD, Suffredini AF, Eichacker PQ, Danner RL. 1994. Selected treatment strategies for septic shock based on proposed mechanisms of pathogenesis. Ann Intern Med 120:771. doi: 10.7326/0003-4819-120-9-199405010-00009. [DOI] [PubMed] [Google Scholar]
- 37.Bian Z, Brauner A, Li Y, Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis 181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 38.Cordeiro MA, Werle CH, Milanez GP, Yano T. 2016. Curli fimbria: an Escherichia coli adhesin associated with human cystitis. Braz J Microbiol 47:414–416. doi: 10.1016/j.bjm.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnqvist A, Olsén A, Pfeifer J, Russell DG, Normark S. 2006. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol 6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 40.Olsén A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y, Smith DR, Hufnagel DA, Chapman MR. 2013. Experimental manipulation of the microbial functional amyloid called curli. Methods Mol Biol 966:53–75. doi: 10.1007/978-1-62703-245-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White AP, Gibson DL, Kim W, Kay WW, Surette MG. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol 188:3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.