Abstract
Pancreatic cancer (PC) is one of the most common forms of malignant tumors and causes of tumor-related death worldwide. The current prognosis of PC still remains poor due to the lack of effective early detection method. Recently, there is strong support that circulating miRNAs can be used as biomarkers for early detection of various cancers, including PC. The purpose of this review is to provide an overview of previous published studies on circulating miRNAs in plasma/serum for early detection of PC and summarize their diagnostic value. PubMed, Embase and Web of Science were systematically searched for eligible studies on circulating miRNAs for PC detection. Overall, 29 studies published between 2009 and 2018 evaluating 51 individual miRNAs (no P-value exceeding 0.05) and 13 miRNAs panels were included. Generally, the diagnostic performance of circulating miRNAs for PC detection was strong, with both the sensitivity and specificity of 36% individual miRNAs and 40% miRNAs panels exceeding 80%. Moreover, two promising miRNA panels were discovered and verified externally with all AUC values exceeding 0.95. Therefore, circulating miRNAs may hold potential to be used as noninvasive diagnostic biomarkers for PC, but large-scale studies are still needed to validate the promising miRNAs and optimize the miRNA panels. Since, the tremendous heterogeneity of studies in this field hampers translating miRNA markers into clinical practice, miRNA analytical procedures are also needed to be standardized in the future.
Keywords: pancreatic cancer, early detection, circulating microRNAs
Introduction
Pancreatic cancer (PC) is one of the most malignant tumors worldwide. The morbidity is projected to grow at a rate of 3% per year in males in the United States,1 and is predicted to become the second leading cause of total cancer-related death before 2030.2 Currently, radical resection is always the most effective curative option for patients with localized and regional PCs.3 However, most PC patients are diagnosed with major vascular invasion or distant metastasis when radical resection is usually not available.4 Consequently, early diagnosis and effective screening of high-risk populations for PC is a valid approach to improve prognosis. Traditional PC imaging tests have drawbacks that are often not suitable for PC screening: computed tomography (CT) has radiation exposure and a high false positive rate;5,6 magnetic resonance (MR) is expensive and prone to misdiagnosis because of its thicker scanning layer;7,8 endoscopic ultrasound (EUS) is generally less tolerant, has certain risks, and is limited by technical difficulties.9 In clinical, several serological biomarkers are widely used for PC diagnosis and prognosis evaluation, such as CA199, CA50, CEA, and CA242, but are usually negative in smaller pancreatic tumors, and show poor specificity for PC detection due to being overexpressed in many other diseases, such as gastroenteric tumors, bile duct cancer, and pancreatitis.10–13
In recent years, liquid biopsy based on microRNAs (miRNAs) has become a popular research field for the early diagnosis of malignancies. MicroRNAs are highly conserved, small noncoding RNA species of 17–25 nucleotides in length14 and remarkable stable in tissue, saliva, urine, serum, plasma, and exosomes.15 Approximately 50% of miRNAs are located in tumor-related regions.16 Aberrantly expressed miRNAs profiles were found in plasma/serum of PC patients and many PC-related circulating miRNA candidates/panels, have been identified for PC detection with high diagnostic efficiency. Several studies have even identified abnormally expressed exosomal miRNAs in plasma specimens of PC patients, suggesting that exosomal miRNA may also be useful for PC diagnosis.17–19 Two recent prospective studies20,21 demonstrated that the closer the recruitment time to PC occurrence, then the higher the diagnostic value of miRNAs, which offers evidence for circulating miRNAs as noninvasive diagnostic markers for early stage PC. The purpose of this systematic review is to provide an overview of published studies on circulating miRNAs for early detection of PC, and to summarize their diagnostic performance.
Methods
This review was implemented in accordance with a predefined protocol, and follows the PRISMA statement for systematic reviews and meta-analysis of priority reporting items.22
Literature search strategy
A systematic literature search was performed to identify studies assessing circulating miRNAs as biomarkers for detection of PC. We searched PubMed, ISI Web of Knowledge, and EMBASE databases for eligible articles until June 28, 2018. The combination keywords were as follows: ([pancreatic OR pancreas] AND [cancer OR carcinoma OR neoplasm OR tumor OR malignancy OR adenocarcinoma OR adenoma] AND [microRNA* OR miRNA* OR miR*] AND [detection OR diagnosis OR biomarker OR marker OR sensitivity OR specificity OR area under the curve] AND [blood OR serum OR plasma]). Duplicate publications were removed.
Eligibility criteria
Only articles written in English were included in this review. Non-original articles such as reviews and conference abstracts were excluded because of insufficient information reported regarding the diagnostic performance of miRNA markers. We required studies that reported relevant information on the diagnostic performance of miRNA markers for human PC detection as well as the sample sizes used in the studies. Studies using treated cases before sampling or disease controls were further excluded.
Data extraction and statistical analysis
Two investigators (EJ and HY) independently filtered the relevant studies against the above-mentioned criteria. Information on first author, publication year, country, sample size, mean or median age, male proportion, specimen type, PC stage, miRNA and/or miRNAs panels investigated, diagnostic related indicators (sensitivity, specificity, AUC), and P-value were extracted by the two investigators independently. MicroRNAs with P-value greater than 0.05 were ruled out. Any inconsistency was resolved by further review and discussion among the authors. MiRbase was used to check and unify the same miRNA with different names (http://www.mirbase.org/). Mean or median age, and male proportion of included studies were calculated using statistical software R (version 3.4.3, R Foundation, Vienna, Austria) if relevant information was not reported but raw data was available.
Quality assessment of the included studies
The two investigators independently assessed the quality of the included studies using QUADAS-2 (quality assessment tool for diagnostic accuracy studies)23 included in the Review Manager software (version 5.3.5, Cochrane Collaboration, Copenhagen, Denmark) package.24 QUADAS-2 is used to evaluate the risk level of bias, which mainly consists of four components: (1) patient selection; (2) index test; (3) reference standard; and (4) flow and timing. The first three components also evaluate clinical applicability. Based on the answers to signaling questions included in each component, the risk level of bias is judged as “low”, “high” or “unclear”, and the clinical applicability is judged as “low”, “high” or “unclear”. Any disagreement, such as inconsistent answers to the questions, was settled by further discussion between the two investigators.
Results
Literature search result
The initial literature search yielded 903 articles according to the aforementioned retrieval strategy (Figure 1). After removing 294 duplicates, we looked through the titles and abstracts of the remaining 609 articles and further excluded 557 articles based on the exclusion criteria. The remaining 52 articles went through full-text reading, of which 23 articles were excluded for the following reasons: 13 using disease controls, three recruiting treated cases before specimen collection, and seven studies not reporting sensitivity, specificity or AUC values. Finally, 29 studies17-21,25–48 published between 2009 and 2018 were eligible for inclusion in this systematic review, and used to evaluate the diagnostic performance of circulating miRNAs for PC.
Figure 1.
Overview of the literature search process (up to 28th of June 2018).
Study quality and characteristics
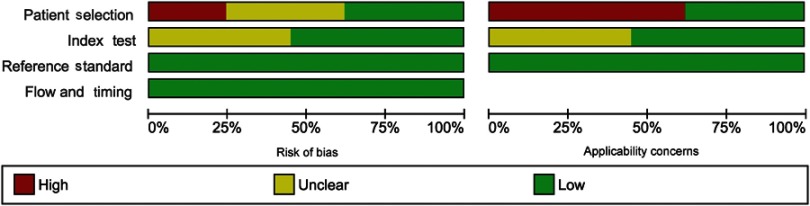
QUADAS-2 was carried out for the 29 included studies for quality assessment (Figures S1 and S2). High risk bias was found in seven studies (24%) in the patient selection domain, and unclear risk bias was found in 13 studies (45%) in the index test domain. For applicability concerns, 18 studies (62%) displayed high concerns in patient selection domain, and 13 studies (45%) displayed unclear concerns in the index test domain.
Of the 29 included studies, 18 were from East Asia,17,25–28,30–33,35,37–40,42,44,46,47 nine studies were from Europe and North America,18–21,34,41,43,45,48 one from Africa,29 and one from South America.36 The majority of the included studies were cross-sectional studies, and only two were nested case-control studies20,21 in which blood samples were taken before diagnosis. The median number (range) of included cases and controls was 56 (9–303) and 30 (6–600), respectively. Among the 29 included studies, four cross-sectional studies17,18,34,43 reported the diagnostic value of miRNAs for early stage (stage I and II) PC, and two nested case-control studies reported the predicted value of miRNAs for PC risk (Tables 1 and 2).
Table 1.
Diagnostic performance of miRNA markers in pancreatic cancer
| Study | Country | Cases vs Controls | Specimen | Stage | miRNA | SEN | SPE | AUC | P-value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age(y) | Male (%) | |||||||||
| Goto, 201817 | Japan | 32/22 | 64/58 | 53/64 | Exosomes | I-IV I-IIa II-IV |
miR-191 miR-21 miR-451a miR-191 miR-21 miR-451a miR-191 miR-21 miR-451a |
72 81 66 67 67 67 79 86 70 |
84 81 86 84 81 86 79 81 81 |
0.79 0.83 0.76 0.75 0.74 0.74 0.80 0.86 0.77 |
0.001 <0.001 0.002 0.032 0.004 0.044 0.001 <0.001 0.002 |
| Hua, 201730 | China | 103/50 | / | 60/NA | Serum | I-IV | miR-373 | 81 | 84 | 0.85 | / |
| Imamura, 201728 | Japan | 100/80 | / | 52/NA | Plasma | / | miR-107 | 82 | 69 | 0.85 | <0.0001a |
| Xu, Y, 201718 | USA | 15/15 | 67/48 | 53/28 | Exosomes | I-IIa | miR-1246 miR-196a miR-196b |
/ / / |
/ / / |
0.73 0.81 0.71 |
0.019 <0.001 0.033 |
| Yu, 201725 | China | 31/28 | 49/45 | 65/64 | Plasma | / | miR-21 | / | / | 0.85 | 0.000a |
| miR-210 | / | / | 0.69 | 0.013a | |||||||
| miR-155 | / | / | 0.82 | 0.002a | |||||||
| miR-196a | / | / | 0.79 | 0.000a | |||||||
| miR-20a | / | / | 0.88 | 0.000a | |||||||
| miR-25 | / | / | 0.76 | 0.000a | |||||||
| Qu, 201726 | China | 56/15 | 52/NA | 61/NA | Serum | I-IV | miR-21 | 77 | 80 | 0.78 | / |
| Li, F, 201727 | China | 87/48 | / | 58/NA | Plasma | I-IV | miR-221 | / | / | 0.69 | / |
| Lai, 201719 | USA | 29/6 | 67/NA | 52/NA | Plasma | / | miR-10b | 100 | 100 | 1.00 | <0.001 |
| miR-21 | 86 | 100 | 0.95 | <0.001 | |||||||
| miR-30c | 100 | 100 | 1.00 | <0.001 | |||||||
| miR-106b | 97 | 100 | 0.98 | <0.001 | |||||||
| miR-20a | 93 | 100 | 0.99 | <0.001 | |||||||
| miR-181a | 97 | 100 | 0.97 | <0.001 | |||||||
| miR-let7a | 93 | 100 | 0.99 | <0.001 | |||||||
| miR-122 | 100 | 67 | 0.89 | 0.003 | |||||||
| Exosomes | miR-10b | 100 | 100 | 1.00 | <0.001 | ||||||
| miR-21 | 100 | 100 | 1.00 | <0.001 | |||||||
| miR-30c | 100 | 100 | 1.00 | <0.001 | |||||||
| miR-106b | 62 | 100 | 0.85 | 0.007 | |||||||
| miR-20a | 83 | 100 | 0.95 | <0.001 | |||||||
| miR-181a | 100 | 100 | 1.00 | <0.001 | |||||||
| miR-let7a | 100 | 100 | 1.00 | <0.001 | |||||||
| miR-122 | 93 | 100 | 0.99 | <0.001 | |||||||
| Hussein, 201729 | Egypt | 35/15 | 57/41 | 40/27 | Plasma | Ib-IV | miR-22 | 97 | 93 | 0.94 | <0.001 |
| miR-642b | 100 | 100 | 1.00 | <0.001 | |||||||
| miR-885-5p | 100 | 100 | 1.00 | <0.001 | |||||||
| Duell, 2017b20 | Europec | 29/29 | / | / | Plasma | / | miR-10a | / | / | 0.75 | / |
| miR-10b | / | / | 0.76 | / | |||||||
| miR-21-3p | / | / | 0.74 | / | |||||||
| miR-21 | / | / | 0.79 | / | |||||||
| miR-30c | / | / | 0.77 | / | |||||||
| miR-106b | / | / | 0.74 | / | |||||||
| miR-155 | / | / | 0.74 | / | |||||||
| miR-212 | / | / | 0.73 | / | |||||||
| Yuan, 201631 | China | 82/88 | 59/59 | 57/49 | Plasma | I-IV | miR-21 | / | / | 0.81 | <0.001a |
| miR-25 | / | / | 0.66 | <0.001a | |||||||
| Sun, 201633 | China | 126/47 | 60/61 | / | Serum | I-IV | miR-124 | / | / | 0.98 | <0.001a |
| Xu, J, 201632 | China | 156/65 | / | 61/NA | Plasma | I-IV | miR-938 | 62 | 74 | 0.69 | <0.0001 |
| miR-126 | 62 | 60 | 0.62 | 0.0044 | |||||||
| miR-486 | 75 | 88 | 0.86 | <0.0001 | |||||||
| Deng, 201635 | China | 303/600 | 62/49 | 62/60 | Serum | I-IV | miR-25 | 76 | 93 | 0.92 | / |
| Alemar, 201636 | Brazil | 24/9 | 62/NA | 50/NA | Serum | Ia-IV | miR-21 | 83 | 78 | 0.89 | 0.001 |
| miR-34a | 91 | 78 | 0.87 | 0.002 | |||||||
| Miyamae, 201537 | Japan | 94/68 | / | 55/NA | Plasma | 0-IV | miR-744 | 59 | 90 | 0.83 | <0.0001a |
| Komatsu, 201538 | Japan | 71/67 | / | 58/NA | Plasma | / | miR-223 | 62 | 94 | 0.83 | <0.001a |
| Abue, 201539 | Japan | 32/30 | 71/45 | 69/37 | Plasma | I-IV | miR-483 | / | / | 0.75 | <0.0006a |
| miR-21 | / | / | 0.79 | <0.0001a | |||||||
| Zhang, 201440 | China | 70/40 | / | / | Serum | / | miR-194 | 54 | 58 | 0.57 | / |
| Slater, 201441 | Germany | 9/10 | / | NA/30 | Serum | I-IV | miR-196b | 100 | 78 | 0.86 | / |
| miR-196a | 90 | 89 | 0.97 | / | |||||||
| Lin, 201442 | China | 49/27 | 62/61 | 55/56 | Serum | I-IV | miR-663a | 86 | 80 | 0.87 | <0.05a |
| miR-492 | 76 | 70 | 0.79 | <0.05a | |||||||
| Ganepola, 201443 | USA | 11/11 | 68/46 | 54/54 | Plasma | IIa- IIb | miR-22 | 82 | 82 | 0.86 | 0.004 |
| miR-642b | 82 | 55 | 0.79 | 0.02 | |||||||
| miR-885-5p | 82 | 73 | 0.84 | 0.006 | |||||||
| Zhao, 201344 | China | 70/40 | / | 60/NA | Serum | I-IV | miR-192 | 76 | 55 | 0.63 | / |
| Li, A., 201345 | USA | 41/19 | 65/44 | 61/90 | Serum | I-III | miR-1290 | 88 | 84 | 0.96 | <0.001a |
| miR-744* | 68 | 53 | 0.69 | 0.0187a | |||||||
| miR-628 | 75 | 84 | 0.82 | <0.001a | |||||||
| miR-550 | 73 | 58 | 0.74 | 0.0022a | |||||||
| miR-1825 | 63 | 79 | 0.70 | 0.0012a | |||||||
| miR-24 | 73 | 68 | 0.79 | 0.0003a | |||||||
| miR-134 | 73 | 68 | 0.80 | 0.0002a | |||||||
| miR-146a | 78 | 79 | 0.82 | <0.001a | |||||||
| miR-378 | 76 | 79 | 0.81 | 0.0002a | |||||||
| miR-210 | 73 | 58 | 0.73 | 0.0052a | |||||||
| miR-22 | 71 | 79 | 0.73 | 0.005a | |||||||
| miR-625 | 63 | 53 | 0.66 | 0.0175a | |||||||
| miR-484 | 76 | 63 | 0.78 | 0.0005a | |||||||
| Liu, J., 201247 | China | 138/68 | 62/61 | 64/66 | Plasma | I-IV | miR-16 | / | / | 0.77 | 0.000 |
| miR-21 | / | / | 0.83 | 0.000 | |||||||
| miR-155 | / | / | 0.80 | 0.000 | |||||||
| miR-181a | / | / | 0.86 | 0.000 | |||||||
| miR-181b | / | / | 0.84 | 0.000 | |||||||
| miR-196a | / | / | 0.88 | 0.000 | |||||||
| miR-210 | / | / | 0.80 | 0.000 | |||||||
| Wang, 200948 | USA | 28/19 | / | / | Plasma | / | miR-21 | 46 | 89 | 0.62 | / |
| miR-210 | 42 | 73 | 0.65 | / | |||||||
| miR-155 | 53 | 78 | 0.67 | / | |||||||
| miR-196a | 43 | 84 | 0.69 | / | |||||||
Note: aP-value represents the difference of miRNA levels between cases and controls (all other P-values represent the statistical significance of AUC values; brepresents the incidence of Pancreatic Cancer by follow-up 5 years); c10 European countries including Denmark (Aarhus, Copenhagen), France, Germany (Heidelberg, Potsdam), Greece, Italy (Florence, Turin, Varese, Naples, Ragusa), The Netherlands (Bilthoven, Utrecht), Norway, Spain (Asturias, Granada, Murcia, Navarra, Guipuzcoa), Sweden (Malmo, Umeå) and the United Kingdom (Oxford, Cambridge); SENs, SPEs, and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation).
Abbreviations: SEN, sensitivity; SPE, specificity; AUC, area under the curve; NA, not available.
Table 2.
Diagnostic performance of miRNA panels in pancreatic cancer
| Study | County | Cases vs Controls | Specimen | Stage | miRNA | SEN | SPE | AUC | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Age(y) | Male (%) | ||||||||
| Franklin, 2018a21 | Sweden | 23/22 | 64/62 | 52/55 | Plasma | I-II | Panel A | / | / | 0.96 |
| Duell, 2017b20 | Europec | 29/29 | / | / | Plasma | / | Panel B | / | / | 0.73 |
| Johansen, 201634 | Denmark | 86/44 | 67/55 | 57/50 | Serum | I-IV | Panel C | 85 | 67 | 0.84 |
| Panel D | 85 | 71 | 0.72 | |||||||
| Denmark, Germany | 153/247 | / | / | Serum | I-II | Panel E | 86 | 68 | 0.83 | |
| Panel F | 83 | 73 | 0.86 | |||||||
| Panel C | 82 | 60 | 0.77 | |||||||
| Panel D | 86 | 73 | 0.87 | |||||||
| Denmark, Germany | 11/247 | / | / | Serum | I | Panel E | 73 | 68 | 0.70 | |
| Panel F | 55 | 73 | 0.76 | |||||||
| Panel C | 82 | 60 | 0.74 | |||||||
| Panel D | 64 | 73 | 0.78 | |||||||
| Denmark, Germany | 142/247 | / | / | Serum | II | Panel E | 87 | 68 | 0.84 | |
| Panel F | 85 | 73 | 0.87 | |||||||
| Panel C | 82 | 60 | 0.77 | |||||||
| Panel D | 88 | 73 | 0.88 | |||||||
| Alemar, 201636 | Brazil | 24/10 | 62/NA | 50/NA | Serum | I-IV | −21, −34a | / | / | 0.89 |
| Slater, 201441 | Germany | 9/10 | / | / | Serum | I-IV | −196b, −196a | 100 | 100 | 1.00 |
| Lin, 201442 | China | 49/27 | 62/61 | 55/56 | Serum | I-IV | −663a, −492 | 86 | 80 | 0.87 |
| Ganepola, 201443 | USA | 11/11 | 68/46 | 54/54 | Plasma | II | Panel G | 91 | 91 | 0.97 |
| Liu, R, 201246 | China | 95/81 | / | / | Serum | I-IV | Panel H | 94 | 93 | 0.99 |
| Liu, J, 201247 | China | 138/68 | 62/61 | 64/66 | Plasma | I-IV | −16, −196a | 87 | 74 | 0.90 |
| Wang, 200948 | USA | 28/19 | / | / | Plasma | / | Panel I | 64 | 89 | 0.82 |
Notes: aRepresents the study concluded prospective and cross-sectional study and the data extracted from the part of cross-sectional study; brepresents the incidence of Pancreatic Cancer by follow-up 5 years); c10 European countries including Denmark (Aarhus, Copenhagen), France, Germany (Heidelberg, Potsdam), Greece, Italy (Florence, Turin, Varese, Naples, Ragusa), The Netherlands (Bilthoven, Utrecht), Norway, Spain (Asturias, Granada, Murcia, Navarra, Guipuzcoa), Sweden (Malmo, Umeå) and the United Kingdom (Oxford, Cambridge); SENs, SPEs, and AUCs in bold fonts represent results from validation set (non-bold fonts represent results without validation). Panel A (15 miRs): −106b, −574, −34a, −451a, −130b, −26a, −144, −423, −101, −122,-24, −22-5p, let-7d-3p, −197, −885-5p; Panel B: −10a, −10b, −21, −30c, −106b, −155, −212; Panel C: −16, −27a, −25, −29c, −483-5p; Panel D (12 miRs): −16, −18a, −24, −27a, −30a, −323, −20a, −25, −29c,-191, −345, −483-5p; Panel E: −16, −27a, −30a, −323, −20a, −29c, −483-5p; Panel F: −16, −24, −27a, −30a, −323, −20a, −25, −29c, −483-5p; Panel G: −22, −642b, −885-5p; Panel H: −20a, −21, −24, −25, −99a, −185, −191; Panel I: −21, −210, −155, 196a.
Abbreviations: SEN, sensitivity; SPE, specificity; AUC, area under the curve; NA: not available.
Fifteen studies analyzed plasma samples for miRNA,17,19–21,25,27–29,31,32,37–39,43,47 12 studies analyzed serum samples for miRNA,26,30,33–36,40–42,44–46 and two studies additionally analyzed exosomes samples for miRNA.17,18 Twenty-six studies reported 51 individual miRNAs, 17–20,25–33,35–45,47,48 among which six studies carried out external validation.28,31,32,37,43,45 Ten studies reported 13 miRNAs panels,20,21,34,36,41–43,46–48 of which three studies performed external validation (Tables 1 and 2).34,43,46 All included studies used quantitative real-time polymerase chain reaction (qRT-PCR) to detect miRNAs concentrations. The miRNAs were isolated by different extraction kits among the included studies; six studies21,25,29,31,41,42 used miRNeasy Kit which has been proven to have a higher extraction efficiency.49 The normalization methods for the expression of miRNAs were not uniform, with cel-miR-39, U6 snRNA, miR-16 being the three most common reference standards for data normalization (Table S1).
Table S1.
Protocols of blood miRNA detection
| Ref | Specimen | Centrifugation | Extraction | Normalization |
|---|---|---|---|---|
| Yu, 20171 | Plasma | 1200g for 10min, 12000g for 10min | miRNeasy Serum/Plasma Kit | miRNeasy Serum/Plasma Spike-In Control (miR-39) |
| Qu, 20172 | Plasma | NA | TRIzol LS Reagent | cel-miR-39 |
| Li, 20173 | Plasma | 3500rpm for 10min | mirVana PARIS Kit | U6 snRNA |
| Lai, 20174 | Plasma | 1000g for 30min | NA | NA |
| Exosomes | 1000g for 30min, 10000g for 30min, (thaw)10000g for 30min, 11000g for 2h | Trizol-LS and Direct-zol RNA MiniPrep kit | miR-425-5p | |
| Hussein, 20175 | Plasma | NA | miRNeasy serum/plasma Kit | miR-3196 |
| Franklin, 20176 | Plasma | NA | miRNeasy Serum/Plasma Kit | NA |
| Duell, 20177 | Plasma | NA | Trizol-LS and Direct-zol RNA MiniPrep kit | miR-425-5p |
| Yuan, 20168 | Plasma | NA | miRNeasy Serum/Plasma Kit | cel-miR-39 |
| Xu, 20169 | Plasma | 3000rpm for 10min | mirVana PARIS kit | U6 snRNA |
| Sun, 201610 | Serum | NA | NA | U6 snRNA |
| Johansen, 201611 | Serum | 2500 g for 10 min | TRI Reagent BD | NA |
| Deng, 201612 | Serum | NA | NA | normalized to the serum volume |
| Alemar, 201613 | Serum | 1500rpm for 10 min | mirVana PARIS kit | cel-miR-39 |
| Miyamae, 201514 | Plasma | 1500rpm for 30min, 3000rpm for 5min, 4500rpm for 5min | mirVana PARIS Kit | cel-miR-39 |
| Komatsu, 201515 | Plasma | 1500rpm for 30min, 3000rpm for 5min, 4500rpm for 5min | mirVana PARIS Kit | cel-miR-39 |
| Abue, 201516 | Plasma | 3500rpm for 10min | mirVana PARIS kit | miR-16 |
| Zhang, 201417 | Serum | NA | mirVana PARIS kit | U6 snRNA |
| Slater, 201418 | Serum | NA | miRNeasy Kit | miR-24 |
| Lin, 201419 | Serum | 1500g for 10 min | miRNeasy Kit | cel-miR-39 |
| Ganepola, 201420 | Plasma | NA | TRI Reagent BD | miR-3196 |
| Zhao, 201321 | Serum | NA | mirVana PARIS kit | U6 snRNA |
| Li, 201322 | Serum | NA | mirVana PARIS kit | miR-16 |
| Liu, R., 201223 | Serum | 800g for 10min, 10000g for 15min, 12000g for 10min, 16000g for 20min | TRIzol Reagent | normalized to the serum volume |
| Liu, J., 201224 | Plasma | 1200g for 10min, 12000g for 10min | TRI Reagent BD | cel-miR-39 |
| Wang, 200925 | Plasma | 1300g for 10min, 12000g for 30min | Trizol LS reagent | miR-16 |
| Goto, 201826 | Exosomes | 5000g for 10min | Trizol kit | normalized to the serum volume |
| Hua, 201727 | Serum | 3500rpm for 10min | mirVana PARIS kit | U6 snRNA |
| Imamura, 201728 | Plasma | 1500rpm for 30min, 3000rpm for 5min, 4500rpm for 5min | mirVana PARIS kit | cel-miR-39 |
| Xu, Y., 201729 | Exosomes | 2000rpm for 15min, 10000g for 30min, 10000g for 1h | Trizol reagent | cel-miR-54 |
Diagnostic efficiency of miRNAs
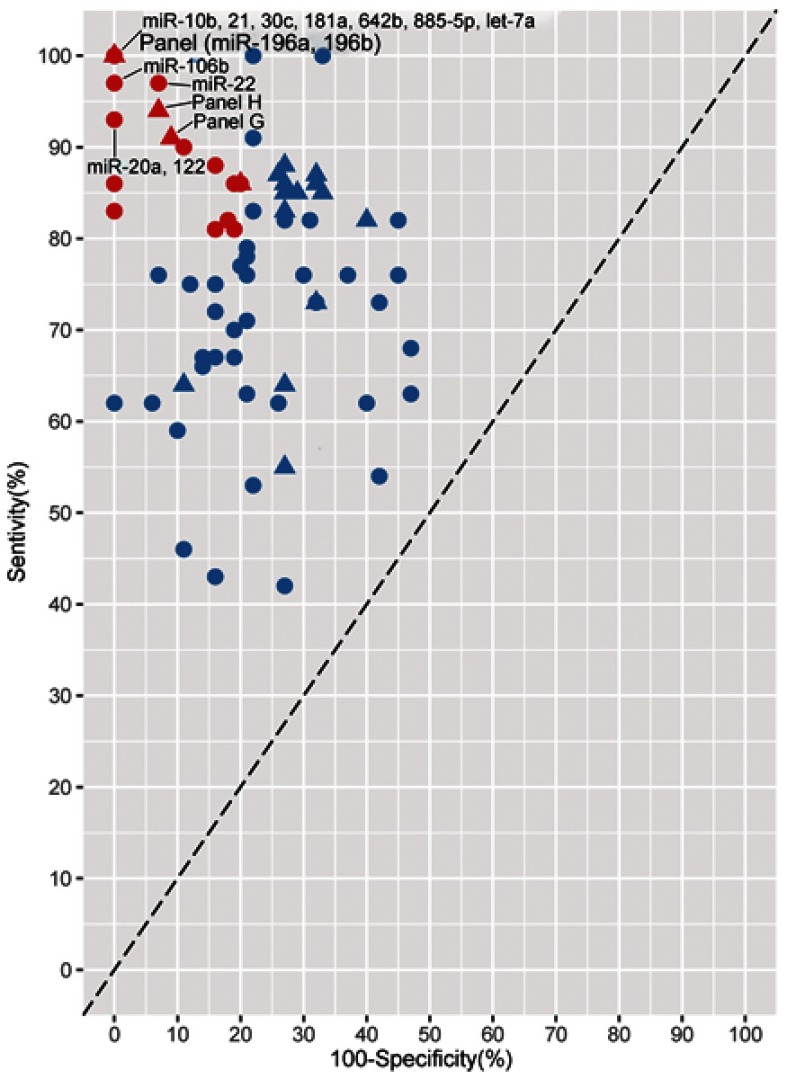
The 29 included studies reported a total number of 68 miRNAs with the diagnostic potential for PC, of which, 21 miRNAs were reported in more than two studies. The reported miRNA panels for PC diagnosis contained the number of miRNAs from 2 to 15, with 10 miRNAs appearing in at least two panels (Table S2). Among studies with reported sensitivity and specificity, both exceeded 80% among 14 individual miRNAs (36%) and 4 miRNAs panels (40%) (Figure 2). Twenty-three individual miRNAs and four miRNA panels were externally validated, and diagnostic performance with ≥0.70 AUC was observed in 18 miRNAs and all the four miRNA panels (Figure 3). MiR-21 is the most frequently reported miRNA (Table 3), whose sensitivity ranged from 46% to 100% (median sensitivity 78%), the specificity ranged from 78% to 100% (median specificity 86%), and the AUC values ranged from 0.62 to 1.00 (median AUC value =0.83). In the study by Lai et al.,19 the sensitivity and specificity of miR-21, miR-10b, miR-30c, miR-181a, and miR-let7a in exosomes all reached 100% (Table 1). Several miRNAs panels showed excellent diagnostic performance for PC;41,46 the AUC values of 7-miRNA panel (miR-20a, −21, −24, −25, −99a, −185, and −191) in Liu R’s study and 2-miRNA panel (miR-196a and −196b) in Slater’s study were 0.99, and 1.00, respectively (Table 2).
Table S2.
Summary of studies reporting diagnostic performance of miRNAs in miRNA panels with pancreatic cancer (only miRNAs that have been reported in ≥2 panels)
| miRNA | Franklin, 2018 6 | Duell, 2017 7 | Johansen, 2016 11 | Alemar, 2016 13 | Slater, 2014 18 | Ganepola, 2014 20 | Liu.R, 2012 23 | Liu.J.Q, 2012 24 | Wang, 2009 25 | Number Of Studies |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-21 | ○↑ | ○↑ | ○↑ | ○↑ | 4 | |||||
| miR-24 | ○↑ | ○- | ○↑ | 3 | ||||||
| miR-196a | ○↑ | ○↑ | ○↑ | 3 | ||||||
| miR-106b | ○↓ | ○↑ | 2 | |||||||
| miR-25 | ○- | ○↑ | 2 | |||||||
| miR-155 | ○↑ | ○↑ | 2 | |||||||
| miR-34a | ○↑ | ○↑ | 2 | |||||||
| miR-191 | ○ | ○↑ | 2 | |||||||
| miR-20a | ○- | ○↑ | 2 | |||||||
| miR-885-5p | ○↑ | ○↑ | 2 |
Notes: ○ represents miRNAs which are part of a panel; ↑ represents upregulation; ↓ represents down-regulation; - represents no difference in overall study population.
Figure 2.
Graphical representation of sensitivity vs specificity of analyzed miRNAs. Sensitivity is plotted on the y-axis while on the x-axis the false-positive rate is presented (100-Specificity). ○ miRNA individual; △ miRNA panel. Plots in red color represent miRNAs or miRNA panels with ≥80% sensitivity and ≥80 specificity. (G): −22, −642b, −885-5p; (H): −20a, −21, −24, −25, −99a, −185, −191.
Figure 3.
Graphical representation of diagnostic performance of the externally validated miRNAs and miRNA panels in PC. (C): −16, −27a, −25, −29c, −483-5p; (D) (12 miRs): −16, −18a, −24, −27a, −30a, −323, −20a, −25, −29c,-191, −345, −483-5p; (G): −22, −642b, −885-5p; (H): −20a, −21, −24, −25, −99a, −185, −191.
Abbreviations: AUC, area under the curve; PC, pancreatic cancer.
Table 3.
Summary of studies reporting diagnostic performance of miRNAs for pancreatic cancer (Only miRNAs that have been reported in ≥2 studies)
| miRNA | Goto, 201817 | Xu, Y., 201718 | Yu, 201725 | Qu, 201726 | Lai, 201719 | Hussein, 201729 | Franklin, 201821 | Duell, 201720 | Yuan, 201631 | Johansen, 201634 | Deng, 201635 | Alemar, 201636 | Abue, 201539 | Slater, 201441 | Ganepola, 201443 | Li, A, 201345 | Liu, R, 201246 | Liu, J, 201247 | Wang, 200948 | Number Of Studies |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-21 | △↑ | △↑ | △ | △↑ | △↑ | △↑ | ○↑ | △↑ | ○↑ | △↑ | ○↑ | 11 | ||||||||
| miR-196a | △↑ | △↑ | △ | ○↑ | ○↑ | ○↑ | 6 | |||||||||||||
| miR-25 | △↑ | △↑ | ○- | △↑ | ○↑ | 5 | ||||||||||||||
| miR-155 | △↑ | △↑ | △ | △ | ○↑ | 5 | ||||||||||||||
| miR-24 | ○↑ | ○- | △↑ | ○↑ | 4 | |||||||||||||||
| miR-210 | △↑ | △↑ | △↑ | ○↑ | 4 | |||||||||||||||
| miR-20a | △↑ | △↑ | ○- | ○↑ | 4 | |||||||||||||||
| miR-885-5p | △↑ | ○↑ | ○↑ | 3 | ||||||||||||||||
| miR-106b | △↑ | ○↓ | △↑ | 3 | ||||||||||||||||
| miR-22 | △↑ | ○↑ | △↑ | 3 | ||||||||||||||||
| miR-191 | △↑ | ○ | ○↑ | 3 | ||||||||||||||||
| miR-34a | ○↑ | ○↑ | 2 | |||||||||||||||||
| miR-642b | △↑ | ○↑ | 2 | |||||||||||||||||
| miR-483 | △ | △↑ | 2 | |||||||||||||||||
| miR-181a | △↑ | △↑ | 2 | |||||||||||||||||
| miR-30c | △↑ | △↑ | 2 | |||||||||||||||||
| miR-10b | △↑ | △↑ | 2 | |||||||||||||||||
| miR-122 | △↓ | ○↑ | 2 | |||||||||||||||||
| miR-16 | ○- | ○↑ | 2 | |||||||||||||||||
| miR-451a | △↑ | ○↓ | 2 | |||||||||||||||||
| miR-196b | △ | ○↑ | 2 |
Notes: ○ represents miRNAs which are part of a panel; △ represents miRNAs which have only been analyzed individually and not as a part of a miRNA panel; ↑ represents upregulation; ↓ represents down-regulation; -represents no difference in overall study population.
For early stage of PC, miR-20a, miR-21, miR-24, miR-25, miR-99a, miR-185, and miR-191 were significantly dysregulated in serum samples of stage I (26 cases) and II (48 cases) PC patients compared to healthy controls in Liu R et al.’s study,46 with positive detection rates of 96% and 91.7%, respectively. Johansen et al.34 evaluated the diagnostic efficiency of four miRNAs panels for stage I and II PC (Table 2), and the results showed AUC values of 0.87, 0.86, 0.77, and 0.83. Ganepola et al.43 investigated a 3-miRNA panel (miR-22, −642b, and −885–5p) for stage II PC and found the sensitivity, specificity, and AUC value were 91%, 91%, and 0.97, respectively. A nested case-control study by Duell et al.20 explored the risk prediction value of a 7-miRNA panel (miR-10a, −10b, −21, −30c, −106b, −155, and −212) in plasma for PC occurring in ≤5 years, ≤8 years, and ≤12 years, and the results of which showed that the AUC values were 0.73, 0.70, and 0.69, respectively. More recently, Franklin et al.21 conducted a study which contained both prospective and cross-sectional (PC stage: I-II) parts. The prospective part indicated that the AUC values of a 15-miRNA panel (miR-106b, −574, −34a, −451a, −130b, −26a, −144, −423, −101, −122, −24, −22-5p, let-7d-3p, −197, and −885-5p) for predicting PC occurring in ≤5 years, 5–10 years, and >10 years were 0.60, 0.55, and 0.65, respectively. The cross-sectional part reported that the AUC value of the above-mentioned miRNA panel for PC at diagnosis was 0.96.
Regulation direction of PC-related miRNA
Of the 21 miRNAs reported more than twice, the dysregulation direction of most miRNAs was consistently upregulated, but the dysregulation direction of three miRNAs (miR-106b, miR-122, and miR-451a) was inconsistent (Table 3). Of which, miR-106b was found to be upregulated in two studies19,20 and downregulated in one study;21 miR-122 and miR-451a were reported upregulated in one study17,21 and downregulated in another study,19,21 respectively. The inconsistent dysregulation direction of the above-mentioned miRNAs was not found to be significantly related to the specimen types or the stage of PC.17,19–21
Discussion
Our systematic review identified a total number of 68 miRNAs from 29 eligible studies evaluating the diagnostic performance of circulating miRNA for PC detection. Ten studies integrated individual miRNAs into miRNA panels (2–15 miRNAs for each panel) (Table 2). Two promising miRNA panels were discovered and verified in two cross-sectional studies,43,46 with AUC values all exceeding 0.95. Only two studies17,34 conducted PC stage subgroup analysis for the diagnostic performance of miRNA. However, due to the lack of sufficient data, stage-specific miRNA for PC is still elusive.
Overall, circulating miRNAs present strong diagnostic value for PC with the sum of sensitivity and specificity of all reported miRNAs or miRNA panels being greater than one (Figure 2). Sensitivity and specificity both exceeded 80% in 36% of individual miRNAs and 40% of miRNA panels (Figure 2). Eleven miRNAs and three panels marked in Figure 2 showed even better diagnostic performance for PC with ≥90% sensitivity and ≥90% specificity. Ganepola et al.43 used a panel composed of miR-22, miR-642b-3p, and miR-885-5 in plasma for the diagnosis of stage II PC, and the AUC value reached 0.97. Another study by Liu R et al.46 used a panel consisting of miR-20a, miR-21, miR-24, miR-25, miR-99a, miR-185, and miR-191 in serum for the diagnosis of stage I-IV PCs, and the AUC value reached 0.99. Moreover, the abovementioned results of the two studies have been externally verified (Figure 3). Two nested case-control studies20,21 showed that circulating miRNAs had certain predictive value for PC occurring in 5 years before diagnosis, but the performance in the PC-free participants is significantly lower compared to the participants being diagnosed with PC. The sample sizes were small in most included studies, and few studies conducted external validation, so the possibility of overestimation cannot be ruled out. Hence, further validation is still indispensable, especially based on large scale PC screening studies.
Some benign diseases and treatment measures may affect the identification of circulating miRNAs. Expression profiles of circulating miRNAs in chronic pancreatitis are different from that of PC, but approximately 4% of chronic pancreatitis cases can develop PC within 20 years.50 Some studies51-53 have demonstrated that antineoplastic drugs and chemical regulators could regulate cell proliferation, apoptosis, and angiogenesis, all of which may impact miRNAs expression profiles. Therefore, in order to avoid the effect of disease and treatment on miRNA concentration, we only included healthy controls and PC cases sampled before any therapy.54–56
The overlap rates of PC-specific miRNAs are low in the current literature reports, and sometimes the regulation expression of the same miRNA in different studies was inconsistent.17,19–21 Consequently, screening of circulating miRNAs for PC detection requires attention. Circulating miRNAs concentration could be influenced by many factors, including: (1) population differences;57 (2) specimen types and volume;58–60 (3) specimen preservation methods and time;61 (4) centrifugation steps;58 (5) miRNA extraction kits;62 (6) normalization methods.58,63 The concentration of intracellular miRNAs is higher than that of extracellular miRNAs in blood, so hemolysis can cause a release of intracellular miRNAs, which may contaminate extracellular miRNAs, and affect the identification of PC-specific miRNAs.64–66 In addition, the blood samples in some studies18,19,34 were processed with only one-step centrifugation (Table S1), so the residual cell debris, containing high concentration miRNAs, may remain in the supernatant and contribute to the total miRNA content. At present, two-step centrifugation procedure is recommended, and the second step requires high-speed with a centrifugal force of >15000 g to remove maximal cell debris to reduce their effect on the quantification of miRNAs in plasma and serum.58,59,67 The miRNeasy kit is recommended as it has a higher miRNAs extraction efficiency compared to other kits,49 but not all studies have applied this extraction kit (Table S1). Different normalization methods could also influence the final quantitative results of circulating miRNAs and could even affect miRNAs expression regulation.58,68–70 Currently, qPCR quantitative standardization methods of miRNAs concentration are not uniform; cel-miR-39, U6 snRNA, and miR-16 are the most used standardization references in the included studies. The concentration of molecules used as the reference should be very stable among individuals, but there are still some references whose concentration varies between cancer cases and healthy controls, and result in a detection bias of miRNA concentration.58,69,71–74
Compared with other types of blood-based markers for PC detection, circulating miRNAs have the following advantages: (1) miRNAs are relatively stable and are insensitive to ribonuclease, acid or alkali environment, long-term room temperature preservation, and repeated freeze-thaw;68,75 (2) it can be repeatedly used as a non-invasive detection method;76,77 (3) it has certain predictive value for high PC risk population;20,21 and (4) the detection of miRNAs is relatively cheap. Other blood markers currently being used to diagnose PC - eg CA199, CA50, and CA242 - are often used to monitor the disease progression,78,79 but their diagnostic value is relatively low (whose sensitivity and specificity are generally lower than 81% and 80%, respectively).80–82 In recent years, circulating tumor DNA (ctDNA), as a novel diagnostic marker for PC, has also shown pretty good diagnostic value, the specificity of which can reach 92.6% or even exceed 99.9% in some studies,83,84 but the sensitivity is usually lower than 75%.84 In addition to identifying more circulating miRNAs for the formation of diagnostically superior miRNA panels for PC, future research should also focus on exploring possibilities of enhancing diagnostic power by combining miRNA makers with other novel laboratory markers, such as ctDNA markers, in a diagnostic model for early detection of PC.
Conclusion
This review indicates that circulating miRNAs hold the potential of being applied as diagnostic markers for PC. Future studies should pay more attention to the standardization of samples processing procedures and miRNA detection protocol. It is also necessary to verify these PC-specific miRNAs in larger scale screening studies, and examine the diagnostic efficiency of circulating miRNA for early stage PC.
Supplementary materials

References
- 1.Yu Q, Xu C, Yuan W, et al. Evaluation of plasma MicroRNAs as diagnostic and prognostic biomarkers in pancreatic adenocarcinoma: miR-196a and miR-210 Could be negative and positive prognostic markers, respectively. Biomed Res Int. 2017;2017:6495867. doi: 10.1155/2017/6495867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu K, Zhang X, Lin T, et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7(1):1692. doi: 10.1038/s41598-017-01904-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, Xu JW, Wang L, Liu H, Yan Y, Hu SY MicroRNA-221-3p is up-regulated and serves as a potential biomarker in pancreatic cancer. Artif Cells Nanomed Biotechnol. 2018;46(3):482–487. doi: 10.1080/21691401.2017.1315429 [DOI] [PubMed] [Google Scholar]
- 4.Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86–93. doi: 10.1016/j.canlet.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussein NA, Kholy ZA, Anwar MM, Ahmad MA, Ahmad SM Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin Oncol. 2017;143(1):83–93. doi: 10.1007/s00432-016-2248-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin O, Jonsson P, Billing O, et al. Plasma micro-RNA alterations appear late in pancreatic cancer. Ann Surg. 2018;267(4):775–781. doi: 10.1097/SLA.0000000000002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duell EJ, Lujan-Barroso L, Sala N, et al. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141(5):905–915. doi: 10.1002/ijc.30790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan W, Tang W, Xie Y, et al. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7(48):80033–80045. doi: 10.18632/oncotarget.12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Cao Z, Liu W, et al. Plasma miRNAs effectively distinguish patients with pancreatic cancer from controls: a multicenter study. Ann Surg. 2016;263(6):1173–1179. [DOI] [PubMed] [Google Scholar]
- 10.Sun B, Liu X, Gao Y, Li L, Dong Z Downregulation of miR-124 predicts poor prognosis in pancreatic ductal adenocarcinoma patients. Br J Biomed Sci. 2016;73(4):152–157. doi: 10.1080/09674845.2016.1220706 [DOI] [PubMed] [Google Scholar]
- 11.Johansen JS, Calatayud D, Albieri V, et al. The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int J Cancer. 2016;139(10):2312–2324. doi: 10.1002/ijc.30291 [DOI] [PubMed] [Google Scholar]
- 12.Deng T, Yuan Y, Zhang C, et al. Identification of circulating MiR-25 as a potential biomarker for pancreatic cancer diagnosis. Cell Physiol Biochem. 2016;39(5):1716–1722. doi: 10.1159/000447872 [DOI] [PubMed] [Google Scholar]
- 13.Alemar B, Izetti P, Gregorio C, et al. miRNA-21 and miRNA-34a are potential minimally invasive biomarkers for the diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2016;45(1):84–92. doi: 10.1097/MPA.0000000000000383 [DOI] [PubMed] [Google Scholar]
- 14.Miyamae M, Komatsu S, Ichikawa D, et al. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113(10):1467–1476. doi: 10.1038/bjc.2015.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komatsu S, Ichikawa D, Miyamae M, et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin Biol Ther. 2015;15(6):773–785. doi: 10.1517/14712598.2015.1029914 [DOI] [PubMed] [Google Scholar]
- 16.Abue M, Yokoyama M, Shibuya R, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46(2):539–547. doi: 10.3892/ijo.2014.2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Zhao CY, Zhang SH, et al. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Rep. 2014;31(3):1157–1164. doi: 10.3892/or.2013.2960 [DOI] [PubMed] [Google Scholar]
- 18.Slater EP, Strauch K, Rospleszcz S, et al. MicroRNA-196a and-196b as potential biomarkers for the early detection of familial pancreatic cancer. Transl Oncol. 2014;7(4):464–471. doi: 10.1016/j.tranon.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin MS, Chen WC, Huang JX, Gao HJ, Sheng HH Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int J Clin Exp Med. 2014;7(12):5226–5234. [PMC free article] [PubMed] [Google Scholar]
- 20.Ganepola GAP, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastro Oncol. 2014;6(1):22–33. doi: 10.4251/wjgo.v6.i1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao C, Zhang J, Zhang S, et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol Rep. 2013;30(1):276–284. doi: 10.3892/or.2013.2420 [DOI] [PubMed] [Google Scholar]
- 22.Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19(13):3600–3610. doi: 10.1158/1078-0432.CCR-12-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Chen X, Du YQ, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–618. doi: 10.1373/clinchem.2011.172767 [DOI] [PubMed] [Google Scholar]
- 24.Liu JQ, Gao J, Du YQ, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422 [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Chen JY, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto T, Fujiya M, Konishi H, et al. An elevated expression of serum exosomal microRNA-191, - 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18(1):116. doi: 10.1186/s12885-018-4242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua Y, Chen H, Wang L, et al. Low serum miR-373 predicts poor prognosis in patients with pancreatic cancer. Cancer Biomarkers. 2017;20(1):95–100. doi: 10.3233/CBM-170231 [DOI] [PubMed] [Google Scholar]
- 28.Imamura T, Komatsu S, Ichikawa D, et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7(1):5708. doi: 10.1038/s41598-017-06137-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu YF, Hannafon BN, Zhao YD, Postier RG, Ding WQ Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget. 2017;8(44):77028–77040. doi: 10.18632/oncotarget.20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T, Wood LD, Itoi T, Takaori K Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi: 10.1016/S0140-6736(16)00141-0 [DOI] [PubMed] [Google Scholar]
- 4.Gillen S, Schuster T, Meyer Zum Buschenfelde C, Friess H, Kleeff J Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010;7(4):e1000267. doi: 10.1371/journal.pmed.1000267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prokesch RW, Chow LC, Beaulieu CF, Bammer R, Jeffrey RB Jr. Isoattenuating pancreatic adenocarcinoma at multi-detector row CT: secondary signs. Radiology. 2002;224(3):764–768. doi: 10.1148/radiol.2243011284 [DOI] [PubMed] [Google Scholar]
- 6.To’o KJ, Raman SS, Yu NC, et al. Pancreatic and peripancreatic diseases mimicking primary pancreatic neoplasia. Radiographics. 2005;25(4):949–965. doi: 10.1148/rg.254045167 [DOI] [PubMed] [Google Scholar]
- 7.Chu LC, Goggins MG, Fishman EK Diagnosis and detection of pancreatic cancer. Cancer J. 2017;23(6):333–342. doi: 10.1097/PPO.0000000000000290 [DOI] [PubMed] [Google Scholar]
- 8.Chen FM, Ni JM, Zhang ZY, Zhang L, Li B, Jiang CJ Presurgical evaluation of pancreatic cancer: a comprehensive imaging comparison of CT versus MRI. AJR Am J Roentgenol. 2016;206(3):526–535. doi: 10.2214/AJR.15.15236 [DOI] [PubMed] [Google Scholar]
- 9.Wiersema MJ, Vilmann P, Giovannini M, Chang KJ, Wiersema LM Endosonography-guided fine-needle aspiration biopsy: diagnostic accuracy and complication assessment. Gastroenterology. 1997;112(4):1087–1095. doi: 10.1016/s0016-5085(97)70164-1 [DOI] [PubMed] [Google Scholar]
- 10.Satake K, Kanazawa G, Kho I, Chung Y, Umeyama K Evaluation of serum pancreatic enzymes, carbohydrate antigen 19-9, and carcinoembryonic antigen in various pancreatic diseases. Am J Gastroenterol. 1985;80(8):630–636. [PubMed] [Google Scholar]
- 11.Ballehaninna UK, Chamberlain RS Biomarkers for pancreatic cancer: promising new markers and options beyond CA 19-9. Tumour Biol. 2013;34(6):3279–3292. doi: 10.1007/s13277-013-1033-3 [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Wang X, Yu F, et al. Combined detection of preoperative serum CEA, CA19-9 and CA242 improve prognostic prediction of surgically treated colorectal cancer patients. Int J Clin Exp Pathol. 2015;8(11):14853–14863. [PMC free article] [PubMed] [Google Scholar]
- 13.Takezako Y, Okusaka T, Ueno H, Ikeda M, Morizane C, Najima M [Tumor markers for pancreatic and biliary tract cancer]. Gan to Kagaku Ryoho Cancer Chemother. 2004;31(9):1443–1446. [PubMed] [Google Scholar]
- 14.Iorio MV, Croce CM MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2017;9(6):852. doi: 10.15252/emmm.201707779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 17.Goto T, Fujiya M, Konishi H, et al. An elevated expression of serum exosomal microRNA-191, - 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18(1):116. doi: 10.1186/s12885-018-4242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu YF, Hannafon BN, Zhao YD, Postier RG, Ding WQ Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget. 2017;8(44):77028–77040. doi: 10.18632/oncotarget.20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86–93. doi: 10.1016/j.canlet.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duell EJ, Lujan-Barroso L, Sala N, et al. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017. doi: 10.1002/ijc.30790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franklin O, Jonsson P, Billing O, et al. Plasma Micro-RNA Alterations Appear Late in Pancreatic Cancer. Annals of Surgery. 2018;267(4):775–781. doi: 10.1097/sla.0000000000002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–1012. doi: 10.1016/j.jclinepi.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 23.Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–U104. doi: 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 24.Bae JM An overview of systematic reviews of diagnostic tests accuracy. Epidemiol Health. 2014;36:e2014016. doi: 10.4178/epih/e2014016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Q, Xu CQ, Yuan W, et al. Evaluation of plasma MicroRNAs as diagnostic and prognostic biomarkers in pancreatic adenocarcinoma: miR-196a and miR-210 could be negative and positive prognostic markers, respectively. Biomed Res Int. 2017. doi: 10.1155/2017/6495867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu K, Zhang X, Lin T, et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7(1):1692. doi: 10.1038/s41598-017-01904-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Xu JW, Wang L, et al. MicroRNA-221-3p is up-regulated and serves as a potential biomarker in pancreatic cancer. Artif Cells Nanomed Biotechnol. 2017;46(3):482–487. doi: 10.1080/21691401.2017.1315429 [DOI] [PubMed] [Google Scholar]
- 28.Imamura T, Komatsu S, Ichikawa D, et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7(1):5708. doi: 10.1038/s41598-017-06137-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussein NA, El Kholy ZA, Anwar MM, Ahmad MA, Ahmad SM Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin Oncol. 2017;143(1):83–93. doi: 10.1007/s00432-016-2248-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua Y, Chen H, Wang L, et al. Low serum miR-373 predicts poor prognosis in patients with pancreatic cancer. Cancer Biomarkers. 2017;20(1):95–100. doi: 10.3233/CBM-170231 [DOI] [PubMed] [Google Scholar]
- 31.Yuan W, Tang W, Xie Y, et al. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7(48):80033–80045. doi: 10.18632/oncotarget.12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Cao Z, Liu W, et al. Plasma miRNAs effectively distinguish patients with pancreatic cancer from controls a multicenter study. Ann Surg. 2016;263(6):1173–1179. [DOI] [PubMed] [Google Scholar]
- 33.Sun B, Liu X, Gao Y, Li L, Dong Z Downregulation of miR-124 predicts poor prognosis in pancreatic ductal adenocarcinoma patients. Br J Biomed Sci. 2016;73(4):152–157. doi: 10.1080/09674845.2016.1220706 [DOI] [PubMed] [Google Scholar]
- 34.Johansen JS, Calatayud D, Albieri V, et al. The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int J Cancer. 2016;139(10):2312–2324. doi: 10.1002/ijc.30291 [DOI] [PubMed] [Google Scholar]
- 35.Deng T, Yuan YZ, Zhang CN, et al. Identification of circulating MiR-25 as a potential biomarker for pancreatic cancer diagnosis. Cell Physiol Biochem. 2016;39(5):1716–1722. doi: 10.1159/000447872 [DOI] [PubMed] [Google Scholar]
- 36.Alemar B, Izetti P, Gregorio C, et al. miRNA-21 and miRNA-34a are potential minimally invasive biomarkers for the diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2016;45(1):84–92. doi: 10.1097/MPA.0000000000000383 [DOI] [PubMed] [Google Scholar]
- 37.Miyamae M, Komatsu S, Ichikawa D, et al. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113(10):1467–1476. doi: 10.1038/bjc.2015.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu S, Ichikawa D, Miyamae M, et al. Malignant potential in pancreatic neoplasm: new insights provided by circulating mir-223 in plasma. J Am Coll Surg. 2015;221(4):e20–e21. doi: 10.1016/j.jamcollsurg.2015.08.351 [DOI] [PubMed] [Google Scholar]
- 39.Abue M, Yokoyama M, Shibuya R, et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46(2):539–547. doi: 10.3892/ijo.2014.2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Zhao CY, Zhang SH, et al. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Rep. 2014;31(3):1157–1164. doi: 10.3892/or.2013.2960 [DOI] [PubMed] [Google Scholar]
- 41.Slater EP, Strauch K, Rospleszcz S, et al. MicroRNA-196a and −196b as potential biomarkers for the early detection of familial pancreatic cancer. Transl Oncol. 2014;7(4):464–471. doi: 10.1016/j.tranon.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin MS, Chen WC, Huang JX, Gao HJ, Sheng HH Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int J Clin Exp Med. 2014;7(12):5226–5234. [PMC free article] [PubMed] [Google Scholar]
- 43.Ganepola GA, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol. 2014;6(1):22–33. doi: 10.4251/wjgo.v6.i1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao C, Zhang J, Zhang S, et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol Rep. 2013;30(1):276–284. doi: 10.3892/or.2013.2420 [DOI] [PubMed] [Google Scholar]
- 45.Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19(13):3600–3610. doi: 10.1158/1078-0432.CCR-12-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R, Chen X, Du Y, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–618. doi: 10.1373/clinchem.2011.172767 [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Gao J, Du Y, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422 [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10(3):203–211. doi: 10.2353/jmoldx.2008.070153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steer ML, Waxman I, Freedman S Chronic pancreatitis. N Engl J Med. 1995;332(22):1482–1490. doi: 10.1056/NEJM199506013322206 [DOI] [PubMed] [Google Scholar]
- 51.Blower PE, Chung JH, Verducci JS, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7(1):1–9. doi: 10.1158/1535-7163.MCT-07-0573 [DOI] [PubMed] [Google Scholar]
- 52.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–1281. doi: 10.1158/0008-5472.CAN-05-3632 [DOI] [PubMed] [Google Scholar]
- 53.Liu H, D’Andrade P, Fulmer-Smentek S, et al. mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol Cancer Ther. 2010;9(5):1080–1091. doi: 10.1158/1535-7163.MCT-09-0965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morimura R, Komatsu S, Ichikawa D, et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105(11):1733–1740. doi: 10.1038/bjc.2011.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kojima M, Sudo H, Kawauchi J, et al. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS One. 2015;10(2):e0118220. doi: 10.1371/journal.pone.0118220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawaguchi T, Komatsu S, Ichikawa D, et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108(2):361–369. doi: 10.1038/bjc.2012.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker N, Lockwood CM Pre-analytical variables in miRNA analysis. Clin Biochem. 2013; 46 (10–11): 861–868. doi: 10.1016/j.clinbiochem.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 58.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57(6):833–840. doi: 10.1373/clinchem.2010.157198 [DOI] [PubMed] [Google Scholar]
- 59.Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One. 2013;8(10):e77963. doi: 10.1371/journal.pone.0077963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim DJ, Linnstaedt S, Palma J, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn. 2012;14(1):71–80. doi: 10.1016/j.jmoldx.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Kowdley KV Method for microRNA isolation from clinical serum samples. Anal Biochem. 2012;431(1):69–75. doi: 10.1016/j.ab.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kroh EM, Parkin RK, Mitchell PS, Tewari M Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petriv OI, Kuchenbauer F, Delaney AD, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci U S A. 2010;107(35):15443–15448. doi: 10.1073/pnas.1009320107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kannan M, Atreya C Differential profiling of human red blood cells during storage for 52 selected microRNAs. Transfusion. 2010;50(7):1581–1588. doi: 10.1111/j.1537-2995.2010.02585.x [DOI] [PubMed] [Google Scholar]
- 66.Rossi RL, Rossetti G, Wenandy L, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12(8):796–803. doi: 10.1038/ni.2057 [DOI] [PubMed] [Google Scholar]
- 67.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6(6):e20769. doi: 10.1371/journal.pone.0020769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiang M, Zeng Y, Yang R, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454(1):210–214. doi: 10.1016/j.bbrc.2014.10.064 [DOI] [PubMed] [Google Scholar]
- 70.Qi R, Weiland M, Gao XH, Zhou L, Mi QS Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology. 2012;55(5):1640–1642; author reply 1642–1643. doi: 10.1002/hep.25558 [DOI] [PubMed] [Google Scholar]
- 71.Fan L, Qi H, Teng J, et al. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol. 2016;37(6):7777–7784. doi: 10.1007/s13277-015-4608-3 [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, Gu J, Roth JA, et al. Pathway-based serum microRNA profiling and survival in patients with advanced stage non-small cell lung cancer. Cancer Res. 2013;73(15):4801–4809. doi: 10.1158/0008-5472.CAN-12-3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diaz-Garcia CV, Agudo-Lopez A, Perez C, et al. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: implications for outcomes and histologic classification. Carcinogenesis. 2013;34(5):1031–1038. doi: 10.1093/carcin/bgt022 [DOI] [PubMed] [Google Scholar]
- 74.Appaiah HN, Goswami CP, Mina LA, et al. Persistent upregulation of U6: SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Research. 2011;13(5):R86. doi: 10.1186/bcr2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grasedieck S, Scholer N, Bommer M, et al. Impact of serum storage conditions on microRNA stability. Leukemia. 2012;26(11):2414–2416. doi: 10.1038/leu.2012.106 [DOI] [PubMed] [Google Scholar]
- 76.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 77.Chevillet JR, Lee I, Briggs HA, He Y, Wang K Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules. 2014;19(5):6080–6105. doi: 10.3390/molecules19056080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y, Gao SG, Chen JM, et al. Serum CA242, CA199, CA125, CEA, and TSGF are biomarkers for the efficacy and prognosis of cryoablation in pancreatic cancer patients. Cell Biochem Biophys. 2015;71(3):1287–1291. doi: 10.1007/s12013-014-0345-2 [DOI] [PubMed] [Google Scholar]
- 79.Zhou G, Niu L, Chiu D, He L, Xu K Changes in the expression of serum markers CA242, CA199, CA125, CEA, TNF-alpha and TSGF after cryosurgery in pancreatic cancer patients. Biotechnol Lett. 2012;34(7):1235–1241. doi: 10.1007/s10529-012-0908-5 [DOI] [PubMed] [Google Scholar]
- 80.Lei XF, Jia SZ, Ye J, et al. Application values of detection of serum CA199, CA242 and CA50 in the diagnosis of pancreatic cancer. J Biol Regul Homeost Agents. 2017;31(2):383–388. [PubMed] [Google Scholar]
- 81.Ni XG, Bai XF, Mao YL, et al. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur J Surg Oncol. 2005;31(2):164–169. doi: 10.1016/j.ejso.2004.09.007 [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int J Clin Exp Med. 2015;8(7):11683–11691. [PMC free article] [PubMed] [Google Scholar]
- 83.Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. doi: 10.1038/ncomms8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28(4):741–747. doi: 10.1093/annonc/mdx004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials