Abstract
Propose
To study CCDC103 expression profiles and understand how pathogenic variants in CCDC103 affect its expression profile at mRNA and protein level.
Methods
To increase the knowledge about the CCDC103, we attempted genotype-phenotype correlations in two patients carrying novel homozygous (missense and frameshift) CCDC103 variants. Whole-exome sequencing, quantitative PCR, Western blot, electron microscopy, immunohistochemistry, immunocytochemistry, and immunogold labelling were performed to characterize CCDC103 expression profiles in reproductive and somatic cells.
Results
Our data demonstrate that pathogenic variants in CCDC103 gene negatively affect gene and protein expression in both patients who presented absence of DA on their axonemes. Further, we firstly report that CCDC103 is expressed at different levels in reproductive tissues and somatic cells and described that CCDC103 protein forms oligomers with tissue-specific sizes, which suggests that CCDC103 possibly undergoes post-translational modifications. Moreover, we reported that CCDC103 was restricted to the midpiece of sperm and is present at the cytoplasm of the other cells.
Conclusions
Overall, our data support the CCDC103 involvement in PCD and suggest that CCDC103 may have different assemblies and roles in cilia and sperm flagella biology that are still unexplored.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01509-7) contains supplementary material, which is available to authorized users.
Keywords: CCDC103, Infertility, Primary ciliary dyskinesia, Situs-inversus-totalis, Cilia, Spermatozoa
Introduction
Cilia, motile or non-motile, are surface cell projections whose skeleton is made of the axoneme. Non-motile cilia are ubiquitous, whereas motile cilia occur in specific tissues, namely in upper respiratory airways and reproductive system [1, 2]. Respiratory cilia are multiple per cell and present a rotational, very fast motion. In contrast, the sperm flagellum is single, longer, and presents a wave-like motion [3]. In common, they possess the same skeletal structure, the axoneme (Ax), which is responsible for motility [2, 4]. In these cells, the Ax is composed of two central single microtubules surrounded by nine peripheral microtubule doublets, forming a 9d+2s pattern. Doublets are linked by a dynein regulatory complex (DRC or nexin links) and contain a pair of projections, named dynein arms (DA), which are nominated by their position as the inner (IDA) and outer (ODA). The coordinated action of DA generates motion and thus provides the force necessary to move fluids or the cell [5]. The two central microtubules are linked by a central bridge and are surrounded by a fibrillar central sheath, which constitutes the central pair complex (CPC). The doublets bind to the CPC by radial projections, called radial spokes (RS) [2].
Cilia play critical roles in physiology and development, and when disrupted cause diseases, known as ciliopathies. Primary ciliary dyskinesia (PCD) is an autosomal recessive ciliopathy caused by anomalies in motile cilia. Patients with PCD present a heterogeneous combination of symptoms [6], including sinusitis, hearing impairment, chronic bronchitis, and subfertility or infertility. Situs-inversus occurs in half of the PCD cases, a clinical situation known as Kartagener syndrome (KS). The diagnosis of PCD is complex, needing, besides strict clinical features selection, the ultrastructural analysis of the axonemes, motion analysis, and genetics tests [7]. Given the sophistication of motile cilia, as well as the high complexity of ciliogenesis [8], the genetic characterization is also very complex, with several genes having been associated to PCD [9, 10].
We previously described a PCD infertile male patient with situs-inversus-totalis, the absence of sperm DA and presence of a novel missense homozygous variant in the CCDC103 (coiled-coil domain containing-103) gene [11]. CCDC103 was identified as a PCD gene in Pakistani PCD families with ODA defects [12]. It was shown to code for an oligomeric coiled-coil domain protein that was found in the sperm axoneme and in cytoplasmic extracts of C. reinhardtii and Zebrafish (D. rerio). However, as CCDC103 was present in mutants lacking ODA components or assembly factors, the authors suggested that those components are not necessary for CCDC103 integration within the sperm axoneme [12]. Later, CCDC103 was shown to specifically bind polymerized microtubules (MT), being critical to stabilize the MT polymeric structure, which suggests that CCDC103 may have a critical role in ODA assembly [13]. Nevertheless, the knowledge about the role of this protein in ciliary biology is still very scarce, with CCDC103 expression in human respiratory cilia and sperm having not yet been studied.
To further investigate the role of CCDC103 in ciliary structure and function, we examined the consequences of CCDC103 pathologic variants in two PCD patients with situs-inversus-totalis, at the ultrastructural, RNA, and protein level. Furthermore, we analyzed the CCDC103 profile in different reproductive cells and somatic cells (nasal cells and white blood cells). Our results revealed that these CCDC103 variants have as consequence the absence of both DA and affected gene and protein expression. Moreover, we observed that CCDC103 was expressed with tissue-specific features, suggesting distinct tissue-specific regulation. Our data corroborate the involvement of CCDC103 in PCD/KS and in infertility and suggest that CCDC103 may have different assemblies and roles in cilia and sperm flagellum biology that are still unexplored.
Materials and methods
Ethics
Ethical guidelines were followed in the conduct of research, with written informed consent obtained before the beginning of the work. For blood and nasal tissue samples, no further ethical or institutional approvals were needed as patient samples and databases are included in the regular clinical assessment of the patients. Surplus sperm, testicular tissue, and oocytes were donated, after assisted reproduction treatments, under the Portuguese National Law on Medically Assisted Procreation (http://data.dre.pt/eli/diario/1/142/2017/0/pt/html) and the Council on Medically Assisted Procreation guidelines (www.cnpma.org.pt), with no further authorizations required. This work did not involve experiments on human or animals. Thus, the provisions of the Declaration of Helsinki as revised in Tokyo 2004 on human experimentation do not apply to this work.
Patients
To understand patient genotype-phenotype correlations, two patients with PCD and situs-inversus-totalis were included in the present study.
Patient 1 is a 45-year-old male from the northern of Portugal. He referred no personal or family consanguinity, chronic respiratory complaints, namely nasal polyps (surgical removal at 2012), sinusitis, rhinitis, and bronchitis, without asthma, and situs-inversus-totalis. As the couple did not achieve a spontaneous pregnancy, they underwent for infertility treatments. The karyotype was normal and there were no Y chromosome microdeletions, cystic fibrosis transmembrane receptor (CFTR) mutations or adverse CFTR polymorphisms (7T/7T). Hormonal values were in the normal range: FSH 1.7 mIU/ml (1.4–18.1), LH 2.7 mIU/ml (1.5–9.3), and total testosterone 15.1 nmol/L (9.1–55.2). The testicular volume was normal (20 ml/20 ml), both epididymides were swollen, without globus-major appearance, and the vas deferens were present. Semen analysis showed a volume of 5.6 ml, a pH 7.8, and the presence of very rare total immotile sperm. The patient undertook testicular sperm extraction (TESE) in November 2006, followed by intracytoplasmic sperm injection (ICSI) with fresh testicular in situ motile sperm. No pregnancy ensued. In 2008, he performed another TESE cycle with fresh sperm but again, no pregnancy was achieved. A third attempt was scheduled with cryopreserved testicular sperm and a successful twin pregnancy was attained with the birth of two male twins, whom at 8 years old perfectly healthy. The genetic analysis by whole-exome sequencing (WES) was formerly published and revealed a novel missense homozygous variant in the CCDC103 gene [11]. Patient 2 is a fertile, 53-year-old female from the northern of Portugal. She reported chronic respiratory complaints, namely bronchiectasis and chronic rhinosinusitis, and situs-inversus totalis.
Sample collection
Genomic DNA was extracted from peripheral blood leukocytes of patient 2, using the salting out protocol [14]. The exome was sequenced using the AmpliSeq strategy on an Ion Proton next-generation sequencing (NGS) platform (Life Technologies, Thermo Fisher Scientific, California, USA) and variant calling was performed as described [11, 15]. All variants were listed in a Variant Call Format (VCF) file that was annotated and filtered using the Ion Reporter Software version 5.2 (http://ionreporter.lifetechnologies.com/) and VarAFT 2.10 (http://varaft.eu) (VarAFT, Aix Marseille University, France). An autosomal recessive disease model was used. Alamut Visual v2.10 software (Interactive Biosoftware, France) assisted variant interpretation. All the suspected variants were manually checked on the Binary Alignment Map (BAM) file through GenomeBrowse version 2.0.2 (Golden Helix, Bozeman, USA).
White blood cell (WBC) samples, used for RNA and protein extraction, from both patients and healthy controls, were collected from peripheral blood with EDTA anticoagulant tubes (VACUETTE, Porto, Portugal). Control white blood cells were obtained from healthy blood donor volunteers.
Nasal cells were obtained by nasal brushing from both patients and healthy controls, using a cytology soft sterile brush (Endobrush, Biogyn SNC, Mirandola, Italy), in both nostrils from the inferior nasal turbinate[16]. Control nasal cells were obtained from healthy university and volunteers. After brushing, cells were placed in fixative for transmission electron microscopy (TEM), or RPMI 1640 Medium (Gibco, Thermo Fisher Scientific, Massachusetts, USA), for RNA and immunofluorescence analysis.
Excedentary testicular tissue from patient 1 was obtained under infertility treatments. Excedentary testicular tissue, used as control, was obtained from men with obstructive azoospermia under infertility treatments (these could be used as controls as they presented conserved spermatogenesis, normal karyotypes, absence of Y microdeletions, and CFTR mutations). Control ejaculated sperm were obtained from normozoospermic men from cases undergoing spermiogram evaluation and otherwise healthy. Excedentary oocytes were obtained under infertility treatments. Sertoli cells were obtained from primary cultures originated from testicle biopsies of men with conserved spermatogenesis [17]. Thereafter, Sertoli cells obtained from primary cultures will be designated culture Sertoli cells (cSC).
Transmission electron microscopy
Nasal samples were fixed with 2.5% glutaraldehyde (Sigma-Aldrich, Missouri, USA) in 0.1 M cacodylate buffer (Merck, Darmstadt, Germany), pH 7.2, 2 h, room temperature (RT) [16], post-fixed with 2% osmium tetroxide (Merck), 2 h, 4 °C, and dehydrated in a graded ethanol series (VWR, Pennsylvania, USA). Samples were then treated with 1% tannic acid (Merck) in 100% ethanol and embedded in epoxy resin (Epon, Sigma-Aldrich). Suitable areas of ciliated cells were selected in semithin sections (1 μm) stained with methylene blue-Azur II (Merck). Ultrathin sections were cut on an LKB-ultramicrotome (Leica Microsystems, Wetzlar, Germany) using diamond knives (Diatome, Pennsylvania, USA), and retrieved on copper grids (Taab, Berks, England). After contrasting with aqueous uranyl acetate (BDH, Poole, England) and lead citrate (Merck), they were observed in a JEOL 100CXII transmission electron microscope (JEOL, Tokyo, Japan), operated at 60 kV.
Patient cilia axoneme ultrastructure was evaluated at high magnifications in transverse sections, and the diagnosis was based on the presence of a systematic defect in any of the axonemal structures [18]. The ultrastructure of the testicular sperm axoneme from patient 1 was previously presented [19].
The ciliary beat axis and ciliary deviation were evaluated in a minimum of 100 transverse sections examined after printing. In printed images, a line was drawn a perpendicular to the central microtubules. A reference line was then chosen based on the main orientation of the lines drawn. The angle of each line to the reference line was calculated and subtracted to the mean. These differences are near zero. The standard deviation (SD) of these differences corresponds to the ciliary beat axis and ciliary deviation [20].
Immunohistochemistry
Testicular tissue from controls was analyzed from formalin-fixed (VWR) paraffin-embedded blocks (Merck). Immunohistochemistry (IHC) was performed in tissue sections of 3 μm thickness attached to adhesive slides (Starfrost, Braunschweig, Germany). Tissue sections were deparaffinized with xylene (VWR) and serially hydrated in a decreasing scale of ethanol followed by a wash in distilled water. Heat antigen retrieval was performed with citrate buffer pH 6 (Merck) in a microwave (600 W) for 20 min. Tissue sections were intensely washed in water. Endogenous peroxidases were inhibited with 3% of hydrogen peroxide (Merck) and non-specific background staining was minimized with 5% of non-fat milk (Nestlé, Vevey, Switzerland) in PBS pH 7.4 (Panreac, Barcelona, Spain), with both treatments being performed for 30 min at RT.
Sections were then incubated with primary antibodies, rabbit polyclonal to-CCDC103-C-terminal (reference ab177558; Lot GR279559-4) from Abcam (Cambridge, UK) and mouse anti-acetylated α-tubulin (Santa Cruz Biotechnology, California, USA) for 1 h at RT. A negative control, throught the omission of the primary antibody, was included. Ultraview universal DAB anti-rabbit and anti-mouse detection kit (Ventana Medical Systems, Arizona, USA) was used to reveal the expression of CCDC103 and acetylated α-tubulin antigens, respectively. After dehydration, sections were mounted on Coverquick 2000 (VWR). Slides were observed under a light microscope BX41 (Olympus, Tokyo, Japan).
Immunocytochemistry
For immunocytochemistry (ICC), smears were made from cell suspensions, containing nasal cells, sperm, testicular cells, and cSC, on adhesive slides (Starfrost). Cell smears were air-dried and directly frozen at − 80 °C until use. Afterward, cells were fixed with 4% paraformaldehyde (Merck) in PBS for 20 min at RT, followed by washes in PBS. For permeabilization, cells were incubated in 0.2% Triton X-100 (Sigma-Aldrich) in PBS for 15 min at RT. Then, washed in PBS and incubated with 5% non-fat milk in PBS for 1 h at RT, to inhibit non-specific binding. Afterward, cells were incubated overnight at 4 °C with the antibodies rabbit anti-CCDC103 (Abcam) and mouse anti-acetylated α-tubulin (Santa Cruz Biotechnology). For each experiment, a negative control, throught the omission of the primary antibody, was included. DyLight-488 anti-rabbit (Biolegend, California, USA) and Texas Red anti-mouse (Santa Cruz Biotechnology) were used as secondary antibodies and applied to cells for 1 h at RT. Then, cells were counterstained with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI: Vector Laboratories, California, USA). Slides were observed under an epifluorescence microscope (Eclipse E400; Nikon, Tokyo, Japan), and also analyzed with a FluoView FV1000 laser scanning confocal microscope (Olympus) to screen an eventual co-localization of the signal.
Western blot
For the extraction of total protein from WBC, peripheral blood was incubated with erythrocyte lysis buffer (ELB) containing 155 mM NH4CL (Sigma-Aldrich), 10 mM KHCO3 (Merck) and 1 mM EDTA (Sigma-Aldrich), for 20 min on ice, and then centrifuged at 2000 rcf (G-force) for 10 min. The cellular pellet was washed twice with ELB and then in cold PBS, centrifuged at 3000 rcf for 5 min at 4 °C and then stored at − 20 °C until protein extraction.
For sperm, testicular tissue cells, and cSC, cells were washed in cold PBS, centrifuged at 3000 rcf for 5 min at 4 °C and ten stored at − 20 °C until protein extraction. In the case of sperm, the pellet was stored in PBS supplemented with a commercial protease inhibitor cocktail (Sigma-Aldrich).
For total protein extraction, the cellular pellet was resuspended in lysis buffer containing 125 mmol/L Tris-HCl, pH 6.8 (Sigma-Aldrich), 2% SDS (Bio-Rad, Hercules, USA), 4 M urea (Sigma-Aldrich), 10% β-mercaptoethanol (Sigma-Aldrich) and a commercial protease inhibitor cocktail (Sigma-Aldrich), and incubated under gentle agitation for 30 min at RT.
Protein concentration was estimated using the Qubit protein assay kit (Life Technologies). After protein measurement, the protein was precipitated using trichloroacetic acid (Merck), washed in acetone (Merck), and stored at − 20 °C until further use. Then, aliquots of total protein (50 to 100 μg) were subjected to electrophoresis using the NuPAGE® system on NuPAGE Bis-Tris midi gels (Life Technologies), according to manufacturer’s instructions. Gels were subsequently transferred overnight at 35 V, 4 °C, to Amersham Protan 0.45 NC nitrocellulose membranes (GE Healthcare, Illinois, USA). Membranes were blocked with 5% non-fat milk with 0.05% Tween-20 (Sigma-Aldrich) in TBS and incubated with rabbit anti-CCDC103 (Abcam) and mouse anti-HSP 70 antibodies (Santa Cruz Biotechnology), overnight at 4 °C. As a negative control, an immunized CCDC103 peptide (ABCAM) is specifically prepared for blocking peptide for anti-CCDC103 antibody—C-terminal. Following incubation with an HRP conjugated goat anti-rabbit or anti-mouse secondary antibody (Santa Cruz Biotechnology), for 2 h at RT. The detection was done using ECL Blotting Substrate (Bio-rad) and the image analyzer LAS3000 (FujiFilm, Tokyo, Japan). The band size estimation was done with the Image Lab software (version 6, Bio-rad).
Gene expression analysis
Total ribonucleic acid (RNA), from cell suspensions containing nasal cells, sperm, testicular cells, cSC, and WBC, was extracted using the PerfectPure RNA Cell and Tissue Kit (5 PRIME GmbH, Hamburg, Germany), according to manufacturer’s instructions. For oocytes, due to the limited number of cells available, the single-cell RNA purification kit was used (Norgen Biotek, Thorold, Canada). The concentration and purity of RNA samples were determined on a Nanodrop spectrophotometer ND-1000 (Version 3.3; LifeTechnologies). Only samples with an A260/A280 ratio between 1.8 and 2.1 were selected as it is indicative of highly purified RNA. RNA was then converted to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, California, USA), according to manufacturer’s instructions and including the optional step of “DNase treatment” that is intended to digest genomic deoxyribonucleic acid (gDNA) to avoid gDNA contamination.
For gene expression analysis of CCDC103 (NM_213607.2), specific PCR primers were designed and amplified by polymerase chain reaction (PCR). The list of primers and conditions are included in Supplementary Table S1. Successful PCR products were enzymatically purified using Illustra ExoStar kit (GE Healthcare, Buckinghamshire, UK) and sequenced with BigDye Terminator v1.1 Cycle Sequencing Kit (LifeTechnologies) according to manufacturer’s instructions and analyzed by high-resolution electrophoresis in a 3130xl genetic analyzer (LifeTechnologies).
Real-time quantitative PCR (qPCR) was performed to evaluate the mRNA expression of CCDC103 in all cells. The B2M and GAPDH genes were used as housekeeping genes to normalize gene expression levels. qPCRs were carried out in a Bio-rad CFX96 (Bio-Rad), with NZY qPCR Green master mix (NZYTech), following manufacture instructions. “Three technical, and, excluding the patient samples, three biological replicates were performed in each PCR assay. Fold variation of gene expression levels was calculated following a mathematical model using the formula 2-ΔΔCt [21]. The statistical significance was determined using the non-parametric statistical test Mann-Whitney test, with alpha < 0.05, Two-tailed p value. Manny-Whitney is the non-parametric counterpart of the t test. Tests were performed in the GraphPad Prism (version 6.01, GraphPad Software, California, USA). When analyzing patient’s data (Fig. 3), patient samples were compared with samples from individuals without PCD/KS. When analyzing the mRNA expression from different control cells types (Fig. 8), samples were compared in pairs (as described in legend from Fig. 8b).
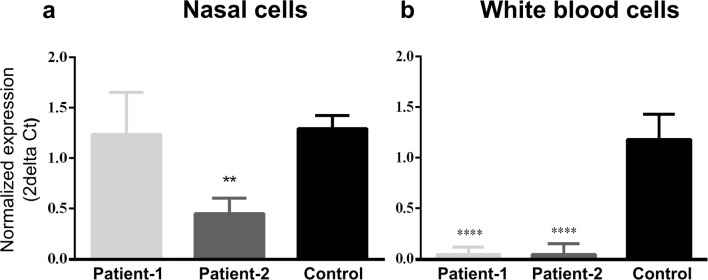
Fig. 3.
CCDC103 mRNA expression levels in a nasal cells and b white blood cells from patients in comparison to cells from healthy individuals (without PCD/KS or other related diseases found). SYBR Green was the fluorescent dye used. Statistical significance was determined using the Mann-Whitney test, with alpha < 0.05. **p < 0.01 and ***p < 0.0001. B2M and GAPDH were used as reference genes, with no statistical significance being observed between the normalizations performed with each locus
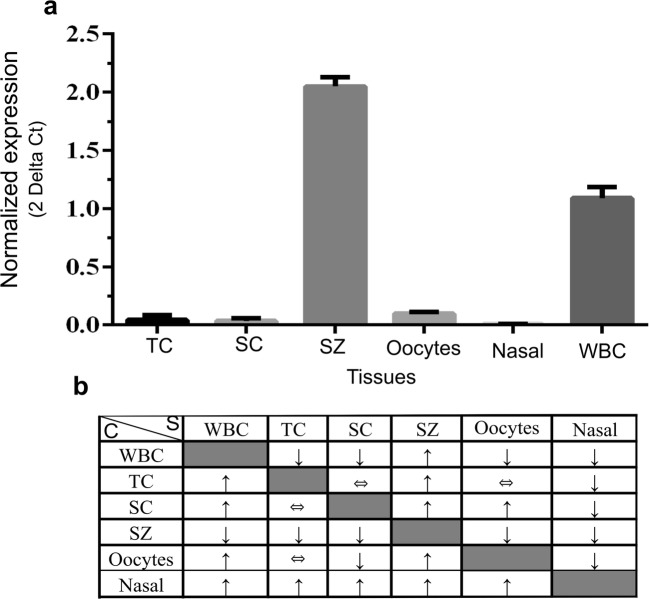
Fig. 8.
CCDC103 RNA expression levels (a) from control testicular cells (TC), culture Sertoli cells (SC), sperm (SZ), oocytes, nasal cells (nasal) and white blood cells (WBC). SYBR Green was the fluorescent dye used. Statistical significance was determined using the Mann-Whitney test, with alpha < 0.05. WBC was used as a calibrator and B2M as the reference gene. Resume of all possible combinations between samples (b). The C (calibrator) column means that this tissue was used as calibrator. For example, using WBC as a calibrator, it was observed that samples (S) from TC, SC, oocytes, and nasal cells present a reduced expression (↓), while SZ has a higher expression (↑) than WBC. ⇔ means that there were no statistically significant differences between the two groups. Controls: for TC cells are samples obtained from men with conserved spermatogenesis that are under infertility treatments; for the remaining cells, are cells from healthy individuals, without PCD/KS or other related diseases found
Results
Patients with CCDC103 disease-causing variants present absence of dynein arms
The main aim of the present study was to analyze the genotype-phenotype correlations between two cases with Kartagener syndrome that presented different variants in the CCDC103 gene. Following clinical studies and transmission electron microscopy, the genetic nature of the diseases was evaluated through WES analysis.
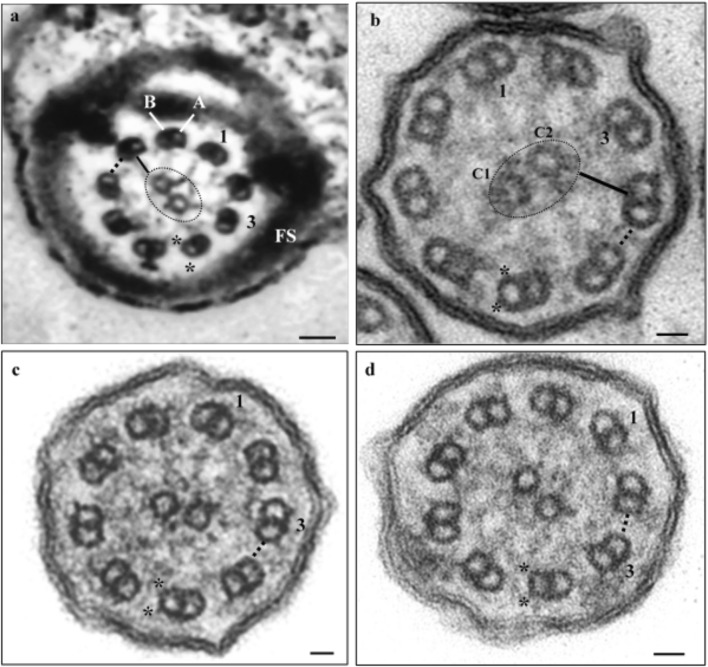
Patient 1 presents chronic respiratory complaints, situs-inversus totalis, and infertility due to the presence of rare total immotile sperm in the ejaculate. The ultrastructure of testicular sperm showed the absence of DA and DRC [19]. Here, we analyzed nasal cilia and found the same ultrastructural axoneme defects observed in sperm (Fig. 1a, b), which suggests that this gene affects both structures indistinctively. Altogether with the high SD value (SD 26.94) obtained from the determination of the ciliary beat axis and ciliary deviation, data establishes the diagnosis of PCD/KS in this patient.
Fig. 1.
Ultrastructure of the axoneme. a Sperm axoneme of patient 1; b axoneme of nasal cilia cells of patient 1; c, d axonemes of nasal cilia cells of patient 2. Note absence of dynein arms (*) and nexin links (dotted line). 1 and 3, numbering of the peripheral duplets; A, microtubule A of a peripheral duplet; B, microtubule B of a peripheral duplet; dotted circle, central sheath; C1 and C2, central microtubule pair; black bar, radial spokes; FS, fibrous sheath. Scale bars, 25 nm
Patient 2 presents chronic respiratory complaints, situs-inversus totalis, but no infertility. The axoneme ultrastructure of nasal cilia revealed the absence of DA and DRC (Fig. 1c, d), and the determination of the ciliary beat axis and ciliary deviation showed a high SD (SD 24.60), which corroborates the diagnosis of PCD/KS in this patient.
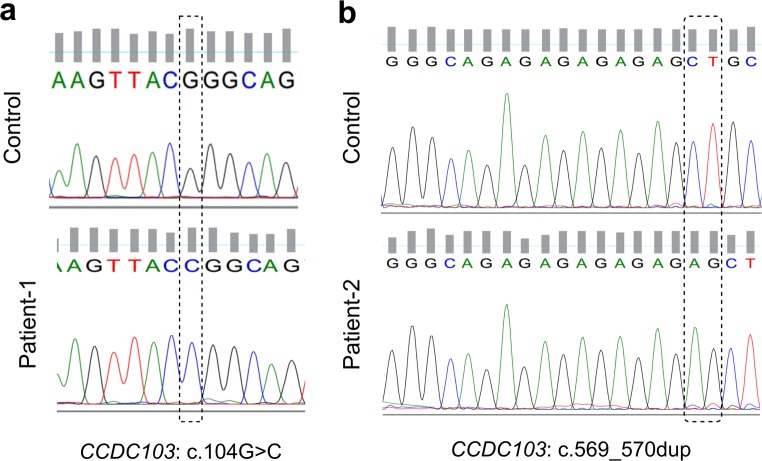
In both patients, WES analysis leads to the identification of novel homozygous, potentially pathogenic, gene variants in CCDC103 (Fig. 2). Patient 1 showed a missense variant, caused by a guanine to an adenine substitution at position 104 (c.104G > A), that corresponds to the replacement of a positively charged arginine (Arg) by a non-polar proline at protein position 35 [11]. According to the American College of Medical Genetics and Genomics (ACMGG) guidelines from 2015 [22], this variant is considered of unknown clinical significance, with a moderate evidence (PM2), and is a recessive variant with a very reduced frequency (0.00001) according to Exome Aggregation Consortium (ExAC) and supporting (PP3) since several bioinformatic programs, namely Polyphen (score = 0.999), SIFT (score = 0.002), FATHMM (score = − 2.69), and MutPred2 (score = 0.896), predicted this variant as damaging with high confident scores. Patient 2 presents a duplication of two nucleotides (AG) between chromosomal position 17:42980013 and 17:42980014 (c.569_570dup), which results in a frameshift variant. This variant was submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), with accession number SUB5094211 (under processing). According to the ACMGG guidelines from 2015 [22], this variant is considered likely pathogenic, with a pathogenic very strong evidence (PVS1), since this variant is expected to affect protein length as a result of the frameshift variant, and with a moderate evidence (PM2), as this variant is absent from variant populational databases.
Fig. 2.
Sequencing electropherograms of variants found in the CCDC103 gene (NM_213607.2). a Variant c.104G>A of patient 1. b Variant c.569_570dup of patient 2
Patients with the CCDC103 variant present reduced mRNA and protein expression
Patient and control cDNA, from WBC and nasal cells, was used to analyze CCDC103 mRNA expression. In WBC of both patients, mRNA expression was significantly reduced (p < 0.0001) of about 0.2-fold (Fig. 3) to controls. In nasal cells of patient 1, no significant differences were observed, whereas, in patient 2, mRNA expression was significantly reduced (p = 0.0013) of about 0.5-fold (Fig. 3) to controls.
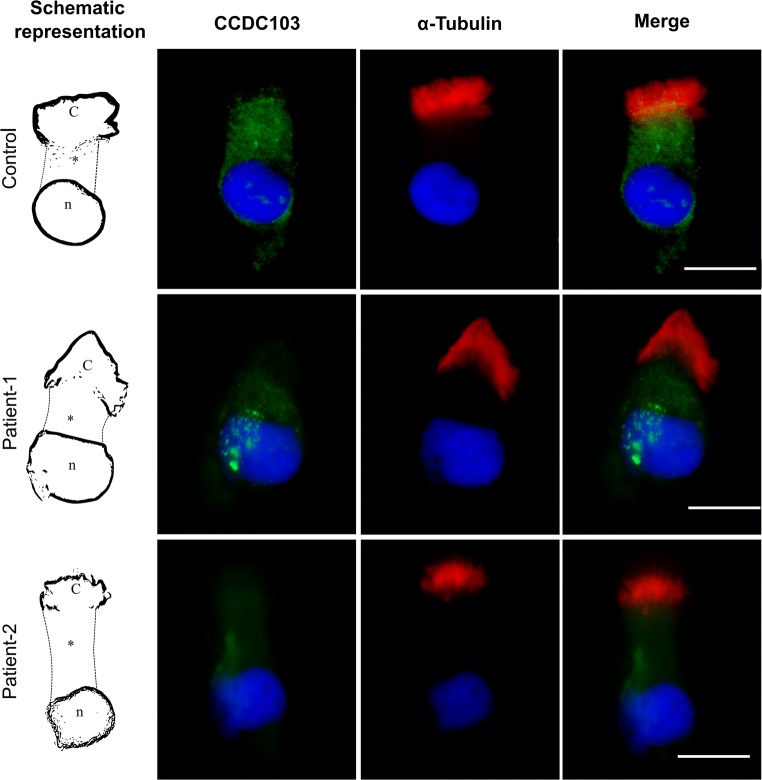
Protein expression was evaluated by ICC in nasal cells of patients and controls. In controls, staining was localized in the cytoplasm and at the base of cilia (Fig. 4). In patients, cytoplasmic staining was strongly reduced, especially in patient 2, and did not invades the base of cilia (Fig. 4), which suggests that CCDC103 pathogenic variants affect protein expression but do not cause protein misallocation.
Fig. 4.
Immunocytochemical detection of CCDC103 (green) and of axoneme-specific acetylated α-tubulin (red), with merged images, in nasal cilia cells of controls (i.e., cells from healthy individuals, without PCD/KS or other related diseases found), patient 1 and patient 2. Nuclei stained with DAPI (blue). C, cilia; *, cytoplasm; n, nucleus. Scale bars, 10 μm
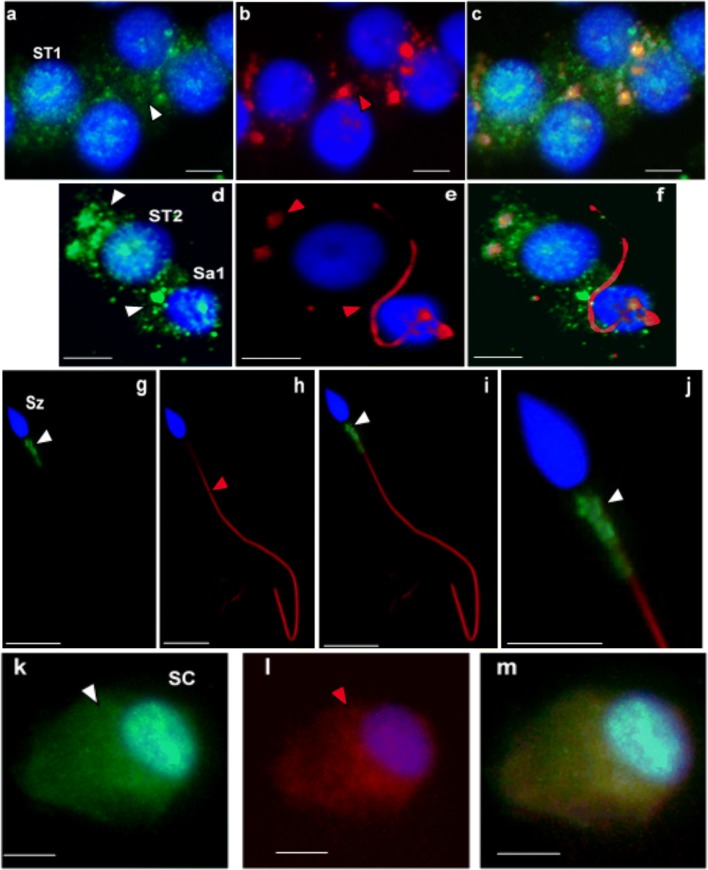
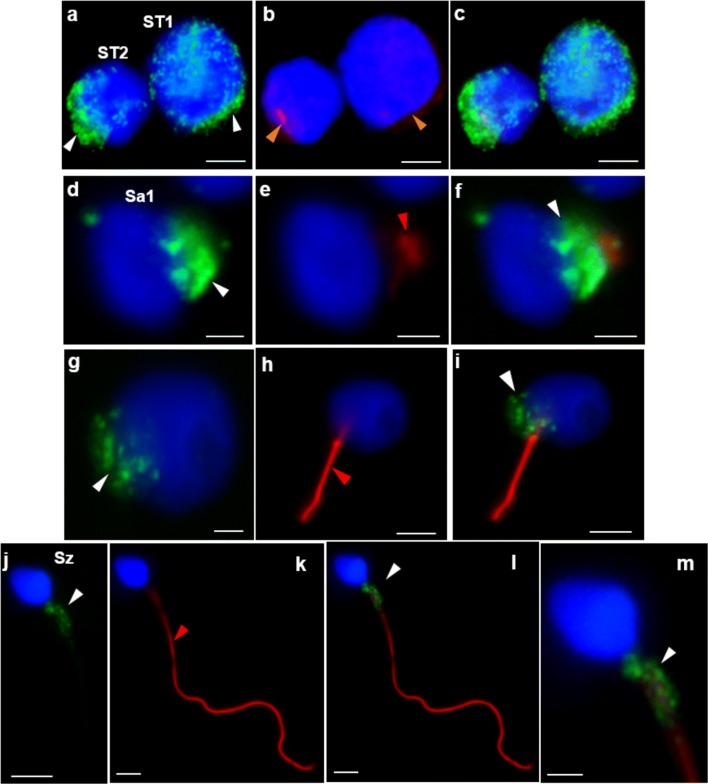
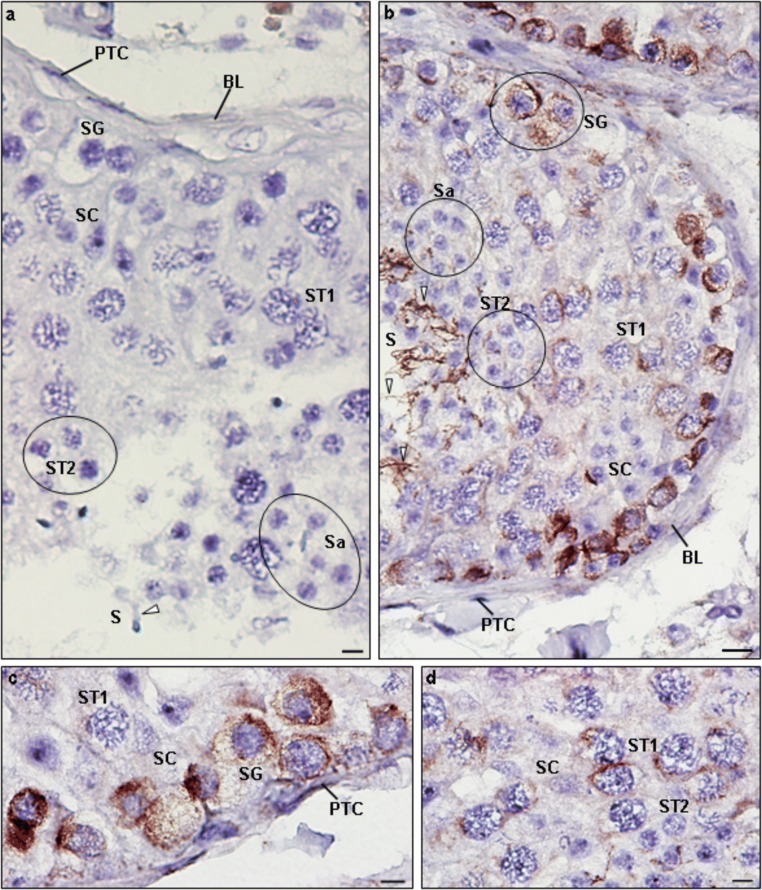
Protein expression was also evaluated by ICC in testicular cells of patient 1 and controls, in ejaculated sperm of controls and in cSC. In controls, CCDC103 was localized in the cytoplasm and perinuclear region of cSC, in the cytoplasm of primary spermatocytes (ST1), secondary spermatocytes (ST2), and round spermatids (Sa), and in the midpiece of sperm (Fig. 5). In patient 1, staining was observed in the cytoplasm and perinuclear region of ST1 and ST2, in the basal nuclear pole of Sa and in the midpiece of sperm (Fig. 6).
Fig. 5.
Immunocytochemical detection of CCDC103 (green) and of axoneme-specific acetylated α-tubulin (red), with merged images, in cells of the seminiferous tubules from controls (i.e., samples obtained from men with conserved spermatogenesis that are under infertility treatments). Staining was observed in the cytoplasm of primary spermatocytes (ST1: a–c), secondary spermatocytes (ST2: d–f), and round spermatids (Sa: d–f), and in the midpiece of sperm (Sz: g–j). In culture Sertoli cells (SC: k–m), staining was observed in the cytoplasm and in the perinuclear region. Nuclei stained with DAPI (blue). White arrowheads-CCDC103 staining, red arrowheads-tubulin staining. Scale bars, 5 μm
Fig. 6.
Immunocytochemical detection of CCDC103 (green) and of axoneme-specific acetylated α-tubulin (red), with merged images, in cells of the seminiferous tubules from patient 1. Staining was observed in the cytoplasm and perinuclear region of primary spermatocytes (ST1: a–c) and secondary spermatocytes (ST2: a–c), at the basal pole of the nucleus preceding axoneme extrusion in early round spermatids (Sa1: d–f) and late round spermatids (Sa2: g–i), and (granular appearance) in the midpiece of sperm (Sz: j–m). Nuclei stained with DAPI (blue). White arrowheads, CCDC103 staining; red arrowheads, tubulin staining. Scale bars: a–c, 4 μm; d–f, 2 μm; h, i, 4 μm; g, 2 μm; j–m, 2 μm
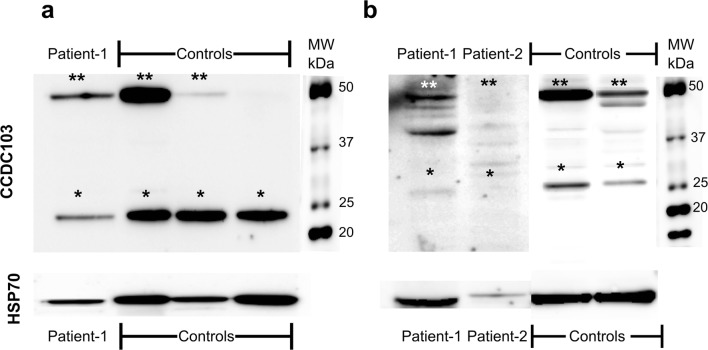
Protein expression by Western blot analysis was evaluated in the WBC from patients and controls. This revealed very challenging due to CCDC103 specific biochemical characteristics. Further, there are no other commercial available monoclonal antibody against CCDC103 and all available commercial antibodies were designed using the same immunogen, a synthetic peptide directed towards the C-terminal region, thus we could not discard the hypothesis of incomplete access to native protein due to the presence of self-association or complex alpha helices in these regions. In our Western blot experiences, we were unable to obtain at least three consistent replicates as we observed variable band intensity and instability of the monomer band, which was not always detected. This occurred despite all protocol procedures being carefully followed. Nevertheless, it was possible to infer that both patients revealed reduced protein expression, especially patient 2 (Fig. 7).
Fig. 7.
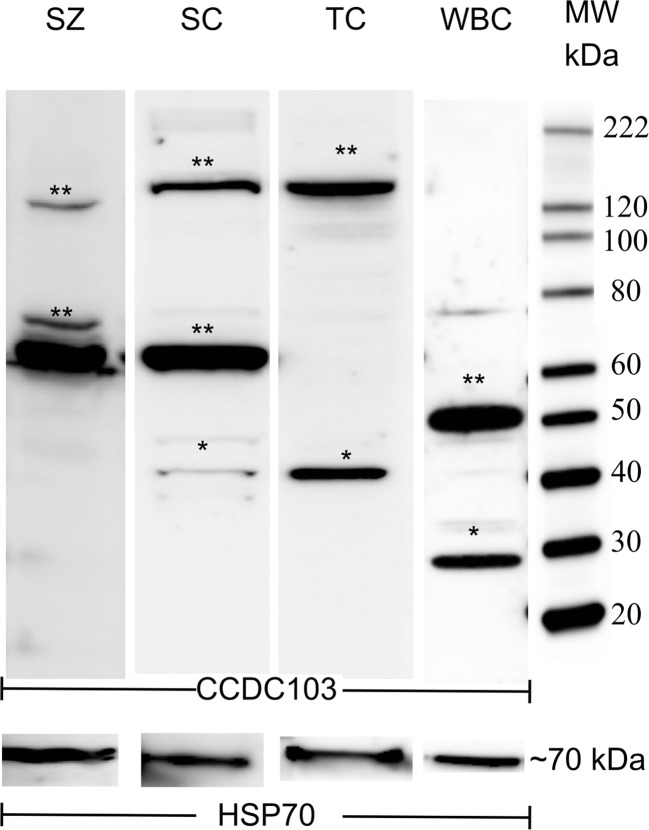
CCDC103 protein detection in white blood cells from controls (i.e., cells from healthy individuals, without PCD/KS or other related diseases found), patient 1 and patient 2 (a, b). *Possibly monomer form; **possibly dimer and/or higher order oligomers. HSP70 (molecular weight (MW) of about 70 kDa) was used as loading control
CCDC103 is present in several cells at different expressions levels
To further increase the knowledge of CCDC103 wild-type expression in human cells, cells from somatic and reproductive tissues obtained from healthy individuals (i.e., individuals without PCD/KS or other related diseases found) and from men with conserved spermatogenesis that are under infertility treatments were compared. Taken WBC as a reference, results showed that mRNA was expressed in all tissues but with different expression levels. Testicular cells, cSC, oocytes, and nasal cells evidenced low expression levels. Ejaculated sperm showed the highest expression levels, about 10-fold more than oocytes and about 2-fold more than WBC, the second more expressed tissue, with nasal cells showing the lowest expression levels (Fig. 8).
Western blot analysis revealed the presence of the protein in ejaculated sperm, testicular cells, cSC, and WBC, with different size bands. This confirms previous findings obtained in model species, which showed that the CCDC103 protein has special biochemical features, namely great oligomerization capabilities [13]. We observed that CCDC103 forms dimers and higher order oligomers whose sizes appear tissue-specific (Fig. 9). The theorical band size of about 27 kDa (monomer) was only observed in WBC, which also presented a dimer of about 50 kDa. In cSC, bands of about 37, 70, and 150 kDa were observed . If in these cells, we assume the 37 kDa band as a cSC-specific monomer, thus the other correctly correspond to dimer and higher order oligomers. Testicular cells presented a similar profile to cSC, but with the absence of the 70 kDa dimer. In ejaculated sperm, the protein was hard to detect despite multiple extraction protocols tested and was only achieved when total protein was protected with proteinase inhibitors before and after pelleted cells. Sperm evidenced the nearly 70 kDa dimer, comparably to cSC, and a higher order oligomer of about 125 kDa.
Fig. 9.
CCDC103 protein detection from control sperm (SZ), culture Sertoli cells (SC), testicular cells (TC), and white blood cells (WBC). *Possibly monomer form. **Possibly dimer and/or higher order oligomers. Polyclonal CCDC103 antibody from ABCAM was used. HSP70 was used as loading control
Total mRNA was sequenced, including the untranslated regions (UTR), to search if the presence of the different isoforms could explain those protein size differences. As no differences in mRNA sequences were found, results suggest that these differences observed in WB experiments might be caused by tissue-specific post-translational modifications or tissue-specific differences in oligomerization.
CCDC103 is differentially located in germ cells
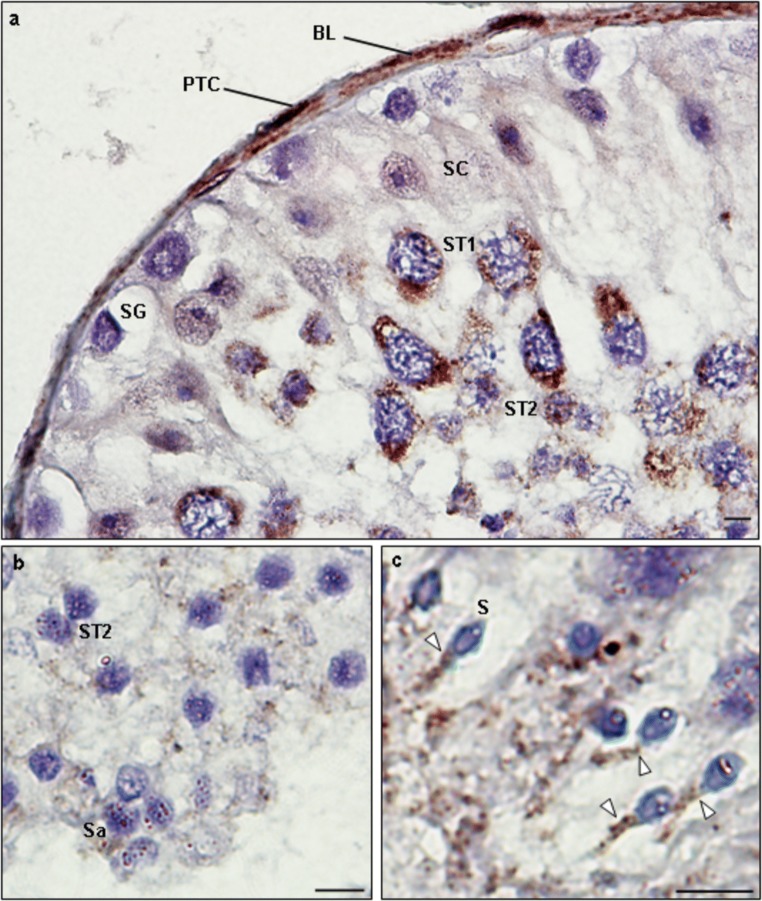
To gain further insights about CCDC103 location in germ cells, IHC detection in paraffin sections of seminiferous tubules was performed. Tubulin staining was observed in the cytoplasm and perinuclear region of cells, being intense in spermatogonia (SG), moderate intense in ST1, weakly intense in ST2, Sa, and Sertoli cells, and intense in the sperm flagellum (Fig. 10). CCDC103 staining was also observed in the cytoplasm and perinuclear region of cells but, in contrast to tubulin, staining was weak in SG, intense in ST1 and ST2, moderate in Sa and Sertoli cells, and intense in the sperm midpiece (Fig. 11).
Fig. 10.
Immunohistochemical detection of tubulin in paraffin sections of control seminiferous tubules. a Negative control (absence of staining). b–d Staining was intense in the cytoplasm and perinuclear region of spermatogonia (SG), moderately intense in the cytoplasm and perinuclear region of primary spermatocytes (ST1), and weakly intense in secondary spermatocytes (ST2), round spermatids (Sa), and Sertoli cells (SC). The flagellum (white arrowheads) of late spermatids (Sd) presents strong staining. PTC, peritubular cells; BL, basal lamina. Scale bars, 10 μm
Fig. 11.
Immunohistochemical detection of CCDC103 in paraffin sections of control seminiferous tubules obtained from men with conserved spermatogenesis that are under infertility treatments (a, b). Staining was weakly intense in spermatogonia (SG), intense in the cytoplasm and perinuclear region of primary spermatocytes (ST1) and secondary spermatocytes (ST2), and moderately intense in the cytoplasm and perinuclear region of Sertoli cells (SC) and round spermatids (Sa). c The midpiece of the elongated spermatids (Sd) showed intense staining (white arrowheads). PTC, peritubular cells; BL, basal lamina. Scale bars, 10 μm
CCDC103 location in sperm was also inspected by laser scanning confocal microscopy. Staining exhibited a helix shape throughout the midpiece region, involving it as a scarf (Supplementary videos S1, S2). No staining were observed by immunogold detection (Supplementary file S1).
Discussion
Patient 1 presents PCD, situs-inversus-totalis, the absence of DA in the sperm axoneme, severely reduced total sperm count, total sperm immobility, and infertility [19]. WES analysis revealed a novel homozygous pathogenic variant in the CCDC103 gene [11]. Panizzi and co-workers have shown that CCDC103 is an essential gene for dynein arm assembly and cilia motility, as in Zebrafish model, an antisense morpholino knockdown of ccdc103, induced curved body axes, left-right asymmetry defects, hydrocephalus and kidney cysts, and caused cilia paralysis [12]. The variant detected in this gene caused a change of an arginine by a proline in the protein α-helix region that contains the dynein attachment factor N-terminus domain. The wild-type arginine is a polar amino acid that contains a side chain consisting of a 3-carbon straight chain ending in a guanidine group that is positively charged. This amino acid is frequent in protein binding sites, as the positive charge enables to interact with negatively charged groups, forming multiple hydrogen bonds [23]. In contrast, the mutant amino acid proline is smaller, has a neutral charge, and is hydrophobic. The side chain of proline is a cyclic structure, named pyrrolidine ring, that confers non-polar characteristics. This characteristic side chain is non-reactive and gives proline an exceptional conformational rigidity. Consequently, proline is not commonly involved in protein function [23], suggesting that this mutation may impact on the protein structure, namely preventing dynein binding [24].
In patient 1, axoneme ultrastructure of both sperm and nasal cilia revealed the absence of DA. In humans, there are no studies that had simultaneously evaluated the effect of PCD gene mutations on the ultrastructure of the axoneme in both respiratory cilia and sperm flagellum. Although the axoneme structure of cilia and sperm are quite similar, some differences are known in the pathways regulating both. It is known that NOTCH signalling plays a crucial role in the differentiation of ciliated cells, with E2F transcription factors being required to activate centriole amplification genes, which are essential to the formation of multiple cilia. In contrast, WNT signalling plays a critical role in sperm flagellum formation, with E2F transcription factors being not so critical as sperm develop a single flagellum, with only one centriole [25]. The present observations that this variant in CCDC103 affected indistinctively both cilia and the sperm flagellum suggest that this variant may act in a shared pathway of DA formation in both cell types.
Patient 2 also presents PCD, situs-inversus-totalis, and absence of DA in respiratory cilia. Interestingly, WES analysis revealed a novel frameshift pathogenic variant also in CCDC103. This variant causes duplication of nucleotides AG at positions 569 and 570 in exon 4. The mutant protein presents a Glu187Argfs*22 change at the RPAP3-Cter domain. Presently, only three genes, RPAP3, SPAG1, and CCDC103 have been found to code for an RPAP3 domain and, curiously, all those genes are directly (CCDC103 and SPAG1) or indirectly (RPAP3) involved to cilia assemble and formation [26]. Although its function is not fully understood, the RPAP3 domain was suggested to participate in protein oligomerization and to be critical for maintaining ATPases in a conformation suitable for Client binding [26]. Client is the designation of a limited set of substrate proteins whose maturation is assisted by the heat-shock protein 90 (Hsp90), a molecular chaperone [27]. Thus, the variant found in patient 2 could affect the ATPases cycle and consequently DA assembly.
Expression studies of CCDC103 have only been performed in Chlamydomonas and Zebrafish. Studies showed that CCDC103 is an oligomeric coiled-coil domain protein present in the sperm Ax. This protein was found to bind to MT, forming linear arrays along MT with spacing consistent with the ODA repeat distance. Further, CCDC103 was shown to be critical to stabilize the MT polymeric structure [13].
As far as we know, no human studies regarding mRNA and protein expression of CCDC103 in normal and pathologic conditions have yet been performed, and to get further insights on the role of CCDC103 variants in cilia biology, we studied CCDC103 gene expression in both patients at mRNA and protein levels.
Quantification of mRNA revealed that CCDC103 is expressed in WBC and nasal cilia cells. In WBC, patients exhibited significantly reduced mRNA expression. In nasal cells, no significant differences to controls were observed for patient 1, whereas a significant reduction in mRNA expression was found for patient 2. We believe that the observed differences between both patients are due to the mutation type, with the frameshift variant having a higher potential to affect the phenotype than the missense variant. However, as in nasal cells, the wild-type expression of CCDC103 was much reduced compared with CCDC103 expression in the other cell types, specifically in blood cells (Fig. 7), this could alternatively explain the observed differences of CCDC103 expression in patient nasal and blood cells. In this explanation, as the effect of the missense variant (P1) could be small, consequently, the differences to controls could not be statistically significative. Another alternative hypothesis to explain the observed differences in the two cell types is that CCDC103 may be differently regulated in blood cells and in nasal cells due to alternative splicing mechanisms, and thus, the same variant could affect differently both cell type [28]. For instance, the binding site of the spliceosome may be distinct in different cell types.
As referred above, the missense variant found in patient 1 is predicted by several bioinformatic tools, with high confident scores, as deleterious. It is highly conserved as the proline replacement is not found in any animal homologues of CCDC103. Further, this variant shows a very reduced frequency in population databases. Altogether, it supports a presumed pathogenic role of this sequence variant. However, the mechanism underlying the reduction of CCDC103 expression observed in blood cells of patient 1 was not elucidated and beyond the scope of this study. Nonetheless, we can speculate some gene regulatory mechanisms that could explain such effect. One hypothesis is that the observed reduction of gene expression is due to the regulation of transcriptions factors. Several studies show that the FOXJ1 and RFX transcription factors regulate a multiple sets of genes involved in motile ciliogenesis [25, 29]. A study from Geremek and co-workers had suggested the existence of a coordinated regulation among the ciliary genome, and that the presence of a mutated ciliary gene may interfere with the FOXJ1 and RFX transcription factors and thus down-regulate the expression of a part of ciliary genes that act together [30]. Consequently, it is possible that the missense variant causing a defective CCDC103 protein presented may activate such regulatory mechanism, leading to a down-regulation of ciliary genes, including CCDC103 itself. Another possible explanation is the involvement of micro RNAs (miRNAs). Recent bioinformatics experiments suggest that up to 30% of human genes may be regulated by micro RNAs (miRNA) [31] and, interestingly, according to TargetScan (a web-based tool that predicts biological targets of miRNAs), CCDC103 includes a small sequence region that is well conserved and is complementary to the miRNA-140. Although, the variant found in P1 is not placed in this sequence, this may suggest that CCDC103 may be regulated by miRNA and the variant may interfere in the cascade of regulation. miRNA are still largely unknown, but several lines of evidence are beginning to recognize miRNAs as important cytoplasmic regulators of gene expression that may act as post-transcriptional regulators of their mRNA targets commanding the mRNA degradation and/or translational repression [32]. Further, recent studies have revealed the role of miRNA, namely miR-34/449, in ciliogenesis of motile cilia in vertebrates [33, 34]. Therefore, if we assume that TargetScan database is accurate and CCDC103 is regulated by miRNA, the missense variant presented may be a signal to miRNA-140 to activate a cascade of post-transcriptional regulators that will lead to down-regulation of CCDC103 gene.
We are aware that further studies using animal models or culture of ciliated cells are needed to evaluate the hypothesis here presented. Further, culture of ciliated cells, by ALI-culture, which is already known as a valuable tool for PCD research [35, 36], may be also an alternative to increase statistical power, particularly in the cases where the cells are rare and repeated nasal brushing should be avoided. However, this recent technique still needs further studies to guarantee that the genetic information is not affected by the culture process.
Immunofluorescence staining revealed the presence of CCDC103 in the cytoplasm of cilia cells. Patients exhibited reduced staining, but again, the reduction in patient 2 was more notorious, corroborating the hypothesis that the frameshift variant could have a more deleterious impact on the phenotype. Further studies on animal models are important to understand if those differences in patients’ phenotypes are related to the type of variant or the variant location itself. Immunocytochemistry also revealed CCDC103 staining in the cytoplasm and perinuclear region of cSC and in control and patient 1 germ cells, and in the sperm midpiece. As labelling was reduced in patient 1, results indicate that the CCDC103 pathogenic variant can affect reproductive cells.
Cytoplasmic staining of CCDC103 was previously observed in the cell body of C. reinhardtii [13]. It was proposed that CCDC103 may assemble during axonemal growth, providing a high-affinity track along doublets to allow the association between the ODA/docking complex [35]. Our observation of CCDC103 expression in cells without motile cilia, altogether with its cytoplasmic staining, supports previous findings that CCDC103 act as cytoplasmatic dynein assemble factor. However, in contrast with what was described in the sperm of C. reinhardtii [13], we did not observe CCDC103 staining along the whole flagellum length of human sperm, but only in the sperm midpiece. C. reinhardtii flagella have been proved to be a suitable model species to study the molecular components of axonemes and the ciliogenesis process [36], and the CCDC103 gene is well conserved. Nevertheless, the protein sequence of both species only shares 32% identity, which could explain those location differences. Moreover, both species may certainly have different post-translational modifications that can likely regulate protein location. The present observations clearly indicate that CCDC103 has a broad distribution, being present both in somatic tissues, namely WBC and nasal cilia cells, and in reproductive cells (Sertoli cells, spermatogonia, spermatocytes, spermatids, and sperm). As cytoplasmic dyneins are present in nearly all animal cells [37], our present data corroborates that CCDC103 is a cytoplasmic dynein arm assembly factor and suggests that the CCDC103 gene may be present in an initial pathway of the ciliogenesis process and/or that its function is not exclusive to axonemal DA assemble.
The present results showed that CCDC103 mRNA expression in control nasal cells was very low in comparison to WBC. A quantitative proteomic analysis of human airway cilia also observed that CCDC103 expression was about 0.0015-fold lower than that found in DNAH5 and DNAH9, two well-known cilia genes [38]. In contrast to cilia cells, we observed that sperm presented the highest expression levels, 10-fold more than oocytes and about 2-fold more than WBC, which further confirms that CCDC103 may have a special role in reproductive function and corroborates findings in reproductive cells of patient 1. We here also firstly describe the presence of mRNA CCDC103 transcripts in human cSC and oocytes.
Western blot experiments in control somatic and germ cells showed that the protein CCDC103 forms monomers, dimers, and higher order oligomers, whose size appeared tissue-specific, corroborating previous findings that state oligomerization as a property of CCDC103 [13, 39]. These results show the heterogeneity of this protein and likely reflect different functions or interactions. Earlier studies of CCDC103 in Chlamydomonas and Zebrafish presented that CCDC103 can homodimerize and migrate as a dimer or higher order oligomer following SDS-PAGE [12, 13]. Further, these studies showed that CCDC103 exhibited extraordinary biophysical properties and highlighted the difficulties in obtaining the linear protein structure, as it refold to its native conformation even following heating at 100 °C in the presence of detergent (SDS) [13]. We obtained a great variability among experiences, with the monomer bands not always being observed, which corroborates its strong dimerization proprieties. Therefore, the observed variability was probably due to these biophysical proprieties that made difficult to disrupt the protein native conformation into a linear structure, leading to the absence of a consistently detected in Western blot experiments. Additionally, these results may suggest that this protein may have a high turnover rate, making hard to have good reproducibility. Nevertheless, the great variability among experiments precluded us to comparatively quantify the protein in these experiments.
To infer about the observed size differences among cells, we sequenced the full mRNA of CCDC103, including UTR, which are known to play crucial roles in the post-transcriptional regulation of gene expression [40]. As no differences were detected between cells, data indicate that the differences could be explained by external factors that can regulate the dimerization process. For instance, post-translational modifications, such as tyrosine phosphorylation, were described as having a role in dimerization of the RACK1A protein [41]. According to the NetPhos 3.1 Server (http://www.cbs.dtu.dk/cgi-bin/), several phosphorylation sites are predicted for the CCDC103 sequence, and thus, it is possible that tissue-specific protein regulation could explain differences in the dimerization process. Besides post-translational modifications, the alternative splicing mechanism can also be a possible explanation to the observed CCDC103 protein size differences among different cell types. Although we did not find differences in the sequence of the mRNA, we can not exclude elementary alternative splicing events that do not necessarily affect the mRNA sequence [37]. For instance, cassette exons are splicing events in which an exon from the mature mRNA sequence can be either included or skipped in order to generate two distinct protein isoforms [38].
Moreover, another possibility could be the regulation by microRNAs. It is still a largely unknown mechanism, but several evidences showed that microRNA have an important role in the regulation of mRNA and could thus also have a role in the generation of protein isoforms [39]. Further studies are needed to be performed in cell cultures or animal models to better understand those mechanisms.
We further report, for the first time, CCDC103 staining in testicular germ cells, showing the presence of CCDC103 throughout spermatogenesis, in a cell-stage specific manner. Cytoplasmic and perinuclear staining was observed in spermatogonia (weak), spermatocytes (intense), spermatids (moderate), and Sertoli cells (moderate). Sperm showed intense staining in the midpiece. This data suggests that also exists a cell-stage regulation of CCDC103. This localization is similar to ZMYND10. ZMYND10 is another gene known to be involved in the assembly of DA, which was shown to be strongly expressed in the cytoplasm of mouse primary spermatocytes and spermatids [42]. This suggests that DA assembly could start in spermatocytes.
Conclusions
In conclusion, here we describe that CCDC103 is expressed differently in different reproductive cells and in WBC and that the protein forms dimers whose sizes are tissue specific. We also described for the first time the presence of CCDC103 in WBC and in testicular germ cells, with higher staining at the spermatocyte-stage and local staining in the midpiece of both testicular and ejaculated sperm. Further, we could show that the pathogenic variants found in the CCDC103 gene lead to an absence of DA and to a significant reduction of gene expression and protein expression that was also tissue-specific.
Our work gives a genotype-phenotype correlation of CCDC103 pathogenic variants in PCD patients with situs-inversus totalis and firstly characterizes the expression profiles of CCDC103 in human cells, thus increases the knowledge regarding its expression and subcellular localization. Our data corroborate the involvement of CCDC103 in PCD and suggest also a role in infertility. We believed that CCDC103 may have more unknown functions in cilia biology and male reproduction. Thus, these results added an additional piece to the complex puzzle of the axonemal dynein assembly process and ultimately may help to further understand the PCD pathophysiology.
Electronic supplementary material
Immunocytochemical detection of CCDC103 from control sperm (SZ) obtained from a confocal Z-stack. CCDC103 is present, with a helix-like shape, at sperm midpiece (green). Sperm flagella stained with axoneme-specific acetylated α-tubulin (red) and nuclei stained with DAPI (blue). (AVI 23 kb)
(AVI 27 kb)
List of primers used in this study. (DOCX 16 kb)
Immunogold staining (DOCX 1360 kb)
Acknowledgments
The authors would like to thank Ana Rita Gonçalves, MSc, Center of Medical Genetics Dr. Jacinto Magalhães (CGMJM-CHUP) for Sanger sequencing samples processing; Ângela Alves, MSc, Institute of Biomedical Sciences Abel Salazar-University of Porto (ICBAS-UP) for technical electron microscopy assistance; Raquel Bernardino, PhD, Tânia Dias, Msc, and Ana Maria, MSc for Sertoli cell culture assistance (ICBAS-UP); Fátima Ferreirinha, MSc (ICBAS-UP) for confocal microscopy assistance; and Rui Fernandes, MSc, Institute of Health Research and Innovation (IBMC/i3S-UP) for immunogold labelling assistance.
Funding
UMIB (Pest-OE/SAU/UI0215/2014) is funded by the National Funds through FCT-Foundation for Science and Technology, and FCT Grant ref.: PD/BD/105767/2014 (R.P.).
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The authors declare that they have followed all the rules of ethical conduct regarding originality, data processing and analysis, duplicate publication, and biological material.
Consent for publication
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
J. Oliveira and M. Sousa contributed equally to this work.
Contributor Information
R. Pereira, Email: ruterpereira@gmail.com
M. E. Oliveira, Email: marcia.oliveira@chporto.min-saude.pt
R. Santos, Email: rosario.santos@chporto.min-saude.pt
E. Oliveira, Email: emoliveira@icbas.up.pt
T. Barbosa, Email: telmab@gmail.com
T. Santos, Email: tiago.santos@chedv.min-saude.pt
P. Gonçalves, Email: pmoreiragoncalves@gmail.com
L. Ferraz, Email: ferrasluis@gmail.com
S. Pinto, Email: soraiapinto.pt@gmail.com
A. Barros, Email: abarros@med.up.pt
J. Oliveira, Email: jorgemsmoliveira@gmail.com
M. Sousa, Email: msousa@icbas.up.pt
References
- 1.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison HM, Valente EM. Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol. 2017;241(2):294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 3.Fawcett D. Cilia and flagella. In: Fawcett D, editor. The cell: biochemistry, physiology, morphology. 2. Phyladelphia, USA: WB Saunders company; 1981. pp. 575–603. [Google Scholar]
- 4.Pereira R, Sá R, Barros A, Sousa M. Major regulatory mechanisms involved in sperm motility. Asian J Androl. 2017;19(1):5–14. doi: 10.4103/1008-682X.167716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King SM. The dynein microtubule motor. Biochim Biophys Acta (BBA) - Mol Cell Res. 2000;1496(1):60–75. doi: 10.1016/S0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- 6.Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, Davis SD, Knowles MR, Zariwala MA. Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet Med. 2009;11:473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson CL, Behan L, Collins SA, Goggin PM, Adam EC, Coles JL, Evans HJ, Harris A, Lackie P, Packham S, Page A, Thompson J, Walker WT, Kuehni C, Lucas JS. Accuracy of diagnostic testing in primary ciliary dyskinesia. Eur Respir J. 2016;47:837–848. doi: 10.1183/13993003.00749-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 9.Pereira R, Oliveira J, Sousa M. A molecular approach to sperm immotility in humans: a review. Med Reprod Embriol Clín. 2014;01(1):15–25. [Google Scholar]
- 10.Kurkowiak M, Ziętkiewicz E, Witt M. Recent advances in primary ciliary dyskinesia genetics. J Med Genet. 2015;52:1–9. doi: 10.1136/jmedgenet-2014-102755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira R, Oliveira J, Ferraz L, Barros A, Santos R, Sousa M. Mutation analysis in patients with total sperm immotility. J Assist Reprod Genet. 2015;32(6):893–902. doi: 10.1007/s10815-015-0474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panizzi JR, Becker-heck A, Castleman VH, Al-mutairi D, Liu Y, Loges NT, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King SM, Patel-King RS. The oligomeric outer dynein arm assembly factor CCDC103 is tightly integrated within the ciliary axoneme and exhibits periodic binding to microtubules. J Biol Chem. 2015;290:7388–7401. doi: 10.1074/jbc.M114.616425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira J, Negrao L, Fineza I, Taipa R, Melo-Pires M, Fortuna AM, et al. New splicing mutation in the choline kinase beta (CHKB) gene causing a muscular dystrophy detected by whole-exome sequencing. J Hum Genet. 2015;60:305–312. doi: 10.1038/jhg.2015.20. [DOI] [PubMed] [Google Scholar]
- 16.Rutland J, Dewar A, Cox T, Cole P. Nasal brushing for the study of ciliary ultrastructure. J Clin Pathol. 1982;35:357–359. doi: 10.1136/jcp.35.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernardino RL, Alves MG, Oliveira PF. Establishment of primary culture of Sertoli cells. In: Alves MG, Oliveira PF, editors. Sertoli Cells. New York, NY: Springer, Humana Press; 2018. pp. 1–8. [DOI] [PubMed] [Google Scholar]
- 18.Afzelius BA, Srurgess JM. The immotile-cilia syndrome: a microtubule-associated defect. Crit Rev Biochem Mol Biol. 1985;19:63–87. doi: 10.3109/10409238509086788. [DOI] [PubMed] [Google Scholar]
- 19.Sousa M, Oliveira E, Alves Â, Gouveia M, Figueiredo H, Ferraz L, et al. Ultrastructural analysis of five patients with total sperm immotility. Zygote. 2015;23(6):1–8. doi: 10.1017/S0967199414000616. [DOI] [PubMed] [Google Scholar]
- 20.De Iongh R, Rutland J. Orientation of respiratory tract cilia in patients with primary ciliary dyskinesia, bronchiectasis, and in normal subjects. J Clin Pathol. 1989;42:613–619. doi: 10.1136/jcp.42.6.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for Geneticists. New York: John Wiley & Sons, Ltd; 2003. pp. 289–316. [Google Scholar]
- 24.Venselaar H, Te Beek T A, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinforma. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014;141(7):1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 26.Maurizy C, Quinternet M, Abel Y, Verheggen C, Santo PE, Bourguet M, C.F. Paiva A, Bragantini B, Chagot ME, Robert MC, Abeza C, Fabre P, Fort P, Vandermoere F, M.F. Sousa P, Rain JC, Charpentier B, Cianférani S, Bandeiras TM, Pradet-Balade B, Manival X, Bertrand E. The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nat Commun. 2018;9(1):2093. doi: 10.1038/s41467-018-04431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wayne N, Mishra P, Bolon DN. Hsp90 and client protein maturation. In: Calderwood SK, Prince TL, editors. Molecular chaperones: methods and protocols. Totowa, NJ: Humana Press; 2011. pp. 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mcmanus CJ, Graveley BR, Treisman J, Richter J. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas J, Morlé L, Soulavie F, Laurençon A, Sagnol S, Durand B. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell. 2010;102(9):499–513. doi: 10.1042/BC20100035. [DOI] [PubMed] [Google Scholar]
- 30.Geremek M, Ziętkiewicz E, Bruinenberg M, Franke L, Pogorzelski A, Wijmenga C, Witt M. Ciliary genes are down-regulated in bronchial tissue of primary ciliary dyskinesia patients. PLoS One. 2014;9(2):e88216. doi: 10.1371/journal.pone.0088216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA Targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi L-E, Cibois M, Robbe-Sermesant K, Jolly T, Cardinaud B, Moreilhon C, Giovannini-Chami L, Nawrocki-Raby B, Birembaut P, Waldmann R, Kodjabachian L, Barbry P. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 34.Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, Kessel M, He L. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510(7503):115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King SM. Axonemal dynein arms. Cold Spring Harb Perspect Biol. 2016;8(11):a028100. doi: 10.1101/cshperspect.a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components Unicellular algae Chlamydomonas. Mol Hum Reprod. 2011;17(8):524–538. doi: 10.1093/molehr/gar034. [DOI] [PubMed] [Google Scholar]
- 37.Neely MD, Boekelheide K. Sertoli cell processes have axoplasmic features: an ordered microtubule distribution and an abundant high molecular weight microtubule-associated protein (cytoplasmic dynein) J Cell Biol. 1988;107(5):1767–1776. doi: 10.1083/jcb.107.5.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackburn K, Bustamante-Marin X, Yin W, Goshe MB, Ostrowski LE. Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J Proteome Res. 2017;16(4):1579–1592. doi: 10.1021/acs.jproteome.6b00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoemark A, Moya E, Hirst RA, Patel MP, Robson E, Hayward J, et al. A high prevalence CCDC103 p.His154Pro mutation causing primary ciliary dyskinesia is associated with normal diagnostic investigations. Eur Respir J. 2017;50(suppl 61):PA1851. doi: 10.1136/thoraxjnl-2017-209999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9(5):563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 41.Sabila M, Kundu N, Deana S, Ullah H. Tyrosine phosphorylation based homo-dimerization of Arabidopsis RACK1A proteins regulates oxidative stress signaling pathways in yeast. Front Plant Sci. 2016;7:176. doi: 10.3389/fpls.2016.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali GR, Yeyati PL, Mizuno S, Dodd DO, Tennant PA, Keighren MA, et al. ZMYND10 functions in a chaperone relay during axonemal dynein assembly. eLife. 2018;7:e34389.10.7554/eLife.34389. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunocytochemical detection of CCDC103 from control sperm (SZ) obtained from a confocal Z-stack. CCDC103 is present, with a helix-like shape, at sperm midpiece (green). Sperm flagella stained with axoneme-specific acetylated α-tubulin (red) and nuclei stained with DAPI (blue). (AVI 23 kb)
(AVI 27 kb)
List of primers used in this study. (DOCX 16 kb)
Immunogold staining (DOCX 1360 kb)