Abstract
Background
Strategies for combatting residual malaria by targeting vectors outdoors are gaining importance as the limitations of primary indoor interventions are reached. Strategies to target ovipositing females or her offspring are broadly applicable because all mosquitoes require aquatic habitats for immature development irrespective of their biting or resting preferences. Oviposition site selection by gravid females is frequently studied by counting early instar larvae in habitats; an approach which is valid only if the number of larvae correlates with the number of females laying eggs. This hypothesis was tested against the alternative, that a higher abundance of larvae results from improved survival of a similar or fewer number of families.
Methods
In a controlled experiment, 20 outdoor artificial ponds were left uncovered for 4 days to allow oviposition by wild mosquitoes, then covered with netting and first and second instar larvae sampled daily. Natural Anopheles habitats of two different types were also identified, and all visible larvae sampled. All larvae were identified to species, and most samples of the predominant species, Anopheles arabiensis, were genotyped using microsatellites for sibling group reconstructions using two contrasting softwares, BAPS and COLONY.
Results
In the ponds, the number of families reconstructed by each software significantly predicted larval abundance (BAPS R2 = 0.318, p = 0.01; COLONY R2 = 0.476, p = 0.001), and suggested that around 50% of females spread larvae across multiple ponds (skip oviposition). From natural habitats, the mean family size again predicted larval abundance using BAPS (R2 = 0.829, p = 0.017) though not using COLONY (R2 = 0.218, p = 0.68), but both softwares once more suggested high rates of skip oviposition (in excess of 50%).
Conclusion
This study shows that, whether in closely-located artificial habitats or natural breeding sites, higher early instar larval densities result from more females laying eggs in these sites. These results provide empirical support for use of early instar larval abundance as an index for oviposition site preference. Furthermore, the sharing of habitats by multiple females and the high skip-oviposition rate in An. arabiensis suggest that larviciding by auto-dissemination of insecticide may be successful.
Keywords: Microsatellites, Anopheles arabiensis, Larval control, Skip-oviposition, Auto-dissemination, Sibship-reconstruction
Background
Concerted efforts towards malaria control and elimination have led to a global decline in malaria cases by about 40% between 2000 and 2015 [1]. Almost 80% of this reduction is attributed to vector control by widespread distribution of insecticide-treated nets (ITNs) and indoor residual spray (IRS) [1]. Despite these successes, malaria reduction in the World Health Organization (WHO) African region has stalled [2], which may in part reflect limitations of ITNs and IRS. These limitations include plasticity in behaviours including early biting and outdoor resting, and feeding on animals allowing adult malaria vectors to avoid exposure to insecticides [3, 4], widespread insecticide resistance [5, 6], and high operational costs of IRS in particular which limit coverage [2]. Implementation of supplementary vector control tools is required to further reduce malaria transmission in a trajectory toward elimination [7, 8].
Larval source management (LSM) is a tool for further development because all mosquitoes need to lay eggs in an aquatic habitat irrespective of their biting or resting preferences [9]. However, the uptake of this intervention is impeded by the management effort required [10] and the lack of knowledge of aquatic habitats that are the most preferred for egg laying, which would allow a more spatially-targeted approach. The oviposition behaviour of Anopheles mosquitoes, and specifically the preference of particular aquatic habitats for egg-laying, has been studied in order to better target LSM and to develop novel attract and kill strategies for vector control [11, 12]. Habitat preferences are frequently inferred from the abundance of early instar larvae in a habitat [13]. This relies on the assumption that higher early instar larval density results from a greater number of gravid females selecting the site for oviposition [11, 12]. However, to date only indirect tests of this assumption exists [14]. Furthermore, whilst there is strong indication from cage experiments that species of the Anopheles gambiae complex frequently distribute their eggs in more than one egg-laying sites (skip-oviposition) [15], this behaviour has not yet been widely accepted in Anopheles due to the few studies providing supporting evidence [14].
An LSM tactic currently being explored is the auto-dissemination of insect growth regulators (IGRs) where adult mosquitoes naturally transfer the IGR from resting to breeding sites [16, 17]. Large numbers of females visiting multiple breeding sites would aid transfer of IGR among habitats to achieve biologically-relevant mortality of the immature stages. However, skip oviposition has been widely accepted to occur habitually in Aedes mosquitos unlike in Anopheles [18–20]. In typical habitats of western Kenya early instar densities vary from 160/m2 in puddles to between 0.3 and 10/m2 in open drainages, cultivated swamps, and river fringes [13, 21]. Anopheles females can lay up to 200 eggs [15], and it is unclear whether the typically-low densities of early instars are a result of few gravid females visiting most habitats or of high mortality of the eggs or larvae from many females.
Here genotype-based family reconstructions methods were used to enumerate the number of female Anopheles arabiensis depositing eggs in single and multiple habitats in relation to overall larval densities. It was hypothesized that relative high number of early instar An. arabiensis larvae in a habitat is an indicator of high number of females laying eggs in a site; therefore, differences in early instar abundance across similar habitat types of comparable size correlate with the number of females laying eggs.
Methods
Collection and rearing of Anopheles arabiensis mosquito families for relatedness testing
Wild blood-fed Anopheles females were collected in May 2015 by aspiration from a cattle-baited trap [22] set up in Kirindo village, Mbita sub-county (0°26′38.46″S 34°15′36.95″E), western Kenya. Anopheles gambiae sensu lato (s.l.) females were identified morphologically, and other mosquito species discarded. Individual females were held in paper cups covered with a fine net. Moist cotton wool covered with filter paper was provided at the base for egg laying. Food was provided ad libitum by a disc of cotton wool soaked with 10% glucose solution on top of the cup. After egg laying, single legs from individual females were genotyped to identify species [23] to ensure that only An. arabiensis families were reared. Anopheles arabiensis eggs from each mother were raised separately in trays and the mothers preserved individually at − 80 °C in Eppendorf tubes containing absolute ethanol. After hatching, larvae were fed twice daily on TetraMin fish food (Tetra, Germany) and pupal stages from each family were separated into females and males prior to emergence to prevent mating. Following emergence, single virgin females and three males from different families were held in 15 × 15 cm mosquito cages for 48 h to copulate. Females were then offered two blood meals and held in paper cups to lay eggs, with resulting offspring reared to early instar stage. All the early instar larvae offspring were preserved at − 80 °C in Eppendorf tubes (20 larvae/tube) containing absolute ethanol for subsequent DNA analyses.
Standardized semi-field evaluation
Semi-field experiments were conducted at the International Centre for Insect Physiology and Ecology (ICIPE), Mbita, western Kenya, between May and June 2016. Twenty artificial ponds were created in a field on campus using plastic tubs (40 cm diameter, 20 cm deep) fully buried in the ground and filled with 50 l of untreated water originating from Lake Victoria to mimic typical Anopheles mosquito breeding habitats. The ponds were set up in a 4 × 5 pond grid with neighboring ponds in each gridline 4 m apart. Ponds were left open for 4 days to allow for natural colonization by egg-laying wild female mosquitoes and then covered with netting on day 5 to prevent further egg laying. All visible larvae were sampled exhaustively daily for a further 6 days to allow time for most of the eggs to hatch. Larvae from each habitat were preserved at − 80 °C in 15 ml Falcon tubes containing absolute ethanol for further analysis.
Field survey of natural aquatic mosquito larval habitats
Field surveys and larval sampling from natural aquatic habitats was conducted between March and May 2017 within a rice irrigation scheme (00°28.473′S 034°32.786′E) (see Additional file 1). Two common mosquito larval habitat categories in the area—puddles and drainage ditches—were identified. Six puddles and five ditches of a perimeter size between 0.64 and 11 m2 were the selected, and exhaustively sampled using larval sweep nets. The estimated pairwise distance between the habitats ranged from 2 m between drainage 2 (D2) and puddle 7 (P7) to 1264 m between puddle 1 (P1) and puddle 8 (P8) Anopheles larvae were separated from the collections morphologically and preserved at − 80 °C in 15 ml Falcon tubes containing absolute ethanol.
DNA extractions and species identification
Genomic DNA was extracted from: (i) the two An. arabiensis families (Family A—adults n = 4, larval offspring n = 46; Family B—adults n = 4, larval offspring n = 36) collected from cattle-baited traps; (ii) the An. gambiae s.l. larvae collected from the artificial pond habitats (n = 466); and (iii) the natural field habitats (n = 702). Samples were extracted using the Nexttec DNA isolation kit (Biotechnologie, GmbH) following manufactures instructions. Following PCR-based species identification [23], but prior to further analysis, An. arabiensis samples from different artificial ponds and natural habitats were randomized on DNA extraction plates to mitigate any bias from plate batch effects.
Microsatellite genotyping
Fifteen An. arabiensis microsatellite DNA markers located on chromosomes 2 and 3 were used in this study (Table 1). Markers were allocated into four multiplexes based on compatible primer annealing temperatures and non-overlapping expected allele size ranges using Multiplex Manager software [24]. Forward primers were fluorescently labelled with either NED, VIC, 6-FAM, or PET dyes. Amplification was carried out using the Qiagen Type-it microsatellite PCR kit with each reaction consisting of 6.25 μl Type-it master mix, 1.25 μl primer mix, 3 μl water, and 2 μl DNA template. The thermocycling conditions were as follows: initial activation at 95 °C for 5 min, 30cycles of denaturation at 95 °C for 30 s, annealing at 57–63 °C for 90 s, extension at 72 °C for 30 s with a final extension at 60 °C for 30 min. The PCR fragments were separated on an ABI 3730XL (Applied Biosystem) sequence analyser using the GeneScan™–500LIZ™ size standard. The allele sizes were scored using GeneMarker software v.2.6.7 (SoftGenetics) with each allele size score checked manually.
Table 1.
Details of microsatellite primer sequences used for the genotyping
| Plex | Marker (label) | Concentration (μM) | Chromosome | Genetic location | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|---|---|---|---|
| 1 | CDC675-FAM [33] | 1.75 | 2L | 24C | TCAAACTCGAACTCCTCAAC | TTTCCGTCGATAGTTTTCTG |
| CDC22-FAM [33] | 1.75 | 2L | 22D | GGGCAAAGAGAAAGCAA | AGCTGTGTGGCAGGTTT | |
| CDC46-NED [33] | 2.5 | 3L | 45C | GTGGTTGACCGATTTGTAAG | ATTTATTCACTCGCCAAGAA | |
| 2 | CDC18-FAM [33] | 1.9 | 2R | 18C | CAGGAAGCGATGTGAAAGT | GGAGTGTTGTCGTTCATCTT |
| CDC28C-FAM [33] | 2.3 | 2L | 28C | TGTGCCGGTTGAGAGAGA | GGGCGAGAACATTAACAA | |
| Ag2:79-NED [34] | 1.9 | 2R | 11B | CGGGTAGCGCTAGAAGTATG | AGAGAAATGTGCCGAAGGGG | |
| 2RiS5-PET [35] | 1.9 | 2R | 12C | TTCTCGAAAGACTGCTGCTG | ATTGGATCGAAAACGGTCTG | |
| 3 | CDC40B-FAM [33] | 1.9 | 3L | 40B | ATGCATGCAAATCGGTAT | TATCGAGGCAAATCGGTA |
| CDC44-VIC [33] | 2.3 | 3L | 44B | ATGCATGCAAATCGGTAT | TATCGAGGCAAATCGGTA | |
| CDC32-NED [33] | 1.9 | 3R | 32A | GTTTGCTTGCTTGTTGTTGT | GTGCTCAACGCCTACAAAT | |
| CDC34-PET [33] | 1.9 | 3R | 34B | AAAACTTTTCCCTCCCATTC | AAGTGCAGCAATTGACGAG | |
| 4 | Ag3:128-FAM [34] | 1.9 | 3R | 29C | CGGGACGGCTAGATAAAGCG | CCGGGCGACATAACCCACCC |
| Ag2:143-VIC [34] | 1.5 | 2L | 25D | CGTACGAGTGAGTGAGTTGG | CAAAAATAGCATCACGGCCG | |
| Ag2:46-NED [34] | 2.7 | 2R | 7A | CGCCCATAGACAACGAAAGG | TGTACAGCTGCAGAACGAGC | |
| Ag3:249-NED [34] | 1.9 | 3R | 30B | ATGTTCCGCACTTCCGACAC | GCGAGCTACAACAATGGAGC |
The markers were grouped into four PCR multiplexes
Family group assignment
Genotyping errors possibly due to null alleles, large allele dropouts and mis-scoring of stutter peaks were identified using MICRO-CHECKER v.2.2.3 [25].
The capacity of the markers to determine sibships within known pedigrees was first tested using the two An. arabiensis families, A and B, with known parents. Family reconstruction was performed using two programs, COLONY [26] and BAPS 6 [27], that differ fundamentally in their methodologies and thus provide independent analyses. COLONY implements a likelihood algorithm in which individuals are simultaneously inferred into groups with the aim of finding the most likely grouping. In COLONY a full-likelihood (FL) analysis algorithm was used without a sibship prior and using a pool of allele frequencies from the natural habitats as a reference population. BAPS implement a Bayesian clustering algorithm treating both sample allele frequencies and the number of larval family groups as random variables when inferring clusters. In BAPS an individual clustering analysis was performed with the number of possible clusters ranging from 5 to 100.
Statistical analysis
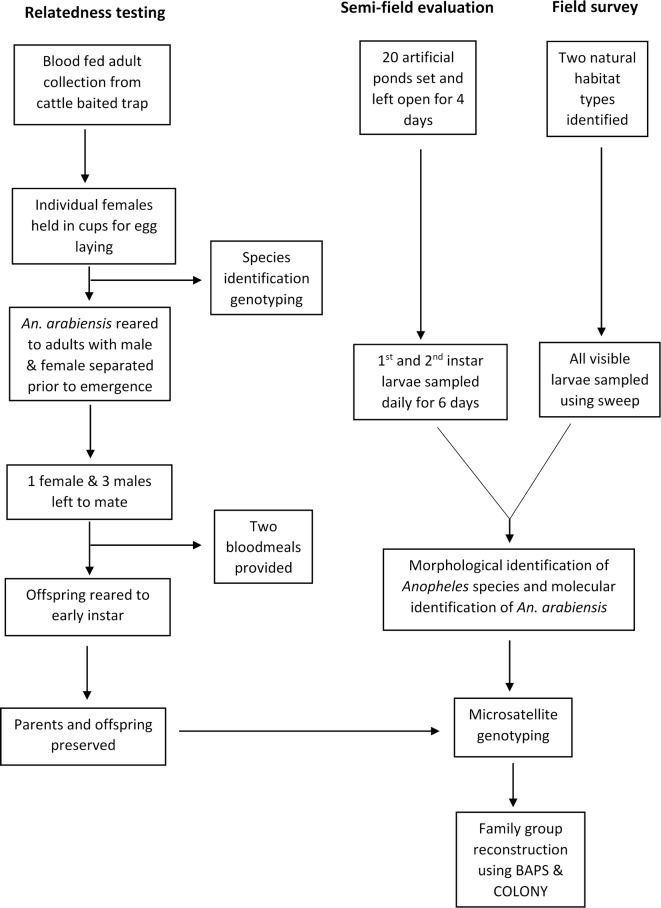
A Kolmogorov–Smirnov test was used to check for normality of the distributions for numbers of larvae and reconstructed families, followed by a Grubbs test to check for outliers in the data, which were removed if detected. Pearson correlation and linear regression were used to test the relationship between the number of families and larval abundance. A partial Mantel test was used to test whether large families were more likely to be split across multiple habitats and if split families were more likely to occur in closer habitats in the software ZT [28]. For this test, a dummy (1/0) variable indicated whether habitats were direct neighbours or not, whilst controlling for sample size (as the minimum sample size per pair of habitats). The different experimental designs and analysis methods used in this study have been summarized in Fig. 1.
Fig. 1.
Flowchart of the experimental design and analysis methods used in the study
Results
Marker testing using known sibship
The capacity of 15 microsatellite markers, developed for An. arabiensis, to determine relatedness within known pedigrees was tested using two families of known full-siblings (A: n = 46 offspring; B: n = 36 offspring). The genotype from the mothers in Family B were inconclusive because of lack of informativeness of the loci, with some genotypes missing and therefore, only genotypes from family A were used for determination of parent–offspring Mendelian segregation but both families were used for sibship analysis. Twelve of the 15 markers yielded segregation patterns between the parents and amongst full-sibs in family A or amongst full sibs in family B consistent with Mendelian inheritance (see Additional file 2), and these markers were retained for subsequent analyses. Clustering analysis using either COLONY or BAPS provided entirely concordant results and correctly identified a single cluster of siblings within each family with no cross-family assignment.
Relatedness among the standardized artificial ponds
Eighteen of the twenty artificial ponds were colonized, with a total of 466 Anopheles larvae collected. The total number of larvae collected from each pond ranged from 4 to 62 (median = 16.5, inter-quartile range = 31) (Table 2). From the species identification genotyping, 96.8% (n = 451) of the larvae were identified as An. arabiensis and were subsequently genotyped using microsatellites, 1.9% (n = 9) An. gambiae sensu stricto (s.s.), with 1.3% (n = 6) failed reactions.
Table 2.
The number of mosquito families estimated using BAPS and COLONY in the artificial ponds
| POND | N | n | BAPS | COLONY |
|---|---|---|---|---|
| A | 42 | 37 | 6 | 13 |
| C | 5 | 4 | 1 | 1 |
| D | 40 | 40 | 5 | 6 |
| E | 8 | 6 | 5 | 5 |
| F | 10 | 10 | 3 | 9 |
| G | 17 | 14 | 5 | 6 |
| H | 33 | 26 | 2 | 7 |
| I | 5 | 4 | 3 | 3 |
| J | 59 | 44 | 6 | 9 |
| K | 41 | 36 | 5 | 11 |
| L | 52 | 23 | 3 | 4 |
| M | 62 | 60 | 6 | 10 |
| N | 4 | 4 | 2 | 2 |
| O | 39 | 34 | 2 | 5 |
| P | 13 | 9 | 2 | 4 |
| Q | 16 | 16 | 4 | 5 |
| R | 10 | 8 | 5 | 6 |
| T | 10 | 10 | 3 | 5 |
N is the total number of An. arabiensis larvae from each pond, and n is the number successfully genotyped
COLONY estimated on average more females per pond compared to BAPS (Table 2), but the results were well correlated across families (r = 0.715, p < 0.001). Overall, BAPS estimated a total of 28 gravid females to have laid eggs across the 18 ponds while the COLONY analysis suggested 70 gravid females. Despite the difference in the total number of families estimated, 95% of the pairwise assignments of individuals as siblings in COLONY were also identified by BAPS, but the latter then appears to reconstruct many additional family pairings within its much larger clusters. In both clustering methods, the total number of larvae in each pond significantly predicted the number of families reconstructed (BAPS R2 = 0.318, p = 0.01; COLONY R2 = 0.476, p < 0.001) (Fig. 2). This corroborated the hypothesis that a relatively higher number of larvae in a pond is indicative of a higher number of females laying eggs in the pond as compared to a pond with a lower number of larvae in close vicinity.
Fig. 2.
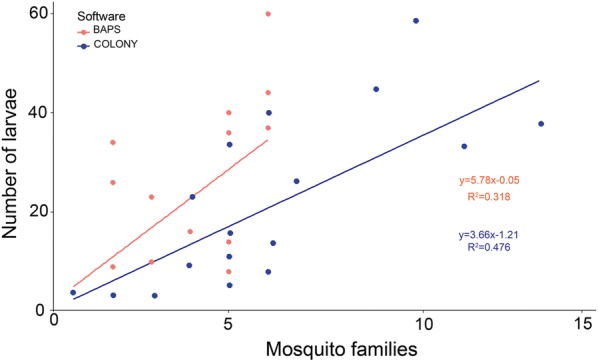
Relationship between mosquito family clusters inferred using COLONY and BAPS with the total number of larvae from each artificial pond habitat
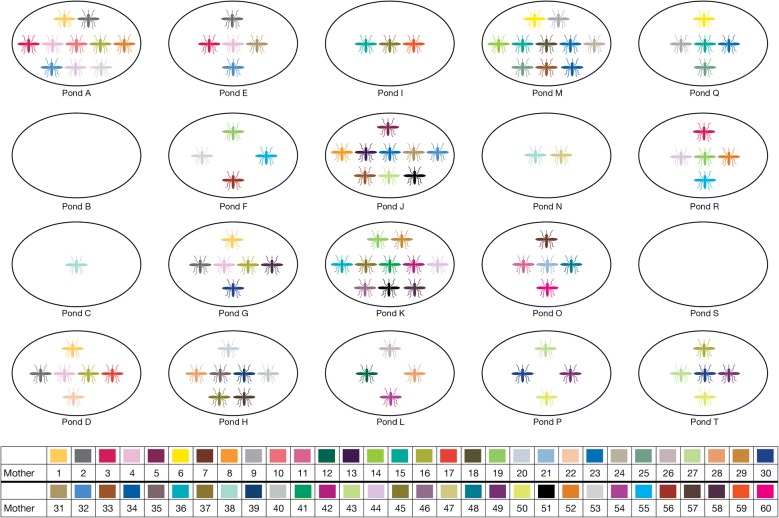
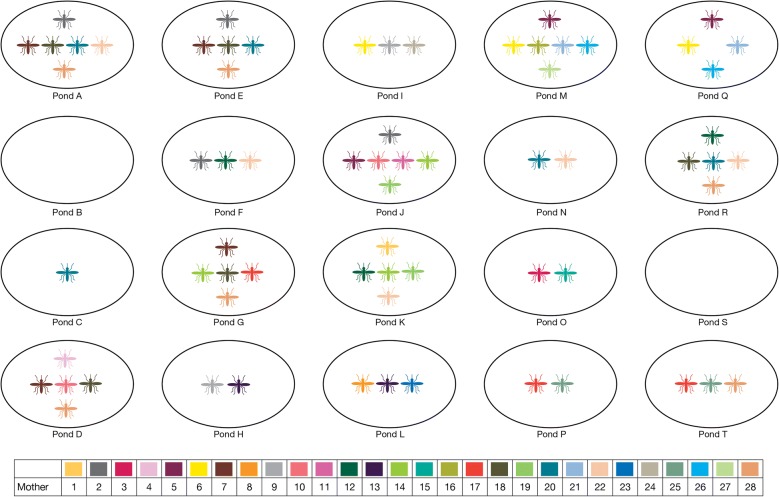
Average numbers of larvae per family were quite low but very variable (median, IQR-BAPS 4.3, 4.9; COLONY 3.0, 2.9) suggesting high mortality of unsampled first instar larvae or eggs. Overall, BAPS estimated that 57% (95% CI 39, 75) females to have deposited larvae in multiple ponds and COLONY 43% (95% CI 30, 56), suggesting a high frequency of skip-oviposition by the mothers (Figs. 3, 4). Given the high skip-oviposition estimates, two linked questions were asked: are larger families more likely to be split across ponds, and are split families more likely to occur in closer ponds? There was a significant correlation between the number of larvae from each mosquito family and the number of ponds in which they were found using either estimator (BAPS r = 0.46, p = 0.015; COLONY r = 0.35, p = 0.003). Dependence of family sharing on pairwise proximity of pond pairs, while controlling for the minimum sample size per pair of ponds, was also evaluated in a partial Mantel test. There was a non-significant result for BAPS (r = 0.107 p = 0.11) and a marginally significant correlation for COLONY (r = 0.15 p = 0.04). This suggests that over the small spatial scale examined in the experimental pond design (Figs. 3, 4), larger families were more likely to be split but the preference of females to lay their eggs in physically closer habitats was weak.
Fig. 3.
Pond setup and mosquito family distribution across the 18 ponds as inferred using BAPS. The colours represent families from a single mother
Fig. 4.
Pond setup and mosquito family distribution across the 18 ponds as inferred using COLONY. The colours represent families from a single mother with only families with two or more offspring visualized (n = 60)
Relatedness in natural habitats
A total of 702 larvae were sampled from across 11 natural habitats with the total number of larvae in each habitat ranging from 28 to 109 (median = 41, IQR = 41) (Table 3); 79% (n = 556) were identified as An. arabiensis, which were genotyped using microsatellites, 9.7% (n = 68) An. gambiae s.s., and 11.1% (n = 74) were failed reactions, some of which may have been non-An. gambiae s.l. Owing to much greater variability in genotyping success in the natural habitat samples, mean family size (number of larvae genotyped divided by number of families) was used as a metric for analysis rather than the number of mosquito families as in the ponds. Larval counts from habitat 8 was found to be an outlier compared to the other 11 natural habitats (p < 0.05) and was excluded from the analysis. After this exclusion all the larval counts from all habitats were normally distributed, Kolmogorov–Smirnov test p > 0.05.
Table 3.
The number of mosquito families estimated using BAPS and COLONY in the natural habitats
| Habitat | Habitat size (M) | N | n | BAPS | COLONY | Mean family size | |
|---|---|---|---|---|---|---|---|
| BAPS | COLONY | ||||||
| P1 | 0.6 | 28 | 18 | 9 | 13 | 2.0 | 1.4 |
| P5 | 0.3 | 36 | 22 | 13 | 20 | 1.7 | 1.1 |
| P6 | 1.4 | 37 | 25 | 14 | 19 | 1.8 | 1.3 |
| P7 | 1.3 | 66 | 10 | 6 | 9 | 1.7 | 1.1 |
| P8 | 6.9 | 191 | 97 | 21 | 75 | 4.6 | 1.3 |
| P9 | 2.6 | 55 | 51 | 17 | 37 | 3.0 | 1.4 |
| D1 | 11.0 | 33 | 30 | 16 | 25 | 1.9 | 1.2 |
| D2 | 5.8 | 109 | 60 | 19 | 42 | 3.2 | 1.4 |
| D3 | 1.9 | 41 | 15 | 9 | 12 | 1.7 | 1.3 |
| D9 | 3.7 | 33 | 24 | 14 | 18 | 1.7 | 1.3 |
N is the total number of An. arabiensis larvae from each habitat, and n is the number successfully genotyped. In the habitat column, prefix P is puddle habitats while D is drainage ditches
Once again COLONY estimated many more families (N = 171) compared to BAPS (N = 41) but the results for mean family sizes estimated using each method were marginally correlated (r = 0.634, p = 0.049). Despite this correlation, only 56% of the pairwise sibship assignments in COLONY were also detected by BAPS suggesting a more moderate agreement between the two software than in the pond experiment. Mean family sizes from the BAPS estimation predicted larval abundance but those estimated by COLONY did not (BAPS R2 = 0.829, p = 0.017; COLONY R2 = 0.218, p = 0.68) (Fig. 5). Similarly, only results from the BAPS analysis supported the hypothesis that a higher number of larvae in one habitat compared to another at the same time is indicative of a higher number of females choosing to lay eggs. Nevertheless, both estimators suggested once again a high frequency of skip-oviposition (BAPS = 73%; 95% CI 59, 87; COLONY = 60%; 95% CI 53, 67.
Fig. 5.
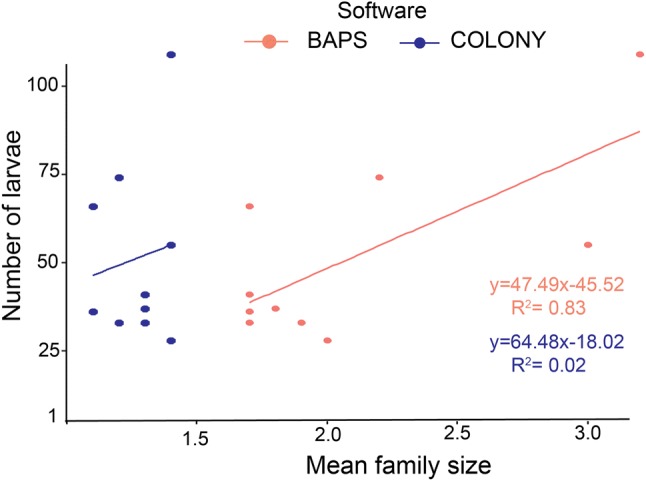
A regression curve comparing relationship between mosquito families inferred using COLONY and BAPS with the total number of larvae from natural habitats
Analysis to answer the same two linked questions as before, found that the mean family size was significantly positively correlated with the number of habitats among which they were distributed using BAPS (r = 0.902, p < 0.001) or COLONY (r = 0.451, p < 0.001). The tests of whether family sharing was dependent on proximity of habitat pairs were both non-significant (BAPS partial Mantel test r = 0.104, p = 0.22; COLONY r = − 0.027, p = 0.43).
Discussion
Understanding mosquito oviposition behaviour is important to inform the development and implementation of breeding habitat-targeted mosquito control strategies. However, direct observational studies of oviposition behaviour are challenging due to its nocturnal occurrence. As such, limited evidence exists to support phenomena such as habitat preference, variations in larval densities across breeding habitats and frequency of skip oviposition by An. gambiae complex females.
In this study, genetic clustering (BAPS) and sibship reconstruction (COLONY) methods were applied to enumerate the number of female An. arabiensis depositing eggs in single and multiple habitats in relation to overall larval densities. The findings show that relative high number of early instar larvae in a breeding habitat is indicative of an increased number of ovipositing females. This supports the hypothesis that early instar larval abundance of An. arabiensis is an indicator of the number of females laying eggs in a habitat and can hence be used to investigate oviposition preferences. This finding is consistent with a study that previously found a positive correlation between the number of mosquito families and number of larvae in a breeding site [29]. The family sizes detected were generally low, concordant with previous studies [14, 29], and the discrepancy between potential an actual family sizes maybe attributable to low egg hatchability and first instar survival rates [30].
This study also found that An. arabiensis females often display skip oviposition, distributing their eggs in multiple habitats. On average, more than half of the families investigated from the experimental and natural habitat surveys resulted from skip-oviposition. The ponds and natural habitats were in a small spatial scale, which could have contributed to the high skip oviposition rates. Since gravid Anopheles females tend to hover over habitats before deciding to lay eggs, it was not surprising that skip-oviposition occurred over the entire range. This finding is similar to a study on An. gambiae s.s., which found that 57% of females had skip-oviposited in habitats [14]. In contrast, a much lower skip-oviposition frequency of 26% was observed in two-choice cage assays on An. gambiae s.s. oviposition in the laboratory [15]. The low frequencies observed in this cage assay could have been due to the individual mosquitoes responding to a similar oviposition substrate reducing preference to skip oviposit. There was also a relationship between mosquito family size and the number of habitats in which they were distributed in both the artificial ponds and natural habitats implying that females that laid more eggs were more likely to exhibit skip oviposition.
Frequent skip-oviposition behaviour could contribute positively to the success of An. arabiensis mosquito control methods using auto-dissemination of insect growth regulators (IGRs) where skipping adult females naturally transfer the IGR from resting to breeding sites. Anopheles arabiensis have been shown to experimentally transfer pyriproxyfen from their resting sites to aquatic habitats leading to significantly reduced larval emergence [16, 17]. Our results suggest that skip oviposition behaviour should also be considered as an important factor in such studies and in the ecology of these vectors.
The number of families estimated using BAPS were consistently much lower than COLONY both in the artificial ponds and natural habitats. This could been in part due to a tendency of COLONY to over-split large families as previously noted in simulation studies [31, 32]. Also, whilst both estimators supported the relationship that high larval density in a breeding habitat is indicative of an increased number of ovipositing females in the ponds, only BAPS did so in the natural habitats. However, despite contrasting family sizes between the two software, individual pairwise sib-ship assignments made by both agreed strongly in the ponds, although the correspondence was more moderate in the natural habitats. This may reflect a problem with ‘self-referencing’ in COLONY, whereby in the natural habitats the same genotype set was used as those assigned, rather than a wider set fully representing genetic diversity of the population. A similar observation was made in the two families of known full-siblings and in the pond samples when ‘self-referencing’ with COLONY.
Conclusion
The current study demonstrates the potential use of An. arabiensis larval abundance in breeding habitats as an indicator of oviposition site preference and provides empirical evidence of frequent skip-oviposition behaviour, which should be considered when studying their ecology and applying larval control methods. These findings support previous findings for An. gambiae s.s., but at present little is known about oviposition behavior in the other major East African malaria vector Anopheles funestus, and studies are now warranted.
Supplementary information
Additional file 1. Schematic view of larval sampling sites at Oluch-Kimira rice irrigation scheme in Homa-Bay county, western Kenya. In the site labels, prefix P is puddle while D is drainage ditch.
Additional file 2. Mendelian test for allele inheritance for parents and offspring in family A. The markers with p values in bold indicate consistency with Mendelian allele segregation rule. N/A represent markers that did not amplify, and n/a represent homozygous alleles whose p values could not be calculated.
Acknowledgements
Thanks to Elizabeth Masinde, Paul Ouma, and Rose Imelda for assistance with the field collection of adult and larval mosquitoes, setting up the artificial ponds, and morphological identification of the mosquito larvae.
Abbreviations
- LSM
larval source management
- IGRs
insect growth regulators
- CBT
cattle baited traps
- ITN
insecticide-treated net
- IRS
indoor residual spray
Authors’ contributions
UF, JO, DKM and DW conceived the study, secured funding, and designed the experiments. JO and EJR performed the laboratory analysis and collected all data. Statistical analysis and interpretation were performed by JO, UF and DW. JO drafted the manuscript which was critically revised by UF and DW. All authors read and approved the final manuscript.
Funding
This work was supported by the Wellcome Trust International Masters Fellowships, Grant # 106450/Z/14/Z to JO. JO was also partly supported by a DELTAS Africa Initiative Grant # DEL-15-011 to THRiVE 2.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-019-2917-5.
References
- 1.Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. World Malaria Report. Geneva: World Health Organization; 2018. https://www.who.int/malaria/publications/world-malaria-report-2018/report/en/. Accessed 5 Dec 2018.
- 3.Govella NJ, Ferguson H. Why use of interventions targeting outdoor biting mosquitoes will be necessary to achieve malaria elimination. Front Physiol. 2012;3:199. doi: 10.3389/fphys.2012.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Killeen GF. Characterizing, controlling and eliminating residual malaria transmission. Malar J. 2014;13:330. doi: 10.1186/1475-2875-13-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranson H, Edi CVA, Koudou BG, Jones CM, Weetman D. Multiple-insecticide resistance in Anopheles gambiae mosquitoes, southern Côte d’Ivoire. Emerg Infect Dis. 2012;18:1508–1511. doi: 10.3201/eid1809.120262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranson H, Lissenden N. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 2016;32:187–196. doi: 10.1016/j.pt.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Killeen GF, Tatarsky A, Diabate A, Chaccour CJ, Marshall JM, Okumu FO, et al. Developing an expanded vector control toolbox for malaria elimination. BMJ Glob Health. 2017;2:e000211. doi: 10.1136/bmjgh-2016-000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams YA, Tusting LS, Hocini S, Graves PM, Killeen GF, Kleinschmidt I, et al. Expanding the vector control toolbox for malaria elimination: a systematic review of the evidence. Adv Parasitol. 2018;99:345–379. doi: 10.1016/bs.apar.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Fillinger U, Lindsay SW. Larval source management for malaria control in Africa: myths and reality. Malar J. 2011;10:353. doi: 10.1186/1475-2875-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fillinger U, Kannady K, William G, Vanek MJ, Dongus S, Nyika D, et al. A tool box for operational mosquito larval control: preliminary results and early lessons from the Urban Malaria Control Programme in Dar es Salaam, Tanzania. Malar J. 2008;7:20. doi: 10.1186/1475-2875-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera-Varela M, Lindh J, Lindsay SW, Fillinger U. Habitat discrimination by gravid Anopheles gambiae sensu lato—a push–pull system. Malar J. 2014;13:133. doi: 10.1186/1475-2875-13-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eneh LK, Fillinger U, Borg Karlson AK, Kuttuva Rajarao G, Lindh J. Anopheles arabiensis oviposition site selection in response to habitat persistence and associated physicochemical parameters, bacteria and volatile profiles. Med Vet Entomol. 2018;33:56–67. doi: 10.1111/mve.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ndenga BA, Simbauni JA, Mbugi JP, Githeko AK, Fillinger U. Productivity of malaria vectors from different habitat types in the western kenya highlands. PLoS ONE. 2011;6:e19473. doi: 10.1371/journal.pone.0019473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Fillinger U, Yan G. Oviposition behavior of female Anopheles gambiae in western Kenya inferred from microsatellite markers. Am J Trop Med Hyg. 2006;75:246–250. doi: 10.4269/ajtmh.2006.75.246. [DOI] [PubMed] [Google Scholar]
- 15.Okal MN, Lindh JM, Torr SJ, Masinde E, Orindi B, Lindsay SW, et al. Analysing the oviposition behaviour of malaria mosquitoes: design considerations for improving two-choice egg count experiments. Malar J. 2015;14:250. doi: 10.1186/s12936-015-0768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mbare O, Lindsay SW, Fillinger U. Pyriproxyfen for mosquito control: female sterilization or horizontal transfer to oviposition substrates by Anopheles gambiae sensu stricto and Culex quinquefasciatus. Parasit Vectors. 2014;7:280. doi: 10.1186/1756-3305-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lwetoijera D, Harris C, Kiware S, Dongus S, Devine GJ, McCall PJ, et al. Effective autodissemination of pyriproxyfen to breeding sites by the exophilic malaria vector Anopheles arabiensis in semi-field settings in Tanzania. Malar J. 2014;13:161. doi: 10.1186/1475-2875-13-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caputo B, Ienco A, Cianci D, Pombi M, Petrarca V, Baseggio A, et al. The “Auto-Dissemination” approach: a novel concept to fight Aedes albopictus in urban areas. PLoS Negl Trop Dis. 2012;6:e1793. doi: 10.1371/journal.pntd.0001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unlu I, Suman DS, Wang Y, Klingler K, Faraji A, Gaugler R. Effectiveness of autodissemination stations containing pyriproxyfen in reducing immature Aedes albopictus populations. Parasit Vectors. 2017;10:139. doi: 10.1186/s13071-017-2034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suman DS, Wang Y, Faraji A, Williams GM, Williges E, Gaugler R. Seasonal field efficacy of pyriproxyfen autodissemination stations against container-inhabiting mosquito Aedes albopictus under different habitat conditions. Pest Manag Sci. 2018;74:885–895. doi: 10.1002/ps.4780. [DOI] [PubMed] [Google Scholar]
- 21.Minakawa N, Mutero CM, Githure JI, Beier JC, Yan G. Spatial distribution and habitat characterization of anopheline mosquito larvae in western Kenya. Am J Trop Med Hyg. 1999;61:1010–1016. doi: 10.4269/ajtmh.1999.61.1010. [DOI] [PubMed] [Google Scholar]
- 22.Njoroge MM, Tirados I, Lindsay SW, Vale GA, Torr SJ, Fillinger U. Exploring the potential of using cattle for malaria vector surveillance and control: a pilot study in western Kenya. Parasit Vectors. 2017;10:18. doi: 10.1186/s13071-016-1957-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 24.Holleley CE, Geerts PG. Multiplex Manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. Biotechniques. 2009;46:511–517. doi: 10.2144/000113156. [DOI] [PubMed] [Google Scholar]
- 25.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 26.Jones OR, Wang J. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour. 2010;10:551–555. doi: 10.1111/j.1755-0998.2009.02787.x. [DOI] [PubMed] [Google Scholar]
- 27.Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnet E, Van de Peer Y. zt: a software tool for simple and partial Mantel tests. J Stat Softw. 2002;7:20197. doi: 10.18637/jss.v007.i10. [DOI] [Google Scholar]
- 29.Chen H, Minakawa N, Cui L, Yan G. Conspecific sharing of breeding sites by anopheline female mosquitoes (Diptera: Culicidae) inferred from microsatellite markers. J Insect Behav. 2008;21:24–33. doi: 10.1007/s10905-007-9101-4. [DOI] [Google Scholar]
- 30.Munga S, Minakawa N, Zhou G, Githeko AK, Yan AG. Survivorship of immature stages of Anopheles gambiae s.l. (Diptera: Culicidae) in natural habitats in Western Kenya Highlands. J Med Entomol. 2007;44:758–764. doi: 10.1093/jmedent/44.5.758. [DOI] [PubMed] [Google Scholar]
- 31.Almudevar A, Field C. Estimation of single-generation sibling relationships based on DNA markers. J Agric Biol Environ Stat. 1999;4:136–165. doi: 10.2307/1400594. [DOI] [Google Scholar]
- 32.Butler K, Field C, Herbinger CM, Smith BR. Accuracy, efficiency and robustness of four algorithms allowing full sibship reconstruction from DNA marker data. Mol Ecol. 2004;13:1589–1600. doi: 10.1111/j.1365-294X.2004.02152.x. [DOI] [PubMed] [Google Scholar]
- 33.Howell P. Genetic components of insecticide resistance in Anopheles arabiensis. Liverpool: University of Liverpool; 2018. [Google Scholar]
- 34.Zheng L, Benedict MQ, Cornel AJ, Collins FH, Kafatos FC. An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae. Genetics. 1996;143:941–952. doi: 10.1093/genetics/143.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witzig C, Parry M, Morgan JC, Irving H, Steven A, Cuamba N, et al. Genetic mapping identifies a major locus spanning P450 clusters associated with pyrethroid resistance in kdr-free Anopheles arabiensis from Chad. Heredity (Edinb). 2013;110:389–397. doi: 10.1038/hdy.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Schematic view of larval sampling sites at Oluch-Kimira rice irrigation scheme in Homa-Bay county, western Kenya. In the site labels, prefix P is puddle while D is drainage ditch.
Additional file 2. Mendelian test for allele inheritance for parents and offspring in family A. The markers with p values in bold indicate consistency with Mendelian allele segregation rule. N/A represent markers that did not amplify, and n/a represent homozygous alleles whose p values could not be calculated.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.